Abstract
Significance:
Glioblastoma is an aggressive and devastating brain tumor characterized by a dismal prognosis and resistance to therapeutic intervention. To support catabolic processes critical for unabated cellular growth and defend against harmful reactive oxygen species, glioblastoma tumors upregulate the expression of wild-type isocitrate dehydrogenases (IDHs). IDH enzymes catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), NAD(P)H, and CO2. On molecular levels, IDHs epigenetically control gene expression through effects on α-KG-dependent dioxygenases, maintain redox balance, and promote anaplerosis by providing cells with NADPH and precursor substrates for macromolecular synthesis.
Recent Advances:
While gain-of-function mutations in IDH1 and IDH2 represent one of the most comprehensively studied mechanisms of IDH pathogenic effects, recent studies identified wild-type IDHs as critical regulators of normal organ physiology and, when transcriptionally induced or down regulated, as contributing to glioblastoma progression.
Critical Issues:
Here, we will discuss molecular mechanisms of how wild-type IDHs control glioma pathogenesis, including the regulation of oxidative stress and de novo lipid biosynthesis, and provide an overview of current and future research directives that aim to fully characterize wild-type IDH-driven metabolic reprogramming and its contribution to the pathogenesis of glioblastoma.
Future Directions:
Future studies are required to further dissect mechanisms of metabolic and epigenomic reprogramming in tumors and the tumor microenvironment, and to develop pharmacological approaches to inhibit wild-type IDH function. Antioxid. Redox Signal. 39, 923–941.
Keywords: glioma, isocitrate dehydrogenases, metabolism, cancer immunology, small molecule inhibitor
Introduction
The reprogramming of cellular metabolism disrupts normal organ physiology and contributes to or causes many human diseases, including cancer (Hanahan and Weinberg, 2011; Yin et al., 2017). Cancer cells undergo extensive metabolic rewiring to support unabated tumor growth and progression. In 1902, Theodor Boveri hypothesized that human cancers are caused by genomic aberrations converting normal cells into incessantly dividing tumor cells. By extension, in the 1920s, Otto Warburg proposed that tumor cells exhibit defects in mitochondrial energy metabolism to adapt to the increased demands of limitless proliferation (Pavlova et al., 2022).
Warburg observed that even in the presence of abundant oxygen, cancer cells rely on enhanced glucose uptake and conversion to pyruvate (aerobic) or lactic acid (anaerobic) to fuel macromolecular synthesis and generate energy in the form of adenosine 5′-triphosphate (ATP). Boveri's concepts of cell cycle checkpoints, oncogenes, and tumor suppressors and Warburg's idea of metabolic adaptation to cancerous transformation ignited the field of cancer genetics and metabolism to elucidate the molecular underpinnings of metabolism-driven tumor initiation and progression. Today, 100 years after the seminal discoveries of Boveri and Warburg, the idea that tumorigenesis and altered metabolism are intertwined (“metabolic transformation”) is more relevant than ever.
Cancer cells can efficiently take up and utilize nutrients for energy production and biomass expansion. Such metabolic adaption, in turn, profoundly affects gene expression and associated differentiation processes. In the past decade, cancer metabolism witnessed a renaissance and renewed interest. Research revealed that cancer metabolic rewiring, particularly in glioblastoma, is far more complex than initially postulated, and extends beyond changes in glycolytic flux.
Research over the past decade added immensely to our understanding of cancer metabolism and identified many changes in different metabolic pathways, including enhanced glutamine catabolism, heightened glucose oxidation through the tricarboxylic acid (TCA) cycle, and increased lipid biosynthesis, as contributing to or driving malignant phenotypes (Tommasini-Ghelfi et al., 2019; Vander Heiden and DeBerardinis, 2017).
The identification of neomorphic mutations in the isocitrate dehydrogenase (IDH) family members IDH1 and IDH2 over a decade ago, together with recent studies that identified the transcriptional upregulation of wild-type IDHs as a tumor-promoting event, helped further fuel cancer metabolism research (Yan et al., 2009; Yang et al., 2012). As wild-type IDHs play a fundamental role in energy metabolism, redox balance, and anaplerosis, metabolism research has begun to investigate the role of the IDH family of enzymes more comprehensively, unraveling their contribution to cancer initiation, progression, and therapy resistance.
Among the many functions of wild-type IDHs, the regulation of redox homeostasis has been identified as one of the enzymes' most critical protumorigenic activities. Cancer cells present with heightened levels of reactive oxygen species (ROS) (Liou and Storz, 2010). These ROS are derived from oxygen and exist as highly reactive free radicals. ROS are capable of oxidizing molecules and macromolecular structures either to the benefit or to the detriment of cancer cell survival (Glasauer and Chandel, 2013; Sies et al., 2022).
Tumor cells are, therefore, under constant pressure to maintain an auspicious equilibrium between the tumor-promoting and tumor-antagonizing effects of oxidative stress. Due to the high basal metabolic rate and fatty acid content in many different cancer types, in particular those that grow within the central nervous system (CNS), including glioblastoma, changes in redox balance are especially critical for their pathogenesis and progression (Belanger et al., 2011; Gopal et al., 1963).
Research in the past few years pointed to wild-type IDHs as metabolic enzymes that initiate, propagate, and protect glioblastoma through their effect on oxidative stress responses. In this review, we will discuss the ROS scavenging properties of wild-type IDHs, the effects of wild-type IDH overexpression on gene expression and key metabolic pathways, including de novo lipid biosynthesis and one-carbon metabolism, and provide an outlook on future research strategies to fully understand and therapeutically target oncogenic mechanisms of wild-type IDHs.
Grade 4 Malignant Glioma: Phenotypes, Genotypes, and Mechanisms of Progression
Glioblastoma is a devastating, invariably lethal brain tumor characterized by poor prognosis and resistance to therapeutic intervention (Lukas et al., 2019). Glioblastoma is the most common subset of a heterogeneous group of brain tumors known as gliomas, their name reflecting a presumed glial progenitor cell-of-origin (McKinnon et al., 2021). Classified as a World Health Organization (WHO) grade 4 astrocytoma, the median survival time for glioblastoma is a mere 14–16 months after diagnosis (McKinnon et al., 2021; Tan et al., 2020). Glioblastoma presents with high mitotic indices, vascular proliferation, diffuse invasion into the normal brain parenchyma, extensive intratumoral necrosis, and resistance to extant therapies (Olar and Aldape, 2014).
Glioma-initiating cells as drivers of glioblastoma phenotypes
The most critical factor impacting patient survival is the invariable recurrence of glioblastoma after surgical resection and therapeutic intervention, typically within 2–3 cm of the initial tumor site (Campos et al., 2016; Jue and McDonald, 2016). Recurrent glioblastoma is thought to arise from a multipotent population of glioblastoma stem or stem-like cells, which are characterized by heightened therapy resistance (Bao et al., 2006). Also known as glioma-initiating cells (GICs), they have acquired the stem-like properties of multilineage differentiation and self-renewal capabilities.
GICs were first isolated based on the expression of the neural stem cell (NSC) surface marker CD133 (Singh et al., 2003). Upon orthotopic implantation into immunocompromised mice, CD133+ GICs give rise to tumors with phenotypic hallmarks of the human disease (Lathia et al., 2015), including resistance to chemotherapy and radiation therapy (RT) (Bao et al., 2006; Chen et al., 2012). Differentiation of GICs along astroglial and neuronal axes induced by bone morphogenetic proteins resulted in decreased proliferation and suppression of tumor growth, demonstrating that differentiation reduces the tumor-propagating potential of GICs (Piccirillo et al., 2006).
Dedifferentiation of GICs into induced pluripotent cells followed by differentiation into specific cellular lineages alters their tumorigenic potential, suggesting that developmental programs and genetic alterations define the fundamental properties of GICs in gliomas (Stricker et al., 2013). Further underscoring the importance of neurodevelopmental reprogramming in GICs, by expressing a core set of neurodevelopmental transcription factors (SOX2, OLIG2, POU3F2, and SALL2), differentiated glioblastoma cells can be fully reprogrammed into GICs (Suva et al., 2014).
Badhuri et al. (2020) created a glioblastoma tumor cell atlas using single-cell transcriptomics of cancer cells. When mapped onto a reference framework of the developing and adult human brain, the study identified multiple GIC subtypes that coexist within a single tumor, including a highly infiltrative subpopulation that recapitulates the hallmarks of outer radial glia (oRG) cells (Bhaduri et al., 2020). The oRG-like identity was controlled by the expression of the oRG cell surface marker protein tyrosine phosphatase receptor type Z1 (PTPRZ1) (Pollen et al., 2015).
Orthotopic transplantation of PTPRZ1-positive cells resulted in glioblastoma formation in mice, with tumor-derived oRG-like cells undergoing mitotic somal translocation. As mitotic somal translocation is not known to occur outside CNS development, the presence of oRG-like GICs highlights the importance of reactivating neurodevelopmental pathways during glioblastoma tumorigenesis that are typically inactive in adulthood.
Hypoxic conditions are necessary for the maintenance and propagation of normal stem cell populations within the brain (Pistollato et al., 2007) and similarly, fuel the propagation, maintenance, and transdifferentiation of GICs (Lathia et al., 2015). The role of hypoxia in propagating glioblastoma progression was first suggested by the presence of microvascular hyperplasia and palisading necrosis (Monteiro et al., 2017). Pseudopalisades, or hypercellular zones, include hypoxic cells that, though less proliferative than adjacent astrocytoma cells, are highly motile and migrate away from hypoxic centers (Brat et al., 2004).
Secreted factors from the pseudopalisading cells, such as vascular endothelial growth factors (VEGFs), promote the recruitment of endothelial cells to support microvascular proliferation and cellular invasion; however, the vasculature is often abnormal and consists of endothelial cells displacing the pericytes that provide mechanical stability and support to the microvasculature (Carmeliet and Jain, 2011; Kaur et al., 2005). This localized disruption of the microvasculature interferes with oxygen distribution, and thus extends areas of hypoxia and necrosis.
As a result, hypoxia-inducible factor 1 (HIF-1) expression is induced within palisading cells, which, in turn, transcriptionally controls oxidative stress response and proangiogenic genes (Kaur et al., 2005). In addition to promoting tumor angiogenesis, GIC-associated expression of HIF-1 activates the phosphatidylinositol 3-kinase-Akt and extracellular signal-regulated kinase 1 and 2 pathways to drive CD133+ GIC expansion (Soeda et al., 2009).
Strikingly, HIF-1 activation within normoxic glioma cells is sufficient to drive the expression of CD133 and promotes a more stem-like state (Bar et al., 2010). Conversely, sustained expression of HIF-1 elicits transdifferentiation of GICs into endothelial cells via enhanced VEGF signaling, underscoring the delicate dichotomy between GIC maintenance and differentiation (Soda et al., 2011; Xiong et al., 2021). Indeed, recent studies have identified hypoxia as a potent intrinsic factor that promotes GIC plasticity, enabling GICs to transition from one cellular state to another (Dirkse et al., 2019). Whereas HIF-1 expression is not restricted to GICs, HIF-2 is preferentially expressed within GIC populations to regulate their tumorigenic properties (Li et al., 2009).
Mechanistically, HIF-2 induces the expression of specific cancer stem cell-related gene signatures, including components of the Notch and calcineurin pathways (Seidel et al., 2010). Expression of HIF-1 and HIF-2 allows GICs to effectively respond to oxidative stress, and facilitates their maintenance and expansion.
Glioblastoma invasion
One of the predominant clinical hallmarks of glioblastoma tumors is extensive infiltration into the adjacent brain parenchyma (Claes et al., 2007). These invasive properties of glioblastoma were first described in a 1938 publication by Scherer (1938).
