ABSTRACT
Hemipteran insects are well-known for their ancient associations with beneficial bacterial endosymbionts, particularly nutritional symbionts that provide the host with essential nutrients such as amino acids or vitamins lacking in the host’s diet. Therefore, these primary endosymbionts enable the exploitation of nutrient-poor food sources such as plant sap or vertebrate blood. In turn, the strictly host-associated lifestyle strongly impacts the genome evolution of the endosymbionts, resulting in small and degraded genomes. Over time, even the essential nutritional functions can be compromised, leading to the complementation or replacement of an ancient endosymbiont by another, more functionally versatile bacterium. Herein, we provide evidence for a dual primary endosymbiosis in several psyllid species. Using metagenome sequencing, we produced the complete genome sequences of both the primary endosymbiont “Candidatus Carsonella ruddii” and an as yet uncharacterized Enterobacteriaceae bacterium from four species of the genus Cacopsylla. The latter represents a new psyllid-associated endosymbiont clade for which we propose the name “Candidatus Psyllophila symbiotica.” Fluorescent in situ hybridization confirmed the co-localization of both endosymbionts in the bacteriome. The metabolic repertoire of Psyllophila is highly conserved across host species and complements the tryptophan biosynthesis pathway that is incomplete in the co-occurring Carsonella. Unlike co-primary endosymbionts in other insects, the genome of Psyllophila is almost as small as the one of Carsonella, indicating an ancient co-obligate endosymbiosis rather than a recent association to rescue a degrading primary endosymbiont.
IMPORTANCE
Heritable beneficial bacterial endosymbionts have been crucial for the evolutionary success of numerous insects by enabling the exploitation of nutritionally limited food sources. Herein, we describe a previously unknown dual endosymbiosis in the psyllid genus Cacopsylla, consisting of the primary endosymbiont “Candidatus Carsonella ruddii” and a co-occurring Enterobacteriaceae bacterium for which we propose the name “Candidatus Psyllophila symbiotica.” Its localization within the bacteriome and its small genome size confirm that Psyllophila is a co-primary endosymbiont widespread within the genus Cacopsylla. Despite its highly eroded genome, Psyllophila perfectly complements the tryptophan biosynthesis pathway that is incomplete in the co-occurring Carsonella. Moreover, the genome of Psyllophila is almost as small as Carsonella’s, suggesting an ancient dual endosymbiosis that has now reached a precarious stage where any additional gene loss would make the system collapse. Hence, our results shed light on the dynamic interactions of psyllids and their endosymbionts over evolutionary time.
KEYWORDS: psyllids, symbiosis, endosymbionts, comparative genomics
INTRODUCTION
Numerous insects maintain long-lasting associations with heritable bacterial endosymbionts that provide the host with essential nutrients lacking in its diet (1). Plant sap-feeding and blood-feeding insects in particular are well-known to harbor nutrient-providing endosymbionts in specialized cells called bacteriocytes, which may form a tissular structure called a bacteriome (2, 3). These so-called primary endosymbionts are obligatory for host survival and reproduction, as they provide essential amino acids and/or vitamins that the host cannot produce or obtain from its food source (4 – 9). Hence, these bacteria have been crucial for the evolutionary success of numerous insects, enabling the exploitation of nutritionally unbalanced food sources such as vertebrate blood and plant sap.
In turn, the host-associated lifestyle has a strong impact on the genome evolution of the endosymbionts. Their strictly intracellular environment, small effective population size, and frequent bottlenecks due to vertical transmission result in genomic decay through the accumulation of deleterious mutations (Muller’s ratchet) and the loss of genes that are no longer needed (10 – 12). Over evolutionary time, this has produced some of the smallest bacterial genomes known to date (6), enriched in genes involved in the production of nutrients required by the host. However, eventually even these pathways can be degraded, leading to either the complementation or the replacement of the ancient endosymbiont by another, more functionally versatile, bacterium or fungus (13 – 19).
This dynamic can be observed in several plant sap-feeding hemipterans that rely on more than one primary endosymbiont to produce all necessary nutrients. Notably, the Auchenorrhyncha (cicadas, planthoppers, spittlebugs) are well-known for their ancient dual endosymbiotic consortia, where two co-primary endosymbionts jointly produce the complete set of essential nutrients required by the host, resulting in an intricate metabolic interdependence between the different partners (20). Nonetheless, multiple endosymbiont replacements occurred over time to compensate for the extreme genome erosion of the ancient symbionts (6, 16, 21 – 26). A similar pattern occurs in aphids (Sternorrhyncha). While most species harbor a single primary endosymbiont, Buchnera aphidicola, which provides the host with the 10 essential amino acids and the vitamins biotin and riboflavin (27), dual-endosymbiotic systems have evolved repeatedly in multiple aphid lineages to compensate for lost pathways in B. aphidicola (13, 14, 18, 28 – 30).
Similar dual primary endosymbioses may be widespread in psyllids (Hemiptera: Psylloidea), a species-rich group of phloem-feeding jumping plant lice. Like other plant sap-feeding insects, psyllids harbor a bacteriocyte-associated primary endosymbiont (“Candidatus Carsonella ruddii”, hereafter Carsonella), which provides the host with essential amino acids (31 – 34). Carsonella is present in all investigated psyllid species and exhibits strict host-symbiont co-divergence, suggesting a single infection of a common ancestor of all extant psyllids (31, 35, 36). Its genome is extremely streamlined and figures among the smallest bacterial genomes known to date (157–175 Kbp) (32). Due to this extreme genome reduction, some Carsonella strains are no longer able to produce the full complement of essential amino acids, questioning their ability to fulfill their symbiotic function without compensation from host genes or co-occurring symbiotic bacteria (34, 37).
Additional endosymbionts have indeed been observed to co-inhabit the bacteriome with Carsonella in several species (3, 38 – 40). In these cases, Carsonella is located in bacteriocytes surrounding the bacteriome, while a second bacterium occurs in the syncytium at the center of the bacteriome. Importantly, the taxonomy of the co-primary endosymbiont varies depending on the psyllid species. While the syncytium-symbiont (Y-symbiont) of the mulberry psyllid Anomoneura mori is an uncharacterized Enterobacteriaceae bacterium (Gammaproteobacteria) (38) whose symbiotic role is unknown, the citrus psyllid Diaphorina citri harbors “Candidatus Profftella armatura” (Betaproteobacteria). The latter is a defensive and nutritional endosymbiont that produces vitamins, carotenoids, and a polyketide toxin, i.e., metabolites that are not provided by Carsonella (40, 41). In contrast, the psyllid species Ctenarytaina eucalypti and Heteropsylla cubana harbor symbionts closely related to the insect endosymbionts “Ca. Moranella endobia” and Sodalis, whose genomes precisely complement several amino acid biosynthesis pathways missing from the co-occurring Carsonella strains (34). Despite typical hallmarks of vertically transmitted intracellular bacteria, the genomes of both endosymbionts are less reduced (>1 Mbp), suggesting a more recent acquisition relative to Carsonella, presumably to compensate for lost functions in the latter. In addition, numerous psyllid microbiome studies revealed highly abundant but yet uncharacterized Enterobacteriaceae bacteria in diverse species from several psyllid families (42 – 47), suggesting that dual primary endosymbioses may be more widespread in psyllids than previously thought.
Herein, we aim to elucidate the evolutionary and metabolic relationships between psyllids of the genus Cacopsylla (Psyllidae) and their Enterobacteriaceae endosymbionts. Many Cacopsylla species have indeed been shown to harbor highly abundant Enterobacteriaceae endosymbionts that are closely related to the Y-symbiont co-inhabiting the syncytium of the bacteriome in A. mori (43, 46, 47). Furthermore, these symbionts were present in all tested individuals of a given species, suggesting that they may represent co-primary endosymbionts widespread in this genus. In this study, we produced the complete genome sequences of both Carsonella and the Enterobacteriaceae endosymbionts of four Cacopsylla species (C. melanoneura, C. picta, C. pyri, and C. pyricola) known to harbor closely related endosymbionts from our previous metabarcoding studies (46, 47). Fluorescent in situ hybridization confirmed the co-localization of both endosymbionts in the bacteriome. Comparative genomic analyses revealed that the Enterobacteriaceae endosymbionts represent a psyllid-associated clade among other insect endosymbionts. Its genome is almost as small as that of Carsonella and complements the tryptophan biosynthesis pathway that is compromised in the co-occurring Carsonella.
