ABSTRACT
Although gut microbiota alteration and related blood microbe profiles in human immunodeficiency virus (HIV)-infected individuals are associated with the disease progression, how abnormal blood microbe profiles influence the inflammation and immune restoration are not fully understood. To address these issues, this study enrolled 24 healthy controls (HCs) and 91 HIV-infected individuals, including 30 treatment-naïve individuals (TNs), 31 immunological non-responders (INRs), and 30 immunological responders (IRs); subsequently, we analyzed blood microbe profiles using metagenomic sequencing and Olink proteomics technology, and identify inflammation-related proteins in peripheral blood samples of these individuals. The results showed increased translocation of microbes in the blood of TNs. This translocation did not return to normal level in either IRs or INRs who received antiretroviral therapy. In addition, Porphyromonas gingivalis significantly increased in TNs, IRs, and INRs compared to HCs. P. gingivalis was inversely associated with CD4+ T-cell counts, CD4/CD8 ratio, latency-associated peptide transforming growth factor-β1, and tumor necrosis factor-related activation-induced cytokine (TRANCE) and positively associated with HIV reservoirs. Burkholderia multivorans and Bacillus thuringiensis significantly decreased in TNs, IRs, and INRs compared to HCs and were positively associated with CD4+ T-cell counts and the CD4/CD8 ratio and negatively associated with HIV reservoir size and pro-inflammatory factors. We identified several species of microbes that were associated with CD4+ T-cell restoration on antiretroviral therapy, Prevotella sp. CAG:5226, Eubacterium sp. CAG:251, Phascolarctobacterium succinatutens, Anaerobutyricum hallii, Prevotella sp. AM34-19LB, and Phocaeicola plebeius were positively associated with HIV reservoir size and pro-inflammatory proteins. Another group of bacteria, B. multivorans, B. thuringiensis, Vibrio vulnificus, and Acinetobacter baumannii, which were negatively associated and pro-inflammatory proteins. In conclusion, different microbes within blood of HIV-infected individuals were found to be closely associated with persistent inflammation and immune restoration, suggesting the blood microbe profiles of HIV-infected individuals also influence disease progression.
IMPORTANCE
The characteristics of blood microbiota in HIV-infected individuals and their relevance to disease progression are still unknown, despite alterations in gut microbiota diversity and composition in HIV-infected individuals. Here, we present evidence of increased blood microbiota diversity in HIV-infected individuals, which may result from gut microbiota translocation. Also, we identify a group of microbes, Porphyromonas gingivalis, Prevotella sp. CAG:5226, Eubacterium sp. CAG:251, Phascolarctobacterium succinatutens, Anaerobutyricum hallii, Prevotella sp. AM34-19LB, and Phocaeicola plebeius, which are linked to poor immunological recovery. This work provides a scientific foundation toward therapeutic strategies targeting blood microbiota for immune recovery of HIV infection.
KEYWORDS: HIV, blood microbiota, inflammation, immune recovery, HIV reservoir
INTRODUCTION
Although antiretroviral therapy (ART) can effectively suppress viral replication and improve the immune status of the human immunodeficiency virus (HIV)-infected patients (1, 2), it is unable to fully solve the problems of residual immune activation and persistent inflammation caused by HIV infection (3) and is linked to several leading causes of morbidity and mortality of HIV infection (4). In fact, after successful ART, patients with HIV infection still have higher levels of interleukin (IL)-6, D-dimers, C-reactive protein, and soluble CD14 (sCD14), predictors of morbidity and mortality in patients on long-term ART (5 – 8). Persistent inflammation is also associated with increased cardiovascular events, accelerated liver disease, impaired immunological recovery, and mortality (5, 9, 10). However, the mechanism underlying HIV-related immune activation and inflammatory response is not fully understood.
Gut microbiota dysbiosis and microbial translocation are important causes of circulating inflammation in both treated and untreated individuals infected with HIV. Several studies have shown altered gut bacterial composition in HIV-infected individuals compared to healthy controls (HCs), with an increase in pro-inflammatory and potentially pathogenic bacteria and a decrease in beneficial bacteria (11 – 13). Particularly, these observations include an increased abundance of Enterobacteriaceae, which induce host inflammation upon infection and can utilize the by-products of this inflammation, namely reactive oxygen species from neutrophils and macrophages, to stimulate an increase in pro-inflammatory Enterobacteriaceae, which in turn exacerbates intestinal inflammation (14, 15). Studies have demonstrated that some species of Prevotella, which are abundant in individuals infected with HIV, enhance T helper (Th)17-mediated mucosal inflammation, have a pro-inflammatory effect, and are positively correlated with immune activation, while Bacteroides, which are reduced in these patients, can produce anti-inflammatory cytokines and have a protective effect against inflammation (12, 16).
Microbial translocation is another important cause of chronic inflammation. In particular, lipopolysaccharide, a component of the cell wall of gram-negative bacteria, is an indicator of microbial translocation and is significantly increased in individuals chronically infected with HIV and simian immunodeficiency virus-infected rhesus monkeys (17). Microbial translocation is also associated with increased activation of CD8+ T-cells and persistent failure of CD4+ T-cell reconstitution in patients receiving ART (18). In contrast, high inflammatory state of the intestinal mucosa and apoptosis of intestinal epithelial cells further promote intestinal microbial translocation (19).
However, most earlier studies have concentrated on how gut microbes affect inflammation and the course of the disease, ignoring the potential direct contribution of the blood microbiota to inflammation in individuals infected with HIV. A recent study found that the diversity of microbial fractions in the blood increases in the late stages of HIV infection, and ART ameliorates this blood microbiota perturbation (20). A longitudinal study revealed that the composition of translocated microbes in the blood affects the degree of recovery of CD4+ T-cells and the presence of persistent systemic inflammation (21). In addition, the characteristics of the blood microbiota at various disease stages of HIV infection and their relationship with inflammation have not been studied. It is unclear which specific microbial species in the blood are linked to HIV disease status and immune recovery, the potential connection between translocated microbiota and gut microbiota, whether blood microbes associated with disease progression are altered in the stool, and the extent of immune recovery after ART in persistent inflammation.
In the present study, we used shotgun metagenomic sequencing to elucidate the characteristics and functions of blood microbiota fractions in individuals infected with HIV at different disease stages and investigated their relationship with systemic inflammation and disease progression. In addition, gut microbiota composition in paired stool samples was investigated to determine whether the blood microbiota associated with disease progression is predominantly derived from gut microbial translocation.
RESULTS
Characteristics of the study population and study design
A total of 115 male subjects participated in the study, including 24 non-HIV-infected individuals as HCs. They were divided into four groups: treatment-naïve individuals (TNs), immunological non-responders (INRs), immunological responders (IRs), and HCs (Fig. 1). Peripheral blood and corresponding stool samples were collected simultaneously. The duration of ART was not significantly different between the IR and INR groups. No differences were observed in body mass index among the four groups (Table 1). The median CD4+ T-cell counts were 341, 317, 820, and 921 cells/µL in TNs, INRs, IRs, and HCs, respectively. Compared to IRs, INRs had significantly lower CD4+ T-cell and CD8+ T-cell counts, and CD4/CD8 ratio.
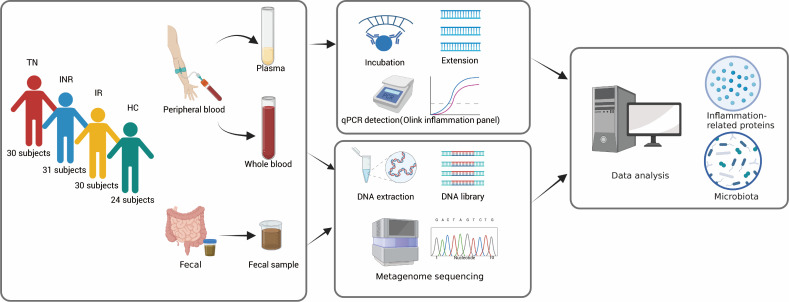
Fig 1.
Study design. Peripheral blood and fecal samples were collected from TNs (n = 30), INRs ( n = 31), IRs (n = 30), and HCs (n = 24). The Olink inflammation panel was used to measure inflammation-related proteins in the plasma. Metagenome sequencing was used to detect the microbiota in the peripheral blood and feces. Data analysis of the microbiota and inflammation-related proteins was performed to explore their correlation and relevance to clinical parameters.
TABLE 1.
| TNs (n = 30) | INRs (n = 31) | IRs (n = 30) | HCs (n = 24) | P value | |
|---|---|---|---|---|---|
| Age (years, IQR) | 30 (26, 37) | 44 (35, 51) | 37 (30, 42) | 32 (27, 43) | < 0.001 a , 0.030 b |
| Male gender (no.,%) | 30 (100) | 31 (100) | 30 (100) | 24 (100) | – |
| BMI (kg/m2, IQR) | 22.62 (20.65, 25.95) | 22.31 (21.45, 25.28) | 22.47 (20.93, 25.93) | 22.98 (21.52, 24.35) | 0.937 a |
| Viral load (log10 copies/mL, IQR) | 4.20 (3.62, 4.91) | <LDL | <LDL | – | – |
| CD4+ T-cell count (cells/μL, IQR) | 341 (183, 500) | 317 (265, 332) | 820 (637, 1,063) | 921 (608, 1,156) | < 0.001 a , < 0.001 b |
| CD8+ T-cell count (cells/μL, IQR) | 1,036 (718, 1,356) | 787 (501, 991) | 975 (705, 1,275) | 790 (673, 1,039) | 0.040 a , 0.024 b |
| CD4/CD8 ratio (IQR) | 0.29 (0.21, 0.50) | 0.38 (0.29, 0.48) | 0.85 (0.67, 1.29) | 1.14 (0.78, 1.50) | < 0.001 a , <0.001 b |
| Duration of ART (years, IQR) | – | 7 (4, 8) | 6 (5, 8) | – | 0.931 c |
Kruskal-Wallis test.
