Abstract
A system was developed for the detection of denitrifying bacteria by the amplification of specific nitrite reductase gene fragments with PCR. Primer sequences were found for the amplification of fragments from both nitrite reductase genes (nirK and nirS) after comparative sequence analysis. Whenever amplification was tried with these primers, the known nir type of denitrifying laboratory cultures could be confirmed. Likewise, the method allowed a determination of the nir type of five laboratory strains. The nirK gene could be amplified from Blastobacter denitrificans, Alcaligenes xylosoxidans, and Alcaligenes sp. (DSM 30128); the nirS gene was amplified from Alcaligenes eutrophus DSM 530 and from the denitrifying isolate IFAM 3698. For each of the two genes, at least one primer combination amplified successfully for all of the test strains. Specific amplification products were not obtained with nondenitrifying bacteria or with strains of the other nir type. The specificity of the amplified products was confirmed by subsequent sequencing. These results suggest the suitability of the method for the qualitative detection of denitrifying bacteria in environmental samples. This was shown by applying one generally amplifying primer combination for each nir gene developed in this study to total DNA preparations from aquatic habitats.
Denitrification is a dissimilatory process of bacteria in which oxidized nitrogen compounds are used as alternative electron acceptors for energy production. The gaseous end products NO, N2O, and N2 are released concomitantly. In the environment, denitrification is responsible for the release of fixed nitrogen into the atmosphere in form of N2 (13). It causes major nitrogen losses in agricultural soils to which fertilizers are applied. Accumulation of the greenhouse gases NO and N2O leads to the destruction of the ozone layer (3, 13). Also, denitrifying bacteria cause the removal of nitrogen compounds from waste water, where denitrification is coupled to the nitrification process (13). Bioremediation of environmental pollutants can be achieved under denitrifying conditions (5, 10, 33).
Denitrifying bacteria are phylogenetically diverse. They belong to all major physiological groups except for the Enterobacteriaceae, obligate anaerobes, and gram-positive bacteria other than Bacillus spp. (34). Defined as a physiological group, these facultative anaerobes can switch from oxygen to nitrogen oxides as terminal electron acceptors when kept under anoxic conditions. Nitrite reductase is the key enzyme in the dissimilatory denitrification process. The reduction of nitrite to NO can be catalyzed by the products of two different nitrite reductase genes: one product contains copper (the nirK product), and the other contains cytochrome cd1 (the nirS product). The two genes seem to occur mutually exclusively in a given strain, but both types have been found in different strains of the same species (4). Although structurally different, both enzyme types are functionally and physiologically equivalent (9, 35). nirS is more widely distributed; nirK is found in only 30% of the denitrifiers studied so far. However, nirK is found in a wider range of physiological groups (4). Several different approaches were used to determine the type of nitrite reductase in laboratory pure cultures. Diethyldithiocarbamate has been used to identify nirK-containing denitrifiers (21). Very specific detection, mostly at the strain level, could be achieved with antisera against dissimilatory nitrite reductase (dNirS [4, 24, 29] and dNirK [4, 17]). Another approach was the use of gene probes for nirK (12, 32) or nirS (12, 15, 24, 29), which were generally specific for the strains investigated. Weak reactivity also occurred for the nirK gene probe with DNA from some of the other nir-type denitrifiers (32); the nirS probe, on the other hand, hybridized with a more limited variety of strains (24, 30). A PCR method with one primer pair to target the nirS nitrite reductase gene showed higher specificity than hybridization experiments (30).
In the present study, we report on the application of new primer systems for both types of nitrite reductase genes. We used several different primer pairs to determine the nir type of denitrifying strains. Using samples from aquatic habitats, we amplified nir fragments and used the most reliable primer pairs for nirK or nirS, respectively, to successfully detect, in these aquatic samples, different populations of denitrifying bacteria.
MATERIALS AND METHODS
Bacteria and growth conditions.
A variety of denitrifying and nondenitrifying bacterial strains (see Tables 2 and 3) were used to evaluate the specificity of designed PCR primers. All strains were grown aerobically at 27°C. For genomic DNA isolation, Pseudomonas, Alcaligenes, Ochrobactrum, Paracoccus, and Azospirillum strains and the denitrifying isolate IFAM 3698 were grown on nutrient broth (NB; Merck, Darmstadt, Germany). Rhizobium strains were grown on yeast extract medium (YEM [27]). Hyphomicrobium zavarzinii IFAM ZV-622T was grown on 337-B1 medium (7) with 0.5% (vol/vol) methanol. Rhodobacter sphaeroides f. sp. denitrificans was grown on trypticase soy broth (TSB; Difco Laboratories, Detroit, Mich.), Roseobacter denitrificans was grown on oligotrophic medium (PYGV [25]) supplemented with 25‰ artificial seawater (16), and Blastobacter denitrificans was grown on peptone yeast extract glucose medium, i.e., PYGV without vitamins. Nondenitrifying strains of the Enterobacteriaceae were grown on Luria broth (LB [19]).
TABLE 2.
Results of PCR amplifications with the different sets of primers for nirKa
| Species or strainb | Source or referenceb | Denitrificationc | nirKc | PCR productsd determined with primers:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| nirK1F-nirK3R | nirK1F-nirK4R | nirK1F-nirK5R | nirK2F-nirK3R | nirK2F-nirK4R | nirK2F-nirK5R | ||||
| Alcaligenes xylosoxidans NCIMB 11015 | NCIMB | + | + | + | + | + | − | − | − |
| Hyphomicrobium zavarzinii IFAM ZV-622T (ATCC 27496) | ATCC | + | + | + | − | + | − | − | − |
| Ochrobactrum anthropi LMG 2136 | LMG | + | + | + | + | + | + | + | + |
| Rhizobium meliloti Rm1021 | C. Elmerich, Institut Pasteur, Paris, France | + | + | + | − | + | + | − | + |
| Rhizobium meliloti 20115 | C. Elmerich, Institut Pasteur, Paris, France | + | + | + | − | + | + | − | + |
| Rhodobacter sphaeroides f. sp. denitrificans | Satoh et al. (20) | + | + | + | − | + | +e | − | + |
| Alcaligenes sp. strain (DSM 30128) | DSM | + | NDg | + | + | + | +e | − | + |
| Alcaligenes xylosoxidans subsp. denitrificans DSM 30026 | DSM | + | ND | + | + | + | − | − | − |
| Blastobacter denitrificans IFAM 1005 (DSM 1113) | DSM | + | ND | + | − | + | − | − | + |
| Enterobacter agglomerans 339 | W. Klingmüller, Universität Bayreuth, Bayreuth, Germany | − | − | −f | − | − | −f | −f | −f |
| Enterobacter cloacae NCIMB 11463 | NCIMB | − | − | − | − | − | −f | − | − |
| Escherichia coli K-12 (DSM 498) | DSM | − | − | −f | − | − | −f | −f | −f |
| Klebsiella pneumoniae subsp. pneumoniae DSM 681 | DSM | − | − | −f | − | − | −f | − | − |
Touchdown reaction from 45 to 40°C used.