RESULTS
All four Cacopsylla species harbor two endosymbionts with tiny genomes
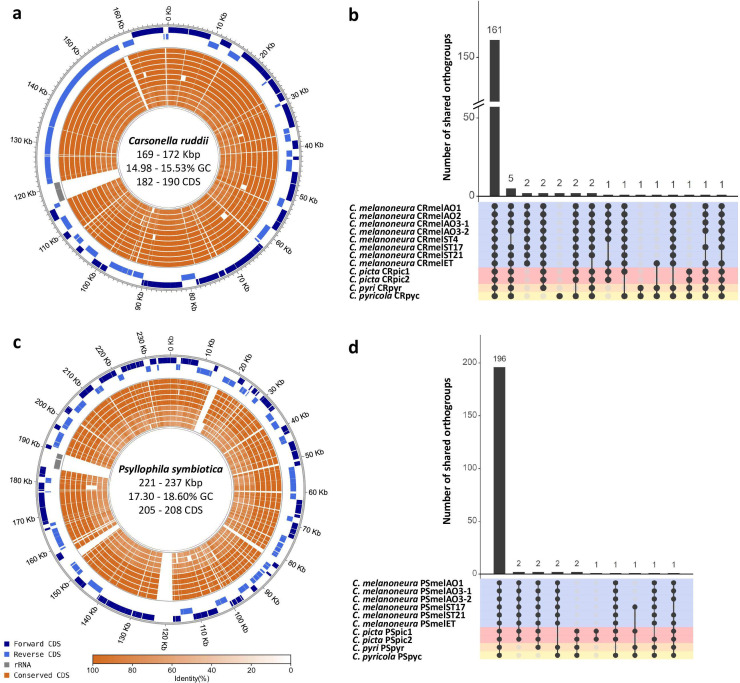
To investigate endosymbiont genetic diversity across different Cacopsylla species and genotypes, 12 insect metagenomes were sequenced. These metagenomes encompassed four different host species: C. melanoneura and C. picta, which complete their development on apple (and also on hawthorn in the case of C. melanoneura) and the pear psyllids C. pyri and C. pyricola. Multiple metagenomes were sequenced for C. melanoneura (N = 8) and C. picta (N = 2), covering different Cytochrome Oxidase I (COI) haplotypes and, in the case of C. melanoneura, different regions of origin (Aosta Valley vs. South Tyrol, Italy), and different host plants (apple vs. hawthorn) (Table 1). As expected, the majority of the Nanopore reads belonged to the insect genome, and only about 5% of the reads (range: 2.94%–9.80%) corresponded to non-host reads. Nonetheless, circular genomes of the primary endosymbiont Carsonella could be assembled from all metagenomes (Table 1) with coverages of 85–408×. Genome size ranged from 169,917 to 171,920 bp with 14.98%–15.53% GC content, similar to previously sequenced Carsonella genomes from other psyllid genera (32, 34, 40, 41). The genomes encoded 182–190 protein-coding genes, 1 ribosomal rRNA operon, and 26–27 tRNAs (Table 1). Synteny and gene content were highly conserved across all genomes, with 161 out of 184 orthogroups (87.5%) shared across all 12 genomes (Fig. 1a and b).
TABLE 1.
Properties of the complete endosymbiont genomes obtained in this study g
| Host species | Cacopsylla melanoneura | Cacopsylla picta | Cacopsylla pyricola | Cacopsylla pyri | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COI haplotype | mel01 a | mel01 a | mel01 a | mel01 a | mel01 a | mel01 a | mel21 b | mel09 c | pic18 d | pic01 e | pyc01 f | ND |
| Origin | Aosta Valley (IT) | Aosta Valley (IT) | Aosta Valley (IT) | Aosta Valley (IT) | South Tyrol (IT) | South Tyrol (IT) | South Tyrol (IT) | South Tyrol (IT) | Trentino (IT) | Trentino (IT) | Starý Lískovec (CZ) | Litenčice (CZ) |
| Host plant | Apple | Apple | Hawthorn | Hawthorn | Apple | Apple | Apple | Apple | Apple | Apple | Pear | Pear |
| Sequencing method | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Nanopore + Illumina | Illumina only |
| Carsonella strain | CRmelAO1 | CRmelAO2 | CRmelAO3-1 | CRmelAO3-2 | CRmelET | CRmelST4 | CRmelST17 | CRmelST21 | CRpic1 | CRpic2 | CRpyc | CRpyr |
| Length (bp) | 169,165 | 169,273 | 169,079 | 169,081 | 169,051 | 169,046 | 169,064 | 169,056 | 171,569 | 171,920 | 168,917 | 169,399 |
| %GC | 15.52 | 15.53 | 15.52 | 15.52 | 15.52 | 15.52 | 15.52 | 15.52 | 15.42 | 15.40 | 14.98 | 15.19 |
| Genes | 216 | 216 | 215 | 215 | 215 | 216 | 215 | 214 | 212 | 211 | 220 | 214 |
| CDS | 186 | 186 | 185 | 185 | 185 | 183 | 185 | 184 | 183 | 182 | 190 | 184 |
| Pseudogenes | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| rRNA | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| tRNA | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 26 | 26 | 27 | 27 |
| Psyllophila strain | PSmelAO1 | PSmelAO3-1 | PSmelAO3-2 | PSmelET | PSmelST17 | PSmelST21 | PSpic1 | PSpic2 | PSpyc | PSpyr | ||
| Length (bp) | 237,114 | 236,266 | 235,918 | 231,767 | 236,072 | 235,744 | 225,730 | 225,865 | 222,755 | 221,413 | ||
| %GC | 18.26 | 18.29 | 18.33 | 18.60 | 18.32 | 18.34 | 17.80 | 17.79 | 17.36 | 17.30 | ||
| Genes | 239 | 240 | 239 | 239 | 242 | 243 | 240 | 240 | 241 | 240 | ||
| CDS | 205 | 206 | 205 | 205 | 207 | 208 | 207 | 207 | 207 | 205 | ||
| Pseudogenes | 2 | 2 | 2 | 2 | 3 | 3 | 1 | 1 | 2 | 3 | ||
| rRNA | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | ||
| tRNA | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | 27 | ||
| ncRNA | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||
Identical to accession no. KM206163.
Accession no. OQ631077.
Identical to accession no. KM206167.
Accession no. OQ631078.
Identical to accession no. KM206174.
Accession no. OQ631079.
IT, Italy; CZ, Czech Republic; ND, not determined. Empty columns for Psyllophila indicate that the genome could not be assembled due to insufficient coverage.
FIG 1.
Endosymbiont genomes are highly conserved across Cacopsylla host species. (a and c) Circular genome plots of the 12 Carsonella genomes (a) and the 10 Psyllophila genomes (c) produced in this study. The three outer-most circles represent forward coding sequences (CDS), reverse CDS, and the ribosomal RNA operon of a reference genome (CRmelAO1 and PSmelAO1, respectively). The inner circles represent the conserved genes in all other genomes of the same taxon (the order is identical to b and d), with the shading indicating the degree of sequence similarity compared to the reference genome. (b and d) Intersection plots showing the number of shared orthogroups across all Carsonella (b) and Psyllophila (d) genomes. The matrix lines are colored according to host species.
