Abstract
Objective
Outcomes are important clinical study information. Despite progress in automated extraction of PICO (Population, Intervention, Comparison, and Outcome) entities from PubMed, rarely are these entities encoded by standard terminology to achieve semantic interoperability. This study aims to evaluate the suitability of the Unified Medical Language System (UMLS) and SNOMED-CT in encoding outcome concepts in randomized controlled trial (RCT) abstracts.
Materials and Methods
We iteratively developed and validated an outcome annotation guideline and manually annotated clinically significant outcome entities in the Results and Conclusions sections of 500 randomly selected RCT abstracts on PubMed. The extracted outcomes were fully, partially, or not mapped to the UMLS via MetaMap based on established heuristics. Manual UMLS browser search was performed for select unmapped outcome entities to further differentiate between UMLS and MetaMap errors.
Results
Only 44% of 2617 outcome concepts were fully covered in the UMLS, among which 67% were complex concepts that required the combination of 2 or more UMLS concepts to represent them. SNOMED-CT was present as a source in 61% of the fully mapped outcomes.
Discussion
Domains such as Metabolism and Nutrition, and Infections and Infectious Diseases need expanded outcome concept coverage in the UMLS and MetaMap. Future work is warranted to similarly assess the terminology coverage for P, I, C entities.
Conclusion
Computational representation of clinical outcomes is important for clinical evidence extraction and appraisal and yet faces challenges from the inherent complexity and lack of coverage of these concepts in UMLS and SNOMED-CT, as demonstrated in this study.
Keywords: natural language processing, Unified Medical Language System, SNOMED-CT, evidence-based medicine
Background and significance
The PICO framework was developed by physicians to formulate clinical questions around 4 elements: Population, Intervention, Comparison, and Outcome.1 This framework is often applied to the randomized controlled trial (RCT) literature to aid in evidence retrieval and to improve evidence-based medicine practice. Not only is efficient evidence search a necessity for clinicians to practice evidence-based medicine without undue burden, but it is also a necessity for modernizing systematic reviews.2 Automated methods or named entity recognition (NER) models for extracting PICO elements from text require gold standard, annotated corpora for training and testing purposes. Often the outcome elements (O in PICO framework) annotated in these corpora have poor inter-annotator agreement compared to other elements or are dropped entirely from the annotation process due to poor inter-annotator agreement overall.3,4 This indicates that outcome elements in RCT abstracts are highly variable.
Two published NER systems provide both open-source annotations of a corpus of RCT abstracts and an annotation guideline to inform outcome element annotation.3,5 However, we are interested in covering a wide range of disease domains, while existing work focuses only on specific domains.3,5 The EBM-NLP corpus places its focus on cardiovascular diseases, cancers, and autism, and the corpus from Sanchez-Graillet et al contains only abstracts on glaucoma and type 2 diabetes.3,5 Other NER systems have taken an automated approach to outcome recognition, which can result in a loss of outcome information in the case where only structured RCT abstracts with a section labeled as “outcomes” are annotated for outcome elements.6,7
Even as NER models improve in outcome element extraction, there will still be a “missing link” preventing efficient evidence retrieval for meta-analysis or systematic reviews. This “missing link” is concept normalization. Once the outcome elements are extracted from RCT abstracts, they cannot be leveraged to the full extent without normalization. Consider, for example, a PubMed abstract (PMID 31941795) that discusses the effect of the intervention N-acetylcysteine on 15-F2t isoprostane concentration.8 An NER model can extract “15-F2t isoprostane concentration” as an outcome. However, if a researcher were to aggregate additional RCT studies with the same extracted outcome, their search may not return all applicable RCTs. Without normalization, this aggregation would not return the synonym “8-iso-PGF2alpha concentration.” Normalization of outcome information can help identify studies using different terms to describe the same outcomes and hence assist with evidence synthesis. Furthermore, normalization can help initiatives such as the Patient-Centered Outcomes Research Institute (PCORI) better evaluate whether or not the outcomes evaluated are patient-centric, and Core Outcome Measures in Effectiveness Trials (COMET), evaluate whether trials are meeting their defined core outcome set.9,10 The information loss due to a lack of normalization can have large consequences on meta-analysis studies or systematic reviews that inform clinical practice guidelines. Additionally, Bai et al11 created a COVID-19 trial knowledge graph by extracting PICO elements from existing clinical trial registries and normalizing them to the UMLS. Their work can provide insight into the impact of the normalization of PICO elements as it presents a computable and standardized representation of COVID-19 evidence that can be queried for downstream evidence synthesis.