Kruskal-Wallis test, INRs vs IRs.
Mann-Whitney U test.
Continuous variables are expressed as median (IQR), and categorical variables are expressed as the number of cases (%).
"–”, not applicable.
HCs, healthy controls; BMI, body mass index; LDL, lower than detectable level.
HIV infection and ART affect the distribution and composition of blood microbiota
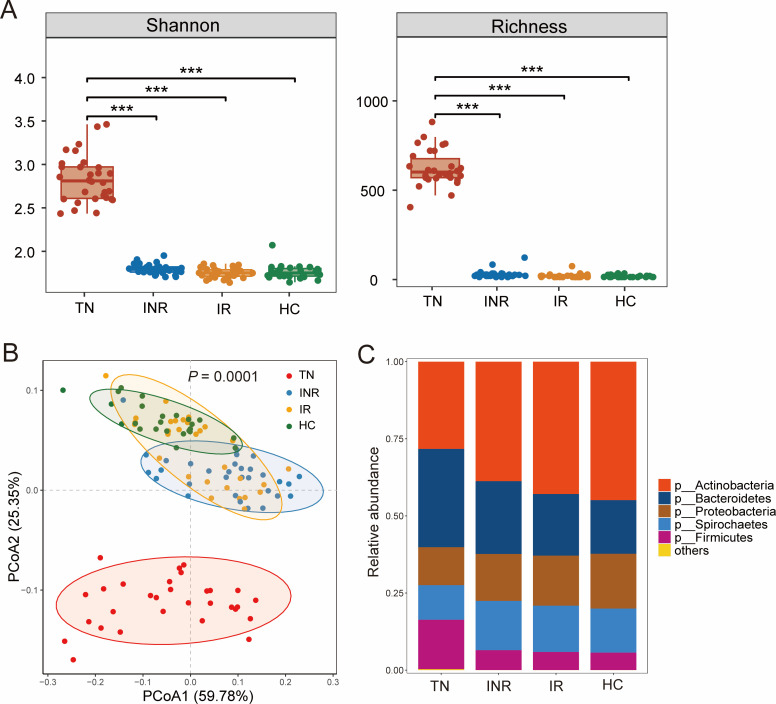
Alpha diversity was used to measure the abundance and evenness of the bacterial taxa within a community. We found that TN individuals exhibited significantly higher alpha diversity of blood microbiota than controls, as shown by the Shannon index (richness and evenness) and Richness index (richness only). ART significantly reduced alpha diversity in IRs and INRs (Fig. 2A). In contrast, TNs, INRs, and IRs (individuals infected with HIV) displayed lower Shannon and Richness indices in stool microbiota than controls, although there was no statistical difference (Fig. S1A).
Fig 2.
Taxonomic analysis of the blood microbiota of study subjects. (A) The Shannon index and richness analysis of blood microbiota in TNs, INRs, IRs, and HCs. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Principal coordinates analysis (PCoA) of blood microbiota composition using Bray-Curtis dissimilarities in TNs, INRs, IRs, and HCs (P = 0.0001, analysis of similarity). (C) Average relative abundances of microbial phyla detected in blood from TNs, INRs, IRs, and HCs. Taxa are merged into the “Others” category if less abundant.
To understand the impact of HIV infection on the distribution of blood microbiota, a principal coordinates analysis (PCoA) comparing samples based on Bray-Curtis differences revealed significantly different blood microbial compositions at the species level between the TN and HC groups. Interestingly, we also observed a significant gradient between TNs, INRs, IRs, and HCs [P = 0.0001, analysis of similarity (ANOSIM); Fig. 2B], with most IR samples overlapping with HC samples, while INR samples overlapped less with HCs and were closer to TNs. This grading suggests that the blood microbial distribution in patients with INRs is still relatively different from that in HCs.
Subsequently, we explored the composition of the blood microbiota and the effect of HIV infection. A graphical representation of the relative proportions of units at the phylum taxonomic level present in the study samples was performed. Blood was mainly composed of Actinobacteria, Bacteroidetes, Proteobacteria, Spirochaetes, and Firmicutes. Compared to HCs, Actinobacteria and Proteobacteria decreased, whereas Bacteroidetes and Firmicutes increased in the TNs. ART reduced this difference but did not restore it to normal levels, and the INRs differed more from that of HCs (Fig. 2C). Stacked bar plots represent the relative proportions of taxonomic units at other taxonomic levels (class, order, family, genus, and species in Fig. S2A through E, respectively). Interestingly, the gut microbiota in feces consisted mainly of the phyla Bacteroidetes, Firmicutes, and Proteobacteria, with a smaller proportion of Actinobacteria. The relative abundance of Bacteroidetes increased and Firmicutes decreased in individuals infected with HIV (Fig. S1B). Therefore, the increased relative abundance of the Bacteroidetes and Firmicutes phyla in the blood indicates that the intestinal lumen may be a source of these bacteria, which are then translocated into the blood.
Identification of specific species associated with disease state and immune response
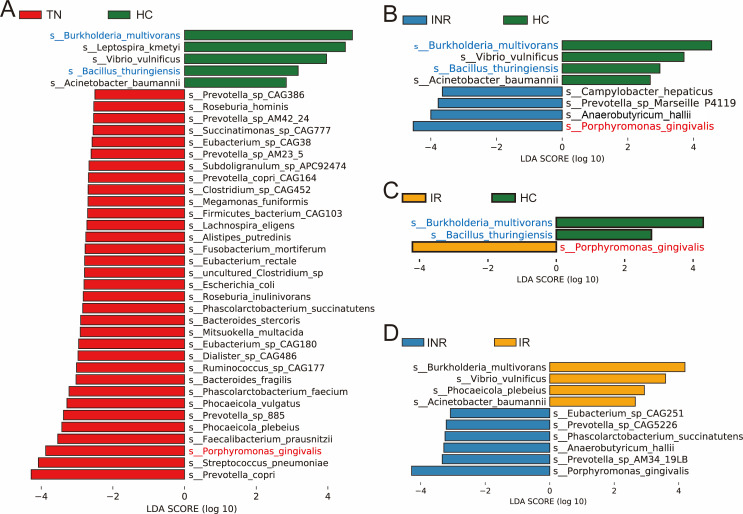
To identify specific species associated with disease states, we performed linear discriminant analysis (LDA) and effect size (LEfSe) analysis (P < 0.05, LDA >2.5). At the species level, the histograms showed that Prevotella copri, Streptococcus pneumoniae, Porphyromonas gingivalis, Faecalibacterium prausnitzii, Phocaeicola plebeius, Prevotella sp. 885, Phocaeicola vulgatus, Phascolarctobacterium faecium, Bacteroides fragilis, Ruminococcus sp. CAG177, and 23 other species were enriched in TNs compared to HCs, while Burkholderia multivorans, Leptospira kmetyi, Vibrio vulnificus, Bacillus thuringiensis, and Acinetobacter baumannii were absent (Fig. 3A). Among the 33 species of bacteria enriched in TNs, 31 (approximately 94%) species belonged to the phyla Bacteroidetes and Firmicutes (Table S1). After long-term ART, this type of biological abnormality was attenuated, and most of the microbiota in IRs and INRs could be restored to normal levels. However, P. gingivalis, Anaerobutyricum hallii, Prevotella sp. Marseille P4119, and Campylobacter hepaticus were enriched, whereas B. multivorans, V. vulnificus, B. thuringiensis, and A. baumannii were absent in INRs compared with HCs (Fig. 3B). In addition, compared with HCs, P. gingivalis was enriched, whereas B. multivorans and B. thuringiensis were deficient in IRs (Fig. 3C). As a result, we found that P. gingivalis was consistently relatively enriched, while B. multivorans and B. thuringiensis were consistently relatively deficient in individuals infected with HIV (TNs, INRs, and IRs) compared to HCs.
Fig 3.
Differential enrichment of microbes in every two groups. Linear discriminant analysis effect size analysis of discriminant taxa in the blood microbiota of pairwise combinations of TNs compared to HCs (A), INRs compared to HCs (B), IRs compared with HCs (C), and INRs compared to IRs (D). Compared to HCs, bacterial species with enriched abundance in individuals infected with HIV are marked in red, and enriched species in HCs are marked in blue. P < 0.05; LDA score >2.5.
