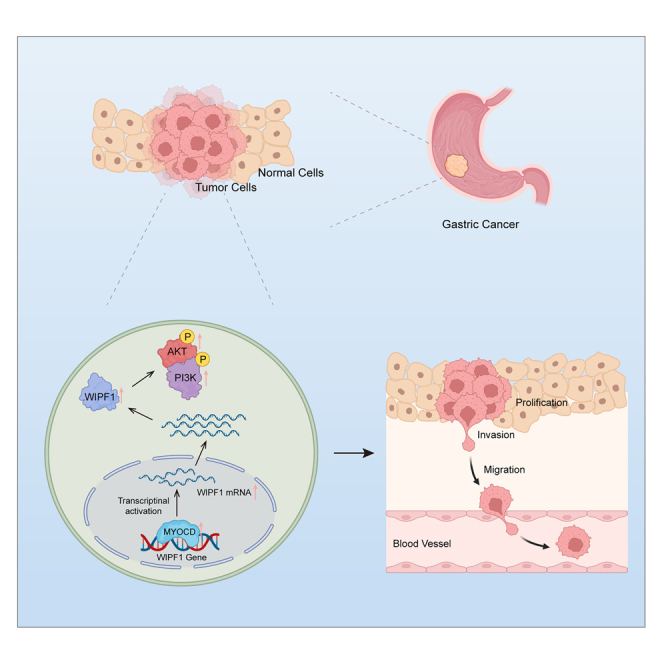
Summary
Wiskott–Aldrich syndrome protein-interacting protein family member 1 (WIPF1) is associated with malignant tumor progression. However, molecular links between WIPF1 and gastric cancer (GC) remain elusive. The expression of WIPF1 was detected in GC tissues and cells. WIPF1 was overexpressed in GC tissues and cells and high expression of WIPF1 was an independent risk factor for a poor prognosis in patients with GC. Further experiments indicated that WIPF1 promoted the proliferation, invasion, and migration of GC cells in vivo and in vitro. WIPF1-regulated genes were closely related to cell proliferation and migration in GC, and silencing WIPF1 significantly repressed PI3K/AKT signaling pathway activation. WIPF1 was activated by myocardin (MYOCD) translation. Rescue experiments confirmed that MYOCD promotes the proliferation, invasion, and migration of GC cells in a WIPF1-dependent manner and activates the PI3K/AKT signaling pathway. MYOCD may transactivate WIPF1 and facilitate GC cell growth and metastasis by activating the PI3K/AKT signaling pathway.
Subject areas: Molecular biology, Cancer, Model organism
Graphical abstract
Highlights
-
•
WIPF1 is highly expressed in GC and is associated with patient prognosis
-
•
WIPF1 promotes tumor malignant biological behavior in vivo and in vitro
-
•
WIPF1 activated the PI3K/AKT signaling pathway in GC
-
•
MYOCD transcriptional activation WIPF1 in GC
Molecular biology; Cancer; Model organism
Introduction
Gastric cancer (GC) is the fourth leading cause of cancer-related death worldwide. There were 1.08 million new cases of GC worldwide in 2021, accounting for 5.6% of all new cancers, and there were about 0.76 million deaths due to GC, accounting for 7.7% of all cancer deaths.1,2 Due to a lack of diagnostic markers for early GC, a large proportion of patients are diagnosed with advanced GC. Although treatment can be improved by surgery, targeted therapy, chemotherapy, radiotherapy, and immunotherapy, the 5-year survival rate is only 10–30% in patients with advanced GC.3,4 Therefore, it is important to study the molecular mechanisms underlying GC tumorigenesis and progression to identify candidate molecular biomarkers for the diagnosis and treatment of GC.
Wiskott–Aldrich syndrome protein-interacting protein family member 1 (WIPF1) plays an oncogenic role in the regulation of cancer invasion and metastasis of cancer.5 WIPF1 is upregulated in breast, glioma, thyroid, and colorectal cancer.6,7 Low WIPF1 expression is associated with a better prognosis in colorectal cancer and glioma.6 One study reported that the long noncoding RNA HCG18 upregulates WIPF1 by inhibiting miR-141-3p in GC.8 However, the specific mechanisms underlying WIPF1-mediated GC progression remain unclear.
Multiple studies have shown that gene set enrichment analysis (GSEA) can be used to identify functional pathways.9,10 Increasing evidence indicates that the PI3K/AKT signaling pathway participates in cell proliferation, metastasis, apoptosis, autophagy, epithelial-mesenchymal transition (EMT), and chemoresistance in different cancer types.11 Epigenetic regulatory mechanisms may contribute to the regulation of PI3K/AKT signaling.12,13 Myocardin (MYOCD) functions as a potent transcription factor in the myocardium and smooth muscles. It promotes the activation of cardiac reporter vectors and enhances the expression of cardiac-specific genes.14 Upregulation of MYOCD increases EMT and cell invasion induced by TGF-β in non-small cell lung cancer.15 However, the potential role and mechanism of action of MYOCD in GC tumorigenesis and progression requires further clarification.
In this study, it was demonstrated that WIPF1 increases the proliferation and migration of GC cells in vitro and promotes GC growth and metastasis in vivo by activating PI3K/AKT signaling. Moreover, bioinformatics analysis and subsequent experiments confirmed that MYOCD transactivated WIPF1 expression. Additionally, WIPF1 is necessary for MYOCD to promote the malignant phenotype of GC cells by activating the PI3K/AKT signaling pathway, suggesting that the MYOCD/WIPF1/PI3K/AKT axis may contribute to the onset and development of GC.
Results
A proteomics study identified Wiskott–Aldrich syndrome protein-interacting protein family member 1 as an oncogenic protein in gastric cancer
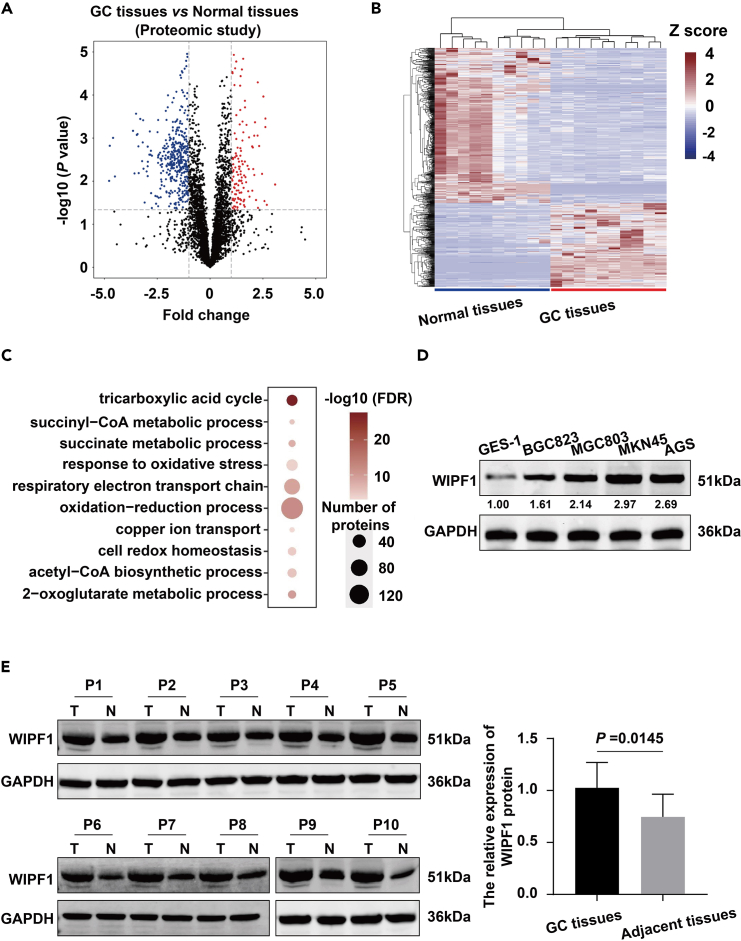
To unveil the potentially pivotal oncogenic proteins that play important roles in the tumorigenesis and progression of GC, proteomic data were carefully analyzed (Figures 1A–1C). Significantly upregulated proteins in GC and adjacent normal tissues are shown in red and blue, respectively, in a volcano plot (Figure 1A). A heatmap shows the 741 significantly differentially expressed proteins (DEPs). Blue represents downregulated proteins while red represents upregulated proteins in the GC tissues (Figure 1B). Gene Ontology (GO) analysis showed that the top 20 DEPs were related to metabolic pathways, mainly focusing on oxidation-reduction (Figure 1C). The most upregulated protein, WIPF1, was selected for further analysis. Although several studies have reported the essential role of WIPF1 in GC, its precise molecular and cellular regulatory networks remain poorly understood. Here, it was found that WIPF1 was highly expressed in GC cell lines and GC tissues compared to GES-1 cells or the corresponding adjacent tissues (p < 0.05; Figures 1D and 1E). These results demonstrate that WIPF1 may act as an oncogene in GC.
