Abstract
Strains of Pseudomonas syringae pv. syringae were isolated from healthy and diseased stone fruit tissues sampled from 43 orchard sites in California in 1995 and 1996. These strains, together with P. syringae strains from other hosts and pathovars, were tested for pathogenicity and the presence of the syrB and syrC genes and were genetically characterized by using enterobacterial repetitive intergenic consensus (ERIC) primers and PCR. All 89 strains of P. syringae pv. syringae tested were moderately to highly pathogenic on Lovell peach seedlings regardless of the host of origin, while strains of other pathovars exhibited low or no pathogenicity. The 19 strains of P. syringae pv. syringae examined by restriction fragment length polymorphism analysis contained the syrB and syrC genes, whereas no hybridization occurred with 4 strains of other P. syringae pathovars. The P. syringae pv. syringae strains from stone fruit, except for a strain from New Zealand, generated ERIC genomic fingerprints which shared four fragments of similar mobility. Of the P. syringae pv. syringae strains tested from other hosts, only strains from rose, kiwi, and pear generated genomic fingerprints that had the same four fragments as the stone fruit strains. Analysis of the ERIC fingerprints from P. syringae pv. syringae strains showed that the strains isolated from stone fruits formed a distinct cluster separate from most of the strains isolated from other hosts. These results provide evidence of host specialization within the diverse pathovar P. syringae pv. syringae.
Bacterial canker and blast of stone fruit trees, caused by Pseudomonas syringae pv. syringae, affects all commercially grown Prunus species in California including peach (Prunus persica), European plum and French prune (P. domestica), Japanese plum (P. salicina), sweet cherry (P. avium), apricot (P. armeniaca), and almond (P. dulcis). Losses can result from a direct reduction in yield due to cold-induced blast or death of buds and flowers or from tree decline and death due to the development of cankers in branches and major scaffold limbs (20).
P. syringae pv. syringae is unique among most P. syringae pathovars in its ability to cause disease in over 180 species of plants in several unrelated genera (1). Strains of P. syringae pv. syringae are identified on the basis of biochemical and nutritional tests and symptom expression in host plants. In many cases, strains of P. syringae that are found infecting a previously unreported host and are biochemically similar to P. syringae pv. syringae strains have been placed in this pathovar without establishment of a common host range (34).
The relationship between P. syringae pv. syringae strains infecting Prunus species and strains that infect other crops such as tomato, cereals, citrus, and kiwi fruit is unknown and needs to be elucidated. Biochemical tests are not reliable for differentiating strains at or below the pathovar level (12, 25), and pathogenicity tests in greenhouses are not reliable indicators of natural host preferences (2). Peach seedlings (22) and cowpea leaves (14) were found to be susceptible to P. syringae pv. syringae strains from various hosts. There is, however, evidence of host specificity among P. syringae pv. syringae strains infecting beans (26, 27) and grasses (10) based on the results of pathogenicity tests.
Molecular analysis of genomic variability has been used to differentiate and classify bacterial strains below the level of species. Analysis of restriction fragment length polymorphisms (RFLP) of the chromosomal DNA of P. syringae strains detected differences between and within the pathovars (5, 11, 16). More recently, enterobacterial repetitive intergenic consensus (ERIC) sequences and repetitive extragenic palindromic (REP) sequences, which are short repetitive DNA sequences with highly conserved central inverted repeats that are dispersed throughout the genomes of diverse bacterial species (32), have been used to design universal PCR primers that generate highly reproducible, strain-specific fingerprints that can differentiate bacterial strains below the level of species or subspecies (4, 19).
The objective of this study was to identify and characterize strains of P. syringae pv. syringae isolated from various Prunus species and other plant hosts by using pathogenicity testing and RFLP and ERIC-PCR analyses.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. Many of these strains have been well characterized in previous pathogenicity, biochemical, and genetic studies (6, 9, 23). Strains were maintained in 15% glycerol at −80°C and subcultured on King’s medium B (KB) (13) as needed.
TABLE 1.
