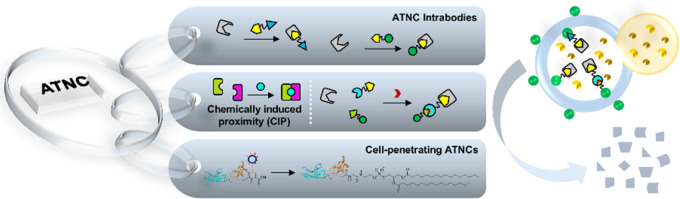
Abstract
Targeted protein degradation (TPD) through the autophagy pathway displays broad substrate scope and is gaining increasing interest in biology and medicine. However, current approaches using small-molecule degraders have limitations due to the lack of versatility, modularity, and ease of implementation and are restricted to addressing only ligandable proteins. Herein, we report a nonsmall molecule-based autophagy-targeting nanobody chimera (ATNC), or phagobody, for selective degradation of intracellular targets, which overcomes these limitations. The core of an ATNC features a nanobody for recruiting proteins as well as an autophagic pathway-directing module. ATNC turns out to be a general, modular, and versatile degradation platform. We show that ATNC can be versatilely implemented in different ways including expressed ATNC intrabodies for ease of use, chemically induced proximity (CIP)-operated logic-gated conditional and tunable degradation, and cyclic cell-penetrating peptide-tethered cell-permeable phagobodies that selectively degrade the undruggable therapeutically relevant HE4 protein, resulting in effective suppression of ovarian cancer cell proliferation and migration. Overall, ATNC represents a general, modular, and versatile targeted degradation platform that degrades unligandable proteins and offers therapeutic potential.
Introduction
Targeted protein degradation (TPD) strategies are rapidly emerging as new modalities in drug discovery.1,2 As a promising therapeutic strategy, TPD shows advantages over traditional inhibitors that depend on occupancy-driven pharmacology, while degraders enable durable knockdown of protein levels.3 Moreover, TPD approaches are able to address challenging targets for traditional drugs, such as proteins with resistance mutations.4 Mechanistically, TPD can be categorized into ubiquitin-proteasome system (UPS)-based and lysosome-associated degradations; the latter can be further classified into the autophagy pathway5 and endosome–lysosome system-based degradation approaches.6,7 Proteolysis targeting chimera (PROTAC), a representative UPS-based approach, uses bifunctional small-molecule degraders to recruit target proteins to E3 ligases for proteasomal clearance.8 On the other hand, lysosome-mediated degradation approaches display broad substrate scope ranging from cell surface and extracellular proteins6,9 to various intracellular targets that are inaccessible for proteasomal clearance.10−12 For example, macroautophagy (hereafter, autophagy)-based degradation methods have been introduced in recent years using bifunctional small molecules5,10−12 for degradation of not only soluble proteins but also aggregated proteins, nonproteinous biomolecules, and organelles.10,13 Autophagy relies on autophagosome encapsulation of cellular debris followed by fusion with lysosomes to give autolysosomes that subsequently accomplish degradation.14 However, these autophagy-based approaches have limitations such as the lack of modularity and versatility, the requirement of multistep organic synthesis, and the limitation to degrading proteins with intracellular ligandable domains. Additionally, bivalent small-molecule degraders often exhibit the so-called “hook effect,”15 which refers to the reduced degradation efficiency resulting from the increased formation of binary complexes accompanied by a reduced ratio of ternary complexes at higher degrader concentrations. The large molecular weight and complicated chemical structure may affect the drug-likeness16,17 profile of these bivalent small-molecule degraders.
In this study, we introduced general, modular, and versatile autophagy-targeting nanobody chimeras (ATNCs) for the selective degradation of intracellular targets. Compared to recently introduced small-molecule-based18 autophagy-targeting degraders, the modular feature of ATNC allows degradation of alternative targets without compromising the degradation efficacy. In addition, ATNC is easy to implement, can address unligandable targets, and shows broad substrate scope including nonproteinous ones like organelles. We also integrated chemically induced proximity (CIP) with ATNC to establish a CIP-operated logic-gated degradation system that enables conditional and tunable degradation. Many therapeutically relevant proteins do not have the right small-molecule binders or are undruggable. For this, we employed a cyclic cell-penetrating peptide-based nonendocytic delivery system19 and designed cell-permeable ATNC drugs, or cell-permeable phagobodies, for selective degradation of the undruggable HE4 protein, which allows efficient suppression of ovarian cancer cell proliferation and migration.
Results
Design and Evaluation of ATNC Degradation Systems
First, we aimed to identify an appropriate autophagy-targeting module. LC3-interacting region (LIR) motifs are short peptide sequences that bind with the LC3 protein, a key marker in autophagy that localizes to preautophagosomal structures (PAS) during autophagosome formation.20 We envisioned that nanobody-LIR chimeras might bring a protein for autophagic clearance (Figure S1A). We successfully identified an LIR sequence DSEDEDFEILSL, derived from the C-terminus of the ATG4B protein,21 that serves as a good candidate showing clear colocalization with LC3B-labeled autophagic puncta (Figure S1B,C). Hence, we designed the GBP-LIR chimera for degradation of the widely used unligandable green fluorescent protein (GFP) and its fusion chimeras. The GFP binding protein (GBP) is a high-affinity nanobody against GFP and its variants, and it binds with GFP with Kd = 1.4 nM.22 We were happy to find that the expressed GBP-LIR chimera efficiently degrades the enhanced GFP (EGFP) protein according to confocal microscopic visualization (Figure S1D,E) and western blot (WB) validation (Figure S1F,G).
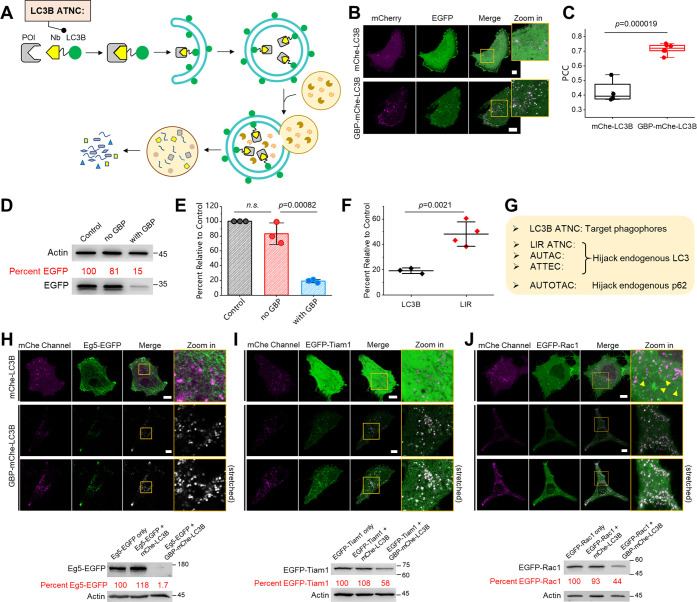
Next, we envisioned that direct fusion of a nanobody with the LC3B protein may result in an even better degradation profile, since LC3B directly localizes to phagophores (Figure 1A). Hence, we designed the GBP-LC3B chimera and were delighted to find that EGFP could also be degraded by the GBP-LC3B chimera in a more complete fashion according to both confocal microscopy (Figure 1B,C) and WB analysis (Figure 1D,E). We compared LC3B- and LIR-based ATNCs and found that the LC3B-based ANTC indeed provides a stronger degree of degradation (Figure 1F). Hence, in the following section, we focus on LC3B-based ATNC for a more detailed study. Compared to recently reported small-molecule degraders that work via autophagic clearance, nanobody-LC3B ATNC relies on a supplement of extra LC3B chimeras that localize to phagophores/autophagosomes without hijacking endogenous LC3 or p62 proteins that are necessary for endogenous functions23 (Figure 1G).
