Abstract
Tumor-specific antigens (TSAs) are crucial for tumor-specific immune response that reduces tumor burden and thus serve as important targets for immunotherapy. Identification of novel TSAs can provide new strategies for immunotherapies. In this study, we demonstrated that the upstream open reading frame (uORF) of RNF10 encodes an antigenic peptide (RNF10 uPeptide), capable of eliciting a T cell-mediated anti-tumor immune response. We initially demonstrated the immunogenicity of the RNF10 uPeptide in a CT26 tumor mouse model, by showing that its epitope was specifically recognized by CD8+ T cells. Vaccination of mice with the long form of the RNF10 uPeptide conferred strong anti-tumor activity. Next, we proved that the human RNF10 uORF could be translated. In addition, we predicted the binding of an RNF10 uPeptide epitope to HLA-A∗02:01 (HLA-A2). This HLA-A2-restricted epitope of the RNF10 uPeptide induced a potent specific human T cell response. Finally, we showed that an HLA-A2-restricted cytotoxic T cell (CTL) clone, derived from a pancreatic cancer patient, recognized the RNF10 uPeptide epitope (RLFGQQQRA) and lysed HLA-A2+ pancreatic carcinoma cells expressing the RNF10 uPeptide. These results indicate that the RNF10 uPeptide could be a promising target for pancreatic carcinoma immunotherapy.
Keywords: MT: Regular Issue, upstream ORF, tumor antigen, tumor vaccine, RNF10, immunotherapy
Graphical abstract
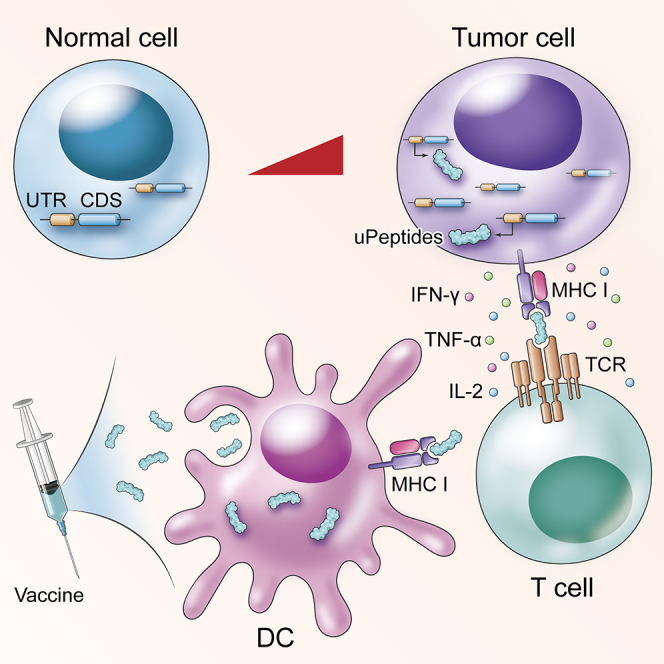
The upstream ORF of RNF10 encoded immunogenic peptides that hold potential as targets for cancer immunotherapy. The RNF10 uPeptides could be used as a peptide vaccine in a murine cancer model; these immunogenic peptides may increase the number of candidates for use as therapeutic vaccines.
Introduction
Tumor-associated antigens (TAAs) and neoantigens play vital roles in cancer immune response and have immense potential to be used in cancer immunotherapy.1 These proteins are usually translated from the CDS region of mRNAs. Recent studies have shown that protein or peptides can be encoded through noncanonical translation of long non-coding RNA and circular RNAs.2 Combining ribosome profiling, proteomics, and immunopeptidomics, researchers have confirmed the widespread translation of cryptic peptides from non-coding regions, including the 5′-UTRs and 3′-UTRs, non-coding RNAs, and intronic, intergenic, and out-of-frame regions, and provided the first insights into their presentation on human leukocyte antigen (HLA) class I molecules.3 Identification of novel TAAs and neoantigens could benefit for developing strategies for immunotherapies.
It is reported that approximately 40% of human mRNAs have upstream open reading frames (uORFs),4,5 and some of them are capable of encoding peptides. Previous studies have revealed their versatile functions such as translational regulators, protein kinase inhibitors, and stress responses.6,7 However, only very few uORF-encoded peptides have been functionally studied.8 Remarkably, uORF-encoded peptides play important roles in rapid and reversible changes upon cell stress. A well-known example is uORF encoded by ATF4 mRNA, which is regulated by cellular stress and plays crucial roles in endoplasmic reticulum (ER) stress responses.9 Precancerous or cancer cells encounter various stress during both tumorigenesis and tumor progression, which could induce expression of numerous uORFs. So, these hidden peptides could be big source of potential TAAs or neoantigens. However, whether these uORF-encoded peptides are capable of activating T cells remains largely unknown.
The RING finger (RNF) family proteins are conserved from yeast to humans. It is estimated that over 600 ubiquitin E3 ligases are encoded by the mammalian genome, surpassing the 518 protein-kinase-encoding genes.10 Since so many of the RNF proteins are ubiquitin E3 ligases,11 and therefore the primary determinants of substrate specificity in ubiquitylation, it has become evident that many RNF ubiquitin E3 ligases are implicated in malignancy. Ubiquitin E3 ligases regulate crucial cellular functions, such as cell cycle control, DNA repair, cell signaling, and the response to hypoxia.12 The therapeutic success of the proteasome inhibitor bortezomib demonstrates the value of targeting the ubiquitin-proteasome system.13 A question of great importance is whether RNF family proteins and the related genes can be used in the development of targeted therapeutics with greater specificities than those of proteasome inhibitors.
In this study, we found RNF family RNF10, which mediates ubiquitination of the 40S ribosomal subunit and coordinates ribosome turnover in response to cellular stress,14 was highly upregulated in tumor cells compared with normal tissues. Remarkably, we identified uORF-encoded peptide of RNF10 and showed that it encoded immunogenic peptides, which could be targeted by cancer immunotherapy. We then used the RNF10 uORF peptide (uPeptide) as a peptide vaccine in a murine cancer model to demonstrate that RNF-protein-related genes can act as precise therapeutic targets for tumor eradication. Additionally, studying these immunogenic peptides may increase the number of candidates for use as therapeutic vaccines.
Results
uORF of RNF10 encoded a peptide in CT26 cells
The CT26 colon carcinoma cell line is commonly used in drug development. Therefore, we initially use CT26 cells to determine the expression levels of RNF-protein-encoding genes. To screen for RNF-protein-encoding genes in CT26 cells, we retrieved relevant RNA sequencing (RNA-seq) data from published studies; CT26 and BALB/cJ NGS fastq reads are available from the European Nucleotide Archive as PRJEB5320 (https://www.ebi.ac.uk/ena/browser/view/PRJEB5320).15 RNF-protein-encoding candidates were selected based on the following standards: (1) expression fold change >2; (2) reads per kilobase per million mapped read (RPKM) >10; (3) uORF presence; and (4) unique peptides with a good major histocompatibility complex (MHC) class I binding rank (“low rank” <2). From these candidates, we further narrowed down the list by selecting genes encoding abundant RNF proteins. Eventually, we selected the most abundant RNFs—RNF10, which was upregulated in CT26 cells (Figures 1A and S1). The expression of RNF10 in CT26 cells was also validated by qRT-PCR (Figure S2).