Bacterial strains used in this study
| Strain | Host of origin | Location collected | Source |
|---|---|---|---|
| P. syringae pv. syringae | |||
| 61 | Wheat | Delaware | D. Cooksey |
| 142 | Beet | California | E. Little |
| 321, 408 | Tomato | California | E. Little |
| 82-12 | Tomato | Georgia | R. Gitaitis |
| 728A | Bean | Wisconsin | S. Lindow |
| B18 | Millet | ? | J. DeVay |
| B37 | Rose | California | J. DeVay |
| B42 | Lemon | California | J. DeVay |
| B39 | Corn | Nebraska | J. DeVay |
| B40 | Foxtail grass | ? | J. DeVay |
| 84-160 | Kiwi fruit | California | K. Conn |
| B36 | Peach | New Zealand | J. DeVay |
| B3A | Peach | California | J. DeVay |
| B15 | Almond | California | J. DeVay |
| B301 | Pear | England | J. DeVay |
| B21 | Apricot | ? | J. DeVay |
| 32 strains | Almond | California | This study |
| 19 strains | Peach | California | This study |
| 13 strains | Cherry | California | This study |
| 12 strains | Prune plum | California | This study |
| 8 strains | Apricot | California | This study |
| 3 strains | Japanese plum | California | This study |
| 072, 073 | Geranium sp. | California | This study |
| 070, 071 | Malva sp. | California | This study |
| P. syringae pv. coriandricola | |||
| 269 | Cilantro | California | E. Little |
| P. syringae pv. morsprunorum | |||
| B28 | Cherry | ? | J. DeVay |
| 048 | Cherry | California | E. Little |
| 150 | Cherry | California | E. Little |
| P. syringae pv. tomato | |||
| 320 | Tomato | California | E. Little |
Strain isolation.
In 1995 and 1996, samples of both healthy and diseased tissues from stone fruit trees were collected from orchard sites in the Sacramento and San Joaquin valleys of California. Samples included healthy flowers, healthy and diseased dormant buds, diseased leaves, twigs, and branches. In addition, samples of weeds were collected during the winter of 1996 from orchards with a history of bacterial canker. Healthy tissues were washed in 0.01 M potassium phosphate buffer (PB) with 0.02% Tween 20 (ca. 3 g of flowers or 5 g of dormant twigs/25 ml of PB; 5 g of weed leaf tissue/100 ml of PB) on a platform shaker at 250 rpm for 30 min, and 100 μl of the wash liquid was spread onto KB plates containing 50 μg of cycloheximide per ml. Three to five healthy buds were ground in 2 ml of PB in a Pyrex tissue grinder, and 100 μl of either undiluted or 1:10-diluted (in PB) wash liquid was plated onto KB or KB with 50 μg of cycloheximide per ml. Diseased tissues were surface sterilized in 0.5% sodium hypochlorite for 1 min, rinsed in sterile water, and ground in a small amount of PB, and the liquid suspension was spread onto KB. The plates were incubated for 3 days, and then blue fluorescent colonies were counted, purified, and tested for the oxidase reaction, the ability to rot potato slices, the presence of arginine dihydrolase, levan production, and tobacco hypersensitivity (17).
Pathogenicity tests.
Bacterial cells grown for 24 h on solid KB at 24°C were suspended in PB to a concentration of ∼5 × 107 CFU/ml. Bacterial suspensions (∼0.1 ml) were injected into the stems of 10- to 12-week-old Lovell peach seedlings by using a 22-gauge needle inserted tangentially under the cambium. PB was injected as a control. The plants were maintained in a greenhouse at 28°C and rated after 10 days for disease development on a scale of 0 to 3 as follows: 0, light necrosis associated with wounding at the area of inoculation; 1, dark, water-soaked necrosis confined to the immediate area of inoculation, with some streaking in the cambium; 2, streaking in the cambium extending away from the site of inoculation, necrosis around the wound up to 2 mm above and below the wound with gumming; and 3, necrotic lesion and streaking involving the entire stem, often with girdling and death of distal portions and extensive gumming. Each seedling was inoculated in three places with a strain, and an average pathogenicity rating for each strain was used to determine the mean and standard deviation of the pathogenicity for all strains isolated from a particular host.
DNA preparation.
