Abstract
Objective
Use heuristic, deep learning (DL), and hybrid AI methods to predict semantic group (SG) assignments for new UMLS Metathesaurus atoms, with target accuracy ≥95%.
Materials and Methods
We used train-test datasets from successive 2020AA–2022AB UMLS Metathesaurus releases. Our heuristic “waterfall” approach employed a sequence of 7 different SG prediction methods. Atoms not qualifying for a method were passed on to the next method. The DL approach generated BioWordVec and SapBERT embeddings for atom names, BioWordVec embeddings for source vocabulary names, and BioWordVec embeddings for atom names of the second-to-top nodes of an atom’s source hierarchy. We fed a concatenation of the 4 embeddings into a fully connected multilayer neural network with an output layer of 15 nodes (one for each SG). For both approaches, we developed methods to estimate the probability that their predicted SG for an atom would be correct. Based on these estimations, we developed 2 hybrid SG prediction methods combining the strengths of heuristic and DL methods.
Results
The heuristic waterfall approach accurately predicted 94.3% of SGs for 1 563 692 new unseen atoms. The DL accuracy on the same dataset was also 94.3%. The hybrid approaches achieved an average accuracy of 96.5%.
Conclusion
Our study demonstrated that AI methods can predict SG assignments for new UMLS atoms with sufficient accuracy to be potentially useful as an intermediate step in the time-consuming task of assigning new atoms to UMLS concepts. We showed that for SG prediction, combining heuristic methods and DL methods can produce better results than either alone.
Keywords: unified medical language system, semantic network, artificial intelligence, heuristic reasoning, deep learning
BACKGROUND AND SIGNIFICANCE
Professional societies, government institutions, and other organizations have separately maintained terms within 222 sources and contributed them to the UMLS Metathesaurus.1 The Metathesaurus is the largest component of the UMLS. The UMLS facilitates interpretation of biomedical meanings across disparate electronic information and data sources in systems serving scientists, health professionals, and the public.2 Every 6 months, UMLS editors at NLM complete the arduous task of incorporating several hundred thousand new atoms (source-specific text strings) into the Metathesaurus. A critical part of that activity involves consolidating like-meaning atoms into UMLS concepts, each assigned a permanent concept unique identifier (CUI). The 2022AB UMLS Metathesaurus embodies 16 857 345 atoms—each assigned an atom unique identifier (AUI), and grouped into 4 553 796 unique concepts.
The biannual updating task requires substantial human time and expertise. Deciding which, if any, of 4 million concepts a new atom matches is a daunting task that carries a risk of misclassification. As the number of unique UMLS concepts grows, both the level of effort required to classify new atoms and the possibility of misclassifications will presumably increase. Automated assistance with assigning new atoms to UMLS concepts could alleviate some of the burden on UMLS editors and reduce the risk of errors.
Previous NLM work showed that deep learning (DL) methods could predict synonymy between pairs of atoms.3 Our group also found that the addition of semantic information about each atom could improve DL prediction accuracy.4 Our previous work used SGs as the indicator of the semantics of an atom. However, the SG is only known after an atom is incorporated into the UMLS (ie, when UMLS editors assign semantic types to UMLS concepts). Each new UMLS concept is assigned to at least 1 of 127 semantic types that categorize the functional meaning/usage of a concept, for example, “disease or syndrome,” “gene or genome,” “laboratory procedure.”5 Once a semantic type is assigned, it can be rolled up to 1 of 15 sematic groups (SGs).6 For example, the SG “DISO” encompasses disorder, cellular dysfunction, sign, or symptom etc. The SG “GENE” encompasses semantic types gene, molecular sequence, amino acid sequence, and others. If SGs are to be useful in assigning new atoms to UMLS concepts, a new atom’s SG must be predicted with high accuracy. This was the motivation of this study.
The current study compares 2 artificial intelligence (AI) computational approaches to predict the SG for new atoms: heuristic reasoning and DL. Heuristic reasoning refers to the use of programs or algorithms that “gain their power from qualitative, experiential judgments, codified in so-called rules of thumb or heuristics, in contrast to numerical calculation programs whose power derives from the analytical equations used. The heuristics focus the attention of the reasoning program on the parts of the problem that seem most relevant. They also guide the application of the domain knowledge.”7
Due to their ability to learn high-level representations from raw text data, DL methods have gained increasing utilization in text analysis and classification tasks. Large language models pretrained on large corpora of biomedical texts have become the work horse of many biomedical natural language processing (NLP) research efforts. BioWordVec, which was trained on PubMed articles, MeSH terms, and clinical notes, has been shown to outperform other general-purpose word embedding models for various NLP tasks.8 The recent success of the self-attention mechanism has led to a flurry of transformer-based language models such as BERT,9 GPT,10 and Chinchilla.11 Of particular relevance to our study is SapBERT trained with the transformer architecture, augmented by PubMed articles and UMLS synonyms.12
The specific contributions of this study have been:
1) Demonstration that AI methods can predict SG assignments for new UMLS atoms with sufficient potential accuracy to be useful in the larger task of assigning new atoms to CUIs.