Figure 1.
Design and evaluation of LC3B-based ANTC: (A) Schematic overview of the working mechanism of LC3B-based ATNC that recruits protein of interest to the autophagic degradation pathway. (B) Representative confocal microscopy images of live HeLa cells coexpressing EGFP and GBP-mCherry-LC3B or mCherry-LC3B showed that EGFP was degraded only in the presence of GBP-mCherry-LC3B with clear autophagic puncta formation. (C) PCC colocalization analysis between EGFP and mCherry channels (n = 4/6 cells for mCherry-LC3B/GBP-mCherry-LC3B groups). (D) Representative WB result (48 h) of the HeLa cell sample for control (EGFP only), no GBP group (EGFP + mCherry-LC3B), and GBP group (EGFP + GBP-mCherry-LC3B). (E) Statistical quantification of WB degradation results (n = 3 experiments). (F) Statistical comparison between LIR- and LC3B-based ATNCs shows that LC3B-based ATNC resulted in a higher degradation degree. (G) Mechanistic difference between LC3B-based ANTC, LIR-based ANTC, and other autophagic small-molecule degradation tools is shown. One-sided Student’s t-test was used in this figure; n.s.: nonsignificant; bar graphs denote mean ± standard deviation (SD); mChe: mCherry. (H–J) LC3B-based ATNC is a general tool for the degradation of EGFP-tagged proteins; in (I), Tiam1 indicates the DHPH functional domain of Tiam1. Confocal micrographs of live HeLa cells coexpressing the EGFP-fused protein and mCherry-LC3B or GBP-mCherry-LC3B showed the entrapment of the EGFP-fused protein into autophagic puncta; stretched (brightness enhanced) images were provided for the better clarity of the degradation panel; and WB analysis (48 h) confirmed degradation of these EGFP fusions. Scale bar: 10 μm.
ATNC is a General Tool for Targeted Degradation of Tagged Proteins
Motivated by the above results, we envisioned that proteins carrying an EGFP tag could be degraded, turning ATNC into a general and robust selective degradation tool for the widely used EGFP-tagged proteins (Figure 1H–J). To test this hypothesis, we used expressed GBP-mCherrry-LC3B for degradation of the EGFP-fused Eg5 protein. Eg5 is a large-molecular-weight motor protein responsible for proper bipolar spindle assembly during cell division, and it is also a prognostic biomarker and a potential therapeutic target for hepatocellular carcinoma.24 Confocal microscopy images showed that Eg5-EGFP was targeted to autophagic puncta (Figure 1H, up) and WB analysis revealed near-complete degradation (Figure 1H, down). We next tested degradation of another protein of therapeutic relevance, T-cell lymphoma invasion, and metastasis-inducing protein 1 (Tiam1).25 Delightfully, both confocal microscopy images (Figure 1I, up) and WB analysis revealed efficient degradation of EGFP-Tiam1 (Figure 1I, down). Finally, we used this general degradation system to study the cellular processes (Figure S2A). For this, we aimed to degrade the Rac1 protein, a key player upstream of the lamellipodia signaling cascade, whose activity might be highly relevant to cell sizes.26 As can be seen from both confocal microscopy images (Figure 1J, up) and WB analysis (Figure 1J, down), EGFP-Rac1 was efficiently degraded. In parallel, we found that cell areas were significantly reduced (Figure S2B,C), suggesting that the presence of Rac1 promotes the expansion of cells. This result is consistent with the critical role of Rac1 for downstream actin polymerization that eventually induces lamellipodia formation.26 Hence, GBP-mCherry-LC3B ATNC is a general tool for degradation of proteins that are fused with a nanobody binding tag, such as the widely used fluorescent protein EGFP.
ATNC is a Modular System for Targeted Degradation of Either Tagged or Endogenous Proteins
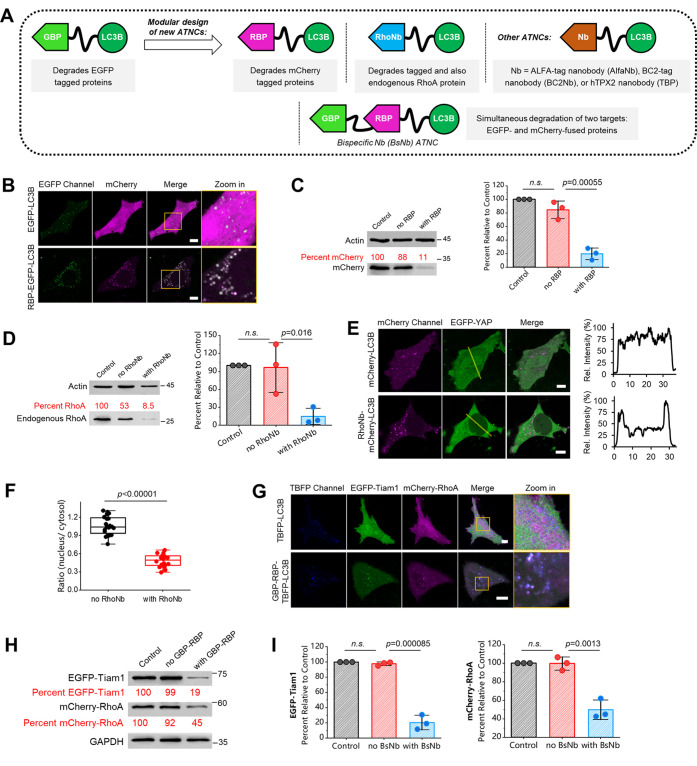
Next, we investigated whether ATNC can be extended to degrade alternative antigenic targets via the modular design of new nanobody-LC3B chimeras (Figure 2A). Therefore, we first chose the nanobody against mCherry red fluorescent protein binding protein (RBP). RBP strongly and specifically interacts with mCherry, another widely used fluorescent protein.27 We designed the RBP-LC3B chimera for selective degradation of mCherry. As seen from both confocal microscopy images (Figure 2B) and WB validation (Figure 2C), mCherry was efficiently degraded by RBP-LC3B chimera. Hence, the modular feature of ATNC allows an easy extension to degrade different targets simply via alternating the nanobody module. Next, we tested a third degradation chimera, RhoNb-LC3B (RhoNb: RhoA nanobody). We chose a nanobody against RhoA, a small GTPase that regulates cell morphology, motility, and transformation.28 Also, RhoA is a disease-relevant small GTPase overexpressed in many cancer cells; for example, it was suggested that downregulation of RhoA could provide a new avenue in reversing the malignant phenotype of gastric cancer.29 For this purpose, we first tested EGFP-RhoA, which carried an EGFP tag to facilitate the visualization of degradation. Confocal microscopy images showed that EGFP-RhoA was targeted to autophagic puncta (Figure S3A), and WB analysis revealed efficient degradation of EGFP-RhoA (Figure S3B).