Figure 1.
Screening and identification of WIPF1
(A) WIPF1 were identified in primary gastric tumors and adjacent normal tissues through a proteomics study of 10 paired primary gastric tumors and adjacent normal tissues (volcano plot).
(B) heatmap.
(C) The results of GO analysis using DEPs in primary gastric tumor and adjacent normal tissues are shown. The size of the circles represents the protein numbers in each GO term and the color represents the –log10 p value.
(D) The protein level of WIPF1 was detected in GC cell lines (BGC823, MGC803, MKN45, and AGS) and GES-1 by Western blot assay.
(E) WIPF1 expression was detected in 10 pairs of GC and adjacent normal tissues by Western blot assay.
Independent validation of the prognostic value of Wiskott–Aldrich syndrome protein-interacting protein family member 1 in gastric cancer by tissue microarray-based immunohistochemistry
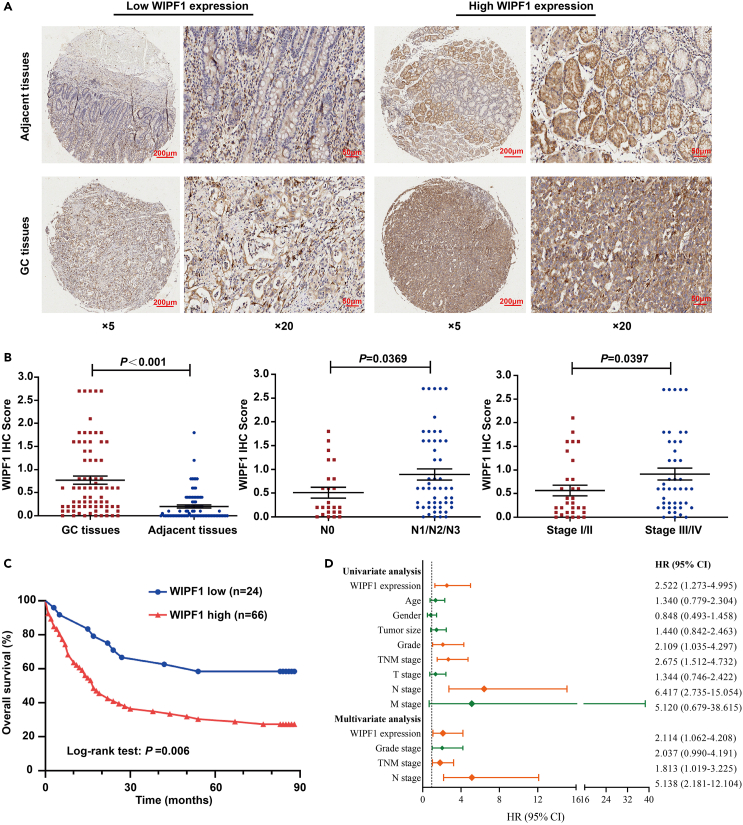
To explore the prognostic value of WIPF1, tissue microarray (TMA)-based immunohistochemistry (IHC) was performed on 90 paired GC and adjacent tissues. The IHC results showed that WIPF1 was mainly localized in the cytosol of GC cells (Figure 2A). IHC scores revealed that WIPF1 expression was higher in GC tissues than in adjacent tissues. It was also found that WIPF1 expression was lower in stage N0 than in stages N1/N2/N3, and was lower in stages I/II than in stages II/IV (Figure 2B). High WIPF1 expression was closely associated with lymph node status and TNM stage in patients with GC (Table 1). High expression of WIPF1 was correlated with poor overall survival in patients with GC (p = 0.006, Figure 2C). Univariate and multivariate analyses indicated that WIPF1 was a significant independent prognostic factor of GC (Figure 2D). Taken together, the results showed that WIPF1 is highly upregulated in GC, and its high expression is closely associated with a poor prognosis.
Figure 2.
High expression of WIPF1 was related to an unfavorable outcome in GC
(A) WIPF1 expression was detected by IHC assay in GC and paired adjacent tissues.
(B) The IHC score of WIPF was statistically constructed in GC tissues and adjacent tissues, the N0 and N1/N2/N3 stage, and Stage I/II and Stage III/IV of GC.
(C) Curves of overall survival in GC. The relationship between WIPF1 high-/low-expression and overall survival in GC patients (log rank test: p = 0.006).
(D) Cox regression model as well as univariate and multivariate analysis of independent risk factors in patients with GC.
Table 1.
Correlation between WIPF1 expression and clinicopathological characteristics
| Variables | WIPF1 expression |
Total | χ2 | p-value | |
|---|---|---|---|---|---|
| Low (n = 24) | High (n = 66) | ||||
| Age (year) | 0.417 | 0.518 | |||
| <60 | 9 (37.5) | 20 (30.3) | 29 | ||
| ≥60 | 15 (62.5) | 46 (69.7) | 61 | ||
| Sex | 1.023 | 0.312 | |||
| Female | 10 (41.7) | 20 (30.3) | 30 | ||
| male | 14 (58.3) | 46 (69.7) | 60 | ||
| Tumor size | 0.877 | 0.349 | |||
| <5cm | 9 (37.5) | 18 (27.3) | 27 | ||
| ≥5cm | 15 (62.5) | 48 (72.7) | 63 | ||
| Grade stage | 1.790 | 0.181 | |||
| I/II | 3 (12.5) | 17 (25.8) | 20 | ||
| III/IV | 21 (87.5) | 49 (74.2) | 70 | ||
| TNM stage | 4.574 | 0.032 | |||
| Ι/II | 11 (45.8) | 15 (22.7) | 26 | ||
| III/IV | 13 (54.2) | 51 (77.3) | 64 | ||
| T stage | 0.395 | 0.530 | |||
| T1/T2/T3 | 17 (70.8) | 51 (77.3) | 68 | ||
| T4 | 7 (29.2) | 15 (22.7) | 22 | ||
| N stage | 5.448 | 0.020 | |||
| N0 | 12 (50.0) | 16 (24.2) | 28 | ||
| N1/N2/N3 | 12 (50.0) | 50 (75.8) | 62 | ||
| M stage | 0.368 | 0.544 | |||
| M0 | 24 (100.0) | 65 (98.5) | 89 | ||
| M1 | 0 (0) | 1 (1.5) | 1 | ||
Wiskott–Aldrich syndrome protein-interacting protein family member 1 knockdown suppressed the growth and metastasis of gastric cancer cells in vitro
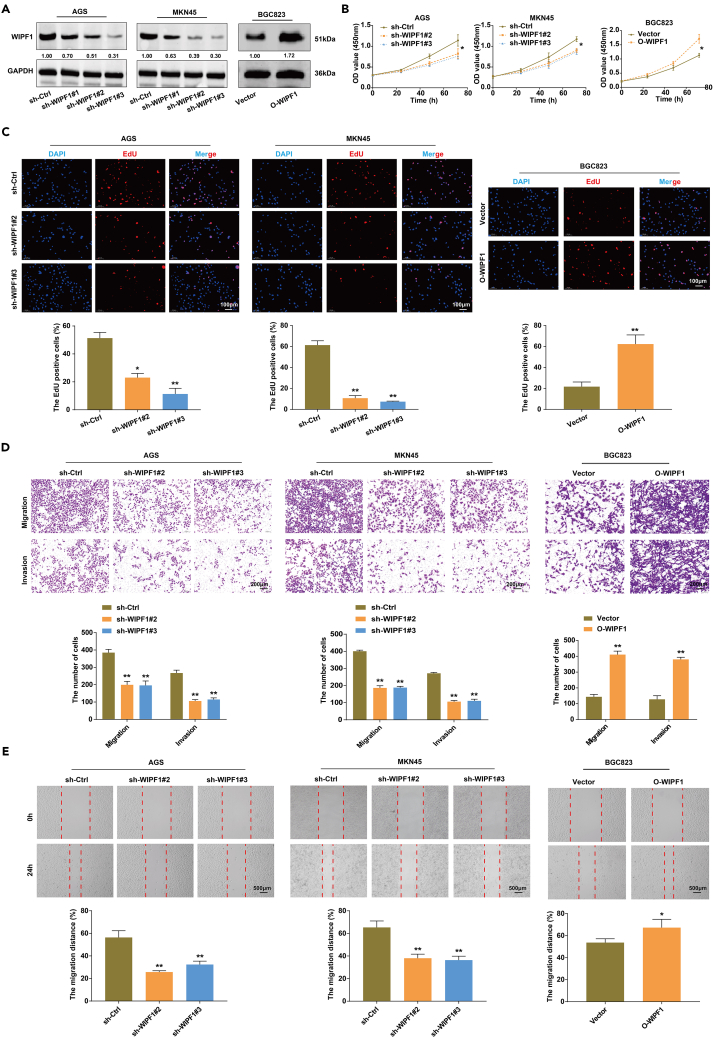
To identify the biological effects of WIPF1 on the characteristics of GC cells in vitro, the expression of WIPF1 was knocked down using RNAi in AGS and MKN45 cells and the expression of WIPF1 was upregulated by transfecting BGC823 cells with overexpressed WIPF1 (O-WIPF1) or the constructed vector. Transfection efficiency was confirmed by western blotting. The results indicated that sh-WIPF1#2 and sh-WIPF1#3 effectively inhibited the expression of WIPF1, while O-WIPF1 markedly upregulated the expression of WIPF1 (Figure 3A). Under these knockdown or overexpression conditions, the proliferation, invasiveness, and migration of AGS, MKN45, and BGC823 cells were evaluated using the CCK8 assay, 5-ethynyl-2′-deoxyuridine (EdU) staining, Transwell assay, and wound-healing assay, respectively. The CCK8 assay indicated that WIPF1 knockdown significantly attenuated the proliferative ability of AGS and MKN45 cells, whereas overexpression of WIPF1 promoted the proliferative ability of BGC823 cells (Figure 3B). EdU staining indicated that WIPF1 knockdown significantly reduced the proportion of Edu-positive AGS and MKN45 cells, whereas WIPF1 overexpression increased the proportion of Edu-positive BGC823 cells (Figure 3C). Furthermore, WIPF1 knockdown suppressed the invasiveness and migration of AGS and MKN45 cells compared to sh-Ctrl, whereas WIPF1 overexpression markedly promoted the invasion and migration of BGC823 cells (Figures 3D and 3E). In summary, these results indicated that WIPF contributes to GC progression in vitro.