Scherer (1938) described gliomas migrating along defined brain structures and infiltrating neighboring tissue. He further noted that glioma cells tend to change morphologically, adapting their physical shape to assume that of the brain region they infiltrate. Since his seminal work, investigators have demonstrated that glioblastoma migrate away from tumor bulk hypoxic niches, traveling along white matter tracts and basement membranes of blood vessels into the brain parenchyma (Cuddapah et al., 2014; Westphal and Lamszus, 2011).
Contrary to other high-grade cancers, which intravasate into the blood and lymphatic vessels, glioblastoma cells travel along such scaffolding. This process is reminiscent of radial glia asymmetric self-renewal and migration during cortical development (Hansen et al., 2010; Hatoum et al., 2019).
Indeed, several genes typical of neurodevelopment are repurposed for invasion. Myosin II, for example, is implicated in somal translocation within radial glia and plays critical roles during brain development by relocating microtubules toward the leading edge of protrusions (Tsai et al., 2007). Myosin II is similarly upregulated during glioblastoma invasion, facilitating leading-edge migration and invasion into neighboring brain regions (Osswald et al., 2015).
With the recent identification of an oRG-like population of cells that persists in adult glioblastoma patients (Bhaduri et al., 2020), mentioned above, it is conceivable that a radial glia-like population of cell extends protrusions into neighboring brain regions and drive glioma infiltrative properties (Qin et al., 2017; Shi et al., 2017).
Grade 4 Gliomas: A Disease of IDH1 Expression and Mutation
Historically, the neuro-oncology field classified glioblastomas into two major subtypes: primary (de novo) and secondary glioblastoma. With a more detailed understanding of the importance of IDHs in the pathogenesis and molecular diagnosis of gliomas, the WHO has recently reclassified these two manifestations of grade 4 gliomas as IDH-wildtype glioblastoma and grade 4 IDH-mutant astrocytoma (Louis et al., 2016). Wild-type IDH1 and IDH2 enzymes catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG), NADPH, and CO2.
Mutations in IDH1 and IDH2 are neomorphic gain-of-function mutations, which affect cofactor binding affinity and the conformation of the enzymes' active center. Arg100 and Arg132 in IDH1, and Arg140 and Arg172 in IDH2 form hydrogen bonds with the α-carboxyl and β-carboxyl groups of isocitrate. When mutated, the enzymes' binding affinity to isocitrate decreases, while affinity to NADPH increases.
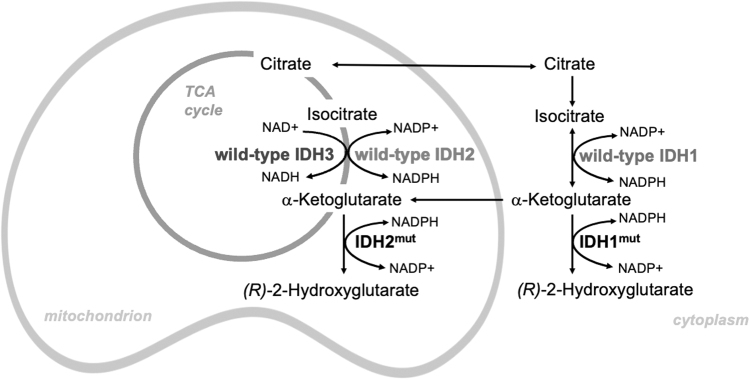
As a consequence, mutations abrogate the IDH forward reaction; that is, the oxidative decarboxylation of isocitrate to α-KG (Xu et al., 2004), and due to the ensuing conformational change of the active center, result in the catalysis of a partial reverse reaction, in which α-KG is reduced to (R)-2-hydroxyglutarate [(R)-2HG], but not further carboxylated (Dang et al., 2009). All neomorphic IDH-mutant enzymes produce (R)-2HG (Fig. 1).
FIG. 1.
Subcellular localization and chemical reactions catalyzed by wild-type IDH and tumor-derived IDH mutant. IDH, isocitrate dehydrogenase. Color images are available online.
Wild-type IDH glioblastoma account for ∼90% of all grade 4 astrocytoma cases. These tumors present as a grade 4 glioma without previous indication of a lesser malignant tumor, and are typically diagnosed in an older patient population (Ohgaki and Kleihues, 2005). Expanding the understanding of pathogenic wild-type IDH1 function, two recent studies revealed that 2/3 of high-grade glioma (HGG) overexpress wild-type IDH1 mRNA, as defined by having a fold change >1.5 in comparison with normal brain tissue (Calvert et al., 2017; Wahl et al., 2017).
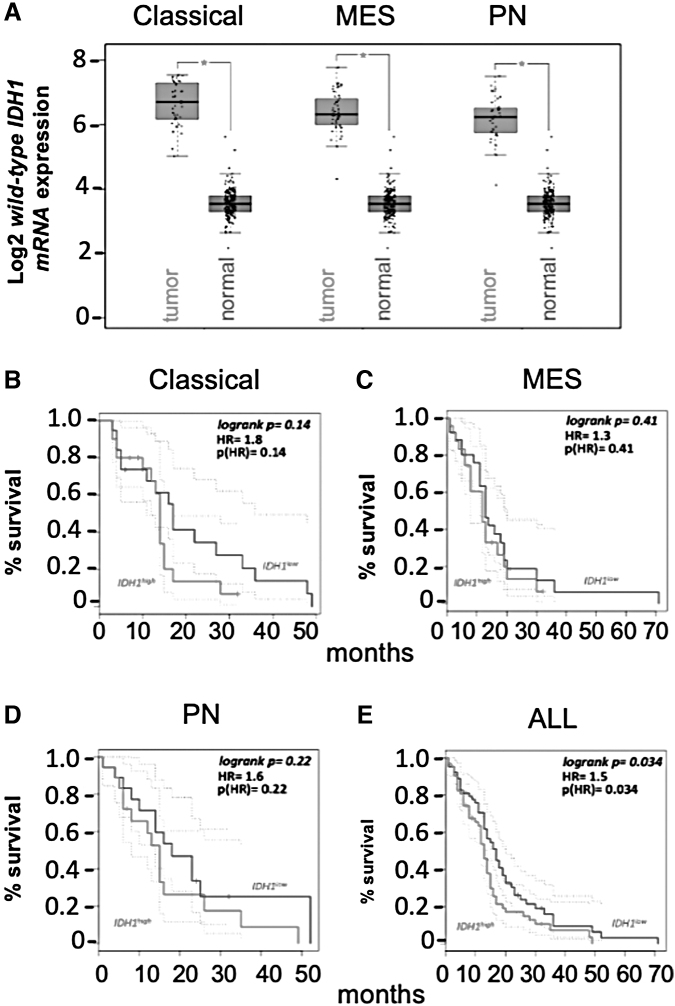
IDH1 point mutation and wild-type IDH1 transcriptional upregulation were mutually exclusive in this analysis (Calvert et al., 2017). Wild-type IDH1 overexpression was observed across different genetic subclasses of glioblastoma (Fig. 2A) and inversely correlated with patient survival (Fig. 2B–E). Levels of wild-type IDH1 mRNA varied according to tumor cellular differentiation, transcriptome-defined subclassification, and tumor grade, with grade 4 gliomas showing the most robust wild-type IDH1 expression.
FIG. 2.
Wild-type IDH1 is overexpressed in glioblastoma. (A) TCGA analysis of wild-type IDH1 expression in classical, MES, and PN glioblastoma tumors. *p < 0.05. (B–E) Glioblastoma patient survival as a function of wild-type IDH1 expression. The tumors were stratified based on median wild-type IDH1 expression, with cutoff high and low percentage set at 50%. The dotted lines represent the 95% confidence intervals for the high group and low groups. HR, hazard ratio; MES, mesenchymal; PN, proneural; TCGA, The Cancer Genome Atlas. Color images are available online.
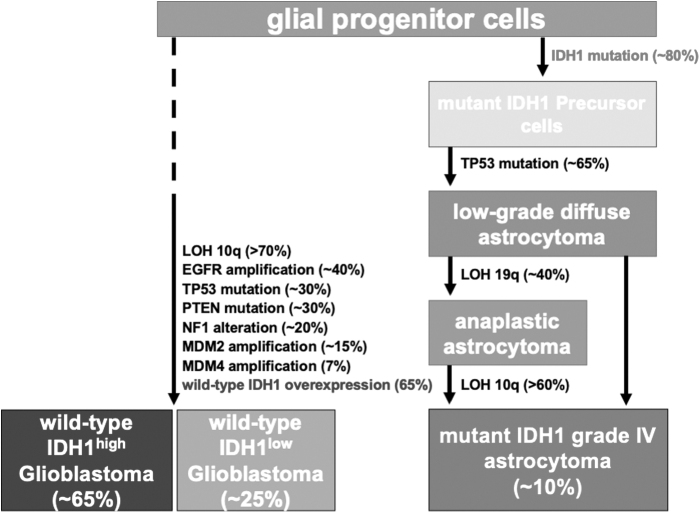
Reflecting increases in wild-type IDH1 transcript level, elevated wild-type IDH1 protein expression was also evident through immunohistochemical analysis (Calvert et al., 2017). These findings point to distinct subtypes of HGG: low and high expressers of wild-type IDH1 (wild-type IDH1low, wild-type IDH1high, respectively) and mutant IDH1 grade 4 astrocytoma (Fig. 3) (Calvert et al., 2017).
FIG. 3.
The genetics of wild-type-IDH glioblastoma versus mutant IDH grade 4 astrocytoma. Major subtypes of adult gliomas. Based upon the IDH mutational status, adult diffuse gliomas are classified into two subgroups: wild-type and mutant IDH gliomas. LOH, loss of heterozygosity. Color images are available online.
Wild-type IDH tumors present later in life and display widespread anatomical distribution within the brain (Brennan et al., 2013; Parsons et al., 2008; Wang et al., 2018). Grade 4 IDH-mutant astrocytomas, on the contrary, arise from a lower grade glioma in younger patients and account for the remaining 10% of cases. These grade 4 gliomas are less aggressive and have a median survival time of 7.8 months without intervention, compared with 4.7 months reported for wild-type IDH glioblastoma (Ohgaki and Kleihues, 2005). Due to their distinct clinical and pathological presentation, wild-type IDH glioblastoma and grade 4 IDH-mutant astrocytoma tumors are considered separate disease entities (Fig. 3) (Cloughesy et al., 2014; Ohgaki and Kleihues, 2013).
IDH-mutant grade 4 gliomas are predominantly located in frontal lobe structures and maintain a gene signature associated with the proneural subtype, as discussed below. In contrast, wild-type IDH tumors show a more widespread anatomical distribution and display genetic features common to all transcriptionally defined subtypes of high-grade glioma (Lai et al., 2011). While region-specific differences in the tumor microenvironment likely influence tumor phenotypic and molecular characteristics, the concept of distinct progenitor cell-of-origin, as a contributing factor underlying differences between IDH1 wild-type and mutant tumors, has gained traction (Alcantara Llaguno et al., 2015; Zong et al., 2015).
The restricted phenotypic and spatial presentation of mutant IDH1 gliomas, combined with similarities these tumors share with fetal and adult brain parenchyma, is consistent with their origin from a lineage-committed precursor with limited differentiation potential. Such cells are abundant at a specific stage and location in forebrain development (Lai et al., 2011).
Together with the presence of oligodendroglial histologic features, the findings suggest that oligodendrocyte precursor cells (OPCs) may represent the cell-of-origin for mutant IDH1 grade 4 astrocytoma (Lai et al., 2011). Although IDH mutation and associated CpG island methylator phenotype together with p53 mutation may occur in quiescent NSCs (cell-of-mutation), tumors can arise from a lineage-committed progeny (cell-of-origin), such as OPCs, after proliferative expansion related to forebrain maturation.
Mosaic analysis with double markers in a genetically engineered murine glioblastoma model confirmed that mutations initially induced NSCs can lie dormant and only trigger malignant transformation after differentiation into OPCs. These findings suggest that mutation may initially occur in either NSCs or OPCs, and only OPCs provide the proper cellular context needed for transformation (Liu et al., 2011).
