Abstract
Objective
Generation of automated clinical notes has been posited as a strategy to mitigate physician burnout. In particular, an automated narrative summary of a patient’s hospital stay could supplement the hospital course section of the discharge summary that inpatient physicians document in electronic health record (EHR) systems. In the current study, we developed and evaluated an automated method for summarizing the hospital course section using encoder-decoder sequence-to-sequence transformer models.
Materials and Methods
We fine-tuned BERT and BART models and optimized for factuality through constraining beam search, which we trained and tested using EHR data from patients admitted to the neurology unit of an academic medical center.
Results
The approach demonstrated good ROUGE scores with an R-2 of 13.76. In a blind evaluation, 2 board-certified physicians rated 62% of the automated summaries as meeting the standard of care, which suggests the method may be useful clinically.
Discussion and conclusion
To our knowledge, this study is among the first to demonstrate an automated method for generating a discharge summary hospital course that approaches a quality level of what a physician would write.
Keywords: natural language processing, machine learning, abstractive summarization, automated clinical notes, automated patient summary, clinician burnout
Objective
Physicians spend approximately 2 h in the electronic health record (EHR) for every 1 h of patient care.1 The time required for recording, reviewing, and summarizing information in EHRs has imposed complex and burdensome workflows on physicians, which has contributed to burnout.2,3 To alleviate documentation burden,4 multiple efforts have pursued automated summary of the hospital patient record through natural language processing (NLP).5–7
When a patient is discharged from a hospital, a physician authors a discharge summary, a transition of care document that summarizes the patient’s hospital stay and is sent to providers who continue the patient’s care in other settings.8 While the discharge summary is both required and valuable, clinical workflow can delay its availability,9 which can increase the risk of rehospitalization10 and medication errors.11 In the United States, the content of this document can only include information that has already been documented within the EHR.12 In generating a discharge summary, physicians spend most of their time manually writing the hospital course section, a textual narrative that describes the progress of treatment for the patient from admission to discharge. Automating this section could potentially save time for physicians, as note templates have automated other sections of the discharge summary13 but not the hospital course section.14
In a prior study, we demonstrated the feasibility of automating the hospital course section of a discharge summary.15 However, the study employed the MIMIC-III dataset,16 which was limited to intensive care unit (ICU) patients and did not cover the full hospital stay. Furthermore, the prior study only measured textual overlap between the automated and reference summaries but not quality nor factuality. In the current study, we developed a novel method for generating clinical summaries using EHR data from inpatient neurology hospitalizations. We evaluated performance with state-of-the-art benchmarks and physician experts.
Background and significance
Text summarization can be categorized into 2 subdomains, extraction and abstraction. Extraction identifies key terms and phrases and concatenates them to form a summary, whereas abstraction generates new sentences to synthesize a summary. Generally, abstraction is more fluent and coherent. Until about 2017, clinical text summarization was mainly through extraction17; abstraction required substantial time and domain expertise with unpromising results.5 Abstractive text summarization has accelerated due to applications of deep learning models called transformers, especially Bidirectional Encoder Representations from Transformers (BERT)18 and Bidirectional and Auto-Regressive Transformers (BART).19
BERT has a bidirectional language representation structure that overcomes restrictions with unidirectional language representation models.18,20 With BERT, the entire input sequence is fed in at once unlike previous deep learning models, such as recurrent neural networks (RNNs), which read text sequentially.
BART was created specifically for abstractive text summarization.19 BART uses a BERT autoencoder with noisy masked input data so as to force a decoder to denoise and reconstruct the original text. Encoder-decoder sequence-to-sequence models, such as BART, are very effective for sequence generation tasks such as text summarization.21 BART is a well performing open source model for text summarization on the CNN/Daily Mail and XSum datasets.22
In the domain of clinical summarization, Yalunin et al23 presented a Longformer encoder and BERT decoder for an abstractive summary of patient hospitalization histories. Additional studies used physician notes rather than the entirety of structured and unstructured data available in the EHR: Shing et al6 demonstrated that automation of discharge summaries was possible but noted issues of factuality, and Cai et al24 showed an approach for automating a patient-facing After-Visit Summary when provided a clinical note summary. In a related effort, Gao et al25 demonstrated that encoder-decoder sequence-to-sequence transformers can summarize a patient’s primary diagnostic problems from a current progress note. Despite these studies demonstrating summarization of physician notes, physicians may desire a summary note generated from their prior notes and not from their current note that is being authored.
Krishna et al26 demonstrated the ability to generate Subjective, Objective, Assessment and Plan (SOAP) notes through an extractive-abstractive summarization pipeline. In a similar paper, Joshi et al27 showed that a pointer generator network with a penalty can be used to summarize medical conversations with 80% of relevant information captured.
While nearly all EHR summarization studies have used automated measures such as ROUGE scores to evaluate performance, Zhang et al28 engaged physician experts to measure clinical validity of summaries. Because automated metrics do not address grammar and consistency, physician evaluations using a Likert-scale can improve understanding of automated summary quality, readability, factuality, and completeness.
This study employs techniques from the previous works: BERT and BART models as well as both automated and physician scoring. We expand on the prior research by applying a generative text method to automate the discharge summary hospital course section using EHR data from a real-world setting and a clinical assessment measuring the validity of the method.