2) Demonstration that for SG prediction, heuristic methods, and DL methods can be combined to produce better results than either approach alone.
MATERIALS AND METHODS
Overview of methods
Our goal has been to eventually assist UMLS editors algorithmically when they are identifying CUIs for new atoms. We recognized that any future CUI prediction algorithm would itself be imperfect. For SG prediction to serve as a useful intermediate step in that context, we set a target accuracy of at least 95% for SG prediction. Because extreme accuracy was critical, we designed the current study’s heuristic and DL algorithms to be “self-aware”—that is, able to estimate accuracy for each SG prediction they made. This ability also enabled us to determine when a hybrid algorithm combining both approaches might outperform either one alone. Specifically, if one AI method could assign an atom’s SG with greater predicted accuracy than the other algorithm, the hybrid system could use the better of the 2.
Figure 1 gives an overview of our methods. Both AI methods used data from prior UMLS releases to derive information useful for predicting the SG of a new atom. The content of UMLS updates has been known to vary in important ways (eg, which sources contributed what numbers of new atoms/concepts, the distribution of semantic types of new atoms, the number of new atoms with novel names). To lessen concerns about idiosyncratic release-to-release variability, our analysis covered 6 consecutive UMLS releases: 2020AA, 2020AB, 2021AA, 2021AB, 2022AA, and 2022AB. The project restricted UMLS contents under review to the current 184 English language sources. We further excluded 25 inactive English language sources (ie, those designated by NLM as not updated in recent years). Finally, to avoid unwanted ambiguities and redundancies, the study excluded atoms that the UMLS editors had deemed suppressible. Suppressible terms have uncertain meanings or lack face validity. For example, the term “pancreas” in a hierarchical list of primary cancers in a source vocabulary would be suppressible since the term refers to pancreatic cancer instead of the anatomical entity. For each AI method, a new model was trained on one base UMLS release version and tested on the new atoms added to the UMLS in the immediate next release (eg, train on 2020AA, test using 2020AB). Altogether, the study analyzed 5 train-test UMLS release pairs.
Figure 1.
Overview of the heuristic approach and deep learning approach.
Both AI methods utilized training and test data extracted from UMLS release files MRCONSO (access to CUI, SCUI, AUI, atom name, and source vocabulary), MRSTY (semantic types enabling derivation of SGs), and MRHIER (source hierarchy).13 Extracted information used by one or both methods included:
source vocabulary
name (spelling) of atom
SG assigned in training dataset—for the small number (<0.1%) of atoms with multiple SGs, one SG was chosen at random; SG from test dataset only used to verify predictions
hierarchical context of atom in source vocabulary—in sources that had a hierarchical structure, we used the second top node as a proxy for the atom’s hierarchical context. This is because the top node is often the source vocabulary and not useful. For example, for the atom “Myocardial infarction (disorder)” from SNOMED CT, the second top node is “Clinical finding (finding),” and the top node is “SNOMED CT concept.”
Source asserted synonymy—some source vocabularies assert synonymy between its atoms by assigning a source concept unique identifier (SCUI). UMLS editors generally honor source synonymy and do not split source-synonymous atoms into different CUIs.
We tracked the amount of effort (human and computer) utilized in developing and deploying each AI approach.
Heuristic approach
Our heuristic approach was structured as a “waterfall” of different customized methods (steps) to classify each new atom. If one of the methods (STEP 1–STEP 5) below could not assign a new atom’s SG with at least 95% confidence (based on statistics derived from the training release—eg, the frequency with which a given UMLS training release source was associated with training release terms categorized by a specific SG), that atom was passed on to the next method. When an atom met eligibility criteria for a given step, that step-predicted SG was assigned to the atom. The atom was not processed in subsequent steps. We determined the sequence of steps to be employed in the waterfall by first using each waterfall prediction method (independently of the others) on all training data. The waterfall steps were then arranged in descending order of each step’s previously established SG prediction accuracy.
Using the following methods, the heuristic algorithms could estimate (statistically, as noted above) whether the SG prediction of an atom was likely to meet or exceed the target 95% accuracy threshold:
STEP 1 (HiPPv SRCs—high positive predictive value sources): Use the frequency that an atom’s source in the previous UMLS release matched a specific SG. For example, 99% of atoms from sources RXNORM and USP mapped to SG CHEM, and 98% of atoms from source CPT matched SG PROC.
STEP 2 (HiPPv SCUIs—high positive predictive value SCUIs): Use a combination of an atom’s source and its SCUI to predict SGs based on how often the combination did so in the training dataset (only useful if a source provided SCUIs).
STEP 3 (Matched Atom Names): Use the frequency that a new (test) atom name exactly matched a training set atom name; if ≥95%, use the SG of the matched atom. Approximately 15% of new atoms from the subsequent UMLS release had identical names with atoms in the previous release. Often, independent UMLS sources selected the same wordings to describe the same phenomena.