Figure 2.
ANTC is a modular degradation system: (A) ATNC is a modular tool that can degrade different targets simply by exchanging the nanobody module. (B) Confocal microscopy images of live HeLa cells coexpression of mCherry and EGFP-LC3B or RBP-EGFP-LC3B. (C) Representative WB result (48 h) and statistical comparison of the degradation of the control sample (mCherry only), no RBP group (mCherry + EGFP-LC3B), and with RBP group (mCherry + RBP-EGFP-LC3B) (n = 3 experiments). (D) Representative WB result (48 h) and statistical comparison of the degradation result of the control sample (blank), no RhoNb group (mCherry-LC3B), and with RhoNb group (RhoNb-mCherry-LC3B) (n = 3 experiments). (E) Confocal microscopy images of live HeLa cells coexpressing EGFP-YAP and mCherry-LC3B or RhoNb-mCherry-LC3B revealed export of EGFP-YAP from the nucleus to the cytosol for cells expressing RhoNb-mCherry-LC3B; line profile analysis was drawn from higher left to lower right. (F) Box plot analysis of the fluorescence ratio of EGFP-YAP in the nucleus versus cytosol (n = 20 cells for both). (G) Representative confocal microscopy images of live HeLa cells coexpressing EGFP-Tiam1, mCherry-RhoA, and TBFP-LC3B (up, control) or GBP-RBP-TBFP-LC3B (down, experiment) for 24 h. (H) Representative WB result (48 h) of the degradation of the control group (EGFP-Tiam1 + mCherry-RhoA), no GBP-RBP group (EGFP-Tiam1 + mCherry-RhoA + TBFP-LC3B), and with GBP-RBP group (EGFP-Tiam1 + mCherry-RhoA + GBP-RBP-TBFP-LC3B). (I) Statistic analysis of the WB results (n = 3 experiments); BsNb: bispecific nanobody GBP-RBP. One-sided Student’s t-test was used for related subfigures; n.s.: nonsignificant; see Methods section for description of box plots; bar graphs denote mean ± SD in this figure; mChe: mCherry; TBFP: TagBFP2.
Then, without overexpressing EGFP-RhoA, we found that endogenous RhoA could still be efficiently degraded according to the WB analysis (Figure 2D). Since RhoA regulates cellular contractility and coordinates with nuclear localization of the transcriptional factor Yes-associated protein (YAP),30 we used the RhoNb-LC3B ATNC degrader to study the downregulation of RhoA on cellular translocation of YAP. After degradation of endogenous RhoA, we found that EGFP-YAP was exported from the nucleus to the cytosol (Figure 2E,F). This result is in agreement with previous findings that translocation of YAP between the cytoplasm and nucleus responds to sustained increased traction forces regulated by RhoA.30,31 Hence, the ATNC system is an easy-to-implement and valuable tool for degradation of endogenous proteins for studying related signaling cascades, for example, mechanosensitive signaling pathways and related transcriptional changes, as shown in this study, without the need to overexpress target proteins.
We further showcased the modularity of ANTC by degrading alternative protein targets. We chose three proteins of wide interest, including ALFA-tag32 and BC2-tag,33 which were recently developed as next-generation protein tags in biotechnology, as well as hTPX2 protein34,35 that exhibits strong nucleus distribution due to the presence of the nucleus localization signal (NLS) in its sequence. All of the new ATNCs carrying corresponding nanobodies enable robust degradation of their cognate cellular targets (Figure S4A–C). It is noteworthy that even hTPX2, which shows strong nucleus distribution, was also degraded, which might be attributed to the presence of nucleus transporting proteins that help to shuttle the nucleus proteins between the cytosol and the nucleus.34
Finally, we demonstrated that the modularity of ATNC could be further extended for simultaneous degradation of two protein targets using bispecific nanobody (BsNb)-containing ATNC. Confocal microscopy images revealed that EGFP-Tiam1 and mCherry-RhoA could be simultaneously recruited to autophagic puncta in the presence of GBP-RBP-TBFP-LC3B BsNb ANTC but not TBFP-LC3B (control; Figure 2G). WB analysis further confirmed the successful simultaneous degradation of EGFP-Tiam1 and mCherry-RhoA proteins in the presence of GBP-RBP-TBFP-LC3B (Figure 2H,I). Simultaneous dual-target degradation would be invaluable to produce synergetic pharmacological effects in drug development.36
Characterization and Mechanistic Elucidation of the ATNC Degradation System
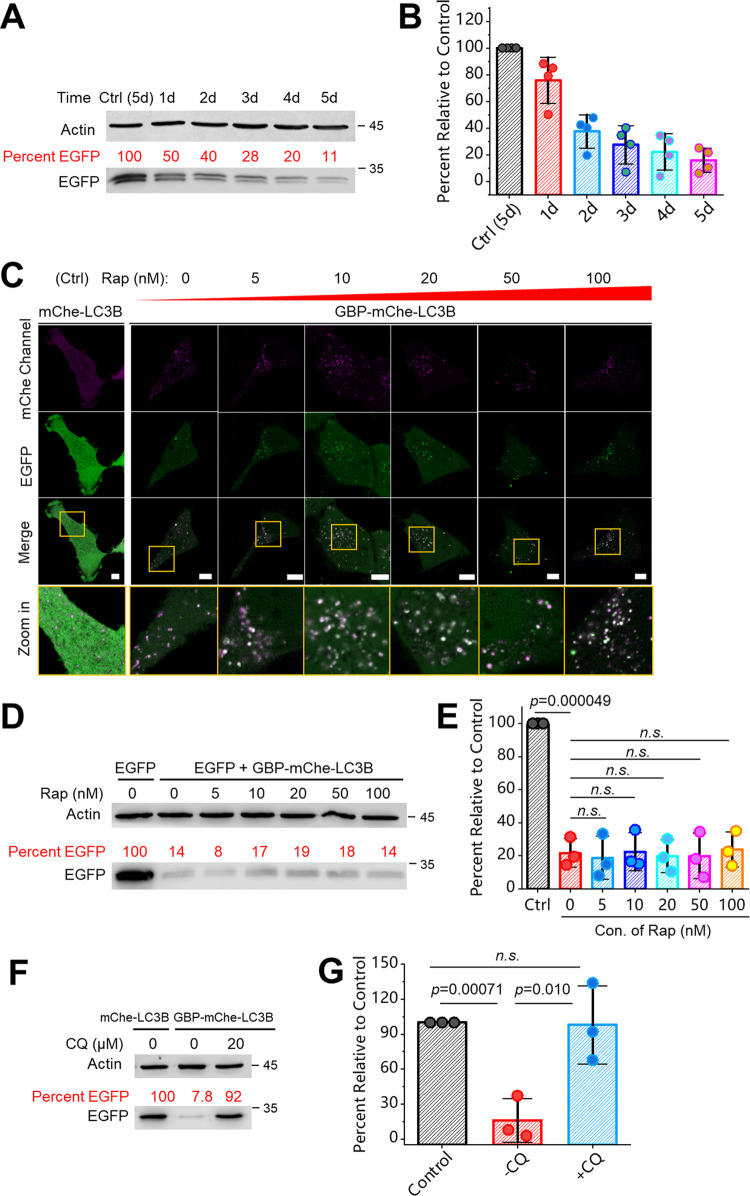