Materials and methods
Abstractive pipeline summarization approach
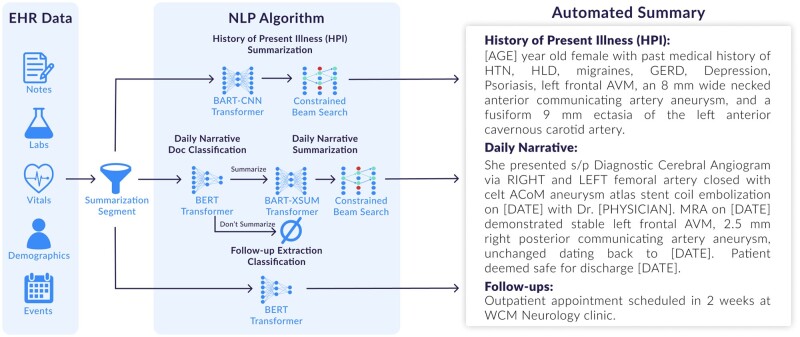
When using transformers with smaller datasets, the recommended best approach is to fine-tune a pretrained model.18 Pretrained transformers generally have a maximum input token length of 512 or 1024, which is challenging to summarize the full patient record.29 To overcome this limitation, 3 different strategies have traditionally been used: (1) truncating the document by, for example, taking only the first 512 tokens as input,30 (2) employing a neural network that scales sequentially, such as a RNN with a transformer output or a transformer with attention that scales with sequence length,31 or (3) summarizing or extracting individual sections first and then performing another layer of summarization during the merging of those sections.32,33 The preferred strategy in the medical domain is to summarize smaller individual sections and combine those sections with a second layer of summarization.6,14 This approach is preferred since it more closely resembles the physicians’ current workflow where they extract salient information first and then synthesize it into a narrative summary.34 Similar to this ensemble strategy, we employed a “day-to-day approach”15 that overcomes the limitations of long-form documents in transformers by summarizing individual clinical notes per day and concatenating them to form a clinical narrative summary (see Figure 1). Each of these 3 parts ingests only specific types of clinical notes as a means to limit the total amount of input words into the transformer models (see “Designing the Day-to-Day Method” presented in the Appendix).
Figure 1.
Data flow that shows how EHR data is segmented into 3 separate sections through the following transformer models referred to as the “day-to-day approach”: (1) HPI summarization, (2) daily narrative document classification and summarization, and (3) follow-up extraction classification. The automated summary is constructed by chronologically assembling the results.
Data collection
From an institutional repository containing data from EHR and other source systems,35 we obtained a dataset consisting of 6600 hospital admissions from 5000 unique patients admitted to the inpatient neurology unit at NewYork-Presbyterian/Weill Cornell Medical Center, a 2600-bed quaternary-care teaching hospital in New York City affiliated with Weill Cornell Medicine of Cornell University. We focused on neurology patients because the speciality is known to exhibit higher clinical complexity as compared to a general inpatient patient; neurology patients have 20% more interactions with physicians, 27% more comorbidities, spend 81% more days in the hospital, and have a 16% higher mortality rate than the general inpatient patient.36 All patients had a hospitalization with a length of stay of at least 48 hours between the years of 2010 and 2020. The dataset contained at least one admit note and one discharge summary per patient, which is a regulatory requirement for each hospital admission.12,37 Each record of the dataset contained a combination of demographics and clinical details (primarily free text documents) as described in Table 1.
Table 1.
Data types used from the dataset.
| Type of data | Description |
|---|---|
| Descriptive | Age, sex, marital status, race, mortality status |
| Encounters | Admission date, admission diagnoses (ICD-10 code and description), discharge date, discharge disposition |
| Free text documents | Admission notes, emergency department provider notes, progress notes, consult notes, operative reports, pathology reports, radiology reports, discharge summaries |
| Measurements | Laboratory results (LOINC), vital signs |
For each discharge summary, we extracted the hospital course section using a regular expression. We only used the hospital course sections as our labels since EHR note templates have automated the other discharge summary sections.13 The corpus of hospital course sections served as the gold standard for model development and evaluation. We created train, validation, and test datasets with a ratio of 80:10:10. Dataset statistics can be seen in Table S6 in the Appendix.
Based on abstractive pipeline summarization strategy using the “day-to-day” approach (see Figure 1), we further segmented the dataset into 3 parts: (1) history of present illness (HPI) summarization which is primarily constructed from the admission note, (2) daily narrative document classification and summarization which chronologically details the patient’s full course of treatment, and (3) follow-up extraction classification which identifies any follow-ups to occur at a subsequent outpatient encounter that are documented in any clinical notes within 72 hours of discharge.15
For the HPI summarization dataset, we used the 6600 admission notes as the source data. We appended the corresponding patient descriptive data (see Table 1), admission date, and admission diagnoses to the beginning of the admit note. We used the first sentence of the hospital course section as the labels.
For the daily narrative summarization dataset, we used 71 115 clinical notes as the source data (note types are in Table 1 and author types are in Table S7 in the Appendix). Likewise, we appended the corresponding patient descriptive data. For the labels, we extracted 7200 sentences from the hospital course sections, excluding the first sentence, that included a date in various formats such as 10/25, 10-25, Oct 25, etc. We found the best corresponding source note for that same patient and date using the highest ROUGE-L score. We created a separate daily classification dataset of the 71 115 clinical notes where the 7200 matching notes were labeled with a 1 signifying that the document had relevant content captured in the discharge summary; the other 63 915 were labeled with a 0.
Lastly, we split the 6600 hospital course sections into 64 747 sentences. Using a BERT-model trained on CLIP,38 we created the follow-up dataset by classifying the sentences with either a 1 or 0 signifying if the sentences were a follow-up for future plans of care or not. Furthermore, 2 researchers with backgrounds in clinical informatics and NLP reviewed all the sentences manually and made any obvious labeling corrections. The researchers were informed that clinical follow-ups sentences contain commands, advice, or vital instructions that need to be performed after the current hospitalization.