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung von Mikroorganismen; IFAM, Institut für Allgemeine Mikrobiologie, Universität Kiel, Kiel, Germany. LMG, Laboratorium voor Mikrobiologie, Universiteit Gent, Ghent, Belgium; NCIMB, National Collections of Industrial and Marine Bacteria, Aberdeen, United Kingdom.
Data from literature.
+, PCR product of the expected size; −, no PCR product.
Expected PCR products and weaker products of unexpected and nonspecific size.
No expected PCR products, only products of unexpected and nonspecific size.
ND, not determined.
TABLE 3.
Results of PCR amplifications with the different sets of primers for nirSa
| Strainb | Source or referenceb | Denitrificationc | nirSc | PCR productsd determined with primers:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nirS1F-nirS3R | nirS1F-nirS5R | nirS1F-nirS6R | nirS2F-nirS3R | nirS2F-nirS5R | nirS2F-nirS6R | nirS3F-nirS5R | nirS3F-nirS6R | nirS4F-nirS5R | nirS4F-nirS6R | ||||
| Alcaligenes eutropus H16 (DSM 428) | DSM | + | + | + | + | + | + | − | + | − | − | − | − |
| Alcaligenes faecalis A15 | H. Bothe, Universität Köln, Cologne, Germany | + | + | + | − | + | + | − | + | + | + | − | + |
| Azospirillum brasilense Sp7 (DSM 1690) | DSM | + | + | − | − | + | − | − | + | − | + | − | + |
| Paracoccus denitrificans ATCC 19367 | ATCC | + | + | + | − | + | + | + | + | + | − | + | + |
| Pseudomonas aeruginosa DSM 6195 | DSM | + | + | − | − | +e | − | + | − | − | − | − | + |
| Pseudomonas stutzeri ATCC 14405 | ATCC | + | + | + | + | + | + | + | + | + | + | + | + |
| Roseobacter denitrificans ATCC 33942T | ATCC | + | + | + | + | + | + | + | +e | + | + | − | − |
| Alcaligenes eutrophus DSM 530 | DSM | + | NDg | + | + | + | + | − | + | − | − | − | − |
| Denitrifying isolate IFAM 3698 | IFAM | + | ND | − | − | + | + | + | +e | − | + | − | + |
| Escherichia coli K-12 (DSM 498) | DSM | − | − | − | −f | − | −f | −f | −f | −f | −f | − | − |
| Enterobacter agglomerans 339 | W. Klingmüller, Universität Bayreuth, Bayreuth, Germany | − | − | − | − | − | − | −f | −f | − | −f | − | − |
| Enterobacter cloacae NCIMB 11463 | NCIMB | − | − | − | − | − | − | − | −f | −f | − | − | − |
| Klebsiella pneumoniae subsp. pneumoniae DSM 681 | DSM | − | − | − | − | − | − | − | − | − | − | − | − |
Touchdown reaction from 45 to 40°C used.
See Table 2, footnote b.
Data from literature.
+, PCR product of the expected size; −, no PCR product.
Expected PCR products and weaker products of unexpected and nonspecific size.
No expected PCR products, only products of unexpected and nonspecific size.
ND, not determined.
Extraction of genomic DNA.
Genomic DNA was obtained from pure cultures by lysozyme-proteinase K-sodium dodecyl sulfate (SDS) treatment followed by phenol-chloroform extraction and subsequent ethanol precipitation (8). The purity and concentration of the DNA preparations were determined spectrophotometrically.
Preparation of total DNA from an enrichment culture and four environmental samples.
DNA was prepared from five samples. (i) A 500-ml volume of medium 337-B1 with 0.5% (wt/vol) KNO3 for the enrichment of denitrifying methylotrophic bacteria was inoculated with 100 μl of activated sludge from a sewage treatment plant near Plön (Schleswig-Holstein, Germany). After 4 weeks at 28°C under anaerobic conditions, 500 μl of the enrichment was again inoculated and kept under the same growth conditions. Six months later, cells were harvested by centrifugation (6,000 × g for 60 min at 4°C) and resuspended in 400 μl of double-distilled water. The DNA was extracted with Chelex 100 (28).
(ii) A 1.5-ml volume of activated sludge from a sewage treatment plant in Plön (Schleswig-Holstein, Germany) was pelleted (13600 × g for 10 min at 4°C), and the pellet was air dried and resuspended in 0.85% NaCl solution. DNA extraction (8) was followed by an additional hexadecyltrimethylammonium bromide (CTAB; Sigma Aldrich, Steinheim, Germany) precipitation step (1) to remove humic acids and carbohydrates.
(iii) Surface water (30 liters) from Lake Kleiner Plöner See (Schleswig-Holstein, Germany; collected in April 1996) was filtered through a cellulose filter (Sartorius, Göttingen, Germany) to remove particles larger than 100 μm and then through a fiberglass filter (pore size, 3 μm; Millipore, Bedford, Mass.), and cells were collected on a Durapore filter (pore size, 0.22 μm; Millipore). Bacterial cells were removed from the filter by shaking it (100 rpm for 5 h at 4°C) in 100 ml of filtered lake water (pore size, 0.22 μm) containing 0.1 mM EDTA. The cells were then harvested by centrifugation (8,000 × g for 45 min at 4°C). The air-dried pellet was resuspended in 10 ml of SET buffer (5% sucrose, 50 mM EDTA, 50 mM Tris-HCl [pH 7.6]). The cells were lysed by the method of Smalla (22) with modifications suggested by Gliesche et al. (8). The suspension was frozen (20 min at −20°C) and thawed (5 min at 30°C) and kept on ice with 1 volume of chilled acetone for 30 min. The pellet (after centrifugation at 4000 × g for 10 min) was dried, resuspended in 5 ml of SET buffer containing 5 mg of lysozyme, and incubated at 37°C for 1 h. DNA extraction and purification were performed by the method of Gliesche et al. (8).