STEP 4 (HiPPv Trigrams—high positive predictive value trigrams): Derive from an atom’s name its individual words (ASCII strings separated by spaces), bigrams (pairs of adjacent words with preserved word order), and trigrams (triplets with preserved order). Determine how often each would match individual SGs. In STEP 4, predict SGs for any new atom containing a trigram when the training trigram-SG association frequency was at least 95%. For example, trigram “acute transverse myelitis” occurred 14 times in atom names from UMLS 2020AA; 100% of those atoms belonged to SG “DISO.”
STEP 5 (SRC-Parens rules—source parenthesis rules): Derive rules based on the idiosyncratic nuances that each source used to construct atom names. We predicted the likelihood of an SG match based on whether a rule was satisfied. Many UMLS sources embedded unique-for-that-source parenthetical expressions within their atom names. For example, in previous UMLS releases, when a SNOMEDCT_US atom name contained “(dose form)” then its SG was always CHEM.
As noted, each of the above methods could be configured to predict a new atom’s SG with ≥95% certainty based on information derived from the training dataset. For atoms not classified by STEP 1–STEP 5, we used less accurate ad hoc algorithms. Atoms became eligible for STEP 6 if they contained 3 or more words. The STEP 6 (SRCs, Trigrams, Words) algorithm first identified all trigrams that could be derived from an atom’s name and added each trigram’s predictive accuracy for each specific SG (as a percentage) to a running total. To candidate SG running totals, STEP 6 also added the percentage association of an atom’s source with the SG. If one SG then had the highest score, STEP 6 assigned the SG to the atom. Otherwise, STEP 6 repeated the same procedure but scored an atom’s words instead of its trigrams. If no SG had a top score, the atom was passed on to the last waterfall step. STEP 7 (SRCs, Bigrams, Words) was similar to STEP 6, but the atoms it processed were composed of only 1 or 2 words. Parallel to STEP 6, STEP 7 used bigrams and individual words to predict SG candidates. For remaining atoms for which STEP 7 had not yet selected an SG, STEP 7 arbitrarily assigned the SG “DISO.” Previous analysis had indicated that for the most common UMLS SGs, LIVB, and CHEM, STEPS 1–6 could select SGs reasonably well based on their unique characteristics (especially sources and atom spellings); this was less so the case for new atoms corresponding to disease names and finding names whose SG would be DISO.
Deep learning approach
To feed text data into our DL models, we converted it into numerical vectors. We chose BioWordVec8 and SapBERT12 as language models based on their performance during earlier testing.14 We generated BioWordVec and SapBERT embeddings for atom strings (200 and 768 dimensions, respectively), a BioWordVec embedding for the name of the source vocabulary (200 dimensions), and a BioWordVec embedding for the string of the second top atom of the source hierarchy (200 dimensions). The reason for using both SapBERT and BioWordVec embeddings of the atom string was that we noticed an increase in performance of up to 5% compared to using either embedding alone, presumably because SapBERT additionally incorporated information in UMLS synonymy. If the second top atom was not available (eg, source with no hierarchy), we used an all-zero vector. We concatenated the 4 embeddings and fed the result into a fully connected multilayer neural network. The output layer had 15 nodes, one for each SG. We split the data from the base UMLS version into training and validation sets (80% and 20%, respectively). We used the validation set to fine-tune the hyperparameters of the neural network.
To develop a method to estimate the probability that a prediction would be correct, we hypothesized that the difference (delta) between the DL’s score for the top SG and second-to-top predicted SG for an atom would be correlated with the accuracy of the DL’s prediction. Intuitively, we expected that a higher delta would be associated with a higher probability of a correct prediction. Since DL scores varied between 0 and 100, we separated the delta into intervals of 1 and computed the accuracy for the atoms in each delta interval for each dataset. We used the microaverage across the 5 datasets as an indicator of the overall expected accuracy for each delta interval.
Hybrid approach
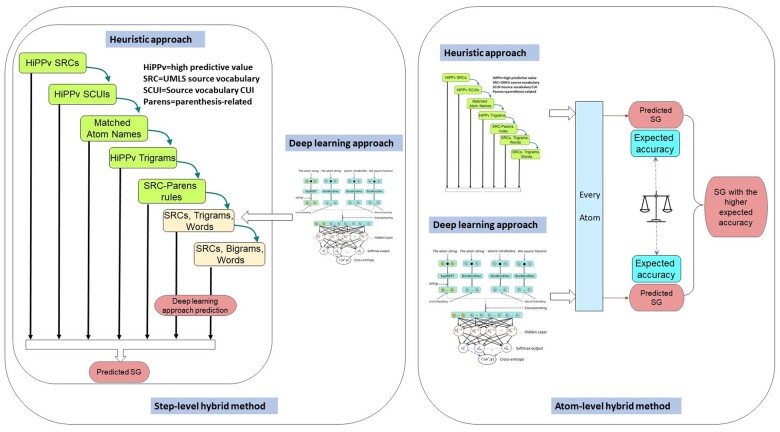
Assuming that our DL and heuristic methods were complementary orthogonal approaches, combining them could theoretically yield better results. Since we have developed methods for each approach to estimate its expected accuracy for SG predictions, we explored 2 methods of combining them into a hybrid model. The “step-level hybrid method” started with STEP 1 of the heuristic method, following each step until the estimated accuracy of the next step was expected to be lower than the DL method, and then switched to DL to process the rest of the atoms. The “atom-level hybrid method” generated SG predictions for all atoms by both the heuristic and DL methods. The results were then combined. When the predictions for a specific atom did not concur, the prediction from the method that had the higher expected accuracy was used. Figure 2 gives an overview of the 2 hybrid methods.