Subsequently, we searched for differentially abundant species in the blood microbiota of INRs and IRs. An LEfSe analysis was performed (P < 0.05, LDA >2.5), and eight differentially abundant species were found in INRs compared to IRs. As shown in Fig. 3D, there were six species enriched in INRs and four species enriched in IRs. P. gingivalis, Prevotella sp. AM34 19LB, A. hallii, Phascolarctobacterium succinatutens, Prevotella sp. CAG5226, and Eubacterium sp. CAG251 were enriched, and B. multivorans, V. vulnificus, P. plebeius, and A. baumannii were deficient in INRs compared with IRs. We found that approximately 50% (3/6) of these species-enriched INRs belonged to the phylum Bacteroidetes and the others belonged to the phyla Firmicutes. B. multivorans, V. vulnificus, and A. baumannii enriched in IRs were all classified as Proteobacteria at the phylum level (Table S2).
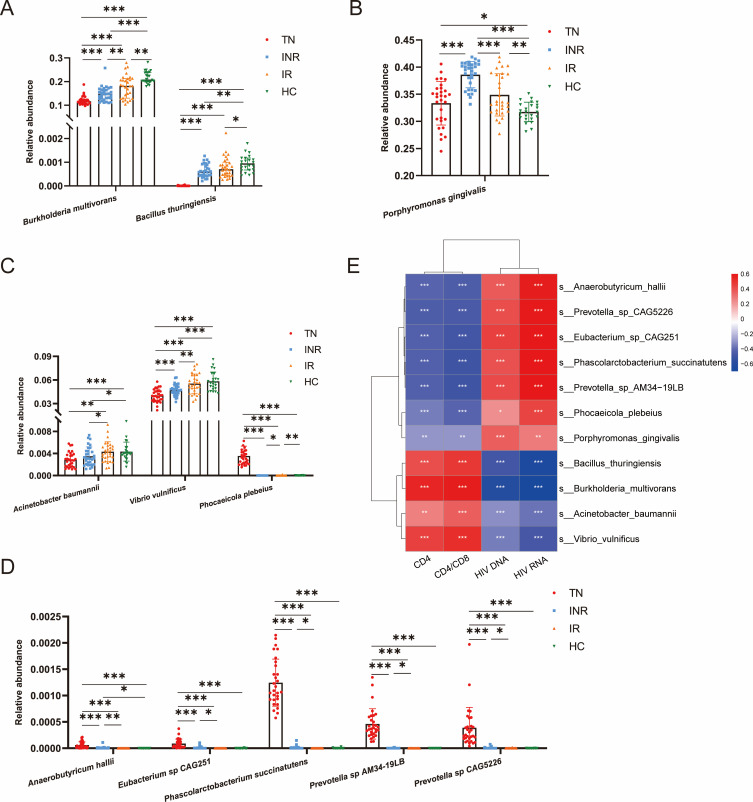
We then visualized the relative abundance of the above 11 bacterial species in the four groups. The bar plots indicated that two species were consistently enriched in HCs (Fig. 4A), and the one species was enriched in individuals infected with HIV (Fig. 4B). The relative abundances of B. multivorans and B. thuringiensis were highest in HCs and lowest in TNs, whereas those in IRs were higher than those in INRs. However, P. gingivalis showed the opposite trend. Among the differential bacteria in INRs and IRs, three species were enriched in IRs (Fig. 4C), and five species were enriched in INRs (Fig. 4D), except B. multivorans and P. gingivalis. The relative abundances of A. baumannii and V. vulnificus were lowest in TNs and highest in HCs and were lower in INRs than in IRs. Interestingly, the relative abundances of P. plebeius, A. hallii, E. sp. CAG251, P. succinatutens, P. sp. AM34-19LB, and P. sp. CAG5226 were high in TNs but very low in INRs, IRs, and HCs, some even close to zero, probably from translocation. Analysis of these 11 bacterial species in fecal samples did not reveal the same alteration in abundance as in blood (Fig. S3A through D).
Fig 4.
Differential bacterial species in blood associated with HIV disease status and immune recovery. (A and B) Species associated with disease states. Relative abundance of the two species enriched in HCs (A) and one species enriched in individuals infected with HIV (B). (C and D) Species associated with immune recovery. Relative abundance of the three species enriched in IRs (C) and five species enriched in INRs (D). Each dot represents a participant. *P < 0.05, **P < 0.01, ***P < 0.001. (E) Heatmap showing Spearman correlations between differential species selected above and clinical parameters and HIV reservoir indicators. Red and blue indicate positive and negative associations, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
To assess the possible impact of blood bacterial fractions on disease progression, we performed correlation analysis based on Spearman correlation with clinical indicators (CD4+ T-cell counts, CD4/CD8 ratio, HIV DNA, and RNA) for 11 bacteria selected from LEfSe analysis. We found that B. multivorans, B. thuringiensis, V. vulnificus, and A. baumannii showed significant positive correlations with the CD4/CD8 ratio and CD4+ T-cell counts, and significant negative correlations with HIV DNA and RNA. In addition, all six INRs-enriched bacteria and P. plebeius were negatively correlated with CD4+ T-cell counts and the CD4/CD8 ratio and positively correlated with HIV DNA and RNA (Fig. 4E).
Plasma inflammation-related protein analysis
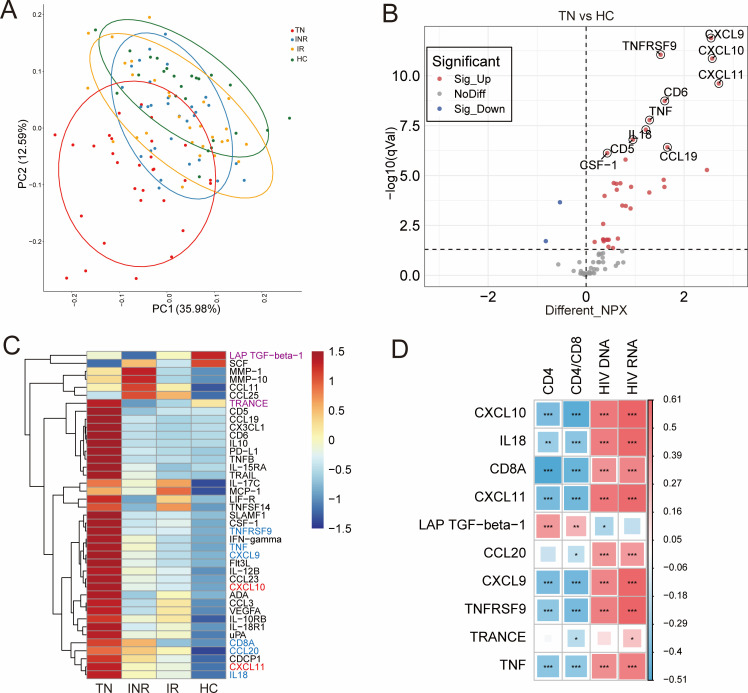
To investigate the clinical significance of plasma inflammation-related proteins in HIV-infected patients, we measured 92 different inflammation-related proteins using the Olink multiplex inflammation assay panel. The expression profiles based on all proteins were visualized using the principal component analysis (PCA) downscaling method, which showed that the protein distribution was different and distinguishable between the four groups (Fig. 5A). We calculated the mean difference in the expression of each protein in the four groups of samples and compared their statistical significance based on statistical analysis. Thirty-seven inflammation-associated proteins were significantly different in TNs compared to HCs; 35 proteins were elevated, and 2 proteins were decreased (Fig. 5B; Table S3). Fourteen inflammation-related proteins differed in INRs compared to HCs; 12 proteins were elevated, and 2 proteins were decreased (Fig. S4A; Table S4). Compared to HCs, three inflammation-related proteins were different, and all were elevated in IRs (Fig. S4B; Table S5). As a consequence, a total of 41 inflammation-related proteins were significantly different in individuals infected with HIV compared to HCs (including TNs versus HCs, IRs versus HCs, and INRs versus HCs). Of these, two differential inflammation-related proteins were included in INRs and IRs (Table S6). In addition, the NPX expression levels of these inflammation-associated proteins were represented by a visual heatmap. Compared with HCs, CXCL10 and CXCL11, marked in red in Fig. 5C, are elevated in individuals infected with HIV (TNs, INRs, and IRs). Box plots showed that they were lowest in HCs and then increased in the order of IRs, INRs, and HCs (Fig. S5A). LAP TGF-β1 and tumor necrosis factor-related activation-induced cytokine (TRANCE) marked in purple are decreased, and TNFRSF9, TNF, CXCL9, CD8A, CCL20, and IL18 marked in blue are elevated in INRs, but there was no significant difference in IRs (Fig. 5C). Box plots revealed the expression of NPX in the four groups (Fig. S5B and C).
Fig 5.
Alterations in inflammation-related proteins and correlation with clinical parameters in study design. (A) Principal component analysis showing the distribution of inflammation-related proteins in plasma in TNs (red), INRs (blue), IRs (yellow), and HCs (green). (B) Volcano plot of differentially expressed inflammation-related proteins in TNs compared to HCs. The red dots and blue dots represent the proteins with significantly higher and lower expression (adjusted P < 0.05) in the TN group, respectively. (C) Heatmap of differentially expressed inflammation-related proteins between the two different groups. Differential proteins that are elevated in TNs, INRs, and IRs compared to HCs are labeled in red, proteins that are elevated in INRs but not in IRs are labeled in blue, and proteins that are decreased in INRs but not in IRs are labeled in purple. (D) Heatmap showing Spearman correlations between differential inflammation-related proteins and clinical parameters and HIV reservoir indicators. Red and blue indicate positive and negative associations, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, to explore the relationship between inflammation-related proteins and clinical indicators, we correlated the 10 proteins most associated with the above disease progression with CD4+ T-cell counts, CD4/CD8 ratio, HIV DNA, and HIV RNA. The results showed that CXCL10, IL18, CD8A, CXCL11, CXCL9, TNFRSF9, and TNF were negatively correlated with CD4+ T-cell counts and the CD4/CD8 ratio but positively correlated with HIV DNA and RNA. CCL20 and TRANCE were weakly negatively correlated with the CD4/CD8 ratio and positively correlated with HIV RNA levels. Unlike other inflammation-related proteins, LAP TGF-β1 was positively correlated with CD4+ T-cell counts and the CD4/CD8 ratio and negatively correlated with HIV DNA (Fig. 5D).
