ABSTRACT
Nucleic acid-based assays, such as polymerase chain reaction (PCR), that amplify and detect organism-specific genome sequences are a standard method for infectious disease surveillance. However, challenges arise for virus surveillance because of their genetic diversity. Here, we calculated the variability of nucleotides within the genomes of 10 human viral species in silico and found that endemic viruses exhibit a high percentage of variable nucleotides (e.g., 51.4% for norovirus genogroup II). This genetic diversity led to the variable probability of detection of PCR assays (the proportion of viral sequences that contain the assay’s target sequences divided by the total number of viral sequences). We then experimentally confirmed that the probability of the target sequence detection is indicative of the number of mismatches between PCR assays and norovirus genomes. Next, we developed a degenerate PCR assay that detects 97% of known norovirus genogroup II genome sequences and recognized norovirus in eight clinical samples. By contrast, previously developed assays with 31% and 16% probability of detection had 1.1 and 2.5 mismatches on average, respectively, which negatively impacted RNA quantification. In addition, the two PCR assays with a lower probability of detection also resulted in false negatives for wastewater-based epidemiology. Our findings suggest that the probability of detection serves as a simple metric for evaluating nucleic acid-based assays for genetically diverse virus surveillance.
IMPORTANCE
Nucleic acid-based assays, such as polymerase chain reaction (PCR), that amplify and detect organism-specific genome sequences are employed widely as a standard method for infectious disease surveillance. However, challenges arise for virus surveillance because of the rapid evolution and genetic variation of viruses. The study analyzed clinical and wastewater samples using multiple PCR assays and found significant performance variation among the PCR assays for genetically diverse norovirus surveillance. This finding suggests that some PCR assays may miss detecting certain virus strains, leading to a compromise in detection sensitivity. To address this issue, we propose a metric called the probability of detection, which can be simply calculated in silico using a code developed in this study, to evaluate nucleic acid-based assays for genetically diverse virus surveillance. This new approach can help improve the sensitivity and accuracy of virus detection, which is crucial for effective infectious disease surveillance and control.
KEYWORDS: nucleic acid-based assays, PCR assays, norovirus, virus surveillance, in silico analysis, virus mutations
INTRODUCTION
Tracking pathogens is a fundamental public health intervention to control communicable diseases (1). Nucleic acid-based assays that analyze organism-specific genome sequences, such as polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), and clustered regularly interspaced short palindromic repeats (CRISPR)-based assays (2), are employed widely as a standard method for virus surveillance. However, challenges arise in virus surveillance utilizing these nucleic acid-based assays because virus genomes constantly mutate (3). The viral genome mutation rate is typically orders of magnitude faster than that of the other types of biological units, such as bacteria, which enables viruses to evolve quickly, resulting in genetic variation within the virus population (4). For instance, noroviruses (Caliciviridae) are composed of 10 genetically variable genogroups (from GI to GX) (5). Rotaviruses (Reoviridae) have been reported to have eight distinct strains (from A to H) (6), and human adenoviruses (Adenoviridae) are subdivided across seven separate species (from A to G) (7). As the viral sequence is a crucial determinant for the sensitivity and specificity of nucleic acid-based assays (8), genome diversity and stability must be considered in the design of nucleic acid-based assays for viruses.
The design of nucleic acid-based assay reagents, such as primers and probes for PCR, typically involves two stages: in silico sequence analysis and in vitro verification. For example, six human rotavirus sequences (G2P4, G3P14, G8P6, G12P6, and G1P8) were used to design primer and probe sequences (9). The authors then used 121 human fecal samples collected from 2004 to 2006 in Slovenia to verify their PCR assays. Another study used 178 human rotavirus A VP6 gene sequences to design primers and a probe (10). The authors then used 266 human samples collected between 2009 and 2012 from Western India for assay verification. Similarly, 18 human rotavirus sequences that target G1, G2, G3, G4, G9, G12, P[4], P[6], and P[8] were employed to evaluate RT-qPCR assays in silico, while 775 clinical samples collected from 2010 to 2014 in Sweden were used to validate the RT-qPCR assays (11). Because these reverse transcriptase qPCR (RT-qPCR) assays were designed with different rotavirus sequences and verified with different clinical samples, they may not show the same performance in a certain scenario, for example, where rotavirus surveillance is conducted in the United States in 2023. Indeed, there is a lack of research evaluating the impacts of different nucleic acid-based assays on virus surveillance.
We hypothesize that a nucleic acid-based assay targeting a specific genome sequence would show different genome amplification efficiencies based on the genome diversity of a virus population, and this would ultimately affect virus surveillance. To test this hypothesis, we first carried out in silico analysis to determine the level of nucleotide diversity across the genomes of 10 viral species. Our results showed that endemic virus surveillance could be significantly impacted by the target sequences of nucleic acid-based assays. We then conducted in vitro experiments using multiple RT-qPCR assays for norovirus on clinical and environmental samples. These experiments revealed that an RT-qPCR assay could exhibit a substantially reduced amplification efficiency when compared to other assays, depending on the norovirus genotypes. Based on these findings, we propose a straightforward approach for evaluating the performance of nucleic acid-based assays for virus surveillance in advance.
RESULTS
Viral genetic variability leads to a wide range of probability of detection of PCR assays
We analyzed genome sequences of 10 viral species, including norovirus (Norwalk-like virus or NLV), rotavirus (RV), respiratory syncytial virus (RSV), influenza A virus (InFA), adenovirus (AdV), severe acute respiratory syndrome coronavirus 1 and 2 (SARS-CoV-1 and SARS-CoV-2, respectively), middle east respiratory syndrome (MERS), Ebola (which caused the 2013 outbreak), and monkeypox virus (Mpox; which caused the 2022 outbreak), to cover a wide range of viral genomic characteristics from an evolutionary perspective. For example, NLV, RV, RSV, InFA, SARS-CoV-1, SARS-CoV-2, MERS, and Ebola are RNA viruses, while AdV and Mpox are DNA viruses. These viruses also caused zoonotic outbreaks in humans at different times. Mpox has been causing outbreaks in humans for decades, most recently in 2022 (12). SARS-CoV-1 caused human disease in 2002 (13, 14), while MERS and Ebola viruses caused human outbreaks in 2012 and 2013 (15–18), and SARS-CoV-2 started infecting humans in 2019 (19–21), while the other viruses, such as NLV, RV, RSV, InFA, and AdV, were reported earlier and have been causing outbreaks (22–25).
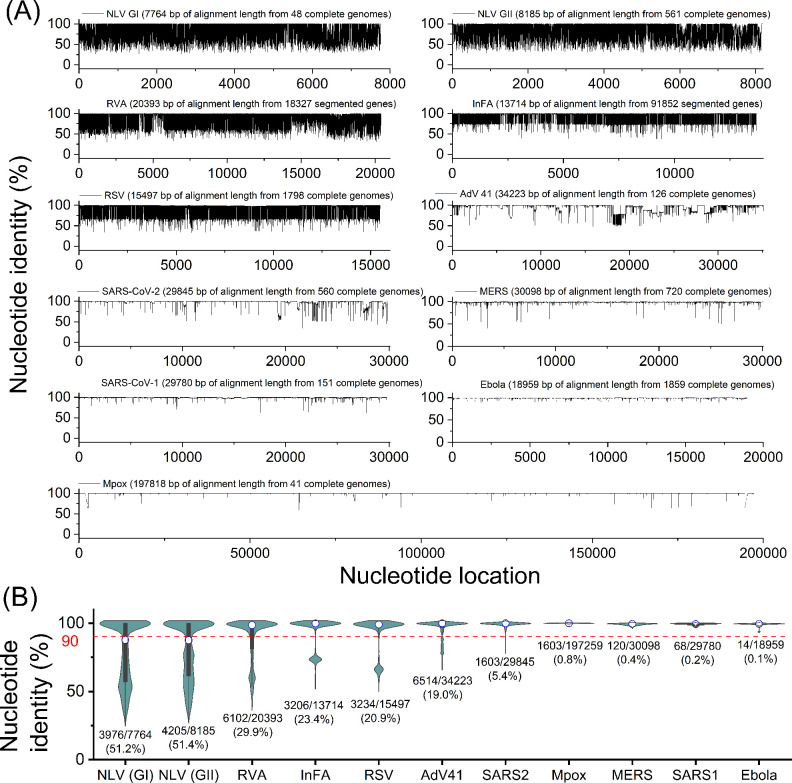
We compared a group of viral sequences to their alignment sequence and determined nucleotide identity (i.e., the percentage of the consensus nucleotide) for the whole viral genomes in Fig. 1A. As viruses mutate and the mutations are passed on to subsequent generations, the viral genomic sequence becomes increasingly diverse, and the nucleotide identity decreases (26, 27). Thus, nucleotide identity has been used to describe genomic diversity (28, 29). In this study, we defined a variable nucleotide as a nucleotide with less than 90% of nucleotide identity to evaluate the level of genome diversity. We found that variable nucleotides, below the red lines in Fig. 1A, are distributed throughout the entire viral genomes (Fig. 1A). Next, we summarized the nucleotide identity for each viral species in a violin chart and determined the percent variable nucleotides that vary substantially depending on viral species (Fig. 1B). We found the percentage of variable nucleotides. Specifically, NLV (51.2% for GI and 51.4% for GII), RV (29.9%), InFA (23.4%), RSV (20.9%), and AdV (19.0%) contain more than 10% variable nucleotides. By contrast, SARS-CoV-2 (5.3%), Mpox (0.8%), MERS (0.4%), SARS-CoV-1 (0.2%), and Ebola (0.1%) represented relatively low values of variable nucleotide. Note that the percentage of variable nucleotides was calculated using only one strain for some viral species, including NLV (GI and GII genogroups), RV (rotavirus A strain), InFA (influenza A virus), and AdV (adenovirus type 41) because the alignment with all strains could not be created due to the genome complexity. Thus, the actual variable nucleotides for these viral species are likely higher than the values shown in Fig. 1. Interestingly, despite the unprecedented COVID-19 pandemic, which has infected more than 673 million cases globally as of February 2023 since the first case was confirmed in 2019, the variable nucleotides of SARS-CoV-2 were fewer than the endemic viruses that were reported earlier and have caused outbreaks. The high level of genetic diversity for endemic viruses suggests that it may be challenging to find a conserved region that can be targeted by nucleic acid-based assay, such as (RT-)qPCR.
Fig 1.
Genetic diversity analysis of 10 viral species. (A) Percent nucleotide identity of the alignment for each viral species with nucleotide locations. (B) Summary of nucleotide identity in violin charts for each viral species. The figures below each violin chart indicate the number of variable nucleotides/the number of total nucleotides (a percentage of variable nucleotides). Any two viral species showed significantly different nucleotide identities (Mann–Whitney U-test, P < 0.05).
We conducted in silico analysis to calculate the probability of detection that previously published primer sets for (RT-)qPCR assays would detect these genome variants (Table 1 and Table S3). We found that the probability of detection significantly varied depending on RT-qPCR assays. For example, RT-qPCR assays for RSV designed by three separate studies only detected 2%, 23%, and 46% of known RSV variants, respectively, when applied to sequences we obtained from Genbank (30–32). Similarly, qPCR assays for AdV type 41 by three groups present probabilities of 43%, 77%, and 100%, respectively, for sequences we obtained from Genbank (33–35). The probability of detection for RV by the other three studies was calculated to be 3%, 16%, and 81%, respectively (9, 10, 36). The variations in the probability of detection observed in Table 1 and Table S3 indicate that the (RT-)qPCR assays targeting different viral sequences may present significantly different probabilities of detection. This finding suggests that different (RT-)qPCR assays could miss the presence of viral variants, which would negatively impact virus surveillance efforts.