Although there is great focus on NER of PICO elements from free text, including RCT studies, our review of the literature found no existing studies that map PICO elements extracted from a diverse range of RCT studies to standardized terminologies or ontologies with the goal of evaluating these terminologies or ontologies. Little knowledge is available regarding whether existing terminologies are suitable for representing PICO elements. Before these outcome elements are normalized, we must first ensure that there are tools to support their accurate and complete normalization. The Unified Medical Language System (UMLS) is a valuable semantic knowledge resource to assess the suitability for normalization as its Metathesaurus contains over 100 biomedical source vocabularies, including clinical documentation vocabularies such as SNOMED-CT and medication-specific vocabularies such as RxNorm.12 Additionally, existing studies have shown that normalization by the UMLS can improve search engine retrieval.13 As the number of studies implementing the SNOMED-CT terminology for concept representation has been increasing since 2012, SNOMED-CT will also be a valuable candidate terminology to assess its suitability for outcome element representation.14
Objective
The goal of this study is to evaluate the UMLS’ and SNOMED-CT’s suitability in representing outcome elements from RCT abstracts. The evaluation will consist of 2 critical metrics, coverage and complexity, modeled after the analysis performed by Friedlin and Overhage.15 This evaluation can point to gaps, if any, in the UMLS Metathesaurus, which, in being filled, will better enable patient-centric outcomes evaluation and evidence-based medicine practice.
Materials and methods
Abstract selection
Five hundred RCT abstracts were selected from PubMed using the following selection criteria. First, all clinical trials from 2010 to 2021 that were Phase 2 or Phase 3, completed interventional studies with results, and from the United States were selected from ClinicalTrials.gov. This resulted in 9338 studies with unique NCT numbers, which were then linked to PubMed articles. The PubMed Entrez Programming Utilities was used to search for PubMed articles with the NCT number in the Title/Abstract or Secondary Source ID and “Randomized Controlled Trial” in the Publication Type.16 If there were multiple PubMed IDs (PMIDs) for a given NCT number, the largest (indicating most recent publication date) PMID was selected. This search returned 3066 NCT numbers linked to 2772 unique PMIDs. Of these 2772 PMIDs, 500 were then randomly selected. During the annotation, if the article was found to be a study protocol or trial design, the abstract was replaced. The disease domains represented by the 500 RCT abstracts were extracted using a previously published ontology-based categorization of the listed ClinicalTrials.gov “Conditions” for their corresponding NCT numbers.17
Abstract annotation
The outcome elements in each abstract were identified by 3 annotators, who followed an annotation guideline developed by us, which is included as Supplementary Material. The primary annotator annotated all 500 abstracts, while the 2 secondary annotators annotated 250 (disjoint) abstracts each. The first 140 abstracts were used for discussion and revision of the annotation guideline, while the remaining 360 abstracts were used to evaluate the annotation guideline. After the annotation process, 3 key changes were made to the annotation guideline. These changes included an important revision allowing for more flexibility with terms such as “increase” and “improvement,” and clearer rules for the annotation of outcomes referred to at baseline and for the conjunction “or” when used as a math symbol (ie, “grade 3 or worse”). All 3 of these improvements represented areas of confusion and disagreement while using the old guideline. All 500 abstracts were updated per the changes, and 140 additional abstracts were annotated to assess the new annotation guideline. These 140 additional abstracts were randomly selected from the EBM-NLP corpus restricted by structured RCTs with Results and Conclusions sections that were linked to an NCTID, and excluding study protocols or trial designs.3 The corresponding outcomes from these 140 additional abstracts are not considered in our evaluation of the UMLS.