In addition to Carsonella, a second circular genome could be assembled from 10 out of the 12 metagenomes (Table 1) with coverages of 19–431×. These genomes belonged to the uncharacterized Enterobacteriaceae endosymbiont previously identified through 16S rRNA gene metabarcoding (46, 47). Contigs of this symbiont were also present in the two remaining metagenomes (both from C. melanoneura), but the coverage was insufficient to assemble complete genomes. The 10 complete chromosomes of the Enterobacteriaceae endosymbiont ranged from 221,413 bp in C. pyri to 237,114 bp in a strain from C. melanoneura from Aosta Valley (strain PSmelAO1; Table 1). GC content varied from 17.30% to 18.60%. Despite the variations in genome size, synteny and gene content were highly conserved across all Enterobacteriaceae genomes (Fig. 1c and d). They contained 205–208 protein-coding genes, 1–3 pseudogenes, 1 ribosomal rRNA operon, 27 tRNAs, and 2 ncRNAs (Table 1). Moreover, 196 out of 209 orthogroups (93.78%) were shared across all 10 genomes (Fig. 1d), indicating that the functional repertoire is highly similar across all four host species. Taken together, all four Cacopsylla species harbor two endosymbionts with typical hallmarks of a long intracellular symbiotic lifestyle, such as extremely small genomes and low GC content.
The Enterobacteriaceae symbionts represent a new clade of insect endosymbionts
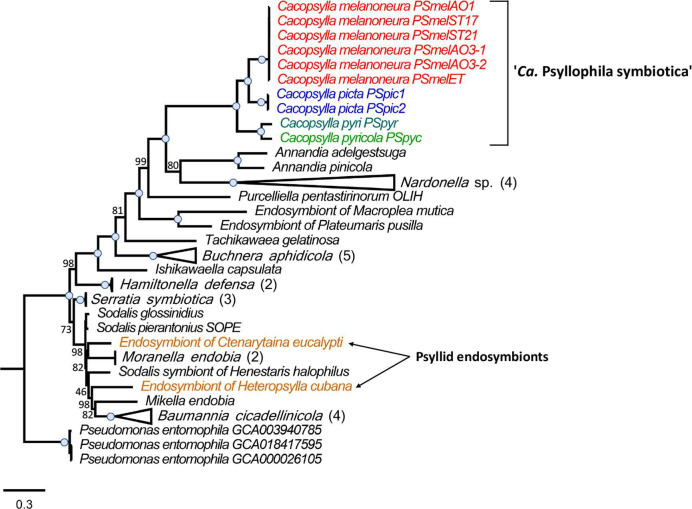
To determine the phylogenetic position of the newly sequenced psyllid endosymbionts, we performed a maximum-likelihood phylogenomic analysis based on 67 single-copy genes present in 46 genomes, namely, the 10 Enterobacteriaceae endosymbionts of Cacopsylla spp., 33 insect endosymbionts from Gammaproteobacteria, and 3 Pseudomonas entomophila strains as outgroups (Fig. 2). The insect endosymbionts included the two previously sequenced endosymbionts of the psyllid species C. eucalypti and H. cubana, as well as both obligate and facultative endosymbionts of diverse hemipterans (aphids, adelgids, leafhoppers, mealybugs, and stinkbugs), beetles (reef beetles and weevils), and the tsetse fly (Table S1). Interestingly, the Enterobacteriaceae endosymbionts of Cacopsylla spp. were not closely related to the previously sequenced endosymbionts of the psyllids C. eucalypti and H. cubana (34) (Fig. 2). Instead, they formed a clade with full bootstrap support that was most closely related to “Ca. Annandia adelgestsuga” and “Ca. Annandia pinicola,” nutritional endosymbionts of adelgids (48) and “Ca. Nardonella sp.,” and ancient endosymbionts of weevils (49) (Fig. 2). Hence, the Enterobacteriaceae endosymbionts of Cacopsylla spp. represent a new psyllid-associated clade of insect endosymbionts for which we propose the name “Candidatus Psyllophila symbiotica” (hereafter Psyllophila).
FIG 2.
The Enterobacteriaceae symbiont represents a new psyllid-associated genus. Maximum-likelihood tree based on the concatenated amino acid sequence alignment of 67 single-copy orthologous genes from 46 genomes, namely, the 10 Enterobacteriaceae endosymbionts of Cacopsylla spp., 33 insect endosymbionts from Gammaproteobacteria, and 3 Pseudomonas entomophila strains as outgroups. The Enterobacteriaceae endosymbionts of Cacopsylla spp. are color-coded based on host species. Branch support is based on 1,000 bootstrap iterations. Blue dots on branches indicate full bootstrap support.
Both Cacopsylla endosymbionts are localized in the bacteriome
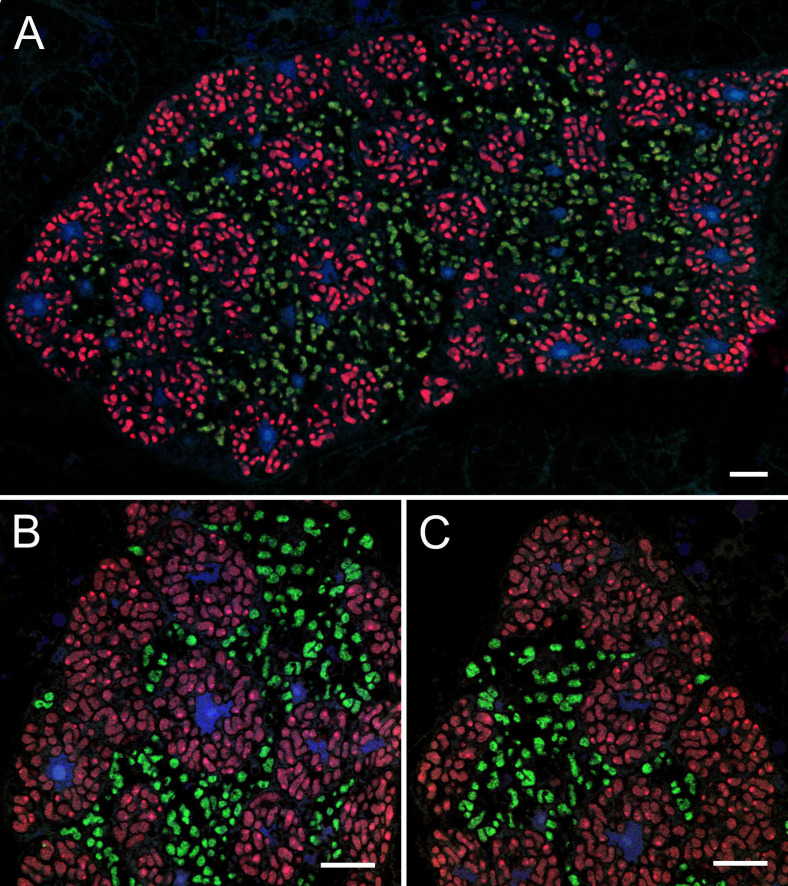
Fluorescence in situ hybridization with Carsonella- and Psyllophila-specific probes revealed that adults of all Cacopsylla species exhibit the same pattern of endosymbiont co-localization in the same bacteriome (Fig. 3). The bacteriomes are large, paired organs localized in the insect’s abdomen. A single bacteriome contains two distinct parts: central and peripheral. Uninucleated bacteriocytes filled with Carsonella are located in the peripheral zone of the bacteriome (Fig. 3), whereas the central part is occupied by a multinucleated syncytium filled with Psyllophila cells as well as some bacteriocytes containing Carsonella (Fig. 3).
FIG 3.
Both Cacopsylla endosymbionts are localized in the bacteriome. Fluorescent in situ hybridization of Carsonella (Cy3, red) and Psyllophila (Cy5, green) symbionts in the bacteriomes of Cacopsylla pyri (a and b) and C. melanoneura (c). Blue represents DAPI. Scale bar = 10 µm.