TABLE 1.
Detailed information about RT-qPCR assays for norovirus GI and GII genogroups
| Target | Assay | Type | Sequence (5′ to 3′) | Degeneracy | Location | Amplicon size (bp) | Length | Gc (%) | Tm (°C)a | Annealing temperature (°C) | Probability of detection |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GIk | A1g | For | GCHATRTTYCGYTGGATG | 24 | 5283–5300b | 93 | 18 | 49.1 | Min: 53.6 Mean: 57.9 Max: 63.6 |
53 | 48/48 (100%) |
| Probe | TGGACAGGRGAYCGCRATCT | 8 | 5322–5341 | 20 | 57.5 | Min: 63.5 Mean: 66.1 Max: 69.7 |
|||||
| Rev | TTAGACGCCATCATCATT | 1 | 5358–5375 | 18 | 38.9 | 56.2 | |||||
| A2h | For | GCYATGTTCCGYTGGATGC | 4 | 5283–5301b | 97 | 19 | 57.9 | Min: 61.5 Mean: 63.4 Max: 66.4 |
57 | 27/48 (56%) |
|
| Probe | TCGGGCAGGAGATYGCGRTCYC | 8 | 5329–5350 | 22 | 65.9 | Min: 68.1 Mean: 70.4 Max: 73.5 |
|||||
| Rev | GTCCTTAGACGCCATCATCATT | 1 | 5358–5379 | 22 | 45.5 | 62.3 | |||||
| A3i | For | GGAGATCGCRATCTCCTGCC | 2 | 5312–5331d | 103 | 20 | 62.5 | Min: 64.8 Mean: 65.1 Max: 66.4 |
60 | 4/48 (8%) |
|
| Probe | GGGGCGTCCTTAGACGCCATCATCATTTA | 1 | 5335–5363 | 29 | 52.7 | 70.6 | |||||
| Rev | CTCYGGTACCAGCTGGCC | 2 | 5397–5414 | 18 | 69.4 | Min: 64.2 Mean: 64.9 Max: 66.7 |
|||||
| GIIl | B1g | For | TTYAGRTGGATGAGRTTYTC | 16 | 5012–5030f | 86 | 19 | 43.9 | Min: 52.7 Ave: 55.8 Max: 60.3 |
53 | 542/561 (97%) |
| Probe | ACDTGGGAGGGYGATCRCAAT | 12 | 5042–5061 | 20 | 55 | Min: 63.3 Ave: 65.3 Max: 68.3 |
|||||
| Rev | YMGAYGCCATCHTCATTC | 24 | 5080–5097 | 18 | 49.1 | Min: 56.3 Ave: 57.5 Max: 60.5 |
|||||
| B2j | For | ATGTTCAGRTGGATGAGRTTCTCWGA | 8 | 5009–5034f | 89 | 26 | 42.3 | Min: 64.2 Mean: 65.3 Max: 67.4 |
60 | 174/561 (31%) |
|
| Probe | AGCACGTGGGAGGGCGATCG | 1 | 5039–5058 | 20 | 70.0 | 70.3 | |||||
| Rev | TCGACGCCATCTTCATTCACA | 1 | 5077–5097 | 21 | 47.6 | 64.1 | |||||
| B3i | For | GTGGGATGGACTTTTACGTGCCAA | 1 | 4974–4997f | 129 | 24 | 50.0 | 66.9 | 60 | 91/561 (16%) |
|
| Probe | AGCCAGATTGCGATCGCCCTCC | 1 | 5051–5072 | 22 | 63.6 | 70.2 | |||||
| Rev | CGTCAYTCGACGCCATCTTCATTCA | 2 | 5078–5102 | 25 | 50.0 | Min: 67.3 Mean: 67.4 Max: 68.4 |
Melting temperatures were calculated with the qPCR parameter sets of OligoAnalyzer (Integrated DNA Technology), which consider 0.2 μM of oligo concentration, 50 mM of Na+ concentration, 3 mM of Mg2+ concentration, and 0.8 mM of dNTPs concentration.
Reference sequence (Genbank ID: KX396056.1).
Reference sequence (Genbank ID: MW305482.1).
Reference sequence (Genbank ID: MW243609.1).
Reference sequence (Genbank ID: MW305489.1).
Reference sequence (Genbank ID: MW661246.1).
Developed in this study.
Adopted from Wolf et al. (2010) (35).
Adopted from Liu et al. (2020) (37).
Adopted from Loisy et al. (2005) (38).
The sequence of a standard sample for the ORF1 and ORF2 genes of norovirus GI genogroup (Integrated DNA Technologies, USA): 5′-CAGAAAAATCTCCAGTAAAGTTATACAGGAGATAAGGACCGGTGGCTTAGAAATGTATGTACCAGGTTGGCAGGCCATGTTCCGCTGGATGCGCTTCCATGATCTCGGATTGTGGACAGGAGATCGCAATCTCCTGCCCGAATTCGTAAATGATGATGGCGTCTAAGGACGCTACACCAAGCGCAGATGGCGCCACTGGCGCCGGCCAGCTGGTACCGGAGGTTAATACAGCTGACCCTATACCCATTGACCCTGTGGCTGGCTCCTCTACAGCCCTTGCCACTGCGGGCCAAGTTAATT-3′ (GenBank accession number: MW305499.1).
The sequence of a standard sample for the ORF1 and ORF2 genes of norovirus GII genogroup (Integrated DNA Technologies, USA): 5′-ACCCCACTCTCAAAGACCCATACAGCTCATGGCACTGCTTGGTGAGGCCTCCCTTCACGGACCCTCTTTCTACAGCAAAATCAGTAAATTGGTCATAACTGAACTCAAAGAAGGTGGGATGGACTTTTACGTGCCAAGGCAGGAACCCATGTTCAGGTGGATGAGGTTCTCTGACTTGAGCACGTGGGAGGGCGATCGCAATCTGGCTCCCAGTTTTGTGAATGAAGATGGCGTCGAATGACGCCGCTCCATCTACTGATGGTGCAGCCGGCCTCGTGCCAGAAAGTAACAGTGAGGTCA-3′ (GenBank accession number: MW661246.1).
Application of RT-qPCR assays to clinical samples reveals a failure of virus detection by an assay with a low probability of detection
We hypothesized that the probability of detection for target viral species or strains could serve as an indicator of nucleic acid-based assay performance in virus surveillance. To validate this hypothesis, we first prepared RT-qPCR assays for norovirus with varying levels of probability of detection. We designed RT-qPCR assays with a high probability of detection; the A1 assay with a 100% probability of detection for GI genogroup and the B1 assay with a 97% probability of detection for GII genogroup. Other RT-qPCR assays, such as the A2 (56%) and A3 (8%) for GI and B2 (31%) and B3 (16%) for GII, were adopted from previously published studies (Table 2). Norovirus was selected for the in vitro experiment due to its significant genetic variability, as demonstrated in Fig. 1, and its importance for public health (39).
TABLE 2.
Norovirus sequences of clinical samples
| Sample # | Confirmed viral sequencesa | Noteb |
|---|---|---|
| GI-1 | - | - |
| GI-2 |
5’-CAACAGATATAGAATTTGACCCAATCAAACTGACACAAATACTGAAGGAATATGGTTTGAAA
CCCACAAGACCTGACAAAACTGATGGCCCAATTATAGTCAGACAGCAAGTG GATGGC CTGGTCTT CCTCCGGCGCACCATCTCTAAGGATGCTATTGGATACCAGGGACGGCTCGATCGCAATTCCATTG AAAGACAGCTATGGTGGACTCGCGGGCCAAATCACG ATGACCCGTTT GAGACACTGGTCCCGCA TTCACAGAGGAAGGTCCAATTAGTATCTCTGCTTGGTGAAGCAGCACTTCATGGTGAAAAGTTCT A CAGAAAGATAGCCGGCAGAG TTATTCAAGAAGTCAAAG AGGGGGGGCTTGAAATCTACATTCCC GGCAGGCAGGCCATGTTCCGCTGGATGCGCTTTCATGATCTGAGTTTGTGGACAGGGGACCGC G ATCTC CTGCCCGATTATGTAAATGATGATG GCGTCTAAGGACGCCCCAACAAACATGGATGGCAC CAGTGGTGCCGGCCAGCTGGTACCAGAGGCAAATACAGCTGAGCCTATATCAATGG AGCCTGTG G CTGGGGCAGCGACAGCTGCC GCAACCGCTGGCCAAGTTAAT-3’ |
616 out of 621 (99%) nucleotides match to MT031988.1 |
| GI-3 | 5′-CCGATCATTGTAAGGCAACAGGTTGATGGCTTGGTTTTCCTCCGGCGCACCATCTCAAAGGAT GCCATCGGGTACCAGGGCCGGCTTGACCGTAATTCCATTGAAAGACAGCTCTGGTGGACCCGG GGGCCCAACCATGATGATCCATTTGAAACCTTGGTCCCACACCCGCAGAGGAAGGTCCAACTGA TATCCCTGCTGGGTGAAGCTGCACTCCATGGTGAGAAGTTCTACAGAAAGATAGCCAGTAGGGT GATCCAGGAGGTTAAAGAAGGAGGATTGGAAATTTACATCCCTGGGTGGCAGGCCATGTTCCGC TGGATGCGATTCCATGATTTGAGCTTGTGGACAGGAGACCGCGATCTCTTGCCCGATTATGTAAA TGATGATGGCGTCTAAGGACGCCCCAACAAACATGGATGGCACCAGTGGTGCCGGTCAGCTGG TACCAGAGGCAAATACAGCTGAACCTATATCAATGGATCCAGTAGCTGGAGCCGCAACAGCGGTT GCAACTG-3′ |
518 out of 518 (100%) nucleotides match to MN922735.1 |
| GI-4 | 5′-AACCATGATGATCCCTTTGAGACATTAATACCCCATCAACAAAGAAAGATTCAATTGATTTC CTTACTTGGTGAGGCTGCGCTCCACGGAGAGAAATTCTATAGAAAGATTGCCAACAGAGTCATAC AGGAAGTCAAAGAAGGGGGCCTTGAGCTCTATATACCAGGTTGGCAGGCCATATTCCGCTGGAT GCGTTTCCATGACTTGAGCTTGTGGACAGGAGATCGCAATCTCCTGCCCGATTATGTAAATGATG ATGGCGTCTAAGGACGCCCCCTCAAACATGGATGGCACTAGTGGTGCCGGTCAGCTGGTTCCA GAGGTTAATGCAGCTGAACCCCTACCCCTTGAGCCGGTGGTGGGTGCCGCAACTGCGGTGGC CACTGCTGGGCAAGTTAA-3′ |
397 out of 398 (99%) nucleotides match to LC646334.1 |