Semi-automated mapping
The outcomes extracted from the annotation process were used as input to the semi-automated mapping process via the strict version of MetaMap 2018, which is the default for semantic NLP.18 The python wrapper for MetaMap, pymetamap, was used to facilitate the mappings.19 Any characters in the outcome string not recognized by MetaMap were written out explicitly (ie, “≥” written as “greater than or equal to”). All parameters were kept in their default settings except for word sense disambiguation, which returned only the best mapping for each input concept.20 No score threshold was used in the mapping. In the case of multiple equivalent “best mappings,” the researcher selected their preferred mapping. The mappings were manually classified into the 3 categories: ie, fully mapped, partially mapped, and unmapped. The following terms were defined to be non-substantive and thus not considered in the mapping designation of an outcome concept: overall, total, score(s), measure(s), level(s), concentration(s), estimated, parameter(s), adjudicated, test, titers, incidence, outcome(s), documented, index or descriptive statistics (“mean,” “median,” “mode,” “rate,” “average,” etc.). The fully mapped category was selected if the UMLS mapping was fully representative of the substantive portion of the outcome concept. The partially mapped category was selected for one of the following 4 reasons: (1) the UMLS mapping returned represented some, but not all, of the substantive outcome concept, (2) the UMLS mapping returned was broader than the outcome concept, (3) the UMLS mapping returned was more specific than the outcome concept, or (4) the UMLS mapping returned was related, but of a different semantic type than the outcome concept. Lastly, the unmapped category was selected if the UMLS mapping did not represent the outcome concept. Terms like change and baseline were required to be correctly mapped for the outcome to be considered fully mapped but did not constitute a partial mapping designation if no other part of the outcome concept was correctly mapped. During the mapping process, partially mapped concepts were tagged with 1 or more of their corresponding reason(s) for partial mapping, shown in Table 1. Unmapped concepts were additionally tagged with “-ab” if an abbreviation was present.
Table 1.
Partial mapping tags
| Tag | Reason | Example |
|---|---|---|
| −t | (i) Temporal component aspect missing |
|
| −v | (i) Numeric value aspect missing |
|
| −nt | (i) Non-temporal, non-numeric aspect missing |
|
| −ntab | (i) Non-temporal, non-numeric aspect that is an abbreviation missing |
|
| −b | (ii) Too broad |
|
| −s | (iii) Too specific |
|
| −sts | (iv) Incorrect semantic type |
|
Semi-automated mapping analysis
Prior to analysis, outcome concepts were preprocessed by conversion into lower case, singularized via the pattern.en python module, and punctuation removed via the string python module.21,22 There were 13 instances, in which the same altered outcomes had different MetaMap mappings. Thus, altered outcome concepts that had the same MetaMap mapping were 1 unique outcome concept. Coverage was calculated as the percent of unique outcomes that were fully mapped to the UMLS.15 Complexity was calculated as the percent of fully mapped unique outcomes that were mapped to 2 or more UMLS concepts.15 Of the returned unique CUIs from the UMLS full mappings, the percentage with SNOMED-CT in the source was calculated. The sources of the atoms belonging to each CUI were determined using the UMLS API with the version 2018 AB to ensure that each returned CUI was not deprecated in the UMLS Metathesaurus.23
Error analysis
Further analysis was performed on certain unmapped outcomes to determine if the error in mapping was caused by the UMLS itself or the MetaMap mapping process. A manual search was performed via the UMLS browser for each unmapped outcome concept of interest to determine whether the UMLS contained the necessary concept(s) to represent the unmapped outcome concept, and these concepts could be evoked using some or all of the outcome concept string. If so, the error was classified as a MetaMap error. Otherwise, further search was performed to determine if the necessary concept(s) were present in the UMLS but lacked a synonymy relationship with the outcome concept (UMLS synonymy error), or if the necessary concept(s) were simply absent from the UMLS (UMLS absence error). Thus, 3 types of mapping errors were established. During this analysis, we required that abbreviations have a one-to-one mapping with their corresponding UMLS concept. For example, “ADAU” stands for “average daily accelerometer units.” Both this abbreviation and its full phrase do not return any results via a UMLS browser search, indicating the concept is absent from the UMLS though this concept could be made up of the UMLS concepts “average,” “daily,” “accelerometer,” and “units” (1 to many). The overall workflow is shown in Figure 1.
Figure 1.
Workflow of methods.
Results
Abstract selection
The 500 randomly selected RCT abstracts were linked to 540 NCT numbers, which were used as input to the ontology-based categorization of disease domains.17 Of the 540 clinical trials, 5 failed in concept mappings of their conditions to SNOMED and 9 trials were not categorized. The 526 categorized clinical trials (and thus the RCT abstract corpus) represented 30 diverse disease domains, as shown in Figure 2. The top 3 disease domains were Metabolism and Nutrition, Endocrinology, and Cardiology/Vascular Diseases. Twenty-one of the 30 disease domains were represented in more than 100 RCT abstracts.