Figure 2.
Overview of the step-level and atom-level hybrid methods.
We used the McNemar test15 to evaluate statistically the differences in performance.
RESULTS
Heuristic approach
Table 1 shows step-by-step results for the heuristic 7-step waterfall SG prediction method. Table 1 provides overall totals as well as individual results for each of the 5 train-test UMLS release pairs. The aggregate of all STEPs in the waterfall approach correctly assigned SGs to 1 475 160 (94.3%) of the 1 563 692 AUIs processed for the 5 UMLS release pairs. By contrast, for STEP 1–STEP 5 inclusive, the heuristic SG prediction accuracy was 98.8% for 1 321 353 AUIs (85% of all test AUIs in the study). Per Table 1, only in STEP 6 and STEP 7 did the waterfall predictive accuracies fall (to 71% and 65%, respectively).
Table 1.
Heuristic semantic group predictions by waterfall step and UMLS dataset release
| UMLS release: train | 2022AA | 2021AB | 2021AA | 2020AB | 2020AA | OVERALL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UMLS release: test | 2022AB | 2022AA | 2021AB | 2021AA | 2020AB | |||||||
| Number of new AUIs in test releases | 275 844 | 175 990 | 455 510 | 226 211 | 430 137 | 1 563 692 | ||||||
| STEP 1: HiPPv SRCs | ||||||||||||
| AUIs processed (% of original) | 140 014 | 51% | 4763 | 3% | 133 684 | 29% | 26 710 | 12% | 290 962 | 68% | 596 133 | 38% |
| AUIs correct (% of processed) | 139 490 | 99.6% | 4621 | 97% | 133 368 | 99.8% | 26 242 | 98% | 289 877 | 99.6% | 593 598 | 99.6% |
| STEP 2: HiPPv SCUIs | ||||||||||||
| AUIs processed (% of original) | 30 443 | 11% | 36 896 | 21% | 213 319 | 47% | 28 398 | 13% | 24 842 | 6% | 333 898 | 21% |
| AUIs correct (% of processed) | 30 346 | 99.7% | 36 731 | 99.6% | 212 659 | 99.7% | 28 283 | 99.6% | 24 673 | 99.3% | 332 692 | 99.6% |
| STEP 3: matched spellings | ||||||||||||
| AUIs processed (% of original) | 19 201 | 7% | 59 935 | 34% | 36 190 | 8% | 95 521 | 42% | 19 086 | 4% | 229 933 | 15% |
| AUIs correct (% of processed) | 18 633 | 97% | 58 952 | 98% | 35 151 | 97% | 94 909 | 99% | 18 243 | 96% | 225 888 | 98% |
| STEP 4: HiPPv trigrams | ||||||||||||
| AUIs processed (% of original) | 23 929 | 9% | 16 950 | 10% | 22 446 | 5% | 28 833 | 13% | 36 258 | 8% | 128 416 | 8% |
| AUIs correct (% of processed) | 23 111 | 97% | 16 379 | 97% | 20 732 | 92% | 27 833 | 97% | 34 344 | 95% | 122 399 | 95% |
| STEP 5: SRC-Parens rules | ||||||||||||
| AUIs processed (% of original) | 7182 | 3% | 6403 | 4% | 6941 | 2% | 5684 | 3% | 6763 | 2% | 32 973 | 2% |
| AUIs correct (% of processed) | 6667 | 93% | 6110 | 95% | 6531 | 94% | 5464 | 96% | 6413 | 95% | 31 185 | 95% |
| STEP 6: SRCs, trigrams, words | ||||||||||||
| AUIs processed (% of original) | 44 712 | 16% | 41 802 | 24% | 32 969 | 7% | 28 475 | 13% | 38 971 | 9% | 186 929 | 12% |
| AUIs correct (% of processed) | 34 753 | 78% | 29 094 | 70% | 22 843 | 69% | 20 053 | 70% | 26 815 | 69% | 133 558 | 71% |
| STEP 7: SRCs, bigrams, words | ||||||||||||
| AUIs processed (% of original) | 10 363 | 4% | 9241 | 5% | 9961 | 2% | 12 590 | 6% | 13 255 | 3% | 55 410 | 4% |
| AUIs correct (% of processed) | 7130 | 69% | 5648 | 61% | 6092 | 61% | 8523 | 68% | 8447 | 64% | 35 840 | 65% |
| TOTALs: STEP 1–STEP 7 | ||||||||||||
| AUIs processed (% of original) | 275 844 | 100% | 175 990 | 100% | 455 510 | 100% | 226 211 | 100% | 430 137 | 100% | 1 563 692 | 100% |
| AUIs correct (% of processed) | 260 130 | 94% | 157 535 | 90% | 437 376 | 96% | 211 307 | 93% | 408 812 | 95% | 1 475 160 | 94.3% |
HiPPv: high predictive value; SRC: UMLS source vocabulary; SCUI: Source vocabulary CUI; Parens: parenthesis-related.