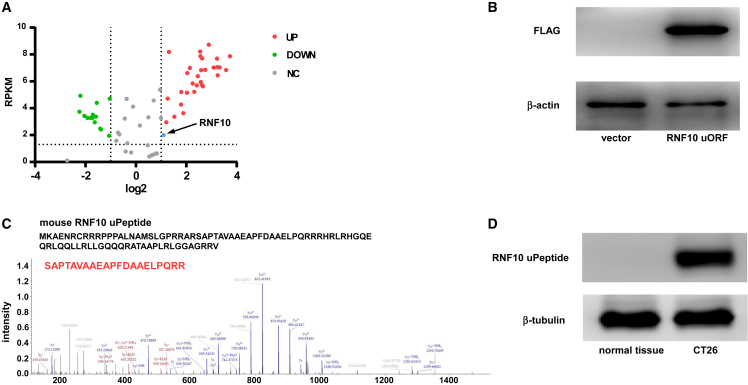
Figure 1.
uORF of RNF10 encoded a peptide in mice CT26 cells
(A) A volcano plot, showing the green, red, and black points represent significantly downregulated (green) or upregulated (red) RING finger (RNF)-protein-encoding genes in CT26 cells and normal colon tissue; black represents genes with no significant expression change. x axis: log2 ratio of RNA expression levels between normal and tumor tissues. y axis: the false discovery rate value (−log10 transformed) of RNF-protein-encoding genes. The blue dot indicates RNF10. (B) An anti-FLAG tag antibody was used to detect RNF10 uPeptide expression in HEK293T cells transfected with the above vectors. (C) Total protein, from HEK293T cells transfected with p-uORF or the control plasmid, was separated via SDS-PAGE. Individual gel bands (10–15 kDa) were extracted and subjected to liquid chromatography with tandem mass spectrometry (LC-MS/MS); the RNF10 uPeptide-specific peptide was identified (lower). (D) RNF10 uPeptide expression was detected in CT26 cells or normal colon tissue of BALB/c mice.
We revealed that a 261-nt ORF was present in the 5′-UTR of RNF10. To evaluate the coding ability of the RNF10 uORF, we used the pcDNA3.1 expression vector. The resulting p-uORF vector contained the RNF10 uORF and a 3×FLAG-coding sequence immediately upstream of the STOP codon. We observed successful plasmid transfection through fluorescence microscopy (Figure S3). The following experiments were carried out in HEK293T cells. The results indicate that, in comparison with mock-treated cells, p-uORF gave rise to an FLAG protein (Figure 1B). We next used liquid chromatography-mass spectrometry (LC-MS) to characterize the amino acid sequences of RNF10 uPeptide in p-uORF-transfected HEK293T cells. We identified distinctive amino acids in the uPeptide sequence, which further suggested that the RNF10 uPeptide was encoded by the uORF (Figure 1C). We then produced and characterized a polyclonal antibody against the amino acid sequence (MKAENRCRRRPPPA) of the RNF10 uPeptide. This antibody detected a specific 10 kDa protein inside CT26 cells, which was not present in the normal tissue (Figure 1D).
Peptides encoded by uORF of RNF10 are immunogenic
Moreover, transfer of CT26 cells into BALB/c mice provides a syngeneic in vivo model system, which is frequently used for developing and testing immunotherapeutic concepts. We therefore chose this model to assess the immunogenicity of the RNF10 uPeptide. RNF10 uPeptide epitopes were prioritized according to their MHC class I binding ability, which was predicted by the consensus method (version 2.5) of the Immune Epitope Database (http://www.iedb.org) (Figure 2A). We used long peptides (27 amino acids in length, with the uPeptide epitope positioned centrally) as antigens to test the immunogenicity of the RNF10 uPeptides. Poly(I:C), which promotes cross-presentation and increases vaccine efficacy, was used as an adjuvant for peptide immunization.16 In the vaccination experiments, the BALB/c mice were vaccinated twice and the immunogenicity of the uPeptides was assessed at 7 days after the last vaccination.
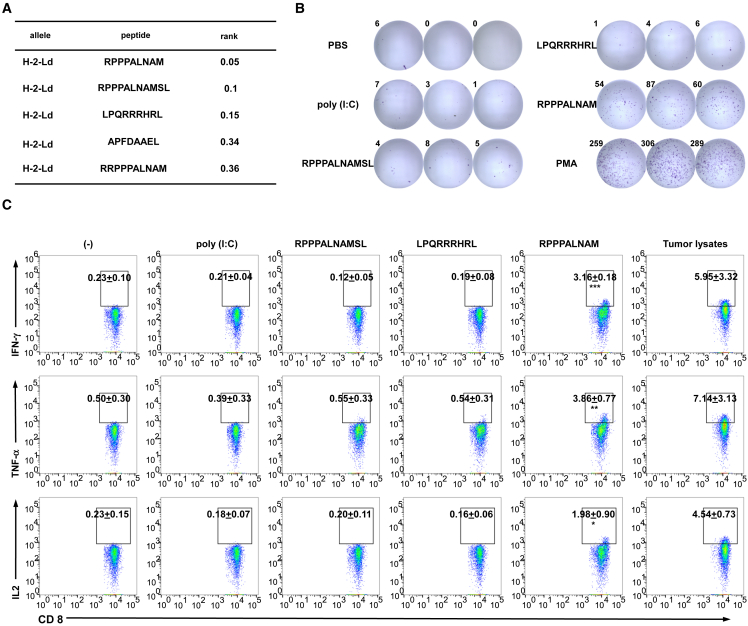
Figure 2.
uPeptides of RNF10 are immunogenic
(A) The binding predictions for RNF10 uPeptides to H2-d MHC class I using the Immune Epitope Database (version 2.5) analysis resource NetMHCpan (ver. 4.1) algorithm. (B) Splenocytes of vaccinated mice were restimulated with the RNF10 uPeptide antigens used for vaccination. The specificity of splenocytes to the RNF10 uPeptide was assess by IFN-γ ELISpot. Representative ELISpot images are shown; spot count per 5 × 105 cells is presented. (C) Splenocytes of mice injected with CT26 cells and vaccinated with peptides and the polyinosinic:polycytidylic acid (poly[I:C]) adjuvant (CT26, n = 3) were tested for recognition of mutated peptides by flow cytometry. Intracellular cytokine staining of CD8+ T cells, evaluated by flow cytometry. Percentages of cytokine-producing T cells are shown (mean ± SD, n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 compared with untreated T cells (−). p-values were determined by two-tailed one-way ANOVA with Dunnett’s multiple-comparisons test.
We found that the RNF10 uPeptide elicited an immune response in the immunized mice. The ELISpot results revealed a strong epitope-specific immune response within the splenocyte population of vaccinated mice (Figure 2B). We further confirmed the immunogenicity of the RNF10 uPeptide epitope by measuring the production of interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and interleukin (IL)-2 by CD8+ T cells using flow cytometry (gating strategy, Figure S4), ELISA, and qRT-PCR (Figure S5). We showed that CD8+ T cells produced cytokines specifically in response to the RNF10 uPeptide (Figure 2C). These data confirm the immunogenicity of the RNF10 uPeptide.
The RNF10 uPeptide antigens elicited antitumoral activity in vitro
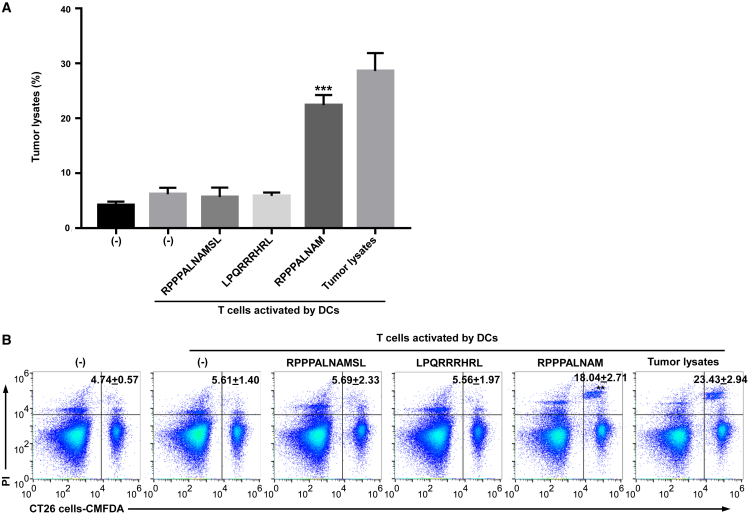
We next set out to investigate whether the RNF10 uPeptide could elicit an RNF10-uPeptide -specific cytotoxic lymphocyte (CTL) response in the immunized mice. CD8+ T cells were sorted from immunized mice and incubated with dendritic cells (DCs) pulsed with each peptide. After three stimulations, the cytotoxicity of CD8+ T cells against CT26 cells was examined using the lactate dehydrogenases (LDH) release assay (Figure 3A). We showed that CTLs primed with RNF10 uPeptide epitopes, but not control peptides, effectively killed CT26 cells. We also examined the capacity of these RNF10-uPeptide-primed CTLs to kill CT26 cell lines by flow cytometry. As shown in Figure 3B, RNF10-uPeptide-reactive CTLs stimulated with the RNF10 uPeptide exhibited effective cytotoxicity against CT26 cells. Collectively, these results indicate that the RNF10 uPeptide successfully primed CTLs and that the RNF10 uPeptide epitopes were naturally processed and expressed on the surface of tumor cells in the context of MHC I molecules.