Figure 3.
WIPF1 knockdown suppressed the growth and metastasis of GC cells in vitro
(A) Western blot analysis of the transfection efficiency of sh-WIPF1#1/#2/#3 in AGS and MKN45 cell lines and the transfection efficiency of overexpressed (O)-WIPF1 in BGC823.
(B) The proliferative ability of AGS and MKN45 cells infected with sh-Ctrl or sh-WIPF1 and BGC823 cells infected with empty vector O-WIPF1 were determined by CCK8 assay respectively.
(C) Detection of AGS, MKN45, and BGC823 cell proliferation in each group using EdU staining. The EdU-labeled cell is shown with red fluorescence and DAPI staining is shown with blue fluorescence in the cell nucleus.
(D) The cell invasion ability of AGS and MKN45 cells infected with sh-Ctrl or sh-WIPF1, and of BGC823 cells infected with the empty vector O-WIPF1, was evaluated by Transwell assay.
(E) A wound-healing assay measured the migration of AGS, MKN45, and BGC823 cells at 0 and 24 h. ∗p < 0.05, ∗∗p < 0.01.
Wiskott–Aldrich syndrome protein-interacting protein family member 1 knockdown suppressed the tumor growth of gastric cancer cells in vivo
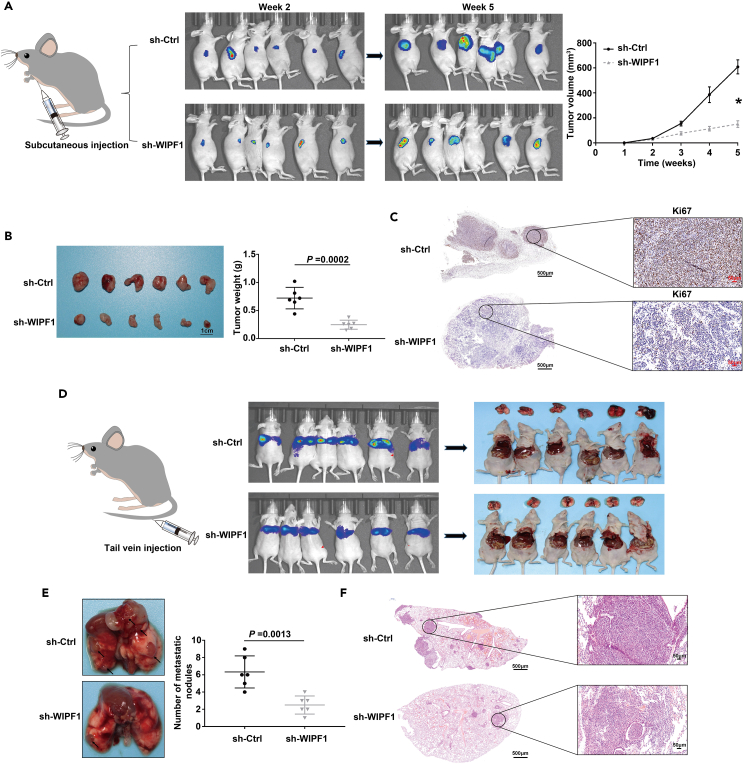
To further investigate the potential function of WIPF1 in vivo, MKN45 cells infected with sh-Ctrl or sh-WIPF1 lentivirus were subcutaneously injected into the armpits of nude mice to construct an orthotropic xenograft model of GC. Chemiluminescent imaging results showed that WIPF1 knockdown significantly inhibited tumor growth at weeks two and five, and that it significantly reduced tumor volume and weight compared to sh-Ctrl (Figures 4A and 4B). Similarly, IHC results further validated that WIPF1 knockdown reduced Ki67 protein expression in mouse GC tumor tissues (Figure 4C). Next, a nude mouse model of lung metastasis was established by tail vein injection of tumor cells into the sh-Ctrl and sh-WIPF1 groups (Figure 4D). Chemiluminescent imaging results showed that WIPF1 knockdown significantly inhibited tumor metastasis compared to the sh-Ctrl-treated group (Figures 4D and 4E). Hematoxylin and eosin (H&E) staining of tumor nodules confirmed tumor metastasis (Figure 4F). Based on these results, it was concluded that WIPF1 knockdown exerted repressive effects on the tumor growth and lung metastasis of GC in vivo.
Figure 4.
WIPF1 knockdown suppressed the tumor growth of GC cells in vivo
(A) The nude mouse model and subcutaneous tumors in the sh-Ctrl group (upper panel) and sh-WIPF1 group (lower panel) at 2 and 5 weeks. The tumor volume was statistically analyzed from weeks one to five.
(B) The tumor weight was statistically analyzed and plotted in columns.
(C) IHC analysis was performed to determine the protein expression of Ki67 in tissues of the sh-Ctrl and sh-WIPF1 groups.
(D) The nude mouse model of lung metastasis was established by the tail vein injection of tumor cells in the sh-WIPF1 group.
(E) The number of tumor nodules on the lung surface was statistically analyzed in the sh-Ctrl and sh-WIPF1 groups.
(F) H&E staining of tumor nodules. ∗p < 0.01.
Wiskott–Aldrich syndrome protein-interacting protein family member 1 promoted GC progression through the PI3K/AKT signaling pathway
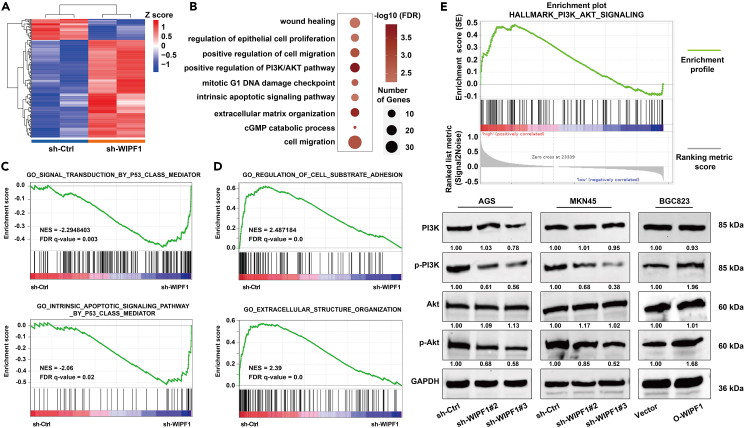
To elucidate the regulatory network mediated by WIPF1 in the malignant process of GC, GO analysis was performed and a heatmap was constructed to interpret the differentially expressed genes (DEGs) upon WIPF1 knockdown in MKN45 cells. Upregulated genes are shown in red and downregulated genes are shown in blue (Figure 5A). GO was used to analyze the biological processes highly associated with these DEGs, which were mainly involved in cell migration and extracellular matrix organization (Figure 5B). Moreover, GSEA results showed that signal transduction by the tumor suppressor p53 was most significantly enriched upon WIPF1 knockdown compared to the vector control (Figure 5C). Genes with low expression upon WIPF1 knockdown were mainly enriched in cell-substrate adhesion and extracellular structure organization (Figure 5D). These results further confirmed that WIPF1 is closely related to the malignancy of GC. Furthermore, WIPF1 was enriched in the HALLMARK PI3K/AKT signaling pathway, and Western blot analysis verified that WIPF1 activated the PI3K/AKT signaling pathway (Figure 5E). Based on these results, it was preliminarily elucidated that WIPF1 promotes GC by activating the PI3K/AKT signaling pathway.