Deep learning approach
In the DL validation phase, we determined that best performance was achieved with the following: one hidden layer of 2048 neurons, one dropout layer (rate = 0.2), one batch normalization layer, the rectified linear unit (ReLU) activation function for the hidden layers, the cross-entropy loss function for computing the loss between the predicted output of the neural network and the true labels of the data, learning rate of 0.00002, and epochs number of 100.
The accuracy of the DL approach in each of the 5 datasets varied from 91.6% to 97.1% (first row, Table 2). The overall microaveraged accuracy across the 5 datasets was 94.3%. Table 2 also shows the accuracy corresponding to each best-to-second-best score delta range. Note that the number of atoms in each range varied in different datasets. The total accuracy for a particular dataset can be interpreted as a weighted average across all ranges. In line with our expectation, the accuracy increased monotonically with delta. Average accuracy of over 95% was achieved with delta of ≥8. Delta scores ≥8 pertained to 78.1% of all AUIs across the 5 datasets.
Table 2.
Performance of deep learning approach and accuracy by top score delta range
| Accuracy |
Proportion of AUIs | ||||||
|---|---|---|---|---|---|---|---|
| Top score delta range a | 2020AA–2020AB | 2020AB–2021AA | 2021AA–2021AB | 2021AB–2022AA | 2022AA–2022AB | Microaverage | |
| Overall | 97.1% | 93.2% | 94.5% | 91.6% | 92.7% | 94.3% | 100% |
| [0, 2) | 52.5% | 58.9% | 52.4% | 50.9% | 52.2% | 53.5% | 4.8% |
| [2, 4) | 73.1% | 79.5% | 71.8% | 68.1% | 64.5% | 72.3% | 4.9% |
| [4, 6) | 87.3% | 90.4% | 85.6% | 81.2% | 75.6% | 84.9% | 5.5% |
| [6, 8) | 95.0% | 94.9% | 93.5% | 90.2% | 83.9% | 92.5% | 6.7% |
| [8, 10) | 98.0% | 96.8% | 96.6% | 94.9% | 90.1% | 96.6% | 7.9% |
| [10, 12) | 99.2% | 97.7% | 98.3% | 97.2% | 93.8% | 97.9% | 9.3% |
| [12, 14) | 99.6% | 98.4% | 99.3% | 98.6% | 96.4% | 98.9% | 10.1% |
| [14, 16) | 99.7% | 98.9% | 99.6% | 99.2% | 97.9% | 99.3% | 10.5% |
| [16, 100] | 99.9% | 99.5% | 99.5% | 99.6% | 99.5% | 99.7% | 40.3% |
Difference of model output score for top 2 SG predictions.
Hybrid approach
Table 3 compares the results of the heuristic and DL approaches. Overall, they agreed for 92.5% of all AUIs processed across the 5 datasets (both correct or both incorrect). The proportion in which one approach was correct while the other was incorrect was: heuristic correct 3.8%, DL correct 3.7%. A small set of examples in each category is available in Supplementary Table S1. The first 5 steps of the heuristic approach outperformed DL (98.8% accuracy vs 94.3%). Therefore, in the step-level hybrid method, we replaced STEP 6 (71% accuracy) and STEP 7 (65% accuracy) of the heuristic approach with DL.
Table 3.
Contingency table for the performance of the 2 approaches
| 2020AA–2020AB | 2020AB–2021AA | 2021AA–2021AB | 2021AB–2022AA | 2022AA–2022AB | Overall | |
|---|---|---|---|---|---|---|
| Both approaches are correct | 402 503 (93.6%) | 199 520 (88.2%) | 418 887 (92.0%) | 149 218 (84.8%) | 245 713 (89.1%) | 1 415 841 (90.5%) |
| Both approaches are incorrect | 6162 (1.4%) | 5003 (2.2%) | 6675 (1.5%) | 6407 (3.6%) | 5648 (2.1%) | 29 895 (1.9%) |
| HE is correct and DL is incorrect | 6280 (1.5%) | 11 906 (5.3%) | 18 494 (4.1%) | 8349 (6.83%) | 14 421 (5.2%) | 59 450 (3.8%) |
| DL is correct and HE is incorrect | 15 192 (3.5%) | 9782 (4.3%) | 11 454 (2.5%) | 12 016 (4.7%) | 10 062 (3.7%) | 58 506 (3.7%) |
HE: heuristic approach; DL: deep learning approach.
Table 4 shows the performance of the heuristic, DL, and the 2 hybrid methods. The atom-level hybrid method achieved the overall best SG prediction accuracy of 96.6%, followed by the step-level hybrid of 96.4%. The full set of results including the gold standard and each method’s predictions is available as Supplementary Material16 (https://doi.org/doi:10.5061/dryad.dfn2z356z). The pairwise differences between the individual approaches and the hybrid methods were all statistically significant (see Supplementary Table S2 for full statistical results).