With the results obtained above, we were motivated to further characterize and mechanistically understand this degradation system. First, we found that cells coexpressing GBP-mCherry-LC3B and EGFP showed time-dependent degradation of EGFP over time (Figure 3A,B); the longer the time, the more complete the degradation. Then, we studied whether rapamycin, an immunosuppressant that activates autophagy via the mTOR pathway,37 affects the degradation or not. Gradient concentrations of rapamycin were added to the cell culture, and it seems that the degree of degradation was not significantly affected (Figure 3C–E). Hence, ATNC generally works in a rapamycin-insensitive fashion, and efficient degradation can be achieved even without adding autophagy-activation drugs, for example, rapamycin that possesses additional side physiological effects. Finally, we used an autophagy inhibitor, chloroquine (CQ),38 to treat the cells and found that degradation of EGFP was almost completely inhibited (Figure 3F,G). This result further validated that nanobody-LC3B ATNC induced degradation through the autophagic pathway.
Figure 3.
Further characterization and mechanistic elucidation of ATNC: (A) WB analysis of live HeLa cells coexpressing EGFP and GBP-mCherry-LC3B was performed and the cell lysate samples were collected 1, 2, 3, 4, and 5 days after transfection; a control cell lysate sample only expressing EGFP was collected at the 5th day after transfection. (B) Statistical quantification of the WB analysis (n = 4 experiments). (C) Gradient concentration of Rap (0, 5, 10, 20, 50, and 100 nM) was added to live HeLa cells coexpressing EGFP and GBP-mCherry-LC3B, and imaging was performed 22 h after drug addition (control: cells coexpressing EGFP and mCherry-LC3B). (D) WB analysis (46 h) of the cell samples corresponding to (C). (E) Quantification of WB results (n = 3 experiments). (F) Live HeLa cells coexpressing EGFP and mCherry-LC3B (control) or GBP-mCherry-LC3B with or without adding CQ (46 h) were subjected to WB analysis. (G) Statistical quantification of WB results (n = 3 experiments). One-sided Student’s t-test was used in this figure; n.s.: nonsignificant; see Methods section for description of box plots; bar graphs denote mean ± SD; see Methods section for more details about drug treatment; mChe: mCherry.
Autophagy-mediated degradation has a key advantage over proteosome-mediated degradation because of its significantly wider substrate scope. Not only can it degrade soluble proteins, but it can also target other cellular components such as organelles and aggregated proteins. To demonstrate this advantage, we tested the degradation of nonproteinaceous targets such as mitochondria organelles, which offer potential for treating diseases related to mitochondria damage.10,39 We used EGFP-mito (mito: mitochondria targeting sequence) to label mitochondrial surfaces in live HeLa cells. Representative confocal microscopy images showed that mitochondria were targeted to autophagic puncta (Figure S5A,B), and western blot analysis confirmed the degradation of EGFP-mito (Figure S5C,D). We further analyzed the Tom20 protein, a key mitochondrial membrane marker, and the western blot also revealed strong degradation of this marker protein (Figure S5C,E). Tau is a prone-to-aggregate microtubule-binding protein, whose clearance is recognized as a route to cure some neuronal degenerative diseases.40 We showed that EGFP-Tau(P301L) was also strongly degraded using the GBP-mCherry-LC3B chimeric degrader (Figure S6). Hence, ATNC is a degradation tool that covers broad substrate scope.
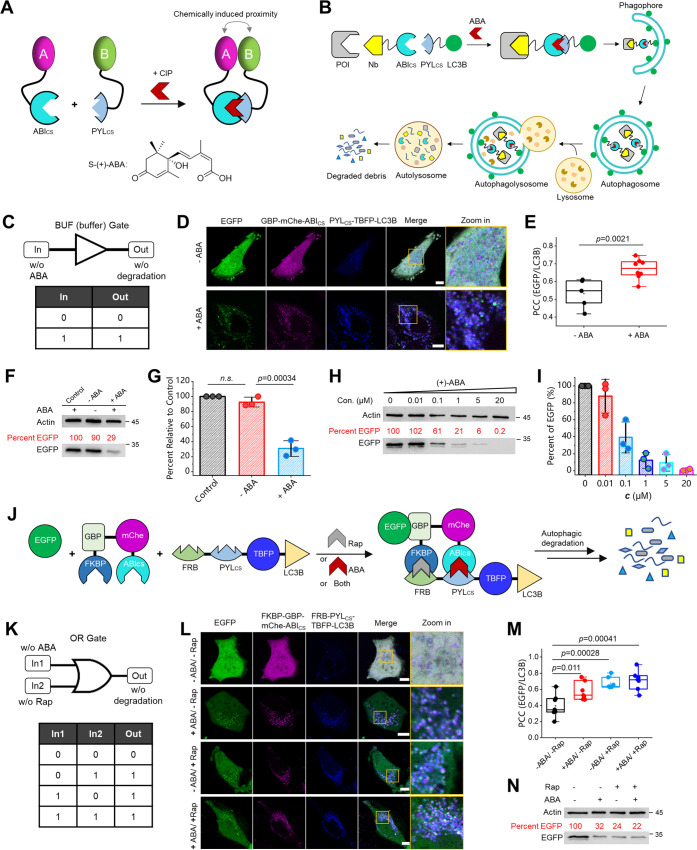
Generation of a Conditional and Tunable Logic Gated Degradation System by Integrating Chemically Induced Proximity with ATNC
The degradation experiments above rely on intracellularly expressed nanobody-LIR/LC3B chimeras or in the form of so-called expressed intrabodies.41 This approach wins in terms of simplicity and ease of use. However, it lacks conditional control and turnability regarding the degree of degradation. Hence, we elaborated ATNC by combining it with chemically induced proximity (CIP)42 or molecular glue43 to create a CIP-controlled conditional degradation system. We chose (+)-abscisic acid (ABA)-based CIP, which enables the dimerization between PYLcs (PYL: pyrabactin resistance-like; cs: complementary surface) and ABIcs (ABI: ABA insensitive) in the presence of ABA, a molecular glue-like dimerizer44 (Figure 4A). PYLcs itself does not interact with ABIcs; however, after PYLcs first binds with ABA, a conformational change of PYLcs occurs, which creates an extensive binding surface that leads to a high affinity to ABIcs.44
Figure 4.