Total genomic DNA was extracted from 10 ml of 24-h shake cultures of bacterial cells. After centrifugation at 10,000 × g for 10 min, the bacterial pellet was resuspended in 1.5 ml of buffer (100 mM Tris-HCl [pH 7.5], 100 mM EDTA [pH 8.0]). Freshly prepared lysozyme (Sigma, St. Louis, Mo.) was added to a final concentration of 25 μg/ml, the volume of the solution was brought to 3 ml with sterile distilled water, and the mixture was incubated on ice for 10 min. Sodium dodecyl sulfate and proteinase K (Gibco BRL, Gaithersburg, Md.) were added to final concentrations of 1% and 200 μg/ml, respectively. The suspension was incubated for 1 h at 50°C and extracted four times with 5 ml of phenol-chloroform (1:1). The nucleic acids were precipitated with 0.1 volume of 3 M sodium acetate (pH 5.2)–1 volume of isopropanol, washed in 70% ethanol, and resuspended overnight in 200 μl of TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]) with 30 μg of RNase (Amresco, Solon, Ohio) per ml. The DNA solution was extracted again with an equal volume of phenol-chloroform (1:1) followed by an equal volume of chloroform-isoamyl alcohol (24:1). The nucleic acids were precipitated with 0.1 volume of sodium acetate–2 volumes of 100% ethanol, rinsed in 70% ethanol, and resuspended in 200 μl of TE buffer. DNA concentrations were determined with a TKO 100 fluorometer (Hoefer Scientific Instruments, San Francisco, Calif.).
RFLP analysis.
Approximately 1 μg of total genomic DNA was digested at 37°C overnight with EcoRI (Pharmacia Biotech, Uppsala, Sweden), and nucleic acid fragments were electrophoresed in 1% agarose gels at 45 V for 5 to 6 h with TAE (0.04 M Tris acetate, 0.001 M EDTA). The DNA was transferred to Nytran (Schleicher & Schuell, Keene, N.H.) nylon membranes, and Southern hybridization analysis was performed as previously described (8) with a [32P]dATP-labeled 7-kb HindIII fragment containing the syrB and syrC genes from plasmid p601D, which was kindly provided by D. Gross (23). The size of restriction fragment(s) that hybridized with the probe was estimated relative to the mobility of 1-kb DNA standards (Gibco BRL).
Oligonucleotide primers and PCR conditions.
ERIC oligonucleotide primers (ERIC1R [5′-ATGTAAGCTCCTGGGGATTCAC-3′] and ERIC2 [5′-AAGTAAGTGACTGGGGTGAGCG-3′]) were purchased from Oligos Etc. (Wilsonville, Oreg.). The PCR conditions were as previously described (21, 32). Bacterial strains were streaked onto plates of KB and incubated for 2 days at 25°C. A small portion of a single colony was transferred to 25 μl of a PCR mixture containing 50 pmol of each primer, 1.25 mM each deoxynucleoside triphosphate, 10% dimethyl sulfoxide, 4 μg of bovine serum albumin (Boehringer Mannheim, Indianapolis, Ind.), 2 U of AmpliTaq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.), 16.6 mM ammonium sulfate, 67 mM Tris HCl (pH 8.0), 6.7 mM magnesium chloride, 6.7 μM EDTA, and 30 mM β-mercaptoethanol. The mixture was overlaid with silicone oil (Aldrich Chemicals, Milwaukee, Wis.), and PCR was performed in a no. 480 DNA thermal cycler (Perkin-Elmer Cetus) under the following conditions: 1 cycle at 95°C for 6 min; 35 cycles at 94°C for 1 min, 52°C for 1 min, and 65°C for 8 min; and a final extension cycle at 68°C for 16 min. Aliquots (8 μl) of the reaction mixture were electrophoresed on 1.5% TAE agarose gels at room temperature at 5 V/cm for 4 h. The DNA fragments in the gel were visualized by staining with ethidium bromide.
Data analysis.
The amplified fragments of each strain were scored as 1 (present) or 0 (absent), and pairwise comparisons were made of each unique pattern by using the Jaccard similarity coefficient (30) and the NTSYS program (Exeter Software, Setauket, N.Y.). A similarity matrix was generated by using the unweighted pair-group method with averages. Phenograms were constructed with the tree display option (TREE). A cophenetic value matrix was calculated by using the COPH option and compared with the original similarity matrix by using the MXCOMP option to test the goodness of fit of the cluster analysis.
RESULTS
Strain collection and identification.