(iv) Cells from 10 liters of lake water (Lake Plussee, Schleswig-Holstein, Germany; collected at a depth of 9 m in August 1996) were concentrated by tangential-flow filtration (31). To 100 μl of the cell suspension was added 200 μl of MilliQ water; DNA was extracted with Chelex 100 (28) and further purified with CTAB (1).
(v) DNA from sediment (Lake Kleiner Plöner See; collected in April 1996) was isolated by the method of van Elsas and Smalla (26) with an additional proteinase K treatment (50 μl of a 20-mg ml−1 solution) after the incubation with SDS.
PCR amplification of the nir genes.
PCR amplifications from pure cultures and environmental samples were performed in a total volume of 50 μl containing 5 μl of 10× PCR buffer (500 mM KCl, 25 mM MgCl2, 200 mM Tris-HCl [pH 8.4], 0.1% Triton X-100), 200 μM each deoxyribonucleoside triphosphate, 1.0 U of Taq polymerase (5 U μl−1; Appligene Oncor, Illkirch, France), 25 pmol (for genomic DNA) or 35 pmol (for total DNA from environmental samples) of both primers (5 pmol μl−1 each), and DNA (10 to 100 ng). After a denaturation step of 5 min at 95°C, a “touchdown” PCR was performed (Thermocycler 2400; Perkin-Elmer, Branchburg, N.J.). This consisted of a denaturation step of 30 s at 95°C, a primer-annealing step of 40 s, and an extension step of 40 s at 72°C. After 30 cycles, a final 7-min incubation at 72°C was performed. During the first 10 cycles, the annealing temperature was decreased by 0.5°C every cycle, starting at 45°C until it reached a touchdown at 40°C. The additional 20 cycles were performed at an annealing temperature of 43°C. The amplification products were analyzed by electrophoresis on 2% (wt/vol) agarose gels (Boehringer, Ingelheim, Germany) followed by a 15-min staining with ethidium bromide (0.5 mg liter−1).
Sequencing of amplified nir products.
For DNA sequencing, amplified PCR products from pure cultures were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany) as specified by the manufacturer. DNA sequences were determined by direct sequencing of purified PCR products with the cycle-sequencing kit (GATC, Konstanz, Germany) and Thermosequenase 2.0 (Amersham, Braunschweig, Germany) as specified by the manufacturers. Labeling was performed by terminating the polymerization with biotin-labeled dideoxynucleoside triphosphates. After a denaturing step of 4 min at 94°C, 30 cycles of denaturation for 30 s at 94°C and primer annealing and extension for 30 s at 55°C were performed, followed by an additional extension step of 6 min at 60°C. The sequencing products were blotted with a direct blotting apparatus (GATC) onto a nylon membrane. The separated products were visualized by an enzyme-linked streptavidin-biotin coupling assay with a streptavidin-alkaline phosphatase conjugate (GATC) and NBT/X-phosphate (Boehringer, Mannheim, Germany) as specified by the manufacturers. The sequences obtained were compared with published nirK and nirS sequences in the EMBL Nucleotide Sequence Database by FASTA analysis of the HUSAR program package based on the Genetics Computer Group sequence analysis package (6).
Hybridization analysis of nir products from total DNA of environmental samples.
Approximately 100 ng (pure cultures) or 250 ng (environmental samples) of PCR product was analyzed on an agarose gel (2%, wt/vol). After electrophoresis, the DNA was transferred onto a positively charged nylon membrane (QIAbrane Nylon Plus; Qiagen) by capillary transfer (24). The DNA was cross-linked to the membrane with UV light (45 s at 302 nm).
Products generated with the primer combination nirK1F-nirK5R from genomic DNA from Alcaligenes xylosoxidans NCIMB 11015 and with the combination nirS1F-nirS6R from genomic DNA from Pseudomonas stutzeri ATCC 14405 were used as probes for nirK and nirS, respectively, to determine the specificity of nir products amplified from total environmental DNA. The nir products were purified by eluting the bands from an agarose gel by using the QIAquick gel extraction kit (Qiagen) as specified by the manufacturer. The probes were labeled randomly with digoxigenin by using the digoxigenin DNA labeling and detection kit (Boehringer) as specified by the manufacturer.
The membrane was prehybridized in 20 ml of DIG-Easy Hyb solution (Boehringer) for 2 h at 42°C. Hybridization was performed with 10 ml of DIG-Easy Hyb solution containing the specific probe (25 ng ml−1) and by incubation overnight at 42°C. After hybridization, the membrane was washed twice for 5 min at room temperature in 100 ml of a solution containing 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% (wt/vol) SDS and twice at 45°C for nirK and 46°C for nirS for 15 min with 100 ml of a solution containing 0.5 × SSC and 0.1% (wt/vol) SDS. Subsequently, the hybridization of the digoxigenin-labeled probe was detected by an enzyme-linked immunoassay with nitroblue tetrazolium/X-phosphate as the substrate as specified by the manufacturer (Boehringer).
Nucleotide sequence accession numbers.
The nirK and nirS sequences have been deposited in the EMBL nucleotide sequence databases under accession no. AJ224902 through AJ224913.
RESULTS
Design of PCR primers.
