Summary
Low protein diets promote health and longevity in diverse species. Restriction of the branched-chain amino acids (BCAAs) leucine, isoleucine and valine recapitulates many of these benefits in young C57BL/6J mice. Restriction of dietary isoleucine (IleR) is sufficient to promote metabolic health and is required for many benefits of a low protein diet in C57BL/6J males. Here, we test the hypothesis that IleR will promote healthy aging in adult, genetically heterogenous UM-HET3 mice. We find that IleR improves metabolic health in young and old HET3 mice, promoting leanness and glycemic control in both sexes, and reprograms hepatic metabolism in a sex-specific manner. IleR reduces frailty and extends the lifespan of male and female mice, but to a greater degree in males. Our results demonstrate that IleR increases healthspan and longevity in genetically diverse mice and suggests that IleR, or pharmaceuticals that mimic this effect, may have potential as a geroprotective intervention.
Keywords: isoleucine, lifespan, frailty, mice, branched-chain amino acids
Graphical Abstract
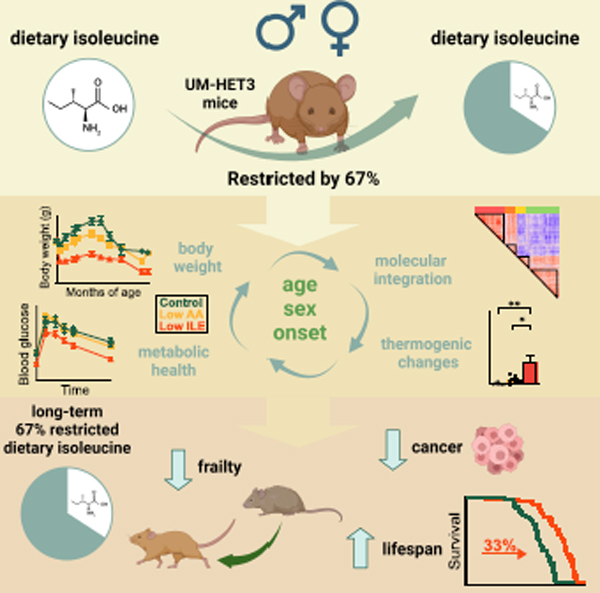
eTOC
Green et al. find that dietary isoleucine is a key regulator of metabolic health and lifespan in genetically heterogeneous mice. Restriction of isoleucine improves metabolic health, reduces frailty, and increases the lifespan of both male and female mice, with greater benefits for males.
Introduction
Dietary interventions such as calorie restriction (CR) extend the lifespan and healthspan of diverse species, including rodents and non-human primates1–5. As adhering to an abstemious CR diet can be difficult, there is great interest in developing interventions that mimic the benefits of a CR diet without reduced calorie intake. Dietary composition has a strong influence on longevity and health, and the role of dietary protein has been the subject of significant investigation.
Beneficial effects of high protein diets for metabolic health have been found by several studies, and these diets are often recommended to the elderly to fight frailty and sarcopenia6,7. In contrast, retrospective and prospective clinical trials have found that lower protein consumption is associated with a decreased risk of age-related diseases and mortality8–11, and a recent twin study found that higher protein diets are paradoxically associated with sarcopenia12. Two short-term clinical trials have found that protein restriction (PR) improves multiple markers of metabolic health, reducing adiposity and improving insulin sensitivity13,14. In rodents, PR improves metabolic health, and the consumption of low protein (LP) diets promotes longevity15–21.
While the mechanisms that mediate the beneficial effects of CR and PR are unknown, both interventions lower consumption of dietary essential amino acids (EAAs). Restriction of the nine EAAs is required for the benefits of CR22, suggesting that one or more of these EAAs regulates lifespan. While significant attention has focused on methionine23–25, we have shown that the branched-chain amino acids (BCAAs) leucine, isoleucine, and valine are of unique metabolic importance. Restriction of all three BCAAs by 67% improves metabolic health in C57BL/6J males and females, and in males extends lifespan by over 30%, indistinguishable from the effect of restricting all 20 common dietary AAs by the same amount19.
While the BCAAs are usually considered as a group, each BCAA has distinct molecular and metabolic effects; this may be due to differential sensing of the BCAAs, for instance by the mammalian target of rapamycin (mTOR) protein kinase, or due to the distinct intermediate and final products of each BCAA. For example, a valine-specific catabolite regulates trans-endothelial fatty acid transport and glucose uptake26,27. We recently showed that the beneficial metabolic effects of protein or BCAA restriction are principally mediated by restriction of isoleucine (IleR). IleR promotes glucose tolerance, reduces adiposity, and is necessary for the metabolic benefits of PR28. In humans, dietary isoleucine levels are associated with body mass index, while blood levels of isoleucine correlate with increased mortality28,29.
Here, we investigated the hypothesis that IleR would extend the healthspan and lifespan of genetically heterogenous UM-HET3 (HET3) mice, which are extensively used in aging studies30–33 as they better reflect the genetically diverse human population than any single inbred strain. Using HET3 mice also increases the likelihood that any findings will be generalizable and robust, and not related to a strain-specific genetic defect or cause of death. We find that short-term IleR improves metabolic health in young HET3 mice of both sexes, reducing weight, adiposity, and improving glucose homeostasis. Lifelong IleR started in 6-month-old adult HET3 mice promotes lifelong leanness and glycemic control, and reprograms hepatic metabolism in a way distinct from a diet in which all dietary AAs are restricted. Finally, IleR reduces frailty in both sexes, extends median and maximum lifespan in male mice, and extends median lifespan more modestly in females. In conclusion, our results demonstrate that reducing dietary levels of isoleucine promotes healthy aging in mice, is more robust than PR in its ability to promote healthy aging, and may be a uniquely potent method to promote healthy aging without requiring reduced calorie consumption.
Results
IleR improves metabolic health in young HET3 mice
In an initial study, we placed male and female 9-week-old HET3 mice (HET3 mice are the F2 progeny of (BALB/cJ × C57BL/6J) mothers and (C3H/HeJ × DBA/2J) fathers, and have segregating alleles from all four parental strains) on one of three AA-defined diets28. Briefly, our Control diet contains all twenty common AAs; the diet composition reflects that of a natural chow in which 21% of calories are derived from protein. We also utilized diets in which all AAs (Low AA) or isoleucine (Low Ile) was reduced by 67%. All three diets are isocaloric, with identical levels of fat; in the Low AA diet, carbohydrates were used to replace calories from AAs, while in the Low Ile diet, non-EAAs were increased to keep the calories derived from AAs constant (Fig. 1A, Table S1).
Figure 1: IleR reduces body weight and improves glycemic control in young HET3 mice.
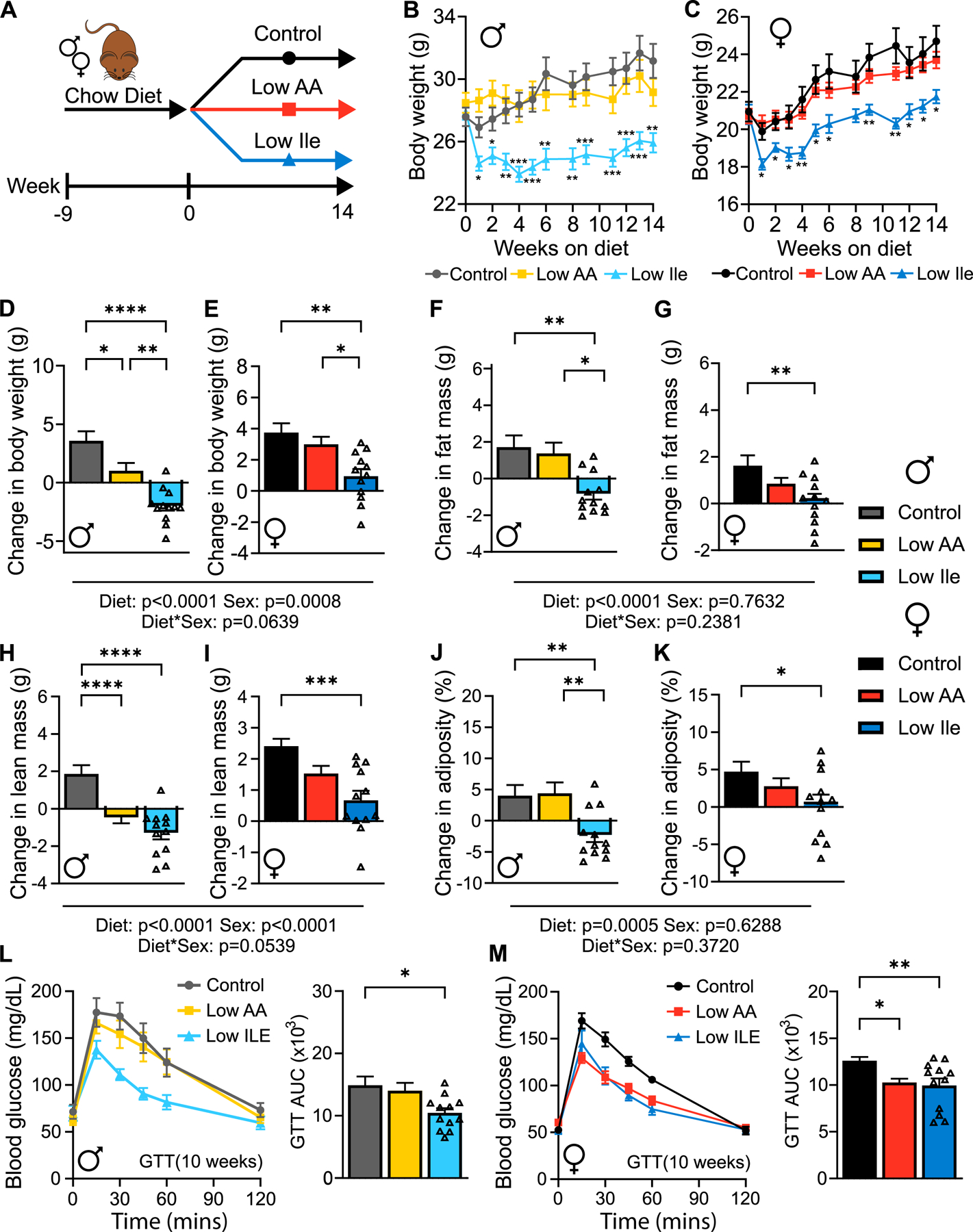
(A) Experimental scheme. (B-C) Male (B) and female (C) mouse weight. (D-K) Change in weight (D-E), fat mass (F-G), lean mass (H-I) and adiposity (J-K) over 14 weeks. (L-M) Glucose tolerance of male (L) and female (M) mice after 10 weeks on the indicated diets. (B-M) n=11–12 mice/group. (B-C) Tukey test following residual maximum-likelihood (REML) analysis model with Geisser-Greenhouse correction. (D-K) Tukey test following 2-way ANOVA. (L, M) Tukey test following ANOVA. (B-M) *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. P-values for the overall effect of Sex, Diet and the interaction represent the significant p-values from the two-way ANOVA. Data represented as mean ± SEM. See also Figure S1.
Both male (Fig. 1B) and female (Fig. 1C) mice on Control and Low AA diets gained weight; however Low Ile-fed mice of both sexes initially lost a significant amount of weight (~12% in males and ~9% in females), which female mice gradually regained. The change in weight was sex and diet-dependent; Low Ile-fed males and females, as well as Low AA-fed males, gained significantly less weight than their Control-fed counterparts (Figs. 1D–E).
Body composition was determined at the beginning and end of the experiment. A Low Ile diet had significant effects on both fat and lean mass, with the effect on lean mass being sex-dependent (Figs. 1F–I). Low Ile-fed males lost both fat and lean mass, while Low AA-fed males accreted a similar amount of fat mass as Control-fed males but lost lean mass (Fig. 1F, H). Low Ile-fed females accreted significantly less fat and lean mass than Control-fed females (Fig. 1G, I). Overall, a Low Ile diet reduced adiposity in both sexes, while a Low AA diet did not reduce adiposity in either sex (Figs. 1J–K and S1A–B).
We expected that a Low Ile and a Low AA diet would improve glycemic control, and we performed glucose tolerance tests (GTTs) after 3 (Figs. S1C–D) and 10 weeks on diet (Figs. 1L–M). The effects were dependent on both diet and sex, with Low Ile-fed males showing significant improvements at both times, while Low Ile-fed females showed improved glucose tolerance only after ten weeks. This was independent of improvements in insulin sensitivity, as measured through an insulin tolerance test (ITT) after either 4 or 11 weeks on diet (Figs. S1E–H), although Low AA-fed females had improved insulin sensitivity at 4 weeks relative to Low Ile-fed mice. Performing an alanine tolerance test (ATT), we found that a Low Ile diet appeared to improve suppression of hepatic gluconeogenesis in both sexes (Figs. S1I–L). Overall, we found that a Low Ile diet was able to reduce weight gain and improve glycemic control in young HET3 mice and was more effective at promoting metabolic health than reducing all AAs.
IleR increases food intake and energy expenditure in young HET3 mice
Despite the effects of a Low Ile diet on body weight and composition, Low-Ile and Low AA-fed males and females ate significantly more calories each day than Control-fed mice (Figs. 2A–B). As a result, both sexes of Low Ile-fed mice consumed significantly more AAs than Control-fed mice (Figs. S2A–B), while consuming significantly less isoleucine (Figs. 2C–D). Notably, Low AA-fed mice of both sexes consumed a similar amount of isoleucine as the Low Ile-fed mice.
Figure 2: IleR alters energy balance in young HET3 mice.
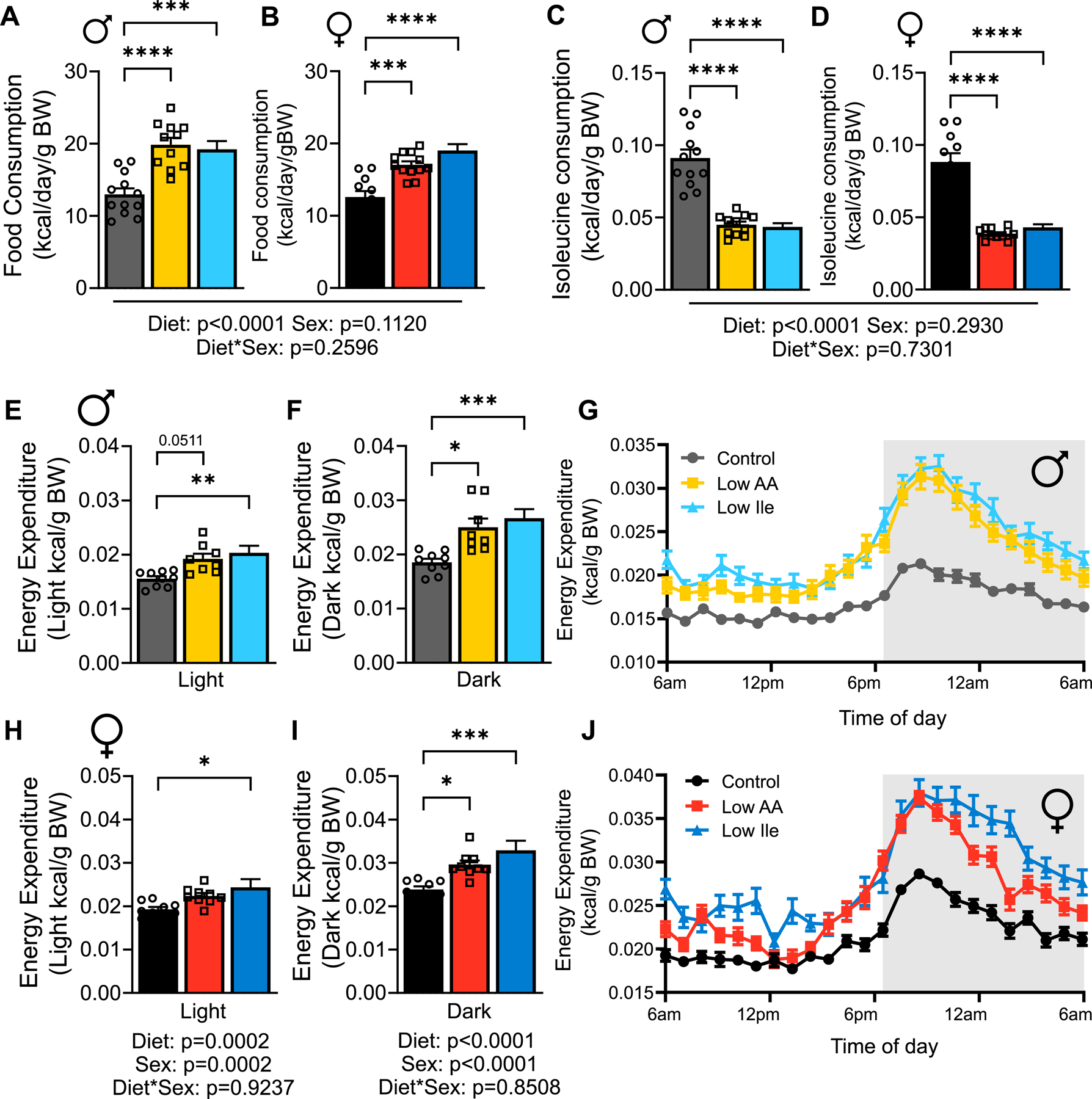
(A-B) Daily food consumption was measured in home cages after 3 weeks on diet in males (A) and females (B). (C-D) Calculated daily isoleucine intake for males (C) and females (D). (A-D) n=11–12 mice/group. (E-J) Energy expenditure measure in males (E-G) and females (H-J); n=8–10 mice/group. (A-F, H-I) Tukey test following 2-way ANOVA, *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. P-values for the overall effect of Diet, Sex and the interaction represent the significant p-values from the two-way ANOVA. Data represented as mean ± SEM. See also Figure S2.