Constrained beam search for medical summarization
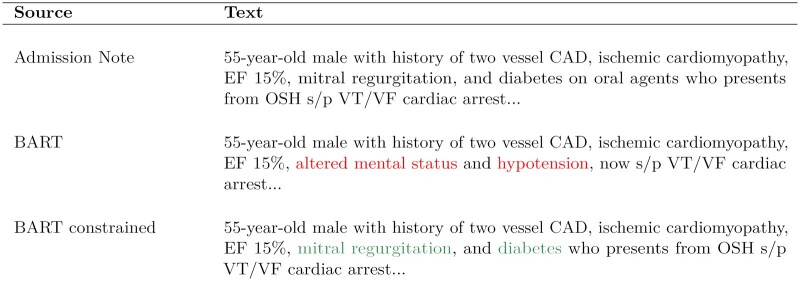
An ongoing research concern of abstractive summarization models is that they can hallucinate text that is not consistent with the source documents which creates factuality problems.39 Abstractive summarization models are trained at the word-level to minimize cross entropy compared with a reference summary, but this is not functionally equivalent to maximizing factuality. Researchers in the field of NLP have investigated some of the following approaches to improve factuality: measuring the inconsistencies through a question answering metric,40 ranking summary correctness through textual entailment predictions,41 using graph-based attention,42 rewarding factuality through reinforcement-learning,28 and constraining beam search during model inference.43 We implemented the latter approach, constrained beam search, as a means to improve factuality in our models. By constraining beam search, inconsistent medical terms are reduced which is the most concerning hallucination to correct for a clinical summary as seen in the example in Figure 2.
Figure 2.
A motivational example of how clinical summaries can hallucinate. Inconsistent medical terms are highlighted in red font. In this example, the proposed BART model with constrained beam search for medical terminology removes the clinical inconsistencies from the HPI summary.
At inference, sequence to sequence transformer models are generally paired with the heuristic algorithm of beam search.44 Beam search allows for multiple candidate summaries for comparison based on a beam width and conditional probabilities. The best beam is then chosen based on a logarithmic score. Our approach constrains traditional beam search by penalizing the logarithmic score of any words from a set of banned words (Algorithm 1). For constructing our set of banned words, we first used a custom medical dictionary derived from SNOMED CT.45 For any medical words and synonyms that intersect both the source document x and , we permit them during beam search; otherwise all other words in the medical dictionary are banned in the generated sequence (the set of banned words) and the next best alternative word is selected as the output. Similar approaches have been found to provide an improvement in the factuality of the generated summary with only a slight decrease in its fluency.43
Algorithm 1.
Constrained beam search approach
for in beam width do
while enddo
ifthen
return
end if
end while
end for
return such that
Models
Our approach is coupled with 2 state-of-the-art NLP models, BERT and BART. We use BERT for classification and BART for text summarization. Additionally, in our evaluation, we used TextRank algorithm46 as a baseline method for comparison with the transformers.
We fine-tuned 2 BERT models on 2 separate datasets—free text documents the model should or should not summarize and the follow-up sentences—with a maximum input length of 512 tokens respectively for 3 epochs.
We used the pretrained BART-CNN and BART-XSum transformer models from HuggingFace. We fine-tuned the models on the HPI and daily narrative summarization datasets for 3 epochs with a maximum input token length of 1024.
Evaluation
We measured the performance of the summarization tasks (HPI summarization, daily narrative summarization, and discharge summary hospital course summarization) and the classification tasks (daily narrative document classification, follow-up extraction classification) as seen in Figure 1. Additionally, we measured physician perception of quality, readability, factuality, and completeness, with a description of each criteria listed in Table S5 in the Appendix.
Summarization tasks
For each of the summarization tasks, we compared BART, BART with beam search constrainment, and TextRank models through ROUGE scores, word count, and error rates. Of note, for the final task of summarizing the discharge summary hospital course, we compared TextRank, the day-to-day approach without beam search constrainment, and the day-to-day approach with beam search constrainment (Figure 1).
ROUGE recall scores measure the textual overlap between the automated and physician-written summary.6,14,19,28 We reported ROUGE scores on a scale from 0 to 100 where a higher score indicates better summarization performance. For longform document summarization tasks such as our study, state-of-the-art ROUGE recall scores are within the ranges of 39-51 for ROUGE-1 (R-1), 10-24 for ROUGE-2 (R-2), and 36-46 for ROUGE-L (R-L).22,47
We measured conciseness through the average word count and standard deviation (SD) of the automated summary in comparison to the physician-written summary.48,49 Conciseness was important since if the summaries were too long, they could lose relevance to downstream outpatient providers.
To understand the effectiveness of constraining medical terminology during beam search for improving factuality, we used a dependency arc entailment (DAE) model that was pretrained on XSum.50 The DAE is an Arc-Factuality model which makes independent factuality judgments, at a word level, then a sentence level, and finally at a summary level, for the generated summary. For each independent dependency arc in the generated summary, the DAE model predicts whether the relationship exists in the input document. It then uses these “arc-level” decisions to extract summary level decisions. If any dependency arc is nonfactual, the generated summary is labeled nonfactual as seen in Figure 3. We generated sentence level summaries for both the HPI and daily narrative summarization tasks and DAE was used to calculate both word and sentence level error rates (the fraction that was determined to be nonfactual). Of note, since hospital charts have many words, we were not able to calculate word and sentence level error rates for discharge summary hospital course summarization due to limits with the DAE model.
Figure 3.