Ninety-one strains of P. syringae pv. syringae collected from 43 almond, prune, plum, peach, apricot, and cherry orchard sites in the San Joaquin and Sacramento valleys were used in this study. Each strain was collected from separate tissue samples within an orchard site. The bacterium was detected in diseased samples and as an epiphyte on apparently healthy twigs, flowers, and buds. In addition, P. syringae pv. syringae was washed from the leaves of two weeds, a Geranium sp. and a Malva sp., that were growing in a prune orchard with trees showing symptoms of bacterial canker. All P. syringae pv. syringae strains used in this study were negative for oxidase, potato rotting, and arginine dihydrolase and positive for levan production and the hypersensitive response on tobacco.
A total of 76 strains of P. syringae pv. syringae isolated in 1995 and 1996 from Prunus hosts were tested for pathogenicity on Lovell peach seedlings. In addition, four strains from orchard weeds, nine strains from nine other hosts, and five strains of three other P. syringae pathovars were tested. All of the P. syringae pv. syringae strains were moderately to highly pathogenic on peach, as evidenced by a pathogenicity rating of 2 or more, except for wheat strain 61, which had a rating of 1.0. The stone fruit strains, together with the bean and lemon strains, had pathogenicity ratings in the range of 2.6 to 3.0, while the grass, millet, pear, tomato, and weed strains had ratings of 2.0. The rose and kiwi strain ratings were 2.5 and 2.3, respectively. P. syringae pv. tomato, morsprunorum, and coriandricola were of low virulence on peach (0.5, 1.1, and 1.0 disease rating, respectively), and each incited only a mild necrotic reaction around the site of inoculation.
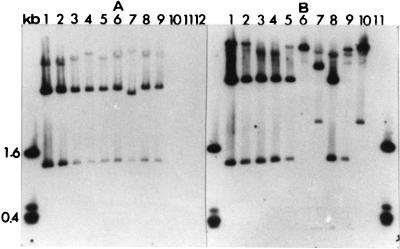
A total of 23 strains, including 19 strains of P. syringae pv. syringae and 4 strains of four other pathovars, were tested for the presence of the syrB and syrC genes. DNA isolated from all of the P. syringae pv. syringae strains, but not the DNA from the other pathovars, hybridized with the syrB and syrC probe (Fig. 1). The kiwi (84-160), rose (B37), Geranium (073), tomato (321), and beet (142) strains and all of the stone fruit strains except for the peach strain from New Zealand (B36) had a similar RFLP pattern.
FIG. 1.
Southern hybridization of EcoRI-digested total genomic DNA of strains of P. syringae pv. syringae and other P. syringae pathovars probed with a 32P-labeled 7-kb HindIII fragment containing the syrB and syrC genes from plasmid p601D. Lanes: kb, the 1-kb molecular marker; A1, B3 peach; A2, B15 almond; A3, 040 almond; A4, B301 pear; A5, 728a bean; A6, B18 millet; A7, B36 peach (New Zealand); A8, 408 tomato; A9, 142 beet; A10, P. syringae pv. maculicola 533; A11, P. syringae pv. coriandricola 269; A12, P. syringae pv. morsprunorum B28; B1, 092 prune; B2, 073 Geranium sp.; B3, B21 apricot; B4, 036 peach; B5, 84-160 kiwi; B6, 61 wheat; B7, 321 tomato; B8, B37 rose; B9, B39, corn; B10, B42 lemon; B11, P. syringae pv. tomato 320 (A and B in the lane designations refer to panels A and B, respectively).
ERIC analysis.
The DNA fingerprints of 104 strains isolated in 1995 and 1996 from 43 orchard sites, including 4 epiphytic weed strains of P. syringae and strains obtained from other hosts and/or sources (Table 1), were determined by using ERIC-PCR. The stone fruit strains (except for strain B36, isolated from peach in New Zealand), rose strain B37, kiwi strain 84-160 and pear strain B301 each generated 1 of 11 distinct ERIC genomic fingerprint patterns, which all shared four fragments of similar mobility (Table 2). These 11 patterns could be differentiated by polymorphisms in one or more of the other amplified DNA fragments (Fig. 2). Ninety-three percent of the stone fruit strains isolated in this study produced either pattern 2, 3, 5, or 6 (Table 2). Pattern 10 was represented by the epiphytic Geranium sp. weed strains and by an epiphytic strain recovered from a healthy prune bud, each from a different orchard site with a history of bacterial canker disease. The Malva sp. weed strains generated a unique pattern, which did not contain the four fragments shared by the stone fruit strain patterns. A strain from a healthy prune flower isolated in the same orchard as the weed strains was the only strain to generate pattern 11. However, 15 other strains isolated from apparently healthy tissues collected in various orchards each generated one of the four most common stone fruit fingerprint patterns.