As Low Ile-fed mice weighed less despite significantly increased calorie intake, we investigated other components of energy balance using metabolic chambers. Male mice fed a Low Ile diet had drastically increased energy expenditure as assessed via indirect calorimetry; the effect was particularly prominent during the “active” dark phase, when Low AA fed mice also had significantly upregulated energy expenditure (Figs. 2E–G). Low Ile and Low AA-fed females displayed a similar pattern of increased energy expenditure, particularly during the dark phase (Figs. 2H–J). The average respiratory exchange ratio (RER) was not affected by IleR (Figs. S2C–F); however, there was an effect on timing, with Low Ile-fed mice, particularly females, shifting from low RER to high RER several hours later than Control or Low AA-fed mice (Figs. S2G–H). These alterations in RER and energy expenditure were not associated with altered activity (Figs. S2I–N). In summary, the lower weight of Low Ile and Low AA-fed mice is not due to decreased food intake; consumption of these diets is instead associated with increased food consumption and energy expenditure.
IleR improves metabolic health in older HET3 mice
To determine if IleR could promote healthy aging, 6-month-old male and female HET3 mice raised on chow were placed on either Control, Low AA, or Low Ile diets; at 24 months of age, a pre-selected group of animals was euthanized and tissues collected, while the remaining mice continued aging (Fig. 3A). Tracking weight, we found that Low Ile-fed mice (Figs. 3B–E) weighed significantly less than Control AA and Low AA-fed mice. Much of the difference in Low Ile-fed males was due to a rapid loss of fat mass (~50%) during the first month (Figs. 3F–G), while much of the difference in females was due to fat gain by Control-fed females (Figs. 3H–I). Differences in lean mass contributed less, with Low Ile-fed males dropping ~10% of their lean mass during the first month which was maintained until late life, and Low Ile-fed females accreting less lean mass than Control-fed females (Figs. 3J–M). All groups of males lost adiposity between 6 and 24 months of age, and while there was no significant difference between groups at these ages, over their life Low Ile-fed males had lower adiposity than Control-fed males, as did Low Ile and Low AA-fed females relative to Control-fed females (Figs. 3N–O, S3A–B).
Figure 3: IleR reduces body weight and improves glycemic control in HET3 mice when started in mid-life.
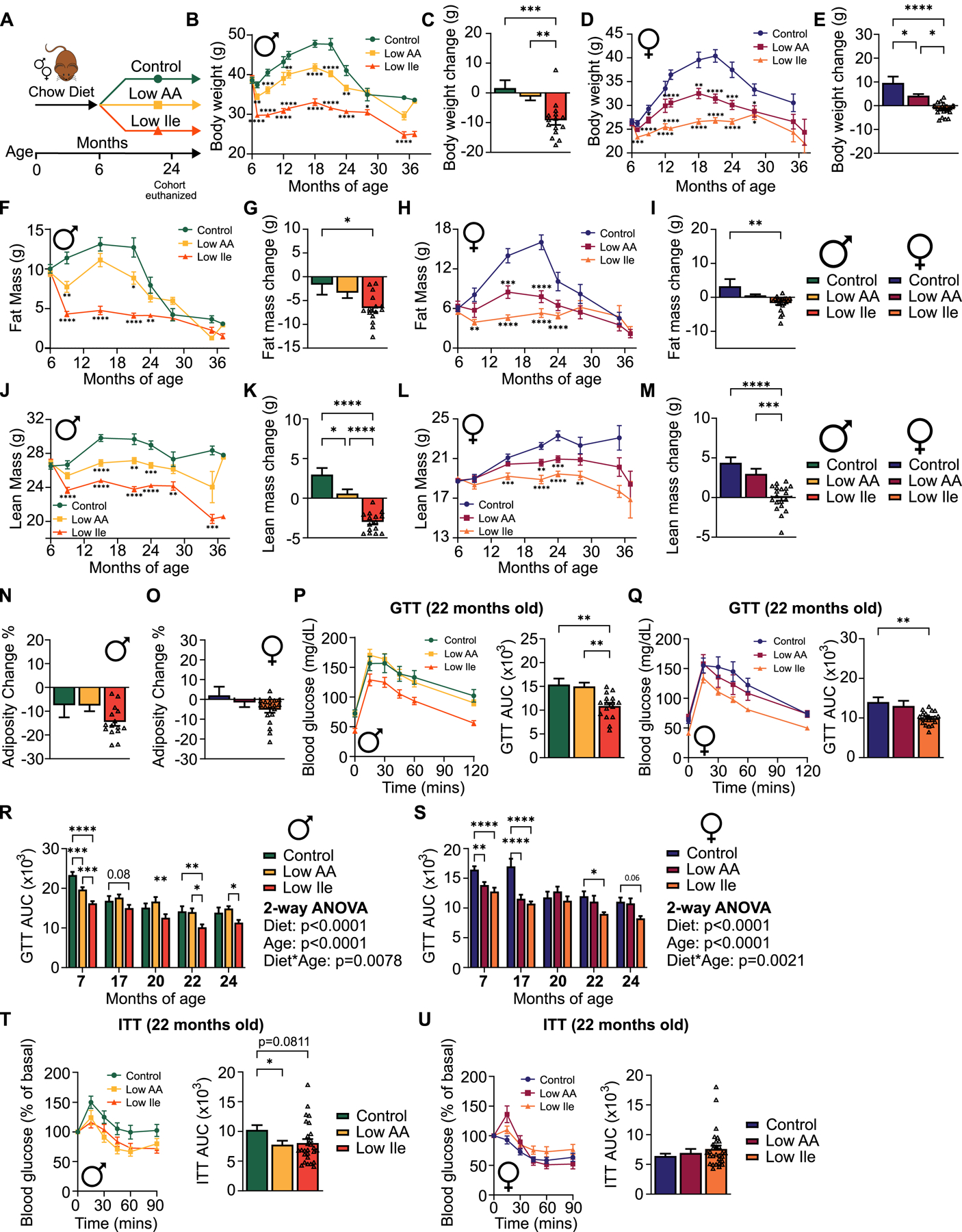
(A) Experimental scheme. At 24 months of age, a pre-selected cohort of mice in each group was euthanized for molecular analysis. (B-E) Male (B-C) and female (D-E) body weight and change from 6–24 months of age. (B, D) n varies by month; maximum 47–53 mice/group. (C) n=9–15 mice/group. (E) n=11–19 mice/group. (F-O) Body composition over the course of the lifespan and change from 6–24 months of age. (F, H, J, L) n varies by month; maximum 47–53 mice/group. (G, K, N) n=9–15 mice/group. (I, M, O) n=11–19 mice/group. (P-Q) Glucose tolerance after 16 months (22 months of age) on the indicated diets in male (P) and female (Q) mice; n=14–16 mice/group for males, 15–20 mice/group for females. (R-S) GTT area-under-the-curve (AUC) for male (R) and female (S) mice over the course of the experiment. Initial n=30–33/group in males, n=30–32 mice/group in females. (T-U) Insulin tolerance after 16 months on diet (22 months of age) on the indicated diets in male (T) and female (U) mice; n=19–27 mice/group for males, 19–29 mice/group for females. (B, D, F, H, J, L) Mixed-effects model with Geisser-Greenhouse correction followed by Dunnett’s multiple comparison correction test, (C, E, G, I, K, M-Q, T,U) Tukey test following ANOVA. (R, S) Tukey test following 2-way ANOVA. (B-U) *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. P-values for the overall effect of Diet, Age and the interaction represent the significant p-values from the two-way ANOVA. Data represented as mean ± SEM. See also Figure S3.
We performed glucose and insulin tolerance tests up to 24 months of age. At 22 months of age (after 16 months on the indicated diets), we found that a Low Ile diet, but not a Low AA diet improved glucose tolerance in both sexes (Figs. 3P–Q). Overall, Low AA and Low Ile diets improved glucose tolerance earlier in life, and this effect persisted in Low Ile-fed mice as they aged (Figs. 3R–S). Low AA-fed males had improved insulin sensitivity relative to Control-fed mice at 22 months of age (Fig. 3T) and trended towards improved insulin sensitivity at earlier timepoints (Fig. S3C); Low Ile-fed males also trended towards improved insulin sensitivity at 22 months of age. In contrast, Low AA-fed females showed improved insulin sensitivity only at 17 months of age, despite a later improvement in glucose tolerance (Figs. 3U, S3D). Overall, IleR powerfully promotes leanness and glycemic control in aging HET3 mice of both sexes, with significantly greater impact than a Low AA diet.
IleR increases food intake and energy expenditure in older HET3 mice
Low AA and Low Ile-fed males and females initially consumed significantly more calories, but significantly less isoleucine, than Control-fed mice; as the mice aged, Low Ile and Low AA-fed mice always ate at least as many calories as Control-fed mice (Figs. 4A–F). Due to the prominent differences in weight and body composition, we used metabolic chambers to investigate how energy balance was impacted by Low AA and Low Ile diets at 9, 14, and 24 months of age (Figs. 4G–L, S3E–T, and S4A–JJ). At all timepoints, Low Ile-fed mice of both sexes had significantly increased energy expenditure relative to Control-fed mice during both the light and dark phases; we also observed increased energy expenditure in Low AA-fed males and females at all ages (Figs. 4G–L, S3O–P, S4A–D and S4S–V). This was not accompanied by significant changes in either respiratory exchange ratio (RER) or activity, although in females at all ages we observed a right-shift in the RER curve of Low Ile-fed mice suggesting an alteration in temporal fuel utilization patterns (Figs. S3E–N, S4G–R, S4Y–JJ). While energy expenditure, RER and activity remained fairly constant over time, 14-month-old females had increased RER on Low AA and Low Ile diets (Fig. S3Q–T).
Figure 4: IleR alters energy balance in HET3 mice when started in mid-life.
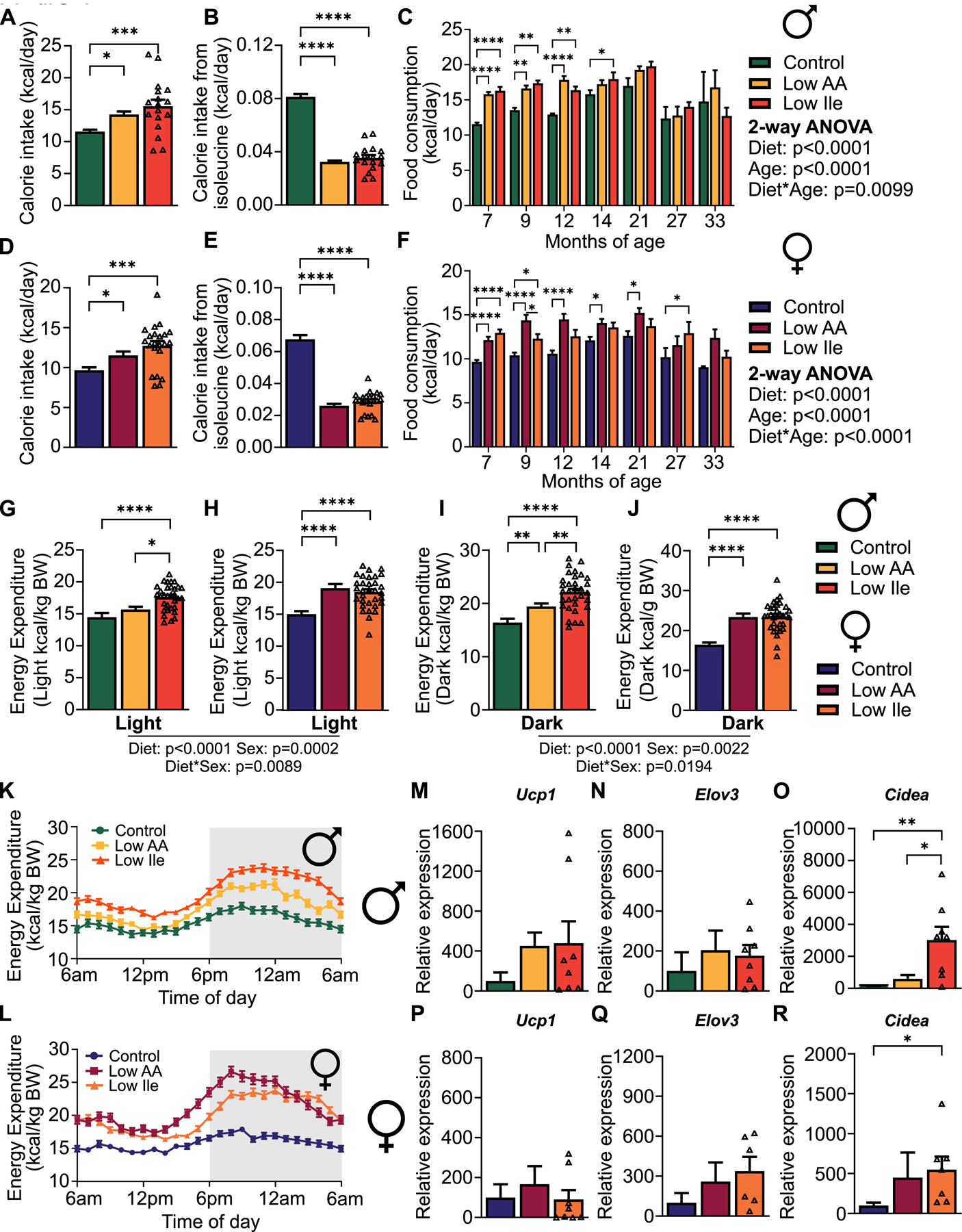
(A-C) Food and isoleucine consumption in male mice after 1 month on diet (A-B) and over the course of the experiment (C). (D-F) Food and isoleucine consumption in female mice after 1 month on diet (D-E) and over the course of the experiment (F). (A, B) n=17–18 mice/group. (C) n varies by month; maximum 47–51 mice/group (D, E) n=20–22 mice/group. (F) n varies by month; maximum 51–53 mice/group. (G-L) Energy expenditure in males and females after 18 months on diet. (G, I, K) n=23–32 mice/group. (H, J, L) n=24–32 mice/group. (M-R) Expression of the indicated genes in the iWAT of 24-month old male (M-O) and female (P-R) mice fed the indicated diets. n=4–8 mice/group. (A,B,D-E,G-J,M-R) Tukey test following ANOVA. (C, F) Tukey test following 2-way ANOVA. (A-R) *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. P-values for the overall effect of Diet, Age and the interaction represent the significant p-values from the two-way ANOVA. Data represented as mean ± SEM. See also Figure S4.
We have previously observed that in C57BL/6J males, a Low Ile diet increases circulating levels of FGF21 and induces hepatic Fgf21, promoting the expression of Ucp1 and other thermogenic, lipogenic, and lipolytic genes in inguinal white adipose tissue (iWAT)28. Consistent with iWAT beiging and the increased energy expenditure we observed, we found that expression of Ucp1 and Elov3 was increased in the iWAT of Low AA and Low Ile-fed males, we did, however, observe a significant increase in the expression of Cidea in Low Ile-fed males (Figs. 4M–O). In females, expression of iWAT Ucp1 was not affected by diet, and although Elov3 levels were increased, it did not reach statistical significance; however, Cidea was significantly upregulated by IleR (Figs. 4P–R). In summary, at all timepoints Low AA and Low Ile-fed mice of both sexes ate at least as many calories as Control-fed mice, yet weighed less and had reduced fat mass, likely due in part to increased energy expenditure associated with increased Cidea expression in iWAT.
Changes in metabolic health relative to isoleucine intake vary with sex and age
We used multivariate analysis to identify age and sex-dependent responses to reduced dietary isoleucine. We correlated the isoleucine intake of each individual mouse on all three diets with 26 phenotypic measurements obtained from each animal for each age and sex, and plotted these in a clustered heatmap (Fig. 5A). For the most part, old and young males and females had similar phenotypic responses to reduced isoleucine intake. In general, isoleucine intake correlates negatively with energy expenditure and RER, and positively with lean and fat mass, body weight, and glucose area under the curve (poor glycemic control). The strength of the changes varies with age, with old male mice having weaker correlations of isoleucine intake with the measured parameters than young males, and vice versa for females (Table S2).