Establishment of logic-gated and tunable degradation systems by integrating CIPs with ATNC: (A) Schematic representation of the ABA-based CIP system and the chemical structure of the CIP inducer (+)-ABA. (B, C) Schematic representation of CIP-integrated ANTC that enables buffer (BUF) gated degradation solely depending on the input of ABA. (D) Live HeLa cells coexpressing EGFP, GBP-mCherry-ABIcs, and PYLcs-TBFP-LC3B (TBFP: mTagBFP2) with or without adding ABA (20 μM, 1.5 h, then wash) were imaged 22.5 h after ABA addition. (E) PCC colocalization analysis between EGFP and PYLcs-TBFP-LC3B with (n = 8 cells) or without (n = 5 cells) adding ABA. (F) WB analysis (48 h) of the control (EGFP only) sample and other cell lysate samples with or without adding ABA (20 μM, 1.5 h, then wash). (G) Statistical quantification of WB results (n = 3 experiments). (H) Live HeLa cells coexpressing EGFP, GBP-mCherry-ABIcs, and PYLcs-TBFP-LC3B were added with gradient concentrations of ABA for 48 h, and the cell lysate was subjected to WB analysis. (I) Statistical quantification of WB results (n = 3 experiments). (J, K) The design of an OR gated degradation system using the mutually orthogonal ABA (ABIcs/PYLcs) and Rap (FKBP/FRB)-based CIPs, which enables OR degradation in the presence of ABA (20 μM), Rap (10 nM), or both. (L) Representative confocal micrographs (24 h) of OR gated degradation. (M) PCC colocalization analysis (n = 7 cells). (N) Representative WB results (48 h). One-sided Student’s t-test was used; n.s.: nonsignificant; see Methods section for description of box plots; bar graphs: mean ± SD; mChe: mCherry.
In this CIP-controlled degradation system, a nanobody fused with ABIcs binds with the target protein, while PYLcs-fused LC3B is localized to phagophores/autophagosomes. Hence, the addition of the ABA molecule will dimerize ABIcs and PYLcs and recruit the protein for degradation (Figure 4B). As a result, a conditional degradation system will be established solely depending on the input of the ABA signal (Figure 4C). We found that ABA indeed induces the dimerization between GBP-mCherry-ABIcs and PYLcs-TBFP-LC3B (Figure 4D,E; TBFP: mTagBFP2 fluorescent protein), subsequently resulting in degradation of EGFP (Figure 4F,G) in an ABA-dependent fashion. Since the degree of dimerization is dependent on the concentration of ABA,44 we envisioned that the degree of degradation could be fine-tuned simply by applying varied concentrations of ABA. Therefore, gradient concentrations of ABA were added to the cell culture, and WB analysis revealed that the degree of degradation was readily fine-tuned by applying different concentrations of ABA (Figure 4H,I).
Next, we were motivated to establish a more advanced OR gated degradation system by integrating two mutually orthogonal CIPs (ABA and Rap) with the ATNC (Figure 4J,K). Rap is a CIP that induces the heterodimerization between FKBP (FK506 binding protein) and FRB (FKBP12-rapamycin-binding) domains.45 In the OR gated system, the GBP nanobody is fused with both FKBP and ABIcs, while the LC3B module is fused with both FRB and PYLcs. Hence, the presence of ABA, Rap, or both will trigger the dimerization and subsequently the degradation. Confocal micrographs (Figure 4L,M) and WB analysis (Figure 4N) revealed successful OR gated degradation.
Development of Cell-Permeable Phagobody Drugs for Degradation of Therapeutically Relevant Undruggable Proteins
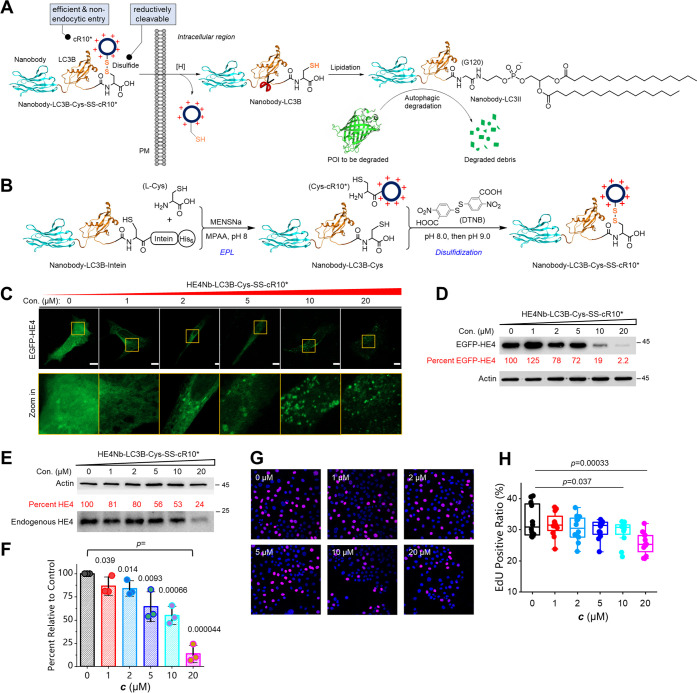
Inspired by the above successes, we envisioned that ATNC could be further extended to create autophagy-based degrader drugs via the generation of cell-penetrating ATNCs or cell-permeable phagobodies for degradation of therapeutically relevant undruggable proteins. To test this possibility, we first designed a cell-permeable ATNC drug called GBP-LC3B-Cys-SS-cR10* (or GLR) for degradation of EGFP (Figure 5A, left). GLR features a GBP nanobody for recruiting EGFP, an LC3B protein for localizing to autophagic structures, and a cyclic cell-penetrating decaarginine (cR10*) peptide (Figure S7) for nonendocytic delivery into a live cell,19,35 and a disulfide bridge (–SS–) that allows efficient cleavage and then removal of the cR10* module inside the reducing intracellular environment (Figure 5A, right). GLR was assembled from the GBP-LC3B-intein precursor via two major steps. The first step was expressed protein ligation (EPL) to cleave the intein tag while installing a C-terminal cysteine residue. The second step was disulfidization to attach the Cys-cR10* module onto GBP-LC3B-Cys to create GBP-LC3B-Cys-SS-cR10* (Figure 5B). GBP-LC3B-intein was expressed in high yield, and the final GLR product was readily obtained (Figure S8). Gratefully, GLR enabled the dose-dependent degradation of EGFP according to microscopic analysis (Figure S9A) and western blot validation (Figure S9B). Hence, cell-permeable phagobodies were generated that can be used to degrade a target protein simply via drug treatment in a dose-dependent fashion.