Metabolic complementarity between Carsonella and Psyllophila
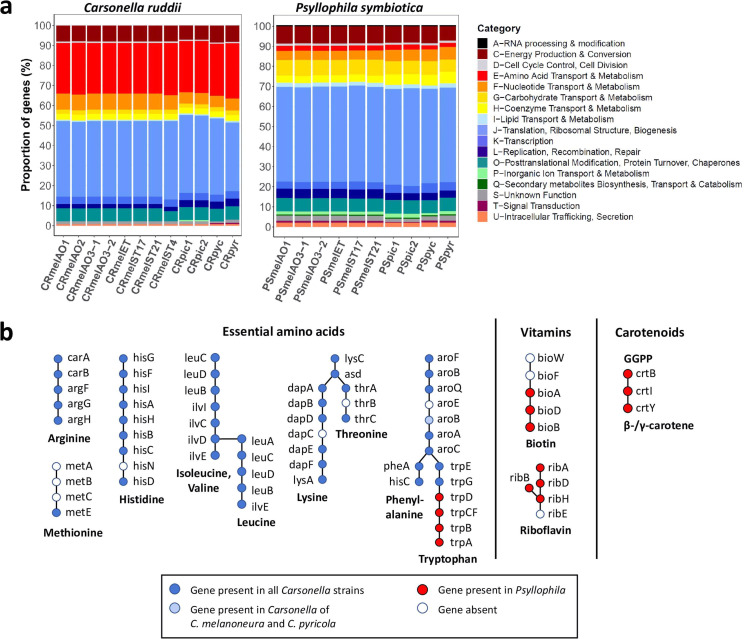
The Cluster of Orthologous Gene (COG) category “amino acid transport and metabolism” was enriched in all sequenced Carsonella genomes (Fig. 4a), in line with its role as a nutritional symbiont. Indeed, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation, the biosynthesis pathways for 8 of the 10 essential amino acids are complete or almost complete in all 12 Carsonella strains from the 4 Cacopsylla species (Fig. 4b). Most of the missing functions (hisN in the histidine pathway, dapC in the lysine pathway, thrB in the threonine pathway, and aroE in the Shikimate pathway, Fig. 4b) are also missing in all previously sequenced Carsonella genomes (Tables S2 and S3). The same applies to the methionine biosynthesis pathway, for which only the last reaction (metE) is present in all sequenced Carsonella genomes from this and previous studies (Fig. 4b; Table S3). The only difference between the Cacopsylla-associated Carsonella strains was the absence of aroB in the stains from C. picta and C. pyri, whereas this gene is present in all Carsonella strains from C. melanoneura and C. pyricola (Fig. 4b; Table S3). Interestingly, the tryptophan biosynthesis pathway was incomplete in all 12 Carsonella strains from Cacopsylla spp., in that only trpE and trpG were present, whereas the rest of the pathway was missing (Fig. 4b).
FIG 4.
Metabolic complementarity between Carsonella and Psyllophila. (a) COG functional categories for the Carsonella and Psyllophila genomes show different proportions of genes involved in amino acid transport and metabolism (red). (b) Schematic representation of the metabolic complementarity between the two symbionts for the biosynthesis of essential amino acids, vitamins, and carotenoids. Genes present in Carsonella genomes are shown in blue, and genes present in Psyllophila are shown in red.
In contrast to Carsonella, the genomes of Psyllophila have lost almost all genes involved in amino acid synthesis (Fig. 4a). Only four genes were retained, and these precisely complement the incomplete tryptophan biosynthesis pathway in Carsonella, namely, trpD, trpCF, trpB, and trpA (Fig. 4b). The four genes were arranged consecutively in the genomes. In addition, all Psyllophila genomes encoded partial biosynthesis pathways for the vitamins biotin (bioA, bioB, bioD) and riboflavin (ribA, ribB, ribD, ribH), as well as all necessary genes for the biosynthesis of carotenoids (crtB, crtI, crtY) (Fig. 4b).
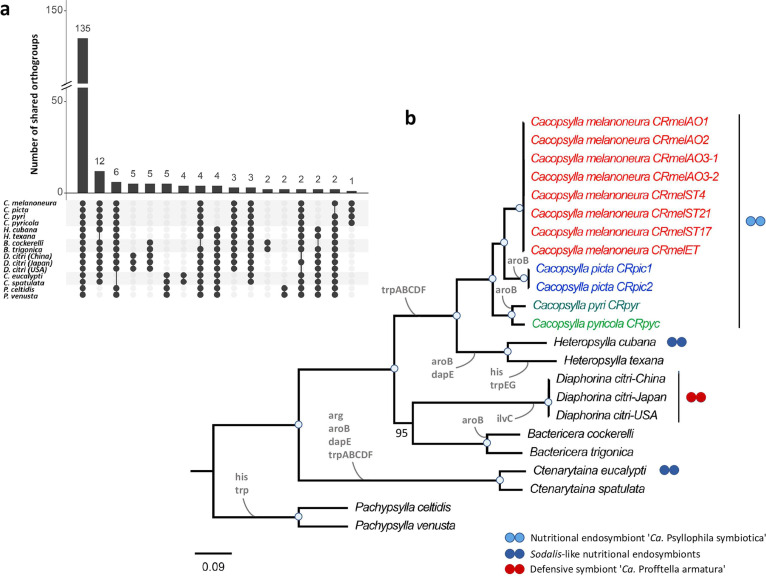
Repeated gene losses throughout Carsonella evolution
Apart from the Carsonella genomes presented herein, complete genome sequences are available for 11 Carsonella strains from 9 psyllid species representing 5 genera and 3 families (Aphalaridae, Psyllidae, and Triozidae) (Table S2). The functional repertoire of these genomes is quite conserved, since 135 out of 197 orthogroups (68.5%) were shared across all 23 genomes and specific orthogroups occurring only in strains from particular host species or genera were rare (14/197) (Fig. 5a). In contrast, host lineage-specific losses of orthogroups were more common. For instance, 12 orthogroups were specifically absent from the 3 Carsonella strains from Heteropsylla texana, Pachypsylla celtidis, and Pachypsylla venusta (Fig. 5a). Similarly, six orthogroups were specifically absent from the Carsonella strains from Ctenarytaina spp., four orthogroups were absent from strains from Cacopsylla spp., and three orthogroups were absent from strains from Pachypsylla spp. (Fig. 5a). These differences are also reflected in repeated losses of genes or entirely new pathways involved in essential amino acid biosynthesis across the Carsonella phylogeny (Fig. 5b). Notably, the tryptophan pathway has been lost at least three times independently, as it is incomplete or missing in all Carsonella strains associated with the genera Cacopsylla and Heteropsylla (Psyllidae), as well as Ctenarytaina and Pachypsylla (Aphalaridae) (Fig. 5b; Table S3). In contrast, this pathway is complete in the Carsonella strains from Bactericera spp. (Triozidae) and Diaphorina citri (Liviidae) (Fig. 5b; Table S3). Other repeatedly lost functions include the histidine biosynthesis pathway as well as the genes aroB and dapE, implicated in the Shikimate and lysine pathways, respectively (Fig. 5b; Table S3). Concomitantly, co-primary endosymbionts complementing the missing amino acid biosynthesis pathways have been identified in several species, i.e., complementing tryptophan in Cacopsylla spp. (this study) and H. cubana (34) and both tryptophan and arginine in C. eucalypti (34) (Fig. 5b).
FIG 5.
Repeated gene losses throughout Carsonella evolution. (a) Intersection plot showing the distribution of orthogroups across 23 Carsonella genomes depending on host species. (b) Maximum-likelihood tree based on the concatenated amino acid sequence alignment of 119 single-copy orthologous genes present in all 23 Carsonella genomes. The Carsonella strains of Cacopsylla spp. are color-coded based on host species. Branch support is based on 1,000 bootstrap iterations. Blue dots on branches indicate full bootstrap support. Losses of genes or pathways involved in the biosynthesis of essential amino acids are indicated on the branches (see Table S3 for a detailed list of genes identified based on KEGG pathway annotation). The presence of known co-primary endosymbionts is indicated using blue dots for amino acid-providing nutritional co-primary endosymbionts and red dots for the defensive and nutritional endosymbiont “Ca. Profftella armatura”.
