ABSTRACT
Disruption of blood glucose utilization may lead to diabetes mellitus, which has complex genetic and environmental aspects, and free fatty acid receptors (FFARs) may bridge the genetic and dietary aspects. FFAR4 has been identified as a new target for diabetes treatment, and it is essential to investigate how FFAR4 affects glucose homeostasis. FFAR4 knockout mice have been shown to cause severely impaired glucose tolerance under high-fat feeding conditions; however, the findings in FFAR4 knockout mice under chow diet conditions seem to be contradictory. Blood glucose utilization in mice under chow diet conditions is thought to show diurnal rhythmicity. In this study, we aim to investigate the role of FFAR4 in glucose utilization rhythm and explore the possible mechanism. Total Ffar4 and gut-specific Ffar4 knockout mice both showed a clear diurnal rhythm in glucose tolerance. However, deletion of total Ffar4 and gut-specific Ffar4 both deteriorate glucose tolerance at the daily light to dark transition (ZT12) in mice. We show that intestinal FFAR4 deficiency leads to significant changes in fecal microbiota at different ZTs. 16S rRNA sequencing results suggested that Akkermansia muciniphila was the main target of intestinal FFAR4 at ZT12. Akkermansia muciniphila supplementation significantly improved the impaired glucose tolerance at ZT12 in mice with gut-specific Ffar4 deletion and was accompanied by recovery of serum hormone level and transcripts of key genes in glucose metabolism.
IMPORTANCE
Alterations in the intestinal environment are associated with various diseases, and FFAR4 is abundantly enriched in the intestine, where it has been shown to have the ability to regulate intestinal hormone secretion and intestinal microbiota; here, we confirmed previous reports. Meanwhile, we found that intestinal FFAR4 regulates glucagon-like peptide 1 secretion by decreasing Akkermansia muciniphila abundance and show that such change is associated with the level of glucose utilization at ZT12 in mice. Intestinal FFAR4 deficiency leads to severely impaired glucose tolerance at the ZT12 moment in mice, and Akkermansia muciniphila supplementation ameliorates the abnormal glucose utilization at the ZT12 moment caused by FFAR4 deficiency, which is very similar to the dawn phenomenon in diabetic patients. Collectively, our data suggest that intestinal Ffar4 deteriorates glucose tolerance at the daily light to dark transition by affecting Akkermansia muciniphila.
KEYWORDS: FFAR4, glucose tolerance, Akkermansia muciniphila, GLP-1
INTRODUCTION
The worldwide epidemic of type 2 diabetes has become a major challenge for public health (1). The most significant concern in the prevention and treatment of type 2 diabetes is the daily management of blood glucose (2). In healthy individuals, insulin sensitivity is significantly higher at the time of awakening than during sleep, which leads to increased glucose tolerance while awakening (3 – 6). Patients with diabetes frequently experience the “dawn phenomenon” (DP), wherein blood glucose or insulin demand increases spontaneously at dawn despite their efforts to lower blood glucose levels (7 – 9). One possible mechanism is impairment in the rhythm of glucose tolerance (9, 10). Little is known about its precise mechanism and the key players governing the glucose utilization rhythm. ZT12 would be the equivalent of dawn for mice since mice are nocturnal. Therefore, the change of glucose tolerance in mice at ZT12 is an important node in the study of dawn phenomenon (9).
G protein-coupled receptors play an important role in regulating glucose metabolism and have been implicated in the development and progression of diabetes (11 – 13). Among them, the free fatty acid receptor (FFAR) FFAR1 (GPR40), FFAR2 (GPR43), FFAR3 (GPR41), and FFAR4 (GPR120) may bridge the genetic and environmental aspects of diabetes (14 – 18). FFAR2 and FFAR3 are activated by short-chain fatty acids (SCFAs) (19), whereas FFAR1 and FFAR4 are activated by medium-chain and long-chain fatty acids (LCFAs) (20, 21). Fatty acids, especially the LCFAs, are major components of the human diet.
FFAR4 is highly expressed in the intestine and primarily serves as the receptor for LCFAs (22 – 24). Studies show that LCFAs regulate the peripheral clock (25). In addition, the circulating level of FGF21, a key downstream regulator of FFAR4, also participates in the rhythm of glucose metabolism (26 – 28). The therapeutic effect of FGF21 on insulin sensitivity has been widely reported. Thus, it is reasonable to hypothesize that FFAR4 may participate in the rhythm of glucose tolerance. Therefore, the role of FFAR4, if any, in improving the rhythm of glucose metabolism deserves attention.
In this study, we found FFAR4 expression shows diurnal oscillations in mice colons, and intestinal Ffar4 deficiency significantly reduced the level of glucose utilization at the daily light to dark transition. Moreover, we found that the impaired glucose tolerance at the daily light to dark transition induced by Ffar4 deletion was mediated by Akkermansia muciniphila.
RESULTS
Ffar4 deficiency impairs the glucose tolerance at certain moments
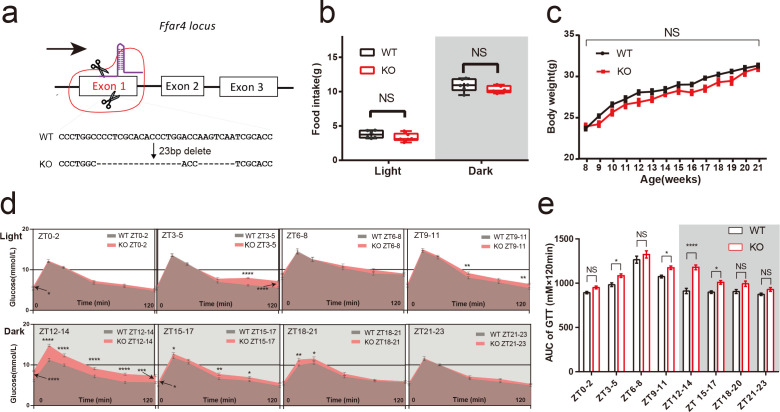
Glucose uptake showed a clear circadian rhythm, which is directly due to glucose-induced differences in insulin release from the islet β cells or the daily variation in tissue sensitivity to insulin (3). FFAR4 has gained considerable recent attention due to its antidiabetic ability (29, 30). To reveal the specific role of Ffar4 in the rhythm of glucose uptake, a mouse model of total Ffar4 knockout (KO) was established to study whether Ffar4 deletion affected the rhythm of glucose uptake (Fig. 1a; Fig. S1a). KO mice showed no effect on their food intake or body weight compared to wild-type (WT) mice under normal chow diet (Fig. 1b and c). On 24-hour glucose detection, there was no difference between WT and KO mice, except for a decrease in blood glucose at the ZT0 moment in the KO mice (P < 0.01) (Fig. S1c). Analysis of glucose tolerance test (GTT) at eight time points revealed that glucose tolerance of both WT and KO mice showed significant diurnal rhythmicity [cosine analysis was performed using areas under curves (AUC): acrophase ZT6.94, bathyphase ZT18.94 (P cosinor < 0.05); acrophase ZT8.27, bathyphase ZT20.27 (P cosinor < 0.001), respectively] (Fig. S1b, d, and e). Notably, KO mice showed a slight impairment of glucose tolerance at ZT3–5; ZT9–11; ZT15–17 (P < 0.05), a robust impairment at ZT12–14 compared with WT mice (P < 0.0001), which is the main cause of the delay of the acrophase and the bathyphase (Fig. 1d and e; Fig. S1d and e). Additionally, analysis of serum metabolism-related hormones revealed that KO mice did not show an altered rhythmicity in their levels of blood insulin, glucagon, and glucocorticoid (GC) but had different levels of growth hormone (GH) at certain ZTs, compared to wild-type mice (S1f-i). Notably, glucagon-like peptide 1 (GLP-1) levels decreased significantly in KO mice at ZT0, ZT6, ZT12, and ZT18 (all P < 0.01) (S1j).
FIG 1.