Further supporting OPC as a contributor to the pathogenesis of grade 4 mutant IDH1 astrocytoma, OPCs can generate protoplasmic astrocytes (Nishiyama et al., 2009), which may allow bipotent OPCs to give rise to brain tumors with astrocytic features. As a result of OPC transformation, low-grade gliomas develop, and through acquiring additional genomic aberrations these tumors progress into grade 4 astrocytomas.
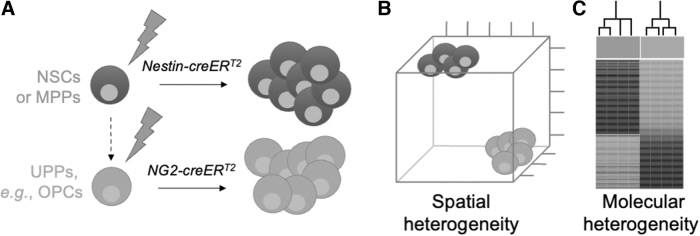
Compared with mutant IDH1 tumors, wild-type IDH1 glioblastoma show greater variability in anatomical location, histopathologic presentation, and genetic profiles, which, together with their molecular resemblance to murine NSCs, point to NSCs as the cell-of-origin (Lai et al., 2011). Using Nestin-creERTM, Ascl1-creERTM, and neuron-glial antigen 2 (NG2)-creERTM transgenes that drive recombination selectively in NSCs, or lineage-restricted CNS progenitor cells, Parada and colleagues demonstrated that distinct glioblastoma-initiating cell populations, in the setting of identical driver mutations, give rise to different subtypes that can be distinguished on histopathologic and molecular levels.
The tumors showed associations with varying types of transcriptomically defined glioblastoma. Furthermore, they had a distinct histopathologic presentation, including varying degrees of necrosis and proliferation, and maintained gene expression profiles specific to the progenitor cell type from which they originate (Alcantara Llaguno et al., 2015). These studies support the idea that glioblastoma tumors have a “cell-of-origin memory” that defines tumor pheno- and genotypes (Fig. 4) (Alcantara Llaguno and Parada, 2016).
FIG. 4.
Progenitor cell-of-origin defines spatial and molecular presentation of glioblastoma. (A) Using inducible Cre recombinases driven by progenitor-specific promoter elements, such as nestin (NSC-selective) or NG2 (OPC-specific), progenitor cell-type–specific expression of wild-type IDHs, in the setting of identical driver mutations, may give rise to different high-grade glioma subtypes. (B, C) These tumors can be distinguished on histopathologic and molecular levels, including spatial presentation and the tumors' association with different transcriptomically defined subtypes. These studies support the idea that glioblastoma tumors have a “cell-of-origin memory” that defines tumor pheno- and genotypes (Alcantara Llaguno and Parada, 2016). Modified from figures 1 and 2 in reference Alcantara Llaguno et al. (2016). NG2, neuron-glial antigen 2; NSC, neural stem cell; OPC, oligodendrocyte precursor cell. Color images are available online.
When expressed in p53-deficient murine NSCs, mutant IDH1 inhibits progenitor cell growth in vitro, reduces glioma formation in vivo, and increases overall animal subject survival (Chen et al., 2014). IDH1R132H growth-inhibitory effect is due to diversion of α-KG from wild-type IDH1, and reduced carbon flux from glucose or glutamine into lipids. Replenishment of α-KG through glutaminolysis compensates for these growth and flux deficiencies (Chen et al., 2014), suggesting that mutant IDH1 tumors require a specialized metabolic niche characterized by elevated glutamate flux for growth and expansion.
Such a niche is provided by frontal lobe neocortical structures, where the majority of IDH-mutant tumors occur. Wild-type IDH1 glioblastoma, on the contrary, are unable to sustain high glutamine flux to support α-KG production and lipid biogenesis (Mashimo et al., 2014). To support anaplerosis in the absence of efficient glutamate catabolism, increased levels of wild-type IDH1 may help sustain macromolecular synthesis via enhanced α-KG and NADPH production, and in doing so, support glioblastoma growth.
Reflecting distinct metabolic wiring in mutant IDH1 versus wild-type IDH1 high-grade tumors, NSC and OPC progenitor cells also exist in specific niches within the brain, and demonstrate metabolic adaptability to specific and changing microenvironments (Roth and Nunez, 2016; van Tilborg et al., 2018). OPCs represent the most abundant and widely distributed population of cycling cells in the adult brain. OPCs differentiate into oligodendrocytes, which produce myelin in the CNS, and express glutamate receptors, the activation of which influences numerous cellular processes.
Glutamate regulates OPC migration, proliferation, and differentiation; maintains progenitor pools; and ensures sustained neuronal myelination by regulating OPC differentiation into mature oligodendrocytes (van Tilborg et al., 2018). In contrast, NSCs are confined to hypoxic subventricular and subgranular zones. NSCs are highly adapted to hypoxia by maintaining low oxidative stress, and show increased expression and activity of rate-limiting enzymes implicated in lipogenesis to sustain anaplerosis that supports stem cell expansion (Bernal and Arranz, 2018).
Recent single-cell RNA-sequencing (scRNA-Seq) studies suggest that IDH-mutant gliomas arise from neural progenitor cells (NPCs) capable of differentiating into astrocytes and oligodendrocytes (Suva and Tirosh, 2020). Although the precise cell-of-origin continues to be subject to debate, studies found that increasing neural lineage restriction effectively prevents gliomagenesis, indicating stem-like cellular states as most susceptible to tumorigenesis and differentiated cells as least vulnerable (Alcantara Llaguno et al., 2019).
Glioblastoma Genetic Subtypes
Besides IDH mutations, grade 4 gliomas can be classified based on their genomic and transcriptional profiles. There are three transcriptionally defined molecular subtypes of grade 4 gliomas—classical, mesenchymal, and proneural (Verhaak et al., 2010). These subtypes are enriched for specific genetic events. Classical grade 4 tumors harbor the most common genetic mutations: chromosome 10 amplification and chromosome 7 deletions, resulting in epidermal growth factor receptor (EGFR) amplification and phosphatase and tensin homolog (PTEN) loss. EGFR amplification is associated with high expression of the neuronal development-related Notch and sonic hedgehog (SHH) signaling pathways.
Intriguingly, the classical subtype may show more pronounced responsiveness to RT than the other two subtypes (Verhaak et al., 2010). By comparison, the mesenchymal subtype is associated with a high percentage of necrosis and related inflammation, and is characterized, on a molecular level, by frequent mutation/deletion of neurofibromatosis type 1 (NF1) and elevated expression of CHI3L1 and MET (Phillips et al., 2006; Verhaak et al., 2010). Defining features of the proneural subtype include platelet-derived growth factor receptor A (PDGFRA) amplification and TP53 mutations, as well as genes that drive the oligodendroglial axis during early tumor formation (Jackson et al., 2006; Verhaak et al., 2010).
Despite gliomas being classified into one predominant subtype, multiregional sampling has revealed that individual tumors are comprised of populations of diverse molecular subtypes (Patel et al., 2014), with longitudinal molecular profiling demonstrating the propensity of tumor cells to change subtype over time (Wang et al., 2018).
Advances in single-cell genomics have enhanced our understanding of intratumoral heterogeneity and tumor cell plasticity. Full-length scRNA-Seq sequencing of 20 adult and 9 pediatric glioblastoma samples identified transcriptional states that drive glioblastoma malignant cell heterogeneity (Neftel et al., 2019). The study demonstrated that glioblastoma tumor cells exist in four distinct cellular states that reflect different gene expression signatures reflective of specific neural cell types; that is, NPC-like, OPC-like, astrocyte cell (AC)-like, and mesenchymal (MES) cell-like states.
The distribution of these cell types varied drastically between each sampled tumor. Each cell type was associated with the key molecular alterations; that is, EGFR amplification in AC-like, NF1 mutations in MES-like, CDK4 amplification in NPC-like, and PDGFRA amplification in OPC-like states. Importantly, any cell state can give rise to tumors comprised of all four states, highlighting the high degree of plasticity of glioblastoma tumor cells.
The identification of glioma molecular subtypes also provided important insight into our understanding of how grade 4 gliomas reappropriate prototypical neurodevelopment pathways. For example, grade 4 mutant IDH astrocytoma are predominantly of the proneural subtype and often have amplification of PDGFRA as previously discussed. PDGFRA is a critical marker of OPCs, and its ligand, PDGF, drives OPC proliferation (Calver et al., 1998; Pringle and Richardson, 1993), further supporting the notion that OPCs may represent the cell of origin of grade 4 mutant IDH astrocytomas. In contrast, classical glioblastoma upregulate nestin, the prototypical marker of NSC and NPC populations.
Glioblastoma of the classic subtype also exhibits activation of the SHH pathway, which, in the context of neural development, is responsible for patterning the brain and spinal cord (Dahmane and Ruiz i Altaba, 1999) and regulating adult NPC populations postnatally (Lai et al., 2003). Similarly, activated Notch signaling is essential for maintaining NSC populations in the developing and adult brain (Imayoshi et al., 2010).
The Roles of Wild-Type IDH Enzymes in Glioblastoma Development
In eukaryotes, three IDH paralogs catalyze the oxidative decarboxylation of isocitrate to α-KG for overlapping but nonredundant metabolic cellular processes (Tommasini-Ghelfi et al., 2019). Subcellular localization, cofactor requirements, structural organization, catalytic mechanism, and allosteric regulation differ between paralogs (Fig. 1). IDH1 localizes to both cytosol and the peroxisomes, while IDH2 and IDH3, as part of the TCA cycle, are found within the mitochondrial matrix. IDH1 and IDH2 function as homodimers, use NADP+ as an electron acceptor, and require binding to a divalent metal ion, typically Mn2+ or Mg2+.
IDH3 is a heterotetramer composed of two 37-kDa α subunits (IDH3α), one 39-kDa β subunit (IDH3β), and one 39-kDa γ subunit (IDH3γ) (Bzymek and Colman, 2007). IDH3 catalyzes an irreversible and rate-limiting step of the TCA cycle, which is tightly regulated through substrate availability (citrate, ADP, isocitrate, NAD+, Mg2+/Mn2+), product inhibition (NADH, α-KG), and competitive feedback inhibition (ATP), to avoid unnecessary depletion of isocitrate and accumulation of α-KG (Bzymek and Colman, 2007; Qi et al., 2008).
Wild-type IDH1 promotes glioblastoma progression and therapy through effect on redox balance and de novo fatty acid biosynthesis
To define the impact of wild-type IDH1 on glioblastoma progression, Calvert et al. (2017) conducted a series of gain-of-function experiments using cDNA complementation and RNAi-mediated loss-of-function experiments in patient-derived GICs and murine NSCs. Knockdown of wild-type IDH1 reduced the proliferation of GICs (Calvert et al., 2017), and knockdown cells grow more slowly as orthotopic xenografts compared with cells without a wild-type IDH1 knockdown after intracranial injection in NOD-SCID mice (Calvert et al., 2017). Conversely, overexpression of wild-type IDH1 in luciferase-expressing murine NCS null for p53 and PTEN increased tumor formation and growth after intracranial injection, and reduced animal survival (Calvert et al., 2017).
Wild-type IDH1 promotes lipid biosynthesis
Studies in liver and adipose cells and tissue revealed that wild-type IDH1 controls lipid metabolism due to its ability to produce cytosolic NADPH, the rate-limiting factor for lipogenesis (Koh et al., 2004; Shechter et al., 2003). During de novo fatty acid synthesis, eight acetyl-CoA molecules are converted into palmitic acid in a series of reactions that utilize 14 molecules of NADPH (Carta et al., 2017). The saturated fatty acid palmitic acid is converted into monounsaturated fatty acids (MUFAs) via stearoyl-CoA desaturase (Enoch et al., 1976).