Figure 5: Correlation analysis identifies diet and age dependent and independent physiological and metabolic responses to a Low Ile diet.
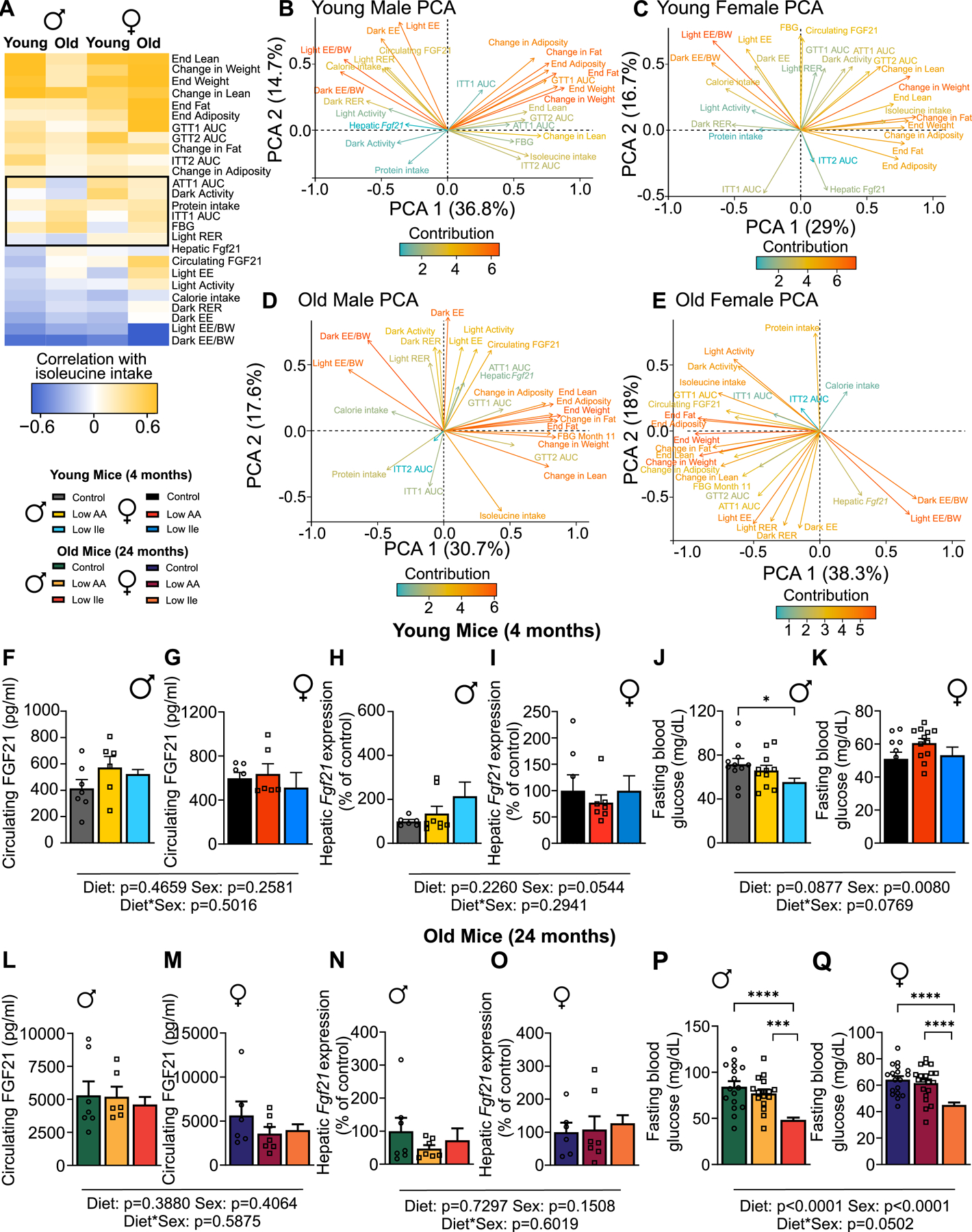
(A) Phenotypic measurements correlated with consumption of isoleucine (kcals) in each mouse (Pearson’s correlation) and clustered (hierarchical clustering). Phenotypic measurements that do not cluster as well appear in the middle of the correlation plot surrounded by a black box. (B-D) Phenotypic measurements from PCA of young and old mice of both sexes were visualized; positively correlated variables point to the same side of the plot, negatively correlated variables point to opposite sides of the plot. Length and color of arrows indicate contribution to the principal components. Young n=11–12 mice/group, old n=17–22 mice/group. (F-Q) Selected phenotypic measurements. Circulating FGF21 n=6–7 mice/group, Hepatic Fgf21 n=6–8 mice/group, FBG n=11–21 mice/group. Tukey test following 2-way ANOVA; *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. P-values for the overall effect of Sex, Diet and the interactions represent the significant p-values from the two-way ANOVA. Data represented as mean ± SEM. See also Table S2. See also Figure S5.
In young and old males, differences between groups are particularly driven by changes in energy expenditure and changes in weight and adiposity, which contributed the most (and oppositely) to the differences in phenotypic variation as shown by principle component analysis (PCA), with isoleucine intake correlating closely with weight, and oppositely with energy expenditure normalized to body weight (Figs. 5B & D). In females, energy expenditure normalized to body weight likewise strongly contributed to differences in phenotypic variation (Figs. 5C & E). Surprisingly, there was a strong contribution and correlation of energy expenditure and hepatic Fgf21 only in aged females (Fig. 5E). When looking across diet and ages groups, energy expenditure appears to contribute to the separation of all groups of Low Ile-fed mice (Figs. 5B–E and S5C–F).
The increased energy expenditure of mice on a PR or IleR diet may be mediated by induction of FGF2117,18,28,34. Circulating FGF21 (Figs. 5F–G & L–M) and hepatic Fgf21 expression (Figs. 5H–I & N–O) did not have a consistent trend across groups. While there was a trend towards increased blood levels of FGF21 and greater hepatic Fgf21 expression in young males, this was not the case in females or in old mice of either sex, nor in the iWAT of old mice (Figs. S5A–B). In contrast, fasting blood glucose was consistently lower in Low Ile-fed mice of both age groups and in aged females (Figs. 5J–K & P–Q).
Using PCA of the variables from Fig. 5A for young, old, male and female mice, we investigated variation across diet groups (Figs. S5C–F). Due to the heterogeneous nature of our study species, there was overlap for all groups, especially as mice age. Interestingly, across all groups, the Low Ile and Control-fed mice had the greatest separation, and Low AA-fed overlapping with both of these groups, consistent with our general observation that a Low Ile diet has greater metabolic and physiological impact than a Low AA diet. Overall, while the correlation between isoleucine consumption and individual measures of metabolic health tend to trend in the same direction regardless of age or sex, the magnitude of the relationship is highly dependent on these variables.
IleR and reducing all dietary amino acids have distinct molecular effects
Due to the central role of the liver in maintaining metabolic homeostasis, we conducted a detailed molecular analysis of the livers of young and old mice of both sexes, consuming either the Control, Low AA, or Low Ile diet. We performed transcriptional profiling as well as metabolomic and lipidomic analysis. After analyzing the effects of diet on each of these parameters separately, we integrated these analyses with phenotypic data.
Low Ile-fed young males showed a distinct pattern of gene expression relative to Control-fed counterparts; interestingly, at this age 56% of the significantly differentially expressed (SDE) genes altered by a Low AA diet are also altered by a Low Ile diet (Fig. 6A, Table S3A–4). In contrast, when we examined the top 50 SDEs altered in a Low Ile diet in 24-month-old males, Low AA-fed males appeared more similar to Control-fed males, and there was no overlap in the SDE genes altered by Low AA and Low Ile diets (Fig. 6B). Conversely, while in young females there were relatively few SDE changes induced by either diet and little overlap in those few genes that were SDE, in old females, many more genes were SDE, and 35% of the genes SDE by a Low Ile diet were also altered by a Low AA diet (Fig. 6C–D, Table S3C).
Figure 6: IleR and Low AA diets have sex- and age-dependent molecular impacts.
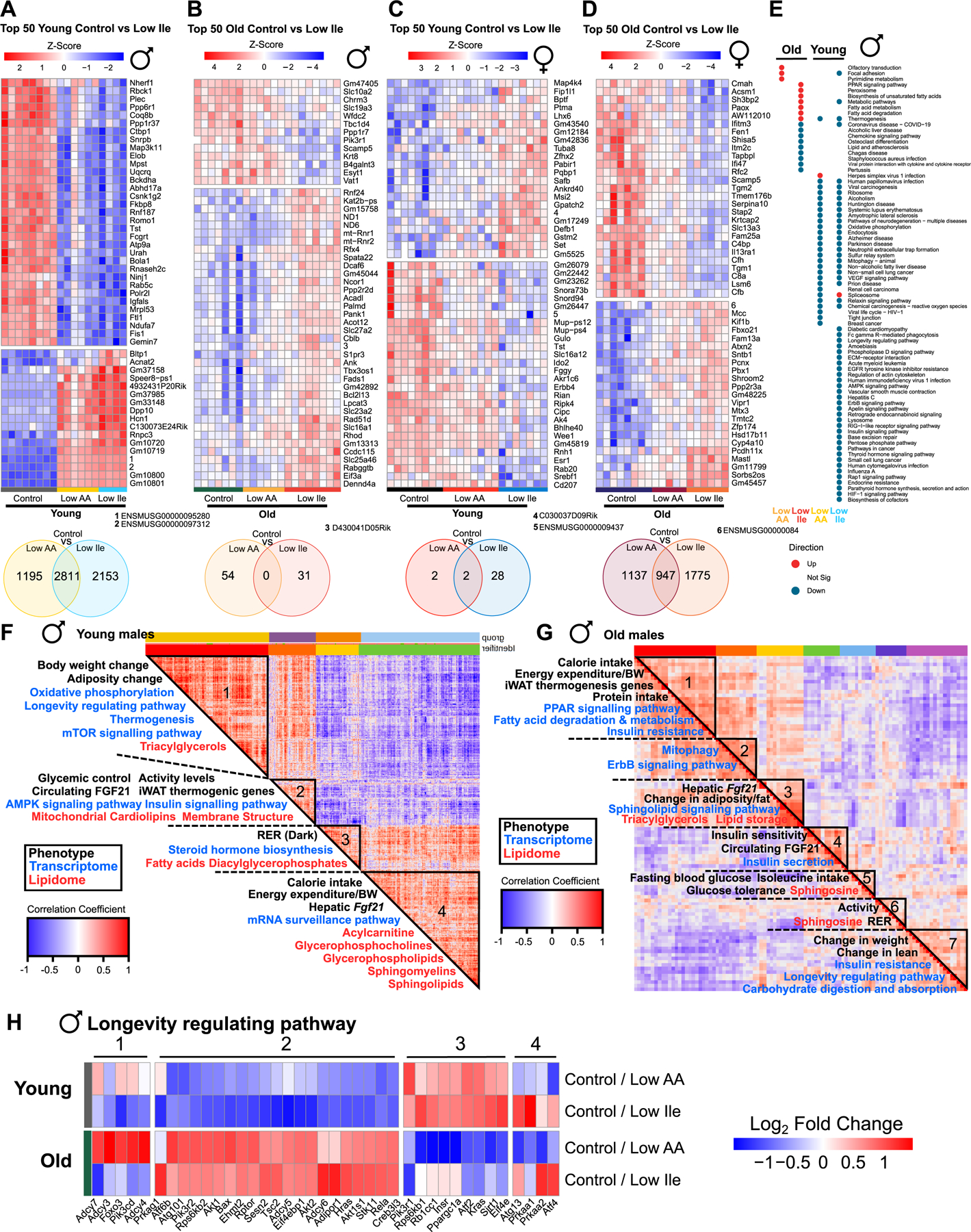
(A-D) Top 50 significantly differentially expressed (SDE) genes for the indicated comparisons, with hierarchical clustering across all groups in each figure. Venn Diagrams below each heatmap indicating significant gene overlap for the comparisons, unadjusted p<0.05. n=4–8 mice/group. (E) Enriched transcriptomic pathways across all male groups (red = upregulated, blue = downregulated, grey = not significant). n=6–8 mice/group. (F-G) Spearman’s rank order correlation matrix of phenotypic, transcriptomic, metabolomic and lipidomic changes across young (F) and old (G) male Control vs Low Ile mice. Mega-clusters identified by hierarchical clustering are outlined in black (Table S5D). N=6–8 mice/group. (H) Genes from KEGG “Longevity regulating pathway” significantly altered in old and young male mice on Low AA or Low Ile diets. n=6–8 mice/group. See also Tables S3A–S5D. See also Figure S6.
We performed KEGG analysis to identify pathways altered by each diet (Figs. 6E, S6A and Table S3D). Interestingly, many more pathways were altered by IleR than a Low AA diet in both young and old males, and more pathways were altered by both diets in young than in old males (Table S3B, S3D). In young males, there was substantial overlap in the pathways altered by Low AA and Low Ile diets; in aged males, there was no overlap in the altered pathways (Fig. 6E). In young males a Low Ile diet downregulated multiple pathways linked to aging, including “Insulin signaling”, “Lysosome”, “Ribosome”, and “AMPK signaling”. In old Low Ile-fed males, upregulated pathways included “PPAR signaling”, “Fatty acid metabolism” and “Thermogenesis” while downregulated pathways included pathways related to immune function, including “Chemokine signaling” and “COVID-19.” Few pathways were altered by diet in young females, while there were numerous pathways with significant overlap between the effects of the Low Ile and Low AA diet in old females (Fig. S6A). In aged females, many of the downregulated pathways on Low AA and Low Ile diets were shared, including pathways associated with the immune system, including the TNF signaling pathway, cytokine, interleukin and MAPK signaling, while upregulated pathways were associated with fatty acid metabolism, including fatty acid degradation and PPAR signaling (Fig. S6A, Table S3D).
We performed untargeted metabolomics and investigated significant changes between the livers of Control and Low AA-fed or Low Ile-fed mice across sexes and ages (Tables S4A–S4B). We did not see many significant changes, likely reflecting the genetic diversity of our studied mice and their variable metabolomic profiles (Table S3B). Relaxing the significance criteria to investigate pathway changes, we found aged Low Ile-fed males had altered carbohydrate and AA metabolism (Table S4B), including BCAA metabolism; they also showed changes in other AA pathways, including glutamine/glutamate, arginine, and histidine metabolism (Fig. S6B). In young Low AA-fed males, only 2 metabolites changed significantly, including glycine which was significantly downregulated, and we observed a similar pattern in young Low Ile-fed males (Table S4A). In females, with relaxed significance criteria, we identified changes in fatty acid metabolism pathways in old Low Ile and Low AA-fed mice and AA pathway changes in young Low AA-fed mice (Table S4C).
To investigate potential long-term diet-induced changes in AAs, we performed metabolomics in the plasma of fasted old mice. Despite the restriction of all AAs by 2/3rds in Low AA-fed mice, we observed significant increases in the blood levels of proline, lysine, arginine and isoleucine in Low AA-fed males and increased levels of isoleucine in Low AA-fed females (Fig. S6C & Table S4D). In contrast, we observed no changes in plasma AA levels in Low Ile-fed males, while in Low Ile-fed females proline, phenylalanine, methionine, leucine and valine were significantly increased (Table S4D). Hepatic levels of isoleucine did not significantly change in any group, but trended upwards in young Low AA-fed males and downwards in old Low Ile-fed mice of both sexes (Figs. S6D–E & Tables S4A, S4C).
We also performed untargeted lipidomics on livers from all groups (Tables S5A–S5B). Similar to our transcriptomics analysis, we found the greatest number of changes in young Low Ile-fed males, and the fewest in young females, with little overlap between Low AA and Low Ile-fed mice for each age and sex (Table S3B, Figs. S6F–G). In young Low AA and Low Ile-fed males, levels of the omega-9 fatty acid oleic acid, which is thought to prevent heart disease and reduce cholesterol, was upregulated (Table S5A).
Using LION, we found that except in aged females fatty acylcarnitines, which play a key role in the regulation of sugar and lipid metabolism, were enriched (Table S5C). In young Low AA and Low Ile-fed male, and Low AA-fed females, there were many changes in fatty acid and triglyceride pathways. In aged Low AA and Low Ile-fed males, there were changes involved in lipid bilayer thickness and diffusion, whereas in aged females consuming these diets, there was an effect on lipid storage and triglycerides. This suggests that these diets induce sex-specific changes in fatty acid metabolism that become more pronounced with age (Table S5C). Looking at the top 50 most significantly altered lipids between Control and Low Ile groups, we observed that Low AA and Low Ile diets appear to have rejuvenating qualities on the lipidomic profile of aged males, which look more similar to young males than to aged Control-fed males; this was less clear in females (Figs. S6F–G).