Ffar4 knockout significantly impaired glucose tolerance at ZT12-14. CRISPR–Cas9 gene editing strategy for total Ffar4 KO mice. Genomic sequencing identified the deletion of 16- and 7-bp nucleotides from the first exon of Ffar4 (a). Day and night food intake (WT/KO mice, n = 6) (b), body weight per week (WT/KO mice, n = 10) (c) between WT and KO mice were recorded. Blood glucose at each time point in GTT (d), AUCs at each ZT (e) between WT and KO mice were performed using two-way ANOVA (WT/KO mice, n = 13 per ZT). Data are expressed as the mean ± standard error of the mean. P < 0.05 was considered statistically significant using two-way ANOVA. * represents P < 0.05;** represents P < 0.01;*** represents P < 0.001;**** represents P < 0.0001, ANOVA, analysis of variance; KO, knockout; NS, not significant; WT, wild type.
Intestinal loss of Ffar4 recapitulates the attenuation of glucose tolerance at ZT12
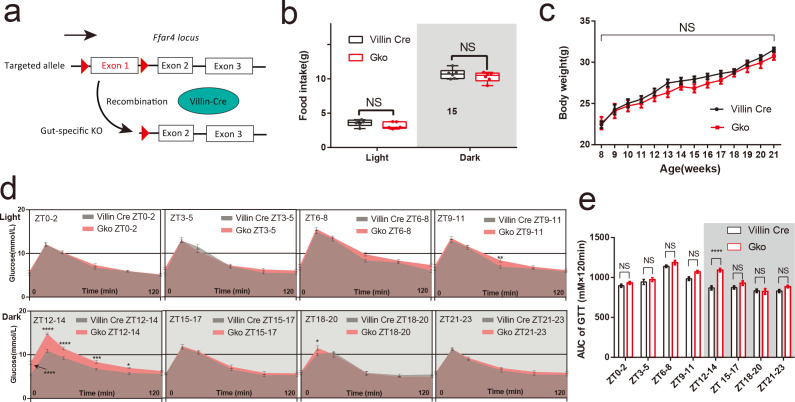
To determine whether intestinal Ffar4 is the main contributor to impairment glucose tolerance, a Gko mouse model was established (Fig. 2a; Fig. S3a). Consistent with the findings in the Ffar4 total KO mice, there were no significant differences in daily food intake, body weight, and 24-hour glucose between Gko mice and Villin-Cre mice (Fig. 2b and c; Fig. S3d). Gko mice also exhibited impaired tolerance at ZT12–14 (Fig. 2d and e). Moreover, glucose tolerance of both Villin Cre and Gko mice showed significant diurnal rhythmicity [cosine analysis was performed using AUC: acrophase ZT6.36, bathyphase ZT18.36 (P cosinor < 0.05); acrophase ZT7.71, bathyphase ZT19.71 (P cosinor < 0.001), respectively] (Fig. S3b and c). Gko mice did not have significant difference levels in blood insulin, glucagon, GC, and GH at any ZTs compared to Villin Cre mice; the difference in GLP-1 levels between Villin Cre and Gko mice was still the most prominent (Fig. S3e and i). These results indicate that loss of intestinal FFAR4 is primarily responsible for the attenuation of the glucose tolerance at ZT12.
FIG 2.
Gut-specific knockout of Ffar4 can result in aberrant glucose tolerance at ZT12–14. Gene targeting strategy for gut-specific Ffar4 knockout (Gko). Mice with floxed exon 1 of Ffar4 were generated by homologous recombination. Gko mice were obtained by mating with Villin-Cre mice (a). Day and night food intake (Villin-Cre/Gko mice, n = 6) (b), body weight per week (Villin-Cre/Gko mice, n = 6) (c). Blood glucose at each time point in GTT (d), AUCs at each ZT (e) between Villin-Cre and Gko mice were performed using two-way ANOVA (Villin-Cre/Gko mice, n = 8 per ZT). Data are expressed as the mean ± standard error of the mean. P < 0.05 was considered statistically significant using two-way ANOVA; * represents P < 0.05;** represents P < 0.01;*** represents P < 0.001;**** represents P < 0.0001; NS, not significant.
FFAR4 deficiency causes changes in the intestinal environment of mice
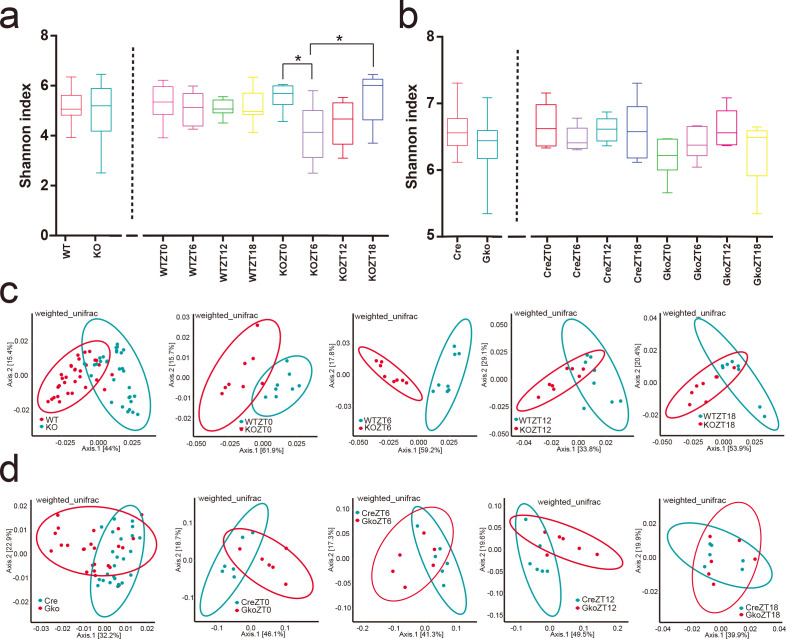
FFAR4 is abundantly distributed in the gut and has been shown to regulate the composition of the intestinal microbiota (31, 32). A variety of gut microbes have been reported to improve glucose metabolism in mice (33, 34). To investigate why FFAR4 deficiency causes impaired glucose tolerance, we analysed the fecal fractions of four genotypes of mice at ZT0, ZT6, ZT12, and ZT18. The alpha diversity of KO mice shows a significant change at the different time points. However, 16S rRNA sequencing analysis from a combined or single ZT both showed no significant differences in microbiota alpha diversity in WT/KO mice (Fig. 3a; Fig. S4c and d) and Villin Cre/Gko mice (Fig. 3b; Fig. S4e and f). In contrast, beta diversity analysis showed that both groups of mice (WT and KO mice; Villin Cre and Gko mice) had a clear separation either individually or combined (Fig. 3c and d). In addition, beta diversity did not change over time at any mouse genotype (Fig. S4g and h). Moreover, mice fecal SCFAs showed that the levels of acetic acid in KO mice increased compared to wild-type mice at ZT12, but no significant differences in propionic and butanoic acid levels at ZT12 (Fig. S4a). The levels of acetic acid, propionic acid, and butyric acid in the feces of Gko mice were not significantly different from those of Villin Cre mice at ZT12 (Fig. S4b).
FIG 3.
FFAR4 deletion induces changes in the alpha and beta diversity of mice intestinal microbes. The alpha diversity (shannon Index) of WT, Villin-Cre, KO, Gko mice were shown in (a and b), * represents P < 0.05. Beta diversity analysis (weighted unifrac) is shown in panels b–d (WT/KO mice, n = 8 per ZT; Villin-Cre/Gko mice, n = 6 per ZT).