DISCUSSION
Herein, we present the complete genome sequences of both Carsonella and the uncharacterized Enterobacteriaceae endosymbionts of four Cacopsylla species from different host plants. The Enterobacteriaceae endosymbionts represent a psyllid-associated clade among other insect endosymbionts, for which we propose the name “Ca. Psyllophila symbiotica”. Both endosymbionts co-occur within the bacteriome, exhibiting the same co-localization pattern (Carsonella in peripheral bacteriocytes, Psyllophila in the central syncytium) as for other dual endosymbioses in D. citri and A. mori (38 – 40). In combination with a small and AT-rich genome, the bacteriome localization confirms that Psyllophila is a co-primary endosymbiont widespread within the genus Cacopsylla. Interestingly, unlike co-occurring endosymbionts in other psyllid species (34, 40, 41), the Psyllophila genome is almost as small as the genome of Carsonella, indicating an ancient dual endosymbiosis rather than a recent acquisition of a more versatile symbiont to rescue a degrading primary endosymbiont.
Despite having a tiny and functionally limited genome, Psyllophila has retained the necessary genes to complement the tryptophan biosynthesis pathway that is compromised in the co-occurring Carsonella. This appears to be a recurring theme across Carsonella evolution since the tryptophan pathway is the most frequently lost amino acid biosynthesis pathway based on the genomes available to date. Specifically, this pathway has been lost multiple times independently, namely, in the Carsonella strains associated with species from the genera Pachypsylla and Ctenarytaina (Aphalaridae) and the psyllid lineage leading to both Heteropsylla and Cacopsylla (Psyllidae). Apart from tryptophan, the arginine and histidine pathways have also been lost in specific Carsonella strains, albeit less frequently (34). However, the currently available Carsonella genomes cover only a few branches on the psyllid tree of life; therefore, we do not know how frequent (or rare) these gene losses actually are. Concomitantly, co-primary endosymbionts complementing the missing amino acid biosynthesis pathways have been identified in several species, namely, Psyllophila and a Sodalis-like symbiont complementing tryptophan in Cacopsylla spp. (this study) and H. cubana (34), respectively, and another Sodalis-like symbiont complementing both tryptophan and arginine in C. eucalypti (34). Intriguing cases in this context are the psyllid species H. texana, P. celtidis, and P. venusta, whose Carsonella strains have lost both the histidine and tryptophan pathways, but no co-primary endosymbionts have been observed to date (34). Possible alternative scenarios are that the missing genes are encoded by the host, e.g., after horizontal transfers of bacterial genes to the host genome, or that the amino acids in question are present in sufficient quantities in the phloem sap of the psyllid’s host plant. Although numerous genes of bacterial origin have indeed been identified in the genome of P. venusta, they do not restore the missing amino acid pathways (37).
Based on its genome sequence, Psyllophila not only rescues tryptophan biosynthesis but also encodes partial biosynthesis pathways for the vitamins biotin and riboflavin, as well as all necessary genes for the synthesis of carotenoids, pigments that may protect against oxidative damage to DNA (50). This represents a striking convergence with “Ca. Profftella armatura,” the co-primary endosymbiont in several Diaphorina species, which has an almost identical gene set for these pathways as Psyllophila (41). As in Psyllophila, the last step in the riboflavin pathway is missing in “Ca. Profftella armatura,” but the relevant gene has been detected in the genomes of the psyllids D. citri and P. venusta, likely due to a horizontal transfer from an unknown bacterium (37). Likewise, we identified a similar riboflavin synthase gene encoded in the genomes of all four Cacopsylla species via blast searches of the D. citri gene against preliminary assemblies of the insect genomes from our metagenomic data sets. Hence, it is likely that riboflavin can be jointly synthesized by Psyllophila and its psyllid hosts, just like in the symbiosis of D. citri and Profftella. In any case, the functional similarity between two distantly related psyllid endosymbionts highlights the importance of these metabolites for the psyllid hosts and/or the endosymbionts.
Taken together, our data shed light on the dynamic interactions of psyllids and their endosymbionts over evolutionary time. Notably, the tiny and highly eroded genome of Psyllophila indicates a long-lasting dual endosymbiosis of Carsonella and Psyllophila within the genus Cacopsylla. Based on fossil records, extant psyllids (superfamily Psylloidea) evolved relatively recently, about 48 mya (51). The Psyllidae family is likely younger (52), with some fossils of Cacopsylla spp. estimated to date back 16–23 mya, but this has not been formally validated. In addition, recent microbiome studies confirm the presence of other Enterobacteriaceae symbionts in psyllid species from other families, notably Triozidae and Aphalaridae (42 – 44, 53), suggesting that related co-primary endosymbionts may be more widespread across psyllid families than currently thought. However, in the absence of genomic data for these Enterobacteriaceae symbionts, it is unknown whether they belong to the same genus or share a common ancestor with Psyllophila.
In any case, the Carsonella-Psyllophila dual endosymbiosis in Cacopsylla spp. has likely reached a highly precarious state since no functional redundancy exists between the two endosymbionts and any additional gene loss would destabilize the symbiotic system. Considering the diversity of predominant psyllid-associated bacteria revealed by previous studies (35, 42 – 47, 54), it is likely that the ancient endosymbiont Psyllophila has already been replaced by younger symbionts in some psyllid lineages. For instance, this may have been the case in H. cubana and C. eucalypti, which harbor more recently acquired co-primary endosymbionts with larger genomes (34). It is tempting to speculate that species that are not known to harbor co-primary endosymbionts today (e.g., P. venusta) may have harbored a similar dual endosymbiosis in the past, but the co-symbiont was lost without replacement, maybe because its functions were no longer required after a change in ecological conditions (e.g., change of host plant, evolution of gall-forming behavior). This could also explain why the Carsonella strains in these species have lost similar genes and pathways as the strains existing in dual endosymbiotic systems today.
This raises the question of whether these pathways were lost before or after the establishment of the dual primary endosymbiosis. According to the Black Queen Theory on the evolution of dependencies within bacterial communities (55), it is advantageous for bacteria to lose costly metabolic functions (i.e., to streamline their genomes), as long as another species within the community still produces these metabolites as “common goods.” Applying this concept to a community with two partners would imply that any essential pathway can be lost in only one of them but has to be retained in the other to maintain all essential functions in the system. Hence, in most psyllid dual endosymbioses, Carsonella may have lost the tryptophan pathway since it was encoded by its symbiotic partner. In turn, the co-primary endosymbionts lost all other genes involved in amino acid biosynthesis since these were maintained in Carsonella, thus establishing the existing metabolic complementarities in different psyllids. An exception to this occurs in Diaphorina spp., whose co-primary endosymbiont Profftella is very similar to Psyllophila in its metabolic repertoire except for the tryptophan pathway, which is complete in the co-occurring Carsonella strains (41). It is therefore conceivable that an ancestral co-primary endosymbiont was replaced by Profftella before Carsonella lost the tryptophan pathway in these species. However, since only a few psyllid endosymbionts have been characterized at the genomic level, more studies across the psyllid tree of life will be necessary to obtain a more complete picture of the evolutionary dynamics of psyllids and their primary endosymbionts.
MATERIALS AND METHODS
Psyllid samples
Genomic data were obtained from four psyllid species: the apple psyllids Cacopsylla melanoneura and C. picta as well as the pear psyllids C. pyri and C. pyricola. All four species are vectors of plant pathogens, namely, “Ca. Phytoplasma mali” and “Ca. Phytoplasma pyri,” respectively, causing apple proliferation and pear decline (56, 57). Remigrants (i.e., adults that return to their host plants for reproduction after overwintering on shelter plants) of C. melanoneura were captured in various apple orchards in two Italian regions (Aosta Valley and South Tyrol) in March 2020 and March 2021. Additional C. melanoneura specimens were sampled on hawthorn (Crataegus sp.) in a single location (Aosta Valley) in March 2021. Remigrants of C. picta were captured from apple orchards in Trentino (Italy) in April 2021. Adults in C. pyri and C. pyricola were collected in pear orchards in Litenčice (Czech Republic) in December 2019 and in Starý Lískovec (Czech Republic) in July 2020, respectively. Sampling was done using the beating tray method.