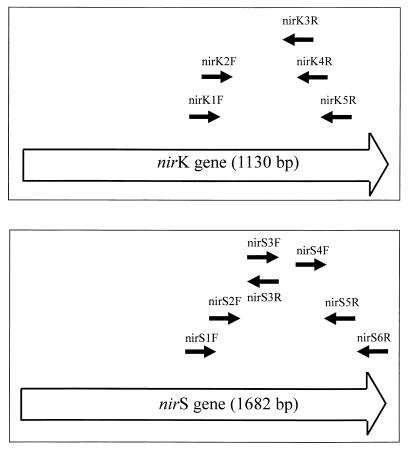
For the nirK gene, six sequences were available from the EMBL database, i.e., from Alcaligenes faecalis S-6 (D13155), Achromobacter cycloclastes (Z48635), Pseudomonas aureofaciens (Z21945), Pseudomonas sp. strain G-179 (M97294), Rhodobacter sphaeroides (U62291), and Rhizobium “hedysari” (U65658). For the nirS gene, six sequences were available, i.e., from Pseudomonas stutzeri JM300 (M80653), Pseudomonas stutzeri (X56813), Pseudomonas aeruginosa (X16452), Paracoccus denitrificans Pd1222 (U05002), Paracoccus denitrificans LMD29.63 (U75413), and Alcaligenes eutrophus H16 (X91394). For each gene, the available sequences were aligned by using the MULTIALIGN program (6). Five conserved regions for nirK and six conserved regions for nirS were chosen to design the primers used in this study. The sequences of the primers are shown in Table 1, and their locations within the nir genes are shown in Table 1 and Fig. 1. Comparison of the chosen primer sequences to all stored sequences in the EMBL database by using the BLASTN program (6) revealed significant similarity only to sequences of the nirK or the nirS gene.
TABLE 1.
Primer sequences and positions used to amplify fragments from nirK and nirS nitrite reductase
| Primera | Positionb | Primer sequence (5′-3′) |
|---|---|---|
| nirK1F | 526–542 | GG(A/C)ATGGT(G/T)CC(C/G)TGGCA |
| nirK2F | 565–581 | GC(C/G)(C/A)T(C/G)ATGGT(C/G)CTGCC |
| nirK3R | 898–918 | GAACTTGCCGGT(A/C/G)G(C/T)CCAGAC |
| nirK4R | 942–959 | GG(A/G)AT(A/G)A(A/G)CCAGGTTTCC |
| nirK5R | 1023–1040 | GCCTCGATCAG(A/G)TT(A/G)TGG |
| nirS1F | 763–780 | CCTA(C/T)TGGCCGCC(A/G)CA(A/G)T |
| nirS2F | 855–874 | TACCACCC(C/G)GA(A/G)CCGCGCGT |
| nirS3F | 1002–1019 | TTCCT(C/G/T)CA(C/T)GACGGCGGC |
| nirS4F | 1317–1336 | TTC(A/G)TCAAGAC(C/G)CA(C/T)CCGAA |
| nirS3R | 1002–1019 | GCCGCCGTC(A/G)TG(A/C/G)AGGAA |
| nirS5R | 1494–1514 | CTTGTTG(A/T)ACTCG(C/G)(C/G)CTGCAC |
| nirS6R | 1638–1653 | CGTTGAACTT(A/G)CCGGT |
FIG. 1.
Diagram showing the positions of primers used to amplify nir fragments: the positions for the nirK primers are correlated with the sequence of the nirK gene from Alcaligenes faecalis S-6 (top), and the positions for the nirS primers are correlated with the sequence of the nirS gene from Pseudomonas stutzeri ZoBell (bottom).
Amplification of nirK and nirS fragments from pure cultures.
The selected primers were used to amplify nirK or nirS fragments from bacterial pure cultures known to denitrify and to contain either nirK or nirS. For nirK, amplification products were obtained with the primer combinations nirK1F-nirK3R (392 bp), nirK1F-nirK4R (433 bp), nirK1F-nirK5R (514 bp), nirK2F-nirK3R (353 bp), nirK2F-nirK4R (394 bp), and nirK2F-nirK5R (475 bp). For nirS, amplification products were obtained with the primer combinations nirS1F-nirS3R (256 bp), nirS1F-nirS5R (751 bp), nirS1F-nirS6R (890 bp), nirS2F-nirS3R (164 bp), nirS2F-nirS5R (659 bp), nirS2F-nirS6R (798 bp), nirS3F-nirS5R (512 bp), nirS3F-nirS6R (651 bp), nirS4F-nirS5R (197 bp), and nirS4F-nirS6R (336 bp). Products of the expected size could be obtained from all selected primer combinations for amplification of nirK fragments with genomic DNA from Ochrobactrum anthropi and for amplification of nirS fragments with genomic DNA from Pseudomonas stutzeri. With the other pure cultures used in this study, not all but several primer combinations generated products of the expected sizes when used in PCR amplification experiments (Tables 2 and 3).
At least one primer combination for each gene which reacted with all pure cultures known to contain either nirK or nirS was found. For nirK, the primer combinations nirK1F-nirK3R and nirK1F-nirK5R amplified with all tested nirK-containing strains. For nirS, amplification with the combination nirS1F-nirS6R generated products of the expected size from all nirS-containing strains.
Whenever weaker products of nonspecific sizes occurred besides the products of the expected sizes (Table 2 and 3), application of a higher temperature for primer annealing eliminated the nonspecific products. By using the selected primer combinations for nirK with DNA from pure cultures of denitrifying strains known to contain the coding sequence for nirS and vice versa, no PCR products were obtained (data not shown). In addition, no products were obtained when applying the nirK or nirS primer combinations to DNA from nondenitrifying strains (Tables 2 and 3). Amplification experiments with eubacterial primers for 16S rRNA genes confirmed the ability to amplify gene fragments from the DNA of the pure cultures (data not shown). The results of the nirK and nirS nitrite reductase PCR experiments are summarized in Tables 2 and 3.
Determination of the type of nir gene of denitrifying bacteria.
Five strains of denitrifying bacteria were tested to determine their type of nitrite reductase. Genomic DNA preparations from these strains reacted with either the nirK or the nirS primer combination in PCR amplification experiments. Blastobacter denitrificans, Alcaligenes xylosoxidans, and Alcaligenes sp. were determined to have the coding sequence for the nirK nitrite reductase as deduced from their amplification products with the nirK primer combinations (Table 2). The coding sequence for the nirS nitrite reductase was identified within genomic DNA from Alcaligenes eutrophus DSM 530 and the denitrifying isolate IFAM 3698 as deduced from their amplification products with the nirS primer combinations (Table 3).
Sequencing of nir gene fragments.
To determine the specificity of the amplified fragments, the PCR products obtained with the primer combinations nirK1F-nirK5R and nirS1F-nirS6R from the pure cultures were partially sequenced. A total of 425 bp of the amplified fragments from nirK were sequenced; the amplified and sequenced regions for nirS were between 774 and 792 bp long. Comparison to all sequences in the EMBL database by using the FASTA program revealed that all sequences obtained were parts of the nirK or the nirS gene, respectively. The levels of homology of the fragments compared to the sequences of the nirK and nirS genes from the EMBL database are summarized in Tables 4 and 5, respectively. The calculations were done with the GAP program for the nucleic acid similarity and the TREE program for the deduced amino acid similarity.