Figure 2.
Disease domains represented in the RCT abstract corpus.
Abstract annotation
The annotation process resulted in moderate agreement amongst annotators for both the 360 abstracts annotated with the original guideline, and the 140 additional abstracts annotated with the “new” guideline, as shown in Table 2.24 Though the average Cohen’s kappa for the new guideline is slightly lower than that for the old version, the new guideline included important changes as referred to in the “Methods” section. Furthermore, this difference could be due, in part, to the greater diversity of outcomes in the EBM-NLP corpus than those previously seen, as the abstracts selected from this corpus were not required to be interventional, Phase II/III trials. The Cohen’s kappa values presented indicate that the outcomes are reasonably independent of the bias of any 1 annotator and the annotators share an understanding of the definition of outcome elements as per the annotation guideline.25 Thus, we could be confident in using these extracted outcome elements for the remainder of our analysis.
Table 2.
Cohen’s kappa scores for annotation batches
| Batch | Cohen’s kappa (w/secondary annotator 1) | Cohen’s kappa (w/secondary annotator 2) | Cohen’s kappa (averaged) |
|---|---|---|---|
| 360 abstracts—old version guideline | 0.747 | 0.780 | 0.763 |
| 140 abstracts—new version guideline | 0.776 | 0.726 | 0.751 |
Mapping analysis
From the 500 randomly selected RCT abstracts, 2731 outcomes were extracted and used as input into MetaMap. Prior to analysis of the mapping results, the outcomes were preprocessed as described in the “Methods” section, resulting in 2617 unique outcomes. As shown in Figure 3, 44% of outcome elements were fully mapped by the UMLS, 36% were partially mapped, and 20% were unmapped. Additionally, of the fully mapped outcome elements, 67% were mapped to 2 or more UMLS concepts. This is within our expectation considering that the average string length for the outcome elements used as input to MetaMap was 26 characters. One example of a complex outcome is the outcome “24-hour ambulatory systolic blood pressure,” which maps to the UMLS concept “24 hour systolic blood pressure” (CUI C1282174) and “Ambulatory—qualifier value” (CUI C0439841). Further examples of each mapping designation are shown in Figure 3.
Figure 3.
MetaMap semi-automated mapping results and examples.
Stratification by disease domain, as shown in Figure 4, revealed no real pattern in the mapping designation for outcome elements, which were grouped into disease domains based on their corresponding PubMed abstract and repeats of the same outcome within 1 PubMed abstract were included. As shown in Figure 4, for those disease domains with greater than 100 abstracts, ie, all those to the left of Obstetrics/Gynecology, the % of full mappings was in the range of 41-56, the % of partial mappings was in the range of 23-37, and the % of unmapped elements was in the range of 17-29. The less represented disease domains of sleep and neonatology had a larger percentage of unmapped elements. However, the unmapped outcome elements in both disease domains originated from only 4 PubMed abstracts.
Figure 4.
MetaMap mapping designation stratified by disease domain of corresponding PubMed abstract.
Further analysis identified that the top 3 causes for partial mapping designation were: (1) a missing aspect of the substantive outcome concept unrelated to the temporal component or numeric values (28%); (2) a missing aspect of the substantive outcome concept unrelated to the temporal component or numeric values that involved an abbreviation (20%); and (3) the UMLS mapping was too specific for the outcome concept (17%). Additionally, 14% of the partial mappings had tags for a missing temporal component, 11% had tags for the incorrect semantic type, 6% had tags for too broad of a returned mapping, and 4% had tags for a missing numeric value.
Of the 20% of outcome elements that were unmapped by MetaMap, 78% of them contained abbreviations. Table 3 shows the most common unmapped terms by count of occurrence in a unique PubMed abstract, labeled by their respective reasons for the designation of “unmapped” (ie, MetaMap extraction error, UMLS synonymy error, or UMLS absence error). As shown in the table, the top 4 unmapped terms were tolerated, safe, AEs, and AE. The most common error type present was MetaMap extraction error.
Table 3.