To examine the relationship between molecular changes and whole-organism physiology and metabolism induced by a Low Ile diet, we constructed a correlation matrix for each age and sex using phenotypic data as well as hepatic transcriptomic (Tables S3A & S3C), metabolomic (Tables S4Aand S4C, S5, S6, S7) and lipidomic data (Tables S5A–S5B). Statistically significant changes in gene expression, metabolite and lipid levels for each group were concatenated with phenotypic data, and Spearman’s correlation was used to calculate coefficients which were then rendered by hierarchical clustering (based on 1 – correlation coefficient between all molecules) to produce 4–7 clusters for each contrast (Table S5D). We identified KEGG-enriched pathways in each cluster and related these to the corresponding cluster phenotypes. Our results are consistent with a broad reprogramming of hepatic metabolism by a Low Ile diet, and link specific molecular changes and metabolic processes in the liver with specific phenotypes.
We previously found in C57BL/6J males IleR induces FGF21 and showed that the effects of a Low Ile diet on energy expenditure and food consumption are partially blocked by deletion of Fgf2128. Interestingly, in young HET3 males FGF21 levels in the blood cluster with the expression of thermogenic genes in iWAT and the inner mitochondrial membrane cardiolipins, while hepatic expression of Fgf21 clusters with energy expenditure and calorie intake (Fig. 6F, clusters 2 and 4). However, in aged males, energy expenditure and iWAT thermogenic gene expression cluster together and do not cluster with FGF21 (Fig. 6G, cluster 1). This suggests that, as we recently found for dietary protein17, FGF21 may be more important in the response to isoleucine in C57BL/6J males than in other genetic backgrounds.
Other associations in aged males related to glycemic control. In addition to containing genes associated with energy expenditure and thermogenesis, Cluster 1 contains genes associated with insulin resistance, fatty acid metabolism and PPAR signaling; this last is known to have a key role in glucose and lipid metabolism (Fig. 6G, cluster 1) 35. We found that changes in genes related to insulin sensitivity and insulin secretion were associated with FGF21 levels (Fig. 6G, cluster 4), while changes in blood glucose levels and glucose tolerance were associated with isoleucine intake and ceramides (Fig. 6G, cluster 5). In aged females Cluster 3, containing genes involved in thermogenesis, was similar to Cluster 1 in old males (Figs. S6H–I).
In both young and old males we found clusters containing genes associated with the KEGG “Longevity regulating pathway (LRP)” (Fig. 6H). In young males, these genes were found in Cluster 1, and were associated with changes in weight and adiposity, as well as transcriptional changes in oxidative phosphorylation, mTOR signaling, and thermogenesis (Fig. 6F, cluster 1). In aged males, the LRP was likewise associated with changes in weight, but clustered with changes in lean mass, insulin resistance, and carbohydrate digestion and absorption (Fig. 6G, cluster 7).
We created clustered heatmaps based on LPR genes altered by diet (Fig. 6H). While some of these clusters appear to be age-dependent and diet-independent, such as Cluster 2 which is associated with mTOR signaling, genes in Cluster 1, associated with the adenylate cyclase activating pathway, retain a more youthful expression profile in Low Ile-fed males and not in Low AA-fed males. In old males, Cluster 3, which contains genes associated with insulin signaling, is less strongly downregulated in Low Ile-fed old males than in Low AA-fed old males. Finally, in cluster 4, genes associated with autophagy/mTOR signaling are downregulated by a Low AA diet and upregulated by a Low Ile diet. In females we saw little evidence of either diet inducing a more youthful expression profile (Fig. S6J).
In summary, we found that consumption of Low AA and Low Ile diets uniquely reprograms hepatic metabolism. In contrast to our initial assumption that Low AA and Low Ile diets would have similar molecular effects on the liver, we found that the effects of the diets were largely distinct. Further, age and sex were extremely important factors in the molecular response to both diets. These effects were also observed in our differential coexpression analysis, where the clustering of molecular changes and phenotypes depended on both sex and age.
IleR increases lifespan and healthspan in HET3 mice
We utilized a recently developed mouse frailty index to examine how diet, age, and sex impact frailty 36. As expected, we observed increased frailty with age in Control-fed mice of both sexes; frailty was significantly lower in Low AA and Low Ile-fed male mice at multiple time points (Fig. 7A), as well as in female mice at 22 and 24 months of age (Fig. 7B). These changes were mainly related to physical frailty in both sexes, and body condition and digestion in males (Figs. S7A–J).
Figure 7: IleR increases lifespan and improves healthspan in HET3 male mice.
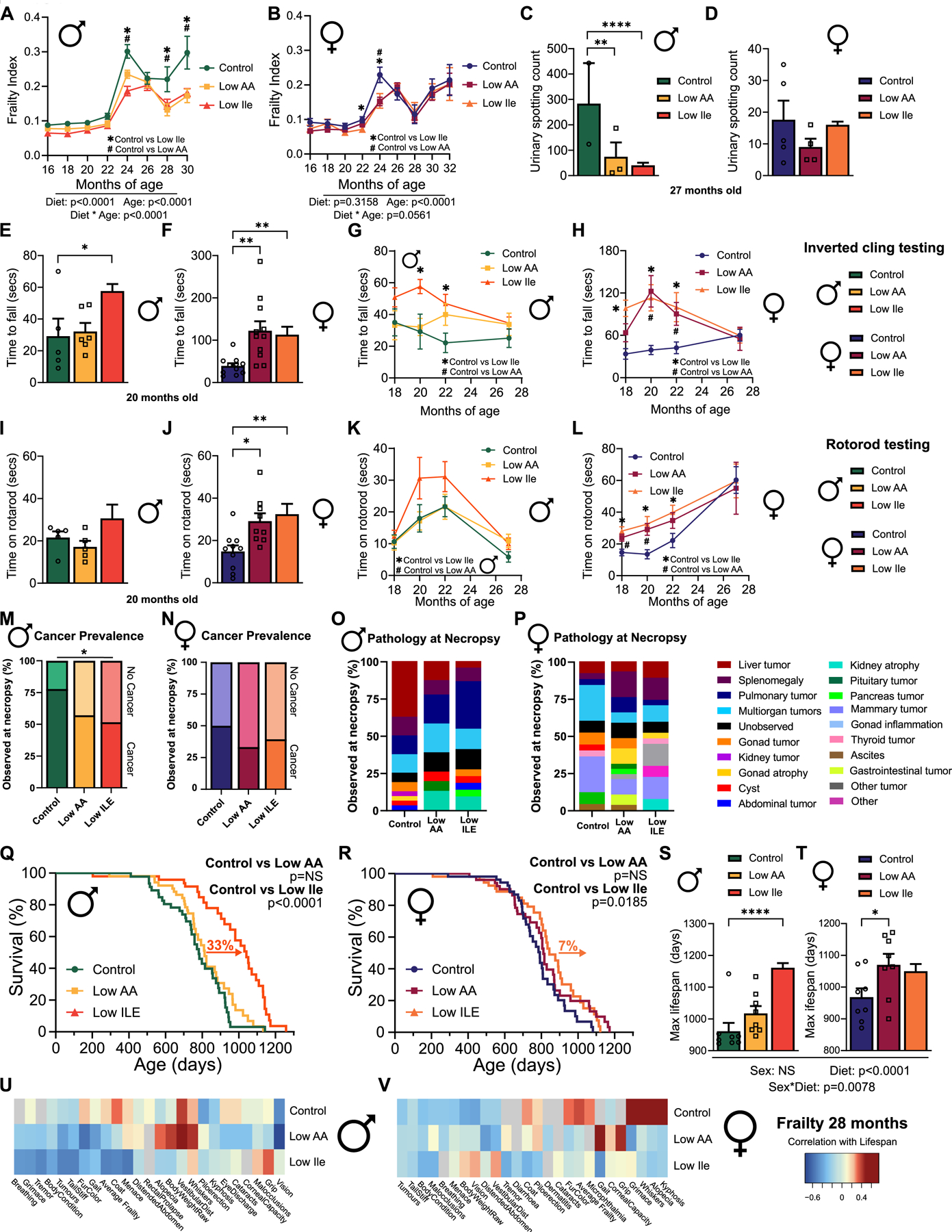
(A-B) Frailty of male (A) and female (B) mice over time. n varies by month; max n=44–49 mice/group. (C-D) Void spot assay conducted in 27-month-old male (C) and female (D) mice. n=2–14 mice/group. (E-H) Inverted cling test in 20-month-old mice and over time. (E-F) n=5–11 mice/group. (G-H) n varies by month; max n=16–22 mice/group. (I-L) Rotarod test in 20-month-old mice and over time. (I-J) n=5–10 mice/group. (K-L) n varies by month; max n=15–22 mice/group. (M-N) Tumor incidence at necropsy; n=31–38 mice/group. (O-P) Pathology observed at necropsy. n=22–32 mice/group. (Q-R) Kaplan-Meier survival curves by diet in male (Q) and female (R) mice. n=47–53 mice/group. (S-T) Average top 10% of lifespans in male (S) and female (T) mice by diet, n=8 mice/group. (U-V) Spearman’s rank correlation of frailty measured at 28 months with final lifespan across male (U) and female (V) mice. n=13–25 mice/group. (A-B, G-H, K-L) Mixed-effects model (REML) for time and diet with post-hoc Tukey testing for multiple comparisons. #P<0.05 Control vs Low AA, *P<0.05 Control vs Low Ile. (C-F, I-J, S-T) Tukey test following ANOVA; *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. (M-N) Two-sided chi-squared test, *P<0.05. (Q-R) Log-rank test for Control vs Low AA and Control vs Low Ile. Outlier removed from (D) using ROUT outlier test. Data represented as mean ± SEM. See also Tables S6A and S6C. See also Figure S7.
We also investigated other age-associated phenotypes. Lower urinary tract dysfunction increases with age, particularly in males37, and urinary spotting was significantly increased in aged Control-fed males relative to Low AA and Low Ile-fed males (Figs. 7C–D). We performed inverted cling and rotarod assays to assess muscle strength, function, and motor coordination. At 20 and 22 months of age, Low Ile-fed males and females were able to cling for longer than Control-fed mice; a similar affect was observed in Low AA-fed females (Figs. 7E–F). In late life, concurrently with the convergence of body weights, we found no significant differences in inverted cling performance between diet groups in either sex (Figs. 7G–H). Similarly, 20 and 22 months of age Low AA and Low Ile-fed females were able to stay on the rotarod longer than their Control-fed counterparts, although this was not the case in males (Figs. 7I–J). As females aged, we no longer saw significant differences in rotarod performance between diet groups, and we saw no significant differences between diet groups in males at any time point (Figs. 7K–L). We observed no diet-induced effects on either short-term or long-term memory during novel object recognition testing in either sex (Figs. S7K–L).
Cellular senescence is a underlying mechanism of age-related pathophysiological decline38. To investigate if a Low Ile diet altered cellular senescence, we measured the expression of p16 and p21 as well as several senescence-associated secretory phenotype (SASP) genes in the livers of aged Low Ile-fed mice relative to Control-fed mice. In males only, p16 trended towards a significant reduction on a Low Ile diet and the inflammatory marker interleukin 1-α (Ila) was downregulated, and no changes were seen in the markers measured in females (Figs. S7M–N).
In HET3 mice, cancer is the major cause of death, with >80% of mice dying of neoplastic lesions 39. At necropsy Low Ile-fed males were less likely to have a tumor present at death than Control-fed males, but in females there was no significant difference between groups (Fig. 7M–N). When looking more broadly at pathologies observed at death, we found differences in the spectrum of pathologies observed. The most common pathology found at necropsy in Control-fed and Low AA-fed males were liver tumors, while Low Ile-fed males had fewer liver tumors (p=0.0083) (Fig. 7O). In Control-fed females the most common pathology at necropsy were mammary and pulmonary tumors (Fig. 7P), and Control-fed females had a greater prevalence of mammary gland tumors relative to Low AA fed mice (p=0.1, Fig. S7O).
In agreement with the significant difference in frailty and decrease in tumors found at necropsy, we found that consumption of a Low Ile diet from 6 months of age significantly increased male lifespan, with a 33% increase in median lifespan relative to Control-fed mice (Fig. 7Q, Table S6A) and a significant increase in maximum lifespan (Wang-Allison p=0.0257, Table S6A). Surprisingly, despite inhibiting frailty a Low AA diet had no effect on male lifespan (Fig. 7Q, Table S6A). A Low Ile diet also increased median lifespan in females, albeit by a more modest 7%, but did not increase maximum lifespan; once again, there was no effect of a Low AA diet on median or maximum lifespan (Fig. 7R, Table S6A). Investigating the survival of the top 10% longest-lived animals in each group we found the top 10% of Low Ile-fed males had a 17% increase in lifespan relative to the top 10% of Control-fed males (Fig. 7S). In females, a Low AA diet increased the lifespan of the 10% longest-lived mice relative to Control-fed mice by 12% (Fig. 7T).
To determine how frailty might interact with lifespan in a diet- and sex-dependent manner, we correlated frailty measures with lifespan in male and female mice (Fig. 7U–V). In Low Ile-fed males, we observed a strong negative correlation between lifespan and frailty measures associated with body condition, including tail stiffness, fur color and tremor occurrence. By contrast, Low AA-fed males had a strong positive correlation between lifespan and alopecia, whisker loss and body weight (Fig. 7U). Females similarly showed different correlation patterns based on diet; in Control-fed females, we found that lifespan was strongly positively correlated with whisker loss, alopecia and kyphosis, while these deficits correlated negatively with lifespan in Low AA and Low Ile-fed females (Fig. 7V).
In summary, a Low Ile diet improves healthspan and reduces frailty in both male and female mice, and robustly extends median and maximum lifespan in males. In females, a Low Ile diet more modestly promotes median but not maximum lifespan. Notably, a Low AA diet reduced in all AAs including isoleucine benefits aspects of healthspan in both sexes, and slows the development of frailty equivalently to the effects of a Low Ile diet, but does not extend lifespan in either males or females.
Discussion
Recently it has become clear that diet macronutrient composition is important for metabolic health and aging. Multiple studies have highlighted dietary protein as a key determinant of human health and suggest that a low protein diet is optimum metabolic health8,9,11,13,14,40. In animals, dietary protein is a key regulator of metabolic health as well as longevity, with mice living longer when fed a low protein diet16,41.
We have hypothesized that many of the beneficial effects of PR may result from the reduced consumption of specific AAs. We have shown that 67% restriction of the three BCAAs is sufficient to recapitulate the benefits of PR and can extend the lifespan of male mice equivalently to restriction of all twenty common AAs19. Conversely, increasing dietary BCAAs promotes hyperphagia, obesity, and shortens lifespan42. While the BCAAs are often considered as a group, it is becoming clear that the three BCAAs do not all have equivalent metabolic effects, and we have shown that isoleucine is the most potent of these in the regulation of metabolic health28. IleR is both necessary for the metabolic benefits of PR, and is sufficient to promote glucose tolerance, hepatic insulin sensitivity, and energy expenditure in mice C57BL/6J males28 Here, we tested the hypothesis that IleR would promote healthy aging in genetically heterogenous HET3 mice, which better model the human population than inbred strains. We found that IleR has dramatic effects on weight, fat mass, glycemic control, and energy expenditure in both young and old mice of both sexes. Interestingly, these effects were distinct – and generally greater in magnitude – than the effects in mice consuming a Low AA diet in which all AAs were restricted. Furthermore, we found that in males, IleR substantially increases both median and maximum lifespan while reducing frailty; IleR had a more modest effect on the median lifespan of female HET3 mice, while surprisingly a Low AA diet did not extend lifespan in either sex.
Our most striking finding was the robust extension of median and maximum lifespan induced in HET3 male mice by IleR. The ability of IleR to extend the lifespan of this heterogeneous population beginning at 6 months of age, as well as the effect of IleR on frailty and maximum lifespan, clearly indicate that IleR is geroprotective. Unlike BCAA restriction19, IleR increases median lifespan not only in males, but also more modestly in females. This type of male-specific or male-biased effect has also been seen before in numerous studies30,43. Our correlation and PCA analyses of the relationship between isoleucine intake and metabolic phenotypes clearly show sex differences, the accrual of which over time may contribute to differences in longevity. Pharmacological or genetic disruption of the AA-sensitive mTORC1 signaling pathway consistently shows larger benefits for female than male longevity44–47. In combination with the fact that isoleucine is not a strong mTORC1 agonist, this suggests that IleR may work through largely mTORC1-independent mechanisms.