Figure 5.
WIPF1 regulated genes related to cell proliferation and migration in GC
(A) The heatmap shows the DEGs in MKN45 cells with or without WIPF1 knockdown.
(B) GO analysis of DEGs upon WIPF1 knockdown. The details of this result were demonstrated by the enrichment analysis.
(C) GSEA in MKN45 cells with WIPF1 knockdown. Genes related to signal transduction by a p53 class mediator and an intrinsic apoptotic signaling-mediated pathway by p53 class mediator were enriched for genes highly expressed upon WIPF1 knockdown.
(D) Genes related to the regulation of cell substrate adhesion and extracellular structure organization showed low expression upon WIPF1 knockdown.
(E) The PI3K/AKT signaling pathway was enriched in the WIPF1 regulated function based on TCGA database analysis. Western blot analysis was used to verify that WIPF1 activated the PI3K/AKT signaling pathway.
Myocardin promotes the transcription of Wiskott–Aldrich syndrome protein-interacting protein family member 1 in gastric cancer
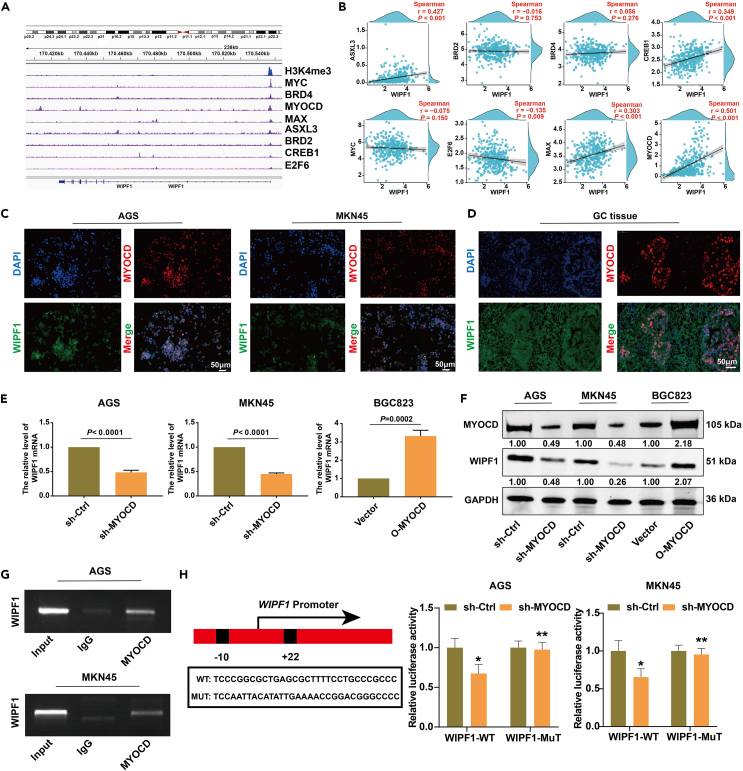
To explore the upstream regulators of WIPF1 in GC, the translation factor of the WIPF1 promoter was analyzed using ChIP-seq. H3K4me3, MYC, BRD4, MYOCD, MAX, ASXL3, BRD2, CREB1, and E2F6 were also predicted (Figure 6A). The correlation between WIPF1 and the expression of the above transcription factors in GC was further analyzed using The Cancer Genome Atlas (TCGA) data. Notably, the correlation between the expression of MYOCD and WIPF1 was highest in GC (Figure 6B). Immunofluorescence (IF) staining indicated that MYOCD was mainly localized in the nuclei of GC cells, and WIPF1 was predominantly localized in the cytoplasm of GC cells (Figures 6C and 6D). qPCR and Western blot assays indicated that MYOCD knockdown reduced WIPF1 expression in GC cells, and overexpression of MYOCD facilitated the expression of WIPF1 in GC cell lines (Figures 6E and 6F). To further validate the binding sites of MYOCD in the proximal promoter of WIPF1, ChIP-qPCR was performed using ChIP-grade MYOCD antibodies in AGS and BGC823 GC cell lines. The results revealed that compared to the isotope IgG control, MYOCD directly bound to the promoter of WIPF1 but did not bind to the negative region (Figure 6G). The results of the dual-luciferase reporter gene assay confirmed that the MYOCD protein directly bound to the WIPF1 promoter region (Figure 6H). Therefore, it was concluded that MYOCD directly bound to the proximal promoter of WIPF1 and activated WIPF1 expression in GC.
Figure 6.
WIPF1 expression was activated by MYOCD translation
(A) The translation factor of the WIPF1 promoter were analyzed by ChIP-seq analysis.
(B) The linear correlation between MYC, BRD4, MYOCD, MAX, ASXL3, BRD2, CREB1, and E2F6 and the mRNA expression of WIPF1 were plotted by Pearson regression analysis.
(C) IF double staining was used to detect the localization of MYOCD and WIPF1 in GC cells.
(D) IF double staining was used to detect the localization of MYOCD and WIPF1 in GC tissues.
(E) The mRNA expression of WIPF1 was detected in AGS and MKN45 cells treated with sh-MYOCD and in BGC823 cells treated with the overexpression of MYOCD.
(F) Western blot analysis of the protein level of MYOCD and WIPF1 in AGS and MKN5 cells treated with sh-MYOCD and in BGC823 cells treated with the overexpression of MYOCD.
(G) A ChIP-PCR assay was used to confirm that MYOCD protein directly bound to the WIPF1 promoter region.
(H) Dual luciferase reporter gene experiment.
Myocardin is necessary for Wiskott–Aldrich syndrome protein-interacting protein family member 1 to promote gastric cancer processes
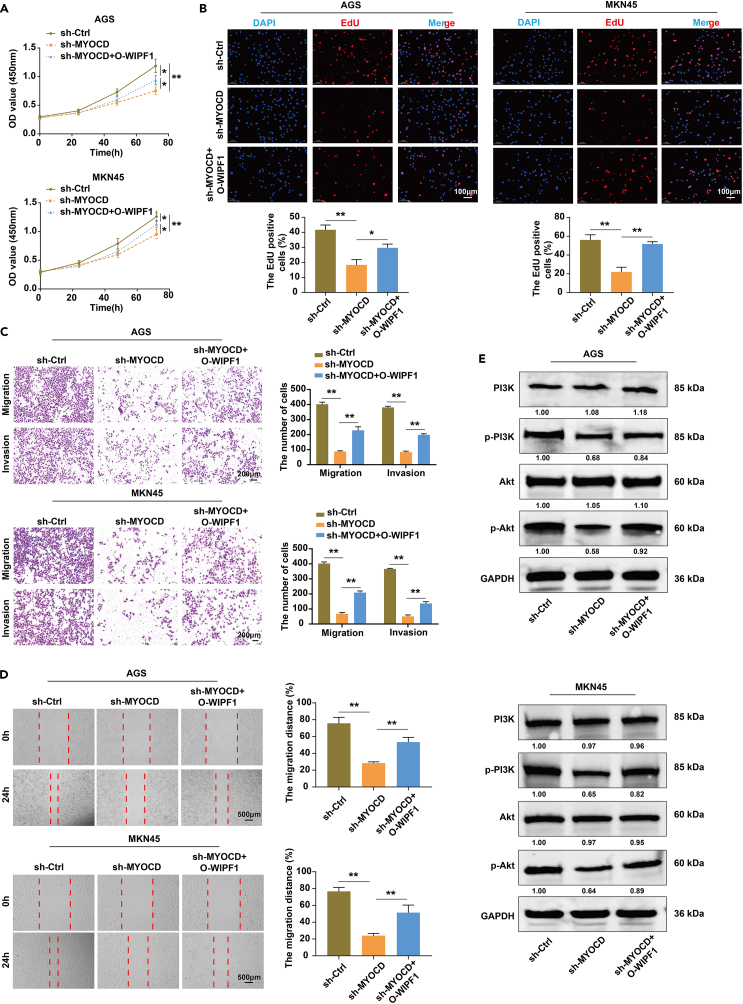
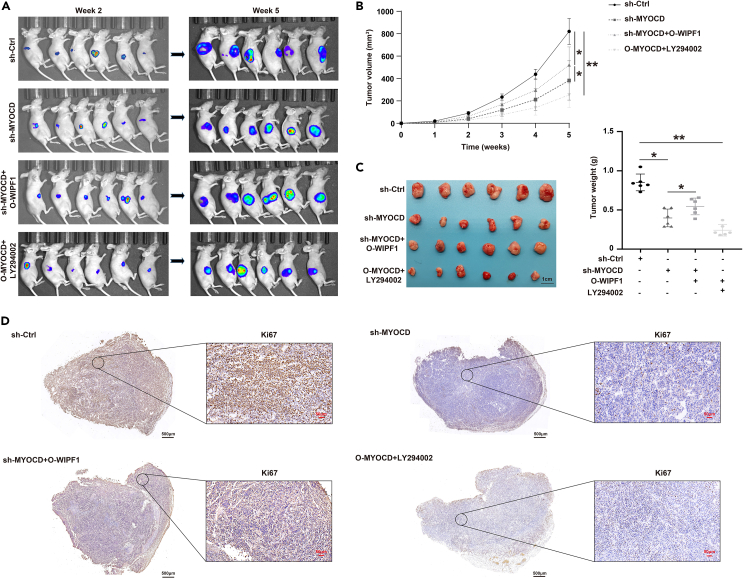
To identify the biological effects of MYOCD on the characteristics of GC cells in vitro, MYOCD expression was knocked down by transfecting AGS and MKN45 cells with sh-MYOCD, sh-MYOCD + O-WIPF1, or the control constructs. The proliferation, invasiveness, and migration of AGS and MKN45 cells were evaluated using the CCK8 assay, EdU staining, Transwell assay, and wound-healing assay, respectively. The CCK8 assay indicated that MYOCD knockdown significantly reduced the proliferative ability of AGS and MKN45 cells, and overexpression of WIPF1 reversed these effects (Figure 7A). EdU staining indicated that MYOCD knockdown significantly reduced the proportion of EdU-positive AGS and MKN45 cells, whereas WIPF1 overexpression reversed the proportion of sh-MYOCD-positive cells (Figure 7B). In addition, MYOCD knockdown suppressed the invasion and migration of AGS and MKN45 cells compared to sh-Ctrl, while WIPF1 overexpression markedly reversed the invasion and migration of sh-MYOCD cells (Figures 7C and 7D). Through in vivo rescue studies, it was also demonstrated that overexpression of WIPF1 could reverse the tumor growth inhibition caused by MYOCD knockdown (Figures 8A–8D). Western blot analysis indicated that MYOCD knockdown suppressed the protein levels of p-PI3K and p-AKT. WIPF1 overexpression markedly reversed the effect of sh-MYOCD on PI3K/AKT signaling pathway protein levels (Figure 7E). To further verify the role of MYOCD in the GC process through the activation of the PI3K/AKT signaling pathway, a subcutaneous tumor formation model of MYOCD overexpression was constructed in nude mice. The tumors of nude mice were then treated with LY294002. Consistent with the expected results, LY294002 partially suppressed excessive tumor growth following MYOCD overexpression (Figures 8A–8D). This indicated that MYOCD activates the PI3K/AKT signaling pathway in GC. Based on the results of the in vivo and in vitro studies, it is possible that WIPF1 promotes GC progression by regulating PI3K/Akt signaling in an MYOCD-dependent manner.
Figure 7.
WIPF1 was necessary for MYOCD to promote GC cell proliferation, invasion, and migration by activating the PI3K/AKT signaling pathway