| GI-5 | 5′-CGCAACTCCATTGAAAGACAATTATGGTGGACCCGGGGCCCAAACCATGATGATCCCTTT GAGACATTAATACCCCATCAACAAAGAAAGATTCAATTGATTTCCTTACTTGGTGAGGCTGCGCTC CACGGAGAGAAATTCTATAGAAAGATTGCCAACAGAGTCATACAGGAAGTCAAAGAAGGGGGCC TTGAGCTCTATATACCAGGTTGGCAGGCCATATTCCGCTGGATGCGTTTCCATGACTTGAGCTTGT GGACAGGAGATCGCAATCTCCTGCCCGATTATGTAAATGATGATGGCGTCTAAGGACGCCCCCTC AAACATGGATGGCACTAGTGGTGCCGGTCAGCTGGTTCCAGAGGTTAATGCAGCTGAACCCCTA CCCCTTGAGCCGGTGGTGGGTGCCGCAACTGCGGTGGCCACTGCTGGGCAAGTTAAT-3′ |
441 out of 442 (99%) nucleotides match to LC646334.1 |
| GI-6 | 5′-CAGGCCATATTCCGCTGGATGCGTTTCCATGACTTGAGCTTGTGGACAGGAGATCGCAATCTC CTGCCCGATTATGTAAATGATGATGGCGTCTAAGGACGCCCCCTCAAACATGGATGGCACTAGTG GTGCCGGTCAGCTGGTTCCAGAGGTTAATGCAGCTGAACCCCTACCCCTTGAGCCGGTGGTGG GTGCCGCAACTGCGGTGGCCACTGCTGGGCAAGTTAAT-3′ |
229 out of 229 (100%) nucleotides match to MN421785.1 |
| GI-7 | - | - |
| GI-8 | 5′-GAAGCATCAAATAGATGGGTTAGTTTTTCTGAGGCGCACTATATCAAAAGATGCTGCTGG CTACCAAGGGCGCTTGGACCGCAACTCCATTGAAAGACAATTATGGTGGACCCGGGGCCCAAA CCATGATGATCCCTTTGAGACATTAATACCCCATCAACAAAGAAAGATTCAATTGATTTCCTTACT TGGTGAGGCTGCGCTCCACGGAGAGAAATTCTATAGAAAGATTGCCAACAGAGTCATACAGGA AGTCAAAGAAGGGGGCCTTGAGCTCTATATACCAGGTTGGCAGGCCATATTCCGCTGGATGCG TTTCCATGACTTGAGCTTGTGGACAGGAGATCGCAATCTCCTGCCCGATTATGTAAATGATGAT GGCGTCTAAGGACGCCCCCTCAAACATGGATGGCACTAGTGGTGCCGGTCAGCTGGTTCCA GAGGTTAATGCAGCTGAACCCCTACCCCTTGAGCCGGTGGTGGGTGCCGCAACTGCGGTGG CCACTGCTG-3′ |
509 out of 510 (99%) nucleotides match to LC646334.1 |
| GI-9 | - | |
| GI-10 | 5′-TAGTCTCAACAGATATTGAATTTGACCCAAACAGGTTAACACAAGTTCTAAGAGAGTATG GCTTAAAGCCCACAAGACCTGACAAGACTGATGGCCCAATCATTGTGAGACAGCAAGTTGATG GCTTGGTTTTCCTCCGGCGCACCATTTCGAAAGATGCCATTGGATACCAGGGACGCCTCGACC GAAATTCCATTGAGAGACAGCTCTGGTGGACTCGTGGGCCCAACCATGATGATCCATTTGAAAC CTTAGTCCCACATACACAGAGAAAGGTTCAGCTAATATCCCTACTAGGTGAAGCTGCACTCCAT GGTGAGAAATTCTACAGAAAGATAGCCAGTAGGGTGATCCAGGAAGTCAAAGAGGGGGGGTTG GAAGTTTACATCCCTGGGTGGCAGGCCATGTTCCGCTGGATGCGATTCCATGATTTGAGCTTGT GGACAGGAGACCGCGATCTCTTGCCCGATTATGTAAATGATGATGGCGTCTAAGGACGCCCCAA CAAACATGGATGGCACCAGTGGTGCCGGTCAGCTGGTACCAGAGGCGAATACAGCTGAACCTAT ATCAATGGATCCAGTGGCTGGAGCCGCAACAGCGGTCGCTACTGCTGGACAAGTTAATA-3′ |
620 out of 628 (99%) nucleotides match to MN922741.1 |
| GII-1 | 5′-CTCTTAGTGCTATGTCTGAGGTCTCTGGTCTTTCCCCTGAGGTTGTGCAAGCCAACTCCT GTTTCTCATTCTATGGGGATGATGAAATAGTCAGCACAGATATAAACCTAGACCCAGAAAAACTCA CCAGGAAACTGAGGGAGTATGGCCTCGTCCCAACAAGGCCAGACAAAACTGAGGGCCCACTTG TGATCACTCAGGATTTGAATGGTCTCACATTCTTGAGGCGAACCATAGTGCGGGACCCCGCAGG TTGGTTTGGAAAATTGGATCGTGATTCCATTCTAAGGCAGTTATACTGGACCAGAGGACCCAATC ATGAGAACCCCTTTGAAAGTATGATTCCCCACTCCCAGAGAGCAACCCAGTTAATGGCCCTTCTT GGGGAAGCCTCGTTGCATGGTCCCCAATTTTACAAGAAGGTGAGTAAAATGGTCATCAGTGAGA TCAAGAGTGGTGGTCTGGAGTTTTACGTGCCCAGACAGGAGGCCATGTTTAGATGGATGAGATT TTCAGACCTCAGCACGTGGGAGGGCGATCGCAATCTGGCTCCCGAGAATGTGAATGAAGATGG CGTCGAATGACGCAGCTCCATCGAATGATGGCGCGGCTGGCCTCGTACCAGAGATCAACCATG AGGTCATGGCCATAGAGCCTGTTGCAGGGGCCTCTCTAGCAGCCCCTGTCGTAGGACAACTTA ATATAATTGATCCCTGGATTAGAAATAATTTTGTACAAGCCCCTGCTGGAGAATTCACTGTTTCGC CTAGAAATGCTCCAGGTGAATTTTTGTTAGATTTAGAGTTAGGTCCAGAATTGAATCCTTATCTTGCA-3′ |
831 out of 833 (99%) nucleotides match to OP686904.1 |
| GII-2 | - | - |
| GII-3 | 5′-TCCTCCGCCGAACAGTCACCCGTGATCCAGCAGGTTGGTTTGGAAAGTTGGACCAAAACTC CATCCTCAGGCAGTTGTACTGGACAAGAGGACCCAACCATGAAGACCCCAGTGAGACCATGAT ACCACACGCACAAAGACCTGTGCAGCTCATGGCACTACTAGGAGAATCCTCCCTACATGGACC CTCATTTTACAGCAAGGTTAGCAAATTAGTCATATCTGAACTTAAAGAGGGAGGAATGGATTTTT ATGTGCCCAGACAAGAGTCAATGTTCAGGTGGATGAGGTTCTCAGATCTAAGCACATGGGAGG GCGATCGCAATCTGGCTCCCAGTTTTGTGAATGAAGATGGCGTCGAATGACGCCGCTCCATC TAATGATGGTGCTGCTGGTCTCGTACCAGAGGGCAACAACGAGACCCTTCCCCTAGAACCAG TTGCGGGCGCAGCTATAGCCGCACCCGTCACTGGCCAAAATAA-3′ |
482 out of 482 (100%) nucleotides match to KT326180.1 |
| GII-4 | 5′-TGATGATGAGATTGTGAGCACAGACATAAAATTGGACCCAGAAAAATTGACCGCAAAG CTCAAAGAATATGGCCTTAAACCCACTCGGCCCGACAAAACTGAGGGGCCGTTGGTGATTAG TGAGGACCTGAATGGGTTGACTTTCCTCCGCCGAACAGTCACCCGTGATCCAGCAGGTTGG TTTGGAAAGTTGGACCAAAACTCCATCCTCAGGCAGTTGTACTGGACAAGAGGACCCAACC ATGAAGACCCCAGTGAGACCATGATACCACACGCACAAAGACCTGTGCAGCTCATGGCACTA CTAGGAGAATCCTCCCTACATGGACCCTCATTTTACAGCAAGGTTAGCAAATTAGTCATATCTG AACTTAAAGAGGGAGGAATGGATTTTTATGTGCCCAGACAAGAGTCAATGTTCAGGTGGATGA GGTTCTCAGATCTAAGCACATGGGAGGGCGATCGCAATCTGGCTCCCAGTTTTGTGAATGAA GATGGCGTCGAATGACGCCGCTCCATCTAATGATGGTGCTGCTGGTCTCGTACCAGAGGGC AACAACGAGACCCTTCCCCTAGAACCAGTTGCGGGCGCAGCTATAGCCGCACCCGTCACTG GCCAAAATAATGTAAT-3′ |
629 out of 631 (99%) nucleotides match to KY905330.1 |
| GII-5 | 5′-GACGGTGACTCGTGACCCAGCTGGCTGGTTTGGAAAACTGGACCAAAGTTCAATTTTG AGGCAGATGTACTGGACTAGAGGACCAAATCATGAAGACCCCAATGAGACAATGATACCCCATT CTCAAAGACCCATACAGCTCATGGCACTGCTTGGTGAAGCCTCTCTTCACGGACCCTCTTTCTA CAGTAGAATCAGTAAATTGGTCATAACTGAACTTAAAGAAGGTGGGATGGACTTTTACGTGCCAA GGCAGGAACCCATGTTCAGGTGGATGAGGTTTTCTGACTTGAGCACGTGGGAGGGCGATCGC AATCTGGCTCCCAGCTTTGTGAATGAAGATGGCGTCGAGTGACGCCAACCCATCTGATGGGTC CGCAGCCAACCTCGTACCAGAGGTCAACAATGAGGTTATGGCTTTG-3′ |
420 out of 422 (99%) nucleotides match to MK752943.1 |
| GII-6 | 5′-CATACAGCTCATGGCACTGCTTGGTGAAGCCTCTCTTCACGGACCCTCTTTCTACAGTAG AATCAGCAAATTGGTCATAACTGGAACTTAAAGAAGGTGGTATGGATTTTTACGTGCCAAGACAG GAACCCATGTTCAGGTGGATGAGGTTTTCTGACTTGAGCACGTGGGAGGGCGATCGCAATCTG GCTCCCAATTTTGTGAATGAAGATGGCGTCGAGTGACGCCAACCCATCTGATGGGTCCGCAGC CAACCTCGTACCAGAGGTCAACAATGAGGTTATGGCTTTGGAGCCCGTTGTTGGTGCCGCTATT GCGGCACCTGTAGCGGGCCAACAAAATGTAATTGACCCCTGGATTAGAAATAATTTTGTACAAGC CCCTGGTGGGGAGTTTACAGTATCCCCTAGAAACGCTCCAGGTGAAATACTATGGAGCGCGCCC CTAGGCCCCGACCTAAACCCCTATCTATCCCATTTGGCCAGAATGTACAATGGTTATGCAGGTGG TTTTGAAGTGCAGGTAATTCTCGCGGGGAACGCGTTCACCGCCGGGAAGGTTATATTTGCAGCA GTCCCACCAAATTTTCCAACTGAAGGCTTAAGTCCTAGCCAGGTCACTATGTTCCCCCATATAATA GTAGATGTTAGACAATTAGAACCTGTGCTAATTCCCTTACCCGATGTTAGGAATAATTTTTATCATTA CAATCAGTCAAATGACTCCACTATTAAGTTGATAGCAATGTTGTATACACCACTTAGGGCTAATAAT GCTGGGGATG-3′ |
774 out of 784 (99%) nucleotides match to MW661260.1 |
| GII-7 | 5′-CGGACCCTCTTTCTACAGTAGAATCAGCAAATTGGTCATAACTGAGCTCAAAGAAGGTGG GATGGACTTTTACGTGCCAAGGCAGGAACCCATGTTCAGGTGGATGAGGTTTTCTGACTTGAGC ACGTGGGAGGGCGATCGCAATCTGGCTCCCAATTTTGTGAATGAAGATGGCGTCGAATGACGC CAACCCATCTGATGGGTCCGCAGCCAACCTCGTACCAGAGGTCAACAATGAGGTTATGGCTTTG GAGCCCGTTGTTGGTGCCGCTATTGCGGCACCTGTAGCGGGCCAACAAAATGTAATTGACCCCT GGATTAGAAATAATTTTGTACAAGCCCCTGGCGGGGAGTTCACAGTATCCCCTAGAAACGCTCC AGGTGAAATACTATGGAGCGCGCCCCTAGGCCCTGACCTAAATCCCTACCTGTCCCATTTGGCC AGAATGTACAATGGTTATGCAGGTGGTTTTGAAGTGCAGGTAATTCTCGCGGGGAACGCGTTCA CCGCCGGGAAGGCGGGGAACGCGTTCACCGCCGGGAAGTTATATTTGCAGCAGTCCCACCAAA TTTTCCAACTGAAGGCCTAAGTCCTAGCCAGGTCACTATGTTCCCCCACATAATAGTAGATGTTAG ACAATTAGAACCTGTGCTAATTCCCTTACCCGATGTTAGGAATAATTTCTATCATTACAATCAATCAA ATGACTCCACTATTAAGTTGATAGCAATGTTGTA-3′ |
703 out of 712 (99%) nucleotides match to MW661278.1 |