Akkermansia muciniphila is a target of Ffar4 deletion-induced aberrant glucose homeostasis
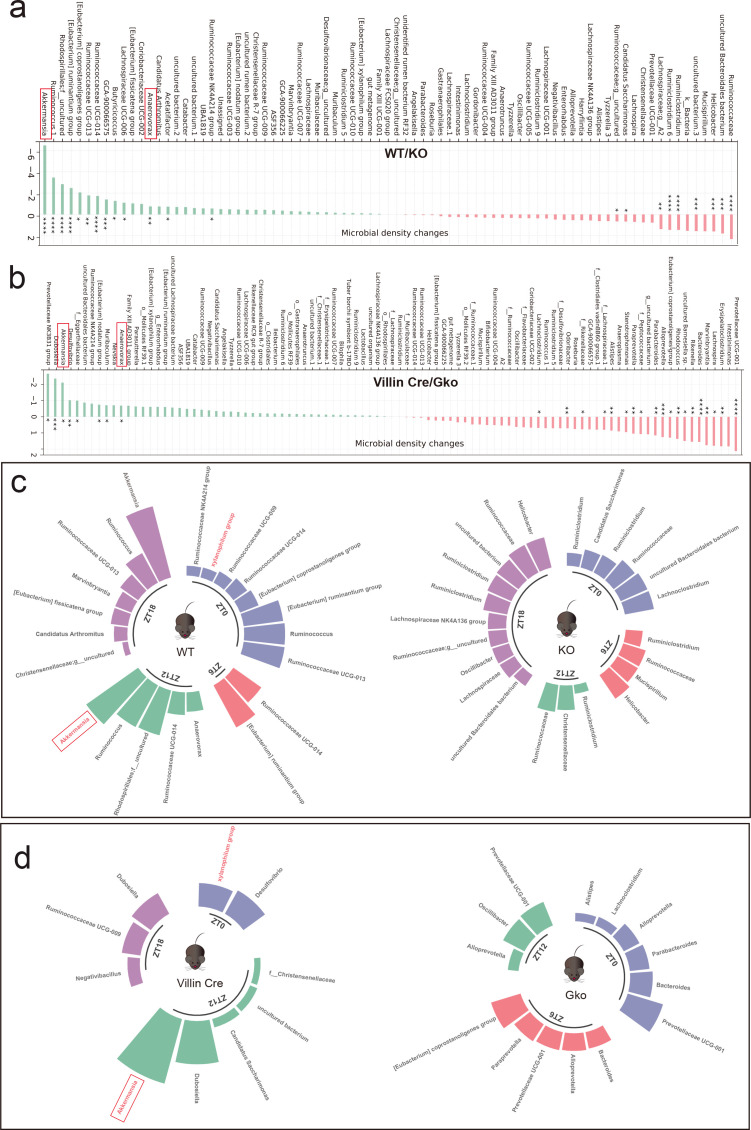
Next, quantification of microbial absolute abundance differences (QMD) was used to identify the dominant microbes at different time points; g_Akkermansia muciniphila was found to be the most dominant groups in the intersection of two groups of mice (Fig. 4a and b). As the microbiome has circadian rhythmicity, it is worth mentioning that g_Akkermansia muciniphila is the most different taxa in WT and KO mice at ZT12 and ZT18 moments (Fig. 4c) and in Villin Cre and Gko mice at ZT12 (Fig. 4d). Moreover, both total Ffar4 deletion and intestinal Ffar4 deletion decreased the abundance levels of Akkermansia muciniphila (A. muciniphila), as detected by quantitative real-time PCR (qPCR) at ZT12 and ZT18 (Fig. S5a and b). Studies by Chiang et al. (35) and Plovier et al. (36) have demonstrated that A. muciniphila is sufficient to improve the metabolic syndromes, such as type 2 diabetes mellitus and obesity (35, 36). Shin et al. (37) found that the oral administration of A. muciniphila to mice reversed high-fat diet-induced obesity and improved glucose tolerance when fed a high-fat diet (37). These results indicate that intestinal Ffar4 deletion significantly reduces the absolute abundance of A. muciniphila, which may be a key gut microbiota in maintaining the glucose homeostasis.
FIG 4.
Deletion of gut Ffar4 led to a decrease in the abundance of Akkermansia muciniphila. QMD was used to show the most significant dominant microbial species in the two groups of mice (a and b) at different ZTs (c and d) (WT/KO mice, n = 8 per ZT; Villin-Cre/Gko mice, n = 6 per ZT). KO, knockout; WT, wild type. * represents P < 0.05;** represents P < 0.01;*** represents P < 0.001;**** represents P < 0.0001.
A. muciniphila supplementation improves abnormal glucose tolerance in Gko mice
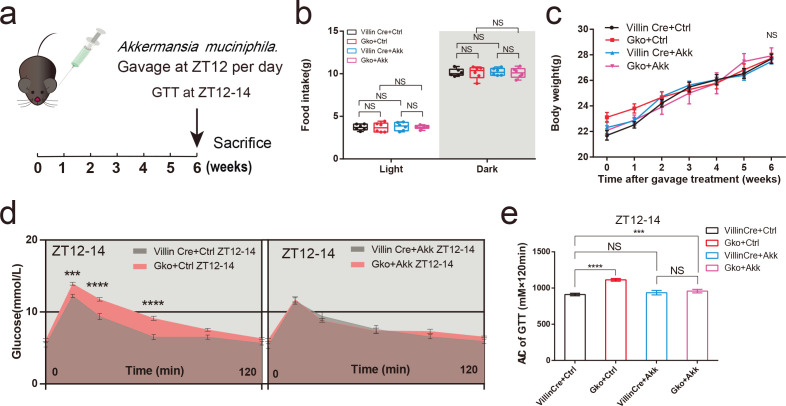
To determine the specific role of A. muciniphila in aberrant glucose tolerance induced by intestinal Ffar4 deletion, Villin Cre and Gko mice were gavaged with a pasteurized solution of A. muciniphila at ZT12 for 6 weeks (Fig. 5a). Within 6 weeks of treatment, there were no significant differences in body weights and the daily food intake of mice compared with those in the control group (Fig. 5b and c). After administration of the oral gavage of pasteurized A. muciniphila solution, Gko mice showed significant improvements in glucose tolerance at ZT12–14 but not in Villin Cre mice. The differences between Gko + Ctrl group and Gko + Akk group were considered significant (P < 0. 001) (Fig. 5d and e). Compared to the control group (Gko + Ctrl), treatment with A. muciniphila significantly increased serum insulin and GLP-1 level (Fig. S6a and e) and decreased glucagon and GH level of Gko mice (Fig. S6b and d) . Insulin levels were upregulated in Villin Cre mice at the ZT12 after pasteurized A. muciniphila gavage compared to the control group (Villin Cre + Ctrl) (Fig. S6a). Although the upregulation of GLP-1 after pasteurized A. muciniphila gavage was not significant, the significant difference in serum GLP-1 levels at ZT12 between Villin Cre and Gko mice was diminished by pasteurized A. muciniphila gavage (Fig. S6e).
FIG 5.
Pasteurized A. muciniphila can improve impaired glucose tolerance at ZT12 in gut-specific Ffar4 knockout (Gko) mice. Gko mice were gavaged with pasteurized A. muciniphila (Akk) for 6 weeks, and the glucose tolerance test was conducted at ZT12–14 in the 6th week (a). Day and night food intake (b), body weight per week (c) between four groups of mice were recorded (Villin Cre + Ctrl/Villin Cre + Akk/Gko + Ctrl/Gko + Akk, n = 7). Blood glucose at each time point in GTT were performed using two-way ANOVA (d), AUCs at ZT12–14 (e) between WT and KO mice were performed using one-way ANOVA (Villin Cre + Ctrl/Villin Cre + Akk/Gko + Ctrl/Gko + Akk, n = 7) (d and e). Data are expressed as the mean ± standard error of the mean. Interaction effects analysis was used to analyze differences between groups (P < 0.05 was considered statistically significant). *** represents P < 0.001;**** represents P < 0.0001; NS, not significant.