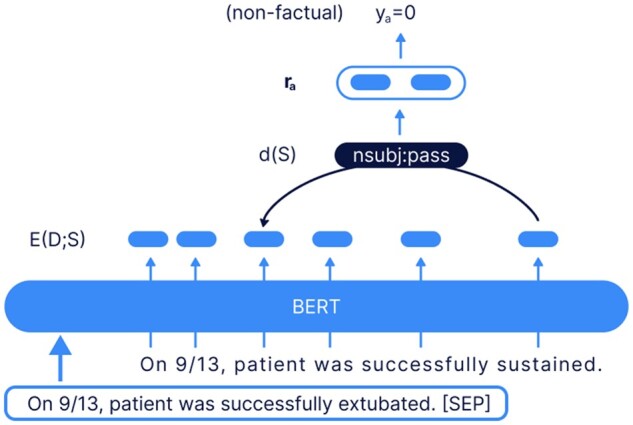
The dependency arc entailment (DAE) model was pretrained on BERT XSum.50 If the arc is nonfactual, then the sentence summary is marked as nonfactual.
Classification tasks
We measured accuracy, recall, precision, and F1-scores for the document classification and follow-up extraction models. Statistics were captured from a binary label of 0 and 1 for both models. The labels measured either (1) correctly identifying the document that should or should not be summarized and (2) correctly selecting the follow-up sentences that should or should not be included in the hospital course summary.
Physician perception
To measure quality, readability, factuality, and completeness of an automated summary,28,51 2 board-certified physicians (M.G.W., an internist; and B.B.N., a neurologist) blindly rated 25 pairs of patient discharge summaries, one generated by the automated method and one written by a physician for a particular hospitalization. The summaries were randomly selected from the test dataset and were ordered randomly so neither physician could ascertain whether they were reviewing the computer- or physician-generated summary. Each physician rated all 50 summaries on a Likert scale of 1-10 where 1 was poor and 10 was excellent with the criteria of each metric listed in Table S5 in the Appendix. Note that our quality measure effectively encompassed a summary being simultaneously concise, readable, factual, and complete. Additionally, both physicians agreed that a Likert score of 7 for quality indicated that the summary met overall sufficient clinical validity.
To measure inter-rater reliability of the scores between the 2 physicians, we used intraclass correlation coefficient (ICC) with a 2-way random effects model and consistency, which measured for the degree of similarity among the 2 reviewers and their ratings; it has a range of 0-1, where 1 represents unanimous agreement and 0 indicates no agreement.52
Results
Summarization tasks
As shown in Table 2, the BART-based approaches had higher ROUGE scores indicative of better performance compared to the TextRank baseline in all 3 subtasks. Given that the HPI task for TextRank had a high ROUGE-2 of 28.94 as a baseline, the implication was that there is high textual overlap between the source notes and produced summary for the HPI segment. Higher textual overlap has been shown in other studies to assist with maintaining factuality.53 Although BART with constrainment had slightly lower ROUGE scores than BART without constrainment, BART with constrainment had lower word and sentence error rates. Stated differently, we observed a tradeoff between ROUGE scores and error rates, which is consistent with the literature; thus, constraining beam search at inference reduces ROUGE scores but lead to an increase in sentence factuality.43 The overall performance of the day-to-day approach with constrainment (Figure 1) had a ROGUE-2 score of 13.76, which was within the lower range of other state-of-the-art longform document summarization models.47
Table 2.
ROUGE recall scores {R-1/R-2/R-L}, word count (WC) mean and standard deviation (SD), and word and sentence error rates (ER) as seen with a dependency arc entailment (DAE) model are presented for our proposed models for comparing automated and physician-written summaries.
| Summarization task | R-1 | R-2 | R-L | Word count mean (±SD) | Word-ER | Sent-ER |
|---|---|---|---|---|---|---|
| History of present illness (HPI) | ||||||
| Baseline: Textrank | 43.30 | 28.94 | 38.40 | 53 (±9) | 0.3 | 0.7 |
| BART | 61.67 | 53.12 | 59.69 | 43 (±9) | 6.3 | 42.4 |
| BART constrained | 61.02 | 52.78 | 59.05 | 44 (±11) | 5.7 | 40.6 |
| Daily narrative | ||||||
| Baseline: Textrank | 9.55 | 1.32 | 8.93 | 20 (±3) | 31.0 | 42.2 |
| BART | 46.59 | 35.03 | 43.95 | 10 (±7) | 9.9 | 28.8 |
| BART constrained | 46.42 | 34.77 | 43.75 | 11 (±13) | 7.2 | 26.7 |
| Discharge summary hospital course | ||||||
| Baseline: Textrank | 15.48 | 4.18 | 8.51 | 511 (±451) | — | — |
| Day-to-day | 37.10 | 14.44 | 19.64 | 444 (±374) | — | — |
| Day-to-day constrained | 35.97 | 13.76 | 18.83 | 421 (±365) | — | — |
Sentences within each summarization section are individually compared (HPI and daily) along with the full hospital course summary.
Hospital charts had 81.5k words on average, which is too large for the DAE model.
The physician-written reference summaries were highly condensed with 591 words on average (SD±597 words) per hospital chart with 81.5k words on average (SD of ±150.3k words). Conciseness results of our model can be seen in Table 2 with the average word count of 421 words for the discharge summary hospital courses; thus, our day-to-day approach had shorter summaries in length than physician-written ones by 170 words on average.
Classification tasks
For the 2 models for the day-to-day approach for classification, results for accuracy, recall, precision, and F1-scores are in Table 3. The daily narrative document classification task had an accuracy of 78.83% at determining which clinical documents should or should not be included for summarization for the discharge summary hospital course. And the follow-up sentences task had an accuracy of 96.11%, which implied the model was very effective at identifying which follow-up sentences should or should not be included in the hospital course section.
Table 3.