(A) The proliferative ability of the AGS and MKN45 cells infected with sh-Ctrl, sh-MYOCD, or sh-MYOCD + O-WIPF1 was determined by CCK8 assay.
(B) Detection of AGS and MKN45 cell proliferation in each group using EdU staining. The EdU-labeled cell showed red fluorescence, and DAPI staining showed blue fluorescence in the cell nucleus.
(C) The cell invasion ability of AGS and MKN45 cells infected with sh-Ctrl, sh-MYOCD, or sh-MYOCD + O-WIPF1 was evaluated by Transwell assay.
(D) A wound healing assay measured the migration of AGS and MKN45 cells at 0 and 24 h.
(E) A Western blot assay was used to verify that MYOCD activated the PI3K/AKT signaling pathway in the form of dependence on WIPF1. ∗p < 0.05, ∗∗p < 0.01.
Figure 8.
WIPF1 was necessary for MYOCD to promote GC tumor growth by activating the PI3K/AKT signaling pathway
(A) The nude mouse model and subcutaneous tumors in the sh-Ctrl, sh-MYOCD, sh-MYOCD + O-WIPF1, and O-MYOCD + LY294002 groups at 2 and 5 weeks.
(B) The tumor volume was statistically analyzed from 1 to 5 weeks.
(C) The tumor weight was statistically analyzed and plotted in columns.
(D) IHC analysis was performed to determine the protein expression of Ki67 in tissues of the sh-Ctrl, sh-MYOCD, sh-MYOCD + O-WIPF1, and O-MYOCD + LY294002 groups. ∗p < 0.01, ∗∗p < 0.001.
Discussion
GC is one of the most common malignant cancers in humans and its incidence ranks second in China and fifth globally.16 With improvements in living standards and acceleration of the pace of life, the incidence and mortality of GC are increasing, especially in younger individuals.17 Therefore, there is an urgent need to identify biomarkers of GC in fundamental and clinical research. In the last few years, bioinformatics analysis, especially proteomics studies, has gained increasing attention in cancer research. Proteomics studies of GC provide insights into the biology of this cancer and suggest opportunities for personalized therapies that target it. In the present study, it was found that upregulated WIPF1 protein was a significant independent prognostic factor for GC and promoted GC growth and progression. Mechanistically, MYOCD transactivated WIPF1 and activated the PI3K/AKT signaling pathway in GC cells. These results establish MYOCD/WIPF1/PI3K/AKT axis involved in the growth and progression of GC.
WIPF1 is involved in actin cytoskeleton organization and polymerization and is related to cell proliferation and invasion.18,19 Although several dysregulated functions of WIPF1 have been identified in GC, further exploration is necessary to uncover the additional mechanisms.8 A previous TCGA analysis showed that WIPF1 acts as a prognostic biomarker in patients with colorectal cancer, breast cancer, and glioma.6 Furthermore, in this study, bioinformatics analysis, including proteomics analysis, revealed that WIPF1 was overexpressed in GC, resulting in deregulated DEPs that were highly associated with metabolic pathways. GO analysis showed that these DEPs almost converged during the oxidation-reduction process. Taken together, these results demonstrate that WIPF1 is an oncogenic protein in GC and participates in many metabolic pathways, especially the oxidation-reduction pathway. However, the clinicopathological function of WIPF1 in GC remains unclear. Therefore, it is important to identify the different features of WIPF1. In this study, TCGA cohort, Western blot assay, and IHC further confirmed the higher expression of WIPF1 in GC tissues than in the adjacent normal tissues. Interestingly, WIPF1 expression was higher in the advanced stages of GC. A previous study found that WIPF1 is significantly associated with overall survival in head and neck squamous cell cancer.20 Univariate and multivariate analyses indicated that WIPF1 is a significant independent prognostic factor in patients with GC. Therefore, the results of this study further demonstrated the oncogenic role of WIPF1 and provided clinicopathological evidence for GC.
Previously, attention has been paid to the pivotal role of WIPF1 in GC because its downregulation and overexpression could lead to harmful effects in vivo and in vitro. The in vitro results showed that WIPF1 knockdown significantly inhibited the proliferation, invasion, and migration of GC cells. However, the determination of the correlation between WIPF1 and GC requires further in vivo investigation. In a xenograft nude mouse model, WIPF1 knockdown resulted in a similar phenomenon in GC, and the tumor volume and weight were significantly decreased. Additionally, WIPF1 knockdown repressed the expression of Ki67 to a very low level and significantly reduced the number of metastatic tumors. It was concluded that WIPF1 knockdown exerted repressive effects on the tumor growth and lung metastasis of GC in vivo. Although these results emphasize the physiological importance of WIPF1 knockdown, the underlying mechanisms remain elusive. In this study, RNA-seq analysis of MKN45 cells indicated that WIPF1 knockdown led to the identification of 215 DEPs related to cell proliferation and migration, among which p53 was the most highly expressed gene according to GSEA. Therefore, it is proposed that WIPF1 participates in the progression and development of GC via the p53 signaling pathway. Further studies are warranted owing to the lack of solid in vivo and in vitro evidence.
The PI3K/AKT signaling pathway promotes the proliferation, invasion, migration, and EMT of cancer cells, as confirmed by previous studies. In this study, the molecular mechanisms by which WIPF1 plays an oncogenic role in GC were investigated. PI3K/AKT was identified as the major downstream signaling pathway underlying the oncogenic role of WIPF1 in GC. Knockdown of WIPF1 inhibited the phosphorylation of PI3K and AKT proteins and downregulated the PI3K/AKT signaling pathway. WIPF1, thus, promotes GC progression by activating the PI3K/AKT signaling pathway.