TABLE 2.
Number of strains of P. syringae pv. syringae generating 1 of 11 distinct ERIC genomic fingerprint patterns
| Host | No. of strainsa with ERIC patternb:
|
Total no. | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| Strains isolated in this study | ||||||||||||
| Almond | 9 | 2 | 14 | 5 | 1 | 1 | 32 | |||||
| Peach | 4 | 9 | 2 | 4 | 19 | |||||||
| Prune | 8 | 2 | 1 | 1 | 12 | |||||||
| Cherry | 1 | 6 | 4 | 1 | 1 | 13 | ||||||
| Apricot | 6 | 2 | 8 | |||||||||
| Plum | 3 | 3 | ||||||||||
| Geranium | 2 | 2 | ||||||||||
| Total | 1 | 36 | 17 | 0 | 16 | 12 | 1 | 1 | 1 | 3 | 1 | 89 |
| Strains characterized previously | ||||||||||||
| Peach (B3) | 1 | 1 | ||||||||||
| Almond (B15) | 1 | 1 | ||||||||||
| Pear (B301) | 1 | 1 | ||||||||||
| Apricot (B21) | 1 | 1 | ||||||||||
| Rose (B37) | 1 | 1 | ||||||||||
| Kiwi (84-160) | 1 | 1 | ||||||||||
| Total | 1 | 1 | 2 | 2 | 6 | |||||||
Number of strains tested that generated the banding pattern.
ERIC fingerprint patterns that all share four fragments of similar mobility.
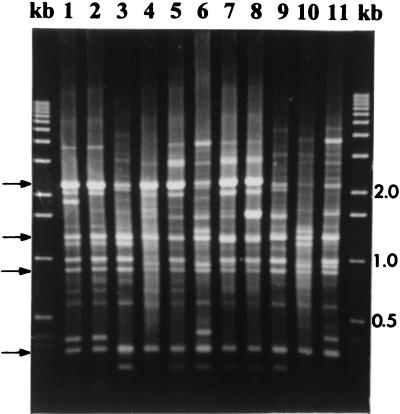
FIG. 2.
The 11 ERIC genomic fingerprint patterns which shared four fragments of similar mobilities generated by 95 of the 104 P. syringae pv. syringae strains tested. Lanes: kb, the 1-kb molecular marker 1 to 11, ERIC fingerprint patterns 1 to 11, respectively. The arrows on the left indicate the four fragments common to the 11 ERIC patterns.
The occurrence of a particular ERIC fingerprint pattern was not host or location specific. In fact, pattern 2 was common to some strains isolated from all Prunus hosts (Table 2). In some cases, strains that generated different patterns were isolated on the same day from separate samples collected in the same orchard. In addition, except for the peach strain from New Zealand, the stone fruit strains from other sources, including B3 and B15, which have been in culture for at least 30 years (6), generated fingerprint patterns similar to those for the strains isolated in this study (Table 2; Fig. 2).
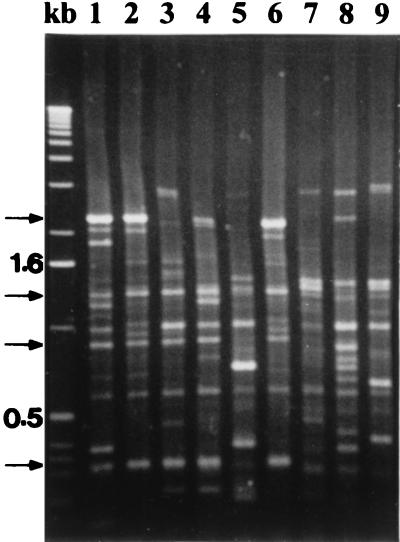
Most P. syringae pv. syringae strains (61, 321, 82-12, B18, B39, B40, and B42) from hosts other than stone fruits, together with the New Zealand peach strain (B36), generated patterns that did not contain any of the four DNA fragments shared by the Prunus strain patterns (Fig. 3). However, the rose (B37) strain generated stone fruit pattern 3 whereas the pear strain (B301) and the kiwi fruit strain (84-160) generated a unique pattern (pattern 4) that contained the four fragments common to the stone fruit patterns (Fig. 2). The bean strain (728a) contained three of the four common bands.