Resultant MUFAs can be further processed into polyunsaturated fatty acids (PUFAs) and phospholipid phosphatidic acid (Koundouros and Poulogiannis, 2020). Conversely, elongation of palmitic acid by fatty acid elongases results in the synthesis of saturated fatty acid stearate to produce more complex lipids (Koundouros and Poulogiannis, 2020; Matsuzaka et al., 2007).
As the de novo fatty acid synthesis requires a vast reservoir of rate-limiting cytosolic NADPH, Calvert et al. (2017) explored the effect of wild-type IDH1 on anaplerotic flux, particularly lipid biosynthesis, by performing targeted metabolomic studies. GICs modified for stable wt-IDH1 knockdown had significantly reduced α-KG and NADPH levels (Calvert et al., 2017). Using liquid chromatography-mass spectrometry–based quantification of free fatty acid species and uniformly 13C6-labeled glucose and 13C2 acetate tracers, the study found the reduction in the NADPH/NADP+ ratio to be associated with diminished de novo saturated fatty acid and MUFA biosynthesis.
Under conditions of hypoxia (Metallo et al., 2012) and anchorage-independent tumor spheroid growth (Jiang et al., 2016), wild-type IDH1 can promote the reductive formation of citrate from glutamine by catalyzing the conversion of α-KG to isocitrate (the “reverse” reaction). Citrate can subsequently be converted to acetyl-CoA (coenzyme A) and then malonyl-CoA, the carbon precursors for de novo lipogenesis.
To determine whether wild-type IDH1, under normoxic conditions, can promote the anaplerotic replacement of acetyl-CoA by stimulating α-KG production (via the “forward reaction”), 13C-label incorporation into acetyl-CoA was assessed. GICs expressing wild-type IDH1 targeting shRNA exhibited elevated levels of 13C2-labeled acetyl-CoA, suggesting that acetyl-CoA accumulates in wild-type IDH1 compromised cells, as it cannot be used for de novo fatty acid synthesis due to limited cytosolic NADPH availability.
Wild-type IDH1 controls redox homeostasis
In addition to supporting de novo lipid biogenesis, cytosolic NADPH is a potent antioxidant playing a pivotal role in combatting oxidative stress and radiation damage. Mice homozygously null for wild-type IDH1 undergo normal pre- and postnatal development, but in contrast to wild-type littermates, are more susceptible to treatment with the endotoxin lipopolysaccharide due to elevated levels of hepatic oxidative stress, pronounced DNA damage leading to apoptosis, and high expression of proinflammatory cytokines. These findings correlated with an increase in the NADP+/NADPH ratio compared with wild-type mice, suggesting that wild-type IDH1's ability to supply cytosolic NADPH is critical for the observed phenotype (Itsumi et al., 2015).
In support of this hypothesis, wild-type IDH1 depletion in vitro in multiple glioblastoma cell lines revealed that cells exhibited heightened sensitivity to ultraviolet radiation and H2O2 due to inefficient recycling of the potent antioxidant reduced glutathione (GSH) and a reduced ability to neutralize ROS (Lee et al., 2002).
In neurons, loss of wild-type IDH1 expression renders cells hypersensitive to oxidative stress due to unabated accumulation of ROS (Yang et al., 2017). When upregulated in glioblastoma, wild-type IDH1 increased the intracellular pool of NADPH and enhanced cellular GSH levels (Calvert et al., 2017). Conversely, grade 4 IDH-mutant astrocytoma, in which mutant IDH1 utilizes NADPH for the production of 2HG, have reduced intracellular NADPH and GSH levels resulting in elevated ROS abundance (Shi et al., 2014).
Wild-type IDH1 controls susceptibility to radiation and targeted therapies
Knockdown of wild-type IDH1 in RT-resistant glioma cell lines with differing p53 mutational status sensitized each cell line to RT with enhancement ratios between 1.3 and 1.6, like the radiosensitization typically induced by temozolomide (Chakravarti et al., 2006; Choi et al., 2014). The radiosensitization conferred by wild-type IDH1 knockdown was mediated by increased ROS as it was nearly entirely reversed by incubation with the antioxidant precursor N-acetyl cysteine (NAC) (Calvert et al., 2017; Wahl et al., 2017). In vivo tumor regression studies using xenograft models carrying a doxycycline-inducible shRNA directed against wild-type IDH1 demonstrated that reduction in wild-type IDH1 expression enhanced RT antitumor effect, as indicated by reduced tumor volume (Wahl et al., 2017).
Inhibition of growth factor receptors, especially EGFR, is known to suppress lipid biosynthesis (Cheng et al., 2018a; Cheng et al., 2018b; Guo et al., 2009) and cooperate with ROS scavengers to reduce GIC survival (Monticone et al., 2014). Analysis of The Cancer Genome Atlas (TCGA) gene expression data showed a significant correlation between wild-type IDH1 and EGFR mRNA expression, suggesting that the combined inhibition of EGFR and wild-type IDH1 may reduce glioma cell survival (Calvert et al., 2017).
To address whether wild-type IDH1, through its impact on lipid biosynthesis and redox balance, modulates cell responses toward erlotinib, Calvert et al. (2017) defined the apoptotic response of GICs with and without amplified EGFR to RNAi-mediated knockdown of wild-type IDH1. Erlotinib treatment increased Annexin V positivity in shIDH1-expressing GICs with amplified EGFR. Treatment of erlotinib-primed cells with cell-permeable α-KG or the fatty acid palmitate plus the cholesterol precursor mevalonate protected cells from the proapoptotic effects of wild-type IDH1 knockdown, suggesting that a reduction of fatty acid and cholesterol biosynthesis is essential for proapoptotic effects of wild-type IDH1 knockdown.
In addition to promoting erlotinib treatment-associated apoptosis by limiting lipid synthesis, wild-type IDH1 knockdown in GICs augmented cellular ROS levels because of a decrease in reduced glutathione and NADPH. Treatment of wild-type IDH1 knockdown cells with the ROS scavenger EUK-134 or NAC reduced effector caspase activation in response to erlotinib treatment, suggesting elevated ROS in GICs with reduced wild-type IDH1 expression contributes to the proapoptotic effects of EGFR inhibition (Calvert et al., 2017).
Wild-type IDHs and the epigenetic control of gene expression
Wild-type IDH1, via production of α-KG, regulates the enzymatic activity of α-KG-dependent oxygenases and, in doing so, influences gene expression and affects a broad spectrum of cellular functions. α-KG-dependent oxygenases catalyze a diverse range of biochemical processes and reactions, including DNA and histone methylation (Tommasini-Ghelfi et al., 2019). These modifications, in turn, regulate a diverse range of cellular processes, including maintenance of pluripotency, hypoxic responses, angiogenesis, inflammation, among many others (Büchler et al., 2003; Gonzalez et al., 2018; Keith and Simon, 2007; Palazon et al., 2014).
Due to their essential roles in regulating cellular homeostasis, α-KG-dependent oxygenase activity, when deregulated, can contribute to various diseases. Histone methylation at Lys and Arg residues results in DNA supercoiling, and modulates gene expression by diminishing accessibility of transcription factors and enhancers to specific chromatin regions (Sullivan and Karpen, 2004). Jumonji C (JmjC) catalytic domain containing α-KG-dependent oxygenases demethylate these histone marks and enhance chromatin accessibility (Tsukada et al., 2006; Walport et al., 2016). These enzymes have proven critical in the regulation of pluripotency.
The JmjC domain protein Jarid2, for example, controls the activity of polycomb repressive complex 2, an important regulator of developmental genes in embryonic stem cells (Peng et al., 2009). In line with this observation, naïve embryonic stem cells exhibit high levels of intracellular α-KG that, in turn, functions to catalyze JmJC-dependent histone demethylation (Carey et al., 2015).
When wild-type IDH1 is silenced, GICs showed increases in trimethylation on H3K4, H3K9, H3K27, and H3K36 (Calvert et al., 2017). Functionally, reduced wild-type IDH1 expression attenuated GIC self-renewal, as evidenced by reduced sphere formation, and resulted in a more differentiated GIC phenotype, as indicated by augmented microtubule-associated protein 2 and glial fibrillary acidic protein expression, markers for neuronal and glial differentiation, respectively.
Chromatin immunoprecipitation-sequencing (ChIP-Seq) experiments, using antibodies recognizing trimethylated H3K27, K36, and K4 proteins, RNA-Seq, together with Spatial Clustering for Identification of ChIP-Enriched Regions and ingenuity pathway analysis identified a tumor suppressor gene signature, including NDUFS1, GNG4, and TNFAIP1, modulated by wild-type IDH1 through its impact on histone methylation (Calvert et al., 2017).
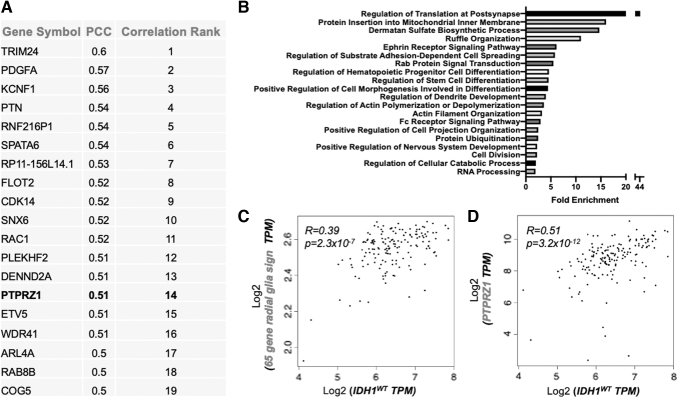
To identify additional gene signatures regulated by wild-type IDH1, we performed an unbiased TCGA analysis to identify the top transcriptional correlates with wild-type IDH1 mRNA expression in glioblastoma tumors (Fig. 5A). Gene ontology pathway analysis of the top 500 wild-type IDH1 correlates identified significant enrichment for pathways involved in neurodevelopmental processes and stem cell maintenance (Fig. 5B).
FIG. 5.
Correlation of wild-type IDH1 with an oRG gene signature. (A) TCGA dataset analysis profiling top mRNA correlates with wild-type IDH1 transcript level in glioblastoma. (B) Gene-ontology analysis of the top 500 TCGA wild-type IDH1 correlates. (C) TCGA dataset analysis of wild-type IDH1 mRNA correlation with oRG gene signature (n = 65 genes). (D) TCGA dataset analysis of wild-type IDH1 transcript correlation with PTPRZ1 mRNA levels. CR, correlation rank; oRG, outer radial glia; PTPRZ1, protein tyrosine phosphatase receptor type Z1; R, correlation coefficient; TPM, transcripts per million. Color images are available online.
Specifically, we observed a high correlation between wild-type IDH1 mRNA expression and an oRG gene signature (as described in Pollen et al. (2015) (Fig. 5C) that included PTPRZ1, the master of oRG identity (Fig. 5D). Together with the published results described above, these studies point to wild-type IDH1 as a critical regulator of progenitor cell fate and will motivate further efforts to fully define the mechanisms, by which wild-type IDHs promote glioblastoma stemness and progression, as detailed below.
Pharmacologically targeting wild-type IDHs
A series of compounds have been developed to inhibit mutant IDH1, mutant IDH2, or both, including GSK321 and GSK864 (GlaxoSmithKline), IDH305 (Novartis), AG-221 and AGI-6780 (Agios). These and other compounds are discussed in detail elsewhere (Tommasini-Ghelfi et al., 2019). For many of these compounds, the mechanism of inhibition has been elucidated at the molecular level by defining cocrystal structures of the inhibitor bound to enzymes by X-ray crystallography. Most inhibitors regulate enzyme activity allosterically by engaging an allosteric pocket, locking the enzyme in an open, catalytically inactive conformation.