| GII-8 | 5′-ACAGTAGAATCAGCAAAATTGGTCATAACTGAGCTCAAAGAAGGTGGGATGGACTTTTAC GTGCCAAGGCAGGAACCCATGTTCAGGTGGATGAGGTTTTCTGACTTGAGCACGTGGGAGGG CGATCGCAATCTGGCTCCCAATTTTGTGAATGAAGATGGCGTCGAATGACGCCAACCCATCTGA TGGGTCCGCAGCCAACCTCGTACCAGAGGTCAACAATGAGGTTATGGCTTTGGAGCCCGTTGT TGGTGCCGCTATTGCGGCACCTGTAGCGGGCCAACAAAATGTAATTGACCCCTGGATTAGAAAT AATTTTGTACAAGCCCCTGGCGGGGAGTTCACAGTATCCCCTAGAAACGCTCCAGGTGAAATAC TATGGAGCGCGCCCCTAGGCCCTGACCTAAATCCCTACCTGTCCCATTTGGCCAGAATGTACAA TGGTTATGCAGGTGGTTTTGAAGTGCAGGTAATTCTCGCGGGGAACGCGTTCACCGCCGGGA AGATTATATTTGCAGCAGTCCCACCAAATTTTCCAACTGAAGGCCTAAGTCCTAGCCAGGTCACT ATGTTCCCCCACATAATAGTAGATGTTAGACAATTAGAACCTGTGCTAATTCCCTTACCCGATGTT AGGAATAATTTCTATCATTACAATCAATCAAATGACTCCACTATTAAGTTGATAGCAATGTTGT-3′ |
688 out of 698 (99%) nucleotides match to MW661278.1 |
| GII-9 | - | - |
| GII-10 | 5′-CACCGACATAAAATTGGACCCTGAGCAGTTAACCGCCAAGTTGAGGGAGTACGGCCTGA AGCCAACCCGCCCAGACAAGACCGAGGGACCCCTGATCATCAGTGAAGACTTGAACGGACTCA CTTTCCTCCGAAGGACGGTGACTCGTGACCCAGCTGGCTGGTTTGGAAAACTGGATCAAAGTT CAATTCTGAGGCAGATGTACTGGACTAGAGGACCAAATCATGAAGACCCCAATGAGACAATGATA CCCCACTCTCAAAGACCCATACAGCTCATGGCACTGCTTGGTGAAGCCTCTCTTCACGGACCCT CTTTCTACAGTAGAATCAGCAAATTGGTCATAACTGAGCTCAAAGAAGGTGGGATGGACTTTTAC GTGCCAAGGCAGGAACCCATGTTCAGGTGGATGAGGTTTTCTGACTTGAGCACGTGGGAGGG CGATCGCAATCTGGCTCCCAATTTTGTGAATGAAGATGGCGTCGAATGACGCCAACCCATCTG ATGGGTCCGCAGCCAACCTCGTACCAGAGGTCAACAATGAGGTTATGGCTTTGGAGCCCGTT GTTGGTGCCGCTATTGCGGCACCTGTAGCGGGCCAACAAAATGTAATTG-3′ |
608 out of 615 (99%) nucleotides match to OM185499.1 |
Bold indicates nucleotides that did not match to the reference sequence. Underlines represent the annealing sites for three RT-qPCR assays (A1, A2, and A3 for GI samples and B1, B2, and B3 assay for GII samples).
The norovirus sequences were blasted to find the reference sequences (Genbank ID).
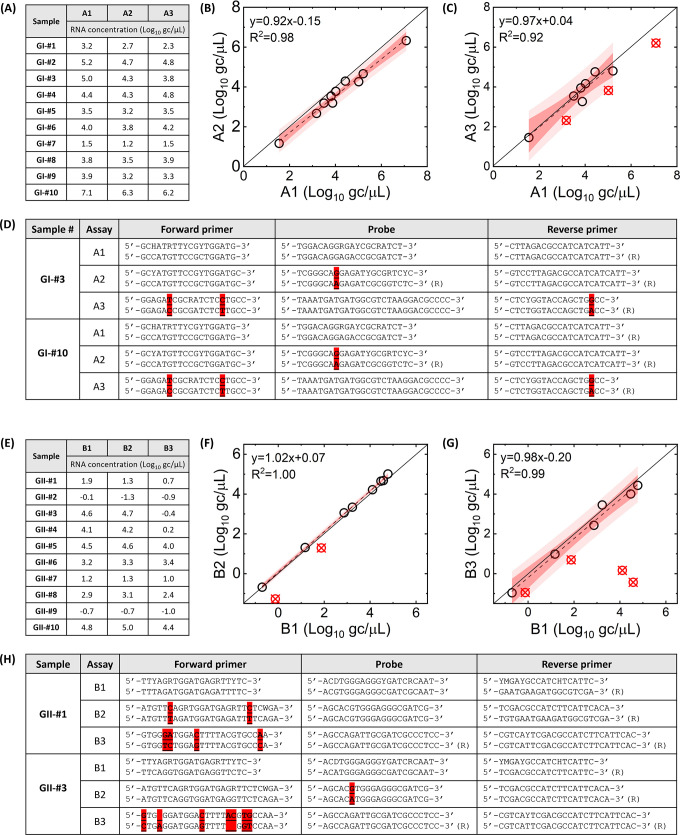
The RT-qPCR assays were then applied to clinical samples (Fig. 2A and E for GI and GII, respectively). We compared the RNA concentration of the RT-qPCR assays with lower probability of detection levels, such as A2, A3, B2, and B3, to those with high probability of detection levels, such as A1 and B1 (Fig. 2B and C for GI and Fig. 2F and G for GII). The comparison results present that some samples (red circle with a cross in Fig. 2C, F, and G, respectively) are located significantly below the regression lines (i.e., the studentized residual was less than −1.5). These outliers indicate that the RT-qPCR assays with a lower probability of detection yielded significantly lower RNA concentrations than those with a high probability of detection. For instance, RNA concentrations of GII-#3 and GII-#4 measured by the B3 assay were 105.0- and 103.9-fold lower than those by the B1 assay, respectively (Fig. 2E). These results indicate that the B3 assays do not amplify specific norovirus samples as effectively as the B1 assay. Excluding these outliers, slopes of regression analysis among RT-qPCR assays were not significantly different from 1 (P > 0.05), meaning that all assays yielded similar RNA concentrations for the rest of the samples.
Fig 2.
Clinical samples analyzed by RT-qPCR assays. (A) and (E) show GI and GII RNA concentrations, respectively. GI RNA concentrations by A1 assay are compared to those by A2 (B) and A3 assay (C). GII RNA concentrations by B1 assay are compared to those by B2 (F) and B3 assay (G). Red circles with a cross indicate outliers that deviated from the regression line (a cutoff studentized residual is −1.5). Dark and pale shades represent 95% confidence and prediction intervals, respectively. Possible annealing sequences for GI-#3 and GI-#10 with A1, A2, and A3 assays are presented in (D). Possible annealing sequences for GI-#1 and GI-#3 with B1, B2, and B3 assays are presented in (H). Sequences of RT-qPCR assay and viral genome are illustrated top and bottom, respectively. Highlighted nucleotides show mismatches.
The reduced amplification efficiencies can be explained by the mismatches between RT-qPCR assay sequences and viral sequences. Potential bindings between the primer/probe of RT-qPCR assays and the annealing sites of the virus genome are illustrated in Tables S4 through S9; Fig. 2D and H. We found that GII-#3 and GII-#4, which showed a significant reduction in RNA concentration by the B3 assay, had seven mismatches with the B3 assay while the B1 assay, which had no mismatches with these two samples (Fig. 2H). Furthermore, we discovered that a lower probability of detection by RT-qPCR, as determined in silico, may suggest a larger number of mismatches between RT-qPCR assays and viral genomes confirmed in vitro. For example, the A1 assay (100% probability of detection for GI) had no mismatches with the 10 GI samples, while the A2 (56% probability of detection) and A3 (13% probability of detection) assays had 0.75 and 2 average mismatches, respectively. Similarly, the B1 assay (97% probability of detection for GII) had no mismatches, whereas the B2 (31% probability of detection) and B3 (16% probability of detection) assays had 1.12 and 2.5 average mismatches, respectively, with the GII samples.
Mismatches between RT-qPCR assays and norovirus genomes explain RNA quantification of a mixture of norovirus sequences in wastewater
As an important epidemiological tool for disease surveillance, we also evaluated the relationship between the detection probability of RT-qPCR assay and results from wastewater-based epidemiology (WBE). We first conducted an experiment in which we spiked local wastewater with known quantities and genotypes of norovirus. In this experiment, we added 2 mg of each clinical sample (i.e., GII-#1, GII-#3, and GII-#10) or mixtures of those samples to 500 mL of local wastewater, from which endogenous norovirus was not detected by the RT-qPCR assays. We then processed the wastewater to obtain the concentrated sludge and quantified norovirus RNA concentrations. As a result, we detected norovirus RNA using the B1 assay, with an average norovirus recovery efficiency of 16.8% (n = 7), which is comparable to those by wastewater surveillance procedures for SARS-CoV-2 (40). This finding demonstrates that our sewage processing method can effectively concentrate norovirus RNA from wastewater. We also found that the B2 assay failed to detect GII-#1 and the B3 assay could not amplify GII-#1, GII-#3, and the mixture of these two samples (Table 3), which agrees with the results from clinical sample analyses. This finding suggests that variation in RNA concentrations of wastewater among RT-qPCR assays can also be explained by the mismatches between RT-qPCR assays and viral genomes.
TABLE 3.