Since psyllids are too small to obtain sufficient DNA for long-read sequencing from a single individual, several specimens need to be pooled, which introduces genetic variation that can hinder genome assembly. We used two different strategies to reduce the genetic variation among the pooled individuals, depending on the host plant of the different species. For C. melanoneura and C. picta collected on apple trees, we applied the same experimental design as in reference (56). In the green house, the field-caught adults were sorted into mating couples, and each couple was caged on a branch of an apple tree (cultivar Golden Delicious) using nylon nets. Once the offspring of the mating couples had reached adulthood, all newly emerged siblings were collected and stored at −20°C. Since the primary endosymbionts are vertically transmitted from mother to offspring, all siblings harbor genetically identical endosymbionts and can therefore be pooled without introducing genetic variation for the endosymbionts. In addition, the Cytochrome Oxidase I (COI) haplotype was determined for two individuals per sibling group according to reference (58), to determine the genetic diversity among the different populations and mating couples. For all psyllids that do not develop on apples (C. melanoneura from hawthorn and the pear psyllids C. pyri and C. pyricola), adults collected in the field were immediately stored at −20°C. Subsequently, the COI haplotype was determined for numerous individuals of each species in order to select individuals with identical COI haplotype for pooling.
DNA extraction
For each sibling group of the apple psyllids C. melanoneura and C. picta selected for long-read metagenome sequencing, DNA was extracted from two pools, each containing four to six whole females. For the field-caught C. melanoneura from hawthorn and C. pyricola from pears, DNA was first extracted from individual females and subjected to COI haplotype determination as outlined above. Subsequently, two pools, each combining the DNA extracts of five females with identical haplotypes, were established for each species. Only females were used since they are larger and hence provide more DNA, and we reasoned that their endosymbiont titers may be higher since the endosymbionts are harbored in two tissues, the bacteriome and the ovaries. DNA extraction was performed using a modified protocol of the PureGene Tissue Kit (Qiagen, Venlo, Netherlands). Whole insects were ground in 100 µL of cell lysis solution and 5 µL of proteinase K solution and incubated at 56°C for 3 h, followed by an incubation with 1.5 µL of RNase A at 37°C for 30 min. Subsequently, proteins were precipitated by adding 35 µL of protein precipitation solution. DNA was then extracted with 1 vol of chloroform/isoamyl alcohol (24:1 vol/vol) and precipitated in 1 vol of isopropanol after overnight incubation at −20°C. The DNA pellet was resuspended in 40 µL of sterile water and incubated at 65°C for 1 h to increase DNA rehydration. For C. pyri, DNA was extracted from a single female using the QIAamp DNA Micro Kit (Qiagen) according to the manufacturer’s instructions.
Metagenome sequencing and assembly
Long-read metagenome sequencing using an Oxford Nanopore-Illumina hybrid approach was performed for C. melanoneura, C. picta, and C. pyricola. For each sample, one pool was used for long-read sequencing on the MinION (Oxford Nanopore Technologies, UK), and the second pool was used for 2× 150 bp paired-end sequencing on an Illumina NovaSeq (Macrogen Europe, Netherlands). About 1.5 µg of DNA was used for library preparation using the Oxford Nanopore Ligation Sequencing Kit SQK-LSK 109 (Oxford Nanopore Technologies, UK). Each library was sequenced on an entire R9.4 flowcell for 43–72 h, depending on pore activity. Basecalling was done using Guppy v5.0.11 (Oxford Nanopore Technologies, UK) in high-accuracy mode. Low-quality (<Q7) and short (<500 bp) reads were discarded, and host reads were removed via mapping against a genome scaffold of C. melanoneura (J.M. Howie & O. Rota-Stabelli, unpublished data) using Minimap2 v2.15 (59). The remaining non-host reads ≥500 bp were assembled using Flye v2.9 (60) with the metagenome option. Contigs belonging to the endosymbionts were identified using blast (61). Reads were mapped back onto the endosymbiont contigs using Minimap2 v2.15, and all mapped reads were assembled again with Flye v2.9 using the same parameters. This produced two circular genomes for most data sets. These genomes were first polished with Nanopore reads using Medaka v1.5.0 (https://github.com/nanoporetech/medaka) and subsequently with Illumina reads using several iterations of Polca, a genome polisher integrated in the MaSuRCa toolkit v4.0.7 (62), until no more errors were found. It is important to note that the two endosymbiont genomes need to be polished together to avoid the introduction of errors in highly conserved regions (e.g., the ribosomal RNA operon) of the endosymbiont genome with lower coverage. In rare cases, two rounds of Flye assemblies did not produce circular endosymbiont genomes. Two of these genomes (CRmelAO2 and PSmelET) could be finished using alternative assembly approaches: (i) assembly with Canu v2.1.1 (63), polishing with Medaka and Polca as outlined above, followed by scaffolding and gap-closing with Redundans v0.14 (64) and (ii) Nanopore and Illumina reads mapping onto the complete endosymbiont genomes were assembled together using SPAdes v3.15.1 (65). Genome coverage was estimated by mapping the Nanopore reads onto the finished genomes during the polishing step with Medaka.
The metagenome of C. pyri was assembled from 42 million 2× 250 paired-end reads from a single female sequenced on an Illumina NovaSeq (University of Illinois, Urbana-Champaign, IL, USA). The metagenome was assembled using SPAdes v3.15.1 (65) with the meta option and default kmers. Endosymbiont contigs were identified based on coverage, which initially produced two contigs for Carsonella and three contigs for the Enterobacteriaceae symbiont. These contigs were ordered based on the complete genomes obtained using long-read sequencing and closed after scaffolding and gap-closing with Redundans v0.14 (64). The completeness of all genomes was assessed using BUSCO (gammaproteobacteria_odb10 data set) (66).
Functional genome analyses
All complete endosymbiont genomes were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) version 2021-07-01.build5508 (67). Circular plots of conserved protein-coding genes were produced using MGCplotter (https://github.com/moshi4/MGCplotter). For each endosymbiont (i.e., Carsonella ruddii and Psyllophila symbiotica), all protein-coding genes were assigned to orthogroups using Orthofinder v2.5.2 (68), and shared orthogroups were plotted using the package UpsetR (69). Cluster of Orthologous Gene (COG) categories were determined using eggNOG-mapper v2.1.7 (70), and KEGG pathway annotations were obtained using BlastKOALA v2.2 (71).
Phylogenomics
Orthofinder v2.5.2 (68) was used to identify single-copy orthologous genes shared (i) between all available Carsonella genomes and (ii) between the Enterobacteriaceae endosymbionts of Cacopsylla spp., 33 other nutritional endosymbionts from Gammaproteobacteria, and 3 Pseudomonas entomophila strains as outgroups. The amino acid sequences of each conserved gene were aligned using Muscle v3.1.31 (72), and the alignments were concatenated into a partitioned supermatrix using the script geneStitcher.py (https://github.com/ballesterus/Utensils/blob/master/geneStitcher.py). IQ-TREE v1.6.1 (73) was used to predict the optimal amino acid substitution model for each gene partition (74, 75) and to produce a maximum-likelihood phylogenetic tree with 1,000 bootstrap iterations. The tree was visualized in FigTree v1.4.4 (https://github.com/rambaut/figtree).
Fluorescence in situ hybridization
Fluorescence in situ hybridization was conducted on adult insects with symbiont-specific probes complementary to their 16S rRNA gene sequences (Carsonella: Probe Carso107 “Cy3-ATACTAAAAGGCAGATTCTTG,” Psyllophila: Probe Psyllo118 “Cy5-TCCATTGAGTAGTTTCCCAG”). For Carsonella, two helper probes were used to increase their signals (helperFCarso107: “AGCGAACGGGTGAGTAATATG,” helperRCarso107: “ACATTTCTATATACTTTCCA”).
Insects preserved in ethanol were rehydrated and then postfixed in 4% paraformaldehyde for 2 h at room temperature. Next, the specimens were dehydrated again by incubation in increased concentrations of ethanol and acetone, embedded in Technovit 8100 resin (Kulzer, Wehrheim, Germany), and cut into semithin sections (1 µm). The sections were then incubated overnight at room temperature in hybridization buffer containing the specific probes at a final concentration of 100 nM. After hybridization, the slides were washed three times in PBS, dried, covered with ProLong Gold Antifade Reagent (Life Technologies, Carlsbad, California, USA), and observed using a Zeiss LSM 900 Airyscan 2 confocal laser scanning microscope.