Accuracy, recall, precision, and F1-score of the classification models of the day-to-day approach.
| Classification task | Accuracy | Recall | Precision | F1 |
|---|---|---|---|---|
| Daily narrative document | 0.7883 | 0.4390 | 0.7500 | 0.5538 |
| Follow-up sentences | 0.9611 | 0.7851 | 0.8364 | 0.8100 |
Physician perception
As shown in Table 4 and Figure 4, the quality of the automated summaries had an average rating of 6.52; 62% met the quality standards of care, defined as a quality score of 7 or higher, compared to 94% of the physician-written summaries. The automated summaries were also highly readable with a mean score of 7 out of 10; in fact, the 2 raters misclassified 34% of the physician-written summaries as being generated by the automated method. The factuality (6.4 vs 8.6) and completeness (6.9 vs 8.7) ratings were slightly lower for the automated as compared with the physician-written summaries.
Table 4.
Mean and standard deviation (SD) of the ratings for quality, readability, factuality, and completeness of the automated and physician-written summaries.
| Automated | Physician-written | |
|---|---|---|
| Quality, mean (±SD) | 6.52 (±1.50) | 8.16 (±1.13) |
| Readability | 7.00 (±1.46) | 8.00 (±1.44) |
| Factuality | 6.44 (±1.55) | 8.60 (±0.88) |
| Completeness | 6.88 (±1.70) | 8.68 (±0.84) |
Figure 4.
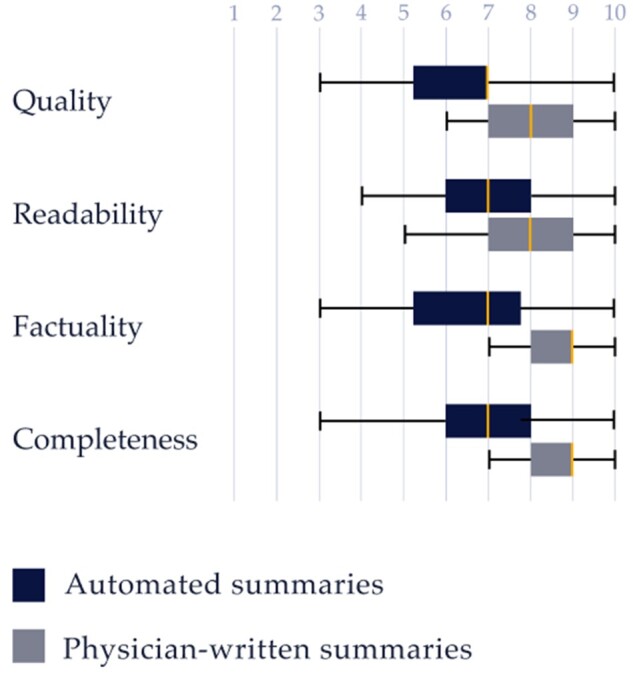
Box plot for the Likert scores for quality, readability, factuality, and completeness of the automated and physician-written summaries by the physician reviewers.
ICCs were 0.64 for quality, 0.59 for readability, 0.73 for factuality, and 0.68 for completeness, which indicate moderate reliability of agreement for the 2 physician reviewers.52
Discussion
We found the automated approach did very well in mirroring the structure of a manually written physician summary (see example in Table S8 in the Appendix). Notably, 62% of randomly sampled automated summaries met the standards of clinical validity, which suggests the approach is effective. Similar to how prior research has demonstrated clinical validity for summarizing the impression section for radiology reports,28 our results demonstrate how transformer models are effective at summarizing a patient’s hospital stay to generate the hospital course section of a discharge summary.
Note that in order to perform well on the quality metric, as our approach did, our technique had to perform well for every day of a patient’s stay in the hospital for conciseness, readability, factuality, and completeness and then select the most salient content within that time interval along with any clinical follow-ups for discharge. Therefore, the very high ROUGE scores for the HPI and daily segments and the high accuracy scores for the document classification and follow-up sentences all came together as an adequate measure for the quality performance of our automated summaries. Another contributing factor was the baseline performance of the TextRank method (a nonsupervised approach) on the dataset; the high ROUGE scores on TextRank implied that our transformer models had high textual overlap, which has been previously shown to assist with factuality for abstractive summarization.53 And although this study did not use an Oracle algorithm that measured the upper-limit for the model’s performance, the study put particular emphasis on the physician evaluation that measured performance of the physician-written summaries as the gold standard.
While we demonstrated the success of our model in emulating a physician summary, we recognize several limitations and areas of future work, especially for factuality and completeness. Of interest is that the physician clinical notes, such as Progress Notes and Consults, were overwhelmingly the primary source of content selected for summarization because of their high textual overlap with content in the discharge summary hospital course section; our model included very little source data from labs and vitals that were not contextualized within a physician note. Thus, if a physician did not include any context with respect to labs and vitals in their notes (such as documenting the implications of an abnormality for lab results), our model assumed that the data was more or less irrelevant. The implication is that our model, as constructed, was not adequate to find, interpret, and synthesize lab and vitals data that may have been missed by physicians in their clinical notes during the course of the patient’s hospital stay.
Given these limitations, we make a few recommendations. The BERT-based document classification model (see Figure 1) had an accuracy of 78.83% and a recall of 43.90%, as it primarily struggled with false negatives. The classification model should be paired with a rule-based structure to rebalance the dataset by always summarizing documents for specific procedures, consults, and events (even if the findings are insignificant) and never summarizing specific documents such as nutritionist progress notes. For example, if a patient is administered a tissue plasminogen activator drug, commonly used for emergency treatment for ischemic stroke, the event should always be included in a hospital course summary. Our BERT-based document classification model approach could then be used for cases where it is too difficult to create such rules. Secondly, we would recommend that the documentation classification dataset should be manually annotated in lieu of our approach of selecting corresponding notes using the highest ROUGE-L score that matched content within the reference hospital course sections.