Several inhibitors that were developed to target mutant IDH1 showed significant inhibitory activity against wild-type IDH1. Treatment of GICs harboring high-level expression of wild-type IDH1 with GSK864, a compound structurally related to GSK321, resulted in a dose-dependent decrease of the NADPH/NADP+ ratio, while cells that have low IDH1 expression, including minimally transformed cortical astrocytes, did not respond to treatment (Calvert et al., 2017). Moreover, when tested in a panel of transformed glioma cells and patient-derived tumor spheres, GSK864 reduced cell viability and stem cell frequency, increased apoptosis when used in combination with inhibitors of receptor tyrosine kinases, and delayed tumor progression in vivo (Calvert et al., 2017).
To develop high-activity small molecule inhibitors specific for wild-type IDH1 for the treatment of wild-type IDH1 cancers, AbbVie performed a high-throughput screen using their proprietary library, which encompasses 750,000 compounds. The research team identified a hit series, centered around compound 1i, which contained a large cluster of α,β-unsaturated enones (Jakob et al., 2018). As indicated by the wild-type IDH1:1i crystal structure, 1i bound to a fully closed wild-type IDH1enzyme and competed with NADPH for access to the wild-type IDH1 active site via the formation of a covalent adduct and the reversible trapping of H315 located within the NAPDH binding site (Jakob et al., 2018).
Covalent engagement with the NADPH pocket resulted in robust enzyme inhibition, as indicated by a cell-free EC50 of 49 nM. Further compound optimization led to the synthesis of 13i. 13i, but not a structurally related, inactive enone analog, 18i, robustly decreased the activity of recombinant wild-type IDH1with an EC50 of 14 nM (Chung et al., 2020).
Furthermore, 13i showed dose-dependent inhibition of cellular wild-type IDH1 activity, as measured by tracking incorporation of 13C from glutamine to citrate (Jakob et al., 2018), and reduced the viability of patient-derived GICs and the clonogenic potential of glioma cells (Chung et al., 2020). Improved potency due to the replacement of a methyl with a phenyl group addition at R1 is consistent with filling a lipophilic groove between K374/L383/A378 observed in wild-type IDH1:inhibitor cocrystal structures and suggested a clear trajectory to the K260 residue of the opposing monomer.
Like adult glioblastoma, wild-type IDH1 is overexpressed in pediatric high-grade gliomas (Chung et al., 2020), as determined by scRNA-Seq analyses of H3K27M DIPG tumors, with high-level expression of wild-type IDH1 detectable across all three major DIPG subtypes. Wild-type IDH1 protein was elevated in H3K27M mutant compared with H3.3 WT tumors, as determined by immunoblot analysis using lysates from patient-derived cells and by query of the Pediatric Brain Tumor Atlas (PedcBioPortal) (Chung et al., 2020). Mirroring the effect of genetic inactivation in adult glioblastoma cells, RNAi-mediated knockdown of wild-type IDH1 or treatment with 13i reduced the growth of H27K27M mutant (wild-type IDH1high) but not H3.3 WT DIPG cells (wild-type IDH1low).
Consistent with the progrowth activity, knockdown of wild-type IDH1 or pharmacological inhibition using 13i reduced the growth rates of H3K27M DIPGs cell lines. In vivo, intraperitoneally administered 13i crossed the blood–brain barrier, as shown by the micromolar accumulation of 13i within the brainstem of nontumor-bearing mice, and reduced tumor progression and increased survival in two H3K27M wild-type IDH1high patient-derived xenograft (PDX) models (Chung et al., 2020).
Wild-type IDH2 regulates mitochondrial homeostasis
Highlighting the overlapping but nonredundant functions of IDH enzymes, the production of mitochondrial NADPH by wild-type IDH2 is critical for inhibiting mitochondrial oxidative stress, mitochondrial dysfunction, and the ensuing execution of cell death. During mitochondrial respiration, superoxide anions are converted to toxic H2O2 by manganese superoxide dismutase and further reduced to H2O by the mitochondrial components of the GSH- and NADPH-dependent antioxidant system.
In response to elevated levels of ROS, wild-type IDH2 expression is induced to facilitate the defense against ROS (Jo et al., 2001; Kong et al., 2018). Reduced expression of wild-type IDH2 leads to ROS accumulation, DNA fragmentation, lipid peroxidation, and mitochondrial damage; conversely, overexpression of wild-type IDH2 depleted ROS levels and promoted cell survival (Jo et al., 2001).
Under aerobic conditions, mitochondrial α-KG feeds forward into the TCA cycle to support oxidative phosphorylation and the generation of ATP (Martinez-Reyes and Chandel, 2020). Like wild-type IDH1, wild-type IDH2 catalyzes the reductive carboxylation of α-KG into isocitrate at the expense of NADPH. This process allows cancer cells growing in hypoxic conditions or under conditions of diminished oxidative phosphorylation capabilities to maintain anaplerosis. Here, mitochondrial α-KG generated through glutaminolysis is reductively carboxylated by wild-type IDH2 to generate citrate, which is then exported from the mitochondria into the cytosol to support de novo lipid biogenesis (Wise et al., 2011).
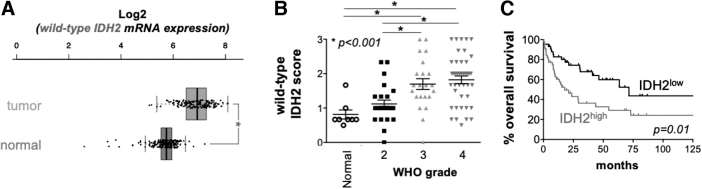
Like wild-type IDH1, wild-type IDH2 levels are elevated in TCGA glioblastoma patient samples (Fig. 6A), with the extent of wild-type IDH2 protein overexpression correlating with glioma grade (Fig. 6B) and negatively correlating with patient survival (Fig. 6C). Despite well-defined pathogenic activities of mutant IDH2 across multiple forms of cancer, including grade 4 glioma, molecular studies directly implicating wild-type IDH2 in glioma pathogenesis and progression are presently lacking. It seems plausible that wild-type IDH2, through scavenging mitochondrial ROS and fueling macromolecule synthesis, is essential for the growth and progression of high-grade glioma.
FIG. 6.
Expression analysis of wild-type IDH2 in glioblastoma. (A) TCGA analysis of wild-type IDH2 expression in normal brain and glioblastoma tumors. *p < 0.05. (B) wild-type IDH2 protein expression in gliomas as a function of tumor grade. Tumors were stained with IDH2-specific antibodies, and staining intensity was quantified by a trained neuropathologist. *p < 0.05. (C) Glioblastoma patient survival as a function of wild-type IDH2 protein expression. Stratification was determined by Cutoff Finder: https://molpathoheidelberg.shinyapps.io/CutoffFinder_v1/. Color images are available online.
Wild-type IDH3 regulates mitochondrial bioenergetics and one-carbon metabolism
Wild-type IDH3 is predominantly localized to mitochondria, but unlike the wild-type IDH2 reaction, the conversion of isocitrate to α-KG by wild-type IDH3 is irreversible and represents one of the rate-limiting steps of the TCA cycle. This reaction is tightly regulated by substrate availability, product inhibition, redox state, and competitive inhibition via ATP (Qi et al., 2008). Wild-type IDH3 subunits further control holoenzyme properties. While the α subunit is essential for converting isocitrate into α-KG, the β and γ subunits function to improve substrate and cofactor binding affinity.
Once α-KG is generated by wild-type IDH3, it enters the TCA cycle, and is further metabolized into succinate, then fumarate, facilitating TCA cycle progression and promoting biosynthesis of critical macromolecules such as fatty acids, nucleotides, and hemes (Krebs and Johnson, 1937). In addition, the electrons generated from NADH are transferred to oxygen via a series of enzymatic processes known as the electron transport chain. This exergonic series of reactions is coupled with the endergonic synthesis of ATP (Vander Heiden and DeBerardinis, 2017).
In contrast to its paralogs IDH1 and IDH2, genomic-sequencing studies revealed that glioma-associated IDH3α, IDH3β, and IDH3γ subunits are not mutated (Krell et al., 2011). In the absence of mutation, May et al. (2019) discovered that IDH3α mRNA and protein levels are elevated within glioblastoma patient samples when compared with glial cells in normal brain, with IDH3α abundance most prominent within the leading edge of glioblastoma tumor specimens. Using CRISPR-Cas9 and RNAi loss of function, together with gain-of-function studies in orthotopic glioblastoma tumor models, May et al. (2019) further demonstrated that IDH3α promoted glioblastoma progression.
Reflecting its central role in the TCA cycle, IDH3α ablation reduces TCA cycle turnover and shunts energy metabolism. Intriguingly, IDH3 function is not confined to mitochondria. A conserved nuclear localization signal at amino acid 124 of the α subunit allows wild-type IDH3 to translocate to the cellular nucleus. This finding confirms previous proteomic analyses of isolated cancer cell nuclei and mitochondria, which demonstrated distribution of various TCA cycle enzymes, including IDH3α, to both nucleus and mitochondria (Qattan et al., 2012).
In particular, Nagaraj et al. (2017) demonstrated that multiple TCA enzymes, including IDH3, localize to the cell nucleus while maintaining a mitochondrial pool during embryonic development. Enzymes associated with the cell nucleus are enzymatically active and, as part of the first half of the TCA cycle, contribute to metabolites, such as acetyl-CoA and α-KG, which are essential for epigenetic control of gene expression and activation of the zygotic genome during preimplantation development.
Within the nuclear lamina, IDH3α interacted directly with cytosolic serine hydroxymethyltransferase (cSHMT), promoted thymidylate synthesis, and enhanced nucleotide availability during DNA synthesis (May et al., 2019). Subcellular fractionation revealed this interaction with cSHMT and nuclear lamina localization was cell cycle dependent, occurring during S-phase when DNA replication machinery is active. cSHMT is a rate-limiting enzyme of the de novo thymidylate synthesis pathway (Anderson et al., 2012). It functions as a lamin-binding protein, serves as a scaffold required for the recruitment of other enzymes implicated in thymidylate synthesis, and interacts with components of the DNA replication machinery to support de novo thymidylate synthesis at sites of DNA replication (Anderson et al., 2012).
Like cSHMT loss of function, genetic inactivation of IDH3α, while impairing nucleotide biosynthesis and cellular growth, enhances the cellular methylation potential by increasing methionine cycle pathway utilization, resulting in DNA hypermethylation and the suppression of an oncogenic signature important for cellular growth and differentiation. Through integrative analysis of methylation array and RNA-seq data, followed by pathway enrichment analysis, May et al. (2019) found that cAMP-mediated signaling and the regulation of EMT pathways were deregulated in the setting of IDH3α deletion.
With limited overlap between differentially methylated and differentially expressed genes, and the enrichment of hypermethylated CpG elements in IDH3α-deficient cells within open sea regions, IDH3α, by promoting DNA hypermethylation, may regulate gene expression through additional mechanisms, for example, by regulating the repetitive genome (May et al., 2019).
Together, the identification of a noncanonical extramitochondrial function of IDH3α suggests a previously unrecognized functional interplay between mitochondrial energy and one-carbon metabolism. This finding points to cancer-associated IDH3α expression and IDH3α-controlled one-carbon metabolism as novel metabolic vulnerabilities in glioblastoma. It is plausible that noncanonical function of IDH3α, that is, its interaction with cSHMT and the modulation of one-carbon metabolism, selectively occurs in rapidly dividing cells with high demand for nucleotide biosynthesis. Therefore, targeting the IDH3α-cSHMT signaling axis via small molecules to disrupt their interaction may represent a novel therapeutic strategy for the treatment of glioblastoma.
As cancer cells depend on DNA synthesis to support unabated growth, multiple enzymes implicated in one-carbon metabolism are upregulated in cancer, including cSHMT (Mehrmohamadi et al., 2014). These data point to IDH3α overexpression as a means to regulate cSHMT function and underscore the importance of therapeutic strategies to target components of the thymidylate pathway to halt glioblastoma tumor progression.