Figure 5.
Development of cell-permeable phagobody drugs for degradation of the therapeutically relevant undruggable protein HE4: (A) Schematic view of the working mechanism of the cell-permeable phagobody degrader. (B) Schematic view of the two-step assembly of nanobody-LC3B-Cys-SS-cR10* degrader. (C) Representative confocal micrographs of live HeLa cells expressing EGFP-HE4 treated with gradient concentrations of HE4Nb-LC3B-Cys-SS-cR10* (HLR) for 24 h. (D) WB analysis results of the cell lysate sample 24 h after drug treatment. (E) Representative WB analysis of OVCAR3 ovarian cancer cells treated with gradient concentrations of HLR for 24 h. (F) Statistical quantification of WB results (n = 3 experiments). (G) Representative confocal microscopy images of the OVCAR3 cells treated with different concentrations of HLR (blue: nucleus stained by 5 μg mL–1 Hoechst; magenta: EdU-labeled S-phase cell). (H) Quantification of EdU assay results (n = 12 cells for each). One-sided Student’s t-test was used; see Methods section for description of box plots; bar graphs denote mean ± SD.
Next, we were motivated to design a cell-permeable phagobody for selective degradation of a disease-related undruggable protein, human epididymis protein 4 (HE4). HE4 is a glycoprotein recently identified as a tumor marker overexpressed in several carcinomas such as endometrial cancer and ovarian cancer.46 Recent studies showed that overexpression of HE4 promotes tumor growth;47 hence, it was speculated that off-regulation of HE4 would be a promising approach to cure relevant cancers. Unfortunately, no small molecules have been identified so far to inhibit this glycoprotein or specifically bind with it, which renders HE4 both an undruggable protein and a difficult unligandable target for bivalent small-molecule degraders. In order to circumvent this problem, we designed and generated another phagobody degrader, HE4 nanobody-LC3B-Cys-SS-cR10*, abbreviated as HLR. Similarly, HLR was assembled from the HE4 nanobody-LC3B-intein precursor via two steps including EPL and disulfidization (Figure S10). First, we overexpressed EGFP-HE4 in live HeLa cells to facilitate the visualization of the degradation process. Both confocal microscopy images (Figure 5C) and WB analysis (Figure 5D) showed that EGFP-HE4 was degraded in a dose-dependent manner, and the higher the concentration, the more complete the degradation. We next used an ovarian cancer cell line, OVCAR3, to evaluate the degradation of endogenous HE4 and the resulting biological responses. Delightfully, endogenous HE4 in ovarian cancer cells was also degraded by HLR in a dose-dependent manner (Figure 5E,F). Finally, we studied whether degradation of HE4 will inhibit cancer cell proliferation using an EdU (5-ethynyl-2′-deoxyuridine) cell proliferation assay. As can be seen from the microscopy images (Figure 5G) and statistical analysis (Figure 5H), the EdU-positive OVCAR3 cell or the S-phase cell ratio decreased significantly after adding HLR, and the higher the concentration, the stronger the inhibition. Moreover, we found that cell migration was significantly inhibited after adding the HLR degrader using a scratch-wounding cell migration assay (Figure S11). Therefore, we have created a cell-permeable phagobody of therapeutic potential that downregulates the undruggable and unligandable cancer biomarker HE4, which allows suppression of ovarian cancer cell proliferation and migration.
Conclusions
In summary, we introduced general, modular, and versatile autophagy-targeting nanobody chimeras (ATNCs), or phagobodies, for the selective degradation of intracellular unligandable and undruggable proteins. We first showed the general applicability of this tool for the degradation of various proteins fused with a nanobody binding tag such as the commonly used EGFP tag. We next showed the modular feature of this tool by alternating the nanobody module for the degradation of different antigenic targets, including endogenous ones. We also showed that ATNCs degrade not only soluble proteins but also other substrates such as organelles.
We further elaborated ATNC by integrating it with CIP and cyclic CPP methodologies. Therefore, we have introduced in total three variations to implement ATNC technology, each accompanied by different features and advantages (Figure S12). The first variation is simply expressed intracellular ATNCs or so-called ATNC intrabodies. For this purpose, no more than a plasmid is needed to apply the ATNC technology. The second form is to integrate chemically induced proximity to establish logic-gate-operated conditional and tunable TPD systems. The third form is the generation of cell-permeable phagobodies carrying a disulfide-bridged cyclic cell-penetrating peptide that mediates nonendocytic intracellular delivery. Importantly, we use this approach to create a therapeutically relevant degrader, HE4Nb-LC3B-Cys-SS-cR10*, that allows the degradation of the undruggable and unligandable endogenous target HE4 and subsequently achieves the suppression of ovarian cancer cell proliferation and migration.
Compared to recently reported small-molecule degraders working through the autophagic pathway, ATNC can be easily implemented without the necessity to prepare degraders via multistep organic synthesis, hence particularly benign for nonchemistry biologists48 who are interested in autophagy-based degradation. Compared with small-molecule degraders, cell-permeable ATNCs are able to address unligandable and undruggable therapeutically relevant proteins. The use of a nanobody also potentially enhances the specificity and binding affinity toward targets of interest. On the other hand, ATNC methodology may also be associated with few limitations. Due to the relatively large M.W., it typically requires more quantities of the ATNC degrader to achieve sufficient degradation. Also, generation of nanobodies sometimes may also turn out to be a time-consuming process and needs several trials. In general, given the above-mentioned findings, we envision that ATNC will be a promising degradation tool for not only biological studies but also biomedical applications.