Second, we recommend constructing a method for improving factuality for proper names, dates, numbers, and other similar items. As seen in the example in Table S8 in the Appendix, our approach occasionally misidentified the surgeon’s name who performed a procedure or swapped dates for when 2 procedures were performed. This occurred because our model hallucinated the most common surgeon’s name or dates when either was not included in the clinical notes; the model would infer the dates and provider names as opposed to not including them. Since our approach was only designed to guarantee factuality of medical terminology, this kind of hallucination continues to be a known limitation. While our results demonstrated the improved factuality of the HPI and narrative summarization tasks via reduced word and sentence errors rate through DAE, we recommend for future research a physician evaluation that measures the factuality of our constrained beam search method. Such a study would count the number of hallucinations between an automated and reference clinical summary and categorize them based on type and severity of hallucination.
Lastly, the 2 evaluators commented that the readability of the automated summaries could be improved by reducing the prevalence of acronyms. For background, physicians commonly write medical acronyms, including some that are not common outside their specialty, in their clinical notes. As our model was trained with physician-written discharge summaries as the gold-truth, these same acronyms continued forward in the automated summaries. To address this challenge, acronyms in the dataset could be automatically translated and linked to their Unified Medical Language System (UMLS) terminologies.45 Likewise, clinical text in the automated summary could be extracted and contextualized as a benefit for comprehension to clinicians. This could be implemented through tools such as MetaMap,54 cTAKES,55 or MedCAT.56
Our study revealed several expanded applications of clinical note summarizations. While our dataset was specific to patients admitted to a neurology unit, the prior study that used a similar approach used the MIMIC-III dataset for ICU patients.15 Anecdotally, we found that the inpatient neurology discharge summaries and the clinical notes at NewYork-Presbyterian/Weill Cornell Medical Center were structurally similar to the MIMIC-III dataset. Thus, our day-to-day approach could potentially be adapted in the future to other inpatient clinical specialties if fine-tuned on a different dataset.
Likewise, our summary was constructed chronologically for the hospital stay by abstracting a few sentences each day for the patient and condensing duplicate information and events occurring over multiple days; so as new clinical content became available, the prior summarized sentences remained intact. The implication is that our approach could also be used to create a transfer report for patients moved from one medical unit to another before discharge. Finally, our study demonstrated an automated method for complete automation of the discharge summary hospital course. In practice, a healthcare organization could add a step where physicians review a drafted automated summary and make slight corrections before finalization, a process commonly referred to as “human-in-the-loop.” The 2 evaluators provided the feedback that such a workflow would be anticipated to provide significant benefit to physicians in mitigating physician burnout.
Conclusion
Transformers can perform state-of-the-art NLP tasks such as text summarization. We present an approach of using transformers, enhancing these models for clinical factuality by constraining medical terminology, and then dividing the medical chart into 3 separate segments to automate the hospital course section of the discharge summary. Through our work with an inpatient neurology EHR dataset, we have shown the potential of this approach as a means of constructing an automated patient summary of the hospital chart. Findings from this study could be used by a healthcare organization to determine the potential value of implementing clinical text summarization methods in a real-time production setting.
Supplementary Material
Acknowledgments
We are grateful for the guidance we received from Alexander “Sasha” Rush with respect to our approach of improving clinical factuality in a sequence-to-sequence transformer model. We are thankful for the assistance we received from Rita Giordana Pulpo for the graphical designs in our manuscript.
Contributor Information
Vince C Hartman, Cornell Tech, New York, NY 10044, United States; Abstractive Health, New York, NY 10022, United States.
Sanika S Bapat, Cornell Tech, New York, NY 10044, United States; Abstractive Health, New York, NY 10022, United States.
Mark G Weiner, Department of Medicine, Weill Cornell Medicine, New York, NY 10065, United States; Department of Population Health, Weill Cornell Medicine, New York, NY 10065, United States.
Babak B Navi, Department of Neurology and Feil Family Brain and Mind Research Institute, Weill Cornell Medicine, New York, NY 10065, United States.
Evan T Sholle, Department of Population Health, Weill Cornell Medicine, New York, NY 10065, United States.
Thomas R Campion, Jr, Department of Population Health, Weill Cornell Medicine, New York, NY 10065, United States; Clinical & Translational Science Center, Weill Cornell Medicine, New York, NY 10065, United States.
Author contributions
V.C.H. conceptualized the study, developed the technology, coordinated the evaluation, and generated the manuscript text. S.S.B. developed the technology and contributed to the manuscript text and revisions. M.G.W. and B.B.N. contributed to conceptualization of the study, performed the evaluation, and reviewed/edited the manuscript. E.T.S. coordinated access to data, evaluation, and manuscript edits. T.R.C. oversaw study concept, guided evaluation, and contributed/edited manuscript text.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
We received support from NewYork-Presbyterian and Weill Cornell Medicine, including the Joint Clinical Trials Office and Clinical and Translational Science Center (UL1TR002384).
Conflicts of interest
V.C.H. and S.S.B. have commercial interest in Abstractive Health.
Data and code availability
The data is available with appropriate IRB approval and legal agreements. The code, including our abstractive pipeline approach and constrained beam-search methods, is available to the public upon request to the author V.C.H.