TABLE 4.
Nucleic acid and amino acid similarities of nirK gene fragments from denitrifying bacteria
| Organism (EMBL accession no.) | % Similaritya to:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| 1. Alcaligenes faecalis S-6 (D13155) | 84.4 | 64.5 | 78.7 | 62.4 | 79.4 | 66.0 | 67.4 | 66.7 | 82.3 | 84.4 | 84.4 | 68.8 | 100.0 | 71.6 | |
| 2. Achromobacter cycloclastes (Z48635) | 79.8 | 63.8 | 80.8 | 67.4 | 78.0 | 68.8 | 69.5 | 67.4 | 81.6 | 80.8 | 80.8 | 68.8 | 84.4 | 69.5 | |
| 3. Pseudomonas aureofaciens (Z21945) | 67.3 | 68.7 | 65.2 | 66.0 | 65.2 | 82.3 | 80.8 | 78.7 | 68.1 | 63.1 | 63.1 | 70.9 | 64.5 | 68.1 | |
| 4. Pseudomonas sp. strain G-179 (M97294) | 75.5 | 78.6 | 67.5 | 65.2 | 76.6 | 66.7 | 68.8 | 66.7 | 81.6 | 80.1 | 80.1 | 64.5 | 78.7 | 72.3 | |
| 5. Rhodobacter sphaeroides 2.4.3 (U62291) | 67.3 | 72.5 | 71.5 | 68.5 | 66.7 | 70.2 | 70.2 | 68.1 | 68.1 | 64.5 | 64.5 | 83.0 | 62.4 | 68.1 | |
| 6. Rhizobium “hedysari” HCNT1 (U65658) | 74.6 | 73.2 | 64.7 | 73.9 | 66.3 | 66.7 | 66.7 | 66.0 | 81.6 | 80.1 | 80.1 | 67.4 | 79.4 | 73.8 | |
| 7. Alcaligenes xylosoxidans NCIMB 11015 (AJ224905) | 68.0 | 74.8 | 80.9 | 70.1 | 75.8 | 66.6 | 95.7 | 92.2 | 69.5 | 65.2 | 65.2 | 73.0 | 66.0 | 69.5 | |
| 8. Alcaligenes xylosoxidans subsp. denitrificans DSM 30026 (AJ224904) | 69.6 | 73.9 | 77.2 | 70.1 | 75.5 | 65.6 | 91.3 | 95.0 | 71.6 | 67.4 | 67.4 | 73.0 | 67.4 | 71.6 | |
| 9. Alcaligenes sp. strain DSM 30128 (AJ224903) | 68.7 | 73.4 | 79.3 | 68.2 | 75.1 | 65.6 | 89.9 | 89.9 | 70.9 | 66.7 | 66.7 | 70.9 | 66.7 | 71.6 | |
| 10. Hyphomicrobium zavarzinii IFAM-622T (AJ224902) | 76.9 | 79.8 | 68.7 | 76.5 | 70.6 | 73.9 | 70.6 | 70.8 | 69.9 | 85.1 | 85.1 | 70.2 | 82.3 | 71.6 | |
| 11. Rhizobium meliloti 20115 | 78.3 | 80.9 | 66.8 | 76.2 | 71.7 | 75.8 | 73.6 | 72.5 | 72.7 | 81.4 | 100.0 | 68.1 | 84.4 | 72.3 | |
| 12. Rhizobium meliloti Rm1021 (AJ224909) | 78.3 | 80.9 | 66.8 | 76.2 | 71.7 | 75.8 | 73.6 | 72.5 | 72.7 | 81.4 | 100.0 | 68.1 | 84.4 | 72.3 | |
| 13. Rhodobacter sphaeroides f. sp. denitrificans (AJ224908) | 68.5 | 72.2 | 73.4 | 68.0 | 84.7 | 67.8 | 76.5 | 75.3 | 73.6 | 70.6 | 73.5 | 73.5 | 68.8 | 67.4 | |
| 14. Ochrobactrum anthropi LMG 2136 (AJ2249057) | 99.5 | 80.0 | 67.5 | 75.8 | 67.5 | 74.6 | 68.2 | 69.9 | 68.7 | 77.4 | 78.1 | 78.1 | 68.5 | 71.6 | |
| 15. Blastobacter denitrificans IFAM 1005 (AJ224906) | 73.2 | 73.6 | 70.3 | 70.3 | 70.3 | 68.7 | 76.0 | 76.7 | 74.6 | 74.6 | 74.1 | 74.1 | 70.6 | 73.4 | |
Values in the upper right are levels of amino acid similarity, and those in the lower left are levels of nucleic acid similarity. Values are calculated from data obtained from the EMBL nucleotide sequence database by using the GAP program for the similarity of nucleic acids and by using the TREE program for the similarity of deduced amino acids.
TABLE 5.