MYOCD is a key regulator of a variety of cellular functions and acts as a potent transcriptional factor that regulates gene expression in the myocardium and smooth muscle.21,22 It was found that MYOCD directly bound to the proximal promoter of WIPF1 and activated the expression of WIPF1 in GC. Furthermore, WIPF1 is necessary for MYOCD to promote the malignant phenotype of GC cells by activating the PI3K/AKT signaling pathway.
Limitations of the study
This study had some limitations, and further evidence is needed to confirm the conclusions of the bioinformatics analysis. Here, the oncogenic role of WIPF1 in the initiation, progression, and prognosis of GC was identified. Abundant WIPF1 expression was associated with a poor prognosis and poor survival in patients with GC. It was found that WIPF1 knockdown significantly inhibited the proliferation, invasion, and migration of GC cells. This mechanism was further validated by various bioassays in a xenograft nude mouse model, and WIPF1 knockdown significantly predicted tumor weight and lung metastasis; however, only a slight weight gain was observed according to the findings of this study. WIPF1 promoted GC progression by activating the PI3K/AKT signaling pathway. In the upstream regulatory network, WIPF1 was found to be transactivated by MYOCD in GC cells. The findings of this study suggest that MYOCD-transactivated WIPF1 promotes GC growth and metastasis via the activation of the PI3K/AKT signaling pathway, highlighting WIPF1 as a potential prognostic biomarker in the treatment of GC.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-WIPF1 | Santa Cruz | Cat#sc-390099;RRID:AB_1152991 |
| Mouse monoclonal anti- MYOCD | Santa Cruz | Cat#sc-518132;RRID:AB_11424149 |
| Rabbit monoclonal anti-total Akt | Proteintech | Cat#60203-2-Ig; RRID:AB_91578 |
| Rabbit monoclonal anti-p-Akt | Proteintech | Cat#80455-1-RR;RRID:AB_2315049 |
| Rabbit monoclonal anti-PI3K | Proteintech | Cat#67071-1-Ig;RRID:AB_10008352 |
| Mouse monoclonal anti- p-PI3K | Thermo Scientific | Cat#PA5-104853;RRID:AB_10789447 |
| Rabbit monoclonal anti-Ki67 | Abcam | Cat#ab279653;RRID: AB_2756525 |
| Chemicals, peptides, and recombinant proteins | ||
| BCA | Thermo Scientific | Cat#23227 |
| Lipofectamine 3000 Reagent | Thermo Scientific | Cat#L3000015 |
| Plasmid | RiboBio | Cat#PPRO1001 |
| CCK8 kit | Dojindo | Cat#CK04 |
| EdU reagent | Abbkine | Cat#KTA2031 |
| 4% polymerized formaldehyde solution | Sigma-Aldrich | Cat#50-00-0 |
| DAPI | Sigma-Aldrich | Cat#10236276001 |
| TRIzol reagent | Thermo Scientific | Cat#15596018 |
| Deposited data | ||
| GSM1250896 | GEO - Gastric Cancer | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1250896 |
| GSM2359448 | GEO - Gastric Cancer | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2359448 |
| GSM2825420 | GEO - Gastric Cancer | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2825420 |
| Experimental models: Cell lines | ||
| Gastric cancer cell line AGS | Shanghai Fuheng Biotechnology Co., Ltd | FH0261 |
| MKN45 | Shanghai Fuheng Biotechnology Co., Ltd | FH0260 |
| MGC803 | Shanghai Fuheng Biotechnology Co., Ltd | FH0275 |
| BGC823 | Shanghai Fuheng Biotechnology Co., Ltd | FH0263 |
| Experimental models: Organisms | ||
| Nude mice | Ziyuan Laboratory Animal Science and Technology | N/A |
| Software and algorithms | ||
| SPSS software | SPSS Inc. | Version 18.0 |
| Prism | GraphPad Software | Version 8.0.0 |
| Image J | NIH | RRID:SCR_003070 |
| Biological samples | ||
| Gastric cancer tissue and normal paracancerous tissue | The First Hospital of Lanzhou Universit | N/A |
Resource availability
Lead contact
The relevant experimental reagents, experimental methods and related data of this study can be obtained by contacting Quan-lin Guan.(e-mail:guanql@lzu.edu.cn).
Materials availability
The study did not generate new unique reagents.
Experimental model and study participant details
Clinical samples
The Clinical samples used in the experiment were derived from the First Hospital of Lanzhou university. Experiments with TMA were derived from Shanghai Outdo Biotech Co., In this study, we included a total of 100 patients with gastric cancer, aged 40–85 years, with no restriction on gender, ancestry, race or ethnicity or socioeconomic information, with matched gastric and paracancerous tissue samples, none of whom had received radiotherapy or chemotherapy, and no other cancer co-morbidities. The study was approved by the ethics committee of the First Hospital of Lanzhou Universit (LDYYLL2019-84)and the ethics committee of Shanghai Outdo Biotech Co., Ltd (No. SHYJS-CP-1710001). All the patients provided written informed consent.The study was conducted according to the principles expressed in the Declaration of Helsinki.
Animals
Six-week-old male nude mice weighing 16–18 g were purchased from the Shanghai Laboratory Animals Center (SLAC; Shanghai, China) and cultured in a warm and moist-specific pathogen-free room with a 12 h light/dark cycle as well as ample food and water. This study was approved by the Ethics Committee of the First Hospital of Lanzhou University(LDYYLL2019-84).
Method details
Proteomics study and DIA mass spectrometry
Tissue samples were rinsed with Tris-sucrose, triturated, and lysed in tissue lysis buffer as described previously.23 The tissues were sonicated using a Biorupter Pico sonication device (Diagenode, Liege, Belgium). Total protein was collected via ultracentrifugation at 12,000 × g for 10 min at 4°C, and the protein concentration was measured using a BCA assay (Thermo Scientific, California, USA).
Bioinformatics analysis
Gene expression was determined using HTSeq count, and DEGs were identified using DESeq2. GO analysis was performed using the Database for Annotation, Visualization and Integrated Discovery, and GSEA was conducted using GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp).24,25 Enrichment plots were generated and WIPF1 expression in normal and GC samples was compared. Samples were divided into high- and low-expression groups (median) and survival was analyzed using the log rank test. GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the topGO (version 2.42.0) and KEGGREST (version 1.30.1) packages in R.
Cell culture
Human GC cell lines (AGS, MGC803, MKN45, and BGC823) and the gastric epithelial cell line GES-1 were maintained in Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Corning, New York, NY, USA) and 1% penicillin and streptomycin (Thermo Scientific, California, USA). The cells were incubated in a humidified incubator with 5% CO2 at 37°C. Mycoplasma contamination was removed from all cell lines.
Cell transfection
Cells at 75–80% confluency were transfected with sh-Ctrl, sh-WIPF1#1, sh-WIPF1#2, and sh-WIPF1#3; control vector and WIPF1 expression plasmid; sh-Ctrl and sh-MYOCD; and control vector and MYOCD expression plasmid (RiboBio, Guangzhou, China) using Lipofectamine 3000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, California, USA). Subsequently, the lentivirus was packaged and infected, as previously reported.26 The shRNA-targeting sequence was as follows: sh-WIPF1#1, CCATGTGAAGATGAGTGGGAA; sh-WIPF1#2: CCAATACTGGACAAACCTAAA; and sh-WIPF1#3: CATTCAATCAAGTCCGCACAA.