Table 4.
Comparison of the performance of heuristic, DL, and the 2 hybrid methods
| 2020AA–2020AB | 2020AB–2021AA | 2021AA–2021AB | 2021AB–2022AA | 2022AA–2022AB | Microaverage | |
|---|---|---|---|---|---|---|
| Heuristic | 95.0% | 93.4% | 96.0% | 89.5% | 94.3% | 94.3% |
| Deep learning | 97.1% | 93.2% | 94.5% | 91.6% | 92.7% | 94.3% |
| Step-level hybrid | 97.3% | 96.2% | 97.6% | 94.1% | 94.5% | 96.4% |
| Atom-level hybrid | 97.5% | 96.3% | 97.6% | 93.9% | 95.2% | 96.6% |
Development and run-time data for each approach
Development (design, coding, testing, iterative evolution) of the heuristic approach consumed 2.5 person-months of effort by author (RAM). The computational environment used for the heuristic approach was a virtual Linux machine with 16 CPUs and 24 GB of RAM. To run the heuristic algorithms to produce the results described herein required 4 h for data transformation (from raw UMLS distribution files MRCON, MRSTY, and MRHIER), and 1 h to execute all of the 7 steps of the waterfall approach for all 5 datasets.
The DL approach was developed over 3 months by 2 authors (YM and KWF). We utilized the Biowulf high performance computing cluster at the National Institutes of Health.17 Slurm workload manager was used to submit and parallelize the training, evaluation, and testing jobs.18 The average total time for training a DL model was 6.5 days on a v100x GPU with 32GB of RAM and 300GB of CPU RAM. Each dataset took 4 h to prepare the embeddings and 15 min for testing on an average.
DISCUSSION
Accomplishments of this study
The motivation for this study was to create a SG predictor with high enough accuracy to assist in UMLS editing, ie, to group synonymous new atoms into UMLS concepts. In predicting the SG for new atoms, both the heuristic and DL approaches in this study achieved nearly identical overall accuracies (94.3%) across all datasets. The 2 hybrid approaches gained about 2% in accuracy, with the atom-based hybrid method slightly outperforming the step-based method. Our original design goal of at least 95% accuracy was satisfied.
Relevant prior work
An earlier study by Fan et al19 used distributional similarity of UMLS concepts in a corpus of PubMed citations to classify them into 7 semantic classes. The error rate was 19.8% for the top prediction. A subsequent refinement added lexical features which reduced the error rate to 14.3%.20 Compared with Fan et al, our DL approach did not need an external corpus, nor the hand-crafted rules to extract contextual information from text. Some elements of our heuristic approach resembled their lexical methods, but we also used bigrams and trigrams in addition to single words. Furthermore, our approaches leveraged information derived from the source vocabulary, including source name, source synonymy, and source hierarchy. Overall, our approaches had an error rate of 4% (10% lower than their best results). A more recent study by Kudama and Llavori21 employed conditional random fields to predict the SG of the head word in a UMLS atom. The highest overall precision they achieved was 80%.
Contrasting the 2 AI approaches in this study
The study’s 2 approaches can both be classified as AI techniques. The heuristic approach was derived through applying expert knowledge and analysis of existing UMLS data that led to symbolic reasoning algorithms. The DL approach was less dependent on human knowledge and analysis and relied more on machine learning using large volumes of training data. Each approach has its strength and limitations. The heuristic approach relies on manual derivation of highly specific algorithms that could have limited generalizability to other tasks. However, the advantages of the heuristic approach are its flexibility, short processing time, and superior accuracy in waterfall steps 1–5. Due to the modular nature of the steps, each one could be modified individually as required. It would be relatively simple to add new steps, or to reorder existing steps. On the other hand, the DL approach is a monolithic black box. Any modification will need retraining of the whole model, which may take multiple days and more intensive utilization of computing resources. The strength of the DL approach is minimal human intervention, which provides higher consistency and reproducibility.
Application of SG prediction in UMLS editing
This study created a highly accurate SG predictor with the goal of eventually assisting UMLS editors in assigning new atoms to UMLS concepts. Currently, the UMLS editing platform depends primarily on normalized string matching and source synonymy to suggest to UMLS editors possible grouping of new atoms into existing concepts. A logical extension of this work will be to create a DL-based tool that suggests to which existing concepts a new atom belongs (or that it does not match anything and should form a new concept), which can potentially augment or replace the current workflow. We observed considerable variability in the 5 datasets in our study in terms of the performance of our methods. Therefore, the methods should be regularly revisited to ensure that their performance does not degrade over time. Performance might improve following additional insights gained from applying the methods to more datasets, and the availability of newer techniques.