Nucleic acid and amino acid similarities of nirS gene fragments from denitrifying bacteria
| Organism (EMBL accession no.) | % Similaritya to:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| 1. Pseudomonas stutzeri JM300 (M80653) | 93.5 | 55.5 | 53.9 | 53.1 | 58.4 | 58.4 | 55.5 | 53.9 | 87.4 | 71.9 | 49.6 | 87.0 | |
| 2. Pseudomonas stutzeri ZoBell (X56813) | 92.6 | 57.8 | 56.6 | 56.2 | 61.9 | 61.9 | 57.8 | 56.6 | 92.4 | 73.2 | 51.6 | 92.4 | |
| 3. Pseudomonas aeruginosa (X16452) | 67.3 | 67.2 | 67.3 | 68.5 | 70.7 | 70.7 | 100.0 | 67.3 | 56.2 | 60.5 | 61.1 | 55.9 | |
| 4. Paracoccus denitrificans Pd1222 (U05002) | 66.1 | 66.1 | 72.1 | 97.7 | 64.4 | 64.4 | 67.3 | 100.0 | 54.7 | 59.0 | 72.0 | 55.1 | |
| 5. Paracoccus denitrificans LMD29.63 (U75413) | 64.5 | 64.3 | 71.6 | 94.9 | 64.4 | 64.4 | 68.5 | 97.7 | 54.3 | 59.4 | 72.8 | 54.7 | |
| 6. Alcaligenes eutrophus H16 (X91341) | 69.7 | 70.8 | 76.0 | 73.2 | 71.5 | 100.0 | 70.7 | 64.4 | 60.7 | 64.6 | 62.5 | 60.3 | |
| 7. Alcaligenes eutrophus DSM 530 | 69.7 | 70.8 | 76.0 | 73.2 | 71.5 | 100.0 | 70.7 | 64.4 | 60.7 | 64.6 | 62.5 | 60.3 | |
| 8. Pseudomonas aeruginosa DSM 6195 | 67.3 | 67.2 | 100.0 | 72.1 | 71.6 | 76.0 | 76.0 | 67.3 | 56.2 | 60.5 | 61.1 | 55.9 | |
| 9. Paracoccus denitrificans ATCC 19367 | 66.1 | 66.1 | 72.1 | 100.0 | 94.9 | 73.2 | 73.2 | 72.1 | 54.7 | 59.0 | 72.9 | 55.1 | |
| 10. Alcaligenes faecalis A15 (AJ224913) | 87.5 | 89.4 | 68.5 | 67.4 | 65.8 | 71.8 | 71.8 | 68.5 | 67.4 | 72.9 | 49.6 | 97.3 | |
| 11. Azospirillum brasilense Sp7 DSM 1690 (AJ224912) | 72.3 | 72.6 | 68.6 | 68.2 | 66.8 | 71.9 | 71.9 | 68.6 | 68.2 | 72.7 | 57.0 | 72.9 | |
| 12. Roseobacter denitrificans ATCC 33942T (AJ224911) | 59.5 | 61.8 | 64.2 | 74.2 | 73.7 | 66.9 | 66.9 | 64.2 | 74.2 | 61.1 | 63.9 | 49.6 | |
| 13. Denitrifying isolate IFAM 3698 (AJ224910) | 86.1 | 89.4 | 68.2 | 67.1 | 66.0 | 70.8 | 70.8 | 68.2 | 67.1 | 94.4 | 74.9 | 61.3 | |
Values in the upper right are levels or amino acid similarity, and those in the lower left are levels or nucleic acid similarity. Values are calculated from data obtained from the EMBL nucleotide database by using the GAP program for the similarity of nucleic acids and by using the TREE program for the similarity of the deduced amino acids.
Amplification of nitrite reductase genes from environmental samples.
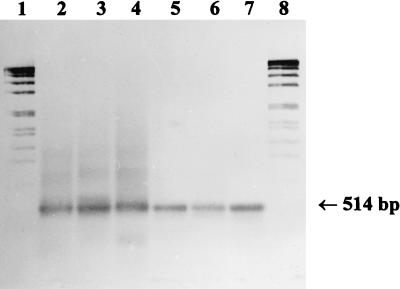
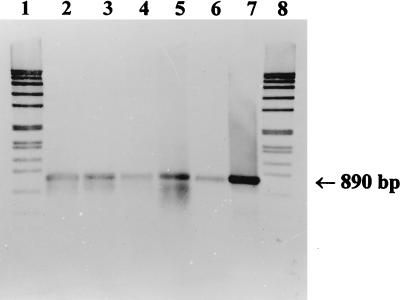
The development of a system of generally amplifying primer combinations for each gene permitted the qualitative detection of denitrifying bacteria in environmental samples. For these experiments, nirK1F-nirK5R or nirS1F-nirS6R were applied to total DNA preparations from an enrichment culture for denitrifying methylotrophic bacteria, from activated sludge, from the water of Lake Kleiner Plöner See, and Lake Plussee, and from sediment of Lake Kleiner Plöner See. For nirK, a weak product of the expected size (514 bp), as well as a smear of nonspecific products, was obtained from the DNA preparations from the water of Lake Kleiner Plöner See and Lake Plussee and from sediment of Lake Kleiner Plöner See. The weak band of 514 bp was extracted and reamplified at annealing temperatures of 65 to 60°C by applying the same primer combination. As a result, distinct bands of 514 bp were obtained (Fig. 2, lanes 2 to 6). For nirS, products of the expected size (about 890 bp) were generated (Fig. 3, lanes 2 to 6). The sizes of the products obtained from the environmental samples were the same as those of the positive controls for nirK of Alcaligenes xylosoxidans (Fig. 2, lane 7) and for nirS of Pseudomonas stutzeri (Fig. 3, lane 7).
FIG. 2.
Southern blot hybridization of nirK fragments obtained from environmental samples with the primer combination nirK1F-nirK5R to the digoxigenin-labeled fragment from Alcaligenes xylosoxidans. Lanes: 1 and 8, digoxigenin-labeled DNA size standard VII (Boehringer); 2, sediment from Lake Kleiner Plöner See (5 μl of PCR product), 3, water from Lake Plussee (7 μl); 4, water from Lake Kleiner Plöner See (7 μl); 5, activated sludge from the sewage treatment plant at Plön (10 μl); 6, enrichment culture for denitrifying methylotrophic bacteria (12 μl); 7, A. xylosoxidans positive control (5 μl).
FIG. 3.
Southern blot hybridization of nirS fragments obtained from environmental samples with the primer combination nirS1F-nirS6R to the digoxigenin-labeled fragment from Pseudomonas stutzeri ZoBell. Lanes: 1 and 8, digoxigenin-labeled DNA size standard VII (Boehringer); 2, sediment from Lake Kleiner Plöner See (15 μl), 3, water from Lake Plussee (12 μl); 4, water from Lake Kleiner Plöner See (20 μl); 5, activated sludge from the sewage treatment plant at Plön (10 μl); 6, enrichment culture for denitrifying methylotrophic bacteria (20 μl); 7, P. stutzeri ZoBell positive control (1 μl).
Hybridization analysis with probes for nirK and nirS.
To confirm the specificity of these amplified nirK and nirS fragments, hybridization experiments were performed. The DIG-labelled PCR product from Alcaligenes xylosoxidans (nirK), obtained with the primer combination nirK1F-nirK5R, and from Pseudomonas stutzeri (nirS), obtained with the primer combination nirS1F-nirS6R, were used as probes. All the amplified fragments hybridized, and the products of the positive controls reacted with the specific probe (Fig. 2 and 3). Previous results had shown that the probes did not hybridize with negative controls (PCR without template), with nonspecific PCR products, or nonlabelled DNA standard (data not shown).