Figure 3.
RNF10 uPeptide antigens elicited antitumoral activity in vitro
(A) The splenic T cells were co-cultured with CT26 cells for 18 h. T-cell-mediated cytotoxicity against CT26 cells was assessed by lactate dehydrogenase (LDH) assay (mean ± SD, n = 3). (B) The splenic T cells were co-cultured with CellTrace CMFDA-labeled CT26 cells for 18 h. Tumor cell death was examined by PI uptake via flow cytometry. Percentages of dead tumor cells are shown (mean ± SD, n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with untreated T cells (−). p-values were determined by two-tailed one-way ANOVA with Dunnett’s multiple-comparisons test.
Injection of RNF10 uPeptide antigens elicited antitumoral activity in vivo
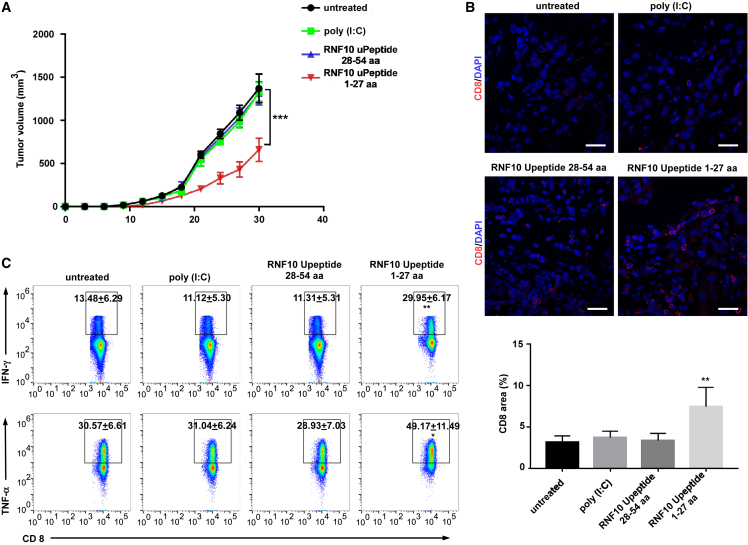
We assessed whether immune responses elicited in vitro translated to antitumoral effects in tumor-bearing mice. Long peptides have many advantages to exact epitopes, as they can be naturally processed and presented on MHC I molecules.17 The therapeutic potential of vaccination with RNF10 uPeptide antigens was explored by immunizing mice with 27 aa peptides (RNF10 uPeptide 1-27aa) containing the uPeptide epitope (RPPPALNAM) in center, after transferring 1.5 × 105 CT26 cells into mice on days 1, 3, 6, 9, and 15. The result showed that the growth of tumors was inhibited in mice vaccinated with the RNF10 uPeptide antigens, compared with the control group (Figures 4A and S6). In addition, the tumors of mice vaccinated with the RNF10 uPeptide contained significantly more infiltrating CD8+ T cells than the tumors of control mice (Figure 4B). Moreover, the CD8+ tumor-infiltrating lymphocytes (TILs) isolated from vaccinated mice produced more IFN-γ and TNF-α than those isolated from control animals (Figure 4C). These results suggest that the immune microenvironment changed after vaccination.
Figure 4.
Induction of efficient tumor control in a CT26 colon cancer mouse model immunized with a long RNF10 uPeptide vaccine
(A) Tumor growth (mean ± SEM) in untreated (control) or RNF10-uPeptide immunized BALB/C mice (n = 7), inoculated subcutaneously (s.c.) with CT26 cells. (B) Upper panel, CD8-stained cancer tissue sections. Scale bar, 20 μm. Lower panel, proportional CD8+ lymphocyte areas in tumor tissue of control (n = 5) or vaccinated (n = 5) animals. (C) Tumor-infiltrating lymphocytes (TILs) or splenocytes were stimulated with PMA and ionomycin to assess the ability of CD8+ T cells to produce IFN-γ and TNF-α. Cytokine production was determined by intracellular cytokine staining and flow cytometry. Data are representative of two independent experiments. Percentages of dead tumor cells are shown (mean ± SD, n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with untreated T cells (−). p-values were determined by two-tailed one-way ANOVA with Dunnett’s multiple-comparisons test.
The uORF of human RNF10 could be translated
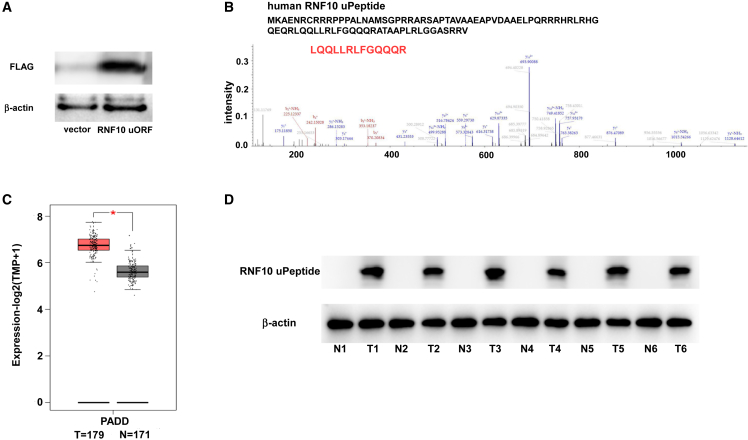
To determine the coding ability of the human RNF10 uORF, a vector containing human RNF10 uORF and a 3×FLAG-coding sequence (immediately upstream of the STOP codon) was transfected into HEK293T cells. The western blotting results indicated that an FLAG protein was expressed in these p-uORF-transfected HEK293T cells (Figure 5A). We next used LC-MS to characterize the amino acid sequences of RNF10 uPeptide in the transfected HEK293T cells (Figure 5B).
Figure 5.
The RNF10 uPeptide is expressed in pancreatic cancer
(A) HEK293T cells were transfected with the p-uORF expression vector carrying an expression cassette for human RNF10 uORF with a 3×FLAG-coding sequence. Immunoblotting for FLAG expression using an anti-FLAG tag antibody in the indicated cells (n = 3). (B) Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis, following SDS-PAGE, was performed to identify epitopes within the human RNF10 uPeptide. (C) Expression (measured and normalized as transcripts per million base pairs [TPM] from the RNA-seq data) of RNF10 in pancreatic carcinoma patient samples from TCGA. The statistical significance of differences was evaluated using the Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. (D) Six randomly selected pancreatic carcinoma patient samples and their paired adjacent normal pancreatic tissues were collected. The expression of human RNF10 uPeptide was detected by immunoblotting.