Although our results show that the frailty index for mice can be successfully employed in genetically heterogeneous mice, we were surprised to find that the Low AA diet uncoupled frailty from lifespan, particularly as a Low AA diet begun early in life extends male C57BL/6J lifespan equivalently to a Low BCAA diet19. In most of the physiological tests we carried out, regardless of age, females on a Low Ile diet had a reduced magnitude of response relative to males. These differences may be due to inherent disparities in AA catabolism between males and females48, or due to sex hormones.
Consistent with our previous findings in C57BL/6J males28, a Low Ile diet did not decrease fasting plasma levels in HET3 mice, although as we previously observed in C57BL/6J males, hepatic levels of isoleucine did trend downwards in old Low Ile-fed HET3 mice. In Low AA-fed mice, plasma levels of isoleucine were actually higher, and trended upwards in male liver, which may help to explain the divergent effects of a Low AA and Low Ile diets on lifespan. This may reflect in part that changes in dietary AA content more directly impact levels of AAs in the portal vein than in the systemic circulation 49. While the precise mechanisms by which circulating AA levels are buffered against changes in dietary Ile are not known, the limited impact of a Low Ile diet on circulating levels of Ile likely reflects in part reduced catabolism of Ile and increased autophagy to preserve this limiting AA for proteogenesis.
The molecular impact of the diets were dependent upon sex and age, with both diets impacting many more pathways in young males and aged females than in either aged males or young females. Notably, the divergent effects of Low AA and Low Ile diets on the lifespan of males was reflected at the transcriptional level, where there was little overlap in the biological pathways affected in males fed these two diets. For example, we observed upregulation of fatty acid metabolism and downregulation of immune pathways in aged Low Ile-fed males, but not in aged Low AA-fed males. In contrast, the biological pathways altered by Low AA and Low Ile-fed diets were very similar in aged females, possibly contributing to the smaller lifespan differences between Low AA-fed and Low Ile-fed females.
We consider it likely that the sex-specific molecular effects of IleR contributes to the greater benefit of IleR on healthy aging in males. We have not investigated the impact of IleR on the immune system, but the downregulation of immune pathways in Low Ile-fed HET3 males suggests that inflammation could be reduced, and reduced inflammation is thought to contribute to the beneficial effects of CR50,51. Effects on IleR on cell senescence could also result in reduced inflammation and immune activation. Future research investigating the effects of IleR on inflammation and cell senescence may shed light on the mechanisms by which IleR extends healthspan and lifespan.
While we and others have shown that PR increases lifespan, these previous studies initiated the diets earlier in life16,19,21, and we may have initiated the Low AA diet too late to extend longevity. This appears paradoxical, as the Low AA-fed mice consumed the same amount of isoleucine as the Low Ile-fed mice. It is possible that the reduction of other EAAs or NEAAs has a negative impact on healthy aging when initiated at 6 months of age. These AAs could include leucine, restriction of which we have found can negatively impact body composition13,28, and serine and glycine, which have important roles in skeletal muscle stem cell function52. The ratio between isoleucine and other dietary AAs may also be important in determining the effects of IleR on healthspan and longevity.
We were concerned about the potential impact of our interventions on sarcopenia53. Comfortingly, we found that a Low Ile diet tended to promote grip strength, and rotarod testing did not show any significant defect induced by either Low AA or Low Ile diets. While grip strength appeared to improve in middle age for Low Ile-fed mice, this is most likely due to reduced body weight. However, we did not examine muscle strength and quality in detail, and more directly assessing the effects of IleR on muscle strength, quality and fiber type will be critical to examine in the future.
IleR significantly improves metabolic health in young HET3 mice of both sexes, mirroring our previous work in Low Ile-fed C57BL/6J males28. Low AA-fed HET3 mice also showed a modest reduction in body weight, but interestingly IleR had a greater impact on glucose tolerance than the Low AA diet at most ages, suggesting that some AAs may have a detrimental effect on glycemic control when reduced. Importantly, when started in 6-month-old mice – roughly equivalent to a 30 year old adult human54, IleR and to a lesser extent a Low AA diet was able to promote leanness in both sexes. While the metabolic benefits of IleR varies by sex and age, the main trend towards improved heath is consistent. All of this highlights the potential translatability of IleR to improve not only longevity, but metabolic health.
FGF21 has been strongly associated with the metabolic health benefits of PR, PR induces FGF21 in C57BL/6J mice, and deletion of Fgf21 blocks many of the benefits of a PR in this strain17,18,21,34,55. However, we saw no statistically significant changes in either circulating FGF21 or hepatic Fgf21 induced by either Low AA or Low Ile diets, and FGF21 levels tended not to correlate or anti-correlate with other phenotypes. These results are in accordance with our recent observation that FGF21 was induced by PR in C57BL/6J mice, but not in HET3 or DBA/2J mice 17. Understanding the role of FGF21 in the benefits of IleR should be a priority for future studies.
Energy expenditure was increased by both Low Ile and Low AA diets; while we expected this to be mediated by induction of the FGF21-UCP1 axis34, we also did not see a clear upregulation of Ucp1 by these diets. Interestingly, we did see a statistically significant increase in Cidea, but not Elov3, both regulators of thermogenesis. The role of Cidea in metabolic health is complex; while Cidea deficient mice are lean and resistant to obesity, it has been suggested that Cidea may regulate lipolysis and thermogenesis through Ucp1 suppression56. In humans, reduced CIDEA expression is associated with high body fat and a low basal metabolic rate and is increased in adipose tissue by CR57. Future work will need to be conducted to determine if induction of Cidea contributes to the metabolic effects of IleR.
In conclusion, we have shown that dietary restriction of a single BCAA isoleucine, can extend both healthspan and lifespan when begun at 6 months of age – roughly equivalent to a human in their 30’s33. This effect is particularly robust in males, but benefits are observed in both sexes; the fact that IleR works in a genetically heterogeneous population suggests that such an intervention may be applicable to humans. IleR improves metabolic health without inducing “binge eating” behavior as seen with CR58, and improves median lifespan in females, which is not seen with BCAA restriction19. Additional research will be required to determine if there are potentially negative effects of IleR, and to examine how optimal levels of isoleucine for lifespan and health span vary with age and sex. In agreement with human data28,29, dietary isoleucine seems to be critically important in metabolic health and aging, and provide new evidence that protein quality – the specific AA composition of dietary protein – is as important, or even more important, than the amount of protein or calories consumed. While additional research will be required to test how isoleucine affects healthy aging in humans, our results support the concept that limiting dietary levels of isoleucine may be a key to healthy aging.
Limitations of study
There are several limitations of the work conducted here. First, we examined only a single level of restriction, and studies of CR as well as PR suggest that different strains and sexes may respond optimally to different levels of restriction. Different levels of IleR may more optimally function to reduce frailty and increase lifespan in females, and examining the graded response to isoleucine may provide new insights into both biological mechanisms as well as translatability.
Other dietary components may also affect longevity. The diets used here are based on the AA profile of whey; however, studies conducted with either whey or casein have found broadly similar effects13,16,18,28,59,60. To keep diets isocaloric, the reduction of dietary AAs in the Low AA diet was balanced by additional carbohydrates, and the reduction of isoleucine from the Low Ile diet was balanced with NEAAs. The protein to carbohydrate ratio, the type of dietary carbohydrate consumed, the precise degree of restriction, and diet-induced changes to the microbiome could all play a role in the responses we observe here16,41,61–63.
Our molecular analysis was largely limited to the liver; while this is the first organ to be exposed to nutrients following their absorption in the intestine, catabolism of isoleucine occurs in many tissues64. We have not yet identified the specific cellular or organismal sensor of isoleucine responsible for the beneficial effects of IleR, and this is a critical area for future research. Notably, we did not observe significant reductions of circulating levels of Ile in IleR mice in this study or in a previous study utilizing young C57BL/6J male mice. We hypothesize that this reflects a dietary adaptation that acts to maintain AA homeostasis by reducing utilization and catabolism of Ile in order to preserve limited Ile for the translation of protein; understanding how this is mediated may provide important insight into understanding the beneficial effects of IleR. The effect of IleR on immune function has not been investigated, and as the mice here were in a specific pathogen-free facility, effects of IleR on immune function may not have revealed themselves.
Surprisingly, PR does not confer the same beneficial effects as reducing isoleucine alone. It is possible that this is due to reduced levels of beneficial AAs; we and others have found that reducing dietary levels of leucine, serine, or glycine may have detrimental effects13,65. In the context of a human diet, this may limit the translation of our findings, as the simplest way to lower dietary isoleucine at the present time is to reduce total protein intake. Finally, while short-term IleR in humans is likely feasible based on recent studies66,67, long-term IleR will likely require novel approaches or the development of pharmaceutical methods to reduce isoleucine uptake or sensing, or the identification of compounds that can mimic the benefits of IleR.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dudley W. Lamming (dlamming@medicine.wisc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The accession number for the hepatic RNAseq gene expression data reported in this paper can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE241904).
This paper does not report original code.
All data required to remake graphs is available in the excel Data S1 file (published at http://data.mendeley.com/preview/dk74jscsww), and where stated is provided in Supplementary Tables.
List of primers used provided in Table S7 and in Key Resources.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Human insulin | Eli Lilly | NDC 0002-8215-17 (Humulin R U-100) |
| TRI reagent | Sigma | T9494 |
| SYBR Green | Thermo Fisher Scientific | 4309155 |
| Superscript III Reverse Transcriptase | Thermo Fisher Scientific | 18080085 |
| Critical commercial assays | ||
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D Systems | Cat# MF2100 |
| Deposited data | ||
| Liver transcriptomics raw and analyzed | This paper | GEO: GSE241904; Tables S3A and S3C. RRID:SCR_017757. |
| Liver metabolomics | This paper | Tables S4A and S4C |
| Plasma metabolomics | This paper | Table S4D |
| Liver lipidomics | This paper | Table S5A and S5B |
| Raw data to recreate figures | This paper | http://data.mendeley.com/preview/dk74jscsww |
| Experimental models: Organisms/strains | ||
| Mouse strain: C57BL6/J (Male) | The Jackson Laboratory | Cat# JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse strain: DBA2/J (Male) | The Jackson Laboratory | Cat# JAX: 000671; RRID: IMSR_JAX:000671 |
| Mouse strain: BALB/cJ (Female) | The Jackson Laboratory | Cat# JAX: 000651; RRID: IMSR_JAX:000651 |
| Mouse strain: C3H/HeJ (Female) | The Jackson Laboratory | Cat# JAX: 000659; RRID: IMSR_JAX:000659 |
| Mouse strain: UM-HET3 (Male and Female) | This paper | F2 progeny of (BALB/cJ x C57BL/6J) mothers and (C3H/HeJ x DBA/2J) fathers |
| Oligonucleotides | ||
| Fgf21: F: CAAATCCTGGGTGTCAAAGC | [13] | N/A |
| Fgf21: R: CATGGGCTTCAGACTGGTAC | [13] | N/A |
| Cidea: F: GAATAGCCAGAGTCACCTTCG | [28] | N/A |
| Cidea: R: AGCAGATTCCTTAACACGGC | [28] | N/A |
| Elov3: F: ATGCAACCCTATGACTTCGAG | [28] | N/A |
| Elov3: R: ACGATGAGCAACAGATAGACG | [28] | N/A |
| Ucp1: F: GCATTCAGAGGCAAATCAGC | [28] | N/A |
| Ucp1: R: GCCACACCTCCAGTCATTAAG | [28] | N/A |
| P16 (Cdkn2a): F: CCCAACGCCCCGAACT | [89] | N/A |
| P16 (Cdkn2a): R: GCAGAAGAGCTGCTACGTGAA | [89] | N/A |
| P21 (Cdkn1a): F: GTCAGGCTGGTCTGCCTCCG | [89] | N/A |
| P21 (Cdkn1a): R: CGGTCCCGTGGACAGTGAGCAG | [89] | N/A |
| Il1a: F: TGCAGTCCATAACCCATGATC | [90] | N/A |
| Il1a: R: ACAAACTTCTGCCTGACGAG | [90] | N/A |
| Il1b: F: TCCTGTGTGATGAAAGACGGCAC | [89] | N/A |
| Il1b: R: GTGCTGATGTACCAGTTGGGGAAC | [89] | N/A |
| Il6: F: CTGGGAAATCGTGGAAT | [89] | N/A |
| Il6: R: CTGGGAAATCGTGGAAT | [89] | N/A |
| Il10: F: ATAACTGCACCCACTTCCCA | [89] | N/A |
| Il10: R: GGGCATCACTTCTACCAGGT | [89] | N/A |
| Mcp1: F: GCATCCACGTGTTGGCTCA | [89] | N/A |
| Mcp1: R: CTCCAGCCTACTCATTGGGATCA | [89] | N/A |
| Actb (β-Actin): F: GATGTATGAAGGCTTTGGTC | [89] | N/A |
| Actb (β-Actin): R: TGTGCACTTTTATTGGTCTC | [89] | N/A |
| Software and algorithms | ||
| edgeR package | [76] | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism |
| MetaboAnalyst | [91] | https://www.metaboanalyst.ca/ |
| R (Version 3.4.2) | N/A | https://www.r-project.org/ |
| MyGene package | N/A | https://www.bioconductor.org/packages/release/bioc/html/mygene.html |
| Factoextra package | [87] | https://cran.r-project.org/web/packages/factoextra/index.html |
| missMDA package | N/A | https://cran.r-project.org/web/packages/missMDA/index.html |
| LION/web | [92] | http://www.lipidontology.com/ |
| Limma package | [77] | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Metabolomics package | N/A | https://cran.r-project.org/src/contrib/Archive/metabolomicsR/ |
| Lipid Annotator | [93] | https://www.agilent.com/en/solutions/omics/lipidomics/lipidomics-data-analysis |
| MassHunter | N/A | https://www.agilent.com/en/promotions/masshunter-mass-spec |
| Emmeans package | N/A | https://cran.rproject.org/web/packages/emmeans/index.html |
| ComplexHeatmap package | [84, 94] | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Cluster package | N/A | https://cran.r-project.org/web/packages/cluster/index.html |
| Other | ||
| Normal Chow | Purina | Cat# 5001 |
| 2019 Teklad Global 19% Protein Extruded Rodent | Teklad | 2019 |
| Teklad Global 2020x | Teklad | 2020X |
| Mouse diets (See Table S1) | This paper | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse information
All procedures were performed in conformance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of the William S. Middleton Memorial Veterans Hospital (Madison, WI, USA). HET3 mice are the F2 progeny of (BALB/cJ x C57BL/6J) mothers and (C3H/HeJ x DBA/2J) fathers; female BALB/cJ (#000651), male C57BL/6J (#000664), female C3H/HeJ (#000659) and male DBA/2J (#000671) were obtained from The Jackson Laboratory and bred to produce heterogeneous HET3 mice. Breeders and their litters prior to weaning were fed 19% protein chow (2019 Teklad Global 19% Protein Extruded Rodent Diet, Table S1). At weaning, mice were switched to Teklad Global 2020x. Approximately one week prior to the start of experiments, mice were switched to a chow diet (Purina 5001) and housed 2–3 per cage. All mice were maintained at a temperature of approximately 22°C, and health checks were completed on all mice daily. The short-term study was started when the mice were 9 weeks of age, and the lifespan study when they were 6 months old (numbers in Table S6B).
At the start of both experiments, mice were randomized to receive either the 22% (Control, TD.140711) or 7% (Low AA, TD.140712) amino acid diet, or 22% amino acid diet with isoleucine reduced by 2/3rds (Low Ile, TD.160734); all diets were obtained from Envigo. Within the diet series, calories from amino acids were replaced by calories from carbohydrates, while calories from fat were held fixed at 20%, making the diets isocaloric (3.6 Kcal/g). For the Low Ile diet, amino acid levels were made up with non-essential amino acids, making the diet isonitrogenous with the Control diet. Full diet descriptions are provided in Table S1. The randomization of mice was performed at the cage level to ensure that all groups had approximately the same initial starting weight and body composition. Mice were housed in a SPF mouse facility in static microisolator cages, except when temporarily housed in a Columbus Instruments Oxymax/CLAMS metabolic chamber system. Mice were housed under a 12:12 h light/dark cycle with free access to food and water, except where noted in the procedures below.
METHOD DETAILS
In vivo Procedures
Glucose, insulin, and alanine tolerance tests were performed by fasting the mice overnight for 16 hours and then injecting glucose (1 g kg−1), insulin (0.75 U kg−1) or alanine (2 g kg−1) intraperitoneally (i.p.) 68,69. Glucose measurements were taken using a Bayer Contour blood glucose meter (Bayer, Leverkusen, Germany) and test strips. Mouse body composition was determined using an EchoMRI Body Composition Analyzer (EchoMRI, Houston, TX, USA). For assay of multiple metabolic parameters [O2, CO2, food consumption, respiratory exchange ratio (RER), energy expenditure] and activity tracking, mice were acclimated to housing in a Oxymax/CLAMS metabolic chamber system (Columbus Instruments) for ∼24 h and data from a continuous 24 h period was then recorded and analyzed. Food consumption in home cages was measured by moving mice to clean cages, filling the hopper with a measured quantity of fresh diet in the morning and measuring the remainder in the morning 3–6 days later. The amount was adjusted for the number of mice per cage, the number of days that passed and the relative weights of the mice (i.e., heavier mice were credited with a larger relative portion of the food intake). Mice were euthanized by cervical dislocation after an overnight (16h) fast and tissues for molecular analysis were flash‐frozen in liquid nitrogen or fixed and prepared as described below.