DISCUSSION
The genetic diversity of denitrifying bacteria in environmental samples can be investigated by different molecular methods. We describe herein the first steps required to detect denitrifying bacteria in aquatic habitats by the use of two distinct PCR systems for the nitrite reductase genes, nirK and nirS. Since denitrifiers are not defined by close phylogenetic relationship, an approach involving 16S rRNA molecules is not suitable for general detection of this physiological group in the environment. The use of rRNA-targeted probes has been successfully applied so far only for strains and specific groups to explore the denitrifying community of activated sludge (18).
A more general approach to the detection of all denitrifying bacteria in environmental samples could be the use of a physiological gene or of an enzyme as a molecular marker. For this purpose, nitrite reductase and its genes have been used by several authors, since this is the key enzyme in the denitrification process. Antisera against the dissimilatory nitrite reductase (dNir) from Pseudomonas stutzeri ATCC 14405 were highly specific and reacted with the immunizing strain and few other closely related bacteria (14, 29). Less specific reactions could be obtained with combinations of antisera against heme-type dNirs from P. stutzeri JM300 and P. aeruginosa (4). When a variety of approximately 150 denitrifying strains of uncharacterized dNir type were screened with this combination and an antiserum against Cu dNir from Alcaligenes cycloclastes, 90% of the strains could be identified as possessing either the heme-type or Cu dNir (4). Due to the inducible nature of the enzyme, antisera could be useful in detecting conditions of active denitrification (29). In contrast, approaches targeting the nirK or nirS gene would detect the denitrifiers irrespective of the denitrifying conditions. Compared to the antisera, a broader response to different pure cultures possessing the nirS gene was achieved by use of the specific gene probes (15, 24). By using hybridization with a gene probe for nirK, this type of nitrite reductase could be always confirmed in pure cultures (32). In enrichment cultures, different populations of denitrifiers could be detected by restriction enzyme-digested preparations (HindIII) of total DNA with a probe for nirS (23).
PCR amplification systems, on the other hand, are limited neither to actively denitrifying cells nor to cultivated strains. Application to pure cultures of a primer pair derived from three nirS sequences flanking a conserved central region of the nirS gene revealed a reactivity broader than that with the use of antisera but not as satisfactory as that with the use of gene probes (30). A strain-specific reaction such as that obtained with the use of antisera was not achieved, although amplification failed with some nirS-containing strains as detected by hybridization. This may be because for PCR amplification only the homology of the primer hybridization region is decisive whereas hybridization of gene probes can be detected if any region of the probe shows sufficient homology. In the present study, a PCR system based on six sequences each for nirK or nirS available from the database promised an even more general approach. Regions that are conserved for both genes could not be found, because the enzymes are structurally different. When the sequences for each of the genes were aligned separately, conserved regions for each became evident. Degenerated primers that flanked regions at the more highly conserved C terminus of the two genes were designed. Primers were not positioned within the nirS region that codes for heme binding, because this is homologous to highly conserved regions in other heme-binding proteins (11). By using different combinations of primers and low-temperature stringency conditions in the PCR assays, amplification of nir fragments from both genes was possible for all denitrifying strains tested. The specificity of the PCR procedure was confirmed, since specific products were not obtained when the primer combinations were used with nondenitrifying strains that could perform assimilatory respiration of NO3− or with strains possessing the gene coding for the other nir type. This is consistent with the finding that the two genes are mutually exclusive in a given strain (4). Combinations containing nirK4R or those containing nirS5R resulted in the smallest number of specific products; this may be due to the degree of conservation in the region where the primer should hybridize. From every pure culture tested, at least two different primer combinations were applied successfully for nirK and at least three were applied successfully for nirS. The specificity of amplifications was confirmed by sequencing the largest products. Generally, sequencing revealed that the products from the pure cultures with the primer pairs nirK1F-nirK5R and nirS1F-nirS6R were specific fragments of the genes coding for copper-containing and cytochrome cd1-containing nitrite reductase, respectively. Calculation of the homology revealed that with these primer pairs, PCR products could be obtained from genes showing homology as low as 65.5% for nirK and 59.5% for nirS to the sequences available for the design of the primers. This indicates that these PCR systems could be reasonable means for a more general detection of denitrifying bacteria.
The fragments of nirS investigated here were more heterogeneous than were those of nirK. In-frame deletions or insertions of up to 18 bp could be observed within the sequenced region of nirS. Since these results are not in agreement with the findings that the Cu dNirs were more heterogeneous than the cd1 dNirs (4), the different molecular weights of the Cu dNir subunits may be a result of processing of the enzyme.
The degrees of conservation of the nir genes are very variable. The nirK fragments from eight strains of Ochrobactrum anthropi were 98.8 to 100.0% homologous (data not shown). On the other hand, the sequence of the fragment from two of these strains was 100% homologous to that of the same fragment from Alcaligenes faecalis S-6 (data not shown). However, both types of nir genes are distributed among closely related Pseudomonas (RNA group I [35]) and Alcaligenes (4) species. Even among strains of Alcaligenes faecalis, both types could be detected. The distribution of nir genes among denitrifying bacteria could be explained in different ways. The occurrence of the same nir type among phylogenetically different groups (34) might be caused by a common denitrifying ancestor. During evolution, the ability to denitrify may have been lost in some branches, resulting in closely related nondenitrifying and denitrifying strains (2). The occurrence of different nir types among the same species could be an indication of a horizontal gene transfer (4).
The PCR systems for the nirK and the nirS genes for nitrite reductase, using one generally amplifying primer combination for each nir gene, could be applied successfully to detect populations of denitrifying bacteria in aquatic systems. Due to higher cell densities, detection of such populations from soils by using these PCR systems should be possible as well.
nirS fragments could be amplified directly from DNA preparations of environmental samples, whereas for nirK a reamplification step was necessary when total DNA from water or sediment of lakes was the target for amplification. This might be in agreement with a numerical preponderance of heme type nitrite reductase, as reported for the distribution of nir types in numerically dominant isolates from soils (4). Further investigations will be necessary to determine the abundance of nirK and nirS type denitrifiers and to obtain more information on the genetic diversity of denitrifying bacteria in the environment.