To assess whether the results of the mouse experiments were applicable to the human setting, we analyzed RNA-seq data from patients with pancreatic cancer, obtained from The Cancer Genome Atlas (TCGA: https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and GEPIA 2 (https://www.gepia2.cancer-pku.cn). We found that the expression of RNF10 differed between pancreatic cancerous and normal tissues (Figures 5C and S7A). The expression of RNF10 in pancreatic cancerous and normal tissues was also validated by qRT-PCR (Figure S7B). Furthermore, western blotting showed that RNF10 uPeptide was overexpressed in pancreatic cancer and pancreatic cancer cell lines (Figures 5D and S7C). Immunohistochemistry confirmed high expression of RNF10 uPeptide in pancreatic cancer tissues (Figure S7C), while it was not detected in some crucial organs (Figure S8).
uORF of RNF10 generates an immunogenic peptide recognized by human autologous T cells
We applied the IEDB algorithm to predict the binding affinities of human RNF10 uPeptide to HLA class I alleles. Based on standard criteria in the field, we considered peptides with a rank <2 as strong binders, a rank of 2–5 as intermediate to weak binders, and a rank >5 as nonbinders. For the RNF10 uPeptide, we found that RLFGQQQRA was predicted to bind strongly to HLA-A2 allele (Figure S9).
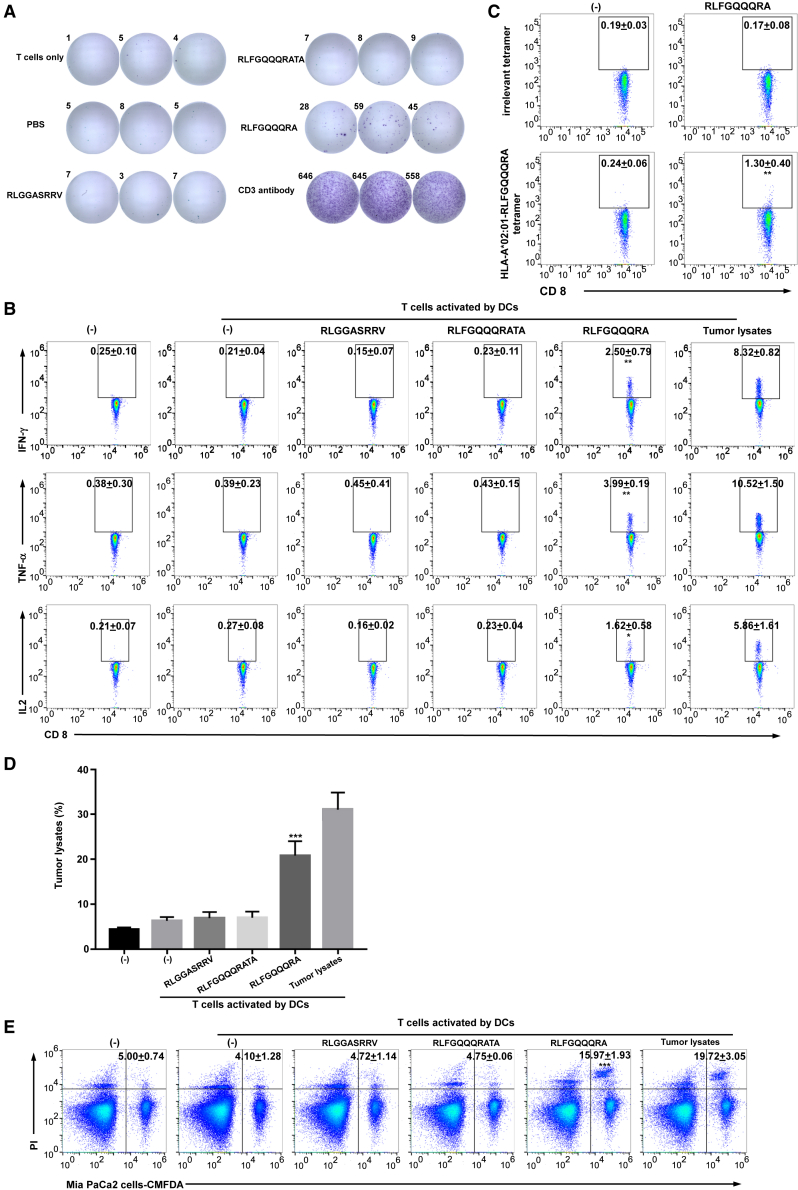
We therefore synthesized the uORF peptides and loaded them onto HLA-A2-positive DCs. The RLFGQQQRA epitope, derived from the RNF10 uPeptide, induced IFN-γ secretion by pancreatic-cancer-patient-derived T cells (Figure 6A), as determined by ELISpot. The ability of patient-derived T cells to specifically produce IFN-γ, TNF-α, and IL-2 in response to DCs pulsed with the RLFGQQQRA peptide, was confirmed by flow cytometry (Figure 6B), ELISA, and qRT-PCR (Figure S10). We further identified the RNF10-uPeptide-specific CD8+ T cells from the HLA-A2-positive pancreatic carcinoma patients (Figure S11). To determine whether peptide-specific T cells can be selectively expanded, T cells were co-cultured with peptide-pulsed DCs for 21 days. Positive staining with the RLFGQQQRA-tetramer-PE demonstrated the successful expansion of peptide-specific CD8+ T cells (Figure 6C). Thus, the RLFGQQQRA epitope from the RNF10 uORF is a strong immunogenic epitope presented on HLA-A2.
Figure 6.
The RNF10 uPeptide generates an immunostimulatory epitope recognized by human autologous T cells
(A) CD8+ T cells were primed by autologous DCs loaded with uPeptide antigens. The CTLs were restimulated with peptides overnight. The uPeptide-specific T cell response was evaluated using an IFN-γ ELISpot. Representative ELISpot images are shown; spot count per 5 × 104 T cells is presented. (B) The primed T cells were restimulated with uPeptide antigens overnight and evaluated by flow cytometry; representative flow cytometry plots of intracellular cytokine staining are shown. Percentages indicate proportions of CD8+ T cells producing different cytokines (mean ± SD, n = 3). (C) CD8+ T cells were primed by autologous DCs loaded with uPeptide antigens. The primed T cells were then stained with the uRNF10 RLFGQQQRA-HLA-A∗02:01 tetramer. Numbers in plots denote percentages of the tetramer+CD8+ T cells (mean ± SD, n = 3). (D) The primed T cells were co-cultured with MIA PaCa-2 cells for 18 h and their cytotoxicity against MIA PaCa-2 cells was assessed by lactate dehydrogenase (LDH) assay (mean ± SD, n = 3). (E) The T cells were co-cultured with CellTrace CMFDA-labeled MIA PaCa-2 cells for 18 h. Tumor cell death was examined by PI uptake via flow cytometry. Percentages of dead tumor cells are shown (mean ± SD, n = 3). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with untreated T cells (−). p-values were determined by two-tailed one-way ANOVA with Dunnett’s multiple-comparisons test.
To investigate whether the RLFGQQQRA epitope could generate RNF10-uPeptide-specific human CTLs, CD8+ T cells sorted from the PBMCs of HLA-A2-positive donors were incubated with autologous DCs pulsed with each peptide. After three stimulations, the cytotoxicity of the CD8+ T cells against MIA PaCa-2 cells that expressed RNF10 uPeptide (Figure S6), was examined using the LDH release assay (Figure 6D) and flow cytometry (Figure 6E). CTLs stimulated with peptides containing the RLFGQQQRA epitope killed the MIA PaCa2 cells, but those stimulated with control peptides did not. These data indicate that the RLFGQQQRA epitope successfully induced peptide-specific human CTLs.