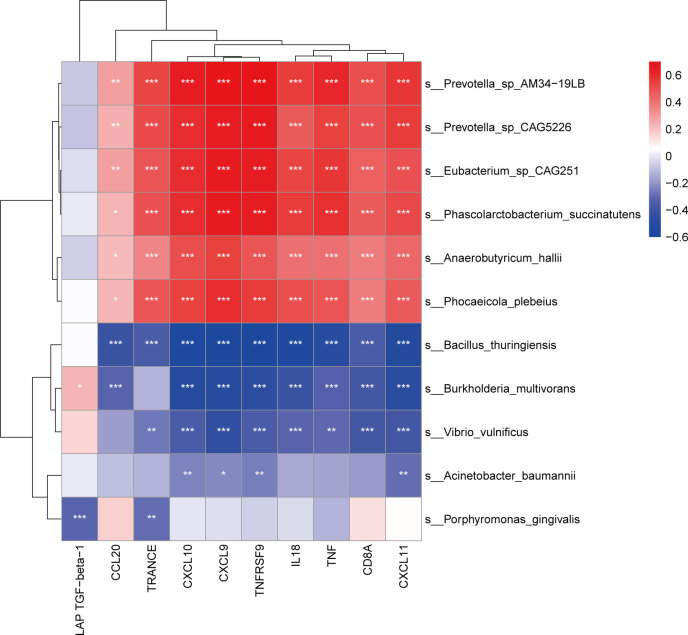
Relationship between blood bacterial fractions and inflammation-related proteins
To investigate the relationship between the blood microbiota associated with disease progression and inflammation-related proteins, we examined 11 bacterial species and 10 inflammation-associated protein correlations based on the Spearman coefficient (Fig. 6). The results demonstrated that inflammation-related proteins, except LAP TGF-β1, were positively correlated with P. plebeius, A. hallii, E. sp. CAG251, P. succinatutens, P. sp. AM34-19LB, and P. sp. CAG5226 and negatively correlated with B. thuringiensis. B. multivorans was positively correlated with LAP TGF-β1 and negatively correlated with other proteins, excluding TRANCE. V. vulnificus was adversely correlated with TRANCE, CXCL10, CXCL9, TNFRSF9, IL18, TNF, CD8A, and CXCL11. A. baumannii was inversely correlated with CXCL10, CXCL9, TNFRSF9, and CXCL11. Interestingly, P. gingivalis was negatively correlated with LAP TGF-β1 and TRANCE. Accordingly, P. gingivalis is more likely to promote disease progression by inhibiting the expression of LAP TGF-β1 and TRANCE.
Fig 6.
Correlations between differential bacterial species and inflammation-related proteins in study subjects. The heatmap shows Spearman correlations between the differentially abundant species and inflammation-related protein-associated disease states and immune recovery. Red and blue indicate positive and negative associations, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
DISCUSSION
In this study, we investigated the characteristics of the blood microbiota and inflammation-related proteins in individuals infected with HIV with different disease states and analyzed the relationship between these microbes and disease progression. We found that alpha diversity in the blood of individuals infected with HIV increased, particularly in TNs, whereas the diversity of gut microbes was reduced in stool samples. Accordingly, community PCoA showed differences in blood microbiota composition among TNs, INRs, IRs, and HCs, especially between TN and other groups, suggesting that ART partly repaired damaged intestinal epithelial integrity and reversed microbial translocation. The overall blood bacterial profile was dominated by the phyla Actinobacteria, Bacteroidetes, Proteobacteria, and Firmicutes. Furthermore, the number of inflammation-related proteins was significantly higher in TNs than in HCs, and INRs had more abnormal inflammation-related proteins than IRs. Among the differential inflammation-related proteins, the level of most inflammation-related proteins was higher in individuals infected with HIV, inversely correlated with CD4+ T-cell counts, and positively correlated with HIV viral reservoir indicators.
Previous studies have shown that reduced gut microbiota diversity is deleterious to the stability of the entire ecosystem and is associated with decreased CD4+ T-cells and increased T-cell activation (22, 23). An increase in gut microbial diversity and abundance is usually considered a more stable microbial community in the gut and more conducive to disease recovery. However, the impact of the increased diversity of blood microbes is different. Although there is rising evidence of the presence of microbes in the blood of healthy individuals, their species and numbers are significantly smaller than those of the gut microbiota (24 – 28). In the early stages of HIV infection, massive depletion of gastrointestinal CD4+ T-cells, especially Th17 cells, and concomitant structural damage to the epithelium lead to a significant increase in intestinal permeability and the entry of microbes and their products into the body circulation through the damaged mucosal barrier (19, 29 – 31). In the study of blood microbial composition, a significant increase was found in the TN group for Bacteroidetes and Firmicutes, which are the main phyla of the gut microbiota composition, suggesting a possible source of blood microbes with intestinal microbiota translocation. Blood microbiota diversity decreased substantially after ART but still did not return to normal levels, suggesting that microbial translocation persisted despite ART, which may be related to the persistence of intestinal damage (32, 33). Increased microbial diversity and altered composition affect the degree of CD4+ T-cell restoration and contribute to persistent inflammation (20, 21). This also responds to the fact that the blood microbiota not only reflects the microbial composition in the gut but also actively participates in the pathogenesis of HIV infection. Recent studies support this conclusion by demonstrating the importance of the blood microbiota in the development of diabetes, colon cancer, alcoholic hepatitis, and major depression in the general population (25, 26, 34, 35), possibly confirming the role of blood microbiota in non-AIDS-related comorbidities (36).
In addition to the increased diversity of the blood microbiota found in patients infected with HIV, the composition of the blood microbiota was also disrupted. Community PCoA highlighted differences in blood microbiota composition between TNs, INRs, IRs, and HCs. Based on metagenomic sequencing, the overall blood bacterial profile was dominated by the phyla Actinobacteria, Bacteroidetes, Proteobacteria, and Firmicutes, which are consistent with the dominant phyla found in most blood studies (21, 34, 37). At the species level, we found that many blood microbes in the TNs were disordered, and most of this disorder was corrected after receiving ART. However, some microbes differed significantly between the INR and IR groups and the HC group. We found that P. gingivalis was consistently enriched in individuals infected with HIV compared to HCs during different disease stages of HIV infection. P. gingivalis is a gram-negative rod-shaped specialized anaerobic bacterium that derives metabolic energy for growth from proteolytic products, heme, and vitamin K (38). It is an oral pathogen and, in addition to being a pathogen of periodontitis, P. gingivalis is involved in various systemic diseases such as cardiovascular disease, Alzheimer’s disease, and rheumatoid arthritis (39 – 41). Xie et al. found that P. gingivalis can mediate receptor-independent HIV-1 entry into the epithelial cells. Invasive bacteria and their outer membrane vesicles that interact with HIV-1 can mediate the transfer of HIV through the mucosa, establish mucosal transmission of HIV-1, and enhance HIV-1 infectivity (42, 43). Analysis of the presence of different bacteria in IRs and INRs, along with the observation that P. gingivalis was enriched in INRs, indicated that this bacterium may also play an important role in immune reconstitution. Interestingly, the other differential bacteria enriched in INRs were mostly present only in samples from the TN group and were absent in most samples from the other groups. These bacteria belong to the phyla Bacteroidetes and Firmicutes and may originate from intestinal microbial translocation. After receiving ART, disruption of the intestinal mucosal barrier was largely restored, and translocations were reduced. Nevertheless, the abundance of translocated bacteria varied owing to different degrees of mucosal recovery, and these translocated bacteria were negatively correlated with CD4+ T-cell counts and positively correlated with viral reservoir size. In particular, several studies have found Prevotella to be strongly associated with disease progression in HIV infection (44 – 46). The negative correlation of these bacteria with CD4+ T-cell counts and the CD4/CD8 ratio and the positive correlation with HIV viral reservoirs were detrimental to disease recovery.
In addition, we found that B. multivorans and B. thuringiensis were enriched in HCs compared to individuals infected with HIV. B. multivorans, A. baumannii, and V. vulnificus were enriched in IRs compared to INRs. B. multivorans kills pests and diseases by inhibiting fungal growth (47). B. thuringiensis (Bt) has become the main microorganism used for biological control. It contains chitinases that have an affinity for polymers and can degrade them, resulting in antimicrobial and immunomodulatory effects in the healthcare field (48). Another promising area is the potential of Bt proteins against cancer cells, with cytotoxic effects on cells altered by certain cancers, such as hepatocellular carcinoma (HepG2) and cervical cancer (HeLa) cells. This indicates the potential of microbes and new opportunities for future applications (49, 50). A. baumannii and V. vulnificus are gram-negative conditional pathogens, and a minor number of bacteria are necessary to maintain microecosystem stability (51, 52). It is worth noting that for these differential bacteria, trends in the feces were not the same as those in the blood. It is not plausible to use bacteria in feces to reflect bacterial changes in the blood, which are substantially less than those in feces, and the host is more sensitive to slight changes in blood bacteria, increasing the impact on disease progression. However, this remains hypothetical and must be tested in appropriately designed studies.