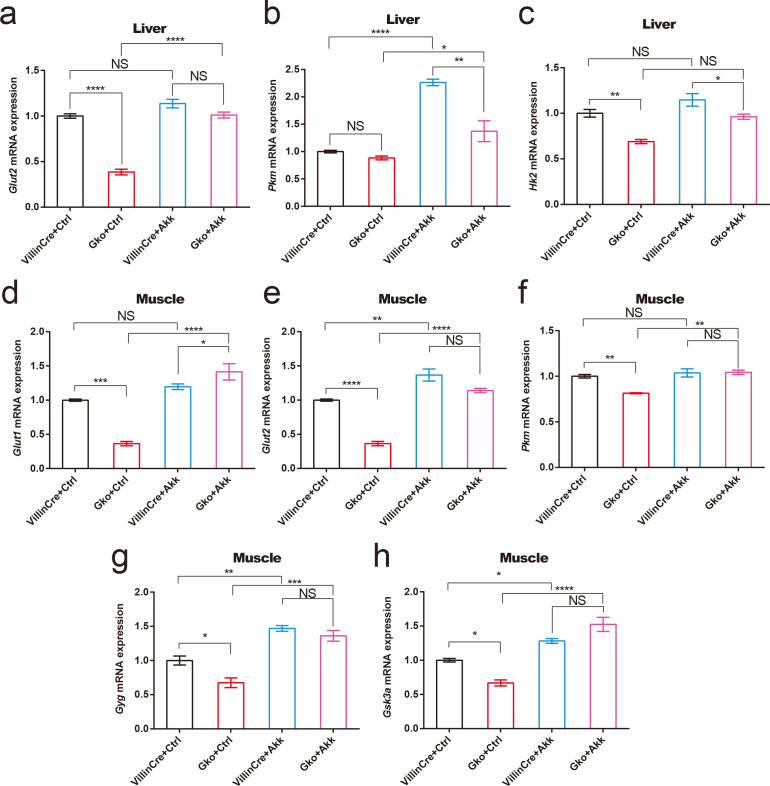
A. muciniphila supplementation improves key transcripts of glucose metabolism at ZT12
In view of the serum hormone changes of mice in each group, we detected the key transcripts of glucose metabolism in muscle and liver. Consistent with the results of impaired glucose tolerance in intestinal FFAR4-deficient mice at the moment of ZT12, compared to Villin Cre control mice, liver Glut2 and Hk2 mRNA levels were significantly downregulated in Gko control mice (Fig. 6a and c); muscle Glut1, Glut2, Pkm, Gyg, and Gsk3a mRNA levels were significantly downregulated in Gko control mice (Fig. 6d and h). These results suggest that intestinal FFAR4 deficiency leads to a downregulation of liver glucose uptake and glycolytic capacity and a downregulation of muscle glucose uptake, glycolysis, and muscle glycogen synthesis capacity at ZT12. After the administration of A. muciniphila solution, liver Pkm mRNA levels and muscle Glut2, Gyg, and Gsk3a mRNA levels were significantly upregulated in Villin Cre mice (Fig. 6b, e, g and h); liver Glut2, Pkm mRNA levels, and muscle Glut1, Glut2, Pkm, Gyg, and Gsk3a mRNA levels were significantly upregulated in Gko mice (Fig. 6a, b, d and h).
FIG 6.
Pasteurized A. muciniphila can improve transcripts of key genes in glucose metabolism at ZT12 in gut-specific Ffar4 knockout (Gko) mice. Glucose metabolism transcript changes in liver (Glut2, Pkm, and Hk2) and muscle (Glut1, Glut2, Pkm, Gyg, and Gsk3a) are shown in panels a–h. Data are expressed as the mean ± standard error of the mean. Interaction effects analysis was used to analyze differences between groups (P < 0.05 was considered statistically significant using one-way ANOVA). * represents P < 0.05;** represents P < 0.01;*** represents P < 0.001;**** represents P < 0.0001; NS, not significant.
DISCUSSION
During the transition from subjective rest to subjective activity, the baseline glucose levels, insulin sensitivity, and glucose tolerance peak gradually in mice, preventing hypoglycemia during sleep and providing energy for activity after waking (3, 8). It has been noted that Ffar3 and Ffar2 mRNA expressions showed a diurnal rhythm in the colonic muscle layer (38). Our data verified the rhythmic expression of FFAR4 from in vivo. Like the diurnal pattern of glucose tolerance in mice, we found that colonic FFAR4 expression was upregulated as mice entered the active phase (Fig. S2). Moreover, both total Ffar4 knockout and gut Ffar4-specific knockout mice severely affected the process of glucose utilization in the waking phase (ZT12). Impaired glucose tolerance at the daily light to dark transition (ZT12) in mice was similar to DP of diabetes in humans (9). Our data are the first to correlate FFAR4 with DP and may lead to a new understanding of the mechanisms of DP occurrence.
Total Ffar4 knockout showed difference in growth hormone and glucagon-like peptide 1 levels at certain ZTs compared to control mice. Compared to control mice, gut-specific knockout mice had severely downregulated serum GLP-1 levels and showed no significant differences in growth hormone, which may be related to the function of FFAR4 in other organs or tissues. It is worth mentioning that FFAR4-specific agonists have been reported to upregulate serum GLP-1 levels in mice, which in turn increases insulin levels (39, 40). However, our data show that the decrease in GLP-1 levels caused by FFAR4 deficiency did not affect the change in serum insulin levels.
Free fatty acid receptor 4 (also known as GPR120) is a specific receptor for n3-polyunsaturated fatty acids (PUFAs). It has been reported that there exist several FFAR4 mutations in diabetic populations (41 – 44); therefore, LCFAs acting as endogenous ligands may influence the risk of DP differently in these populations. Furthermore, we previously found that FFAR4 is not required for n3-PUFA-induced cell growth inhibition and apoptosis in cancer cells (45). FFAR4 may not participate in the regulation of glucose tolerance through LCFA, but may rather directly modulate the dysfunction of glucose tolerance at the daily light to dark transition (ZT12).
This study further suggested that the aberrant glucose tolerance induced by FFAR4 deletion was mediated by A. muciniphila. Ffar4-specific ligands, n3-PUFAs, are influential components of the diet and have been shown to significantly increase A. muciniphila abundance in the gut (46, 47). However, it is worth exploring which key A. muciniphila-derived metabolites affect the process of glucose metabolism. Previous studies have shown that A. muciniphila is an important source of SCFAs (48, 49), and we found that acetic acid abundance at ZT12 was disrupted in total Ffar4 KO mice (P < 0.001) but not in gut Ffar4-specific knockout mice compared to control mice. Additionally, several studies have shown that A. muciniphila can significantly stimulate an increase in serum GLP-1 levels through secretory protein P9, an effective mediator of GLP-1 secretion (50 – 52). Similarly, we found that treatment with A. muciniphila immediately increased GLP-1 and insulin secretion and decreased glucagon and GH secretion in gut-specific knockout of Ffar4 mice. Elevated serum GLP-1 levels in mice have been widely reported to be associated with increased insulin levels, insulin sensitization, and decreased glucagon levels. Serum growth hormone levels have also been reported to be associated with insulin sensitization.