ACKNOWLEDGMENTS
We thank Domagoj Gajski, Michal Štarha, and Valentina Candian for helping collect psyllids in the field, as well as Massimiliano Trenti and Katrin Janik for help with the psyllid rearing in the greenhouse.
This study was funded by a joint project of the province of Bolzano and the Austrian Science Fund FWF (FIGHToplasma, I 4639-B). Anna Michalik was supported by the Polish National Science Centre grant 2017/26/D/NZ8/00799. The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the open access publication charges.
AFTER EPUB
[This article was published on 28 September 2023 with alignment errors in Table 1. The Table 1 alignment was corrected in the current version, posted on 6 October 2023.]
Contributor Information
Jessica Dittmer, Email: jessica.dittmer@inrae.fr.
Hannes Schuler, Email: hannes.schuler@unibz.it.
Jack A. Gilbert, University of California San Diego, La Jolla, California, USA
DATA AVAILABILITY
The genomes produced in this work are accessible in the NCBI database under BioProject accessions PRJNA803426 (endosymbionts of Cacopsylla melanoneura), PRJNA853274 (endosymbionts of Cacopsylla picta), PRJNA853282 (endosymbionts of Cacopsylla pyricola), and PRJNA853726 (endosymbionts of Cacopsylla pyri). New COI haplotypes for C. melanoneura, C. picta, and C. pyricola were deposited under accessions OQ106377–OQ106379.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00578-23.
Insect endosymbiont genomes included in the phylogenomics analysis of Ca. Psyllophila symbiotica.
Previously published Carsonella genomes used for comparative and phylogenomic analyses.
Presence of genes involved in the biosynthesis of essential amino acids in all available Carsonella genomes based on KEGG pathway annotations using BlastKoala.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 2. Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York. [Google Scholar]
- 3. Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. doi: 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- 4. Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A 108:2849–2854. doi: 10.1073/pnas.1013465108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21:1366–1372. doi: 10.1016/j.cub.2011.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bennett GM, Moran NA. 2013. Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol 5:1675–1688. doi: 10.1093/gbe/evt118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32:402–407. doi: 10.1038/ng986 [DOI] [PubMed] [Google Scholar]
- 8. Nováková E, Husník F, Šochová E, Hypša V. 2015. Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl Environ Microbiol 81:6189–6199. doi: 10.1128/AEM.01487-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reis F, Kirsch R, Pauchet Y, Bauer E, Bilz LC, Fukumori K, Fukatsu T, Kölsch G, Kaltenpoth M. 2020. Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles. Nat Commun 11:2964. doi: 10.1038/s41467-020-16687-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- 11. Toft C, Andersson SGE. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11:465–475. doi: 10.1038/nrg2798 [DOI] [PubMed] [Google Scholar]
- 12. Manzano-Marín A, Latorre A. 2016. Snapshots of a shrinking partner: genome reduction in Serratia symbiotica. Sci Rep 6:32590. doi: 10.1038/srep32590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pérez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. 2006. A small microbial genome: the end of a long symbiotic relationship?. Science 314:312–313. doi: 10.1126/science.1130441 [DOI] [PubMed] [Google Scholar]
- 14. Monnin D, Jackson R, Kiers ET, Bunker M, Ellers J, Henry LM. 2020. Parallel evolution in the integration of a co-obligate aphid symbiosis. Curr Biol 30:1949–1957. doi: 10.1016/j.cub.2020.03.011 [DOI] [PubMed] [Google Scholar]
- 15. Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. doi: 10.1111/1462-2920.12121 [DOI] [PubMed] [Google Scholar]
- 16. Koga R, Moran NA. 2014. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8:1237–1246. doi: 10.1038/ismej.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCutcheon JP, Boyd BM, Dale C. 2019. The life of an insect endosymbiont from the cradle to the grave. Curr Biol 29:R485–R495. doi: 10.1016/j.cub.2019.03.032 [DOI] [PubMed] [Google Scholar]
- 18. Manzano-Marín A, Szabó G, Simon J-C, Horn M, Latorre A. 2017. Happens in the best of subfamilies: establishment and repeated replacements of co-obligate secondary endosymbionts within lachninae aphids. Environ Microbiol 19:393–408. doi: 10.1111/1462-2920.13633 [DOI] [PubMed] [Google Scholar]
- 19. Matsuura Y, Moriyama M, Łukasik P, Vanderpool D, Tanahashi M, Meng X-Y, McCutcheon JP, Fukatsu T. 2018. Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci U S A 115:E5970–E5979. doi: 10.1073/pnas.1803245115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ankrah NYD, Chouaia B, Douglas AE. 2018. The cost of metabolic interactions in symbioses between insects and bacteria with reduced genomes. mBio 9:mBio doi: 10.1128/mBio.01433-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCutcheon JP, Moran NA. 2007. Parallel Genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A 104:19392–19397. doi: 10.1073/pnas.0708855104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A 106:15394–15399. doi: 10.1073/pnas.0906424106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 my of evolution. Genome Biol Evol 2:708–718. doi: 10.1093/gbe/evq055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mao M, Yang X, Poff K, Bennett G. 2017. Comparative genomics of the dual-obligate symbionts from the treehopper, Entylia carinata (Hemiptera: Membracidae), provide insight into the origins and evolution of an ancient symbiosis. Genome Biol Evol 9:1803–1815. doi: 10.1093/gbe/evx134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett GM, Mao M. 2018. Comparative genomics of a quadripartite symbiosis in a planthopper host reveals the origins and rearranged nutritional responsibilities of anciently diverged bacterial lineages. Environ Microbiol 20:4461–4472. doi: 10.1111/1462-2920.14367 [DOI] [PubMed] [Google Scholar]
- 26. Michalik A, Castillo Franco D, Kobiałka M, Szklarzewicz T, Stroiński A, Łukasik P. 2021. Alternative transmission patterns in independently acquired nutritional cosymbionts of Dictyopharidae planthoppers. mBio 12:e0122821. doi: 10.1128/mBio.01228-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86. doi: 10.1038/35024074 [DOI] [PubMed] [Google Scholar]
- 28. Lamelas A, Gosalbes MJ, Manzano-Marín A, Peretó J, Moya A, Latorre A. 2011. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet 7:e1002357. doi: 10.1371/journal.pgen.1002357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manzano-Marín A, Simon J-C, Latorre A. 2016. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia Symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol 8:1440–1458. doi: 10.1093/gbe/evw085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yorimoto S, Hattori M, Kondo M, Shigenobu S. 2022. Complex host/symbiont integration of a multi-partner symbiotic system in the Eusocial aphid Ceratovacuna japonica. iScience 25:105478. doi: 10.1016/j.isci.2022.105478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, Baumann P. 2000. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol 66:2898–2905. doi: 10.1128/AEM.66.7.2898-2905.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314:267. doi: 10.1126/science.1134196 [DOI] [PubMed] [Google Scholar]
- 33. Tamames J, Gil R, Latorre A, Peretó J, Silva FJ, Moya A. 2007. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol Biol 7:181. doi: 10.1186/1471-2148-7-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sloan DB, Moran NA. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29:3781–3792. doi: 10.1093/molbev/mss180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall AAG, Morrow JL, Fromont C, Steinbauer MJ, Taylor GS, Johnson SN, Cook JM, Riegler M. 2016. Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ Microbiol 18:2591–2603. doi: 10.1111/1462-2920.13351 [DOI] [PubMed] [Google Scholar]
- 36. Fromont C, Riegler M, Cook JM. 2016. Phylogeographic analyses of bacterial endosymbionts in fig homotomids (Hemiptera: Psylloidea) reveal codiversification of both primary and secondary endosymbionts. FEMS Microbiol Ecol 92:fiw205. doi: 10.1093/femsec/fiw205 [DOI] [PubMed] [Google Scholar]