Lifespan study
Mice were randomized into diet groups and enrolled in the survival study at 6 months of age; deaths prior to randomization and enrollment were not recorded. Mice were euthanized for humane reasons if moribund, if the mice developed other specified problems (e.g. excessive tumor burden), or upon the recommendation of the facility veterinarian. No mice in these studies contracted dermatitis requiring treatment or removal. Mice found dead were noted at each daily inspection and saved in a refrigerator for gross necropsy, during which the abdominal and thoracic cavities were examined for the presence of solid tumors, metastases, splenomegaly, and infection; on the basis of this inspection the presence or absence of observable cancer was recorded. For each necropsied mouse, a “cause of death” was assigned based on the clearest visually detected indication from necropsy and histological analyses. A subset of mice were removed from the study for cross-sectional analysis; these mice were pre-selected on enrollment in the lifespan study and euthanized at 24 months of age. Only mice used for the cross-sectional analysis were used for GTTs and ITTs. Frailty data and chamber data were collected from all mice. Mice were censored as of the date of death if removed for cross-sectional analysis or if death was likely due to experimenter error. The lifespans of all mice, including censored mice, is provided in Table S6C.
Frailty assessment
Frailty was assessed longitudinally in a subset of mice using a list of 28 frailty measures based on the procedures outlined by36. This frailty index reflects an accumulation of deficits associated with aging, akin to Rockwood’s frailty index in humans 70. The items scored are scored 0 (no deficit), 0.5 (mild deficit) or 1 (severe deficit) and included alopecia, loss of fur color, dermatitis, loss of whiskers, coat condition, tumors, distended abdomen, kyphosis, tail stiffening, gait disorders, tremor, body condition score, vestibular disturbance, cataracts, corneal opacity, eye discharge/swelling, microphthalmia, vision loss, menace reflex, nasal discharge, malocclusions, rectal prolapse, vaginal/uterine/penile prolapse, diarrhea, breathing rate/depth, mouse grimace score , and piloerection. Scores for all items are added together, and then divided by the total number of items scored.
Inverted cling and rotarod assays
For inverted cling tests, mice were placed on a wire frame which was carefully inverted until mice were hanging upside down, a timer was started, and the time until the mouse fell was recorded. The average time of two rounds of testing conducted at least 30 minutes apart was calculated. For rotarod testing, mice were trained at a constant speed of 4rpm the day before testing. On the day of testing, mice were put on the rotarod for two rounds, at least 30 minutes apart, and the average time spent on the rotarod and max speed were recorded. For testing, the rotarod started at a speed of 4rpm with an acceleration of 0.5rpm/sec up to a max of 40rpm.
Void Spot Assay
Void spot assays were performed as described previously71. Briefly, mice were individually placed in standard mouse cages with thick chromatography paper (Ahlstrom, Kaukauna, WI). During the 4 hour study period, mice were restricted from water. Chromatography papers were imaged with a BioRad ChemiDoc Imaging System (BioRad, Hercules, CA) using an ethidium bromide filter set and 0.5 second exposure of ultraviolet light. Images were imported into ImageJ and total void spots analyzed with VoidWhizzard72.
Assays and Kits
Blood for circulating FGF21 analysis was obtained following an overnight fast. Blood FGF21 levels were assayed by a mouse/rat FGF-21 quantikine ELISA kit (MF2100) from R&D Systems (Minneapolis, MN, USA).
Quantitative PCR
Liver was extracted with Trireagent (Sigma, St Louis, MO, USA). Then, 1 μg of RNA was used to generate cDNA (Superscript III; Invitrogen, Carlsbad, CA, USA). Oligo dT primers and primers for real‐time PCR were obtained from Integrated DNA Technologies (IDT, Coralville, IA, USA). Reactions were run on a StepOne Plus machine (Applied Biosystems, Foster City, CA, USA) with Sybr Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA). Actin was used to normalize the results from gene‐specific reactions. Primer sequences can be found in Table S7 and in Key Resources.
Transcriptomic Analysis
RNA was extracted from liver as previously described73. The concentration and purity of RNA was determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and RNA was diluted to 100–400 ng/µl for sequencing. The RNA was then submitted to the University of Wisconsin-Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility, and RNA quality was assayed using an Agilent RNA NanoChip. RNA libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation protocol (Illumina, San Diego, CA) with 250ng of mRNA, and cleanup was done using RNA Clean beads (lot #17225200). Reads were aligned to the mouse (Mus musculus) with genome-build GRCm38.p5 of accession NCBI:GCA_000001635.7 and expected counts were generated with ensembl gene IDs74.
Analysis of significantly differentially expressed genes (DEGs) was completed in R version 3.4.375 using edgeR76 and limma77. Gene names were converted to gene symbol and Entrez ID formats using the mygene package. Male and female mice were analyzed separately, and clear outliers were removed following PCA analysis of the raw data. Genes with too many missing values were removed, if genes were present in less than one diet/age group they were removed. To reduce the impact of external factors not of biological interest that may affect expression, data was normalized to ensure the expression distributions of each sample are within a similar range. We normalized using the trimmed mean of M-values (TMM), which scales to library size. Heteroscedasticity was accounted for using the voom function, DEGs were identified using an empirical Bayes moderated linear model, and log coefficients and Benjamini-Hochberg (BH) adjusted p-values were generated for each comparison of interest78. DEGs were used to identify enriched pathways, both Gene Ontology (for Biological Processes) and KEGG enriched pathways were determined for each contrast, enriched significantly differentially expressed genes (FDR cutoff=0.1, FDR cutoff=0.2 old male mice with higher variability). All genes, log2 fold-changes and corresponding unadjusted and Benjamini-Hochberg adjusted p-values can be found in Tables S3A and S3C.
Metabolomic Analysis
Untargeted metabolomics was performed as previously described 79. Samples were kept at −20°C throughout the extractions in an Iso-Therm System (VWR #20901-646). Fifteen milligrams of liver were homogenized in 200 µL of a 3:3:2 solution of acetonitrile:isopropanol:water (MeCN:IPA:H2O) in ceramic bead tubes (1.4 mm, Qiagen #13113-50) using a TissueLyzer II (Qiagen #85300). An additional 800 µL of the extraction solvent was added and samples were centrifuged for 2 min at 14,000xg at RT. Lysate equivalent to 2 mg of tissue was transferred to a new 1.5 mL tube and 100 µL of extraction solvent containing 5 ug succinate-d4 (Sigma-Aldrich #293075) and 1 µg myristate-d27 (CDN Isotopes #D-1711) internal standards was added to each sample to a final volume of 500 µL. 250 µL of extract was transferred to a new tube and dried down in a SpeedVac Plus SC110A, then resuspended in 420 µL 1:1 MeCN:H2O. Samples were centrifuged as before and 400 µL was transferred to a glass autosampler vial with insert and dried down using a SpeedVac. To the dried extract, 10 uL of 20 mg/mL methoxyamine hydrochloride (MP Biomedicals #155405) in pyridine (Millipore PX2012-7) was added and vials were capped, then incubated at 37°C for 1.5h with light flicking every 30 min to mix. 91 µL of MSTFA (Thermo Fisher Scientific #TS-48915) was added to vials and incubation at 37°C with constant shaking for an additional 30 min was done to derivatize samples. Finally, samples were transferred to a GC-MS vial and immediately queued for injection.
Analysis of trimethylsilylated metabolites was performed using an Agilent 5977A Series GC-MS. A Phenomenex ZB-5MSi column (30 m, 0.25 I.D., 0.25 um; #7HG-G018-11) with 5 m column guard was used for chromatographic separation. Splitless, 1 µL sample injections were performed with injector port at 250°C with a 60 s purge at 8.2 psi. Helium was used as carrier gas and kept at 1 mL/min during the GC profile which was as follows: oven at 60°C for 1 min followed by an increase of 10°C/min to 325°C, which was held for 10 min. The MS transfer line was kept at 290°C with a solvent delay of 5.70 min. Ion source was kept at 230°C, quadrupole at 150°C and the acquisition range was from 50–750 m/z collecting 3 spectra/sec. Two quality control samples of pooled liver extracts were run at the beginning and end of each day to monitor for changes in chromatography and to equilibrate the system. A FAME mix was run externally for RI calibration. Collected data was processed in the NIST AMDIS Program using the Fiehn metabolomics library 80 for compound identification and peak integration before being exported to R for normalization to the succinate-d4 internal standard to control for extraction variability. Final data was reported as ion count per 2 mg liver. Data was normalized and analyzed using the metabolomics package in R. Log2 fold-changes and associated p-values for young and old, male and female mice on Low AA or Low Ile diets relative to Control fed mice can be found in Tables S4A and S4C.
For serum metabolomics, 150 µL of methanol was added to 5 µL of serum sample and incubated on ice for 10 min, followed by vortexing and centrifugation at 16,000 x g for 10 min at 4°C. The supernatant (3 µL) was loaded to LC-MS. Samples were analyzed using a quadrupole-orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific, San Jose, CA) operating in negative or positive ion modes, coupled to hydrophilic interaction chromatography via electrospray ionization and used to scan from m/z 70 to 1000 at 1 Hz and 140,000 resolution. LC separation was on a XBridge BEH Amide column (2.1 mm x 150 mm, 2.5 mm particle size, 130A° pore size) using a gradient of solvent A (20mM ammonium acetate, 20mM ammonium hydroxide in 95:5 water: acetonitrile, pH 9.45) and solvent B (acetonitrile). Flow rate was 150 ml/min. The LC gradient was: 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; 30 min, 85% B. Autosampler temperature was 4°C. Data was analyzed using the metabolomics package in R. Log2 fold-changes and associated p-values for 24 month old male and female mice on Low AA or Low Ile diets relative to Control fed mice can be found in Tables S4D.
Lipidomic Analysis
All solutions were pre-chilled on ice. Tissue was transferred to labeled bead-mill tubes (1.4 mm, MoBio Cat# 13113-50) and extracted in a solution of 250 µL PBS, 225 µL MeOH containing internal standards (Avanti SPLASH LipidoMix (Lot#12) at 10 µL per sample) and 750 µL MTBE (methyl tert-butyl ether). The sample was homogenized in two, 30s cycles using the TissueLyzer followed by incubation on ice for 5 minutes. Samples were centrifuged at 16,000g for 10 minutes at 4 °C, and 500 µL of the lipid-containing upper phase was collected in a new tube. Samples were evaporated to dryness using a Savant SpeedVac Plus SC110A (ThermoFischer) at room temperature.
Dried lipids were reconstituted in 150 µL IPA and transferred to an LC-MS vial with insert (Agilent 5182–0554 and 5183–2086) for analysis. A processed blank sample was included with each extraction batch and went through the same procedure except without tissue or internal standard added. Five pooled quality control (QC) samples were prepared by taking equal volumes (~10 µL) from each sample after final resuspension and aliquoting in vials. These were run throughout the LC-MS queue to ensure reproducibility throughout the run. The extraction protocol was based on81.
The Agilent 6546 Accurate Mass Q-TOF dual ESI mass spectrometer equipped with an Agilent HiP 1290 Sampler and Agilent 1290 Infinity II LC system was used for analysis. Samples were run in positive and negative ionization as separate experiments. Samples were injected at either 1µL (positive mode) or 5µL (negative) at a 50x (positive) or 5x (negative) dilution. Samples were separated on an Agilent Poroshell C18 2.1 × 50 mm column maintained at 50°C connected at a sample flow of 0.500 ml/min. Mobile phase A consisted of ACN:H2O (60:40 v/v) in 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consists of IPA:ACN:H2O (90:9:1 v/v) in 10 mM ammonium formate and 0.1% formic acid. The chromatography gradient for both positive and negative modes started at 15% mobile phase B then increases to 30% B over 2.4 min, then to 48% B from 2.4 – 3.0 min, then to 82% B from 3 – 13.2 min, and finally to 99% B from 13.2 – 13.8 min where it was held until 15.4 min and then returned to the initial conditions and equilibrated for 4 min.
For positive ionization, the source gas temperature is set to 250 °C, with a gas flow of 12 L/min and a nebulizer pressure of 35 psig. VCap voltage was 4000 V, fragmentor at 145 V, skimmer at 45 V and Octopole RF peak at 750 V. For negative mode, the source gas temperature was set to 350 °C, with a drying gas flow of 12 L/min and a nebulizer pressure of 25 psig. VCap voltage is set at 4000 V, fragmentor at 200 V, skimmer at 45 V and Octopole RF peak at 750 V. Reference masses in positive mode (m/z 121.0509 and 922.0098) are infused with nebulizer pressure at 2 psig, and negative mode reference masses (m/z 1033.988, 966.0007, and 112.9856) were infused with a nebulizer pressure at 5 psig. Samples were analyzed in a randomized order acquiring with a scan range m/z 100 – 1500. Tandem mass spectrometry was conducted using the same LC gradient at collision energy of 25 V using a pooled sample to create the lipid library.
QC samples and blanks were injected throughout the sample queue to ensure the reliability of acquired lipidomics data. Results from LC-MS experiments were collected using Agilent Mass Hunter (MH) Workstation and analyzed using the software packages MH Qual, MH Quant, Lipid Annotator and Profinder (Agilent Technologies, Inc.) to prepare the data set. In-house R scripts were used for data cleanup and curation (see accompanying code for details). Data was normalized and analyzed using the metabolomics package in R. Data was normalized and analyzed using the metabolomics package in R. Log2 fold-changes and associated p-values for 24-month-old male mice on Low AA or Low Ile diets relative to Control fed mice can be found in Tables S5A and S5B.
Integrative Analysis
Four data types (metabolomics, transcriptomics, lipidomics, phenotypic outcomes) were obtained from experiments with three factors (sex, age (young, old) and diet (Low AA, Low Ile or Control)) from 92 mice (eight old female mice fed Control diet, seven old female mice fed LowAA diet, eight old female mice fed Low Ile diet, seven old male mice fed Control diet, eight old male mice fed Low AA diet, eight old male mice fed Low Ile diet, eight young female mice fed Control diet, eight young female mice fed Low AA diet, eight young female mice fed Low Ile diet, eight young male mice fed Control diet, seven young male mice fed Low AA diet, and seven young male mice fed Low Ile diet). For each of the four contrasts we investigated, young/old male/female Control vs Low Ile, there were different numbers of correlations depending on the number of significant data points.
To identify molecules of interest in each of the -omics datasets, significantly differentially expressed molecules between Control, and Low Ile Groups using an estimated marginal means from a linear model. The Benjamini-Hochberg method was applied to control false discovery rate, selecting those with adjusted p-value < 0.05 (adjusted p-value < 0.2 for metabolites) 78. Metabolomics, transcriptomics, and lipidomics data were preprocessed, log2 transformed, z-scale normalized across molecules and samples for each data type individually. Phenotypic outcome data were similarly z-scale normalized just across phenotypes.
To integrate the data, all four data types were concatenated for each comparison. Correlations were performed between using Spearman’s rank (Young Control x Low Ile Male: 2560 × 2560 = 6553600; Old Control x Low Ile Male: 98 × 98 = 9604; Young Control x Low Ile Female: 42 × 42 = 1764; Old Control x Low Ile Female: 1198 × 1198 = 1435204). Complete hierarchical clustering was used to reorder molecules based on 1 – Spearman correlation between all molecules. The number of clusters were determined by silhouette scores82. However, the silhouette curve was atypical, so the number of clusters were determined based on the 4–7 clusters observed in the heatmap and the consistency of pathways and phenotypes within clusters; heatmaps and correlation plots were generated for all comparisons.
All analyses were performed in R (v. 4.1.0) using using emmeans83 (v. 1.8.4), ComplexHeatmap 84 (v. 2.8.0), cluster 85 (v. 2.1.2). For each cluster, the over representation of KEGG pathways from genes were determined using kegga and the gene ontology terms were determined using goana from limma77 (v. 3.48.3).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analyses
Most statistical analyses were conducted using Prism, version 9 (GraphPad Software Inc., San Diego, CA, USA) and R (version 4.1.0). Tests involving multiple factors used one‐way ANOVA with Diet as the categorical variable followed by a Tukey–Kramer or Dunnett’s post hoc test for multiple comparisons as appropriate. Data distribution was assumed to be normal but was not formally tested. Energy expenditure data did not meet criteria for ANCOVA with body weight as a covariate. Transcriptomics, metabolomics and lipidomics data were analyzed using R (version 3.4.2). All correlations where depicted were produced using Pearson’s correlations and PCA plots were produced by imputing missing data and scaling the data using the R package “missMDA” using the PCA analysis and plots were generated using the R package “factoextra”86,87. Lifespan comparisons were calculated by log-rank test. Maximum lifespan calculations were made by generating a cutoff of the top 10% longest lived animals in each sex, coupled with Boschloo’s Test (Wang-Allison) for significance testing between groups 88. Exact test used and exact n values are provided in each figure legend.