Unmapped outcomes with a count greater than 1 in unique RCT abstracts, ordered by count of occurrence
| Term | Frequency | Reason unmapped | Term | Frequency | Reason unmapped |
|---|---|---|---|---|---|
| tolerated | 95 | a | ACR50 response (at least 50% improvement in American College of Rheumatology response criteria) | 2 | a |
| safe | 38 | s | PVR (pulmonary vascular resistance) | 2 | m |
| AEs (adverse events) | 33 | m | SRI-4 response (at least 4-point reduction in SELENA-SLEDAI score) | 2 | a |
| AE (adverse event) | 33 | m | VTE (venous thromboembolism) | 2 | s |
| well-tolerated | 18 | a | HRs (hazard ratios) | 2 | s |
| TEAEs (treatment-emergent adverse events) | 16 | s | MACE (major adverse cardiovascular events) | 2 | a |
| HbA1c | 15 | m | change from baseline in HbA1c | 2 | m |
| OS (overall survival) | 10 | s | Injection site tenderness | 2 | m |
| Median OS (overall survival) | 8 | s | HbA1c concentration | 2 | m |
| ORR (overall/objective response rate) | 7 | a | SVR (sustained virologic response) | 2 | m |
| LDL-C (low-density lipoprotein cholesterol) | 6 | m | HbA1c concentrations | 2 | m |
|
5 | s | burning | 2 | m |
| Reactogenicity | 4 | m | IGA score of 1 (Investigator’s Global Assessment) | 2 | a |
| HF (heart failure) | 3 | m | IGA score of 0 (Investigator’s Global Assessment) | 2 | a |
| SVR12 (sustained virologic response after 12 weeks post-treatment) | 3 | a | non-HDL-C (nonhigh-density lipoprotein cholesterol) | 2 | m |
| disease progression | 3 | m | HR (heart rate) | 2 | m |
| eGFR (estimated glomerular filtration rate) | 3 | s | SVR12 rates (sustained virologic response after 12 weeks post-treatment) | 2 | a |
|
2 | m | FPG (fasting plasma glucose) | 2 | s |
| PK (pharmacokinetics) | 2 | m | Mean HbA1c | 2 | m |
|
2 | m | BW (body weight) | 2 | s |
| function | 2 | m |
The reason each outcome is unmapped is presented where “m” stands for MetaMap error, “s” stands for UMLS synonymy error, and “a” stands for UMLS absence error. Information on the abbreviations is presented in parentheses.
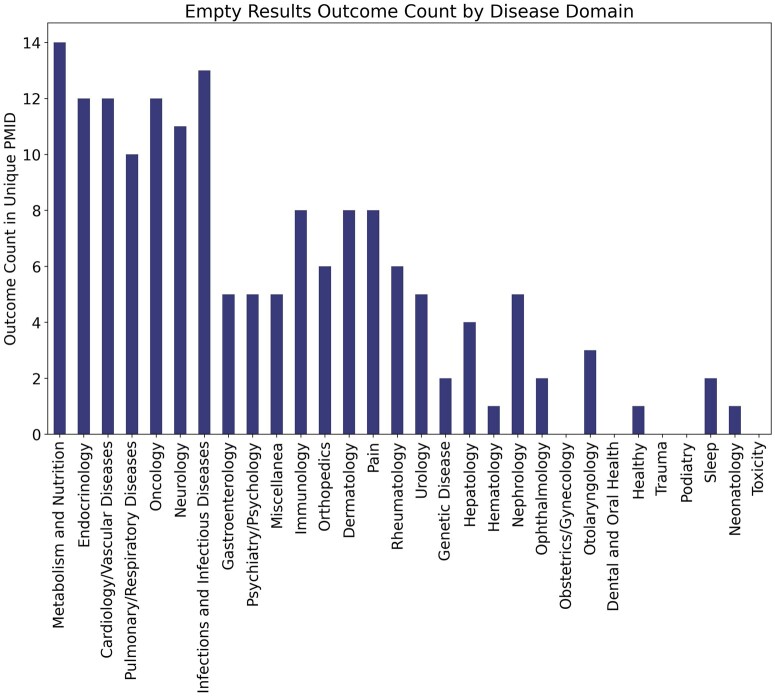
Furthermore, for 80 of the 525 unmapped outcomes (15%), MetaMap did not return any mapping, ie, an empty result (error classification present in Supplementary Material). As most of these empty results involved mapping of abbreviations, which typically belong to a specific clinical sublanguage, the empty results were stratified by disease domain. To inform potential improvements to the UMLS specifically, and not MetaMap, only those unmapped terms due to UMLS Metathesaurus error (either synonymy or absence) were considered, which accounted for 61 of the 80 terms. Additionally, the term “tolerated” was removed from analysis as it had a total term count across disease domains over 6× greater than any other term and is not specific to any disease domain. The results of this analysis are shown in Figure 5 where the count represents the occurrence of an empty result outcome in a unique PubMed abstract belonging to said disease domain. Following the figure, the disease domains of Metabolism and Nutrition, and Infections and Infectious Diseases had the highest count of empty results. These disease domains have a disproportionately high count of empty results returned for outcome concepts compared to domains with similar PubMed abstract counts (Cardiology/Vascular Diseases, Endocrinology, Pulmonary/respiratory diseases). Specific examples of the outcome concepts that result in empty mappings from the Metabolism and Nutrition domain due to UMLS synonymy errors include TEAEs (treatment-emergent adverse events), FPG (fasting plasma glucose), and DMG (dimethylglycine). Those with empty mappings due to UMLS absence errors include AUCGIR (area under the curve for the glucose infusion rate) and SIOGTT (sensitivity index for the oral glucose tolerance test). For the disease domain of Infections and Infectious Diseases, examples of the empty results outcomes due to UMLS synonymy errors include improve VS (viral suppression) and reactogenic while those due to UMLS absence errors include HiSCR75 (75% reduction of Hidradenitis Suppurativa Clinical Response criteria), HiSCR90, and SVR12 (sustained virologic response after 12 weeks post-treatment). Here, it is of note that the broader concepts of HiSCR and SVR are present in the UMLS.
Figure 5.
Count of occurrence of outcome elements with empty MetaMap results due to a UMLS Metathesaurus error in unique PubMed abstracts stratified by disease domain.
Lastly, the MetaMap semi-automated mapping process returned 1160 unique UMLS CUIs for the fully mapped outcome elements, of which 705 (61%) had SNOMED-CT as a source vocabulary.
Discussion
The UMLS had a low coverage of 44% in representing outcome elements from RCT abstracts. Even when considering mappings that were partially mapped solely due to a missing temporal component or numeric value to be full mappings, the UMLS coverage only increases 5%, to 49%. Furthermore, around half of the partial mappings missed a substantive component of the outcome concept (not temporal or numeric value related). Finally, a large percentage of the mappings (20%) were found to be unmapped by MetaMap. This indicates that there is great room for improvement of the UMLS to better represent outcome elements. Instances in which outcome concepts are absent from the UMLS are likely to have the greatest impact on downstream evidence synthesis and indicate improvements are necessary in biomedical source vocabularies that comprise the UMLS.
Of the fully mapped outcome elements, 67% were complex, indicating that there was not a single, atomic UMLS concept to represent the outcome element. Complex outcomes can fall into 2 categories, one of which is known as post-coordination. Post-coordination indicates that the individual UMLS concepts that comprise an outcome concept can be combined to fully capture the outcome concept. An example of post-coordination occurs with the outcome concept “ADHD symptom control” which maps to UMLS concepts “Attention deficit hyperactivity disorder” (C1263846) and “Symptom control (regime/therapy)”(C1274136). However, complex outcomes can also consist of semantic elements that necessitate a more intensive approach than simple combination of their corresponding UMLS concepts.26 For example, the outcome concept “greater than or equal to 20% decreases from the baseline in the lesion area” can be mapped to the UMLS concepts “Greater than” (C0439093), “Equal” (C0205163), “20%” (C3842589), “Decrease” (C0547047), “Baseline” (C1442488), “Lesion” (C0221198), and “Area” (C0205146). However, additional efforts are required to incorporate the UMLS concepts such that they capture the meaning of the outcome concept due to the semantic elements of Boolean and defining connectors.26 Thus, within the overarching designation of “complex” there may be a wide range of UMLS concept manipulation required.
SNOMED-CT was a source in only 61% of the fully mapped MetaMap CUIs, which is not surprising considering its role as a reference terminology for clinical concepts specifically. However, it can serve as a warning to those wishing to standardize PICO elements amongst increasing trends of SNOMED-CT usage for concept representation. While SNOMED-CT certainly has a role in representing outcome concepts, the UMLS, with a Metathesaurus of over 100 source vocabularies is much better suited to handle the variability and complexity of outcome elements.