CCK8 assay
The CCK8 kit (Dojindo, Kumamoto, Japan) was used to measure the proliferation of the AGS, MKN45, and BGC823 cell lines. The cells (2×105) were inoculated into a 6-well plate and transfected. After 48 h, CCK8 solution prepared in advance was gently added to the culture medium of each well. The absorbance was measured at 450 nm using a microplate reader. The proliferative abilities of AGS, MKN45, and BGC823 cell lines were statistically plotted. Each assay was performed in triplicate.27
EdU assay
The GC cells were transfected according to the experimental group after 48 h; The GC cells were cultured in 96-well cell culture plates for 6 h. Next, 10 μmol/L EdU reagent (Abbkine) was added to each well. The cells were incubated at 37°C and 5% CO2 for 6 h. The cells were fixed with a 4% polymerized formaldehyde solution (Sigma-Aldrich, St. Louis, Missouri, USA) for 45 min. The cells were blocked in PBS containing 3% bovine serum albumin (BSA), permeabilized in 0.3% Triton X-100 solution for 15 min, and washed with PBS (5 min/time). Nuclei were stained with 5 mg/L DAPI (Sigma-Aldrich) for 20 min. Finally, staining was observed using fluorescence microscopy. EdU-labeled cells showed red fluorescence and DAPI-stained nuclei showed blue fluorescence. Imaging software was used to analyze cell proliferation.27
Transwell assay
After 48 h of transfection, AGS, MKN45, and BGC823 cells were harvested using trypsin, resuspended in serum-free medium, and seeded onto the upper chamber of an 8-μm pore Transwell plate (Corning, USA) coated with Matrigel. DMEM supplemented with 10% fetal bovine serum was added to the lower chamber. After 24 h, adherent cells in the upper chamber were completely removed using a cell scraper. The invading cells on the opposite side were fixed with 4% formaldehyde for 10 min. After crystal violet staining, images of these cells were recorded using light microscopy (Zeiss Germany, Oberkochen, Germany).27
Wound healing assay
The GC cell lines MKN45, AGS, and BGC823 were seeded in a 6-well plate (Corning, USA) at a concentration of 5×105/mL. Twelve hours later, the wound on the adherent cells was created by scratching them with a 200-μL pipette tip. The culture medium was then replaced immediately, and the cells were maintained for another 24 h. The width of the wound scratch was determined at 0 h and 24 h. The migratory ability of these cells was analyzed routinely.27
Xenograft assay
Six-week-old male nude mice weighing 16–18 g were purchased from the Ziyuan Laboratory Animal Science and Technology (Hangzhou, China) and cultured in a warm and moist-specific pathogen-free room with a 12 h light/dark cycle as well as ample food and water. This study was approved by the Ethics Committee of the First Hospital of Lanzhou University, and all standards were based on the manufacturer’s instructions. MKN45 cells were stably transfected with sh-control, sh-WIPF1, sh-MYOCD, O-MYOCD, or sh-MYOCD + O-WIPF1. In addition, some nude mice were treated with LY294002 (O-MYOCD + LY294002) on days 7, 14, and 21 after MKN45 cells were inoculated, and the dosage of LY294002 was 15 mg/kg.28 LY294002 is a commonly used PI3K inhibitor. LY294002 can permeate cells, inhibit PI3K specifically, and inhibit the PI3K/AKT signaling pathway.29 The xenograft assay was performed as described previously.27
H&E staining and IHC
H&E staining was performed on 5 μm paraffin sections using a standard H&E staining protocol. Tissue samples were fixed, embedded,cut into slices, dewaxed, antigen retrieved, incubated with primary antibody, incubated with secondary antibody, DAB stained, hematoxylin stained, and alcohol denatured.Two experienced pathologists independently reviewed all sections. The IHC score was determined using modified Histo-score (H-score). The staining intensity and percentage of positive cells were scored as described previously. The final immunoreactive score was calculated as the staining intensity score×the percentage of positive cells.The expression level of WIPF1 was considered “low expression” if the immunoreactive score was 0–4 and “high expression” if the score was 5-9.27 The primary antibodies used were anti-WIPF1 (1:200; Santa Cruz Biotechnology) and anti-Ki67 (1:100; Abcam).
RNA sequencing and analysis
Total RNA was extracted from MKN45 cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, California, USA) according to standard procedures. The mRNA-seq library was constructed using the VAHTS mRNA-seq V2 Library Prep Kit from Illumina (Nanjing, China). Briefly, 4 μg of total RNA was combined with mRNA capture beads and bound in a PCR cycler. After capturing the mRNA-bead complex, the mRNA was eluted and incubated at room temperature. Frag/prime Buffer was added to resuspend the complex, and the mRNA was fragmented into 200–300 bp fragments. Next, cDNA strands were synthesized, and a library was constructed and amplified. Sequencing was conducted on the Illumina NovaSeq platform, raw data were deposited, and GO analysis was used to examine the biological processes, cellular components, and molecular functions of the DEGs.
Bioinformatics analyses
The ChIP-seq datasets of MYC, BRD4, MYOCD, and MAX for the WIPF1 regulatory regions in primary GC were obtained from the GEO database (GSM1250896, GSM2359448, and GSM2825420).30,31 Raw data were analyzed using the Fastq-dump algorithm. Bowtie2 software mapped fast files to the hg38 reference genome, and the homer suite generated the visualized files. In addition, RNA-seq data for the CG were obtained from TCGA database to analyze the correlation between the expression of various genes.
qPCR
The expression of WIPF1 in AGS cells, MKN45 cells treated with sh-WIPF1 lentivirus or control, and BGC823 cells treated with O-WIPF1 or vector was evaluated by qPCR. Briefly, total RNA was extracted using TRIzol reagent. qPCR was performed using the first-strand cDNA synthesized using the HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) and AceQ qPCR SYBR Green Master Mix. Each data point was analyzed at least in triplicate, and GAPDH was used for internal normalization.Finally, the average of each data point was analyzed using the 2−ΔΔCt method.27 The forward and reverse primers used were as follows: WIPF1-forward, CGGAGGCGGTGGAAGTTTT and WIPF1-reverse, CCGTGGATCTCAGCTTCGG.
Western blot
Radio immunoprecipitation assay (RIPA, Beyotime, Shanghai, China) supplemented with protease inhibitors (Beyotime) was used to extract total protein from GC cells and tissue samples. Total protein was run on 8-12% SDS/PAGE gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Boston, MA, USA). Subsequently, membranes were blocked with 5% skim milk and incubated overnight at 4°C with the primary antibodies. The next day, PVDF membranes were incubated for 2 h at room temperature with appropriate secondary antibodies, and ECL substrate was applied. Images were visualized using a Tianneng automatic chemiluminescence image processing system (TANO, Shanghai, China).27 The primary antibodies used are summarized in Table S1.
IF staining
Cells or tissues on slides were fixed with paraformaldehyde (4%) and permeabilized with 0.01% Triton X-100 for 15 min. The slides were incubated with primary antibodies anti-PLAUR (1:200, Santa Cruz, sc-13522) or anti-TCF7L2 (1:100, Bioss, bsm-52543R), overnight in a humidified chamber at 37°C in the dark. Subsequently, cell nuclei were stained with DAPI. The fluorescence distribution and intensity of cells and tissues were analyzed using a Zeiss LSM 880 laser microscope (Carl Zeiss AG, Oberkochen, Germany).27 The primary antibodies used were anti-WIPF1 (1:200; Santa Cruz Biotechnology USA) and anti-MYOCD (1:200; Santa Cruz Biotechnology).
ChIP-qPCR
In the logarithmic growth phase, 1 × 107 cells were fixed with 1% formaldehyde and decrosslinked with 2.5 M glycine. After washing and removing the supernatant, cells were lysed and transferred to a centrifuge tube. The nuclear pellet was collected and sonicated, and the supernatant was divided into two samples. MYOCD-specific and control IgG antibodies were added, followed by overnight incubation at 4°C, magnetic bead incubation, and washing steps. Samples were eluted and purified, and 50 μL of purified product was collected for qPCR detection.
Dual luciferase reporter assay
WIPF-MUT and WIPF1-WT luciferase plasmids were co-transfected into cells (AGS and MKN45 cell lines) with sh-MYOCD or sh-ctrl using Lipofectamine 3000. A dual-luciferase reporter assay was performed according to the manufacturer's instructions. After 36 h, the cells were washed and lysed, and the lysates were added to a 96-well plate. Transcriptional activity was measured using a multifunctional reader, and the ratio of luciferin to Revilla fluorescence was used for intergroup comparisons.
Quantification and statistical analysis
All statistical data were analyzed using the SPSS software (version 18.0; SPSS Inc., Chicago, Illinois, USA). Continuous variables are expressed as mean ± standard deviation. All data were normally distributed and the variances were equal. Each in vivo and in vitro assay was performed at least three times. Student’s t-test was used to compare two groups. A p value of less than 0.05 was considered significant.
Acknowledgments
We are grateful to the laboratory staff in the Key Laboratory of Tumor Biological Behavior of Hubei Province and The First Hospital of Lanzhou University for their help. We thank Shanghai Cutseq Bio-medical Technology Co. Ltd. for the DIA-MS guidance. We acknowledge TCGA project, and GEO project for providing their platforms and contributors for uploading their meaningful datasets. This work was supported by funding from: (1) Key R&D Plan of Gansu Provincial Department of Science and Technology (20YF3FA029); (2) ConstructionProject of Clinical Medical Research Center from Gansu Provincial Department of Science and Technology (21JR7RA390); (3) Natural Science Foundation of Gansu Province (21JR7RA386, 22JR5RA909, 22JR5RA918, 22JR5RA899, 21JR7RA382, and 21JR11RA077); (4) Gansu Province Higher Education Innovation Ability Improvement Project (2019B-009,2020B-009, and 2021B-001); (5) Scientific and Technological Development Guiding Plan Project of Lanzhou City (2020-ZD-74); (5) Foundation of the First Hospital of Lanzhou University (ldyyyn2021-1). The authors are responsible for the results and opinions provided by this research, and the sponsor is not responsible for the content published. The authors are responsible for the results and opinions provided by this research, and the sponsor is not responsible for the content published.