Supplementary Material
Supplementary Table 1: Diet Composition.
Diet composition and calorie content for diets used in this study. Related to STAR METHODS Experimental Model and Subject Details.
Supplementary Table 2: The relationship between isoleucine intake and phenotypic changes is dependent on sex and age.
R values for correlations from Figure 5A. Related to Figure 5.
Supplementary Table 3: Changes in the hepatic transcriptome on Low AA and Low Ile diets are dependent on age and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from transcriptomic analysis in males. (B) Significant changes at each omics level (adjusted p-value<0.01 for transcriptomics, p-value=0.05 for metabolomics and lipidomics). (C) Hepatic Log2 fold-changes and related p-values for comparisons of interest from transcriptomic analysis in females. (D) KEGG enriched pathways for significant genes found in Tables S3A and S3C. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 4: Changes in the hepatic and circulating metabolome are dependent of age, diet and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from metabolomic analysis in males. (B) KEGG enriched pathways for metabolites found in Tables S4A and S4C.
(C) Hepatic Log2 fold-changes and related p-values for comparisons of interest from metabolomic analysis in females. (D) Plasma Log2 fold-changes and related p-values for comparisons of interest from amino acid metabolomics analysis in 24 month old males and females. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 5: Changes in the hepatic lipidome on Low AA and Low Ile diets are dependent on age and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from lipidomic analysis in males. (B) Hepatic Log2 fold-changes and related p-values for comparisons of interest from lipidomic analysis in females. (C) LION Lipid Ontology enrichment for pathways of significant lipids (p<0.05) for each of the age/sex/diet groups. (D) Concatenated data for coexpression network for Figure 6F and G, Supplementary Figures 6E and F. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 6: A low isoleucine diet increases median lifespan in both sexes.
(A) Lifespan statistics for Figure 7Q–T. (B) Number of mice for each study cohort. Related to STAR METHODS Experimental Model and Subject Details. (C) Lifespan of all, including censored, mice. Related to Figure 7.
Supplementary Table 7: Primer sequences.
Primer sequences for each gene measured by qPCR. Related to STAR METHODS Quantitative PCR.
Highlights.
Isoleucine restriction (IleR) improves metabolic health in both sexes.
IleR reprograms hepatic metabolism in a sex- and age-dependent manner.
IleR reduces frailty and increases lifespan, with stronger effects on male lifespan.
Amino acid restriction begun at 6 months extends healthspan but not lifespan.
Acknowledgements
The Lamming lab is supported in part by the NIA (AG056771, AG062328, AG081482, and AG084156), the NIDDK (DK125859), and startup funds from UW-Madison. CLG was supported in part by Dalio Philanthropies, a Glenn Foundation Postdoctoral Fellowship, and by Hevolution Foundation award HF-AGE AGE-009. HHP was supported in part by F31AG066311. MET was supported in part by a Supplement to Promote Diversity in Health‐Related Research (R01AG062328-03S1). RB is supported by F31AG081115. MFC is supported by F31AG082504. C-YY was supported by T32AG000213 and by F32AG077916. MMS was supported in part by a Supplement to Promote Diversity in Health‐Related Research RF1AG056771-06S1. TTL was supported by K01AG059899. JS is supported by the NIDDK (R01DK133479), a pilot grant from the Diabetes Research Center at Washington University, P30DK020579, and a UW BIRCWH Scholars Program award to JS (K12HD101368). JS is a HHMI Freeman Hrabowski Scholar and is an American Federation for Aging Research grant recipient. CJ is supported in part by the NIAAA (R01AA029124). IO was supported by UWCCC Support Grant P30 CA014520 and Wisconsin Head and Neck Cancer SPORE CEP P50DE026787. WAR was supported by U54DK104310 and R01DK131175. Support was provided by the UW-Madison OVCRGE with funding from the Wisconsin Alumni Research Foundation. The authors used the UW-Madison Biotechnology Center Gene Expression Center (RRID:SCR_017757), and thank the University of Wisconsin Translational Research Initiatives in Pathology Laboratory, supported by the UW Department of Pathology and Laboratory Medicine and the UWCCC (P30CA014520) for use of its facilities and histopathology services. The Lamming lab was supported in part by the U.S. Department of Veterans Affairs (I01-BX004031), and this work was supported using facilities and resources from the William S. Middleton Memorial Veterans Hospital. Graphical abstract created with BioRender.com. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
DWL has received funding from, and is a scientific advisory board member of, Aeovian Pharmaceuticals, which seeks to develop novel, selective mTOR inhibitors for the treatment of various diseases.
References
- 1.Osborne TB, Mendel LB, and Ferry EL (1917). The Effect of Retardation of Growth Upon the Breeding Period and Duration of Life of Rats. Science 45, 294–295. 10.1126/science.45.1160.294. [DOI] [PubMed] [Google Scholar]
- 2.Weindruch R, Walford RL, Fligiel S, and Guthrie D (1986). The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. The Journal of nutrition 116, 641–654. 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- 3.Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, and Anderson RM (2014). Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nature communications 5, 3557. 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCay CM, Crowell MF, and Maynard LA (1935). The Effect of Retarded Growth Upon the Length of Life Span and Upon the Ultimate Body Size. The Journal of nutrition 10, 63–79. 10.1093/jn/10.1.63. [DOI] [PubMed] [Google Scholar]
- 5.Green CL, Lamming DW, and Fontana L (2022). Molecular mechanisms of dietary restriction promoting health and longevity. Nat Rev Mol Cell Biol 23, 56–73. 10.1038/s41580-021-00411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzuya M (2022). Nutritional Management of Sarcopenia and Frailty-Shift from Metabolic Syndrome to Frailty. J Nutr Sci Vitaminol (Tokyo) 68, S67–S69. 10.3177/jnsv.68.S67. [DOI] [PubMed] [Google Scholar]
- 7.Ribeiro RV, Solon-Biet SM, Pulpitel T, Senior AM, Cogger VC, Clark X, O’Sullivan J, Koay YC, Hirani V, Blyth FM, et al. (2019). Of Older Mice and Men: Branched-Chain Amino Acids and Body Composition. Nutrients 11. 10.3390/nu11081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, et al. (2014). Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19, 407–417. 10.1016/j.cmet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sluijs I, Beulens JW, van der AD, Spijkerman AM, Grobbee DE, and van der Schouw YT (2010). Dietary intake of total, animal, and vegetable protein and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-NL study. Diabetes Care 33, 43–48. 10.2337/dc09-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Nielen M, Feskens EJ, Mensink M, Sluijs I, Molina E, Amiano P, Ardanaz E, Balkau B, Beulens JW, Boeing H, et al. (2014). Dietary protein intake and incidence of type 2 diabetes in Europe: the EPIC-InterAct Case-Cohort Study. Diabetes Care 37, 1854–1862. 10.2337/dc13-2627. [DOI] [PubMed] [Google Scholar]
- 11.Lagiou P, Sandin S, Weiderpass E, Lagiou A, Mucci L, Trichopoulos D, and Adami HO (2007). Low carbohydrate-high protein diet and mortality in a cohort of Swedish women. J Intern Med 261, 366–374. 10.1111/j.1365-2796.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 12.Ni Lochlainn M, Bowyer RCE, Welch AA, Whelan K, and Steves CJ (2023). Higher dietary protein intake is associated with sarcopenia in older British twins. Age Ageing 52. 10.1093/ageing/afad018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. (2016). Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell reports 16, 520–530. 10.1016/j.celrep.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferraz-Bannitz R, Beraldo RA, Peluso AA, Dall M, Babaei P, Foglietti RC, Martins LM, Gomes PM, Marchini JS, Suen VMM, et al. (2022). Dietary Protein Restriction Improves Metabolic Dysfunction in Patients with Metabolic Syndrome in a Randomized, Controlled Trial. Nutrients 14. 10.3390/nu14132670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speakman JR, Mitchell SE, and Mazidi M (2016). Calories or protein? The effect of dietary restriction on lifespan in rodents is explained by calories alone. Exp Gerontol 86, 28–38. 10.1016/j.exger.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19, 418–430. 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green CL, Pak HH, Richardson NE, Flores V, Yu D, Tomasiewicz JL, Dumas SN, Kredell K, Fan JW, Kirsh C, et al. (2022). Sex and genetic background define the metabolic, physiologic, and molecular response to protein restriction. Cell Metab 34, 209–226 e205. 10.1016/j.cmet.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Munzberg H, Hutson SM, Gettys TW, Schwartz MW, and Morrison CD (2014). FGF21 is an endocrine signal of protein restriction. J Clin Invest 124, 3913–3922. 10.1172/JCI74915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson NE, Konon EN, Schuster HS, Mitchell AT, Boyle C, Rodgers AC, Finke M, Haider LR, Yu D, Flores V, et al. (2021). Lifelong restriction of dietary branched-chain amino acids has sex-specific benefits for frailty and lifespan in mice. Nat Aging 1, 73–86. 10.1038/s43587-020-00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green CL, and Lamming DW (2019). Regulation of metabolic health by essential dietary amino acids. Mech Ageing Dev 177, 186–200. 10.1016/j.mad.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill CM, Albarado DC, Coco LG, Spann RA, Khan MS, Qualls-Creekmore E, Burk DH, Burke SJ, Collier JJ, Yu S, et al. (2022). FGF21 is required for protein restriction to extend lifespan and improve metabolic health in male mice. Nature communications 13, 1897. 10.1038/s41467-022-29499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida S, Yamahara K, Kume S, Koya D, Yasuda-Yamahara M, Takeda N, Osawa N, Chin-Kanasaki M, Adachi Y, Nagao K, et al. (2018). Role of dietary amino acid balance in diet restriction-mediated lifespan extension, renoprotection, and muscle weakness in aged mice. Aging Cell 17, e12796. 10.1111/acel.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, and Smith-Wheelock M (2005). Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4, 119–125. 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orentreich N, Matias JR, DeFelice A, and Zimmerman JA (1993). Low methionine ingestion by rats extends life span. The Journal of nutrition 123, 269–274. 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 25.Lee BC, Kaya A, Ma S, Kim G, Gerashchenko MV, Yim SH, Hu Z, Harshman LG, and Gladyshev VN (2014). Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nature communications 5, 3592. 10.1038/ncomms4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. (2016). A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med 22, 421–426. 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop CA, Machate T, Henning T, Henkel J, Puschel G, Weber D, Grune T, Klaus S, and Weitkunat K (2022). Detrimental effects of branched-chain amino acids in glucose tolerance can be attributed to valine induced glucotoxicity in skeletal muscle. Nutr Diabetes 12, 20. 10.1038/s41387-022-00200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu D, Richardson NE, Green CL, Spicer AB, Murphy ME, Flores V, Jang C, Kasza I, Nikodemova M, Wakai MH, et al. (2021). The adverse metabolic effects of branched-chain amino acids are mediated by isoleucine and valine. Cell Metab 33, 905–922 e906. 10.1016/j.cmet.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deelen J, Kettunen J, Fischer K, van der Spek A, Trompet S, Kastenmuller G, Boyd A, Zierer J, van den Akker EB, Ala-Korpela M, et al. (2019). A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nature communications 10, 3346. 10.1038/s41467-019-11311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strong R, Miller RA, Antebi A, Astle CM, Bogue M, Denzel MS, Fernandez E, Flurkey K, Hamilton KL, Lamming DW, et al. (2016). Longer lifespan in male mice treated with a weakly estrogenic agonist, an antioxidant, an alpha-glucosidase inhibitor or a Nrf2-inducer. Aging Cell 15, 872–884. 10.1111/acel.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395. 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. (2011). Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 66, 191–201. 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flurkey K, Astle CM, and Harrison DE (2010). Life extension by diet restriction and N-acetyl-L-cysteine in genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 65, 1275–1284. 10.1093/gerona/glq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill CM, Laeger T, Dehner M, Albarado DC, Clarke B, Wanders D, Burke SJ, Collier JJ, Qualls-Creekmore E, Solon-Biet SM, et al. (2019). FGF21 Signals Protein Status to the Brain and Adaptively Regulates Food Choice and Metabolism. Cell reports 27, 2934–2947 e2933. 10.1016/j.celrep.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contreras AV, Torres N, and Tovar AR (2013). PPAR-alpha as a key nutritional and environmental sensor for metabolic adaptation. Adv Nutr 4, 439–452. 10.3945/an.113.003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitehead JC, Hildebrand BA, Sun M, Rockwood MR, Rose RA, Rockwood K, and Howlett SE (2014). A clinical frailty index in aging mice: comparisons with frailty index data in humans. J Gerontol A Biol Sci Med Sci 69, 621–632. 10.1093/gerona/glt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu TT, Thomas S, McLean DT, Roldan-Alzate A, Hernando D, Ricke EA, and Ricke WA (2019). Prostate enlargement and altered urinary function are part of the aging process. Aging (Albany NY) 11, 2653–2669. 10.18632/aging.101938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeBrasseur NK, Tchkonia T, and Kirkland JL (2015). Cellular Senescence and the Biology of Aging, Disease, and Frailty. Nestle Nutr Inst Workshop Ser 83, 11–18. 10.1159/000382054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehninger D, Neff F, and Xie K (2014). Longevity, aging and rapamycin. Cell Mol Life Sci 71, 4325–4346. 10.1007/s00018-014-1677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergnaud AC, Norat T, Mouw T, Romaguera D, May AM, Bueno-de-Mesquita HB, van der AD, Agudo A, Wareham N, Khaw KT, et al. (2013). Macronutrient composition of the diet and prospective weight change in participants of the EPIC-PANACEA study. PLoS One 8, e57300. 10.1371/journal.pone.0057300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solon-Biet SM, Mitchell SJ, Coogan SC, Cogger VC, Gokarn R, McMahon AC, Raubenheimer D, de Cabo R, Simpson SJ, and Le Couteur DG (2015). Dietary Protein to Carbohydrate Ratio and Caloric Restriction: Comparing Metabolic Outcomes in Mice. Cell reports 11, 1529–1534. 10.1016/j.celrep.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solon-Biet SM, Cogger VC, Pulpitel T, Wahl D, Clark X, Bagley E, Gregoriou GC, Senior AM, Wang QP, Brandon AE, et al. (2019). Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat Metab 1, 532–545. 10.1038/s42255-019-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, et al. (2014). Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell 13, 273–282. 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamming DW (2014). Diminished mTOR signaling: a common mode of action for endocrine longevity factors. SpringerPlus 3, 735. 10.1186/2193-1801-3-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, et al. (2014). Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 13, 468–477. 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, et al. (2008). Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J 22, 807–818. 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 47.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hochmuth L, Körner C, Ott F, Volke D, Cokan KB, Juvan P, Brosch M, Hofmann U, Hoffmann R, Rozman D, et al. (2021). Sex-dependent dynamics of metabolism in primary mouse hepatocytes. Arch Toxicol 95, 3001–3013. 10.1007/s00204-021-03118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap YW, Rusu PM, Chan AY, Fam BC, Jungmann A, Solon-Biet SM, Barlow CK, Creek DJ, Huang C, Schittenhelm RB, et al. (2020). Restriction of essential amino acids dictates the systemic metabolic response to dietary protein dilution. Nature communications 11, 2894. 10.1038/s41467-020-16568-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spadaro O, Youm Y, Shchukina I, Ryu S, Sidorov S, Ravussin A, Nguyen K, Aladyeva E, Predeus AN, Smith SR, et al. (2022). Caloric restriction in humans reveals immunometabolic regulators of health span. Science 375, 671–677. doi: 10.1126/science.abg7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner EM (2005). Caloric restriction decreases survival of aged mice in response to primary influenza infection. J Gerontol A Biol Sci Med Sci 60, 688–694. 10.1093/gerona/60.6.688. [DOI] [PubMed] [Google Scholar]