RNF10 uPeptide elicited antitumoral response in patient-derived xenograft model
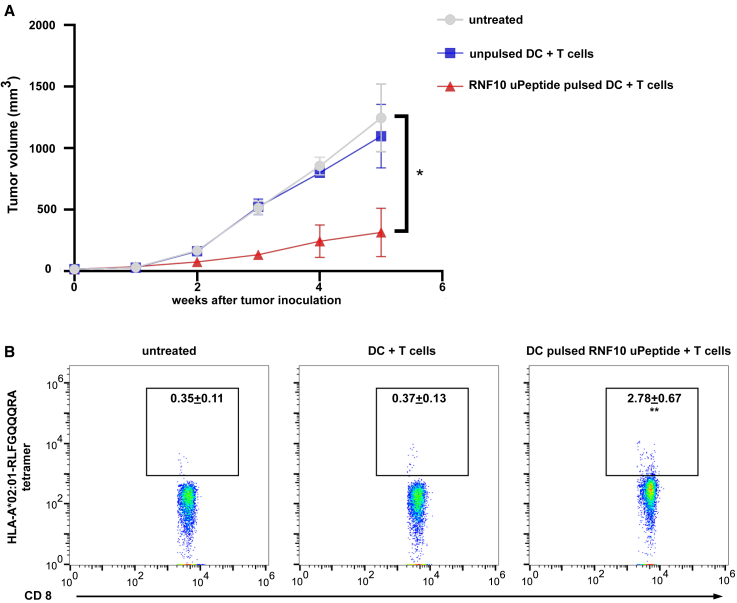
To further explore the immunogenicity of RNF10 uPeptide in vivo, we employed the pancreatic cancer patient-derived xenograft (PDX) model implanted in immunocompromised NOD.SCID mice, followed by intravenous (i.v.) infusion of autologous DCs and T cells. Tumor growth was profoundly retarded when receiving RNF10 uPeptide-loaded DCs, compared with the ones receiving control DCs and the untreated mice, determined by tumor-volume measurement (Figures 7A and S12). We performed flow cytometry and found that in tumors from RNF10 uPeptide-loaded DC-vaccinated mice, RNF10-uPeptide-specific CD8+ T cell infiltrates were significantly more abundant than those in control DC-vaccinated mice (Figure 7B).
Figure 7.
Human RNF10 uPeptide elicites immune response to tumor
(A) Tumor volumes were monitored weekly for 5 consecutive weeks following infusion of untreated, unpulsed, or RNF10 uPeptide pulsed DCs and T cells. (B) Tumor-infiltrating lymphocytes (TILs) were determined by tetramer staining and flow cytometry. Data are representative of two independent experiments. Percentages of dead tumor cells are shown (mean ± SD, n = 5). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with untreated T cells (−). p-values were determined by two-tailed one-way ANOVA with Dunnett’s multiple-comparisons test.
Discussion
Cancer is a multifactorial disease and a major public health concern worldwide.18 Given that RNF proteins are involved in almost all cellular processes, it is not surprising that their dysregulation, caused by genetic alterations, is associated with the development and progression of human cancer, as well as resistance to anti-cancer drugs.19 However, few studies have addressed whether and how certain RNF proteins can contribute to anti-tumor therapy. In this study, we revealed that the uORF of RNF10 encodes a uPeptide, which induces a CD8+ T-cell-mediated anti-tumor immune response. The uPeptide encoded by RNF10 could therefore be a target for use in cancer immunotherapy.
To assess whether RNF proteins could be exploited as therapeutic targets in cancer, we chose the CT26 tumor cell line. Recently, Castle et al. identified the transcriptomic characteristics of the CT26 colorectal carcinoma.15 Many RNF-protein-encoding genes are overexpressed in CT26 cells, compared with normal colon tissue. A few RNF proteins are also involved in cellular stress signaling.20,21,22 Interestingly, uORFs are significantly enriched in stress-response genes and control the translation of certain master regulators of the stress response.23,24 We revealed that the RNF-protein-encoding gene RNF10 contained a uORF and was overexpressed in CT26 cells. Previous studies have shown that an ORF upstream of the 5′-UTR can be translated.25 Here, we identified that the uORF of RNF10 could also be translated and encoded a peptide. This RNF10 uPeptide comprised distinct amino acid sequences, compared with its associated mRNA-translated proteins. We found that the RNF10 uPeptide was expressed in CT26 cells, but could not be detected in the normal colon tissue of BALB/c mice by immunoblotting. This could be explained by the generally low efficiency of uORF translation in normal cells.26,27 Alternative translation initiation at noncanonical uORFs has been demonstrated to be an important mechanism of translational regulation during the stress response or tumor initiation.28,29 For example, BiP encodes a chaperone protein, which is important for ER homeostasis, and contains uORFs. Under normal conditions, canonical eIF2α-mediated translation initiation is responsible for the translation of the BiP CDS. However, during ER stress, elevated eIF2A expression initiates the translation of BiP uORFs.30 Cancer cells are inherently stressed owing to their unprogrammed proliferation, resulting in inadequate blood supply and compromised lymphatic draining, which leads to hypoxia, nutrient deprivation, and chronic toxin exposure.31 Under these conditions, the translation of uORFs may be active. Thus, the RNF10 uPeptide is specifically expressed in tumors.
Recently, it was reported that the frequent expression of uPeptides in benign and malignant human tissues inspire novel approaches in direct molecular as well as immunotherapeutic targeting of cancer-associated uORFs and uPeptides.32 Tumor-associated cryptic peptides from non-coding regions, including the 5′- and 3′-UTR, non-coding RNAs, and intronic, intergenic, and out-of-frame regions, represent highly promising targets for anti-tumor immune surveillance as well as the development of immunotherapeutic approaches.1 In the present study, we proved the immunogenicity of the RNF10 uPeptide, which may be a promising target for cancer immunotherapy.
We demonstrated that the RNF10 uPeptide contained an HLA-A2-restricted epitope, which was recognized by a specific CTL population. These CTLs could effectively kill CT26 cells that overexpressed the RNF10 uPeptide. Many researchers have reported that long peptide vaccines attenuated tumor progression in animal models and cancer patients.33 Our result showed that tumor growth in BALB/c mice injected with CT26 cells was significantly inhibited after vaccination with a 27-aa RNF10 uPeptide epitope. These results revealed that the RNF10 uPeptide vaccine induced strong anti-tumor activity in the CT26 model. In addition, the vaccinated mice had higher numbers of CD8+ TILs in their tumor lesions than unvaccinated controls. Moreover, we found a significant increase in the IFN-γ- and TNF-α-producing capacity of the CD8+ TILs of immunized mice, compared with the control mice. Thus, the vaccination with the RNF10 uPeptide changed the immune microenvironment of the tumor. This approach might be particularly useful for reshaping the tumor microenvironment in patients who lack TILs, and applicable as a standalone or combination therapy to increase the clinical success rate of checkpoint blockade.
To assess whether the findings from the vaccinated mouse model could be replicated in humans, we analyzed RNA-seq data from TCGA. We found that pancreatic adenocarcinoma was among the cancers in which RNF10 was upregulated. We also confirmed that the RNF10 uPeptide was expressed in pancreatic carcinoma tissues.
The immune system has the ability to eradicate cancer cells via the recognition of TSAs and the induction of T-cell-mediated cytotoxicity.34 Moreover, researchers have reported that some patients with pancreatic carcinoma benefit from immunotherapy.35,36 We have uncovered that the human RNF10 uPeptide elicited a T cell response against human pancreatic adenocarcinoma cells. Therefore, the RNF10 uPeptide could be targeted in pancreatic carcinoma immunotherapy. These results illustrate that in the context of pathologically altered cellular processes, the differential translation of uORFs and differential uPeptide processing could produce tumor-specific uORF-derived HLA ligands, which may serve as potent TSAs. Neoantigens are peptides derived from tumor-specific mutations which have not been previously recognized by the body’s immune system.37,38 Most neoantigens are derived from single nucleotide variants (SNVs).34 Unlike the patient-specific nature of SNV neoantigens, uORF-derived antigens may have the advantage of being widely shared by multiple tumors, allowing for the development of universal, off-the-shelf therapies. Furthermore, uORF epitopes may be used as an alternative immunotherapy in patients with low tumor mutational burden and thus few neoantigens. Therefore, studies such as ours may encourage the screening of the entire human genome for uORFs to unravel the whole uORF-derived immunopeptidome landscape in cancer.
There are limitations to this study. We were not able to investigate the expression of RNF10 uPeptides in every mouse and human organ, which may yield more convincing evidence. In addition, we are lacking experiments that directly reflect the situation in human cancer. Further studies will be needed to see whether uORF peptides can be therapeutically targeted in human cancer.