In this study, we found that species enriched in TNs and INRs were positively correlated with HIV DNA and RNA, whereas species enriched in HCs and IRs were negatively correlated with HIV DNA and RNA. This implicates that the HIV reservoir is closely related to blood microbiota. Previous studies have shown that the gut is one of the viral reservoirs (53, 54). A study found that the ratio of Bacteroidales/Clostridiales was inversely correlated with viral reservoir size (55). The relationship between blood microbes and HIV reservoirs has not been reported in current studies. Although there is evidence that the gut and blood microbiota composition have a strong effect on the host immune system (20, 56), it remains worthwhile to investigate how the microbial community interacts with the viral reservoir and whether it can influence the HIV reservoir through immune cell metabolism and proliferation (57 – 59). Inflammation is a hallmark of disease progression in response to HIV infection. We found that the number of inflammatory proteins was significantly higher in the TNs than in the HCs, and this difference did not fully normalize after ART. INRs have more abnormal inflammation-related proteins than IRs do. Among these differential inflammation-associated proteins, the levels of most were higher in individuals infected with HIV, inversely correlated with CD4+ T-cell counts, and positively correlated with HIV viral reservoir indicators. These inflammatory proteins mostly exert pro-inflammatory effects, which are consistent with previous findings. Yin et al. found that increased plasma CXCL9, CXCL10, and CXCL11 measured during primary HIV-1 infection predicted long-term HIV disease prognosis in men who have sex with men (MSM) and has the potential as a novel biomarker for clinical use (60). IL-18 may affect CXCL9, CXCL10, and CXCL11 by driving IFN-γ secretion (61). CCL20 can promote the efficient integration of HIV-1 in resting CD4+ T-cells by binding to CCR6 displayed on these cells (62). Therefore, blocking chemokine pathways, including CCR6/CCL20, maybe a possible route for therapeutic interventions to prevent viral transmission to key immune loci. CD8A is considered a marker molecule for killer T-cells and enhances CD8+ T-cell responses by acting as a co-receptor for TCRs (63). TGF-β1 levels are increased in the plasma and tissues of individuals infected with HIV. The TNF/TNFR pathway is involved in immune activation and viral reservoirs in HIV infection (64). Therefore, modulation of the TNF/TNFR pathway by novel therapeutic approaches could limit immune activation and reduce the size of the HIV reservoir in patients with undetectable viremia treated with ART (65). Notably, LAP TGF-β1 was reduced in individuals infected with HIV and had an anti-inflammatory effect. It was positively associated with CD4+ T-cells and the CD4/CD8 ratio and inversely associated with viral reservoir indicators. TGF-β1 is a homeostatic factor that maintains immune system homeostasis and orchestrates complex tissue repair after organ injury or infection. TGF-β1 also controls the proliferation, survival, and reactivity of naïve CD4+ and CD8+ T-cells in the peripheral blood, which are essential for maintaining peripheral immune homeostasis and T-cells defending against immune change (66). TGF-β1 normally inhibits TCR-mediated T-cell activation, including the suppression of Th1 cells, Th2 cells, and cytotoxic T lymphocytes. In contrast, TGF-β1 supports the differentiation and maintenance of peripheral Tregs, Th9, and Th17 cells (67). TGF-β1 negatively regulates B-cell survival, proliferation, immunoglobulin (Ig) synthesis, and IgG class switching but promotes IgA antibody production and plays an important role in mucosal immunity (68, 69). Interestingly, TRANCE was lower in the INR group than in the other groups. This is consistent with previously reported results (70 – 72). Lower levels of TRANCE were reported to be an independent predictor of nontraumatic fractures, suggesting an effect on osteoclastogenesis (73), affecting bone marrow cell development and leading to a decrease in bone marrow cells, CD34+ hematopoietic progenitor cells, and granulocytes (74).
Translocation of the gut microbiota to the blood is one of the causes of systemic immune activation during HIV infection (20). Our study explored the relationship between blood microbiota and inflammation-related proteins. Blood microbiota was found to be positively correlated with inflammation-related proteins (positively correlated with CD4+ T-cells and negatively correlated with HIV reservoir), suggesting that it may be the blood microbiota that influences disease progression in individuals infected with HIV by causing an inflammatory response that leads to alterations in inflammation-related proteins.
This study had several limitations. First, the frequency of MSM in our HIV group was higher than that in the control group, which may have influenced the results. In addition, the mechanism by which the blood microbiota affects inflammation-related proteins and thus immune recovery needs further analysis, as this association does not necessarily imply causality, and further animal experiments are needed to verify this. Finally, future longitudinal or prospective studies are needed to reveal that comprehensive medical records including lifestyle habits, intestinal inflammation, and intestinal injury may provide further evidence for our hypothesis.
In conclusion, this study characterized blood microbiota abnormalities and inflammation-related proteins in individuals infected with HIV. Blood microbial P. sp. CAG:5226, E. sp. CAG:251, P. succinatutens, A. hallii, P. sp. AM34-19LB, P. plebeius, and P. gingivalis were positively correlated with pro-inflammatory proteins and HIV DNA and RNA and negatively correlated with anti-inflammatory proteins, CD4+ T-cells, and the CD4/CD8 ratio. The opposite was true for the blood microbes B. multivorans, B. thuringiensis, V. vulnificus, and A. baumannii. These results will help identify effective microbial and immunotherapeutic strategies for HIV infection.
MATERIALS AND METHODS
Study population
We recruited 30 TNs with chronic HIV infection, 31 INRs (CD4+ T-cell counts < 350 cells/µL), and 30 IRs (CD4+ T-cell counts > 500 cells/µL) who had successfully received ART for more than 2 years with plasma HIV RNA below detectable levels. Controls were recruited from healthy non-HIV-infected individuals who accompanied patients to the clinic, medical students, or hospital staff to form an age-matched group. Exclusion criteria included co-infection with hepatitis B virus or hepatitis C virus, tuberculosis and other opportunistic infections, pregnancy, use of antibiotics, probiotics, or prebiotics within the past 1 month or diarrhea or digestive symptoms. The detailed characteristics of the participants are listed in Table 1. This study was approved by the Ethics Committee of the Fifth Medical Center of the PLA General Hospital. Each subject signed an informed consent form before enrollment.
Sample collection
Peripheral blood samples were collected under sterile conditions and were immediately processed. The sample was then divided into two aliquots of 4 mL in a biosafety cabinet, and one of the tubes was stored in a −80°C refrigerator until DNA extraction. The other was centrifuged at 400 × g for 10 minutes to isolate the plasma, which was stored frozen at −80°C for further proteomic analysis. Peripheral blood mononuclear cells were isolated using density gradient centrifugation using Ficoll-Paque PLUS (17-1440-03, GE Healthcare).
The researchers distributed disposable sterile potty and tubes to the participants in advance. Participants first discharged the feces in a sterile potty, then washed their hands, put on disposable gloves, and took the middle part of the feces. Fecal samples from patients and HCs were freshly collected and frozen at −80°C within 4 hours after sampling.
DNA extraction
All microbial DNA was extracted from whole blood and stool samples using the QIAamp DNA Blood Mini Kit (51106, Qiagen) and QIAamp PowerFecal Pro DNA Kit (51804, Qiagen), according to the manufacturer’s instructions. The concentration and purity of the extracted DNA were determined using TBS-380 (Turner Biosystems, Sunnyvale, CA, USA) and NanoDrop2000 (Thermo Scientific, Wilmington, DE, USA), respectively. DNA extraction quality was checked on a 1% agarose gel.
DNA library construction and sequencing
Paired-end library was constructed using NEXTflexTM Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). Shotgun metagenomic sequencing was performed on DNBSEQ-T7 platform (MGI Tech Co., Ltd., Guangdong, China) using DNBSEQ-T7RS Reagent Kit (FCL PE150) version 2.0, according to the manufacturer’s instructions. All samples were paired-end sequenced with a 150-bp read length to a targeted data size of 10.0 Gb.
Sequence quality control and genome assembly
The raw reads from metagenome sequencing were used to generate clean reads by removing adaptor sequences, trimming, and removing low-quality reads using fastp (version 0.20.0). The clean reads were mapped to the human hg38 reference genome using BWA (version 0.7.9 a) to identify and remove human host-originated reads.
These high-quality reads were then assembled into contigs using MEGAHIT (version 1.1.2), which use succinct de Bruijn graphs. Contigs with lengths greater than 300 bp were selected as the final assembly result.
Gene prediction and taxonomy
Open reading frames in contigs were identified using MetaGene. A nonredundant gene catalog was constructed using CD-HIT (version 4.6.1) with 90% sequence identity and 90% coverage. Reads after quality control were mapped to the nonredundant gene catalog with 95% identity using SOAPaligner (version 2.21), and gene abundance in each sample was evaluated. Representative sequences of the non-redundant gene catalog were annotated based on the NCBI NR database using BLAST as implemented in DIAMOND version 0.9.19 with an e-value cutoff of 1e−5 using Diamond (version 0.8.35) for taxonomic annotations.