FIG. 3.
ERIC fingerprints of P. syringae pv. syringae strains isolated from various plant hosts, showing strain variability within the pathovar. Lanes: kb, the 1-kb molecular marker; 1, B3 peach (pattern 1); 2, B301 pear (pattern 4); 3, B728a bean; 4, B37 rose (pattern 3); 5, B42 lemon; 6, 84-160 kiwi (pattern 4); 7, B18 millet; 8, B40 foxtail; 9, 321 tomato. Arrows on the left indicate the four fragments common to 95 of the 104 strains tested.
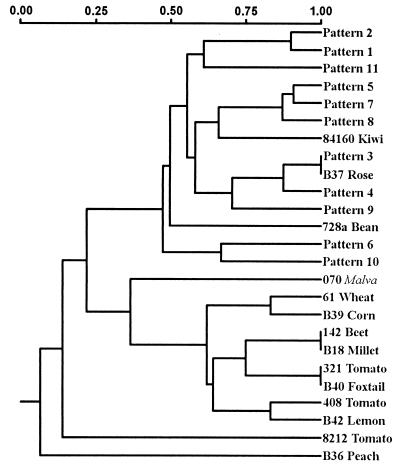
Sixteen bands were scored for the cluster analysis. The resulting dendrogram (Fig. 4) supported the observation that the genomic fingerprints of P. syringae pv. syringae strains from stone fruits had more similarities to each other than to those of most of the strains from other hosts. The P. syringae pv. syringae strains tested formed two clusters. One cluster contained the strains with the 10 stone fruit patterns, together with strains B301 and 84-160 (pattern 4), B37, and 728a. The other cluster contained most of the remaining P. syringae pv. syringae strains from various hosts, with the Malva weed strain 070 being the most divergent strain within this cluster. One tomato strain isolated in Georgia (82-12) and the New Zealand peach strain (B36) were dissimilar from all of the other strains tested and were outliers from the two main clusters. A cophenetic correlation of >0.9 was determined for the similarity matrix, indicating a very high goodness of fit for the cluster analysis.
FIG. 4.
Dendrogram of genetic relatedness of the ERIC fingerprint patterns generated by 104 strains of P. syringae pv. syringae. Cluster analysis was performed by using the Jaccard similarity coefficient (30). Ninety-five of the strains generated 1 of the 11 fingerprint patterns indicated on the dendrogram. The remaining strains are listed with the host from which they were originally isolated. The scale at the top indicates the degree of genetic relatedness between the strains tested.
DISCUSSION
In this study, the P. syringae pv. syringae strains isolated from Prunus hosts in California generated similar genetic profiles in ERIC-PCR whereas most strains of P. syringae pv. syringae isolated from other hosts generated dissimilar patterns. This suggests a host specialization of the stone fruit strains within the heterogeneous pathovar syringae. Specialization of P. syringae pv. syringae strains toward a particular host has been observed in previous studies. Saad and Hagedorn (27) used a bean pod pathogenicity assay and found that strains of P. syringae pv. syringae isolated from beans or as epiphytes from weeds near bean fields, but not strains isolated from other hosts, caused a pathogenic reaction. The same result was observed in other studies of the strains isolated from beans (2, 7, 26), which led Rudolph (26) to propose designating the bean strains P. syringae pv. phaseoli. Legard et al. (16), using RFLP analyses of P. syringae pv. syringae strains from various hosts, found that the bean strains formed a separate cluster within the pathovar, substantiating the results of the greenhouse pathogenicity assays. Gross and DeVay (10) found a tendency for grass strains of P. syringae pv. syringae to be highly virulent on inoculated maize plants and to reach higher populations in maize leaf tissues than did strains isolated from nongrass hosts. In our study, pathogenicity tests with peach seedlings in the greenhouse failed to distinguish between stone fruit strains and strains from other hosts but were useful in differentiating P. syringae pv. syringae strains from strains of other pathovars. Similarly, Otta and English (22) found that P. syringae pv. syringae strains from 30 different hosts induced similar cankers on wound-inoculated peach seedling stems.