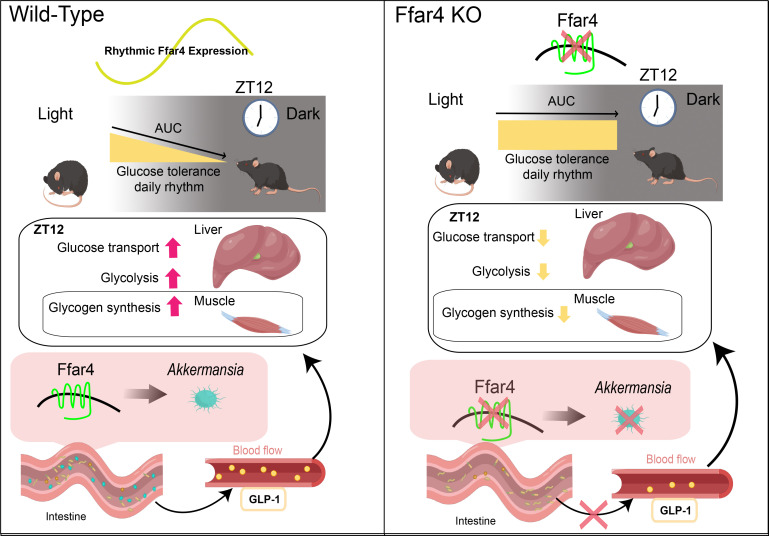
Consistent with our hypothesis, intestinal FFAR4 deficiency, although not leading to the disappearance of the glucose tolerance rhythm in mice, caused a delay in the peak and trough of the glucose tolerance rhythm in mice, which was mainly caused by the impaired glucose tolerance in mice at ZT12 due to intestinal FFAR4 deficiency. Intestinal FFAR4 deficiency resulted in a downregulation of A. muciniphila abundance in the intestine at ZT12, and after pasteurized A. muciniphila supplementation, Gko mice showed improved glucose tolerance at ZT12, accompanied by changes in serum hormones and an increase in peripheral tissue glucose metabolism. In particular, the restoration of serum GLP-1 hormone in Gko mice after A. muciniphila supplementation increased serum insulin levels and improved liver and muscle glucose utilization. GLP-1 receptor is absent in muscle and liver tissue, so changes in glucose utilization in liver and muscle tissue should be mediated by GLP-1-affected insulin. At the same time, a decrease in growth hormone means that the resistance to insulin of mice is weakened, which also facilitates blood glucose utilization in mice. The decrease of serum glucagon level is beneficial to the gluconeogenesis process, which was also demonstrated by upregulation of gluconeogenic transcripts in muscle tissue of A. muciniphila supplemented Gko mice (Fig. 6; Fig. S6). The above results suggest that restoring the FFAR4-A. muciniphila pathway plays a key role in maintaining glucose tolerance levels at ZT12 and even glucose tolerance rhythm in mice (Fig. 7). Moreover, it is necessary to develop new strategies to modulate the activity of the FFAR4-A. muciniphila pathway in vivo as it may serve as a novel intervention for the management of glucose tolerance rhythm in the future.
FIG 7.
Conceptual summary. Intestinal Ffar4 affects hormone secretion such as GLP-1 secretion via Akkermansia muciniphila, which can modify glucose metabolism in the liver and muscle of mice, affecting glucose tolerance at the daily light to dark transition. GLP-1, glucagon-like peptide 1.
MATERIALS AND METHODS
Animals
Ffar4 total KO (RRID: MGI:7256540) mice were provided by Shanghai Bioray Lab. Floxed Ffar4 (fl/fl, RRID: MGI: 7256541) and Villin-Cre mice (RRID: IMSR_JAX: 004586) mouse models were established by Shanghai Biomodel Organism. Tail tips of mice were digested with 100 µL of 50-mM NaOH at 95°C for 20 min. Next, 10 µL of Tris-HCl (pH 8.0) was added to neutralize the mixture. KO mice were genotyped by PCR using tail DNA with the forward primer CCCGGCATGTCCCCTGAGTGT and reverse primer TGGTCGCCCTTGACATCCGAGA at 95°C for 30 s and 63°C for 45 s for 40 cycles, generating an 87-bp fragment for the wild type and a 110-bp fragment for KO mice. Ffar4 (fl/fl) mice were genotyped using the forward primer TGCTCTTTCTGGAGCTGTGT and the reverse primer AGAGATCAGAATGGACAACT at 95°C for 30 s, 58°C for 45 s, and 72°C for 30 s for 40 cycles, generating a 272-bp fragment for fl/fl and a 243-bp fragment for the wild type. Villin-Cre mice were genotyped using the forward primer AGCGATGGATTTCCGTCTCTGG and the reverse primer AGCTTGCATGATCTCCGGTATTGAA at 95°C for 30 s, 58°C for 45 s, and 72°C for 30 s for 40 cycles, generating a 272-bp fragment. Gut-specific Ffar4 knockout mice (Gko) were generated by mating fl/fl mice with Villin-Cre mice. All mice were on a C57BL/6J background. Animals were housed and bred in a sterile-pathogen-free environment, subjected to a 12-hour light/12-hour dark cycle (07:00 lights on, 19:00 lights off), and provided access to water and food ad libitum.
Animal experiments
Adult male mice were used in all experiments. These mice were litter mates or from litters born during the same time period. To test the impact of A. muciniphila on glucose tolerance in Gko mice, the mice were treated with pasteurized A. muciniphila [5 × 1010 colony forming units (CFU)] or the control medium for 6 weeks until the GTT was performed. Mice were euthanised and then tissue sampled after the GTT.
Glucose tolerance test
Mice were fasted for 6 hours before the GTT. Tail blood glucose levels were measured immediately using a glucometer (OneTouch Ultra2), and a glucose injection corresponding to 0.8 g/kg of body weight was administered immediately afterward. Blood glucose levels were measured using glucometers at 15, 30, 60, 90, and 120 min. The tests at ZT12–14, ZT15–17, ZT18–20, and ZT21–23 were performed under dim red light.
Serum hormone assay
After at least 1 week of recovery after GTT, mice blood was collected through orbital puncture in EDTA tubes in non-fasting conditions. Serum insulin, glucagon, corticosterone, and glucagon-like peptide 1 (GLP-1) total levels were measured using an ultra-sensitive mouse insulin enzyme-linked immunosorbent assay (ELISA) kit (90080, Crystal Chem), glucagon ELISA kit (10–1281-01, Mercodia), corticosterone ELISA kit (ADI-900–097, Enzo Life Sciences), mouse GLP-1 ELISA kit (81508, Crystal Chem), and growth hormone ELISA kit (EZRMGH-45K, Sigma), respectively.
Quantitative real-time PCR
According to the manufacturer’s protocol, total RNA was isolated with RNAiso Plus (Takara). RNA was reverse-transcribed using Prime Script RT Master Mix (Takara). mRNA quantification was performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad) and Ultra SYBR Mixture (CWBIO). qPCR included 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 30 s. Animal experiment results were normalized to the housekeeping gene β-actin and were presented as 2−ΔΔCt. Genomic DNA of mice fecal samples collected at different times was extracted using the Fast DNA Spin kit for soil (MP, USA) following the manufacturer’s instructions. An amount of 20 ng of DNA was then used in qPCR reactions for A. muciniphila with specific primers, and for total bacteria for normalisation. The nucleotide sequences of primers used in this experiment are listed in Table 1.
TABLE 1.
Primers used in qRT-PCR
| Primer | Sequence |
|---|---|
| Mus β-actin forward primer | AGGAGTACGATGAGTCCGGC |
| Mus β-actin reverse primer | AGGGTGTAAAACGCAGCTCAG |
| Mus Ffar4 forward primer | AGTCAATCGCACCCACTTCC |
| Mus Ffar4 reverse primer | CCCAGCAGTGAGACGACAAA |
| Mus Glut1 forward primer | TCTGGACAAACCCACTTGTAA |
| Mus Glut1 reverse primer | CCAGCCATTTATATCTGCTTAGGT |
| Mus Glut2 forward primer | GGTGATGGGTCTCATTGGTGA |
| Mus Glut2 reverse primer | ACACGGGAGGCAAACAACTA |
| Mus Pkm forward primer | AGTGCCTGTACCTTGATGGC |
| Mus Pkm reverse primer | TACAAGCGTTGCTGGCCTAA |
| Mus Gyg1 forward primer | GAACCTGTGGTGGGACACTT |
| Mus Gyg1 reverse primer | CCTGAGACATGTTCCATCATTAGA |
| Mus Gsk3a forward primer | CGATGAACTGCGGAGACTCG |
| Mus Gsk3a reverse primer | GGATGGACAGTTCACCAGGAC |
| Mus Hk2 forward primer | ACCCGGGATGTTAGGCAAAG |
| Mus Hk2 reverse primer | ACCATTCTGAAACGCCGACT |
| A. muciniphila forward primer | CAGCACGTGAAGGTGGGGAC |
| A. muciniphila reverse primer | CCTTGCGGTTGGCTTCAGAT |
| UniF340 forward primer | ACTCCTACGGGAGGCAGCAGT |
| UniR514 reverse primer | ATTACCGCGGCTGCTGGC |
Western blotting
Distal colons of mice were lysed in RIPA buffer with 1 × protease inhibitor cocktail. Protein samples were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and electrically transferred to polyvinylidene fluoride membranes. Tris buffered saline with Tween (TBST) containing 5% skim milk was used to block the membranes for 2 hours. The membranes were washed four times with TBST and incubated overnight at 4°C with primary antibodies, including antiβ-ACTIN (Abcam, UK) and anti-FFAR4 (Sigma-Aldrich, USA). The next day, membranes were washed four times with TBST and incubated with horseradish peroxidase-conjugated secondary immunoglobulin G antibody for 2 hours at 25°C. For protein detection, the membranes were washed three times with TBST before imaging.