Norovirus RNA concentrations of the norovirus-spiked wastewater quantified by three RT-qPCR assays
| Spiked sample | B1 assay | B2 assay | B3 assay |
|---|---|---|---|
| GII-#1a | 4.3 ✕ 101 gc/uL | Undetermined | Undetermined |
| GII-#3a | 3.7 ✕ 103 gc/uL | 5.5 ✕ 102 gc/uL | Undetermined |
| GII-#10a | 8.1 ✕ 103 gc/uL | 2.8 ✕ 103 gc/uL | 1.1 ✕ 104 gc/uL |
| GII-#1, GII-#3b | 5.3 ✕ 101 gc/uL | 1.1 ✕ 101 gc/uL | Undetermined |
| GII-#1, GII-#10b | 1.1 ✕ 102 gc/uL | 3.0 ✕ 101 gc/uL | 5.6 ✕ 101 gc/uL |
| GII-#3, GII-#10b | 3.0 ✕ 102 gc/uL | 9.3 ✕ 101 gc/uL | 1.2 ✕ 102 gc/uL |
| GII-#1, GII-#3, GII-#10b | 2.2 ✕ 102 gc/uL | 6.9 ✕ 101 gc/uL | 8.8 ✕ 101 gc/uL |
Two milligrams of each stool sample was added to 500 mL of composite sewage sample. The sewage samples were processed as described in the “Sewage sample collection and processing” chapter, and the 10-fold dilutions of RNA extracts were quantified by RT-qPCR as described in the “RT-qPCR protocol for norovirus quantification” chapter. The figures indicate the concentrations of the 10-fold dilutions of RNA extracts, for which the dilution factor, recovery efficiency, and concentration factor were not considered.
Two milligrams of each stool sample was added to 500 mL of grab sewage sample. The rest of the processing and analyzing procedure was the same as described above.
Application of RT-qPCR assays to wastewater samples corroborates the importance of in silico analysis for virus surveillance
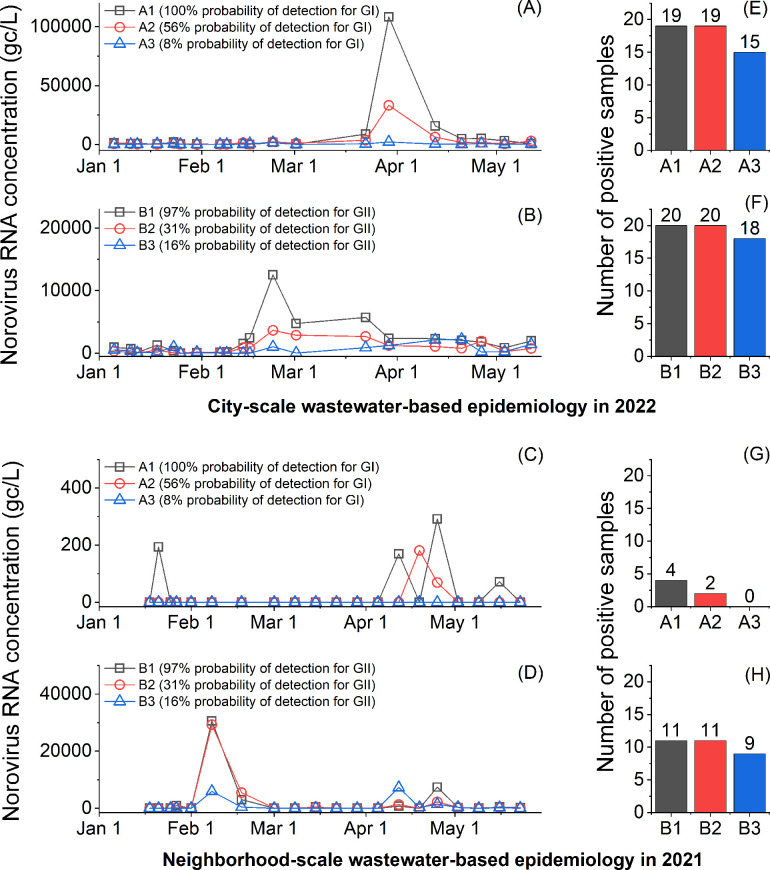
We utilized RT-qPCR assays to detect norovirus RNA in two sets of wastewater samples collected from city-scale and neighborhood-scale sewersheds (Fig. 3). We found that norovirus RNA concentrations of wastewater collected from the city-scale sewershed in 2022 aligned with the percent positive rate of patients by PCR test in Midwestern States (Fig. S5), suggesting the norovirus surveillance results were reliable. Interestingly, the RT-qPCR assays showed varying surveillance results depending on the probability of detection of the assays at a particular monitoring period. For example, from March 2nd to April 21st, 2022, the B1 and B2 assays presented a decreasing tendency in GII RNA concentrations, while the B3 assay, with the lowest probability of detection for GII, demonstrated an increasing tendency. In addition, an RT-qPCR assay with a low probability of detection is more susceptible to false negatives. We found that the number of positive samples by A3 and B3 (the lowest probability of detection for GI and GII, respectively) was lower than those by A1 and B1 (the highest probability of detection for GI and GII, respectively) (Fig. 3E through H). Thus, our findings suggest that caution should be exercised when using an RT-qPCR assay with a low probability of detection to detect or quantify viruses for wastewater surveillance.
Fig 3.
Wastewater samples analyzed by RT-qPCR assays. City-scale and neighborhood-scale wastewater-based epidemiology data are summarized at the top and the bottom, respectively. (A and C) present temporal GI concentrations of 20 samples from a city-scale wastewater treatment plant, and (B and D) show temporal GII concentrations of 20 samples from a manhole. The numbers of norovirus-positive wastewater samples are summarized in (E to H).
DISCUSSION
At the emergence of novel viruses in the human population, primarily from zoonotic spillovers from animal reservoirs (e.g., SARS, Ebola, HIV, MERS, Nipah, and Canine parvovirus), their genome sequences are distinct from closely related viruses (41). For instance, bats were identified as natural hosts of coronaviruses closely related to SARS-CoV-1, which caused the 2002–2004 SARS outbreak (13, 42). This bat virus evolved rapidly in at least two intermediate hosts, such as civets, before being transmitted to humans (43), resulting in a SARS-CoV-1 genome that was distinct from previously known groups of coronaviruses (14). If emerging viruses acquire efficient human-to-human transmission, genetic mutations in the viral genomes can be introduced, causing viruses to diverge from their ancestors, as demonstrated in the influenza virus or SARS-CoV-2 variants (19, 44). Depending on disease pathology (mortality, incubation time, transmissibility) and public health interventions (contact tracing, quarantine, and vaccination), some virus strains may fade out and be contained to a limited number of people or even become extinct. On the other hand, other viruses may establish a stable relationship with humans and become endemic viruses, circulating in the human population and intermittently causing outbreaks. Among the viral species in this study, SARS-CoV-2 (45) and Mpox (46) can be considered as newly emerging viruses, while SARS-CoV-1 (47), MERS (18), and Ebola (17) are considered contained viruses, and NLV, RV, RSV, IAV, and AdV are classified as the endemic viruses (48).
In this study, we discovered that the endemic viruses exhibited a significantly higher level of genome diversity than the emerging viruses and the contained viruses (Mann–Whitney U-test, P < 0.05 in Fig. 1). This finding can be attributed to the evolutionary rate (i.e., the speed of genetic change in a lineage over a specific period) and the time for which the genetic changes are accumulated (23). Table S10 summarizes the previously reported evolutionary rates of various viral species. The evolutionary rates ranged from 4 × 10−4 to 1.2 × 10−2 nucleotide substitutions/site/year (s/s/y) for the RNA viruses (i.e., TV, RVA, IAV, RSV, SARS, MERS, and SARS-CoV-2) and from 5 × 10−6 to 4.1 × 10−5 s/s/y for the DNA viruses (i.e., AdV and Mpox). Interestingly, AdV, a DNA virus with an evolutionary rate of 4.1 × 10−5 s/s/y, presented a higher degree of genome diversity compared to RNA viruses with faster evolutionary rates, such as SARS-CoV-1 (4 × 10−4 s/s/y), SARS-CoV-2 (6.7 × 10−4-3.3 × 10−3 s/s/y), MERS (1.1 × 10−3 s/s/y), and Ebola (1.2 × 10−3 s/s/y). Note that AdV type 41 was estimated to have originated in 1720 (22), allowing the virus a much longer period for the accumulation of mutations. Although the evolutionary rate reflects the rate at which mutations are passed on to descendants, it is not a direct measure of current genome diversity. Instead, the time for which genetic changes accumulate may play a more critical role. This is why endemic viruses are genetically highly variable.
If a viral species has numerous strains with highly variable genome sequences, many of which are currently circulating in the human population, it is challenging to predict which strains would be introduced to a specific location and lead to an outbreak. For example, three geographically adjacent Asian countries suffered from different norovirus genotypes in 2018, showing GII.2 in China (49), GII.4 in Japan (50, 51), and GII.17 in South Korea (52). In this study, we corroborated that RT-qPCR assays for norovirus could have a significantly reduced genome amplification efficiency than the other assays depending on viral sequences present in clinical and environmental samples (Fig. 2 and 3). Therefore, virus surveillance should be conducted with an assay that can cover a wide range of viral sequences that may be introduced to a community. We found that the probability of detection determined in silico with the up-to-date viral sequences could be used to evaluate the likelihood of reduced quantification efficiency for clinical testing or wastewater surveillance.
The calculated probability of detection may not be the perfect parameter to describe the PCR amplification efficiency because it assumes that the perfect match between a PCR assay and a virus genome is a prerequisite for the amplification. The PCR assay could still detect viruses even with mismatches between the genome and primers. The probability of detection does not differentiate the varying impacts of mismatches on the amplification of the target sequence. It is currently challenging to quantitatively evaluate the impact of mismatch because the number, location, and type of mismatch or their combinations have complex impacts on PCR amplification efficiency (53–55). Despite the limitation, the probability of detection would still be helpful in evaluating nucleic acid-based assays because it is better to minimize the number of mismatches. For example, the viral sequences from the databases (e.g., Genbank or GISAID) are probably not perfectly representative of the true viral diversity in reality, meaning that there may be a viral sequence with mismatches on annealing sites that have not been reported in the database yet. Indeed, the Genbank database did not include sequences that are identical to 12 norovirus sequences of our clinical samples (out of 15 sequences). We sequenced norovirus RNA in the clinical samples (from 229 to 833 bases) and the most similar sequences in Genbank showed from 1 to 10 mismatches (Table 2). In addition, mutations frequently occur in viruses, which could eventually lead to the appearance of new mismatches on the annealing sites (37). The unexpected extra mismatch could result in failure of PCR analysis (i.e., false negative).
The current practice for reporting PCR assay results, such as Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines, focuses on the quality control and reproducibility of data and does not demand thorough consideration of the impact of genome diversity on RT-qPCR analysis. For example, the MIQE list requests to present target gene information, but a great level of diversity is also found at a gene level too, as shown in segmented genes of RV and InFA (Fig. 1). This means the target gene may not be enough information to address genome diversity. In this study, we propose that the probability of detection be used to evaluate the performance of nucleic acid-based assays for virus surveillance in advance. The probability of detection of nucleic acid-based assay can be simply calculated in silico, and we published a code that calculates the probability of detection of degenerate RT-qPCR (github.com/Nguyen205/In-silico-analysis-for-degenerate-qPCR-assay). This tool will enable people to easily evaluate their nucleic acid-based assays and improve the reliability of virus surveillance.
MATERIALS AND METHODS
Viral genetic diversity analysis
We obtained complete viral sequences from open-source databases, such as Genbank (ncbi.nlm.nih.gov/genbank) or Global Initiative on Sharing All Influenza Data (GISAID; gisaid.org) to investigate the genetic diversity of viruses. We downloaded all available viral sequences from the databases and, in cases where sufficient sequences were available, we applied an additional filter of collection location and year to complete in silico analysis using an ordinary laptop. The viral sequences were aligned using MUSCLE (v3.8.1551) (56). To expedite the computation, we sliced the complete genome into smaller pieces for viruses with a long sequence length and/or numerous sequences. We compared sequences between the complete alignment and individual viral sequences using Jalview (v.2.11.2.5) (57).