Outlook
Calvert et al. (2017) have shown that except for moderate upregulation of aconitase 1, wild-type IDHs are the only TCA and TCA-associated enzymes that are robustly overexpressed in glioblastoma tumors. This finding suggests that glioblastoma tumors are not characterized by global induction of TCA and TCA-associated anabolic enzymes, and instead supports the hypothesis that IDH induction represents a selective oncogenic mechanism contributing to tumor progression, rather than a passive nonspecific adaptation to increased proliferative rates. To fully characterize and pharmacologically target wild-type IDH oncogenic effects, the following open questions need to be answered.
How do wild-type IDHs control gene expression through chromatin and DNA modifications?
Studies focused on wild-type, and point-mutated IDH1 and IDH2 enzymes have begun unraveling the epigenetic mechanisms by which these metabolic enzymes control gene expression. As a result, novel cellular processes and pathways that contribute to disease pathogenesis and progression have been elucidated. Much work, however, is still needed to fully determine the extent of the epigenetic rewiring in wild-type IDH glioblastoma, the genes and pathways dysregulated, how these epigenetic changes support glioblastoma progression, and whether these pathways can be therapeutically targeted.
Wild-type IDHs, by producing α-KG and activating α-KG-dependent dioxygenases, can modify histones and DNA CpG islands and in doing so, regulate gene expression. The impact of wild-type IDH1 on histone trimethylation marks in glioblastoma, as discussed above, has been linked to the self-renewal capacity of GICs in vitro (Calvert et al., 2017). Future studies need to identify the α-KG-dependent dioxygenase(s) through which wild-type IDHs promote histone trimethylation. Further, to expand upon these results, a global assessment of histone modifications in wild-type IDH glioblastoma, using liquid chromatography-mass spectrometry, will identify additional histone modifications that control gene expression.
In addition, it remains unclear whether wild-type IDH enzymes control DNA methylation. IDH mutations have been linked to a DNA hypermethylation phenotype in grade 4 IDH-mutant astrocytoma. Here, 2HG functions as a competitive inhibitor of α-KG-dependent oxygenases. CpG methylation is often facilitated by α-KG-dependent oxygenases belonging to the ten–eleven translocation methylcytosine dioxygenase (TET) family of DNA demethylases.
In particular, TET2 is involved in the differentiation of embryonic stem cells toward a hematopoietic lineage (Koh et al., 2011; Moran-Crusio et al., 2011). An intracellular pool of α-KG is critical for maintaining TET activity, as direct manipulation of α-KG level disrupts TET activity and associated downstream processes (Carey et al., 2015; Hwang et al., 2016; TeSlaa et al., 2016). Assessing global DNA methylation changes upon wild-type IDH expression will reveal novel mechanisms by which wild-type IDHs control the expression of gene signatures critical for stem-cell like properties of GICs, including oRG-related genes, and if such regulation affect tumor biological features, including dedifferentiation potential and invasion. Notably, a recent study reported that wild-type IDH1 is hyperacetylated at Lys224 in colorectal cancers and liver metastases. Wild-type IDH1 acetylation regulated cellular redox hemostasis promoted colorectal cell invasion and migration in vitro and in vivo (Wang et al., 2020). Future studies will determine whether wild-type IDH1 can regulate glioblastoma invasion through effect on oRG gene signatures, and whether post-translational modifications regulate proinvasive properties of wild-type IDH1.
How do wild-type IDHs affect antitumor immunity?
Studies using a genetically engineered murine mouse model with virally induced overexpression of either IDH1R132H or wild-type IDH1 transgenes and concomitant p53 knockdown and PDGF expression suggested that IDH1 mutation through (R)-2HG–driven epigenetic effect down regulated the expression of leukocyte chemotaxis factors (Amankulor et al., 2017). In particular, the infiltration of immune cells linked to poor prognosis in many cancer types, such as macrophages, microglia, monocytes, and neutrophils, was dampened in mutant IDH1 gliomas (Amankulor et al., 2017).
Consistently, the systemic depletion of neutrophils selectively slowed down disease progression of wild-type IDH1 tumors but had no other effect on the progression of mutant IDH1 tumors (Amankulor et al., 2017). Significant infiltration of wild-type IDH1 gliomas with immune cells, such as macrophages, points to unique immune-related vulnerabilities that can be therapeutically explored, for example, by coinhibiting wild-type IDH1 enzyme activity and macrophage function.
In further support of immune-related differences between wild-type and mutant IDH1 gliomas, in silico analysis using TCGA dataset found that wild-type IDH1 glioma showed enrichment of gene signatures associated with tumor-promoting M2 macrophage and neutrophils (Wang et al., 2018), high-level expression of immunoregulatory molecules, such as programmed death-ligand 1, which correlated with poor patient outcome in wild-type IDH1 as compared with mutant gliomas (Mu et al., 2018), and association between wild-type IDH1 expression level and the extent of immune cell infiltration (Cejalvo et al., 2020).
Future studies will determine the role of wild-type IDHs in shaping the systemic and the tumor-associated immune system, and will define the underlying mechanisms, in particular those mediated by α-KG and the ensuing effect on the regulation of immunogenic factors.
How does wild-type IDH1 by controlling lipid metabolism impact glioblastoma tumor biology?
By generating cytosolic NADPH, wild-type IDH1 promotes the de novo biosynthesis of fatty acids (Calvert et al., 2017). Within tumor cells, increased lipid biosynthesis supports unabated cell growth by providing substrates critical for energy production, plasma membrane composition, and intracellular signaling. Fatty acid esterification to a glycerol moiety generates triacylglycerides, which are utilized during fatty acid oxidation to generate ATP (Snaebjornsson et al., 2020).
In addition, fatty acids are incorporated into the plasma membrane as MUFAs or PUFAs, as glycolipids that function in cell recognition and inflammatory response, or cholesterol, which controls membrane fluidity. Due to the myriad species of fatty acid-derived molecules and their unique roles within normal and tumor cell physiology, identifying global changes in the glioblastoma lipidome as a function of wild-type IDH expression will provide insight into how wild-type IDH1 impacts lipid metabolism, intracellular signaling, and the composition of intracellular and plasma membranes.
In addition to their structural roles as components of the plasma membrane, fatty acid-derived lipids promote ferroptotic cell death when oxidized by iron-dependent lipoxygenase enzymes (Stockwell and Jiang, 2020; Stockwell et al., 2020). Ferroptosis is a recently discovered form of cell death that has rapidly gained recognition as a paradigm-shifting strategy to specifically target cancer cells and persister cells that are resistant to apoptosis induced by a broad spectrum of therapies, including RT and targeted therapies (Stockwell et al., 2020; Viswanathan et al., 2017).
Importantly, recent studies suggest that RT and immune-mediated checkpoint inhibition promote tumor cell ferroptosis. Mechanistically, IFN-γ produced by activated CD8+ T cells in response to immune checkpoint inhibitor treatment, together with ataxia-telangiectasia mutated kinase that becomes activated in tumor cells upon RT treatment synergistically suppressed the expression of SLC7A11, a unit of the glutamate-cystine antiporter xc−. By exchange of glutamate at a 1:1 ratio, system xc− imports cystine, which, upon conversion to cysteine, is used to synthesize GSH by glutathione synthetase. GSH, which is replenished from oxidized glutathione (GSSG) by glutathione reductase, is a cofactor for glutathione peroxidase 4 (GPX4), which converts toxic lipid hydroperoxides to nontoxic lipid alcohols.
Downregulation of system xc− impairs cystine uptake, the rate-limiting substrate for glutathione synthesis, dampens lipid repair mediated by GPX4, and promotes tumor lipid oxidation and ferroptosis. Future studies will test the hypothesis that wild-type IDH1 can inhibit ferroptosis as an important mechanism contributing to RT and checkpoint inhibitor treatment resistance (Lang et al., 2019; Liao et al., 2022). It is plausible that wild-type IDH1 inhibits ferroptosis through its effect on lipid metabolism and lipid repair: PUFAs, upon conversion into phospholipids by ACSL4, are the substrates for lipid peroxidation by iron-dependent lipoxygenase enzymes. Lipid peroxides decompose into reactive derivatives, including aldehydes and Michael acceptors, which can react with proteins and nucleic acids and trigger ferroptosis. Due to increased MUFA synthesis in wild-type IDH1high glioblastoma, exogenous MUFAs, upon incorporation into cellular membranes, can displace oxidizable PUFAs from the plasma membrane and reduce lipid peroxidation (Magtanong et al., 2019).
In addition, it will be important to test the hypothesis if wild-type IDH1 inhibits ferroptosis by promoting lipid repair. Here, wild-type IDH1 increases cellular NADPH level, which is required for GSSG reduction. Beyond providing reducing equivalents necessary for GSH and redox maintenance, wild-type IDH1 may provide carbon necessary for GSH biosynthesis via enhanced α-KG production. Through enzymatic transamination, cytosolic α-KG may fuel glutamate production within glioma cells. This reversible transamination event can be catalyzed by branched-chain amino acid aminotransferase-1 (BCAT1) and BCAT2 at the expense of branched-chain α-ketoacids leucine, isoleucine, or valine (Hall et al., 1993).
Importantly, BCAT1 is upregulated in wild-type IDH1 glioblastoma, likely through an epigenetic and α-KG-dependent mechanism (Tonjes et al., 2013). Derivative glutamate can in turn fuel nucleotide and fatty acid biosynthesis, control production of alanine, aspartate, and serine, or be imported into the mitochondria to support TCA cycle progression (Altman et al., 2016). Glutamate also plays a key role in neutralizing ROS by supporting GSH synthesis. Glutamate contributes to GSH generation via two distinct mechanisms, which are ultimately part of the same pathway.
First, glutamate and cysteine can be converted into gamma-glutamyl cysteine, the immediate precursor to GSH, via glutamate cysteine ligase (Chen et al., 2005). Second, intracellular glutamate can be exported from the cell by antiporter system xc− at a 1:1 ratio coupled with cystine import (Bannai, 1986). Cystine is in turn oxidized into cysteine in an NADPH-dependent manner, thus fueling GSH synthesis, oxidative stress response, and ferroptosis defense (Jiang et al., 2021).
These future studies will help establish the pharmacological inhibition of wild-type-IDH1 as a potent proferroptosis therapy that can amplify therapeutic effects of RT or immunotherapies.
How does the IDH status impact progenitor cell transformation and glioma evolution?
The IDH mutational status defines high-grade gliomas. Molecularly determining how IDH status drives these two distinct disease entities will provide unprecedented insight into the underlying mechanisms of gliomagenesis. Future studies have to determine whether and to what extent wild-type IDH1 expression in different progenitor cells (NSCs or NPCs vs. OPCs) results in distinct tumor molecular and biologic properties and therapy susceptibility. Novel wild-type IDH-gain and -loss-of-function models could define, on a molecular level, the susceptibility of distinct progenitor cell populations to wild-type IDH expression.
As cultured NPCs and OPCs undergo spontaneous differentiation following limited passages, immortalized iPSCs engineered to express tet-inducible wild-type IDH enzymes transdifferentiated into NPCs and OPCs would allow for functional characterization of progenitor cells as a function of wild-type IDH expression. Stereotactic implantation into brain regions specific for glioblastoma and grade 4 IDH-mutant astrocytoma (dorsal striatum and frontal lobe structures, respectively) coupled with tumor progression and survival analysis would provide unprecedented insight into the effect of IDH status on gliomagenesis.
Similarly, inducible Cre alleles driven by NPCs or OPC-specific promoters (Nestin-CreERT2 and NG2-CreERT2) will allow the expression or deletion of wild-type IDHs in specific progenitor cell populations, which together with gross phenotypic and molecular analysis of murine tumors would further define the impact of IDH status on gliomagenesis.
What are the next steps toward pharmacologically targeting wild-type IDH1 in gliomas?
As detailed above, small molecule inhibitors designed to target mutant IDH1 showed significant activity against wild-type IDH1. These include AG-120, AGI-6780, GSK321, GSK864, and IDH305, which inhibit cellular wild-type IDH1 with half maximal inhibitory concentrations (IC50s) of 71 nM, 2.7 μM, 46 nM, 466.5 nM, and 6.14 μM, respectively (Tommasini-Ghelfi et al., 2019). GSK864 is the only mutant IDH1 inhibitor that was tested in wild-type IDH1 glioblastoma cells and derivative orthotopic mouse models.