In conclusion, in this study, we revealed that the uORF of RNF10 encoded immunogenic peptides and that this RNF10 uPeptide was expressed in tumor cells and could be targeted by specific CD8+ T cells. Thus, uORF-associated antigens represent an important source of TSAs. Moreover, uORF-derived epitopes shared between patients may have particular relevance to anti-tumor vaccination.
Materials and methods
Patients and tissues
Tumor tissue and peripheral blood samples were obtained from HLA-A∗02:01+ (HLA-A2 from hereon) patients with pancreatic carcinoma, being treated at the Sun Yat-sen Memorial Hospital, Sun Yat-sen University (Guangzhou, China) between 2020 and 2022. All samples were collected from patients who had provided informed consent, and all related procedures were performed with the approval of the internal review and ethics board of the Sun Yat-sen Memorial Hospital. The HLA-A2+ status of the patients was determined by staining patient peripheral blood mononuclear cells (PBMCs) with a monoclonal anti-HLA-A2 antibody (Cat# 343304, BioLegend), followed by flow cytometry.
Cell lines and mice
Female 8- to 12-week-old BALB/c mice (obtained from the animal center of Guangdong Province) were kept in accordance with the institutional policies on animal research at the Sun Yat-sen University. The CT26 colon cancer cell line, the HPDE6-C7 pancreatic ductal epithelial cell line, the MIA PaCa-2, PANC-1, SW1990 and Capan-2 pancreatic cancer cell line, and HEK293T cells were purchased from the Shanghai Cell Center. Master and working cell banks were generated immediately upon receipt, of which the third and fourth passages were used for the tumor experiments. Cells were tested for mycoplasma every 3 months.
uORF prediction
The ORF finder was used to identify potential ORFs within RNA transcripts and predict the peptides encoded by those ORFs.
Quantitative reverse-transcriptase (qRT)-PCR
qRT-PCR was performed with PrimeScriptTM RT Master Mix (Cat# RR035A, Takara) and SYBR Premix Ex Taq Kit (Cat# RR820A, Takara), according to the manufacturer’s instructions. The primer sequences used are listed in Table S1. Data were collected and analyzed using a LightCycler 480 instrument (Roche).
Plasmids and transfection
To test whether the predicted uORF sequence in the 5′-UTR was translatable, complete uORF sequences of interest (with the 3×FLAG tag immediately upstream of the uORF termination codon and including the HindIII and BamHI restriction site-overhangs for ligation) were amplified by PCR. Amplicons were ligated into the pcDNA3.1 plasmid (+). The plasmids were transfected into HEK293T cells with Lipofectamine 3000 (Cat# L3000015, Invitrogen), according to the manufacturer’s instructions.
Western blotting
Total proteins were extracted from cells or tissues using the radioimmunoprecipitation assay (RIPA) lysis buffer (Cat# 89900, Pierce), supplemented with protease inhibitors (1:100, Cat# ab65621, Abcam). Equal amounts of protein were loaded onto a 12.5% polyacrylamide gel (Cat# PG112, EpiZyme Biotechnology), separated by electrophoresis, and transferred to a PVDF membrane. The membrane was blocked in 5% skim milk solution and hybridized with the following antibodies, depending on the experiment: anti-FLAG antibody (Sigma-Aldrich, Cat# F1804) and anti-β-actin (C4) (Santa Cruz Biotechnology, Cat# sc-47778) to detect actin. A polyclonal antibody against uPeptide was produced by Genscript Biotechnology Co (Nanjing, China).
Liquid chromatography with tandem mass spectrometry analysis of uORF-encoded proteins
Proteins were separated by SDS-PAGE and the relevant protein bands were excised and chopped into 1-mm3 pieces. Peptide mixtures were then extracted from the gel fragments, digested with sequencing-grade trypsin, and dried, prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The peptides were analyzed on a QExactive mass spectrometer (Thermo Fisher, Carlsbad, CA).
Identification of target epitopes for uPeptide
The uPeptide was subjected to MHC class I peptide binding prediction using Immune Epitope Database (http://www.iedb.org) algorithms for 8-mer, 9-mer, 10-mer, and 11-mer, containing the uPeptide sequence.
Synthetic peptides
uORF epitopes were studied in the context of a 27-aa peptide. Peptides were synthesized by the Genscript Biotechnology Co (Nanjing, China).
Immunization of mice with peptides
Research involving animals complied with protocols approved by the Sun Yat-sen University Animal Care and Use Committee of the Sun Yat-sen Memorial Hospital. For the immunogenicity analysis of uPeptide epitopes, age-matched female BALB/c mice were vaccinated on days 1 and 7. Vaccination was performed by subcutaneous (s.c.) injection of 100 μg synthetic peptide and 50 μg polyinosinic:polycytidylic acid (poly [I:C]) (Invivo Gen, Cat# vac-pic) formulated in phosphate-buffered saline (PBS; 200 μL total volume) into the lateral flank. For confirmation of epitope immunogenicity, mice were vaccinated against a single epitope (n = 3). The readout was performed 5 to 6 days after the last immunization (see also enzyme-linked immunospot [ELISpot] and intracellular cytokine staining sections).
For the therapeutic tumor experiments, BALB/c mice were inoculated s.c. with 1.5 × 105 CT26 colon cancer cells and randomly distributed among treatment groups. Tumor volume was measured with a caliper and calculated using the formula A × B2/2, with A as the largest and B as the smallest diameter of the tumor. Tumor growth was assessed by measuring the mean tumor size (with standard error) while disregarding single distant outliers.
DC differentiation and T cell priming
DCs were generated as previously described with modifications.39 DCs were obtained from the peripheral blood of HLA-A2+ patients with pancreatic cancer and cultured for 6 days in DMEM media containing 50 ng/mL GM-CSF (PeproTech, Cat# 300-03-20), 20 ng/mL IL-4 (PeproTech, Cat# 200-04-20), and 10% AB type human serum. The culture medium was replaced every 3 days, and cell differentiation was monitored by light microscopy. DCs were pulsed with 20 μg/mL peptides for 18 h. To generate uPeptide-specific T cells, T cells were isolated from the PBMCs of HLA-A2+ patients with pancreatic cancer using Pan T cell beads (Miltenyi Biotec, Cat# 130-096-535). The T cells were then cultured with antigen-presenting DCs (5:1) in RPMI 1640 media with 25 U/mL interleukin (IL)-2 (PeproTech, Cat# 200-02-50) for 9 days.
ELISpot
T cells were collected from the in vitro DC co-cultures or murine spleens, and were restimulated with peptide-pulsed DCs for 18 h. IFN-γ-secreting T cells were quantified by IFN-γ ELISpot (Mabtech, Cat# 3420-4HST-2 [human], Cat# 3321-4HST-2 [mouse]), according to the manufacturer’s instructions. Developed spots were counted using an ELISpot reader (C.T.L., Shaker Heights, OH).
ELISA
The cell processing is the same as for the ELISpot. Cell supernatants were harvested after restimulation, and quantitative measurements of cytokine were done by ELISA (Dakewe) according to the instructions of the manufacturer. Cytokine concentration was determined spectrophotometrically at 450 nm in combination with a wavelength correction at 570 nm.
Intracellular cytokine staining
A total of 2 × 106 splenocytes were stimulated with 5 μg/mL peptide in the presence of brefeldin A (BioLegend, Cat# 420601). Splenocytes treated with tumor lysates served as a positive control. Cells were incubated for 5 h at 37°C, stained with Zombie Aqua Fixable Viability Kit (BV525, BioLegend, Cat# 423101), anti-mouse CD8 (PerCP, BioLegend, Cat# 100732), and anti-mouse CD3 (APC-Cy 7, BD Pharmingen, Cat# 557596) antibodies for 30 min at 4°C, fixed, and permeabilized using BD Cytofix/Cytoperm soln kit (BD Pharmingen, Cat# 554714), according to the manufacturer’s protocol. Next, the cells were stained with anti-mouse IFN-γ (EFLOUR 450, eBioscience, Cat# 48-7311-82), anti-mouse TNF-α (EFLOUR 450, eBioscience, Cat# 48-7321-82), and anti-mouse IL-2 (EFLOUR 450, eBioscience, Cat# 48-7021-82) antibodies. Cytokine secretion by CD8+ T cells was analyzed by multicolor flow cytometry (n = 3).