ACKNOWLEDGMENTS
We are grateful to P. Hirsch and C. G. Gliesche for critically reading the manuscript. The assistance of M. Beese and B. Albrecht with the photographic work is gratefully acknowledged. We thank H. Bothe and K. Kloos for providing most of the strains investigated, C. G. Gliesche for providing H. zavarzinii IFAM ZV-622T, P. Hirsch for providing B. denitrificans IFAM 1005 and R. denitrificans ATCC 33942T, M. Schloter for providing four strains of O. anthropi, and J. M. Tiedje for providing R. sphaeroides f. sp. denitrificans.
REFERENCES
- 1.Ausubel F M, Brent R, Kinston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 2.Betlach M R. Evolution of bacterial denitrification and denitrifier diversity. Antonie Leeuwenhoek. 1982;48:585–607. doi: 10.1007/BF00399543. [DOI] [PubMed] [Google Scholar]
- 3.Conrad R. Soil organisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO) Microbiol Rev. 1996;60:609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyne M S, Arunakumari A, Averill B A, Tiedje J M. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl Environ Microbiol. 1989;55:2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fries M R, Zhou J, Chee-Sanford J, Tiedje J M. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl Environ Microbiol. 1994;60:2802–2810. doi: 10.1128/aem.60.8.2802-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genetics Computer Group. GCG program manual. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 7.Gliesche C G, Hirsch P. Mutagenesis and chromosome mobilization in Hyphomicrobium facilis B-522. Can J Microbiol. 1992;38:1167–1174. doi: 10.1139/m92-191. [DOI] [PubMed] [Google Scholar]
- 8.Gliesche C G, Menzel M, Fesefeldt A. A rapid method for creating species-specific gene probes for methylotrophic bacteria. J Microbiol Methods. 1997;28:25–34. [Google Scholar]
- 9.Glockner A B, Jüngst A, Zumft W G. Copper-containing nitrite reductase from Pseudomonas aureofaciens is functional in a mutationally cytochrome cd1-free background (NirS−) of Pseudomonas stutzeri. Arch Microbiol. 1993;160:18–26. doi: 10.1007/BF00258141. [DOI] [PubMed] [Google Scholar]
- 10.Hess A, Zarda B, Hahn D, Häner A, Stax D, Höhener P, Zeyer J. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl Environ Microbiol. 1997;63:2136–2141. doi: 10.1128/aem.63.6.2136-2141.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jüngst A, Wakabayashi S, Matsubara H, Zumft W G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991;279:205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- 12.Kloos K, Fesefeldt A, Gliesche C G, Bothe H. DNA-probing indicates the occurrence of denitrification and nitrogen fixation genes in Hyphomicrobium. Distribution of denitrifying and nitrogen fixing isolates of Hyphomicrobium in a sewage treatment plant. FEMS Microbiol Ecol. 1995;18:205–213. [Google Scholar]
- 13.Knowles R. Denitrification. Microbiol Rev. 1982;46:43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Körner H, Zumft W G. Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl Environ Microbiol. 1989;55:1670–1676. doi: 10.1128/aem.55.7.1670-1676.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linne Von Berg K-H, Bothe H. The distribution of denitrifying bacteria in soils monitored by DNA-probing. FEMS Microbiol Ecol. 1992;86:331–340. [Google Scholar]
- 16.Lyman J, Fleming R H. Composition of sea water. J Mar Res. 1940;3:134–167. [Google Scholar]
- 17.Michalski W P, Nicholas D J D. Immunological patterns of distribution of bacterial denitrifying enzymes. Phytochemistry. 1988;27:2451–2456. [Google Scholar]
- 18.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K-H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J E, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1 to 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Satoh T, Hoshino Y, Kitamura H. Rhodopseudomonas sphaeroides forma sp. denitrificans, a denitrifying strain as a subspecies of Rhodopseudomonas sphaeroides. Arch Microbiol. 1976;108:265–269. doi: 10.1007/BF00454851. [DOI] [PubMed] [Google Scholar]
- 21.Shapleigh J P, Payne W J. Differentiation of cd1 cytochrome and copper nitrite reductase production in denitrifiers. FEMS Microbiol Lett. 1985;26:275–279. [Google Scholar]
- 22.Smalla K. Extraction of microbial DNA from sewage and manure slurries. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. p. 1.1.3. [Google Scholar]
- 23.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 25.Staley J T. Prosthecomicrobium and Ancalomicrobium, new prosthecate fresh water bacteria. J Bacteriol. 1968;95:1921–1944. doi: 10.1128/jb.95.5.1921-1942.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Elsas J D, Smalla K. Extraction of microbial community DNA from soils. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. p. 1.3.3. [Google Scholar]
- 27.Vincent J M. A manual for the practical study of root-nodule bacteria. Oxford, United Kingdom: Burgess and Son LTB; 1971. [Google Scholar]
- 28.Walsh P S, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 29.Ward B B, Cockcroft A R, Kilpatrick K A. Antibody and DNA probes for detection of nitrite reductase in seawater. J Gen Microbiol. 1993;139:2285–2293. doi: 10.1099/00221287-139-9-2285. [DOI] [PubMed] [Google Scholar]
- 30.Ward B B. Diversity of culturable denitrifying bacteria. Limits of rDNA RFLP analysis and probes for the functional gene, nitrite reductase. Arch Microbiol. 1995;163:167–175. [Google Scholar]
- 31.Ward B B, Voytek M A, Witzel K-P. Phylogenetic diversity of natural populations of ammonia oxidizers investigated by specific PCR amplification. Microb Ecol. 1997;33:87–96. doi: 10.1007/s002489900011. [DOI] [PubMed] [Google Scholar]
- 32.Ye R W, Fries M R, Bezborodnikov S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA of other denitrifiers. Appl Environ Microbiol. 1992;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Palumbo A, Tiedje J M. Sensitive detection of a novel class of toluene-degrading denitrifiers, Azoarcus tolulyticus, with small-subunit rRNA primers and probes. Appl Environ Microbiol. 1997;63:2384–2390. doi: 10.1128/aem.63.6.2384-2390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zumft W G. The denitrifying procaryotes. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 554–582. [Google Scholar]
- 35.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]