A second potential application of our methods is in the quality assurance of existing semantic type assignment in the UMLS. Previous studies have proposed methods to identify errors in semantic type assignments.22 Several of those methods focused on concepts assigned to multiple semantic types or SGs, which are more likely to contain errors or problems of internal consistency.23–29 Those methods have limited generalizability because only 10% of UMLS concepts have multiple semantic type or SG assignments. The auditing method proposed by He et al30 was restricted to top level semantic types. Another method by Gu et al31 relied on semantic tags which were only available from SNOMED CT terms. In contrast, our approaches can be used to predict the SG of every atom. Another advantage of our method is that we have a built-in metric for the expected accuracy of each prediction. Potential errors detected by any method need to be reviewed by UMLS editors, who can become overwhelmed with false positives if the positive predictive value of the method is not high enough. Using our method, we would recommend that the editors only review potential errors associated with a very high expected accuracy (eg, >98%) to reduce the false positive rate.
Limitations and future work
The hybrid approaches relied on an estimation of the expected accuracy of the prediction by the 2 approaches for each atom. When applying the method to a particular dataset, the expected accuracy could be calculated based on the actual performance of the methods on past datasets. For example, when applying the methods to the 2021AA–2021AB dataset, the expected accuracy should be calculated based on the actual performance of the methods on the 2020AA–2020AB and 2020AB–2021AA datasets. Nevertheless, to simplify the experimental design, we used the microaveraged actual performance of all datasets to derive a single expected accuracy for all datasets, and applied that to evaluate the performance of the hybrid methods over individual datasets. We acknowledge this limitation. However, our analyses across the 5 datasets have led us to believe that the results reported herein are nevertheless largely valid. Another limitation of our study is that, based on the observed variability between UMLS releases, whether our results are generalizable to future releases remains to be seen. That the evaluation model used in this study used training data from the immediately previous UMLS release somewhat mitigates against this concern. Finally, the parentheses-related rules used in STEP 5 of the heuristic method were manually derived by author RAM through direct observations of patterns in the data. That approach may not be fully reproducible for future applications.
We are currently in the final stages of developing and testing an automated approach to derive STEP 5 of the heuristic approach and preliminary results show that the performance is comparable to the hand-crafted rule. We are also experimenting to adapt our SG prediction methods to help with the algorithmic assignment of semantic types in the UMLS editing process, and potentially as an auditing tool for assigned semantic types in the UMLS.
In the future, it may be worthwhile to look at newer versions of DL models and techniques, which are evolving rapidly. As a start, the SapBERT model used in this study, which is based on the 2020AA UMLS, may benefit from retraining using a newer release. Newer and more powerful language models, such as ChatGPT32 and GPT-4,33 may offer additional benefits.
CONCLUSION
Our study demonstrated that AI methods can predict SG assignments for new UMLS atoms with sufficient accuracy to be potentially useful as an intermediate step in the time-consuming task of assigning new atoms to UMLS concepts (CUIs). We showed that for SG prediction, combining heuristic methods and DL methods can produce better results than either alone.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Will Hibbard for his contribution to this work during his internship at NLM. The authors would also like to thank colleagues in the Scientific Computing Branch for their assistance with computing resources to support this work. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov)
Contributor Information
Yuqing Mao, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
Randolph A Miller, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
Olivier Bodenreider, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
Vinh Nguyen, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
Kin Wah Fung, National Library of Medicine, National Institutes of Health, Bethesda, Maryland, USA.
FUNDING
This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
AUTHOR CONTRIBUTIONS
YM, RAM, OB, and KWF conceived and designed the study. RAM developed and implemented the heuristic approach. YM and KWF developed and implemented the DL approach. VN provided helpful suggestions and critiques during development of both methods. YM and RAM performed the data analysis. YM, RAM, and KWF drafted the manuscript and all authors contributed substantially to its revision.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The UMLS data files are available from the NLM website. The full set of results including the gold standard and each method’s predictions is available as online supplementary material.
REFERENCES
- 1. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amos L, Anderson D, Brody S, Ripple A, Humphreys BL.. UMLS users and uses: a current overview. J Am Med Inform Assoc 2020; 27 (10): 1606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nguyen V, Yip HY, Bodenreider O. Biomedical vocabulary alignment at scale in the UMLS Metathesaurus. In: Proceedings of the Web Conference 2021; 2021: 2672–83; Ljubljana, Slovenia. [DOI] [PMC free article] [PubMed]
- 4. Nguyen V, Yip HY, Bajaj G, et al. Context-enriched learning models for aligning biomedical vocabularies at scale in the UMLS Metathesaurus. In: Proceedings of the ACM Web Conference 2022; 2022: 1037–46; Lyon, France. [DOI] [PMC free article] [PubMed]
- 5. Lindberg DA, Humphreys BL, McCray AT.. The unified medical language system. Yearb Med Inform 1993; 2 (1): 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCray AT, Burgun A, Bodenreider O.. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform 2001; 84 (Pt 1): 216–20. [PMC free article] [PubMed] [Google Scholar]
- 7. Clancey WJ, Shortliffe EH.. Readings in Medical Artificial Intelligence: The First Decade. Reading, MA: Addison-Wesley Longman Publishing Co., Inc.; 1984. [Google Scholar]
- 8. Zhang Y, Chen Q, Yang Z, Lin H, Lu Z.. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci Data 2019; 6 (1): 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Devlin J, Chang M-W, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). 2019: 4171–86; Minneapolis, MN.