We defined nucleotide identity as the percentage of the consensus nucleotide of the alignment. When we calculated nucleotide identity using Jalview (58), gaps in each viral sequence that occurred during the genome sequencing process (i.e., undetermined sequence) or were due to different sequence lengths, were ignored. In addition, if a consensus nucleotide was determined with less than 10 viral sequences, those nucleotides were excluded from the alignment. As many studies employ a 90% nucleotide identity threshold to assess the similarity of viral sequences (27, 59–61), we defined nucleotide identity below this threshold as a variable nucleotide, which we utilized to evaluate the level of genome diversity. Further details about sequence download and alignment determination can be found in Supporting Information.
In silico probability of target sequence detection of (RT-)qPCR assays
We determined the impact of viral genome diversity on the performance of (RT-)qPCR by calculating the probability of detection. The probability of detection is defined in this study as the ratio of the number of viral genomes that have identical sequences to those of the (RT-)qPCR assay (e.g., sequences of two primers and one probe) to the total number of viral genomes found databases, such as GenBank or GISAID. Our previous study developed an algorithm that searches for the target sequences within each viral sequence obtained from the database and counts the number of virus genomes that include or exclude the identical target sequence (62). In this study, we improved the algorithm to calculate the probability of detection of (RT-)qPCR assays with degenerate sequences (https://github.com/Nguyen205/In-silico-analysis-for-degenerate-qPCR-assay). The input data for calculating the probability of detection of the (RT-)qPCR assays are viral sequences in a fasta format obtained from the database and sequences of (RT-)qPCR assay.
Degenerate RT-qPCR assay design to detect genetically variable norovirus populations
We collected 637 complete norovirus sequences from Genbank and determined their genogroups and genotypes using the Norovirus Genotyping Tool Version 2.0 (www.rivm.nl/mpf/typingtool/norovirus). The sequences whose genotypes were not determined with this tool were excluded from further analysis. Although noroviruses are genetically diverse, including 10 genogroups (5), the majority of norovirus infections have been caused by the GI and GII genogroups (63, 64). We also found that most of our sequences were either GI (n = 48) or GII (n = 561), while the other genogroups were negligible, showing only three sequences of GIII and eight sequences of GIV. For this reason, we designed two RT-qPCR assays, each targeting GI and GII genogroups, respectively.
Next, we aligned the sequences of each genogroup with MUSCLE (v3.8.1551) (56). The aligned sequences were used to generate degenerate sequences, which contain multiple possible nucleotides at one position, using the DegePrime (65). We selected degenerate sequences that were longer than 18 bases for a primer and 20 bases for a probe, had a degeneracy of less than 24, and an amplicon size between 80 and 200 bases. We further analyzed these degenerate sequences using Oligoanalyzer (Integrated DNA Technologies, USA) to ensure that they met the following requirements. First, the degenerate primers and probes must not have degenerate nucleotides at the last three bases from the 3′ end. Second, the average probes′ melting temperature (Tm) must be 7°C higher than those of primers. Finally, the probe sequence must not start with a guanine from the 5′ end.
Once the candidate primer and probe sequences that satisfied all of the requirements outlined above were determined through in silico analysis, we conducted an in vitro experiment to evaluate the performance of these primers and probes in terms of amplification of the target sequences. These oligonucleotides were synthesized by Integrated DNA Technologies (USA). First, we analyzed melting curves from SYBR-based one-step RT-qPCR analysis and did not detect any obvious evidence for the formation of primer-dimers (Fig. S1). Second, we generated calibration curves with synthetic DNA controls (Integrated DNA Technologies, USA) to confirm that the PCR efficiency fell within the range of 85–110% and that the R2 value was greater than 0.99. Third, we determined the limit of detection (LOD) for the RT-qPCR assay developed in this study, using 20 replicates of serial dilutions of synthetic controls, following a previous study (62). We found that the LODs for GI and GII were 1.1 and 5.7 gc/μL, respectively (Fig. S2). Fourth, we evaluated the specificity of the RT-qPCR to our target sequences. This is an important step, as we designed degenerate PCR assays to cover a wide range of norovirus genotypes. We used clinical samples of GI to test the specificity of the GII assays and vice versa, as these samples contain a high concentration of microbes from humans, including norovirus GI (which is expected to have higher sequence similarity to GII samples). We did not detect fluorescence signals from GI clinical samples when we used GII assays, or vice versa (Fig. S3). Furthermore, in silico analysis showed that the GI and GII assays had a 0% probability of detection for GII (n = 561) and GI viral sequences (n = 48), respectively. These results support that our GI and GII assays specifically amplify target viral sequences. We chose the final primers and probes that showed the highest probability of detection among the candidates, satisfying all in silico and in vitro verification steps (Table 1).
RT-qPCR protocols
The SYBR-based RT-qPCR analysis was conducted to detect the formation of primer dimers. The RT-qPCR mixture for SYBR-based RT-qPCR assay included 3 µL of RNA sample, 0.3 µL of 50 µM forward and reverse primer, 1.275 µL of molecular biology grade water (Corning, NY, USA), 5 µL of 2× iTaq universal SYBR green reaction mix, and 0.125 µL of iScript reverse transcriptase from the iTaq Universal SYBR Green One-Step Kit (1725151, Bio-Rad Laboratories, USA). The PCR cocktail was placed in 96-well plates (4306737, Applied Biosystems, USA) and analyzed by an RT-qPCR system (QuantStudio 3, Thermo Fisher Scientific, USA). The RT-qPCR was performed with a thermocycle of 50°C for 10 min and 95°C for 1 min, and then 40 cycles of denaturation at 95°C for 10 s, annealing at 53°C for 30 s, and extension at 60°C for 30 s. Melting curves were analyzed while the temperature increased from 60°C to 95°C. The SYBR signal was normalized to the ROX reference dye. The cycles of quantification (Cq) were determined by QuantStudio Design & Analysis Software (v1.5.1).
The Taqman-based RT-qPCR assays were conducted for genome quantification. The Taqman-based RT-qPCR started by mixing 2.5 µL of RNA sample, 2.5 µL of Taqman Fast Virus 1-step Master Mix (4444432, Applied Biosystems, USA), and 5 µL of primers/probe mixture to achieve final concentrations of 2,000 nM for primers and 1,000 nM for probes. The PCR cocktail was placed in 96-well plates (4306737, Applied Biosystems, USA) and analyzed by an RT-qPCR system (QuantStudio 3, Thermo Fisher Scientific, USA) with a thermal cycle of 5 min at 50°C, 20 s at 95°C followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 53°C for 30 s, and extension at 60°C for 30 s. When previously designed RT-qPCR assays were used, the annealing temperature was adjusted for each RT-qPCR assay as reported in references (Table 1).
For both SYBR- and Taqman-based assays, at least three replicates were analyzed for serial dilution of synthetic DNA (for a standard curve), nuclease-free water (as a negative control), and samples. All positive samples were positive and negative samples were negative in all RT-qPCR analyses. The linear dynamic range for the serial dilutions of synthetic DNA was between 10° and 105 gc/uL. The PCR efficiencies for RT-qPCR were higher than 85% (R2 >0.99). The details for RT-qPCR assays are summarized in Table S1, in accordance with MIQE guidelines (8).
Clinical samples collection and processing
In all, 20 unidentified stool samples collected from norovirus-infected patients were provided by the Illinois Department of Public Health. Sample collection dates and locations are summarized in Table S2. In total, 10 samples were positive for the GI genogroup, and the other 10 samples were for the GII genogroup. Norovirus RNA was extracted using the following procedure. An amount of 100 mg of stool sample was mixed with 900 µL of deionized water. The mixtures were vortexed for 30 s and centrifuged at 17,000 × g for 10 min. Viral RNA was extracted from 140 µL of the supernatant using a QIAamp Viral RNA mini kit (Qiagen, Germany) following the manufacturer’s protocol (66–68). An inhibition test was conducted by adding the Tulane virus (whose host is a rhesus monkey) to each extract, following our previous protocol (69). We found the impact of any possible inhibitors was negligible (Fig. S4). The RNA extracts were kept at −80°C until downstream analysis.
Sanger sequencing
Clinical samples were analyzed by Sanger sequencing to obtain viral sequences. First, we synthesized complementary DNA (cDNA) from the norovirus genomic RNA using the First Strand cDNA Synthesis Kit (New England BioLabs, USA). An amount of 6 μL of RNA samples was mixed with 10 µL of M-MuLV Reaction Mix, 2 µL of M-MuLV Enzyme Mix, and 2 µL of 10 µM of reverse primers. The mixture was incubated at 42°C for 60 min for cDNA synthesis, followed by 80°C for 10 min for enzyme inactivation. The 3.5 mL of cDNA was then mixed with 0.5 µL of Phusion DNA polymerase, 10 µL of 5X Phusion HF buffer, 2.5 µL of 10 µM forward primer, 2.5 µL of 10 µM reverse primer, 1 µL of 10 mM dNTPs, and 30 µL of nuclease-free water (Phusion High-Fidelity PCR Kit, MA, USA). The forward and reverse primer sequences are summarized in Table 4. This 50 µL of PCR cocktail was incubated at 98°C for 30 s for initial denaturation, 40 cycles of denaturation at 98°C for 10 s, annealing at various temperatures for each primer set (Table 1) for 30 s, extension at 72°C for 30 s, and 72°C for 10 min (final extension). The PCR amplicon was purified using a QIAquick PCR Purification Kit (QIAGEN, Germany) following the manufacturer’s protocol, and the PCR amplicon was eluted in 30 µL of nuclease-free water. In addition, the PCR amplicon was further cleaned up by ExoSAP-IT Express PCR Product Cleanup Reagent (Applied Biosystems, MA, USA) following the manufacturer’s procedure. The double-stranded DNA concentration of the amplicon was determined by a Qubit 2.0 fluorometer (Invitrogen, USA). The Core DNA Sequencing Facility at the University of Illinois Urbana-Champaign analyzed the samples through Sanger sequencing. The norovirus genome sequences were finalized after examining the sequencing chromatogram (i.e., dye terminator peaks, the baseline, and the sequence text) with FinchTV (version 1.4.0).
TABLE 4.
Summary of primers for Sanger sequencing
| Sample ID | Primer type | Sequence (5′ to 3′) | Length (bp) | Tm (°C)a | Gc (%) | Amplicon location (size) |
|---|---|---|---|---|---|---|
| GI_Set3b | Forward primer | TCATTTTATGGTGATGATGAAAT | 23 | 48.5 | 26.1 | 4889–5550 (662 bp) |
| Reverse primer | AGGGGTCAATCATATTAACTTG | 22 | 50.1 | 36.4 | ||
| GI_Set9c | Forward primer | CCTTGCACATCTCAGGTGAATA | 22 | 54.7 | 45.5 | 4703–5572 (870 bp) |
| Reverse primer | TGAGGCCCTAACTGCAAATC | 20 | 55.1 | 50.0 | ||
| GII_Set3d | Forward primer | TTCTATGGTGATGATGAGATTGT | 23 | 51.5 | 34.8 | 4612–5243 (632 bp) |
| Reverse primer | CTAATCCAGGGGTCAATTACAT | 22 | 52.0 | 40.9 | ||
| GII_Set9e | Forward primer | CAATAGCACACTGGATCCTAAC | 22 | 53.1 | 45.5 | 4459–5324 (866 bp) |
| Reverse primer | CTAGCCAGATGTGCAAGATAAG | 22 | 53.1 | 45.5 | ||
| GII_Set11f | Forward primer | CCCATTCTCAAAGACCCATACA | 22 | 54.5 | 45.5 | 4865–5693 (829 bp) |
| Reverse primer | TGAGAACTCGGCACGAAAC | 19 | 55.3 | 52.6 | ||
| GII_Set14g | Forward primer | GATTTGAATGGTCTCACATTCTTG | 24 | 52.4 | 37.5 | 4744–5336 (593 bp) |
| Reverse primer | TTCTGGACCTAACTCTAAATCTAAC | 25 | 51.8 | 36.0 |
Melting temperatures were calculated with the default parameter sets of OligoAnalyzer (Integrated DNA Technology), which consider 0.25 μM of oligo concentration and 50 mM of Na+ concentration.