Syringomycin functions as a nonspecific virulence factor in strains of P. syringae pv. syringae (6, 10). Genes for the synthesis and export of the phytotoxin are found in P. syringae pv. syringae strains but not in several other related pathovars (23). Some other phytotoxin genes are highly pathovar specific and have been used to develop DNA probes to identify coronatine-producing (3) or phaseolotoxin-producing (28) strains. In addition, the production of syringomycin has been used as a determinative characteristic in identifying strains of P. syringae pv. syringae (29, 34). Therefore, the syrB and syrC genes were used as hybridization probes to confirm the identity of a representative group of the P. syringae pv. syringae strains used in this study. The stone fruit strains, except for the New Zealand peach strain (B36), had a similar hybridization pattern to the pear, rose, bean, and kiwi fruit strains (strains which had a similar ERIC pattern), as well as to the strains from millet, beet, and tomato. However, the ubiquitous presence of syringomycin in this pathovar indicates that although strains can be genetically heterogeneous by methods such as ERICs and RFLPs, all of the P. syringae pv. syringae strains tested have the genetic potential to produce syringomycin.
Weed hosts within or near orchards or fields have been hypothesized to provide overwintering sites for P. syringae pv. syringae and to serve as an inoculum source for disease outbreaks (7, 15, 24). In this study, the ERIC patterns of P. syringae pv. syringae strains recovered from weed species were dissimilar to those of strains causing cankers on Prunus hosts. Thus, the role played by P. syringae pv. syringae epiphytes on weeds in the initiation and development of bacterial canker disease of prune in California remains uncertain. Strains from one of the weed species and two epiphytic strains isolated from healthy prune tissues were the only strains to generate two of the ERIC patterns (patterns 10 and 11). Another 15 epiphytic strains generated the same banding patterns as the strains isolated from diseased tissues. Therefore, healthy tissues appear to harbor a heterogeneous population of epiphytic strains, with at least some of these strains being capable of causing bacterial canker in susceptible tissues.
ERIC and REP PCR has been shown to be a rapid and reliable method to differentiate plant-pathogenic bacteria at or below the pathovar level with highly reproducible results (19). In a study which used REP PCR to compare 100 P. syringae pv. syringae strains from ornamental pear trees with 6 strains from peach, wheat, tomato, and maize, all of the ornamental-pear strains clustered into one of two closely related groups while none of the strains from other hosts had any similarities to the pear strains or to each other (31). These results are similar to what was observed in this study when P. syringae pv. syringae strains isolated from stone fruits in California were compared to strains isolated from other hosts and support the theory that some, if not all, strains within the heterogeneous pathovar syringae have adapted genetically to a particular host. In addition, similar to what was observed in this study, previous research has demonstrated a close relationship between strains causing disease on pome fruits, such as pear, and stone fruits (9, 25). Weingart and Völksch (33), however, found few similarities in the ERIC banding patterns of five strains of P. syringae pv. syringae isolated from pear, apple, and cherry trees in Western Europe. This apparent high diversity might be expected in an area with a long history of cultivating Prunus species, where, presumably, the associated microflora would have evolved with and adapted to the various Prunus hosts over time. In our study, a peach strain (B36) isolated in New Zealand generated an ERIC pattern unlike those from all of the other P. syringae pv. syringae strains tested; this strain may be the result of an evolutionary adaptation separate from North American and European P. syringae pv. syringae strains.
Louws et al. (19), using ERIC PCR, found evidence of intrapathovar diversity among strains of Xanthomonas campestris pv. vesicatoria and campestris, pathovars which also have more than one host. Other pathovars with a more restricted host range, such as P. syringae pv. morsprunorum and tomato, had low or no diversity in their ERIC profiles. Additional studies by other genetic characterization methods support the hypothesis that variation was greater among strains from pathovars with wide host ranges, such as P. syringae pv. syringae. Denny et al. (5) used RFLP to analyze six P. syringae pv. syringae strains and found that the strains clustered into two groups which contained strains either from monocots or from dicots whereas strains of P. syringae pv. tomato were less genetically diverse. In another study involving RFLP and randomly amplified polymorphic DNA analyses (18), strains of P. syringae pv. apii, which infect only celery, were more genetically homogeneous than were strains of P. syringae pv. maculicola, which infect a wide range of crucifer hosts. Overall, our results suggest that strains of P. syringae pv. syringae that are adapted to a specialized niche, such as California stone fruits, may be the result of a recent adaption and/or genetic isolation, resulting in the genetically homogeneous population of P. syringae pv. syringae strains from stone fruits observed in this study, which formed a distinct group from strains isolated from other hosts.