16S rRNA gene sequencing and analysis
Genomic DNA of mice fecal samples collected at different times (ZT0, ZT6, ZT12, and ZT18) was extracted using the Fast DNA Spin kit for soil (MP, USA) following the manufacturer’s instructions. Subsequently, the V3–V4 region (forward primer, 5-CCTACGGGNGGCWGCAG-3; reverse primer, 5-GGACTACHVGGGTATCTA AT-3) of the 16S rDNA gene fragments was amplified. The 50-µL reaction system contained 25 µL of premix Taq lus dye (TaKaRa, China), 0.5 µL of the forward primer, 0.5 µL of the reverse primer, 2 µL of the DNA template, and 22 µL of ddH2O. The amplification conditions were as follows: 95°C for 5 min, 40 cycles of 95°C for 30 s, 52°C for 30 s, 72°C for 30 s, and maintaining at 12°C for 10 min. The PCR products were purified after amplification using the QIAquick PCR Purification kit (Qiagen, USA). An Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and Qubit (version 2.0; Applied Biosystems, Carlsbad, CA, USA) were used to verify and quantify the DNA libraries; moreover, paired-end sequencing was performed using the Illumina Miseq platform. After sequencing, Quantitative Insights into Microbial Ecology 2 (version 2019.7) was used to analyze the high-quality sequences (53). A naive Bayes classifier was trained using the SILVA reference database (version 132) (54). DADA2 was used for quality control, to remove low quality sequences, chimeras, and generate OTU tables with 100% similarity (now called feature tables)(55). Phylogenetic trees were constructed de novo; the construction process includes multiple sequence alignment, removal of high variation sites, construction of an unrooted phylogenetic tree, and transformation of the unrooted phylogenetic tree into a rooted phylogenetic tree. Diversity analyses were performed after samples were rarefied to a common sampling depth of 5623. Differential microbial analysis was performed using QMD (a method for quantifying differences in absolute microbial abundance) (56).
Culture and pasteurization of A. muciniphila
A. muciniphila (ATCC BAA-835) was cultured in brain-heart infusion broth containing 50-mg/L L-cysteine and 500-mg/L mucoprotein under strict anaerobic conditions. A representative culture stock was used to determine the CFU per milliliter under anaerobic conditions by plate counting using mucin media containing 1% agarose. This culture was diluted with anaerobic PBS containing 2.5% glycerol to a final concentration of 2.5 × 1010 CFU/mL. Then, A. muciniphila was inactivated by pasteurization for 30 min at 70°C. After pasteurization, no viable A. muciniphila could be recovered in the culture.
Gas chromatography-mass spectroscopy
Fresh feces of mice were collected at ZT12, and 500 µL of saturated sodium chloride solution was added immediately. After shaking treatment, 10% sulfuric acid solution was added for acidification, and the SCFAs were extracted using anhydrous ether. The ether phase was separated and anhydrous sodium sulfate was added to absorb the residual moisture. The ether phase was analyzed using gas chromatography-mass spectroscopy (TRACE1310-TSQ 8000 Evo; Thermo Fisher Scientific, USA). An Rtx-5 MS capillary tube chromatographic column (0.25 mm ×30 m, 0.25 µm) was purchased from Restek, USA, and water for experiments was produced using a Milli-Q8 ultrapure water generator (Millipore Co., USA). Concentrated sulfuric acid, acetic acid, propionic acid, and n-butyric acid solutions of analytically pure grade were purchased from China National Pharmaceutical Group Co., Ltd. (Sinopharm).
Statistical analysis
Cosine analysis was used to analyze the circadian rhythm of the data; we estimated the loss or gain of rhythms and circadian properties including amplitude, phase, and mesor. P < 0.05 was considered statistically significant. Animal data were analyzed using two-way analysis of variance (ANOVA) and one-way ANOVA using SPSS (SPSS Inc.). For sequencing data, the R platform (http://www.r-project.org/) was used for statistical analysis. Differences in the diversity of intestinal flora were compared using the function of the Kruskal–Wallis test of the vegan package of R. All plots are presented as the mean ± standard error of the mean. P < 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (82000808 and 82000685) and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21-2051).
The mechanic diagram was created by Figdraw.
Z.W. and S.L.Z. designed the experiments. Z.W., T.T.Z., W.W., and J.Y.L. conducted the experiments. Z.W., S.Y.C., and S.L.Z. analyzed the data. Z.W., S.L.Z., and Y.Q.C. wrote the paper. All authors read and approved the final manuscript.
The authors declare no conflicts of interest.
Contributor Information
Y. Q. Chen, Email: yqchen@jiangnan.edu.cn.
Sheng long Zhu, Email: shenglongzhu@jiangnan.edu.cn.
Sarah M. Allard, University of California San Diego, La Jolla, California, USA
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request. The 16S rRNA gene sequences are available in the NCBI SRA database under accession no. PRJNA1001966.
ETHICS APPROVAL
All animal procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals of the School of Medicine, Jiangnan University, and approved by the Animal Ethics Committee of Jiangnan University (JN No: 20171030 c0110506).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00573-23.
Identification of mouse tail genotypes.
Ffar4 mRNA and FFAR4 protein expression oscillations showed a circadian rhythm in the colons of mice.
Identification of mouse tail genotypes.
Comparison of SCFA levels in mouse intestinal feces.
Relative abundance of A. muciniphila.
Serum insulin, glucagon, GC, GH, and GLP-1 levels after gavage with pasteurized A. muciniphila.