Microbial community diversity analysis
Each sample’s alpha diversity (Shannon index) was calculated using the R package VEGAN (version 2.5.3) on the relative abundance of species. Species richness for all samples was estimated based on rarefied data. Beta diversities (Bray-Curtis dissimilarities) among samples were calculated using VEGAN based on the relative abundance of species.
Plasma protein profiling
The OLINK inflammation panel of 92 proteins was used, which uses the proximity extension assay technology to obtain normalized protein expression (NPX) values for 92 proteins. (Olink Proteomics, Watertown, MA, USA; see Table S1 for an overview of immune markers, including their full names). Each antibody was labeled separately with unique proximity extension assay oligonucleotide probes, two separate and complementary sequences. NPX was calculated from the Ct values of the PCR readout. The analytical performance of each protein assay included in the panel was carefully validated based on specificity, sensitivity, dynamic range, precision, scalability, endogenous interference, and detectability (http://www.olink.com). After quality control, 67 proteins were included in the analysis.
Detection of HIV DNA and RNA
Total cellular HIV DNA and RNA were extracted from peripheral blood mononuclear cells using QIAamp DNA Blood Mini Kit (51106, Qiagen) and HiPure Total RNA Plus Mini Kits (R4121-01, Magen), respectively. HIV DNA and RNA were quantified using the HIV DNA Quantitative Detection Kit (SUPI-1116, SUPBIO) and HIV RNA Quantitative Detection Kit (SUPI-0103, SUPBIO). Both kits were used according to the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using R Studio version 4.1.0 (R Studio, Boston, MA, USA), GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA), and SPSS version 26.0 (SPASS, Chicago, IL, USA). Continuous variables are expressed as medians (IQRs), whereas categorical variables are presented as numbers (percentages). The Mann-Whitney U test (between two groups) and Kruskal-Wallis test (for more than two groups) were used to compare continuous data. To visualize the microbial community and functional profiles, as well as the protein distribution among the three groups, we calculated the between-sample diversity score (Bray-Curtis distance). PCA was used to visualize the separation. To test the difference in microbial composition between two or more groups, ANOSIM was employed based on Bray-Curtis dissimilarity. LEfSe analysis was used to identify the taxa or functional profiles most likely to explain the differences between two or three groups. An LDA score cutoff of 2.5 indicated a significant difference. Correlations were determined using the Spearman rank correlation test between two continuous variables. P < 0.05 was considered statistically significant. In addition, a Benjamini-Hochberg false-discovery rate-corrected P value was estimated.
ACKNOWLEDGMENTS
The authors thank all the volunteers and colleagues from the Fifth Medical Center of Chinese PLA General Hospital who volunteered to provide samples and help for this study.
This work was supported by the National Natural Science Foundation of China under grants 82272317, 82130019, and 82171732 and the National Key Research and Development Program of China under grant 2022YFA1303600.
The authors report that there are no competing interests to declare.
AFTER EPUB
[This article was published on 12 September 2023 with an error in the order of corresponding authors in the article PDF. The error was corrected in the current version, posted on 26 October 2023.]
Contributor Information
Fu-Sheng Wang, Email: fswang302@163.com.
Lijun Sun, Email: sunlijunkity@sina.com.
Yan-Mei Jiao, Email: jiaoyanmei@sina.com.
Hongwei Zhou, Southern Medical University, Guangzhou, Guandong, China .
DATA AVAILABILITY
Sequences and metadata are available in NCBI under accession number PRJNA953845.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00467-23.
Legends to Fig. S1 to S5.
Taxonomic analysis of the gut microbiota of study subjects.
Taxon relative abundance of blood microbiota at every taxonomic level.
Expression levels of the species in feces corresponding to Figure 4.
Differential inflammation-related proteins in immunological non-responders and immunological responders versus healthy controls.
Inflammation-related proteins in plasma associated with HIV disease status and immune recovery.
Tables S1 to S6.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong M, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, Tiemessen CT, Vandekerckhove L, Lewin SR, International AIDS Society (IAS) Global Scientific Strategy working group . 2021. Research priorities for an HIV cure: international AIDS society global scientific strategy 2021. Nat Med 27:2085–2098. doi: 10.1038/s41591-021-01590-5 [DOI] [PubMed] [Google Scholar]
- 2. Ndung’u T, McCune JM, Deeks SG. 2019. Why and where an HIV cure is needed and how it might be achieved. Nature 576:397–405. doi: 10.1038/s41586-019-1841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lv T, Cao W, Li T. 2021. HIV-related immune activation and inflammation: current understanding and strategies. J Immunol Res 2021:7316456. doi: 10.1155/2021/7316456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neuhaus J, Jacobs DR, Baker JV, Calmy A, Duprez D, La Rosa A, Kuller LH, Pett SL, Ristola M, Ross MJ, Shlipak MG, Tracy R, Neaton JD. 2010. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J Infect Dis 201:1788–1795. doi: 10.1086/652749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, Robinson J, Huang Y, Epling L, Martin JN, Deeks SG, Meinert CL, Van Natta ML, Jabs DA, Lederman MM. 2014. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis 210:1228–1238. doi: 10.1093/infdis/jiu238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, Plants J, Seth A, Wilson CC, Deeks SG, Lederman MM, Landay AL. 2014. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis 210:1248–1259. doi: 10.1093/infdis/jiu254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunt PW. 2017. Very early ART and persistent inflammation in treated HIV. Clin Infect Dis 64:132–133. doi: 10.1093/cid/ciw697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. So-Armah KA, Tate JP, Chang C-CH, Butt AA, Gerschenson M, Gibert CL, Leaf D, Rimland D, Rodriguez-Barradas MC, Budoff MJ, Samet JH, Kuller LH, Deeks SG, Crothers K, Tracy RP, Crane HM, Sajadi MM, Tindle HA, Justice AC, Freiberg MS, VACS Project Team . 2016. Do biomarkers of inflammation, monocyte activation, and altered coagulation explain excess mortality between HIV infected and uninfected people. J Acquir Immune Defic Syndr 72:206–213. doi: 10.1097/QAI.0000000000000954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. doi: 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serrano-Villar S, Sainz T, Lee SA, Hunt PW, Sinclair E, Shacklett BL, Ferre AL, Hayes TL, Somsouk M, Hsue PY, Van Natta ML, Meinert CL, Lederman MM, Hatano H, Jain V, Huang Y, Hecht FM, Martin JN, McCune JM, Moreno S, Deeks SG. 2014. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell Subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. PLoS Pathog 10:e1004078. doi: 10.1371/journal.ppat.1004078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, Cox S, Engen P, Chakradeo P, Abbasi R, Gorenz A, Burns C, Landay A. 2014. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 10:e1003829. doi: 10.1371/journal.ppat.1003829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, Deeks SG, Hunt PW, Lynch SV, McCune JM. 2013. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med 5:193ra91. doi: 10.1126/scitranslmed.3006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zicari S, Sessa L, Cotugno N, Ruggiero A, Morrocchi E, Concato C, Rocca S, Zangari P, Manno EC, Palma P. 2019. Immune activation, inflammation, and non-AIDS co-morbidities in HIV-infected patients under long-term ART. Viruses 11:200. doi: 10.3390/v11030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. 2013. Host-derived nitrate boosts growth of E.coli in the inflamed gut. Science 339:708–711. doi: 10.1126/science.1232467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Deleage C, Schuetz A, Alvord WG, Johnston L, Hao X-P, Morcock DR, Rerknimitr R, Fletcher JLK, Puttamaswin S, Phanuphak N, Dewar R, McCune JM, Sereti I, Robb M, Kim JH, Schacker TW, Hunt P, Lifson JD, Ananworanich J, Estes JD, on behalf of the RV254/SEARCH 010 and RV304/SEARCH 013 Study Groups . 2016. Impact of early cART in the gut during acute HIV infection. JCI Insight 1:e87065. doi: 10.1172/jci.insight.87065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larsen JM. 2017. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151:363–374. doi: 10.1111/imm.12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. doi: 10.1038/nm1511 [DOI] [PubMed] [Google Scholar]