- 37. Sloan DB, Nakabachi A, Richards S, Qu J, Murali SC, Gibbs RA, Moran NA. 2014. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol 31:857–871. doi: 10.1093/molbev/msu004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukatsu T, Nikoh N. 1998. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl Environ Microbiol 64:3599–3606. doi: 10.1128/AEM.64.10.3599-3606.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Subandiyah S, Nikoh N, Tsuyumu S, Somowiyarjo S, Fukatsu T. 2000. Complex endosymbiotic microbiota of the citrus psyllid Diaphorina citri (Homoptera: Psylloidea). Zoolog Sci 17:983–989. doi: 10.2108/zsj.17.983 [DOI] [Google Scholar]
- 40. Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, van Echten-Deckert G, Okamura K, Yamamoto K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima S, Hattori M, Piel J, Fukatsu T. 2013. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23:1478–1484. doi: 10.1016/j.cub.2013.06.027 [DOI] [PubMed] [Google Scholar]
- 41. Nakabachi A, Piel J, Malenovský I, Hirose Y. 2020. Comparative genomics underlines multiple roles of Profftella, an obligate symbiont of psyllids: providing toxins, vitamins, and carotenoids. Genome Biol Evol 12:1975–1987. doi: 10.1093/gbe/evaa175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morrow JL, Hall AAG, Riegler M. 2017. Symbionts in waiting: the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 5:58. doi: 10.1186/s40168-017-0276-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morrow JL, Om N, Beattie GAC, Chambers GA, Donovan NJ, Liefting LW, Riegler M, Holford P. 2020. Characterization of the bacterial communities of psyllids associated with Rutaceae in Bhutan by high throughput sequencing. BMC Microbiol 20:215. doi: 10.1186/s12866-020-01895-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwak Y, Sun P, Meduri VR, Percy DM, Mauck KE, Hansen AK. 2021. Uncovering symbionts across the psyllid tree of life and the discovery of a new Liberibacter species, "Candidatus Liberibacter capsica". Front Microbiol 12:739763. doi: 10.3389/fmicb.2021.739763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakabachi A, Inoue H, Hirose Y. 2022. Microbiome analyses of 12 psyllid species of the family Psyllidae identified various bacteria including Fukatsuia and Serratia symbiotica, known as secondary symbionts of aphids. BMC Microbiol 22:15. doi: 10.1186/s12866-021-02429-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Štarhová Serbina L, Gajski D, Pafčo B, Zurek L, Malenovský I, Nováková E, Schuler H, Dittmer J. 2022. Microbiome of pear psyllids: a tale about closely related species sharing their endosymbionts. Environ Microbiol 24:5788–5808. doi: 10.1111/1462-2920.16180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuler H, Dittmer J, Borruso L, Galli J, Fischnaller S, Anfora G, Rota-Stabelli O, Weil T, Janik K. 2022. Investigating the microbial community of Cacopsylla spp. as potential factor in vector competence of phytoplasma. Environ Microbiol 24:4771–4786. doi: 10.1111/1462-2920.16138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dial DT, Weglarz KM, Aremu AO, Havill NP, Pearson TA, Burke GR, von Dohlen CD. 2022. Transitional genomes and nutritional role reversals identified for dual symbionts of adelgids (Aphidoidea: Adelgidae). ISME J 16:642–654. doi: 10.1038/s41396-021-01102-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anbutsu H, Moriyama M, Nikoh N, Hosokawa T, Futahashi R, Tanahashi M, Meng XY, Kuriwada T, Mori N, Oshima K, Hattori M, Fujie M, Satoh N, Maeda T, Shigenobu S, Koga R, Fukatsu T. 2017. Small genome symbiont underlies cuticle hardness in beetles. Proc Natl Acad Sci U S A 114:E8382–E8391. doi: 10.1073/pnas.1712857114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sloan DB, Moran NA. 2012. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett 8:986–989. doi: 10.1098/rsbl.2012.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drohojowska J, Szwedo J, Müller P, Burckhardt D. 2020. New fossil from mid-cretaceous burmese amber confirms monophyly of Liadopsyllidae (Hemiptera: Psylloidea). Sci Rep 10:17607. doi: 10.1038/s41598-020-74551-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Percy DM, Crampton‐Platt A, Sveinsson S, Lemmon AR, Lemmon EM, Ouvrard D, Burckhardt D. 2018. Resolving the psyllid tree of life: phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst Entomol 43:762–776. doi: 10.1111/syen.12302 [DOI] [Google Scholar]
- 53. Nakabachi A, Inoue H, Hirose Y. 2022. High-resolution microbiome analyses of nine psyllid species of the family Triozidae identified previously unrecognized but major bacterial populations, including Liberibacter and Wolbachia of supergroup O. Microbes Environ 37:ME22078. doi: 10.1264/jsme2.ME22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thao ML, Clark MA, Baumann L, Brennan EB, Moran NA, Baumann P. 2000. Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol 41:300–304. doi: 10.1007/s002840010138 [DOI] [PubMed] [Google Scholar]
- 55. Morris JJ, Lenski RE, Zinser ER. 2012. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corretto E, Trenti M, Štarhová Serbina L, Howie JM, Dittmer J, Kerschbamer C, Candian V, Tedeschi R, Janik K, Schuler H. 2023. Multiple factors driving the acquisition efficiency of apple proliferation phytoplasma in Cacopsylla melanoneura. Journal of Pest Science, in press. doi: 10.21203/rs.3.rs-2646791/v1 [DOI] [PMC free article] [PubMed]
- 57. Jarausch B, Tedeschi R, Sauvion N, Gross J, Jarausch W. 2019. Psyllid vectors, p 53–78. In Bertaccini A, Weintraub PG, Rao GP, Mori N (ed), Phytoplasmas: plant pathogenic bacteria II. Springer, Singapore. doi: 10.1007/978-981-13-2832-9 [DOI] [Google Scholar]
- 58. Oettl S, Schlink K. 2015. Molecular identification of two vector species, Cacopsylla melanoneura and Cacopsylla picta (Hemiptera: Psyllidae), of apple proliferation disease and further common psyllids of northern Italy. J Econ Entomol 108:2174–2183. doi: 10.1093/jee/tov204 [DOI] [PubMed] [Google Scholar]
- 59. Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kolmogorov M, Bickhart DM, Behsaz B, Gurevich A, Rayko M, Shin SB, Kuhn K, Yuan J, Polevikov E, Smith TPL, Pevzner PA. 2020. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat Methods 17:1103–1110. doi: 10.1038/s41592-020-00971-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 62. Zimin AV, Salzberg SL. 2020. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput Biol 16:e1007981. doi: 10.1371/journal.pcbi.1007981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pryszcz LP, Gabaldón T. 2016. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res 44:e113. doi: 10.1093/nar/gkw294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. 2020. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70:e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- 66. Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 67. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Emms DM, Kelly S. 2019. Orthofinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. R Core Team . 2021. R: A language and environment for statistical computing, R foundation for statistical computing, Vienna, Austria. Available from: https://www.R-project.org
- 70. Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. EggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. doi: 10.1093/molbev/msab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and Ghostkoala: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 72. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. Modelfinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol 65:997–1008. doi: 10.1093/sysbio/syw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Insect endosymbiont genomes included in the phylogenomics analysis of Ca. Psyllophila symbiotica.
Previously published Carsonella genomes used for comparative and phylogenomic analyses.
Presence of genes involved in the biosynthesis of essential amino acids in all available Carsonella genomes based on KEGG pathway annotations using BlastKoala.
Data Availability Statement
The genomes produced in this work are accessible in the NCBI database under BioProject accessions PRJNA803426 (endosymbionts of Cacopsylla melanoneura), PRJNA853274 (endosymbionts of Cacopsylla picta), PRJNA853282 (endosymbionts of Cacopsylla pyricola), and PRJNA853726 (endosymbionts of Cacopsylla pyri). New COI haplotypes for C. melanoneura, C. picta, and C. pyricola were deposited under accessions OQ106377–OQ106379.