Supplemental figure legends.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 2. Alvarado MM, Kum H-C, Gonzalez Coronado K, Foster MJ, Ortega P, Lawley MA. 2017. Barriers to remote health interventions for type 2 diabetes: a systematic review and proposed classification scheme. J Med Internet Res 19:e28. doi: 10.2196/jmir.6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saad A, Dalla Man C, Nandy DK, Levine JA, Bharucha AE, Rizza RA, Basu R, Carter RE, Cobelli C, Kudva YC, Basu A. 2012. Diurnal pattern to insulin secretion and insulin action in healthy individuals. Diabetes 61:2691–2700. doi: 10.2337/db11-1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris CJ, Yang JN, Garcia JI, Myers S, Bozzi I, Wang W, Buxton OM, Shea SA, Scheer FAJL. 2015. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci USA 112:E2225–2234. doi: 10.1073/pnas.1418955112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. 2001. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50:1237–1243. doi: 10.2337/diabetes.50.6.1237 [DOI] [PubMed] [Google Scholar]
- 6. Rodríguez-Cortés B, Hurtado-Alvarado G, Martínez-Gómez R, León-Mercado LA, Prager-Khoutorsky M, Buijs RM. 2022. Suprachiasmatic nucleus-mediated glucose entry into the arcuate nucleus determines the daily rhythm in blood glycemia. Curr Biol 32:796–805. doi: 10.1016/j.cub.2021.12.039 [DOI] [PubMed] [Google Scholar]
- 7. Porcellati F, Lucidi P, Bolli GB, Fanelli CG. 2013. Thirty years of research on the dawn phenomenon: lessons to optimize blood glucose control in diabetes. Diabetes Care 36:3860–3862. doi: 10.2337/dc13-2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng F, Li X, Xiao F, Zhao R, Sun Z. 2022. Circadian clock, diurnal glucose metabolic rhythm, and dawn phenomenon. Trends Neurosci 45:471–482. doi: 10.1016/j.tins.2022.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding G, Li X, Hou X, Zhou W, Gong Y, Liu F, He Y, Song J, Wang J, Basil P, Li W, Qian S, Saha P, Wang J, Cui C, Yang T, Zou K, Han Y, Amos CI, Xu Y, Chen L, Sun Z. 2021. REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature 595:763–767. doi: 10.1038/s41586-021-03654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carroll MF, Schade DS. 2005. The dawn phenomenon revisited: implications for diabetes therapy. Endocr Pract 11:55–64. doi: 10.4158/EP.11.1.55 [DOI] [PubMed] [Google Scholar]
- 11. Oliveira de Souza C, Sun X, Oh D. 2021. Metabolic functions of G protein-coupled receptors and beta-arrestin-mediated signaling pathways in the pathophysiology of type 2 diabetes and obesity. Front Endocrinol 12. doi: 10.3389/fendo.2021.715877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimura T, Pydi SP, Pham J, Tanaka N. 2020. Metabolic functions of G protein-coupled receptors in hepatocytes-potential applications for diabetes and NAFLD. Biomolecules 10:1445. doi: 10.3390/biom10101445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J, Yang Z, Ge X-Y, Gao M-X, Meng R, Xu X, Zhang Y-Q, Li R-Z, Lin J-Y, Tian Z-M, Wang J, Ning S-L, Xu Y-F, Yang F, Gu J-K, Sun J-P, Yu X. 2022. Autonomous sensing of the insulin peptide by an olfactory G protein-coupled receptor modulates glucose metabolism. Cell Metabolism 34:240–255. doi: 10.1016/j.cmet.2021.12.022 [DOI] [PubMed] [Google Scholar]
- 14. Tunaru S, Bonnavion R, Brandenburger I, Preussner J, Thomas D, Scholich K, Offermanns S. 2018. 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nat Commun 9:177. doi: 10.1038/s41467-017-02539-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z, Xu X, Huang W, Qian H. 2018. Free fatty acid receptor 1 (FFAR1) as an emerging therapeutic target for type 2 diabetes mellitus: recent progress and prevailing challenges. Med. Res. Rev 38:381–425. doi: 10.1002/med.21441 [DOI] [PubMed] [Google Scholar]
- 16. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. 2012. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61:364–371. doi: 10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Remely M, Aumueller E, Merold C, Dworzak S, Hippe B, Zanner J, Pointner A, Brath H, Haslberger AG. 2014. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 537:85–92. doi: 10.1016/j.gene.2013.11.081 [DOI] [PubMed] [Google Scholar]
- 18. Oh DY, Olefsky JM. 2016. G protein-coupled receptors as targets for anti-diabetic therapeutics. Nat Rev Drug Discov 15:161–172. doi: 10.1038/nrd.2015.4 [DOI] [PubMed] [Google Scholar]
- 19. Kurashima Y, Kiyono H. 2017. Mucosal ecological network of epithelium and immune cells for gut homeostasis and tissue healing. Annu Rev Immunol 35:119–147. doi: 10.1146/annurev-immunol-051116-052424 [DOI] [PubMed] [Google Scholar]
- 20. Hilgendorf KI, Johnson CT, Mezger A, Rice SL, Norris AM, Demeter J, Greenleaf WJ, Reiter JF, Kopinke D, Jackson PK. 2019. Omega-3 fatty acids activate ciliary FFAR4 to control adipogenesis. Cell 179:1289–1305. doi: 10.1016/j.cell.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suckow AT, Polidori D, Yan W, Chon S, Ma JY, Leonard J, Briscoe CP. 2014. Alteration of the glucagon axis in GPR120 (FFAR4) knockout mice: a role for GPR120 in glucagon secretion. J Biol Chem 289:15751–15763. doi: 10.1074/jbc.M114.568683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu S, Zhang J, Jiang X, Wang W, Chen YQ. 2022. Free fatty acid receptor 4 deletion attenuates colitis by modulating treg cells via ZBED6-IL33 pathway. EBioMedicine 80:104060. doi: 10.1016/j.ebiom.2022.104060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gribble FM, Diakogiannaki E, Reimann F. 2017. Gut hormone regulation and secretion via FFA1 and FFA4. Handb Exp Pharmacol 236:181–203. doi: 10.1007/164_2016_46 [DOI] [PubMed] [Google Scholar]
- 24. Miyamoto J, Igarashi M, Watanabe K, Karaki S-I, Mukouyama H, Kishino S, Li X, Ichimura A, Irie J, Sugimoto Y, Mizutani T, Sugawara T, Miki T, Ogawa J, Drucker DJ, Arita M, Itoh H, Kimura I. 2019. Gut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids. Nat Commun 10:4007. doi: 10.1038/s41467-019-11978-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furutani A, Ikeda Y, Itokawa M, Nagahama H, Ohtsu T, Furutani N, Kamagata M, Yang Z-H, Hirasawa A, Tahara Y, Shibata S. 2015. Fish oil accelerates diet-induced entrainment of the mouse peripheral clock via GPR120. PLoS One 10:e0132472. doi: 10.1371/journal.pone.0132472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quesada-López T, Cereijo R, Turatsinze J-V, Planavila A, Cairó M, Gavaldà-Navarro A, Peyrou M, Moure R, Iglesias R, Giralt M, Eizirik DL, Villarroya F. 2016. The lipid sensor GPR120 promotes brown fat activation and FGF21 release from adipocytes. Nat Commun 7:13479. doi: 10.1038/ncomms13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Erickson A, Moreau R. 2016. The regulation of FGF21 gene expression by metabolic factors and nutrients. Horm Mol Biol Clin Investig 30:/j/hmbci.2017.30.issue-1/hmbci-2016-0016/hmbci-2016-0016.xml. doi: 10.1515/hmbci-2016-0016 [DOI] [PubMed] [Google Scholar]
- 28. Yu H, Xia F, Lam KSL, Wang Y, Bao Y, Zhang J, Gu Y, Zhou P, Lu J, Jia W, Xu A. 2011. Circadian rhythm of circulating fibroblast growth factor 21 is related to diurnal changes in fatty acids in humans. Clin Chem 57:691–700. doi: 10.1373/clinchem.2010.155184 [DOI] [PubMed] [Google Scholar]
- 29. Yang L, Lei X-T, Huang Q, Wang T, Sun H-B, Wang H-Y. 2021. A novel GPR120-selective agonist promotes insulin secretion and improves chronic inflammation. Life Sci 269:119029. doi: 10.1016/j.lfs.2021.119029 [DOI] [PubMed] [Google Scholar]
- 30. Sheng R, Yang L, Zhang Y, Xing E, Shi R, Wen X, Wang H, Sun H. 2018. Discovery of novel selective GPR120 agonists with potent anti-diabetic activity by hybrid design. Bioorg Med Chem Lett 28:2599–2604. doi: 10.1016/j.bmcl.2018.06.047 [DOI] [PubMed] [Google Scholar]