- 10. Radford A, Narasimhan K, Salimans T, Sutskever I. Improving Language Understanding by Generative Pre-Training. 2018. https://cdn.openai.com/research-covers/language-unsupervised/language_understanding_paper.pdf. Accessed February 23, 2023.
- 11. Hoffmann J, Borgeaud S, Mensch A, et al. Training compute-optimal large language models. arXiv, arXiv:2203.15556. 2022, preprint: not peer reviewed.
- 12. Liu F, Shareghi E, Meng Z, Basaldella M, Collier N. Self-alignment pretraining for biomedical entity representations. arXiv, arXiv:2010.11784. 2020, preprint: not peer reviewed.
- 13. UMLS ® Reference Manual. 2023. https://www.ncbi.nlm.nih.gov/books/NBK9684/. Accessed June 12, 2023.
- 14. Bajaj G, Nguyen V, Wijesiriwardene T, et al. Evaluating biomedical word embeddings for vocabulary alignment at scale in the UMLS Metathesaurus using Siamese networks. Proc Conf Assoc Comput Linguist Meet 2022; 2022: 82–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947; 12 (2): 153–7. [DOI] [PubMed] [Google Scholar]
- 16. Mao Y, Miller RA, Bodenreider O, Nguyen V, Fung KW.. Data from: two complementary AI approaches for predicting UMLS semantic group assignment: heuristic reasoning and deep learning. Dryad 2023; Dataset. 10.5061/dryad.dfn2z356z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.BIOWULF, High Performance Computing at the NIH. 2023. http://hpc.nih.gov. Accessed February 23, 2023.
- 18. Yoo AB, Jette MA, Grondona M.. Slurm: simple Linux utility for resource management. In: Feitelson D, Rudolph L, Schwiegelshohn U, eds. Job Scheduling Strategies for Parallel Processing: 9th International Workshop, JSSPP 2003. Seattle, WA: Springer; 2003: 44–60. [Google Scholar]
- 19. Fan J-W, Friedman C.. Semantic classification of biomedical concepts using distributional similarity. J Am Med Inform Assoc 2007; 14 (4): 467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan J-W, Xu H, Friedman C.. Using contextual and lexical features to restructure and validate the classification of biomedical concepts. BMC Bioinformatics 2007; 8 (1): 264–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kudama S, Llavori RB. Semantic annotation of UMLS using conditional random fields. In: KDIR. 2014: 335–41; Rome, Italy.
- 22. Zheng L, He Z, Wei D, et al. A review of auditing techniques for the Unified Medical Language System. J Am Med Inform Assoc 2020; 27 (10): 1625–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gu HH, Perl Y, Elhanan G, Min H, Zhang L, Peng Y.. Auditing concept categorizations in the UMLS. Artif Intell Med 2004; 31 (1): 29–44. [DOI] [PubMed] [Google Scholar]
- 24. Gu H, Hripcsak G, Chen Y, et al. Evaluation of a UMLS auditing process of semantic type assignments. In: AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2007: 294; Chicago, IL. [PMC free article] [PubMed]
- 25. Gu HH, Elhanan G, Perl Y, et al. A study of terminology auditors’ performance for UMLS semantic type assignments. J Biomed Inform 2012; 45 (6): 1042–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Halper M, Chen Z, Geller J, Perl Y. A metaschema of the UMLS based on a partition of its semantic network. In: Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001: 234; Washington, DC. [PMC free article] [PubMed]
- 27. Chen Y, Gu H, Perl Y, Halper M, Xu J.. Expanding the extent of a UMLS semantic type via group neighborhood auditing. J Am Med Inform Assoc 2009; 16 (5): 746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Y, Gu HH, Perl Y, Geller J, Halper M.. Structural group auditing of a UMLS semantic type’s extent. J Biomed Inform 2009; 42 (1): 41–52. [DOI] [PubMed] [Google Scholar]
- 29. Morrey CP, Chen L, Halper M, Perl Y.. Resolution of redundant semantic type assignments for organic chemicals in the UMLS. Artif Intell Med 2011; 52 (3): 141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He Z, Perl Y, Elhanan G, Chen Y, Geller J, Bian J.. Auditing the assignments of top-level semantic types in the UMLS semantic network to UMLS concepts. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2017; 2017: 1262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gu H, He Z, Wei D, Elhanan G, Chen Y.. Validating UMLS semantic type assignments using SNOMED CT semantic tags. Methods Inf Med 2018; 57 (1): 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. OpenAI. ChatGPT. 2022. https://openai.com/blog/chatgpt/. Accessed February 23, 2023.
- 33. OpenAI. GPT-4 Technical Report. 2023. https://cdn.openai.com/papers/gpt-4.pdf. Accessed April 6, 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UMLS data files are available from the NLM website. The full set of results including the gold standard and each method’s predictions is available as online supplementary material.