- 52.Gheller BJ, Blum JE, Lim EW, Handzlik MK, Hannah Fong EH, Ko AC, Khanna S, Gheller ME, Bender EL, Alexander MS, et al. (2021). Extracellular serine and glycine are required for mouse and human skeletal muscle stem and progenitor cell function. Molecular metabolism 43, 101106. 10.1016/j.molmet.2020.101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ham DJ, Borsch A, Chojnowska K, Lin S, Leuchtmann AB, Ham AS, Thurkauf M, Delezie J, Furrer R, Burri D, et al. (2022). Distinct and additive effects of calorie restriction and rapamycin in aging skeletal muscle. Nature communications 13, 2025. 10.1038/s41467-022-29714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flurkey K, Currer J, and Harrison D (2007). The Mouse in Aging Research. In The Mouse in Biomedical Research 2nd Edition, JG F, ed. (American College Laboratory Animal Medicine; ), pp. 637–672. [Google Scholar]
- 55.Hill CM, Laeger T, Albarado DC, McDougal DH, Berthoud HR, Munzberg H, and Morrison CD (2017). Low protein-induced increases in FGF21 drive UCP1-dependent metabolic but not thermoregulatory endpoints. Scientific reports 7, 8209. 10.1038/s41598-017-07498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, Yon Toh S, Chen Z, Guo K, Peng Ng C, Ponniah S, Lin S-C, Hong W, and Li P (2003). Cidea-deficient mice have lean phenotype and are resistant to obesity. Nature Genetics 35, 49–56. 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 57.Gummesson A, Jernås M, Svensson PA, Larsson I, Glad CA, Schéle E, Gripeteg L, Sjöholm K, Lystig TC, Sjöström L, et al. (2007). Relations of adipose tissue CIDEA gene expression to basal metabolic rate, energy restriction, and obesity: population-based and dietary intervention studies. J Clin Endocrinol Metab 92, 4759–4765. 10.1210/jc.2007-1136. [DOI] [PubMed] [Google Scholar]
- 58.Mitchell SE, Delville C, Konstantopedos P, Derous D, Green CL, Wang Y, Han JD, Promislow DE, Douglas A, Chen L, et al. (2016). The effects of graded levels of calorie restriction: V. Impact of short term calorie and protein restriction on physical activity in the C57BL/6 mouse. Oncotarget 7, 19147–19170. 10.18632/oncotarget.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maida A, Chan JSK, Sjoberg KA, Zota A, Schmoll D, Kiens B, Herzig S, and Rose AJ (2017). Repletion of branched chain amino acids reverses mTORC1 signaling but not improved metabolism during dietary protein dilution. Molecular metabolism 6, 873–881. 10.1016/j.molmet.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maida A, Zota A, Sjoberg KA, Schumacher J, Sijmonsma TP, Pfenninger A, Christensen MM, Gantert T, Fuhrmeister J, Rothermel U, et al. (2016). A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J Clin Invest 126, 3263–3278. 10.1172/JCI85946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wali JA, Milner AJ, Luk AWS, Pulpitel TJ, Dodgson T, Facey HJW, Wahl D, Kebede MA, Senior AM, Sullivan MA, et al. (2021). Impact of dietary carbohydrate type and protein-carbohydrate interaction on metabolic health. Nat Metab 3, 810–828. 10.1038/s42255-021-00393-9. [DOI] [PubMed] [Google Scholar]
- 62.MacArthur MR, Mitchell SJ, Chadaideh KS, Trevino-Villarreal JH, Jung J, Kalafut KC, Reynolds JS, Mann CG, Trocha KM, Tao M, et al. (2022). Multiomics assessment of dietary protein titration reveals altered hepatic glucose utilization. Cell reports 40, 111187. 10.1016/j.celrep.2022.111187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pak HH, Cummings NE, Green CL, Brinkman JA, Yu D, Tomasiewicz JL, Yang SE, Boyle C, Konon EN, Ong IM, and Lamming DW (2019). The Metabolic Response to a Low Amino Acid Diet is Independent of Diet-Induced Shifts in the Composition of the Gut Microbiome. Scientific reports 9, 67. 10.1038/s41598-018-37177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, Li X, Zhan L, White E, Anthony TG, et al. (2019). Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell Metab 29, 417–429 e414. 10.1016/j.cmet.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Handzlik MK, Gengatharan JM, Frizzi KE, McGregor GH, Martino C, Rahman G, Gonzalez A, Moreno AM, Green CR, Guernsey LS, et al. (2023). Insulin-regulated serine and lipid metabolism drive peripheral neuropathy. Nature 614, 118–124. 10.1038/s41586-022-05637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karusheva Y, Koessler T, Strassburger K, Markgraf D, Mastrototaro L, Jelenik T, Simon MC, Pesta D, Zaharia OP, Bodis K, et al. (2019). Short-term dietary reduction of branched-chain amino acids reduces meal-induced insulin secretion and modifies microbiome composition in type 2 diabetes: a randomized controlled crossover trial. Am J Clin Nutr 110, 1098–1107. 10.1093/ajcn/nqz191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramzan I, Taylor M, Phillips B, Wilkinson D, Smith K, Hession K, Idris I, and Atherton P (2020). A Novel Dietary Intervention Reduces Circulatory Branched-Chain Amino Acids by 50%: A Pilot Study of Relevance for Obesity and Diabetes. Nutrients 13. 10.3390/nu13010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bellantuono I, de Cabo R, Ehninger D, Di Germanio C, Lawrie A, Miller J, Mitchell SJ, Navas-Enamorado I, Potter PK, Tchkonia T, et al. (2020). A toolbox for the longitudinal assessment of healthspan in aging mice. Nature protocols 15, 540–574. 10.1038/s41596-019-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu D, Yang SE, Miller BR, Wisinski JA, Sherman DS, Brinkman JA, Tomasiewicz JL, Cummings NE, Kimple ME, Cryns VL, and Lamming DW (2018). Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J 32, 3471–3482. 10.1096/fj.201701211R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Searle SD, Mitnitski A, Gahbauer EA, Gill TM, and Rockwood K (2008). A standard procedure for creating a frailty index. BMC Geriatr 8, 24. 10.1186/1471-2318-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keil KP, Abler LL, Altmann HM, Bushman W, Marker PC, Li L, Ricke WA, Bjorling DE, and Vezina CM (2016). Influence of animal husbandry practices on void spot assay outcomes in C57BL/6J male mice. Neurourol Urodyn 35, 192–198. 10.1002/nau.22692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wegner KA, Abler LL, Oakes SR, Mehta GS, Ritter KE, Hill WG, Zwaans BM, Lamb LE, Wang Z, Bjorling DE, et al. (2018). Void spot assay procedural optimization and software for rapid and objective quantification of rodent voiding function, including overlapping urine spots. Am J Physiol Renal Physiol 315, F1067–F1080. 10.1152/ajprenal.00245.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cummings NE, Williams EM, Kasza I, Konon EN, Schaid MD, Schmidt BA, Poudel C, Sherman DS, Yu D, Arriola Apelo SI, et al. (2018). Restoration of metabolic health by decreased consumption of branched-chain amino acids. The Journal of physiology 596, 623–645. 10.1113/JP275075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. (2018). Ensembl 2018. Nucleic Acids Res 46, D754–D761. 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Team, R.C. (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- 76.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47–e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benjamini Y, and Hochberg Y (2018). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 79.Fiehn O (2016). Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol 114, 30 34 31–30 34 32. 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, and Fiehn O (2009). FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem 81, 10038–10048. 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, and Schwudke D (2008). Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res 49, 1137–1146. 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rousseeuw PJ (1987). Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. Journal of Computational and Applied Mathematics 20, 53–65. 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 83.Lenth RV (2023). emmeans: Estimated Marginal Means, aka Least-Squares Means
- 84.Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 85.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K (2022). cluster: Cluster Analysis Basics and Extensions
- 86.Lê S, Josse J, and Husson F (2008). FactoMineR: AnRPackage for Multivariate Analysis. Journal of Statistical Software 25, 18. 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 87.Kassambara A, and Mundt F (2020). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.7
- 88.Wang C, Li Q, Redden DT, Weindruch R, and Allison DB (2004). Statistical methods for testing effects on “maximum lifespan”. Mech Ageing Dev 125, 629–632. 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Yousefzadeh MJ, Zhao J, Bukata C, Wade EA, McGowan SJ, Angelini LA, Bank MP, Gurkar AU, McGuckian CA, Calubag MF, et al. (2020). Tissue specificity of senescent cell accumulation during physiologic and accelerated aging of mice. Aging Cell 19, e13094. 10.1111/acel.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonzano S, Crisci I, Podlesny-Drabiniok A, Rolando C, Krezel W, Studer M, and De Marchis S (2018). Neuron-Astroglia Cell Fate Decision in the Adult Mouse Hippocampal Neurogenic Niche Is Cell-Intrinsically Controlled by COUP-TFI In Vivo. Cell Rep 24, 329–341. 10.1016/j.celrep.2018.06.044. [DOI] [PubMed] [Google Scholar]
- 91.Chong J, Yamamoto M, and Xia J (2019). MetaboAnalystR 2.0: From Raw Spectra to Biological Insights. Metabolites 9 10.3390/metabo9030057. [DOI] [PMC free article] [PubMed]
- 92.Molenaar MR, Jeucken A, Wassenaar TA, van de Lest CHA, Brouwers JF, and Helms JB (2019). LION/web: a web-based ontology enrichment tool for lipidomic data analysis. GigaScience 8. 10.1093/gigascience/giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koelmel JP, Li X, Stow SM, Sartain MJ, Murali A, Kemperman R, Tsugawa H, Takahashi M, Vasiliou V, Bowden JA, et al. (2020). Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using A Rapid and User-Friendly Software. Metabolites 10. 10.3390/metabo10030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu Z (2022). Complex heatmap visualization. iMeta 1, e43. 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Diet Composition.
Diet composition and calorie content for diets used in this study. Related to STAR METHODS Experimental Model and Subject Details.
Supplementary Table 2: The relationship between isoleucine intake and phenotypic changes is dependent on sex and age.
R values for correlations from Figure 5A. Related to Figure 5.
Supplementary Table 3: Changes in the hepatic transcriptome on Low AA and Low Ile diets are dependent on age and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from transcriptomic analysis in males. (B) Significant changes at each omics level (adjusted p-value<0.01 for transcriptomics, p-value=0.05 for metabolomics and lipidomics). (C) Hepatic Log2 fold-changes and related p-values for comparisons of interest from transcriptomic analysis in females. (D) KEGG enriched pathways for significant genes found in Tables S3A and S3C. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 4: Changes in the hepatic and circulating metabolome are dependent of age, diet and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from metabolomic analysis in males. (B) KEGG enriched pathways for metabolites found in Tables S4A and S4C.
(C) Hepatic Log2 fold-changes and related p-values for comparisons of interest from metabolomic analysis in females. (D) Plasma Log2 fold-changes and related p-values for comparisons of interest from amino acid metabolomics analysis in 24 month old males and females. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 5: Changes in the hepatic lipidome on Low AA and Low Ile diets are dependent on age and sex.
(A) Hepatic Log2 fold-changes and related p-values for comparisons of interest from lipidomic analysis in males. (B) Hepatic Log2 fold-changes and related p-values for comparisons of interest from lipidomic analysis in females. (C) LION Lipid Ontology enrichment for pathways of significant lipids (p<0.05) for each of the age/sex/diet groups. (D) Concatenated data for coexpression network for Figure 6F and G, Supplementary Figures 6E and F. Related to Figure 6 and Supplementary Figure 6.
Supplementary Table 6: A low isoleucine diet increases median lifespan in both sexes.
(A) Lifespan statistics for Figure 7Q–T. (B) Number of mice for each study cohort. Related to STAR METHODS Experimental Model and Subject Details. (C) Lifespan of all, including censored, mice. Related to Figure 7.
Supplementary Table 7: Primer sequences.
Primer sequences for each gene measured by qPCR. Related to STAR METHODS Quantitative PCR.
Data Availability Statement
The accession number for the hepatic RNAseq gene expression data reported in this paper can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE241904).
This paper does not report original code.
All data required to remake graphs is available in the excel Data S1 file (published at http://data.mendeley.com/preview/dk74jscsww), and where stated is provided in Supplementary Tables.
List of primers used provided in Table S7 and in Key Resources.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Human insulin | Eli Lilly | NDC 0002-8215-17 (Humulin R U-100) |
| TRI reagent | Sigma | T9494 |
| SYBR Green | Thermo Fisher Scientific | 4309155 |
| Superscript III Reverse Transcriptase | Thermo Fisher Scientific | 18080085 |
| Critical commercial assays | ||
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D Systems | Cat# MF2100 |
| Deposited data | ||
| Liver transcriptomics raw and analyzed | This paper | GEO: GSE241904; Tables S3A and S3C. RRID:SCR_017757. |
| Liver metabolomics | This paper | Tables S4A and S4C |
| Plasma metabolomics | This paper | Table S4D |
| Liver lipidomics | This paper | Table S5A and S5B |
| Raw data to recreate figures | This paper | http://data.mendeley.com/preview/dk74jscsww |
| Experimental models: Organisms/strains | ||
| Mouse strain: C57BL6/J (Male) | The Jackson Laboratory | Cat# JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse strain: DBA2/J (Male) | The Jackson Laboratory | Cat# JAX: 000671; RRID: IMSR_JAX:000671 |
| Mouse strain: BALB/cJ (Female) | The Jackson Laboratory | Cat# JAX: 000651; RRID: IMSR_JAX:000651 |
| Mouse strain: C3H/HeJ (Female) | The Jackson Laboratory | Cat# JAX: 000659; RRID: IMSR_JAX:000659 |
| Mouse strain: UM-HET3 (Male and Female) | This paper | F2 progeny of (BALB/cJ x C57BL/6J) mothers and (C3H/HeJ x DBA/2J) fathers |
| Oligonucleotides | ||
| Fgf21: F: CAAATCCTGGGTGTCAAAGC | [13] | N/A |
| Fgf21: R: CATGGGCTTCAGACTGGTAC | [13] | N/A |
| Cidea: F: GAATAGCCAGAGTCACCTTCG | [28] | N/A |
| Cidea: R: AGCAGATTCCTTAACACGGC | [28] | N/A |
| Elov3: F: ATGCAACCCTATGACTTCGAG | [28] | N/A |
| Elov3: R: ACGATGAGCAACAGATAGACG | [28] | N/A |
| Ucp1: F: GCATTCAGAGGCAAATCAGC | [28] | N/A |
| Ucp1: R: GCCACACCTCCAGTCATTAAG | [28] | N/A |
| P16 (Cdkn2a): F: CCCAACGCCCCGAACT | [89] | N/A |
| P16 (Cdkn2a): R: GCAGAAGAGCTGCTACGTGAA | [89] | N/A |
| P21 (Cdkn1a): F: GTCAGGCTGGTCTGCCTCCG | [89] | N/A |
| P21 (Cdkn1a): R: CGGTCCCGTGGACAGTGAGCAG | [89] | N/A |
| Il1a: F: TGCAGTCCATAACCCATGATC | [90] | N/A |
| Il1a: R: ACAAACTTCTGCCTGACGAG | [90] | N/A |
| Il1b: F: TCCTGTGTGATGAAAGACGGCAC | [89] | N/A |
| Il1b: R: GTGCTGATGTACCAGTTGGGGAAC | [89] | N/A |
| Il6: F: CTGGGAAATCGTGGAAT | [89] | N/A |
| Il6: R: CTGGGAAATCGTGGAAT | [89] | N/A |
| Il10: F: ATAACTGCACCCACTTCCCA | [89] | N/A |
| Il10: R: GGGCATCACTTCTACCAGGT | [89] | N/A |
| Mcp1: F: GCATCCACGTGTTGGCTCA | [89] | N/A |
| Mcp1: R: CTCCAGCCTACTCATTGGGATCA | [89] | N/A |
| Actb (β-Actin): F: GATGTATGAAGGCTTTGGTC | [89] | N/A |
| Actb (β-Actin): R: TGTGCACTTTTATTGGTCTC | [89] | N/A |
| Software and algorithms | ||
| edgeR package | [76] | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism |
| MetaboAnalyst | [91] | https://www.metaboanalyst.ca/ |
| R (Version 3.4.2) | N/A | https://www.r-project.org/ |
| MyGene package | N/A | https://www.bioconductor.org/packages/release/bioc/html/mygene.html |
| Factoextra package | [87] | https://cran.r-project.org/web/packages/factoextra/index.html |
| missMDA package | N/A | https://cran.r-project.org/web/packages/missMDA/index.html |
| LION/web | [92] | http://www.lipidontology.com/ |
| Limma package | [77] | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Metabolomics package | N/A | https://cran.r-project.org/src/contrib/Archive/metabolomicsR/ |
| Lipid Annotator | [93] | https://www.agilent.com/en/solutions/omics/lipidomics/lipidomics-data-analysis |
| MassHunter | N/A | https://www.agilent.com/en/promotions/masshunter-mass-spec |
| Emmeans package | N/A | https://cran.rproject.org/web/packages/emmeans/index.html |
| ComplexHeatmap package | [84, 94] | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| Cluster package | N/A | https://cran.r-project.org/web/packages/cluster/index.html |
| Other | ||
| Normal Chow | Purina | Cat# 5001 |
| 2019 Teklad Global 19% Protein Extruded Rodent | Teklad | 2019 |
| Teklad Global 2020x | Teklad | 2020X |
| Mouse diets (See Table S1) | This paper | N/A |