A large percentage of unmapped concepts contained abbreviations. Word sense disambiguation in MetaMap takes into consideration the surrounding text to produce the best possible mappings, but in our case the only text presented to MetaMap was the outcome element itself.27 This led to difficulty in mapping particularly in cases where there was no text surrounding the abbreviation. To better understand the effect of surrounding context, the top 5 unique unmapped abbreviations (AEs, TEAEs, HbA1c, OS, and ORR) were used as input to MetaMap within the first sentence from the most recent PubMed abstract in which they appeared. Sentences that contained both the abbreviation and its full form were not considered. All 3 of the unmapped error designations (MetaMap error, UMLS synonymy error, and UMLS absence error) were present across these 5 abbreviations. Including the surrounding context did not result in full mappings of the abbreviation in any of the 5 cases. Thus, more work needs to be done to discover when including context surrounding an abbreviation improves MetaMap word sense disambiguation and thus mappings. Additionally, oftentimes abbreviations are not expanded within the abstract of a paper. Therefore, our methods may misrepresent the power of MetaMap and the UMLS in cases where the full form is present in the UMLS and the abbreviation is not.
There were several other limitations to this work. First, we allowed for nonsubstantive components of the outcome element to be mapped incorrectly. For example, the outcome element “overall hypoglycemia” mapped to “overall publication type” and “hypoglycemia” and was designated as fully mapped even though the term “overall” was not correctly mapped. This potentially inflated the coverage score for the UMLS. Second, the temporal component was not considered to be fully represented if only the unit of time was present. For example, “day” was not considered to be representative of “day 8.” However, as mentioned above, inclusion of these instances would alter the coverage by less than 5%. Third, the 2018 version of MetaMap was used as it is the most up to date version compatible with a Mac. However, the abstracts were collected from clinical trials up to 2021, so certain abbreviations like COVID-19 were not recognized by MetaMap. Lastly, the mapping was performed by 1 researcher without expertise in the many disease domains represented, and therefore though research was performed to confirm mapping designations, there could be error in the mapping designation of more complex terms.
In addition to exploring the effect of the surrounding context on outcome element phrase mappings, future work includes performing the normalization process on the other PICO elements (PIC) and collaborating with domain experts, particularly in the areas of Metabolism and Nutrition, and Infections and Infectious Diseases, to incorporate missing terms into the UMLS and improve UMLS synonymy of existing concepts.
Conclusion
This work provides insights into the UMLS and SNOMED-CT suitability in representing outcome elements and points to areas of improvement in the UMLS for supporting the normalization of outcome information. The UMLS has great potential for representing PICO elements from RCT abstracts, and improvement of its coverage of these elements will be essential for normalization, which will in turn enable more efficient evidence search and retrieval without loss of information.
Supplementary Material
Acknowledgments
We would like to thank those individuals who provided feedback for our annotation guideline: Noémie Elhadad, Carol Friedman, Soojin Park, Hua Xu, Jordan Nestor, Ali Soroush, Jason Zucker, Steven Shea, Krzysztof Kiryluk, and Elizabeth Park.
Contributor Information
Abigail Newbury, Department of Biomedical Informatics, Columbia University, New York City, NY 10032, United States.
Hao Liu, Department of Biomedical Informatics, Columbia University, New York City, NY 10032, United States.
Betina Idnay, Department of Biomedical Informatics, Columbia University, New York City, NY 10032, United States.
Chunhua Weng, Department of Biomedical Informatics, Columbia University, New York City, NY 10032, United States.
Author contributions
A.N. contributed to conceptualization, methodology, software, data curation, investigation, formal analysis, visualization, and writing—original draft, reviewing, and editing. H.L. and B.I. contributed to methodology and data curation. C.W. contributed to idea initialization, conceptualization, methodology, investigation, research supervision, funding acquisition, writing—reviewing and editing. All authors approved the final manuscript.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work was supported by NLM (grant number R01LM009886).