Acknowledgments
The authors thank the Core Facilities at HCLS and School of Life Science and Technology in HIT. The authors also thank Donglian Wu for sharing additional nanobody vectors and Simin Xia for the assistance in cell culture experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c08843.
Methods and materials; preparation of a cyclic cell-penetrating peptide; design and evaluation of LIR-based ANTC; RhoNb-mCherry-LC3B ATNC degrades overexpressed EGFP-RhoA in live HeLa cell; demonstration of the modularity of ANTC for degradation of alternative cellular targets; degradation of the organelle mitochondria via ATNC; chemical structure and HPLC-MS characterization of Cys-cR10* (PDF)
Source Data of ATNC (XLSX)
Author Contributions
§ H.H. and C.Z. contributed equally to this work. All authors have given approval to the final version of the manuscript.
This work was supported by funding from HIT, Overseas Outstanding Young Talents Program of China, the National Natural Science Foundation of China (Grant No. 32071410 to X.C.), and the Natural Science Foundation of Heilongjiang Province of China (Grant No. YQ2022B004 to X.C.).
The authors declare the following competing financial interest(s): This work was also subjected to invention patent applications.
Supplementary Material
References
- Zhao L.; Zhao J.; Zhong K. H.; Tong A. P.; Jia D. Targeted protein degradation: mechanisms, strategies and application. Signal Transduction Targeted Ther. 2022, 7, 113 10.1038/s41392-022-00966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel R. R.; Lu D.; Chowdhury S. R.; Monroy E. Y.; Wang J. Targeted protein degradation via lysosomes. Biochemistry 2023, 62 (3), 564–579. 10.1021/acs.biochem.2c00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S. X.; Huang W. H.; Zheng X. L.; Cheng L. Y.; Zhang Z. M.; Wang J.; Shen Z. R. Proteolysis targeting chimera (PROTAC) in drug discovery paradigm: Recent progress and future challenges. Eur. J. Med. Chem. 2021, 210, 112981 10.1016/j.ejmech.2020.112981. [DOI] [PubMed] [Google Scholar]
- Jang J.; To C.; De Clercq D. J. H.; Park E.; Ponthier C. M.; Shin B. H.; Mushajiang M.; Nowak R. P.; Fischer E. S.; Eck M. J.; Janne P. A.; Gray N. S. Mutant-selective allosteric EGFR degraders are effective against a broad range of drug-resistant mutations. Angew. Chem., Int. Ed. 2020, 59 (34), 14481–14489. 10.1002/anie.202003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Y.; Wang C.; Wang Z. Y.; Zhu C. G.; Li J.; Sha T.; Ma L. X.; Gao C.; Yang Y.; Sun Y. M.; Wang J.; Sun X. L.; Lu C. Q.; Difiglia M.; Mei Y. N.; Ding C.; Luo S. Q.; Dang Y. J.; Ding Y.; Fei Y. Y.; Lu B. X. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 2019, 575 (7781), 203–209. 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- Banik S. M.; Pedram K.; Wisnovsky S.; Ahn G.; Riley N. M.; Bertozzi C. R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584 (7820), 291–297. 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Han Y.; Yang Y. F.; Lin F.; Li K. X.; Kong L. H.; Liu H. X.; Dang Y. J.; Lin J.; Chen P. R. Covalently engineered nanobody chimeras for targeted membrane protein degradation. J. Am. Chem. Soc. 2021, 143 (40), 16377–16382. 10.1021/jacs.1c08521. [DOI] [PubMed] [Google Scholar]
- Paiva S. L.; Crews C. M. Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 2019, 50, 111–119. 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pance K.; Gramespacher J. A.; Byrnes J. R.; Salangsang F.; Serrano J. A. C.; Cotton A. D.; Steri V.; Wells J. A. Modular cytokine receptor-targeting chimeras for targeted degradation of cell surface and extracellular proteins. Nat. Biotechnol. 2023, 41, 273–281. 10.1038/s41587-022-01456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D.; Moriyama J.; Nakamura T.; Miki E.; Takahashi E.; Sato A.; Akaike T.; Itto-Nakama K.; Arimoto H. AUTACs: Cargo-specific degraders using selective autophagy. Mol. Cell 2019, 76 (5), 797–810. 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- Fu Y. H.; Chen N. X.; Wang Z. Y.; Luo S. Q.; Ding Y.; Lu B. X. Degradation of lipid droplets by chimeric autophagy-tethering compounds. Cell Res. 2021, 31 (9), 965–979. 10.1038/s41422-021-00532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C. H.; Kim H. Y.; Lee M. J.; Heo A. J.; Park D. Y.; Lim S.; Shin S.; Ganipisetti S.; Yang W. S.; Jung C. A.; Kim K. Y.; Jeong E. H.; Park S. H.; Bin Kim S.; Lee S. J.; Na J. E.; Kang J. I.; Chi H. M.; Kim H. T.; Kim Y. K.; Kim B. Y.; Kwon Y. T. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat. Commun. 2022, 13, 904 10.1038/s41467-022-28520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anding A. L.; Baehrecke E. H. Cleaning house: Selective autophagy of organelles. Dev. Cell 2017, 41 (1), 10–22. 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; He S.; Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer 2020, 19 (1), 12 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid R. T. U.; Reichel A. A mechanistic pharmacodynamic modeling framework for the assessment and optimization of proteolysis targeting chimeras (PROTACs). Pharmaceutics 2023, 15 (1), 195 10.3390/pharmaceutics15010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A decade of drug-likeness Nat. Rev. Drug Discovery 2007, 6 (11), 853.