For ex vivo human T cell intracellular cytokine detection, T cells were stimulated with 20 μg/mL peptide in the presence of 10 μg/mL brefeldin A (BioLegend, Cat# 420601) at 37°C overnight. Subsequently, cells were stained with antibodies against surface markers, including human CD8 (PerCP, BioLegend, Cat# 344707), human CD3 (PE/Cyanine7, BioLegend, Cat# 317333) for 20 min at 4°C, followed by fixation and permeabilization. The cells were then stained with anti-human IFN-γ (EFLOUR 450, eBioscience, Cat# 48-7319-42), anti-human TNF-α (EFLOUR 450, eBioscience, Cat# 48-7349-42), anti-human IL-2 (EFLOUR 450, eBioscience, Cat# 48-7029-42) antibodies at 4°C for 1 h to overnight and analyzed by multicolor flow cytometry (Beckman CytoFLEX S).
Detection of RNF10 uPeptide-specific T cells in humans
DCs were pulsed with 20 μg peptides, then irradiated with 50 Gy to serve as antigen-presenting cells (APCs) for T cell expansion. T cells were cultured with the irradiated APCs in the presence of IL-2 (20 U/mL) and IL-15 (10 ng/mL), and analyzed on day 21. The cytokines were replenished every 3 days. RLFGQQQRA-reactive T cells were stained with RLFGQQQRA-tetramer-PE (diluted 1:50 in FACS buffer) for 30 min at room temperature in the dark, followed by staining with antibodies against human CD3 (PE/Cyanine7, BioLegend, Cat# 317333) and CD8 (PerCP, BioLegend, Cat# 344707). Antigen-specific T cells were then analyzed by flow cytometry.
LDH assay
The LDH cytotoxicity assay kit (Beyotime, Cat# C0016) was used to measure T cell cytotoxicity. In brief, 1.5 × 104 target cells/well were seeded into 96-well microplates in a final volume of 100 μL DMEM media (high glucose)/well, and co-cultured with uPeptide-specific CD8+ T cells at 37°C with 5% CO2 overnight. To prepare the maximum LDH activity control sample, 10 μL of 10× lysis buffer was added and incubated under the aforementioned conditions for another 45 min. At the end of the experiment, 50 μL of supernatant from each well was transferred to a new 96-well plate, to which 50 μL of the reaction mixture supplied in the kit was added. The sample was incubated for another 30 min in the dark, after which, 50 μL of stop solution was added. Finally, the absorbance was measured at 490 nm and 680 nm % LDH cytotoxicity was calculated LDH OD readings using the following formula.
Cytotoxicity assays
Target cells, such as CT26 cells and MIA PaCa2 cells, were stained with CMFDA (Thermo Fisher Scientific, C7025) for 15 min at 37°C. RNF10 uPeptide epitope-activated CD8+ T cells, generated by incubation with uPeptide epitope-pulsed APCs as described above, were harvested using CD8 microbeads (Miltenyi Biotec, Cat# 130-116-478 or Cat# 130-045-201). The CD8+ T cells were then co-cultured with the relevant target cells at an effector to target ratio of 10:1 for 12 h at 37°C. After 12 h, all cells were harvested and stained with PI (eBioscience, Cat# 00–6990; 1:500) and immediately analyzed by flow cytometry.
Immunohistochemistry
Paraffin-embedded samples were cut into 4-μm consecutive sections and deparaffinized. Then, sections were stained for RNF10 uPeptide antibody following detection by a horseradish peroxidase-conjugated antibody and the corresponding peroxidase substrate and counterstained with hematoxylin. The sections were captured on an NI-U (Nikon).
Immunofluorescence staining
For immunofluorescence staining, paraffin-embedded samples were cut into 4-μm consecutive sections and deparaffinized. The antigens were then retrieved using a special pressure cooker for 3 min in EDTA buffer (pH 8.0). The tissue sections were incubated with an anti-mouse CD8 antibody (Abcam, Cat# ab17147; 1:50), followed by incubation with an Alexa-Fluor-conjugated secondary antibody (Thermo Fisher Scientific, Cat# A-21206). The sections were then rigorously washed with PBS, counterstained with DAPI (Ca# H21492, Thermo Fisher), and imaged using a confocal laser-scanning microscope (Carl Zeiss) with a core data acquisition system (Applied Precision).
PDX model
Fresh pancreatic adenocarcinoma (PAAD) tissue was obtained with patient consent and approval from our institutional ethics committee. Immediately, the surgical material was subcutaneously transplanted into the flanks of NOD/SCID mice and allowed to grow. Once the tumor burden reached approximately 1,500 mg, the mice were humanely euthanized, and the tumors were carefully collected and divided into pieces. These tumor fragments were then serially propagated in additional mice. T cells were extracted from the peripheral blood of the identical patients. DCs were pulsed with 20 μg/mL peptides, and then co-cultured with autologous T cells at 1:5 ratio for 20 h. When the PDXs were palpable, 2.5 × 106 T cells and 0.5 × 106 DCs were transferred into tumor-bearing NOD/SCID mice through tail vein injection once a week for 2 weeks. Tumor growth was monitored and recorded.
TIL analysis
Tumors were excised and cut into small pieces in 5 mL of RPMI. The samples were then digested in 15 mL RPMI, supplemented with 50 U/mL collagenase IV (Invitrogen, Cat# 17104019), 100 μg/mL hyaluronidase (Macklin, Cat# H822579-10KU), and 20 U/mL DNase (Solarbio, Cat# D8070-15) for 2 h at 37°C, using gentle MACS dissociators (Miltenyi Biotech). Cell suspensions were then filtered through a 40-μM strainer and washed three times with PBS. The lymphocyte population was enriched by Ficoll density gradient centrifugation at 800 × g for 20 min. T cells were purified with mouse or human Pan T MicroBeads (Miltenyi Biotec, Cat# 130-095-130, Cat# 130-096-535). The T cells were eluted from the beads, stained with relevant antibody, and analyzed by flow cytometry.
Statistical analysis
Means were compared using the Student’s t test or one-way ANOVA with Dunnett’s multiple-comparisons test to compare individual treatment and corresponding control groups. Tumor growth among different experimental conditions was compared by calculating the area under the tumor growth curve (AUC) for each mouse. Statistical differences in the medians of two groups were calculated using a nonparametric Mann-Whitney U test. All analyses were two-tailed and were performed using GraphPad Prism 5.03 software. p-values <0.05 were considered as a measure of statistical significance; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data and code availability
All raw data supporting the findings are available from the corresponding authors upon reasonable request.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (grant number: 32000430).
Author contributions
Z.X. and Z.L. designed the study and revised the manuscript. Z.L., Z.W., Z.J., W.J., J.Q., and W.X. conducted the experiments and analyzed the data. Z.X. drafted the manuscript. Z.X. and M.Y. curated the data and edited the manuscript. All the authors listed have made a direct contribution to the work and approved it for publication. The final manuscript has been read by and obtained the approval of all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2023.100737.
Contributor Information
Yaming Meng, Email: 13631397478@163.com.
Xiaofeng Zhu, Email: zhuxf8@mail.sysu.edu.cn.