Author contributions
Q.L.G., M.H. F., F.S., and R.W. X. designed the study. R.C., T.N.Y., and F.S. collated the data, carried out data analyses, and produced the initial draft of the article. X.N.X., F.S., R.C., X.M. H., and D.W.W. conducted experiments. All authors have read and approved the final submitted article.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: October 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108273.
Contributor Information
Quanlin Guan, Email: guanql@lzu.edu.cn.
Maohui Feng, Email: fengmh@whu.edu.cn.
Supplemental information
Data and code availability
-
•
The ChIP-seq data can be found at the following GEO,The accession number is listed in the key resources table. The RNA-seq data can be found at the following SRA database (PRJNA1021699,https://www.ncbi.nlm.nih.gov/sra/PRJNA1021699), and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset(PXD045944,https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD045944).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Park J.Y., Camargo M.C., Lunet N., Forman D., Soerjomataram I. Is gastric cancer becoming a rare disease? A global assessment of predicted incidence trends to 2035. Gut. 2020;69:823–829. doi: 10.1136/gutjnl-2019-320234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Nikšić M., Bonaventure A., Valkov M., Johnson C.J., Estève J., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inokuchi M., Nakagawa M., Tanioka T., Okuno K., Gokita K., Kojima K. Long- and short-term outcomes of laparoscopic gastrectomy versus open gastrectomy in patients with clinically and pathological locally advanced gastric cancer: a propensity-score matching analysis. Surg. Endosc. 2018;32:735–742. doi: 10.1007/s00464-017-5730-7. [DOI] [PubMed] [Google Scholar]
- 5.Pan Y., Lu F., Xiong P., Pan M., Zhang Z., Lin X., Pan M., Huang H. WIPF1 antagonizes the tumor suppressive effect of miR-141/200c and is associated with poor survival in patients with PDAC. J. Exp. Clin. Cancer Res. 2018;37:167. doi: 10.1186/s13046-018-0848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staub E., Groene J., Heinze M., Mennerich D., Roepcke S., Klaman I., Hinzmann B., Castanos-Velez E., Pilarsky C., Mann B., et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J. Mol. Med. 2009;87:633–644. doi: 10.1007/s00109-009-0467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang T., Shen X., Liu R., Zhu G., Bishop J., Xing M. Epigenetically upregulated WIPF1 plays a major role in BRAF V600E-promoted papillary thyroid cancer aggressiveness. Oncotarget. 2017;8:900–914. doi: 10.18632/oncotarget.13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Lin W., Dong Y., Li X., Lin Z., Jia J., Zou W., Pan Y. Long noncoding RNA HCG18 up-regulates the expression of WIPF1 and YAP/TAZ by inhibiting miR-141-3p in gastric cancer. Cancer Med. 2020;9:6752–6765. doi: 10.1002/cam4.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang W., Sun H., Zhang S., Zhou Y., Weng W., Wu B., Ye T., Huang W., Lin Z., Shi L., Shi K. An immunogenomic signature for molecular classification in hepatocellular carcinoma. Mol. Ther. Nucleic Acids. 2021;25:105–115. doi: 10.1016/j.omtn.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J., Zhang C., Jiang G.Q., Jin S.J., Wang Q., Wang A.Q., Bai D.S. Identification of hepatocellular carcinoma-related genes associated with macrophage differentiation based on bioinformatics analyses. Bioengineered. 2021;12:296–309. doi: 10.1080/21655979.2020.1868119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue C., Li G., Lu J., Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct. Target. Ther. 2021;6:400. doi: 10.1038/s41392-021-00788-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fattahi S., Amjadi-Moheb F., Tabaripour R., Ashrafi G.H., Akhavan-Niaki H. PI3K/AKT/mTOR signaling in gastric cancer: Epigenetics and beyond. Life Sci. 2020;262 doi: 10.1016/j.lfs.2020.118513. [DOI] [PubMed] [Google Scholar]
- 13.Ling M., Quan L., Lai X., Lang L., Li F., Yang X., Fu Y., Feng S., Yi X., Zhu C., et al. VEGFB Promotes Myoblasts Proliferation and Differentiation through VEGFR1-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222413352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christoforou N., Chellappan M., Adler A.F., Kirkton R.D., Wu T., Addis R.C., Bursac N., Leong K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong X., Wang S., Lei Z., Li C., Zhang C., Su Z., Liu X., Zhao J., Zhang H.T. MYOCD and SMAD3/SMAD4 form a positive feedback loop and drive TGF-beta-induced epithelial-mesenchymal transition in non-small cell lung cancer. Oncogene. 2020;39:2890–2904. doi: 10.1038/s41388-020-1189-4. [DOI] [PubMed] [Google Scholar]
- 16.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 17.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA. Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 18.Schafer D.A. Coupling actin dynamics and membrane dynamics during endocytosis. Curr. Opin. Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- 19.Donnelly S.K., Weisswange I., Zettl M., Way M. WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr. Biol. 2013;23:999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin W., Zhao C., Jiang L., Pei D., Zhao L., Zhang C. Identification of a Novel Epithelial-Mesenchymal Transition Gene Signature Predicting Survival in Patients With HNSCC. Pathol. Oncol. Res. 2021;27 doi: 10.3389/pore.2021.585192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J.P., He X., Liu F., Yin S., Wu S., Yang M., Zhao J., Dai X., Jiang H., Yu L., et al. YY1 directly interacts with myocardin to repress the triad myocardin/SRF/CArG box-mediated smooth muscle gene transcription during smooth muscle phenotypic modulation. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahvenainen T., Khamaiseh S., Alkodsi A., Mehine M., Nevala R., Äyräväinen A., Bützow R., Vahteristo P. Lung metastases and subsequent malignant transformation of a fumarate hydratase -deficient uterine leiomyoma. Exp. Mol. Pathol. 2022;126 doi: 10.1016/j.yexmp.2022.104760. [DOI] [PubMed] [Google Scholar]
- 23.Su F., Zhou F.F., Zhang T., Wang D.W., Zhao D., Hou X.M., Feng M.H. Quantitative proteomics identified 3 oxidative phosphorylation genes with clinical prognostic significance in gastric cancer. J. Cell Mol. Med. 2020;24:10842–10854. doi: 10.1111/jcmm.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Tan Y., Wang X., Song H., Zhang Y., Zhang R., Li S., Jin W., Chen S., Fang H., Chen Z., Wang K. A PML/RARalpha direct target atlas redefines transcriptional deregulation in acute promyelocytic leukemia. Blood. 2021;137:1503–1516. doi: 10.1182/blood.2020005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang T., Wang B., Su F., Gu B., Xiang L., Gao L., Zheng P., Li X.M., Chen H. TCF7L2 promotes anoikis resistance and metastasis of gastric cancer by transcriptionally activating PLAUR. Int. J. Biol. Sci. 2022;18:4560–4577. doi: 10.7150/ijbs.69933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai J., Qian K., Zuo X., Yue W., Bian Y., Yang J., Wei J., Zhao W., Qian H., Liu B. PLGA nanoparticle-based docetaxel/LY294002 drug delivery system enhances antitumor activities against gastric cancer. J. Biomater. Appl. 2019;33:1394–1406. doi: 10.1177/0885328219837683. [DOI] [PubMed] [Google Scholar]
- 29.Fergusson A.D., Zhang R., Riffle J.S., Davis R.M. Encapsulation of PI3K Inhibitor LY294002 within Polymer Nanoparticles Using Ion Pairing Flash Nanoprecipitation. Pharmaceutics. 2023;15 doi: 10.3390/pharmaceutics15041157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chia N.Y., Deng N., Das K., Huang D., Hu L., Zhu Y., Lim K.H., Lee M.H., Wu J., Sam X.X., et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 31.Rusan M., Li K., Li Y., Christensen C.L., Abraham B.J., Kwiatkowski N., Buczkowski K.A., Bockorny B., Chen T., Li S., et al. Suppression of Adaptive Responses to Targeted Cancer Therapy by Transcriptional Repression. Cancer Discov. 2018;8:59–73. doi: 10.1158/2159-8290.CD-17-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The ChIP-seq data can be found at the following GEO,The accession number is listed in the key resources table. The RNA-seq data can be found at the following SRA database (PRJNA1021699,https://www.ncbi.nlm.nih.gov/sra/PRJNA1021699), and the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset(PXD045944,https://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD045944).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.