Calvert et al. (2017) showed that low micromolar concentrations of GSK864 reduced glioma cell viability and neurosphere formation, did not affect the NADP+/NADPH ratio in minimally transformed astrocytes, and promoted glioma cell apoptosis when combined with receptor tyrosine kinase inhibitors (Calvert et al., 2017). When administered intraperitoneally at 150 mg/kg, GSK864 reduced glioblastoma PDX burden and moderately increased animal subject survival. While these studies established wild-type IDH1 as a targetable oncogenic activity in glioblastoma, GSK864 will require further optimization.
Such efforts should include the comprehensive assessment of GSK864 brain penetrance and pharmacokinetics, the development of optimized dosing regimens and the establishment of optimal drug formulation suitable for intravenous or oral administration. Notably, a recent study found that reducing cellular magnesium levels can enhance potency of AG-120 and GSK321 against wild-type IDH1 (Vaziri-Gohar et al., 2022). Molecularly, Mg2+ interacts with Asp279 in the allosteric pocket of the IDH1 isoforms and competes with small molecule binding.
As wild-type IDH1 has a lower Km for Mg2+ compared with mutant IDH1, Mg2+ may outcompete allosteric inhibitors for binding to Asp279 of the wild-type, but not the mutant, enzyme (Deng et al., 2015). Thus, allosteric mutant IDH1 inhibitors need to be rigorously assessed for wild-type IDH1 engagement under varying Mg2+ conditions. Furthermore, a recent study examining the role of wild-type IDH1 in pancreatic ductal adenocarcinoma (PDAC) demonstrated that nutrient limitation, as observed in the PDAC microenvironment, renders cancer but not normal cells reliant on wild-type IDH1 activity for survival (Vaziri-Gohar et al., 2022).
Consequently, genetic or pharmacological wild-type IDH1 inhibition was associated with minimal systemic toxicity. These findings are supported by the analysis of whole-body constitutive wild-type IDH1 knockout mice. As described above, global depletion of wild-type IDH1 did not affect mouse wellness at baseline but rendered animals susceptible to oxidative liver injury (Itsumi et al., 2015). Together with the favorable safety profile of mutant IDH1 inhibitors, such as FDA-approved AG-120, the therapeutic window is expected to be significant. These findings also point to the glycemic status of the tumor and the tumor microenvironment as a putative biomarker for wild-type IDH1-targeting therapies.
Similar to allosteric mutant IDH1 inhibitors, covalent wild-type IDH1-sepcific inhibitors, such as 13i, also require further preclinical testing, including detailed pharmacokinetic analysis, optimization of dosing, administration and formulation, comprehensive tumor regression analyses in advanced glioma mouse models and in vivo validation of its mechanism of action. 13i efficacy should also be determined as a function of nutrient availability and the level of tumor-associated wild-type IDH1 expression.
Finally, to enhance the antitumor effect of wild-type IDH1 inhibitors, preclinical studies need to investigate vulnerabilities of glioblastoma cells with high wild-type IDH1 expression, to inform combinatorial regimens and to antagonize wild-type IDH1 glioma progression more effectively. A recent study of metabolic dependences in H3K27M DIPGs exemplifies this approach. α-KG serves as a critical cofactor for H3K27M demethylases, and is required for maintaining low levels of global H3K27 trimethylation and sustained growth of H3.3K27M tumor cells (Chung et al., 2020).
Consequently, the combined pharmacologic inhibition of enzymes producing α-KG, that is, glutamate dehydrogenase and wild-type IDH1, resulted in elevation of global H3K27me3, suppression of tumor cell proliferation, reduced tumor burden, and a shift of chromatin accessibility to mainly closed states, consistent with transcriptional repression associated with elevated H3K27me3 (Chung et al., 2020).
Finally, the efficacy of wild-type IDH1 inhibitors in the context of IDH1-mutant gliomas should be investigated. 13i will likely be effective against mutant IDH1 grade IV astrocytomas as the mutant enzyme relies on wild-type IDH1 enzymes for localized supply of the α-KG, which is required for 2HG production (Ward et al., 2013).
Abbreviations Used
- 2HG
2-hydroxyglutarate
- α-KG
α-ketoglutarate
- AC
astrocyte cell
- ACSL4
acyl-CoA synthetase long-chain family member 4
- ASCL1
Achaete-scute homolog 1
- ATP
adenosine 5′-triphosphate
- BCAT
branched-chain amino acid aminotransferase
- CDK4
cyclin-dependent kinase 4
- CHI3L1
chitinase-3–like protein 1
- ChIP-Seq
chromatin immunoprecipitation sequencing
- CNS
central nervous system
- cSHMT
cytosolic serine hydroxymethyltransferase
- EGFR
epidermal growth factor receptor
- GICs
glioma-initiating cells
- GPX4
glutathione peroxidase 4
- GSH
glutathione
- GSSG
oxidized glutathione
- HGG
high-grade glioma
- HIF
hypoxia-inducible factor
- HR
hazard ratio
- IC50
half maximal inhibitory concentrations
- IDH
isocitrate dehydrogenase
- JmjC
jumonji C
- LOH
loss of heterozygosity
- MES
mesenchymal
- MET
MET proto-oncogene, receptor tyrosine kinase
- MUFA
monounsaturated fatty acid
- NAC
N-acetyl cysteine
- NAD(P)H
nicotinamide adenine dinucleotide (phosphate)
- NF1
neurofibromatosis type 1
- NG2
neuron-glial antigen 2
- NPC
neural progenitor cell
- NSC
neural stem cell
- OLIG2
oligodendrocyte transcription factor 2
- OPC
oligodendrocyte precursor cell
- oRG
outer radial glia
- PDAC
pancreatic ductal adenocarcinoma
- PDGF
platelet-derived growth factor
- PDGFRA
platelet-derived growth factor receptor A
- PDX
patient-derived xenograft
- PN
proneural
- POU3F2
POU class 3 homeobox 2
- PTEN
phosphatase and tensin homolog
- PTPRZ1
protein tyrosine phosphatase receptor type Z1
- PUFA
polyunsaturated fatty acid
- ROS
reactive oxygen species
- RT
radiation therapy
- SALL2
spalt-like transcription factor 2
- scRNA-Seq
single-cell RNA sequencing
- SHH
sonic hedgehog
- Sox2
SRY-box 2
- TCA
tricarboxylic acid
- TCGA
The Cancer Genome Atlas
- TET
ten-eleven translocation methylcytosine dioxygenase
- TPM
transcripts per million
- VEGF
vascular endothelial growth factor
- WHO
World Health Organization
Authors' Contributions
K.M.M. and A.H.S. wrote the article and conceptualized the figures. C.H. performed the expression analysis for wild-type IDH2 protein, and edited the article; and K.M.M., C.H., and A.H.S. approved the final version of the article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This research was supported by R01NS129123 (to Alexander H. Stegh), the Northwestern University Brain Tumor SPORE grant (P50CA221747) to Craig Horbinski and Alexander H. Stegh; R01NS118039, R01NS117104, R01NS102669 (to Craig Horbinski), and an NIH T32CA009560 (to Kevin M. Murnan).
References
- Alcantara Llaguno S, Sun D, Pedraza AM, et al. Cell-of-origin susceptibility to glioblastoma formation declines with neural lineage restriction. Nat Neurosci 2019;22(4):545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Parada LF. Cell of origin of glioma: Biological and clinical implications. Br J Cancer 2016;115(12):1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Wang Z, Sun D, et al. Adult lineage-restricted CNS progenitors specify distinct glioblastoma subtypes. Cancer Cell 2015;28(4):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara Llaguno SR, Xie X, Parada LF. Cell of origin and cancer stem cells in tumor suppressor mouse models of glioblastoma. Cold Spring Harb Symp Quant Biol 2016;81:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat Rev Cancer 2016;16(11):749. [DOI] [PubMed] [Google Scholar]
- Amankulor NM, Kim Y, Arora S, et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev 2017;31(8):774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Woeller CF, Chiang EP, et al. Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem 2012;287(10):7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai S. Exchange of cystine and glutamate across plasma membrane of human fibroblasts. J Biol Chem 1986;261(5):2256–2263. [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- Bar EE, Lin A, Mahairaki V, et al. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol 2010;177(3):1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger M, Allaman I, Magistretti PJ. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab 2011;14(6):724–738. [DOI] [PubMed] [Google Scholar]
- Bernal A, Arranz L. Nestin-expressing progenitor cells: Function, identity and therapeutic implications. Cell Mol Life Sci 2018;75(12):2177–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A, Di Lullo E, Jung D, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 2020;26(1):48–63.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brat DJ, Castellano-Sanchez AA, Hunter SB, et al. Pseudopalisades in glioblastoma are hypoxic, express extracellular matrix proteases, and are formed by an actively migrating cell population. Cancer Res 2004;64(3):920–927. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchler P, Reber HA, Büchler M, et al. Hypoxia-inducible factor 1 regulates vascular endothelial growth factor expression in human pancreatic cancer. Pancreas 2003;26(1):56–64. [DOI] [PubMed] [Google Scholar]
- Bzymek KP, Colman RF. Role of alpha-Asp181, beta-Asp192, and gamma-Asp190 in the distinctive subunits of human NAD-specific isocitrate dehydrogenase. Biochemistry 2007;46(18):5391–5397. [DOI] [PubMed] [Google Scholar]
- Calver AR, Hall AC, Yu WP, et al. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron 1998;20(5):869–882. [DOI] [PubMed] [Google Scholar]
- Calvert AE, Chalastanis A, Wu Y, et al. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep 2017;19(9):1858–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos B, Olsen LR, Urup T, et al. A comprehensive profile of recurrent glioblastoma. Oncogene 2016;35(45):5819–5825. [DOI] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, et al. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015;518(7539):413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat Rev Drug Discov 2011;10(6):417–427. [DOI] [PubMed] [Google Scholar]
- Carta G, Murru E, Banni S, et al. Palmitic acid: Physiological role, metabolism and nutritional implications. Front Physiol 2017;8:8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejalvo T, Gargini R, Segura-Collar B, et al. Immune profiling of gliomas reveals a connection with IDH1/2 mutations, tau function and the vascular phenotype. Cancers (Basel) 2020;12(11):3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A, Erkkinen MG, Nestler U, et al. Temozolomide-mediated radiation enhancement in glioblastoma: A report on underlying mechanisms. Clin Cancer Res 2006;12(15):4738–4746. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Kharbanda S, et al. Hominoid-specific enzyme GLUD2 promotes growth of IDH1R132H glioma. Proc Natl Acad Sci U S A 2014;111(39):14217–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shertzer HG, Schneider SN, et al. Glutamate cysteine ligase catalysis: Dependence on ATP and modifier subunit for regulation of tissue glutathione levels. J Biol Chem 2005;280(40):33766–33774. [DOI] [PubMed] [Google Scholar]
- Cheng C, Geng F, Cheng X, et al. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun (Lond) 2018a;38(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Li J, Guo D. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy. Curr Top Med Chem 2018b;18(6):484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EJ, Cho BJ, Lee DJ, et al. Enhanced cytotoxic effect of radiation and temozolomide in malignant glioma cells: Targeting PI3K-AKT-mTOR signaling, HSP90 and histone deacetylases. BMC Cancer 2014;14:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Sweha SR, Pratt D, et al. Integrated metabolic and epigenomic reprograming by H3K27M mutations in diffuse intrinsic pontine gliomas. Cancer Cell 2020;38(3):334–349.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: A guerilla war. Acta Neuropathol 2007;114(5):443–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From molecular pathology to targeted treatment. Annu Rev Pathol 2014;9:1–25. [DOI] [PubMed] [Google Scholar]
- Cuddapah VA, Robel S, Watkins S, et al. A neurocentric perspective on glioma invasion. Nat Rev Neurosci 2014;15(7):455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999;126(14):3089–3100. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G, Shen J, Yin M, et al. Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J Biol Chem 2015;290(2):762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkse A, Golebiewska A, Buder T, et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun 2019;10(1):1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch HG, Catala A, Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem 1976;251(16):5095–5103. [PubMed] [Google Scholar]
- Glasauer A, Chandel NS. ROS. Curr Biol 2013;23(3):R100–R102. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Xie C, Jiang C. The role of hypoxia-inducible factors in metabolic diseases. Nat Rev Endocrinol 2018;15(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal K, Grossi E, Paoletti P, et al. Lipid composition of human intracranial tumors: A biochemical study. Acta Neurochir (Wien) 1963;11:333–347. [DOI] [PubMed] [Google Scholar]
- Guo D, Prins RM, Dang J, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal 2009;2(101):ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TR, Wallin R, Reinhart GD, et al. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J Biol Chem 1993;268(5):3092–3098. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, et al. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010;464(7288):554–561. [DOI] [PubMed] [Google Scholar]