- 18. Marchetti G, Bellistrì GM, Borghi E, Tincati C, Ferramosca S, La Francesca M, Morace G, Gori A, Monforte AD. 2008. Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS 22:2035–2038. doi: 10.1097/QAD.0b013e3283112d29 [DOI] [PubMed] [Google Scholar]
- 19. Marchetti G, Tincati C, Silvestri G. 2013. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev 26:2–18. doi: 10.1128/CMR.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nganou-Makamdop K, Talla A, Sharma AA, Darko S, Ransier A, Laboune F, Chipman JG, Beilman GJ, Hoskuldsson T, Fourati S, Schmidt TE, Arumugam S, Lima NS, Moon D, Callisto S, Schoephoerster J, Tomalka J, Mugyenyi P, Ssali F, Muloma P, Ssengendo P, Leda AR, Cheu RK, Flynn JK, Morou A, Brunet-Ratnasingham E, Rodriguez B, Lederman MM, Kaufmann DE, Klatt NR, Kityo C, Brenchley JM, Schacker TW, Sekaly RP, Douek DC. 2021. Translocated microbiome composition determines immunological outcome in treated HIV infection. Cell 184:3899–3914. doi: 10.1016/j.cell.2021.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Serrano-Villar S, Sanchez-Carrillo S, Talavera-Rodríguez A, Lelouvier B, Gutiérrez C, Vallejo A, Servant F, Bernadino JI, Estrada V, Madrid N, Gosalbes MJ, Bisbal O, de Lagarde M, Martínez-Sanz J, Ron R, Herrera S, Moreno S, Ferrer M. 2021. Blood bacterial profiles associated with human immunodeficiency virus infection and immune recovery. J Infect Dis 223:471–481. doi: 10.1093/infdis/jiaa379 [DOI] [PubMed] [Google Scholar]
- 22. Lu W, Feng Y, Jing F, Han Y, Lyu N, Liu F, Li J, Song X, Xie J, Qiu Z, Zhu T, Routy B, Routy J-P, Li T, Zhu B. 2018. Association between gut microbiota and CD4 recovery in HIV-1 infected patients. Front Microbiol 9:1451. doi: 10.3389/fmicb.2018.01451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun Y, Ma Y, Lin P, Tang Y-W, Yang L, Shen Y, Zhang R, Liu L, Cheng J, Shao J, Qi T, Tang Y, Cai R, Guan L, Luo B, Sun M, Li B, Pei Z, Lu H. 2016. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Infect 5:e31. doi: 10.1038/emi.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLaughlin RW, Vali H, Lau PCK, Palfree RGE, De Ciccio A, Sirois M, Ahmad D, Villemur R, Desrosiers M, Chan ECS. 2002. Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J Clin Microbiol 40:4771–4775. doi: 10.1128/JCM.40.12.4771-4775.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Amar J, Lange C, Payros G, Garret C, Chabo C, Lantieri O, Courtney M, Marre M, Charles MA, Balkau B, Burcelin R, Bayer A, D.E.S.I.R. Study Group . 2013. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: The D.E.S.I.R study. PLoS ONE 8:e54461. doi: 10.1371/journal.pone.0054461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amar J, Serino M, Lange C, Chabo C, Iacovoni J, Mondot S, Lepage P, Klopp C, Mariette J, Bouchez O, Perez L, Courtney M, Marre M, Klopp P, Lantieri O, Doré J, Charles MA, Balkau B, Burcelin R. 2011. Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia 54:3055–3061. doi: 10.1007/s00125-011-2329-8 [DOI] [PubMed] [Google Scholar]
- 27. Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. 2014. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One 9:e105221. doi: 10.1371/journal.pone.0105221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Castillo DJ, Rifkin RF, Cowan DA, Potgieter M. 2019. The healthy human blood microbiome: fact or fiction? Front Cell Infect Microbiol 9:148. doi: 10.3389/fcimb.2019.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gosalbes MJ, Jimenéz-Hernandéz N, Moreno E, Artacho A, Pons X, Ruíz-Pérez S, Navia B, Estrada V, Manzano M, Talavera-Rodriguez A, Madrid N, Vallejo A, Luna L, Pérez-Molina JA, Moreno S, Serrano-Villar S. 2022. Interactions among the mycobiome, bacteriome, inflammation, and diet in people living with HIV. Gut Microbes 14:2089002. doi: 10.1080/19490976.2022.2089002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hunt PW. 2010. Th17, gut, and HIV: therapeutic implications. Curr Opin HIV AIDS 5:189–193. doi: 10.1097/COH.0b013e32833647d9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manzano M, Talavera-Rodríguez A, Moreno E, Madrid N, Gosalbes MJ, Ron R, Dronda F, Pérez-Molina JA, Lanza VF, Díaz J, Moreno S, Navia B, Serrano-Villar S. 2022. Relationship of diet to gut microbiota and inflammatory biomarkers in people with HIV. Nutrients 14:1221. doi: 10.3390/nu14061221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tanes C, Walker EM, Slisarenko N, Gerrets GL, Grasperge BF, Qin X, Jazwinski SM, Bushman FD, Bittinger K, Rout N. 2021. Gut microbiome changes associated with epithelial barrier damage and systemic inflammation during antiretroviral therapy of chronic SIV infection. Viruses 13:1567. doi: 10.3390/v13081567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meyer-Myklestad MH, Medhus AW, Lorvik KB, Seljeflot I, Hansen SH, Holm K, Stiksrud B, Trøseid M, Hov JR, Kvale D, Dyrhol-Riise AM, Kummen M, Reikvam DH. 2022. Human immunodeficiency virus-infected immunological nonresponders have colon-restricted gut mucosal immune dysfunction. J Infect Dis 225:661–674. doi: 10.1093/infdis/jiaa714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puri P, Liangpunsakul S, Christensen JE, Shah VH, Kamath PS, Gores GJ, Walker S, Comerford M, Katz B, Borst A, Yu Q, Kumar DP, Mirshahi F, Radaeva S, Chalasani NP, Crabb DW, Sanyal AJ, TREAT Consortium . 2018. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 67:1284–1302. doi: 10.1002/hep.29623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ciocan D, Cassard A-M, Becquemont L, Verstuyft C, Voican CS, El Asmar K, Colle R, David D, Trabado S, Feve B, Chanson P, Perlemuter G, Corruble E. 2021. Blood microbiota and metabolomic signature of major depression before and after antidepressant treatment: a prospective case-control study. J Psychiatry Neurosci 46:E358–E368. doi: 10.1503/jpn.200159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gelpi M, Vestad B, Hansen SH, Holm K, Drivsholm N, Goetz A, Kirkby NS, Lindegaard B, Lebech A-M, Hoel H, Michelsen AE, Ueland T, Gerstoft J, Lundgren J, Hov JR, Nielsen SD, Trøseid M. 2020. Impact of human immunodeficiency virus–related gut microbiota alterations on metabolic comorbid conditions. Clin Infect Dis 71:e359–e367. doi: 10.1093/cid/ciz1235 [DOI] [PubMed] [Google Scholar]
- 37. Qiu J, Zhou H, Jing Y, Dong C. 2019. Association between blood microbiome and type 2 diabetes mellitus: a nested case-control study. J Clin Lab Anal 33:e22842. doi: 10.1002/jcla.22842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reyes L. 2021. Porphyromonas gingivalis. Trends Microbiol 29:376–377. doi: 10.1016/j.tim.2021.01.010 [DOI] [PubMed] [Google Scholar]
- 39. Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, Nguyen M, Haditsch U, Raha D, Griffin C, Holsinger LJ, Arastu-Kapur S, Kaba S, Lee A, Ryder MI, Potempa B, Mydel P, Hellvard A, Adamowicz K, Hasturk H, Walker GD, Reynolds EC, Faull RLM, Curtis MA, Dragunow M, Potempa J. 2019. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv 5:eaau3333. doi: 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johansson L, Sherina N, Kharlamova N, Potempa B, Larsson B, Israelsson L, Potempa J, Rantapää-Dahlqvist S, Lundberg K. 2016. Concentration of antibodies against Porphyromonas gingivalis is increased before the onset of symptoms of rheumatoid arthritis. Arthritis Res Ther 18:257. doi: 10.1186/s13075-016-1164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kato T, Yamazaki K, Nakajima M, Date Y, Kikuchi J, Hase K, Ohno H, Yamazaki K, D’Orazio SEF. 2018. Oral administration of Porphyromonas gingivalis alters the gut microbiome and serum metabolome. mSphere 3:e00460-18. doi: 10.1128/mSphere.00460-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mantri CK, Chen C, Dong X, Goodwin JS, Xie H. 2014. Porphyromonas gingivalis-mediated epithelial cell entry of HIV-1. J Dent Res 93:794–800. doi: 10.1177/0022034514537647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dong X-H, Ho M-H, Liu B, Hildreth J, Dash C, Goodwin JS, Balasubramaniam M, Chen C-H, Xie H. 2018. Role of Porphyromonas gingivalis outer membrane vesicles in oral mucosal transmission of HIV. Sci Rep 8:8812. doi: 10.1038/s41598-018-27284-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onderdonk AB, Delaney ML, Fichorova RN. 2016. The human microbiome during bacterial vaginosis. Clin Microbiol Rev 29:223–238. doi: 10.1128/CMR.00075-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou J, Zhang Y, Cui P, Luo L, Chen H, Liang B, Jiang J, Ning C, Tian L, Zhong X, Ye L, Liang H, Huang J. 2020. Gut microbiome changes associated with HIV infection and sexual orientation. Front Cell Infect Microbiol 10:434. doi: 10.3389/fcimb.2020.00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Armstrong AJS, Shaffer M, Nusbacher NM, Griesmer C, Fiorillo S, Schneider JM, Preston Neff C, Li SX, Fontenot AP, Campbell T, Palmer BE, Lozupone CA. 2018. An exploration of Prevotella-rich microbiomes in HIV and men who have sex with men. Microbiome 6:198. doi: 10.1186/s40168-018-0580-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmerk CL, Valvano MA. 2013. Burkholderia multivorans survival and trafficking within macrophages. J Med Microbiol 62:173–184. doi: 10.1099/jmm.0.051243-0 [DOI] [PubMed] [Google Scholar]