Reference sequence (Genbank ID: MT031988.1).
Reference sequence (Genbank ID: MW305499.1).
Reference sequence (Genbank ID: OP727614.1).
Reference sequence (Genbank ID: MW305576.1).
Reference sequence (Genbank ID: MZ478141.1).
Reference sequence (Genbank ID: OP686904.1).
Sewage sample collection and processing
We collected wastewater samples from a city-scale and a neighborhood-scale sewershed. Specifically, we obtained 20 influent wastewater samples from the Urbana-Champaign Sanitary District (IL, USA), which serves 144,097 people living in Champaign city, Urbana city, and adjacent areas from January 2022 to May 2022. In addition, we collected 20 samples from a manhole receiving wastewater discharged by 1,675 residents from January 2021 to May 2022. All wastewater samples were obtained using an autosampler (Teledyne ISCO, USA), programmed to collect a 1–2 L of composite sample comprised of samples pumped for 24 h. The composite samples were transferred to sterile sampling bags (14–955-001, Fisher Scientific, USA), and 20 mL of 2.5 M MgCl2 was added to the samples (i.e., final MgCl2 concentrations were from 25 to 50 mM) to coagulate solids including virus particles (69, 70). The samples were transported on ice to a laboratory at the University of Illinois Urbana-Champaign within 3 h. Upon arrival at the laboratory, supernatants from each composite sample were discarded. The remaining 35 mL of sewage, in which solid particles were concentrated, was transferred to a 50 mL tube (12–565-271, Fisher Scientific). The sewage samples were centrifuged at 10,000 × g for 30 min (Sorvall RC 6 Plus, Thermo Scientific, USA). Supernatants were discarded, and a portion of the concentrated sludge (100 µL) was transferred to a sterile 1.5 mL tube (1415–2600, USA Scientific, USA). Nucleic acids were extracted from the sludge with a QIAamp Viral RNA mini kit (Qiagen, Germany) following the manufacturer’s procedure. Sewage collection and processing were conducted on the same day, and the RNA samples were stored at −80°C until RT-qPCR analysis. RNA quantification was conducted with a 10-fold dilution of RNA extracts to lower the impact of the inhibitors to a negligible level (69).
Statistical analysis
Mann-Whitney U-test was conducted to compare the nucleotide identity of two viral species (Fig. 1). Linear regression analysis was conducted to compare norovirus RNA concentrations of clinical samples determined by two RT-qPCR assays (Fig. 2). The slope of the linear regression curve was compared to 1. Samples with a studentized residual of less than −1.5 were defined as outliers and excluded from the linear regression curve to evaluate the potential impacts of mismatch between RT-qPCR assays and norovirus sequences (Fig. 2). Statistical analyses were conducted using OriginPro 2023.
ACKNOWLEDGMENTS
We acknowledge funding support from the JUMP-ARCHES program of OSF Healthcare in conjunction with the University of Illinois, the College of Applied Health Sciences, the Grainger College of Engineering, the VinUni Illinois Smart Health Center, and the EPA grant (R840487). This study has not been formally reviewed by EPA. The views expressed in this document are solely those of Professor Thanh H. Nguyen and do not necessarily reflect those of the Agency. EPA does not endorse any products or commercial services mentioned in this publication.
We thank Brad Bennett and Bruce Rabe at the Urbana-Champaign Sanitary District and Haley Turner and Travis Ramme at the Rantoul Wastewater Treatment Plant for providing us with influent wastewater. The authors also acknowledge Kip Stevenson for sampling deployment, and Yuqing Mao, Matthew Robert Loula, Aashna Patra, Kristin Joy Anderson, Mikayla Diedrick, Hubert Lyu, Hamza Elmahi Mohamed, Jad R Karajeh, Runsen Ning, Rui Fu, and Kyukyoung Kim for sewage sampling and processing. We also acknowledge Dr. Awais Vaid for guidance on sampling site selection.
Contributor Information
Chamteut Oh, Email: co14@illinois.edu.
Nicole R. Buan, University of Nebraska-Lincoln, Lincoln, Nebraska, USA
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00331-23.
Two texts, five figures, and ten tables are included.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Nsubuga P, White ME, Thacker SB, Anderson MA, Blount SB, Broome CV, Chiller TM, Espitia V, Imtiaz R, Sosin D, Stroup DF, Tauxe RV, Vijayaraghavan M, Trostle M. 2006. Public health surveillance: a tool for targeting and monitoring interventions, p 997–1015. In Disease control priorities in developing countries. [PubMed] [Google Scholar]
- 2. Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. 2021. CRISPR-based diagnostics. Nat Biomed Eng 5:643–656. doi: 10.1038/s41551-021-00760-7 [DOI] [PubMed] [Google Scholar]
- 3. Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sanjuán R, Domingo-Calap P. 2021. Genetic diversity and evolution of viral populations. Encyclopedia of Virology 53. doi: 10.1016/B978-0-12-809633-8.20958-8 [DOI] [Google Scholar]
- 5. Chhabra P, de Graaf M, Parra GI, Chan MC-W, Green K, Martella V, Wang Q, White PA, Katayama K, Vennema H, Koopmans MPG, Vinjé J. 2019. Updated classification of norovirus genogroups and genotypes. J Gen Virol 100:1393–1406. doi: 10.1099/jgv.0.001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patton JT. 2012. Rotavirus diversity and evolution in the post-vaccine world. Discov Med 13:85–97. [PMC free article] [PubMed] [Google Scholar]
- 7. Baker AT, Greenshields-Watson A, Coughlan L, Davies JA, Uusi-Kerttula H, Cole DK, Rizkallah PJ, Parker AL. 2019. Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions. Nat Commun 10:741. doi: 10.1038/s41467-019-08599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 9. Gutiérrez-Aguirre I, Steyer A, Boben J, Gruden K, Poljsak-Prijatelj M, Ravnikar M. 2008. Sensitive detection of multiple rotavirus genotypes with a single reverse transcription-real-time quantitative PCR assay. J Clin Microbiol 46:2547–2554. doi: 10.1128/JCM.02428-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joshi MS, Deore SG, Walimbe AM, Ranshing SS, Chitambar SD. 2019. Evaluation of different genomic regions of rotavirus A for development of real time PCR. J Virol Methods 266:65–71. doi: 10.1016/j.jviromet.2019.01.017 [DOI] [PubMed] [Google Scholar]
- 11. Andersson M, Lindh M. 2017. Rotavirus genotype shifts among Swedish children and adults—application of a real-time PCR Genotyping. J Clin Virol 96:1–6. doi: 10.1016/j.jcv.2017.09.005 [DOI] [PubMed] [Google Scholar]
- 12. Happi C, Adetifa I, Mbala P, Njouom R, Nakoune E, Happi A, Ndodo N, Ayansola O, Mboowa G, Bedford T, Neher RA, Roemer C, Hodcroft E, Tegally H, O’Toole Á, Rambaut A, Pybus O, Kraemer MUG, Wilkinson E, Isidro J, Borges V, Pinto M, Gomes JP, Freitas L, Resende PC, Lee RTC, Maurer-Stroh S, Baxter C, Lessells R, Ogwell AE, Kebede Y, Tessema SK, de Oliveira T. 2022. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. PLoS Biol 20:e3001769. doi: 10.1371/journal.pbio.3001769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391 [DOI] [PubMed] [Google Scholar]
- 14. Marra MA, Jones SJM, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YSN, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, Vogrig R, Ward D, Watson B, Brunham RC, Krajden M, Petric M, Skowronski DM, Upton C, Roper RL. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404. doi: 10.1126/science.1085953 [DOI] [PubMed] [Google Scholar]
- 15. Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Al Rabeeah AA, Alhakeem RF, Assiri A, Al-Tawfiq JA, Albarrak A, Barry M, Shibl A, Alrabiah FA, Hajjar S, Balkhy HH, Flemban H, Rambaut A, Kellam P, Memish ZA. 2014. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5:e01062-13. doi: 10.1128/mBio.01062-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holmes EC, Dudas G, Rambaut A, Andersen KG. 2016. The evolution of Ebola virus: insights from the 2013–2016 epidemic. Nature 538:193–200. doi: 10.1038/nature19790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chowell G, Nishiura H. 2014. Transmission dynamics and control of Ebola virus disease (EVD): a review. BMC Med 12:196. doi: 10.1186/s12916-014-0196-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hui DS, Memish ZA, Zumla A. 2014. Severe acute respiratory syndrome vs. the Middle East respiratory syndrome. Curr Opin Pulm Med 20:233–241. doi: 10.1097/MCP.0000000000000046 [DOI] [PubMed] [Google Scholar]
- 19. Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, Wang Y, Qian Z, Cui J, Lu J. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 7:1012–1023. doi: 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang S, Xu X, Wei C, Li S, Zhao J, Zheng Y, Liu X, Zeng X, Yuan W, Peng S. 2022. Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J Med Virol 94:310–317. doi: 10.1002/jmv.27331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaw SM, Tai JH, Chen SL, Hsieh CH, Chang SY, Yeh SH, Yang WS, Chen PJ, Wang HY. 2020. The origin and underlying driving forces of the SARS-CoV-2 outbreak. J Biomed Sci 27:73. doi: 10.1186/s12929-020-00665-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu L, Qian Y, Jia L, Dong H, Deng L, Huang H, Zhao L, Zhu R. 2021. Genetic diversity and molecular evolution of human adenovirus serotype 41 strains circulating in Beijing, China, during 2010–2019. Infect Genet Evol 95:105056. doi: 10.1016/j.meegid.2021.105056 [DOI] [PubMed] [Google Scholar]
- 23. Mugosa B, Vujosevic D, Ciccozzi M, Valli MB, Capobianchi MR, Lo Presti A, Cella E, Giovanetti M, Lai A, Angeletti S, Scarpa F, Terzić D, Vratnica Z. 2016. Genetic diversity of the haemagglutinin (HA) of human influenza a (H1N1) virus in Montenegro: focus on its origin and evolution. J Med Virol 88:1905–1913. doi: 10.1002/jmv.24552 [DOI] [PubMed] [Google Scholar]
- 24. Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. 2010. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 27:2431–2436. doi: 10.1093/molbev/msq137 [DOI] [PubMed] [Google Scholar]
- 25. Yu JM, Fu YH, Peng XL, Zheng YP, He JS. 2021. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci Rep 11:12941. doi: 10.1038/s41598-021-92435-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rubio L, Galipienso L, Ferriol I. 2020. Detection of plant viruses and disease management: relevance of genetic diversity and evolution. Front Plant Sci 11:1092. doi: 10.3389/fpls.2020.01092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harper SJ. 2013. Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4:93. doi: 10.3389/fmicb.2013.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arahal DR. 2014. Whole-genome analyses: average nucleotide identity. Methods Microbiol 41:103–122. doi: 10.1016/bs.mim.2014.07.002 [DOI] [Google Scholar]