Supplemental information
References
- 1.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong C., Müller M., Pak H., Harnett D., Huber F., Grun D., Leleu M., Auger A., Arnaud M., Stevenson B.J., et al. Integrated proteogenomic deep sequencing and analytics accurately identify non-canonical peptides in tumor immunopeptidomes. Nat. Commun. 2020;11:1293. doi: 10.1038/s41467-020-14968-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laumont C.M., Vincent K., Hesnard L., Audemard É., Bonneil É., Laverdure J.P., Gendron P., Courcelles M., Hardy M.P., Côté C., et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aau5516. [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Brunner A.D., Cogan J.Z., Nuñez J.K., Fields A.P., Adamson B., Itzhak D.N., Li J.Y., Mann M., Leonetti M.D., Weissman J.S. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367:1140–1146. doi: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnstone T.G., Bazzini A.A., Giraldez A.J. Upstream ORFs are prevalent translational repressors in vertebrates. EMBO J. 2016;35:706–723. doi: 10.15252/embj.201592759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Wang Y., Lu J. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem. Sci. 2019;44:782–794. doi: 10.1016/j.tibs.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Jayaram D.R., Frost S., Argov C., Liju V.B., Anto N.P., Muraleedharan A., Ben-Ari A., Sinay R., Smoly I., Novoplansky O., et al. Unraveling the hidden role of a uORF-encoded peptide as a kinase inhibitor of PKCs. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2018899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., Wang Y., Wu X., Tang X., Wu C., Lu J. Determinants of genome-wide distribution and evolution of uORFs in eukaryotes. Nat. Commun. 2021;12:1076. doi: 10.1038/s41467-021-21394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J., Wan J., Shu X.E., Mao Y., Liu X.M., Yuan X., Zhang X., Hess M.E., Brüning J.C., Qian S.B. N-Methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell. 2018;69:636–647.e7. doi: 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Bengtson M.H., Ulbrich A., Matsuda A., Reddy V.A., Orth A., Chanda S.K., Batalov S., Joazeiro C.A.P. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipkowitz S., Weissman A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng L., Meng T., Chen L., Wei W., Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 2020;5:11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonneveld P., Chanan-Khan A., Weisel K., Nooka A.K., Masszi T., Beksac M., Spicka I., Hungria V., Munder M., Mateos M.V., et al. Overall survival with daratumumab, bortezomib, and dexamethasone in previously treated multiple myeloma (CASTOR): a randomized, open-Label, phase III trial. J. Clin. Oncol. 2023;41:1600–1609. doi: 10.1200/JCO.21.02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzia A., Meyer C., Tuschl T. The E3 ubiquitin ligase RNF10 modifies 40S ribosomal subunits of ribosomes compromised in translation. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109468. [DOI] [PubMed] [Google Scholar]
- 15.Castle J.C., Loewer M., Boegel S., de Graaf J., Bender C., Tadmor A.D., Boisguerin V., Bukur T., Sorn P., Paret C., et al. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics. 2014;15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dos-Santos J.S., Firmino-Cruz L., da Fonseca-Martins A.M., Oliveira-Maciel D., Perez G.G., Roncaglia-Pereira V.A., Dumard C.H., Guedes-da-Silva F.H., Santos A.C.V., Leandro M.D.S., et al. Immunogenicity of SARS-CoV-2 trimeric spike protein associated to poly(I:C) plus alum. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.884760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bijker M.S., van den Eeden S.J.F., Franken K.L., Melief C.J.M., Offringa R., van der Burg S.H. CD8+ CTL priming by exact peptide epitopes in incomplete Freund's adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J. Immunol. 2007;179:5033–5040. doi: 10.4049/jimmunol.179.8.5033. [DOI] [PubMed] [Google Scholar]
- 18.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 19.Cai C., Tang Y.D., Zhai J., Zheng C. The RING finger protein family in health and disease. Signal Transduct. Target. Ther. 2022;7:300. doi: 10.1038/s41392-022-01152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto T., Imaizumi K., Kaneko M. The role of tissue-specific ubiquitin ligases, RNF183, RNF186, RNF182 and RNF152, in disease and biological function. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21113921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amal H., Gong G., Gjoneska E., Lewis S.M., Wishnok J.S., Tsai L.H., Tannenbaum S.R. S-nitrosylation of E3 ubiquitin-protein ligase RNF213 alters non-canonical Wnt/Ca+2 signaling in the P301S mouse model of tauopathy. Transl. Psychiatry. 2019;9:44. doi: 10.1038/s41398-019-0388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang E., Okumura C.Y.M., Sheffy-Levin S., Varsano T., Shu V.C.W., Qi J., Niesman I.R., Yang H.J., López-Otín C., Yang W.Y., et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sidrauski C., McGeachy A.M., Ingolia N.T., Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015;4 doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andreev D.E., O'Connor P.B.F., Fahey C., Kenny E.M., Terenin I.M., Dmitriev S.E., Cormican P., Morris D.W., Shatsky I.N., Baranov P.V. Translation of 5' leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4 doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingolia N.T., Lareau L.F., Weissman J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Dou S., He F., Luo J., Wei L., Lu J. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2003903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.H., Park S.M., Park J.H., Keum S.J., Jang S.K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova V.V., Shestakova E.D., Nogina D.S., Mishchenko P.A., Prikazchikova T.A., Zatsepin T.S., Kulakovskiy I.V., Shatsky I.N., Terenin I.M. Ribosomal leaky scanning through a translated uORF requires eIF4G2. Nucleic Acids Res. 2022;50:1111–1127. doi: 10.1093/nar/gkab1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H., et al. Translation from unconventional 5' start sites drives tumour initiation. Nature. 2017;541:494–499. doi: 10.1038/nature21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starck S.R., Tsai J.C., Chen K., Shodiya M., Wang L., Yahiro K., Martins-Green M., Shastri N., Walter P. Translation from the 5' untranslated region shapes the integrated stress response. Science. 2016;351:aad3867. doi: 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dersh D., Hollý J., Yewdell J.W. A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat. Rev. Immunol. 2021;21:116–128. doi: 10.1038/s41577-020-0390-6. [DOI] [PubMed] [Google Scholar]
- 32.Nelde A., Flötotto L., Jürgens L., Szymik L., Hubert E., Bauer J., Schliemann C., Kessler T., Lenz G., Rammensee H.G., et al. Upstream open reading frames regulate translation of cancer-associated transcripts and encode HLA-presented immunogenic tumor antigens. Cell. Mol. Life Sci. 2022;79:171. doi: 10.1007/s00018-022-04145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melief C.J.M., van der Burg S.H. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat. Rev. Cancer. 2008;8:351–360. doi: 10.1038/nrc2373. [DOI] [PubMed] [Google Scholar]
- 34.Smith C.C., Selitsky S.R., Chai S., Armistead P.M., Vincent B.G., Serody J.S. Alternative tumour-specific antigens. Nat. Rev. Cancer. 2019;19:465–478. doi: 10.1038/s41568-019-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S., Guo S., Gou M., Pan Y., Fan M., Zhang N., Tan Z., Dai G. A composite indicator of derived neutrophil-lymphocyte ratio and lactate dehydrogenase correlates with outcomes in pancreatic carcinoma patients treated with PD-1 inhibitors. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.951985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K., Tandurella J.A., Gai J., Zhu Q., Lim S.J., Thomas D.L., Xia T., Mo G., Mitchell J.T., Montagne J., et al. Multi-omic analyses of changes in the tumor microenvironment of pancreatic adenocarcinoma following neoadjuvant treatment with anti-PD-1 therapy. Cancer Cell. 2022;40:1374–1391.e7. doi: 10.1016/j.ccell.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott P.A., Hu Z., Keskin D.B., Shukla S.A., Sun J., Bozym D.J., Zhang W., Luoma A., Giobbie-Hurder A., Peter L., et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Löwer M., Bukur V., Tadmor A.D., Luxemburger U., Schrörs B., et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 39.Su S., Liao J., Liu J., Huang D., He C., Chen F., Yang L., Wu W., Chen J., Lin L., et al. Blocking the recruitment of naive CD4 T cells reverses immunosuppression in breast cancer. Cell Res. 2017;27:461–482. doi: 10.1038/cr.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data supporting the findings are available from the corresponding authors upon reasonable request.