- Qin L. A.; Dai H.; Wang J. F. Key considerations in targeted protein degradation drug discovery and development. Front. Chem. 2022, 10, 934337 10.3389/fchem.2022.934337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh-Everiss T.; Laraia L. Small molecule probes for targeting autophagy. Nat. Chem. Biol. 2021, 17 (6), 653–664. 10.1038/s41589-021-00768-9. [DOI] [PubMed] [Google Scholar]
- Park S. E.; Sajid M. I.; Parang K.; Tiwari R. K. Cyclic cell-penetrating peptides as efficient intracellular drug delivery tools. Mol. Pharmaceutics 2019, 16 (9), 3727–3743. 10.1021/acs.molpharmaceut.9b00633. [DOI] [PubMed] [Google Scholar]
- Birgisdottir Å. B.; Lamark T.; Johansen T. The LIR motif - crucial for selective autophagy. J. Cell Sci. 2013, 126 (15), 3237–3247. 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- Rasmussen M. S.; Mouilleron S.; Shrestha B. K.; Wirth M.; Lee R.; Larsen K. B.; Princely Y. A.; O’Reilly N.; Sjottem E.; Tooze S. A.; Lamark T.; Johansen T. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy 2017, 13 (5), 834–853. 10.1080/15548627.2017.1287651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubala M. H.; Kovtun O.; Alexandrov K.; Collins B. M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010, 19 (12), 2389–2401. 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T.; Svenning S.; Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017, 61 (6), 609–624. 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- Shao Y. Y.; Sun N. Y.; Jeng Y. M.; Wu Y. M.; Hsu C.; Hsu C. H.; Hsu H. C.; Cheng A. L.; Lin Z. Z. Eg5 as a prognostic biomarker and potential therapeutic target for hepatocellular carcinoma. Cells 2021, 10 (7), 1698 10.3390/cells10071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens A. E.; Roovers R. C.; Collard J. G. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003, 546 (1), 11–16. 10.1016/S0014-5793(03)00435-6. [DOI] [PubMed] [Google Scholar]
- Nobes C. D.; Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81 (1), 53–62. 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Moustaqil M.; Bhumkar A.; Gonzalez L.; Raoul L.; Hunter D. J. B.; Carrive P.; Sierecki E.; Gambin Y. A split-luciferase reporter recognizing GFP and mCherry tags to facilitate studies of protein-protein interactions. Int. J. Mol. Sci. 2017, 18 (12), 2681 10.3390/ijms18122681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F. M.; Ridley A. J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582 (14), 2093–2101. 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Liu N.; Bi F.; Pan Y.; Sun L.; Xue Y.; Shi Y.; Yao X.; Zheng Y.; Fan D. Reversal of the malignant phenotype of gastric cancer cells by inhibition of RhoA expression and activity. Clin. Cancer Res. 2004, 10 (18), 6239–6247. 10.1158/1078-0432.CCR-04-0242. [DOI] [PubMed] [Google Scholar]
- Dupont S.; Morsut L.; Aragona M.; Enzo E.; Giulitti S.; Cordenonsi M.; Zanconato F.; Le Digabel J.; Forcato M.; Bicciato S.; Elvassore N.; Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474 (7350), 179–183. 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Valon L.; Marin-Llaurado A.; Wyatt T.; Charras G.; Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nat. Commun. 2017, 8, 14396 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzke H.; Kilisch M.; Martinez-Carranza M.; Sograte-Idrissi S.; Rajavel A.; Schlichthaerle T.; Engels N.; Jungmann R.; Stenmark P.; Opazo F.; Frey S. The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 2019, 10, 4403 10.1038/s41467-019-12301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant D.; Traenkle B.; Maier J.; Kaiser P. D.; Bodenhofer M.; Schmees C.; Vojnovic I.; Pisak-Lukats B.; Endesfelder U.; Rothbauer U. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 2018, 9 (1), 930 10.1038/s41467-018-03191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayer G.; Belzil C.; Gruss O. J.; Nguyen M. D. TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 2014, 71 (16), 3027–3047. 10.1007/s00018-014-1582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Zhou C.; Xia S.; Chen X. Small molecule-nanobody conjugate induced proximity controls intracellular processes and modulates endogenous unligandable targets. Nat. Commun. 2023, 14, 1635 10.1038/s41467-023-37237-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschak E.; Stark H.; Merk D. Polypharmacology by design: A medicinal chemist’s perspective on multitargeting compounds. J. Med. Chem. 2019, 62 (2), 420–444. 10.1021/acs.jmedchem.8b00760. [DOI] [PubMed] [Google Scholar]
- Wu L. L.; Feng Z.; Cui S. Y.; Hou K.; Tang L.; Zhou J. H.; Cai G. Y.; Xie Y. S.; Hong Q.; Fu B.; Chen X. M. Rapamycin upregulates autophagy by inhibiting the mTOR-ULK1 pathway, resulting in reduced podocyte injury. PLoS One 2013, 8 (5), e63799 10.1371/journal.pone.0063799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M.; Orhon I.; Rocchi C.; Zhou X. D.; Luhr M.; Hijlkema K. J.; Coppes R. P.; Engedal N.; Mari M.; Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14 (8), 1435–1455. 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti D.; Braidy N.; De Rasmo D.; Signorile A.; Rossi L.; Atanasov A. G.; Volpicella M.; Henrion-Caude A.; Nabavi S. M.; Vacca R. A. Mitochondria as pharmacological targets in Down syndrome. Free Radical Biol. Med. 2018, 114, 69–83. 10.1016/j.freeradbiomed.2017.08.014. [DOI] [PubMed] [Google Scholar]
- Wang Y. P.; Mandelkow E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17 (1), 22–35. 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- Stocks M. R. Intrabodies: Production and promise. Drug Discovery Today 2004, 9 (22), 960–966. 10.1016/S1359-6446(04)03269-6. [DOI] [PubMed] [Google Scholar]
- Stanton B. Z.; Chory E. J.; Crabtree G. R. Chemically induced proximity in biology and medicine. Science 2018, 359, eaao5902 10.1126/science.aao5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K.; Schulman B. A. Molecular glue concept solidifies. Nat. Chem. Biol. 2020, 16 (1), 2–3. 10.1038/s41589-019-0414-3. [DOI] [PubMed] [Google Scholar]
- Liang F. S.; Ho W. Q.; Crabtree G. R. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signaling 2011, 4 (164), rs2 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles S. D.; Diver S. T.; Austin D. J.; Schreiber S. L. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc. Natl. Acad. Sci. U.S.A. 1997, 94 (15), 7825–7830. 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R.; von Horsten H. H.; Lin Y. F.; Mok S. C.; Crum C. P.; Welch W. R.; Hecht J. L. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometriold ovarian carcinomas. Cancer Res. 2005, 65 (6), 2162–2169. 10.1158/0008-5472.CAN-04-3924. [DOI] [PubMed] [Google Scholar]
- Moore R. G.; Hill E. K.; Horan T.; Yano N.; Kim K.; MacLaughlan S.; Lambert-Messerlian G.; Tseng Y. D.; Padbury J. F.; Miller M. C.; Lange T. S.; Singh R. K. HE4 (WFDC2) gene overexpression promotes ovarian tumor growth. Sci. Rep. 2014, 4, 3574 10.1038/srep03574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.; Khoo R.; Peh K. M.; Teo J.; Chang S. C.; Ng S.; Beilhartz G. L.; Melnyk R. A.; Johannes C. W.; Brown C. J.; Lane D. P.; Henry B.; Partridge A. W. bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA). Proc. Natl. Acad. Sci. U.S.A. 2020, 117 (11), 5791–5800. 10.1073/pnas.1920251117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.