- 48. Mizuki E, Ohba M, Akao T, Yamashita S, Saitoh H, Park YS. 1999. Unique activity associated with non-insecticidal Bacillus thuringiensis parasporal inclusions: in vitro cell-killing action on human cancer cells. J Appl Microbiol 86:477–486. doi: 10.1046/j.1365-2672.1999.00692.x [DOI] [PubMed] [Google Scholar]
- 49. Nagamatsu Y, Okamura S, Saitou H, Akao T, Mizuki E. 2010. Three cry toxins in two types from Bacillus thuringiensis strain M019 preferentially kill human hepatocyte cancer and uterus cervix cancer cells. Biosci Biotechnol Biochem 74:494–498. doi: 10.1271/bbb.90615 [DOI] [PubMed] [Google Scholar]
- 50. Yamashita S, Katayama H, Saitoh H, Akao T, Park YS, Mizuki E, Ohba M, Ito A. 2005. Typical three-domain cry proteins of Bacillus thuringiensis strain A1462 exhibit cytocidal activity on limited human cancer cells. J Biochem 138:663–672. doi: 10.1093/jb/mvi177 [DOI] [PubMed] [Google Scholar]
- 51. Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baker-Austin C, Oliver JD. 2020. Vibrio vulnificus. Trends Microbiol 28:81–82. doi: 10.1016/j.tim.2019.08.006 [DOI] [PubMed] [Google Scholar]
- 53. Margolis DM, Archin NM, Cohen MS, Eron JJ, Ferrari G, Garcia JV, Gay CL, Goonetilleke N, Joseph SB, Swanstrom R, Turner A-M, Wahl A. 2020. Curing HIV: seeking to target and clear persistent infection. Cell 181:189–206. doi: 10.1016/j.cell.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neurath MF, Überla K, Ng SC. 2021. Gut as viral reservoir: lessons from gut viromes, HIV and COVID-19. Gut 70:1605–1608. doi: 10.1136/gutjnl-2021-324622 [DOI] [PubMed] [Google Scholar]
- 55. Borgognone A, Noguera-Julian M, Oriol B, Noël-Romas L, Ruiz-Riol M, Guillén Y, Parera M, Casadellà M, Duran C, Puertas MC, Català-Moll F, De Leon M, Knodel S, Birse K, Manzardo C, Miró JM, Clotet B, Martinez-Picado J, Moltó J, Mothe B, Burgener A, Brander C, Paredes R, BCN02 Study Group . 2022. Gut microbiome signatures linked to HIV-1 reservoir size and viremia control. Microbiome 10:59. doi: 10.1186/s40168-022-01247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zheng D, Liwinski T, Elinav E. 2020. Interaction between microbiota and immunity in health and disease. Cell Res 30:492–506. doi: 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alzahrani J, Hussain T, Simar D, Palchaudhuri R, Abdel-Mohsen M, Crowe SM, Mbogo GW, Palmer CS. 2019. Inflammatory and immunometabolic consequences of gut dysfunction in HIV: parallels with IBD and implications for reservoir persistence and non-AIDS comorbidities. EBioMedicine 46:522–531. doi: 10.1016/j.ebiom.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Castellano P, Prevedel L, Valdebenito S, Eugenin EA. 2019. HIV infection and latency induce a unique metabolic signature in human macrophages. Sci Rep 9:3941. doi: 10.1038/s41598-019-39898-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palmer CS, Palchaudhuri R, Albargy H, Abdel-Mohsen M, Crowe SM. 2018. Exploiting immune cell metabolic machinery for functional HIV cure and the prevention of Inflammaging. F1000Res 7:125. doi: 10.12688/f1000research.11881.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yin X, Wang Z, Wu T, Ma M, Zhang Z, Chu Z, Hu Q, Ding H, Han X, Xu J, Shang H, Jiang Y. 2019. The combination of CXCL9, CXCL10 and CXCL11 levels during primary HIV infection predicts HIV disease progression. J Transl Med 17:417. doi: 10.1186/s12967-019-02172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanda N, Shimizu T, Tada Y, Watanabe S. 2007. IL-18 enhances IFN-gamma-induced production of CXCL9, CXCL10, and CXCL11 in human keratinocytes. Eur J Immunol 37:338–350. doi: 10.1002/eji.200636420 [DOI] [PubMed] [Google Scholar]
- 62. Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, Boucher G, Haddad EK, Sekaly R-P, Harman AN, Anderson JL, Jones KL, Mak J, Cunningham AL, Jaworowski A, Lewin SR. 2010. Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci U S A 107:16934–16939. doi: 10.1073/pnas.1002894107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng A-L, Yau T, Furuse J, Park J-W, Boyd Z, Tang HT, Shen Y, Tschaika M, Neely J, El-Khoueiry A. 2020. Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 73:1460–1469. doi: 10.1016/j.jhep.2020.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pasquereau S, Kumar A, Herbein G. 2017. Targeting TNF and TNF receptor pathway in HIV-1 infection: from immune activation to viral reservoirs. Viruses 9:64. doi: 10.3390/v9040064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. International AIDS Society Scientific Working Group on HIV Cure, Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun T-W, Churchill M, Di Mascio M, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud M-C, Peterlin M, Poli G, Routy J-P, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Van Lint C, Verdin E, Woolfrey A, Zaia J, Barré-Sinoussi F. 2012. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12:607–614. doi: 10.1038/nri3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Evans HM, Schultz DF, Boiman AJ, McKell MC, Qualls JE, Deepe GS, Wormley FL. 2021. Restraint of fumarate accrual by HIF-1α preserves miR-27a-mediated limitation of interleukin 10 during infection of macrophages by Histoplasma capsulatum. mBio 12:e0271021. doi: 10.1128/mBio.02710-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gieseck RL, Wilson MS, Wynn TA. 2018. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol 18:62–76. doi: 10.1038/nri.2017.90 [DOI] [PubMed] [Google Scholar]
- 68. Wallace CH, Wu BX, Salem M, Ansa-Addo EA, Metelli A, Sun S, Gilkeson G, Shlomchik MJ, Liu B, Li Z. 2018. B lymphocytes confer immune tolerance via cell surface GARP-TGF-β complex. JCI Insight 3:e99863. doi: 10.1172/jci.insight.99863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), Canadian Interventional Radiology Association (CIRA), Congress of Neurological Surgeons (CNS), European Society of Minimally Invasive Neurological Therapy (ESMINT), European Society of Neuroradiology (ESNR), European Stroke Organization (ESO), Society for Cardiovascular Angiography and Interventions (SCAI), Society of Interventional Radiology (SIR), Society of NeuroInterventional Surgery (SNIS), and World Stroke Organization (WSO), Sacks D, Baxter B, Campbell BCV, Carpenter JS, Cognard C, Dippel D, Eesa M, Fischer U, Hausegger K, Hirsch JA, Shazam Hussain M, Jansen O, Jayaraman MV, Khalessi AA, Kluck BW, Lavine S, Meyers PM, Ramee S, Rüfenacht DA, Schirmer CM, Vorwerk D. 2018. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke 13:612–632. doi: 10.1177/1747493018778713 [DOI] [PubMed] [Google Scholar]
- 70. Babu H, Ambikan AT, Gabriel EE, Svensson Akusjärvi S, Palaniappan AN, Sundaraj V, Mupanni NR, Sperk M, Cheedarla N, Sridhar R, Tripathy SP, Nowak P, Hanna LE, Neogi U. 2019. Systemic inflammation and the increased risk of inflamm-aging and age-associated diseases in people living with HIV on long term suppressive antiretroviral therapy. Front Immunol 10:1965. doi: 10.3389/fimmu.2019.01965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dasgupta Y, Golovine K, Nieborowska-Skorska M, Luo L, Matlawska-Wasowska K, Mullighan CG, Skorski T. 2018. Drugging DNA repair to target T-ALL cells. Leuk Lymphoma 59:1746–1749. doi: 10.1080/10428194.2017.1397662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mora S, Zamproni I, Cafarelli L, Giacomet V, Erba P, Zuccotti G, Viganò A. 2007. Alterations in circulating osteoimmune factors may be responsible for high bone resorption rate in HIV-infected children and adolescents. AIDS 21:1129–1135. doi: 10.1097/QAD.0b013e32810c8ccf [DOI] [PubMed] [Google Scholar]
- 73. Schett G, Kiechl S, Redlich K, Oberhollenzer F, Weger S, Egger G, Mayr A, Jocher J, Xu Q, Pietschmann P, Teitelbaum S, Smolen J, Willeit J. 2004. Soluble RANKL and risk of nontraumatic fracture. JAMA 291:1108–1113. doi: 10.1001/jama.291.9.1108 [DOI] [PubMed] [Google Scholar]
- 74. Geng S-T, Zhang Z-Y, Wang Y-X, Lu D, Yu J, Zhang J-B, Kuang Y-Q, Wang K-H. 2020. Regulation of gut microbiota on immune reconstitution in patients with acquired immunodeficiency syndrome. Front Microbiol 11:594820. doi: 10.3389/fmicb.2020.594820 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends to Fig. S1 to S5.
Taxonomic analysis of the gut microbiota of study subjects.
Taxon relative abundance of blood microbiota at every taxonomic level.
Expression levels of the species in feces corresponding to Figure 4.
Differential inflammation-related proteins in immunological non-responders and immunological responders versus healthy controls.
Inflammation-related proteins in plasma associated with HIV disease status and immune recovery.
Tables S1 to S6.
Data Availability Statement
Sequences and metadata are available in NCBI under accession number PRJNA953845.