- Hatoum A, Mohammed R, Zakieh O. The unique invasiveness of glioblastoma and possible drug targets on extracellular matrix. Cancer Manag Res 2019;111:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IY, Kwak S, Lee S, et al. Psat1-dependent fluctuations in alpha-ketoglutarate affect the timing of ESC differentiation. Cell Metab 2016;24(3):494–501. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, et al. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci 2010;30(9):3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsumi M, Inoue S, Elia AJ, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ 2015;22(11):1837–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, et al. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 2006;51(2):187–199. [DOI] [PubMed] [Google Scholar]
- Jakob CG, Upadhyay AK, Donner PL, et al. Novel modes of inhibition of wild-type isocitrate dehydrogenase 1 (IDH1): Direct covalent modification of His315. J Med Chem 2018;61(15):6647–6657. [DOI] [PubMed] [Google Scholar]
- Jiang L, Shestov AA, Swain P, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016;532(7598):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 2021;22(4):266–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo SH, Son MK, Koh HJ, et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem 2001;276(19):16168–16176. [DOI] [PubMed] [Google Scholar]
- Jue TR, McDonald KL. The challenges associated with molecular targeted therapies for glioblastoma. J Neurooncol 2016;127(3):427–434. [DOI] [PubMed] [Google Scholar]
- Kaur B, Khwaja FW, Severson EA, et al. Hypoxia and the hypoxia-inducible-factor pathway in glioma growth and angiogenesis. Neuro Oncol 2005;7(2):134–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B, Simon MC. Hypoxia inducible factors, stem cells and cancer. Cell 2007;129(3):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh HJ, Lee SM, Son BG, et al. Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem 2004;279(38):39968–39974. [DOI] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011;8(2):200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong MJ, Han SJ, Kim JI, et al. Mitochondrial NADP(+)-dependent isocitrate dehydrogenase deficiency increases cisplatin-induced oxidative damage in the kidney tubule cells. Cell Death Dis 2018;9(5):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer 2020;122(1):4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs HA, Johnson WA. Metabolism of ketonic acids in animal tissues. Biochem J 1937;31(4):645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell D, Assoku M, Galloway M, et al. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PLoS One 2011;6(5):e19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 2011;29(34):4482–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, et al. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci 2003;6(1):21–27. [DOI] [PubMed] [Google Scholar]
- Lang X, Green MD, Wang W, et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov 2019;9(12):1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, et al. Cancer stem cells in glioblastoma. Genes Dev 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Koh HJ, Park DC, et al. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med 2002;32(11):1185–1196. [DOI] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009;15(6):501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P, Wang W, Wang W, et al. CD8(+) T cells and fatty acids orchestrate tumor ferroptosis and immunity via ACSL4. Cancer Cell 2022;40(4):365–378.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010;44(5):479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell 2011;146(2):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of tumors of the central nervous system: A summary. Acta Neuropathol 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- Lukas RV, Wainwright DA, Ladomersky E, et al. Newly diagnosed glioblastoma: A review on clinical management. Oncology (Williston Park) 2019;33(3):91–100. [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ko PJ, To M, et al. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem Biol 2019;26(3):420–432.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 2020;11(1):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Pichumani K, Vemireddy V, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 2014;159(7):1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaka T, Shimano H, Yahagi N, et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat Med 2007;13(10):1193–1202. [DOI] [PubMed] [Google Scholar]
- May JL, Kouri FM, Hurley LA, et al. IDH3alpha regulates one-carbon metabolism in glioblastoma. Sci Adv 2019;5(1):eaat0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon C, Nandhabalan M, Murray SA, et al. Glioblastoma: Clinical presentation, diagnosis, and management. BMJ 2021;374:n1560. [DOI] [PubMed] [Google Scholar]
- Mehrmohamadi M, Liu X, Shestov AA, et al. Characterization of the usage of the serine metabolic network in human cancer. Cell Rep 2014;9(4):1507–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012;481(7381):380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro AR, Hill R, Pilkington GJ, et al. The role of hypoxia in glioblastoma invasion. Cells 2017;6(4):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticone M, Taherian R, Stigliani S, et al. NAC, tiron and trolox impair survival of cell cultures containing glioblastoma tumorigenic initiating cells by inhibition of cell cycle progression. PLoS One 2014;9(2):e90085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20(1):11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L, Long Y, Yang C, et al. The IDH1 mutation-induced oncometabolite, 2-hydroxyglutarate, may affect DNA methylation and expression of PD-L1 in gliomas. Front Mol Neurosci 2018;11:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Sharpley MS, Chi F, et al. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell 2017;168(1–2):210–223.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, et al. Polydendrocytes (NG2 cells): Multifunctional cells with lineage plasticity. Nat Rev Neurosci 2009;10(1):9–22. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol 2005;109(1):93–108. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res 2013;19(4):764–772. [DOI] [PubMed] [Google Scholar]
- Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol 2014;232(2):165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- Palazon A, Goldrath AW, Nizet V, et al. HIF transcription factors, inflammation, and immunity. Immunity 2014;41(4):518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: Still emerging. Cell Metab 2022;34(3):355–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009;139(7):1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9(3):157–173. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006;444(7120):761–765. [DOI] [PubMed] [Google Scholar]
- Pistollato F, Chen HL, Schwartz PH, et al. Oxygen tension controls the expansion of human CNS precursors and the generation of astrocytes and oligodendrocytes. Mol Cell Neurosci 2007;35(3):424–435. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, et al. Molecular identity of human outer radial glia during cortical development. Cell 2015;163(1):55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 1993;117(2):525–533. [DOI] [PubMed] [Google Scholar]
- Qattan AT, Radulovic M, Crawford M, et al. Spatial distribution of cellular function: the partitioning of proteins between mitochondria and the nucleus in MCF7 breast cancer cells. J Proteome Res 2012;11(12):6080–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen X, Beard DA. Detailed kinetics and regulation of mammalian NAD-linked isocitrate dehydrogenase. Biochim Biophys Acta 2008;1784(11):1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin EY, Cooper DD, Abbott KL, et al. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell 2017;170(5):845–859.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth AD, Nunez MT. Oligodendrocytes: Functioning in a delicate balance between high metabolic requirements and oxidative damage. Adv Exp Med Biol 2016;949:167–181. [DOI] [PubMed] [Google Scholar]
- Scherer HJ. Structural development in gliomas. Am J Cancer 1938;34(3):333–351. [Google Scholar]
- Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain 2010;133(Pt 4):983–995. [DOI] [PubMed] [Google Scholar]
- Shechter I, Dai P, Huo L, et al. IDH1 gene transcription is sterol regulated and activated by SREBP-1a and SREBP-2 in human hepatoma HepG2 cells: evidence that IDH1 may regulate lipogenesis in hepatic cells. J Lipid Res 2003;44(11):2169–2180. [DOI] [PubMed] [Google Scholar]
- Shi J, Zuo H, Ni L, et al. An IDH1 mutation inhibits growth of glioma cells via GSH depletion and ROS generation. Neurol Sci 2014;35(6):839–845. [DOI] [PubMed] [Google Scholar]
- Shi Y, Ping YF, Zhou W, et al. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat Commun 2017;8:15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H, Belousov VV, Chandel NS, et al. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat Rev Mol Cell Biol 2022;23(7):499–515. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab 2020;31(1):62–76. [DOI] [PubMed] [Google Scholar]
- Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A 2011;108(11):4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 2009;28(45):3949–3959. [DOI] [PubMed] [Google Scholar]
- Stockwell BR, Jiang X, Gu W. Emerging mechanisms and disease relevance of ferroptosis. Trends Cell Biol 2020;30(6):478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR, Jiang X. The chemistry and biology of ferroptosis. Cell Chem Biol 2020;27(4):365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engstrom PG, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev 2013;27(6):654–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 2004;11(11):1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Rheinbay E, Gillespie SM, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Tirosh I. The glioma stem cell model in the era of single-cell genomics. Cancer Cell 2020;37(5):630–636. [DOI] [PubMed] [Google Scholar]
- Tan AC, Ashley DM, Lopez GY, et al. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin 2020;70(4):299–312. [DOI] [PubMed] [Google Scholar]
- TeSlaa T, Chaikovsky AC, Lipchina I, et al. Alpha-ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab 2016;24(3):485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini-Ghelfi S, Murnan K, Kouri FM, et al. Cancer-associated mutation and beyond: The emerging biology of isocitrate dehydrogenases in human disease. Sci Adv 2019;5(5):eaaw4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjes M, Barbus S, Park YJ, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med 2013;19(7):901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci 2007;10(8):970–979. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006;439(7078):811–816. [DOI] [PubMed] [Google Scholar]
- van Tilborg E, de Theije CGM, van Hal M, et al. Origin and dynamics of oligodendrocytes in the developing brain: Implications for perinatal white matter injury. Glia 2018;66(2):221–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell 2017;168(4):657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri-Gohar A, Cassel J, Mohammed FS, et al. Limited nutrient availability in the tumor microenvironment renders pancreatic tumors sensitive to allosteric IDH1 inhibitors. Nat Cancer 2022;3(7):852–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan VS, Ryan MJ, Dhruv HD, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547(7664):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl DR, Dresser J, Wilder-Romans K, et al. Glioblastoma therapy can be augmented by targeting IDH1-mediated NADPH biosynthesis. Cancer Res 2017;77(4):960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Chowdhury R, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun 2016;7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Ye Y, Yang X, et al. SIRT2-dependent IDH1 deacetylation inhibits colorectal cancer and liver metastases. EMBO Rep 2020;21(4):e48183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 2018;33(1):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Lu C, Cross JR, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem 2013;288(6):3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M, Lamszus K. The neurobiology of gliomas: From cell biology to the development of therapeutic approaches. Nat Rev Neurosci 2011;12(9):495–508. [DOI] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A 2011;108(49):19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Liu H, He C, et al. Hypoxia contributes to poor prognosis in primary IDH-wt GBM by inducing tumor cells MES-like transformation trend and inhibiting immune cells activity. Front Oncol 2021;11:782043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem 2004;279(32):33946–33957. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ye D, Guan KL, et al. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin Cancer Res 2012;18(20):5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kim MJ, Yoon W, et al. Isocitrate protects DJ-1 null dopaminergic cells from oxidative stress through NADP+-dependent isocitrate dehydrogenase (IDH). PLoS Genet 2017;13(8):e1006975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yao J, Brinton RD, et al. Editorial: The metabolic-inflammatory axis in brain aging and neurodegeneration. Front Aging Neurosci 2017;9:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Parada LF, Baker SJ. Cell of origin for malignant gliomas and its implication in therapeutic development. Cold Spring Harb Perspect Biol 2015;7(5):a020610. [DOI] [PMC free article] [PubMed] [Google Scholar]