References
- 1. Arndt BG, Beasley JW, Watkinson MD, et al. Tethered to the EHR: primary care physician workload assessment using EHR event log data and time-motion observations. Ann Fam Med. 2017;15(5):419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Downing NL, Bates DW, Longhurst CA.. Physician Burnout in the Electronic Health Record Era: Are We Ignoring the Real Cause?. American College of Physicians; 2018. [DOI] [PubMed] [Google Scholar]
- 3. Shanafelt TD, Hasan O, Dyrbye LN, et al. Changes in burnout and satisfaction with work-life balance in physicians and the general US working population between 2011 and 2014. Mayo Clin Proc. 2015;90(12):1600-1613. [DOI] [PubMed] [Google Scholar]
- 4. Quiroz JC, Laranjo L, Kocaballi AB, Berkovsky S, Rezazadegan D, Coiera E.. Challenges of developing a digital scribe to reduce clinical documentation burden. NPJ Digit Med. 2019;2(1):114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunter J, Freer Y, Gatt A, et al. Summarising complex ICU data in natural language. In: AMIA Annual Symposium Proceedings. Vol 2008. American Medical Informatics Association; November 8–12, 2008:323; Washington, DC. [PMC free article] [PubMed] [Google Scholar]
- 6. Shing HC, Shivade C, Pourdamghani N, et al. 2021. Towards clinical encounter summarization: learning to compose discharge summaries from prior notes, arXiv, preprint: not peer reviewed.
- 7. Alsentzer E, Kim A.. 2018. Extractive summarization of EHR discharge notes, arXiv, preprint: not peer reviewed.
- 8. Kripalani S, LeFevre F, Phillips CO, Williams MV, Basaviah P, Baker DW.. Deficits in communication and information transfer between hospital-based and primary care physicians: implications for patient safety and continuity of care. JAMA 2007;297(8):831-841. [DOI] [PubMed] [Google Scholar]
- 9. Roughead E, Kalisch L, Ramsay E, Ryan P, Gilbert A.. Continuity of care: when do patients visit community healthcare providers after leaving hospital? Intern Med J. 2011;41(9):662-667. [DOI] [PubMed] [Google Scholar]
- 10. Van Walraven C, Seth R, Austin PC, Laupacis A.. Effect of discharge summary availability during post-discharge visits on hospital readmission. J Gen Intern Med. 2002;17(3):186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergkvist A, Midlöv P, Höglund P, Larsson L, Bondesson Å, Eriksson T.. Improved quality in the hospital discharge summary reduces medication errors—LIMM: Landskrona Integrated Medicines Management. Eur J Clin Pharmacol. 2009;65(10):1037-1046. [DOI] [PubMed] [Google Scholar]
- 12. Kind AJ, Smith MA.. Documentation of mandated discharge summary components in transitions from acute to subacute care. In: Henriksen K, Battles JB, Keyes MA, Grady ML, eds. Advances in Patient Safety: New Directions and Alternative Approaches. Vol 2. Agency for Healthcare Research and Quality; 2008. [PubMed] [Google Scholar]
- 13. Dean SM, Gilmore-Bykovskyi A, Buchanan J, Ehlenfeldt B, Kind AJ.. Design and hospitalwide implementation of a standardized discharge summary in an electronic health record. Jt Comm J Qual Patient Saf. 2016;42(12):555-AP11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Adams G, Alsentzer E, Ketenci M, Zucker J, Elhadad N. What’s in a summary? Laying the groundwork for advances in hospital-course summarization. Vol 2021. Association for Computational Linguistics; June 6–11, 2021:4794. [DOI] [PMC free article] [PubMed]
- 15. Hartman V, Campion TR. A day-to-day approach for automating the hospital course section of the discharge summary. In: AMIA Annual Symposium Proceedings. March 21–24, 2022:216-225; Chicago. [PMC free article] [PubMed]
- 16. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3(1):160035-160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Syed AA, Gaol FL, Matsuo T.. A survey of the state-of-the-art models in neural abstractive text summarization. IEEE Access. 2021;9:13248-13265. [Google Scholar]
- 18. Kenton J, Toutanova LK. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Vol. 1 (Long and Short Papers). June 2–7, 2019:4171-4186; Minneapolis, MA.
- 19. Lewis M, Liu Y, Goyal N, et al. BART: denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics; July 5–10, 2020:7871-7880.
- 20. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: Advances in Neural Information Processing Systems 30 (NIPS 2017). December 4–9, 2017:5998-6008; Long Beach, CA.
- 21. Rothe S, Narayan S, Severyn A.. Leveraging pre-trained checkpoints for sequence generation tasks. Trans Assoc Comput Linguist. 2020;8:264-280. [Google Scholar]
- 22. Rohde T, Wu X, Liu Y. Hierarchical learning for generation with long source sequences. 2021. 10.48550/arXiv.2104.07545. [DOI]
- 23. Yalunin A, Umerenkov D, Kokh V. Abstractive summarization of hospitalisation histories with transformer networks. 2022. arXiv, preprint: not peer reviewed.
- 24. Cai P, Liu F, Bajracharya A, et al. Generation of patient after-visit summaries to support physicians. In: Proceedings of the 29th International Conference on Computational Linguistics (COLING). October 2022; Gyeongju, Republic of Korea.
- 25. Gao Y, Dligach D, Miller T, Xu D, Churpek MM, Afshar M.. Summarizing patients’ problems from hospital progress notes using pre-trained sequence-to-sequence models. Proc Int Conf Comput Ling. 2022;2022:2979-2991. [PMC free article] [PubMed] [Google Scholar]
- 26. Krishna K, Khosla S, Bigham J, Lipton ZC. Generating SOAP notes from doctor-patient conversations using modular summarization techniques. Association for Computational Linguistics; August 1–6, 2021:4958-4972.
- 27. Joshi A, Katariya N, Amatriain X, Kannan A. Summarize: global summarization of medical dialogue by exploiting local structures. Association for Computational Linguistics; November 16–20, 2020:3755-3763.
- 28. Zhang Y, Merck D, Tsai E, Manning CD, Langlotz C. Optimizing the factual correctness of a summary: a study of summarizing radiology reports. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. July 2020:5108-5120.