- 31. Li J, Song J, Li X, Rock SB, Sinner HF, Weiss HL, Weiss T, Townsend CM Jr, Gao T, Evers BM. 2018. FFAR4 is involved in regulation of neurotensin release from neuroendocrine cells and male C57Bl/6 mice. Endocrinology 159:2939–2952. doi: 10.1210/en.2018-00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rubbino F, Garlatti V, Garzarelli V, Massimino L, Spanò S, Iadarola P, Cagnone M, Giera M, Heijink M, Guglielmetti S, Arena V, Malesci A, Laghi L, Danese S, Vetrano S. 2022. GPR120 prevents colorectal adenocarcinoma progression by sustaining the mucosal barrier integrity. Sci Rep 12:381. doi: 10.1038/s41598-021-03787-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Z, Sun B, Yu D, Zhu C. 2022. Gut microbiota: an important player in type 2 diabetes mellitus. Front Cell Infect Microbiol 12:834485. doi: 10.3389/fcimb.2022.834485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang P-P, Li L-L, Han X, Li Q-W, Zhang X-H, Liu JJ, Wang Y. 2020. Fecal microbiota transplantation improves metabolism and gut microbiome composition in dB/dB mice. Acta Pharmacol Sin 41:678–685. doi: 10.1038/s41401-019-0330-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shih CT, Yeh YT, Lin CC, Yang LY, Chiang CP. 2020. Akkermansia muciniphila is negatively correlated with hemoglobin A1C in refractory diabetes. Microorganisms 8:1360. doi: 10.3390/microorganisms8091360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, Loumaye A, Hermans MP, Thissen J-P, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 37. Shin N-R, Lee J-C, Lee H-Y, Kim M-S, Whon TW, Lee M-S, Bae J-W. 2014. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63:727–735. doi: 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- 38. Segers A, Desmet L, Thijs T, Verbeke K, Tack J, Depoortere I. 2019. The circadian clock regulates the diurnal levels of microbial short-chain fatty acids and their rhythmic effects on colon contractility in mice. Acta Physiol (Oxf) 225:e13193. doi: 10.1111/apha.13193 [DOI] [PubMed] [Google Scholar]
- 39. Jaime-Lara RB, Brooks BE, Vizioli C, Chiles M, Nawal N, Ortiz-Figueroa RSE, Livinski AA, Agarwal K, Colina-Prisco C, Iannarino N, Hilmi A, Tejeda HA, Joseph PV. 2023. A systematic review of the biological mediators of fat taste and smell. Physiol Rev 103:855–918. doi: 10.1152/physrev.00061.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji G, Guo Q, Xue Q, Kong R, Wang S, Lei K, Liu R, Wang X. 2021. Novel GPR120 agonists with improved pharmacokinetic profiles for the treatment of type 2 diabetes. Molecules 26:6907. doi: 10.3390/molecules26226907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Codoñer-Alejos A, Carrasco-Luna J, Codoñer-Franch P. 2020. The rs11187533 C>T variant of the FFAR4 gene is associated with lower levels of fasting glucose and decreases in markers of liver injury in children with obesity. Ann Nutr Metab 76:122–128. doi: 10.1159/000506618 [DOI] [PubMed] [Google Scholar]
- 42. Vestmar MA, Andersson EA, Christensen CR, Hauge M, Glümer C, Linneberg A, Witte DR, Jørgensen ME, Christensen C, Brandslund I, Lauritzen T, Pedersen O, Holst B, Grarup N, Schwartz TW, Hansen T. 2016. Functional and genetic epidemiological characterisation of the FFAR4 (GPR120) P. R270H variant in the danish population. J Med Genet 53:616–623. doi: 10.1136/jmedgenet-2015-103728 [DOI] [PubMed] [Google Scholar]
- 43. Bonnefond A, Lamri A, Leloire A, Vaillant E, Roussel R, Lévy-Marchal C, Weill J, Galan P, Hercberg S, Ragot S, Hadjadj S, Charpentier G, Balkau B, Marre M, Fumeron F, Froguel P. 2015. Contribution of the low-frequency, loss-of-function P. R270H mutation in FFAR4 (GPR120) to increased fasting plasma glucose levels. J Med Genet 52:595–598. doi: 10.1136/jmedgenet-2015-103065 [DOI] [PubMed] [Google Scholar]
- 44. Ichimura A. 2017. Polymorphic variation in FFA receptors: functions and consequences. Handb Exp Pharmacol 236:133–158. doi: 10.1007/164_2016_57 [DOI] [PubMed] [Google Scholar]
- 45. Zhu S, Jiang X, Jiang S, Lin G, Gong J, Chen W, He Z, Chen YQ. 2018. GPR120 is not required for Omega-3 pufas-induced cell growth inhibition and apoptosis in breast cancer cells. Cell Biol Int 42:180–186. doi: 10.1002/cbin.10883 [DOI] [PubMed] [Google Scholar]
- 46. Pal A, Sun S, Armstrong M, Manke J, Reisdorph N, Adams VR, Kennedy A, Zu Y, Moustaid-Moussa N, Carroll I, Shaikh SR. 2022. Beneficial effects of eicosapentaenoic acid on the metabolic profile of obese female mice entails upregulation of hepes and increased abundance of enteric Akkermansia muciniphila. Biochimica et Biophysica Acta 1867:159059. doi: 10.1016/j.bbalip.2021.159059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Monk JM, Liddle DM, Hutchinson AL, Wu W, Lepp D, Ma DWL, Robinson LE, Power KA. 2019. Fish oil supplementation to a high-fat diet improves both intestinal health and the systemic obese phenotype. J Nutr Biochem 72:108216. doi: 10.1016/j.jnutbio.2019.07.007 [DOI] [PubMed] [Google Scholar]
- 48. Peng J, Li X, Zheng L, Duan L, Gao Z, Hu D, Li J, Li X, Shen X, Xiao H. 2022. Ban-Lan-Gen granule alleviates dextran sulfate sodium-induced chronic relapsing colitis in mice via regulating gut microbiota and restoring gut SCFA derived-GLP-1 production. JIR 15:1457–1470. doi: 10.2147/JIR.S352863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Szachta P, Bartnicka A, Gałęcka M. 2016. Microbiota - a key to healing the gastrointestinal tract Pomeranian J Life Sci 62:21–24. [PubMed] [Google Scholar]
- 50. Yoon HS, Cho CH, Yun MS, Jang SJ, You HJ, Kim J-H, Han D, Cha KH, Moon SH, Lee K, Kim Y-J, Lee S-J, Nam T-W, Ko G. 2021. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat Microbiol 6:563–573. doi: 10.1038/s41564-021-00880-5 [DOI] [PubMed] [Google Scholar]
- 51. Cani PD, Knauf C. 2021. A newly identified protein from Akkermansia muciniphila stimulates GLP-1 secretion. Cell Metab 33:1073–1075. doi: 10.1016/j.cmet.2021.05.004 [DOI] [PubMed] [Google Scholar]
- 52. Martchenko SE, Martchenko A, Cox BJ, Naismith K, Waller A, Gurges P, Sweeney ME, Philpott DJ, Brubaker PL. 2020. Circadian GLP-1 secretion in mice is dependent on the intestinal microbiome for maintenance of diurnal metabolic homeostasis. Diabetes 69:2589–2602. doi: 10.2337/db20-0262 [DOI] [PubMed] [Google Scholar]
- 53. Hall M, Beiko RG. 2018. 16S rRNA gene analysis with QIIME2. Methods Mol Biol 1849:113–129. doi: 10.1007/978-1-4939-8728-3_8 [DOI] [PubMed] [Google Scholar]
- 54. Liu F, Xu X, Chao L, Chen K, Shao A, Sun D, Hong Y, Hu R, Jiang P, Zhang N, Xiao Y, Yan F, Feng N. 2021. Alteration of the gut microbiome in chronic kidney disease patients and its association with serum free immunoglobulin light chains. Front. Immunol 12:609700. doi: 10.3389/fimmu.2021.609700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mi K, Xu Y, Li Y, Liu X. 2023. QMD: a new method to quantify microbial absolute abundance differences between groups. iMeta 2. doi: 10.1002/imt2.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of mouse tail genotypes.
Ffar4 mRNA and FFAR4 protein expression oscillations showed a circadian rhythm in the colons of mice.
Identification of mouse tail genotypes.
Comparison of SCFA levels in mouse intestinal feces.
Relative abundance of A. muciniphila.
Serum insulin, glucagon, GC, GH, and GLP-1 levels after gavage with pasteurized A. muciniphila.
Supplemental figure legends.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The 16S rRNA gene sequences are available in the NCBI SRA database under accession no. PRJNA1001966.