Conflicts of interest
None declared.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Richardson WS, Wilson MC, Nishikawa J, et al. The well-built clinical question: a key to evidence-based decisions. ACP J Club. 1995;123:A12. [PubMed] [Google Scholar]
- 2. Wallace BC, Dahabreh IJ, Schmid CH, et al. Modernizing the systematic review process to inform comparative effectiveness: tools and methods. J Comp Eff Res. 2013;2(3):273-282. [DOI] [PubMed] [Google Scholar]
- 3. Nye B, Li JJ, Patel R, et al. A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers).Association for Computational Linguistics; 2018:197-207. doi: 10.18653/v1/P18-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zlabinger M, Andersson L, Hanbury A, et al. Medical entity corpus with PICO elements and sentiment analysis. In: Proceedings of the Eleventh International Conference on Language Resources and Evaluation (LREC 2018). European Language Resources Association (ELRA; ); 2018. Accessed December 5, 2022. https://aclanthology.org/L18-1044 [Google Scholar]
- 5. Sanchez-Graillet O, Witte C, Grimm F, et al. An annotated corpus of clinical trial publications supporting schema-based relational information extraction. J Biomed Semantics. 2022;13(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boudin F, Nie J-Y, Bartlett JC, et al. Combining classifiers for robust PICO element detection. BMC Med Inform Decis Mak. 2010;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jin D, Szolovits P. PICO element detection in medical text via long short-term memory neural networks. In: Proceedings of the BioNLP 2018 Workshop. Association for Computational Linguistics; 2018:67-75. doi: 10.18653/v1/W18-2308 [DOI] [Google Scholar]
- 8. Todd JJ, Lawal TA, Witherspoon JW, et al. Randomized controlled trial of N-acetylcysteine therapy for RYR1-related myopathies. Neurology 2020;94(13):e1434-e1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. About PCORI. About PCORI | PCORI. 2021. Accessed October 6, 2022. https://www.pcori.org/about/about-pcori
- 10. COMET Initiative | Home. Accessed December 13, 2022. https://www.comet-initiative.org/
- 11.Bai Y, Sun H, Du J. A PICO-based Knowledge Graph for Representing Clinical Evidence. In: Zhang C, Mayr P, Lu W, et al., eds. Proceedings of the 2nd Workshop on Extraction and Evaluation of Knowledge Entities from Scientific Documents (EEKE 2021) co-located with JCDL 2021, Virtual Event, September 30th, 2021.CEUR-WS.org 2021:58–65. https://ceur-ws.org/Vol-3004/paper8.pdf. [Google Scholar]
- 12. UMLS – Metathesaurus Vocabulary FAQ. Accessed October 6, 2022. https://www.nlm.nih.gov/research/umls/knowledge_sources/metathesaurus/source_faq.html
- 13. Jing X. The Unified Medical Language system at 30 years and how it is used and published: systematic review and content analysis. JMIR Med Inform. 2021;9(8):e20675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang E, Mostafa J.. The use of SNOMED CT, 2013-2020: a literature review. J Am Med Inform Assoc. 2021;28(9):2017-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Friedlin J, Overhage M.. An evaluation of the UMLS in representing corpus derived clinical concepts. AMIA Annu Symp Proc. 2011;2011:435-444. [PMC free article] [PubMed] [Google Scholar]
- 16. Sayers E. E-Utilities Quick Start. National Center for Biotechnology Information; 2018. Accessed December 11, 2022. https://www.ncbi.nlm.nih.gov/books/NBK25500/ [Google Scholar]
- 17. Liu H, Carini S, Chen Z, et al. Ontology-based categorization of clinical studies by their conditions. J Biomed Inform. 2022;135:104235. [DOI] [PubMed] [Google Scholar]
- 18. Demner-Fushman D, Mork JG, Shooshan SE, et al. UMLS content views appropriate for NLP processing of the biomedical literature vs. clinical text. J Biomed Inform. 2010;43(4):587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rios A. pymetamap. 2022. Accessed January 22, 2023. https://github.com/AnthonyMRios/pymetamap
- 20. Word Sense Disambiguation Server (WSD Server). Accessed December 16, 2022. https://lhncbc.nlm.nih.gov/ii/tools/MetaMap/additional-tools/WSDServer.html
- 21. pattern-en. Accessed December 13, 2022. http://digiasset.org/html/pattern-en.html
- 22. String – Common String Operations. Python documentation. Accessed December 13, 2022. https://docs.python.org/3/library/string.html
- 23. UMLS API Home. Accessed December 13, 2022. https://documentation.uts.nlm.nih.gov/rest/home.html
- 24. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22:276-282. [PMC free article] [PubMed] [Google Scholar]
- 25. Craggs R, Wood MM.. Evaluating discourse and dialogue coding schemes. Comput Linguist. 2005;31(3):289-296. [Google Scholar]
- 26. Chen Z, Fang Y, Liu H, et al. Data-driven modeling of randomized controlled trial outcomes. Stud Health Technol Inform. 2022;294:392-396. [DOI] [PubMed] [Google Scholar]
- 27. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.