- 29. Sun C, Qiu X, Xu Y, Huang X. How to fine-tune BERT for text classification? In: China National Conference on Chinese Computational Linguistics. Springer; October 18–20, 2019:194-206; Kunming, China.
- 30. Zhang R, Wei Z, Shi Y, Chen Y. {BERT}-{AL}: {BERT} for arbitrarily long document understanding. 2020. https://openreview.net/forum?id=SklnVAEFDB.
- 31. Beltagy I, Peters ME, Cohan A.. 2020. Longformer: the long-document transformer, arXiv, preprint: not peer reviewed.
- 32. Gehrmann S, Deng Y, Rush AM. Bottom-up abstractive summarization. In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing. October–November 2018:4098-4109; Brussels, Belgium.
- 33. Gidiotis A, Tsoumakas G.. A divide-and-conquer approach to the summarization of long documents. IEEE/ACM Trans Audio Speech Lang Process 2020;28:3029-3040. [Google Scholar]
- 34. Feblowitz JC, Wright A, Singh H, Samal L, Sittig DF.. Summarization of clinical information: a conceptual model. J Biomed Inform. 2011;44(4):688-699. [DOI] [PubMed] [Google Scholar]
- 35. Campion TR Jr, Sholle ET, Pathak J, Johnson SB, Leonard JP, Cole CL.. An architecture for research computing in health to support clinical and translational investigators with electronic patient data. J Am Med Inform Assoc. 2022;29(4):677-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tonelli M, Wiebe N, Manns BJ, et al. Comparison of the complexity of patients seen by different medical subspecialists in a universal health care system. JAMA Netw Open. 2018; 111(7):e184852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Joint Commission: history and physical requirements. https://www.jointcommission.org/standards/standard-faqs/behavioral-health/care-treatment-and-services-cts/000001780/. Accessed January 6, 2023.
- 38. Mullenbach J, Pruksachatkun Y, Adler S, et al. CLIP: a dataset for extracting action items for physicians from hospital discharge notes. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing. Vol 1. August 2021:1365-1378. [Google Scholar]
- 39. Cao Z, Wei F, Li W, Li S. Faithful to the original: fact aware neural abstractive summarization. In: Thirty-Second AAAI Conference on Artificial Intelligence. February 2–7, 2018; New Orleans, LA.
- 40. Durmus E, He H, Diab M. FEQA: a question answering evaluation framework for faithfulness assessment in abstractive summarization. Association for Computational Linguistics; July 5–10, 2020.
- 41. Falke T, Ribeiro LFR, Utama PA, Dagan I, Gurevych I. Ranking generated summaries by correctness: an interesting but challenging application for natural language inference. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics; July 28–August 2, 2019:2214-2220; Florence, Italy.
- 42. Zhu C, Hinthorn W, Xu R, et al. Enhancing factual consistency of abstractive summarization. In: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. June 2021:718-733.
- 43. King D, Shen Z, Subramani N, Weld DS, Beltagy I, Downey D. Don’t say what you don’t know: improving the consistency of abstractive summarization by constraining beam search. In: Proceedings of the 2nd Workshop on Natural Language Generation, Evaluation, and Metrics (GEM). December 2022:555-571; Abu Dhabi, United Arab Emirates.
- 44. Meister C, Vieira T, Cotterell R.. Best-first beam search. Trans Assoc Comput Linguist. 2020;8:795-809. [Google Scholar]
- 45. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267-D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mihalcea R, Tarau P. Textrank: bringing order into text. In: Proceedings of the 2004 Conference on Empirical Methods in Natural Language Processing. July 2004:404-411; Barcelona, Spain.
- 47. Xiong W, Gupta A, Toshniwal S, Mehdad Y, Yih W.. 2022. Adapting pretrained text-to-text models for long text sequences, arXiv, preprint: not peer reviewed.
- 48. Choudhry AJ, Baghdadi YM, Wagie AE, et al. Readability of discharge summaries: with what level of information are we dismissing our patients? Am J Surg. 2016;211(3):631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Myers JS, Jaipaul CK, Kogan JR, Krekun S, Bellini LM, Shea JA.. Are discharge summaries teachable? The effects of a discharge summary curriculum on the quality of discharge summaries in an internal medicine residency program. Acad Med. 2006;81(10 Suppl):S5-S8. [DOI] [PubMed] [Google Scholar]
- 50. Goyal T, Durrett G. Annotating and modeling fine-grained factuality in summarization. In: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics; June 6–11, 2021:1449-1462.
- 51. Bernal JL, DelBusto S, García-Mañoso MI, et al. Impact of the implementation of electronic health records on the quality of discharge summaries and on the coding of hospitalization episodes. Int J Qual Health Care. 2018;30(8):630-636. [DOI] [PubMed] [Google Scholar]
- 52. Gisev N, Bell JS, Chen TF.. Interrater agreement and interrater reliability: key concepts, approaches, and applications. Res Social Adm Pharm. 2013;9(3):330-338. [DOI] [PubMed] [Google Scholar]
- 53. Ladhak F, Durmus E, He H, Cardie C, Mckeown K. Faithful or extractive? On mitigating the faithfulness-abstractiveness trade-off in abstractive summarization. In: Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics. Vol 1. May 2022:1410-1421; Dublin, Ireland. [Google Scholar]
- 54. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proceedings of the AMIA Symposium. American Medical Informatics Association; November 3–7, 2001:17; Washington, DC. [PMC free article] [PubMed]
- 55. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kraljevic Z, Bean D, Mascio A, et al. 2019. MedCAT – medical concept annotation tool, arXiv, arXiv:191210166, preprint: not peer reviewed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available with appropriate IRB approval and legal agreements. The code, including our abstractive pipeline approach and constrained beam-search methods, is available to the public upon request to the author V.C.H.