- 29. Martinez-Hernandez F, Diop A, Garcia-Heredia I, Bobay LM, Martinez-Garcia M. 2022. Unexpected myriad of co-occurring viral strains and species in one of the most abundant and microdiverse viruses on earth. ISME J 16:1025–1035. doi: 10.1038/s41396-021-01150-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamamoto K, Ogasawara N, Yamamoto S, Takano K, Shiraishi T, Sato T, Tsutsumi H, Himi T, Yokota SI. 2018. Evaluation of consistency in quantification of gene copy number by real-time reverse transcription quantitative polymerase chain reaction and virus titer by plaque-forming assay for human respiratory syncytial virus. Microbiol Immunol 62:90–98. doi: 10.1111/1348-0421.12563 [DOI] [PubMed] [Google Scholar]
- 31. Bonroy C, Vankeerberghen A, Boel A, De Beenhouwer H. 2007. Use of a multiplex real-time PCR to study the incidence of human metapneumovirus and human respiratory syncytial virus infections during two winter seasons in a Belgian paediatric hospital. Clin Microbiol Infect 13:504–509. doi: 10.1111/j.1469-0691.2007.01682.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hughes B, Duong D, White BJ, Wigginton KR, Chan EMG, Wolfe MK, Boehm AB. 2022. Respiratory syncytial virus (RSV) RNA in wastewater settled solids reflects RSV clinical positivity rates. Environ Sci Technol Lett 9:173–178. doi: 10.1021/acs.estlett.1c00963 [DOI] [Google Scholar]
- 33. Ding N, Craik SA, Pang X, Lee B, Neumann NF. 2017. Assessing UV inactivation of adenovirus 41 using integrated cell culture real-time qPCR/RT-qPCR. Water Environ Res 89:323–329. doi: 10.2175/106143017X14839994523028 [DOI] [PubMed] [Google Scholar]
- 34. Liu P, Herzegh O, Fernandez M, Hooper S, Shu W, Sobolik J, Porter R, Spivey N, Moe C. 2013. Assessment of human adenovirus removal by qPCR in an advanced water reclamation plant in. J Appl Microbiol 115:310–318. doi: 10.1111/jam.12237 [DOI] [PubMed] [Google Scholar]
- 35. Wolf S, Hewitt J, Greening GE. 2010. Viral multiplex quantitative PCR assays for tracking sources of fecal contamination. Appl Environ Microbiol 76:1388–1394. doi: 10.1128/AEM.02249-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou N, Lv D, Wang S, Lin X, Bi Z, Wang H, Wang P, Zhang H, Tao Z, Hou P, Song Y, Xu A. 2016. Continuous detection and genetic diversity of human rotavirus A in sewage in Eastern China, 2013-2014. Virol J 13:153. doi: 10.1186/s12985-016-0609-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D, Zhang Z, Wu Q, Tian P, Geng H, Xu T, Wang D. 2020. Redesigned duplex RT-qPCR for the detection of GI and GII human noroviruses. Engineering 6:442–448. doi: 10.1016/j.eng.2019.08.018 [DOI] [Google Scholar]
- 38. Loisy F, Atmar RL, Guillon P, Le Cann P, Pommepuy M, Le Guyader FS. 2005. Real-time RT-PCR for norovirus screening in shellfish. J Virol Methods 123:1–7. doi: 10.1016/j.jviromet.2004.08.023 [DOI] [PubMed] [Google Scholar]
- 39. Yu F, Jiang B, Guo X, Hou L, Tian Y, Zhang J, Li Q, Jia L, Yang P, Wang Q, Pang X, Gao Z. 2022. Norovirus outbreaks in China, 2000–2018: a systematic review. Rev Med Virol 32:e2382. doi: 10.1002/rmv.2382 [DOI] [PubMed] [Google Scholar]
- 40. Pecson BM, Darby E, Haas CN, Amha YM, Bartolo M, Danielson R, Dearborn Y, Di Giovanni G, Ferguson C, Fevig S, Gaddis E, Gray D, Lukasik G, Mull B, Olivas L, Olivieri A, Qu Y, SARS-CoV-2 Interlaboratory Consortium . 2021. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ Sci (Camb) 7:504–520. doi: 10.1039/d0ew00946f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woolhouse M, Gaunt E. 2007. Ecological origins of novel human pathogens. Crit Rev Microbiol 33:231–242. doi: 10.1080/10408410701647560 [DOI] [PubMed] [Google Scholar]
- 42. Fields BN, Knipe DM. 2013. Edited by David M. and Howley P. M.. Fields Virology 82 [Google Scholar]
- 43. Wang LF, Eaton BT. 2007. Bats, civets and the emergence of SARS. Curr Top Microbiol Immunol 315:325–344. doi: 10.1007/978-3-540-70962-6_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson KEE, Song T, Greenbaum B, Ghedin E. 2017. Getting the flu: 5 key facts about influenza virus evolution. PLoS Pathog 13:e1006450. doi: 10.1371/journal.ppat.1006450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L, Wang Y, Ye D, Liu Q. 2020. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents 55:105948. doi: 10.1016/j.ijantimicag.2020.105948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thornhill JP, Antinori A, Orkin CM. 2022. Monkeypox virus infection in humans across 16 countries — April–June 2022. N Engl J Med 387:e69. doi: 10.1056/NEJMc2213969 [DOI] [PubMed] [Google Scholar]
- 47. Cleri DJ, Ricketti AJ, Vernaleo JR. 2010. Severe acute respiratory syndrome (SARS). Infect Dis Clin North Am 24:175–202. doi: 10.1016/j.idc.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gerba CP. 2009. Environmentally Transmitted Pathogens. Environmental Microbiology 445–484. [Google Scholar]
- 49. Ji L, Hu G, Xu D, Wu X, Fu Y, Chen L. 2020. Molecular epidemiology and changes in genotype diversity of norovirus infections in acute gastroenteritis patients in Huzhou. J Med Virol 92:3173–3178. doi: 10.1002/jmv.26247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Motoya T, Umezawa M, Saito A, Goto K, Doi I, Fukaya S, Nagata N, Ikeda Y, Okayama K, Aso J, Matsushima Y, Ishioka T, Ryo A, Sasaki N, Katayama K, Kimura H. 2019. Variation of human norovirus GII genotypes detected in Ibaraki, Japan, during 2012-2018. Gut Pathog 11:26. doi: 10.1186/s13099-019-0303-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Misumi M, Nishiura H. 2021. Long-term dynamics of Norovirus transmission in Japan, 2005–2019. PeerJ 9:e11769. doi: 10.7717/peerj.11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jeong MH, Song YH, Ju SY, Kim SH, Kwak HS, An ES. 2021. Surveillance to prevent the spread of norovirus outbreak from asymptomatic food handlers during the Pyeongchang 2018 olympics. J Food Prot 84:1819–1823. doi: 10.4315/JFP-21-136 [DOI] [PubMed] [Google Scholar]
- 53. Lefever S, Pattyn F, Hellemans J, Vandesompele J. 2013. Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clin Chem 59:1470–1480. doi: 10.1373/clinchem.2013.203653 [DOI] [PubMed] [Google Scholar]
- 54. Kamau E, Agoti CN, Lewa CS, Oketch J, Owor BE, Otieno GP, Bett A, Cane PA, Nokes DJ. 2017. Recent sequence variation in probe binding site affected detection of respiratory syncytial virus group B by real-time RT-PCR. J Clin Virol 88:21–25. doi: 10.1016/j.jcv.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brault AC, Fang Y, Dannen M, Anishchenko M, Reisen WK. 2012. A naturally occurring mutation within the probe-binding region compromises a molecular-based West Nile virus surveillance assay for mosquito pools (Diptera: culicidae). J Med Entomol 49:939–941. doi: 10.1603/me11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1–19. doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larminie C, Murdock P, Walhin J-P, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. 2004. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res Mol 122:24–34. doi: 10.1016/j.molbrainres.2003.11.014 [DOI] [PubMed] [Google Scholar]
- 59. Ruiz-Ruiz S, Moreno P, Guerri J, Ambrós S. 2006. The complete nucleotide sequence of a severe stem pitting isolate of citrus tristeza virus from Spain: comparison with isolates from different origins. Arch Virol 151:387–398. doi: 10.1007/s00705-005-0618-6 [DOI] [PubMed] [Google Scholar]
- 60. Zhai L, Dai X, Meng J. 2006. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res 120:57–69. doi: 10.1016/j.virusres.2006.01.013 [DOI] [PubMed] [Google Scholar]
- 61. Amonsin A, Kedkovid R, Puranaveja S, Wongyanin P, Suradhat S, Thanawongnuwech R. 2009. Comparative analysis of complete nucleotide sequence of porcine reproductive and respiratory syndrome virus (PRRSV) isolates in Thailand (US and EU Genotypes). Virol J 6:1–10. doi: 10.1186/1743-422X-6-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oh C, Sashittal P, Zhou A, Wang L, El-Kebir M, Nguyen TH, Elkins CA. 2022. Design of SARS-CoV-2 variant-specific PCR assays considering regional and temporal characteristics. Appl Environ Microbiol 88. doi: 10.1128/aem.02289-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee BE, Pang XL. 2013. New strains of norovirus and the mystery of viral gastroenteritis epidemics. Can Med Assoc J 185:1381–1382. doi: 10.1503/cmaj.130426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Graaf M, van Beek J, Koopmans MPG. 2016. Human norovirus transmission and evolution in a changing world. Nat Rev Microbiol 14:421–433. doi: 10.1038/nrmicro.2016.48 [DOI] [PubMed] [Google Scholar]
- 65. Hugerth LW, Wefer HA, Lundin S, Jakobsson HE, Lindberg M, Rodin S, Engstrand L, Andersson AF. 2014. DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl Environ Microbiol 80:5116–5123. doi: 10.1128/AEM.01403-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tan M, Jin M, Xie H, Duan Z, Jiang X, Fang Z. 2008. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J Med Virol 80:1296–1301. doi: 10.1002/jmv.21200 [DOI] [PubMed] [Google Scholar]
- 67. Richards GP, Watson MA, Meade GK, Hovan GL, Kingsley DH. 2012. Resilience of norovirus GII.4 to freezing and thawing: implications for virus infectivity. Food Environ Virol 4:192–197. doi: 10.1007/s12560-012-9089-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nishimura N, Nakayama H, Yoshizumi S, Miyoshi M, Tonoike H, Shirasaki Y, Kojima K, Ishida S. 2010. Detection of noroviruses in fecal specimens by direct RT-PCR without RNA purification. J Virol Methods 163:282–286. doi: 10.1016/j.jviromet.2009.10.011 [DOI] [PubMed] [Google Scholar]
- 69. Oh C, Zhou A, O’Brien K, Jamal Y, Wennerdahl H, Schmidt AR, Shisler JL, Jutla A, Schmidt AR 4th, Keefer L, Brown WM, Nguyen TH. 2022. Application of neighborhood-scale wastewater-based epidemiology in low COVID-19 incidence situations. Sci Total Environ 852:158448. doi: 10.1016/j.scitotenv.2022.158448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ahmed W, Bertsch PM, Bivins A, Bibby K, Farkas K, Gathercole A, Haramoto E, Gyawali P, Korajkic A, McMinn BR, Mueller JF, Simpson SL, Smith WJM, Symonds EM, Thomas KV, Verhagen R, Kitajima M. 2020. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci Total Environ 739:139960. doi: 10.1016/j.scitotenv.2020.139960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Two texts, five figures, and ten tables are included.