ACKNOWLEDGMENT
This work was supported by a grant from the California Prune Board.
REFERENCES
- 1.Bradbury J F. Guide to Plant Pathogenic Bacteria. Kew, England: CAB International Mycological Institute; 1986. Pseudomonas syringae pv. syringae; pp. 175–177. [Google Scholar]
- 2.Cheng G Y, Legard D E, Hunter J E, Burr T J. Modified bean pod assay to detect strains of Pseudomonas syringae pv. syringae that cause bacterial brown spot of snap bean. Plant Dis. 1989;73:419–423. [Google Scholar]
- 3.Cuppels D A, Moore R A, Morris V L. Construction and use of a nonradioactive DNA hybridization probe for detection of Pseudomonas syringae pv. tomato on tomato plants. Appl Environ Microbiol. 1990;56:1743–1749. doi: 10.1128/aem.56.6.1743-1749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny T P, Gilmour M N, Selander R K. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J Gen Microbiol. 1988;134:1949–1960. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 6.DeVay J E, Lukezic F L, Sinden S L, English H, Coplin D L. A biocide produced by pathogenic isolates of Pseudomonas syringae and its possible role in the bacterial canker disease of peach trees. Phytopathology. 1968;58:95–101. [Google Scholar]
- 7.Ercolani G L, Hagedorn D J, Kelman A, Rand R E. Epiphytic survival of Pseudomonas syringae on hairy vetch in relation to epidemiology of bacterial brown spot of bean in Wisconsin. Phytopathology. 1974;64:1330–1339. [Google Scholar]
- 8.Gilbertson R L, Maxwell D P, Hagedorn D J, Leong S A. Development and application of a plasmid DNA probe for detection of bacteria causing common bacterial blight of bean. Phytopathology. 1989;79:518–525. [Google Scholar]
- 9.Gross D C, Cody Y S, Proebsting E L, Jr, Radamaker G K, Spotts R A. Ecotypes and pathogenicity of ice-nucleation-active Pseudomonas syringae isolated from deciduous fruit tree orchards. Phytopathology. 1984;74:241–248. [Google Scholar]
- 10.Gross D C, DeVay J E. Population dynamics and pathogenesis of Pseudomonas syringae in maize and cowpea in relation to the in vitro production of syringomycin. Phytopathology. 1977;67:475–483. [Google Scholar]
- 11.Henson M, Hildebrand D C, Schroth M N. Relatedness of Pseudomonas syringae pv. tomato, Pseudomonas syringae pv. maculicola and Pseudomonas syringae pv. antirrhini. J Appl Bacteriol. 1992;73:455–464. [Google Scholar]
- 12.Hildebrand D C, Schroth M N, Huisman O C. The DNA homology matrix and non-random variation concepts as the basis for the taxonomic treatment of plant pathogenic and other bacteria. Annu Rev Phytopathology. 1982;20:235–256. [Google Scholar]
- 13.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 14.Lai M, Hass B. Reaction of cowpea seedlings to phytopathogenic bacteria. Phytopathology. 1973;63:1099–1103. [Google Scholar]
- 15.Latorre B A, Jones A L. Evaluation of weeds and plant refuse as potential sources of inoculum of Pseudomonas syringae in bacterial canker of cherry. Phytopathology. 1979;69:1122–1125. [Google Scholar]
- 16.Legard D E, Aquadro C F, Hunter J E. DNA sequence variation and phylogenetic relationships among strains of Pseudomonas syringae pv. syringae inferred from restriction site maps and restriction fragment length polymorphism. Appl Environ Microbiol. 1993;59:4180–4188. doi: 10.1128/aem.59.12.4180-4188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lelliott R A, Billing E, Hayward A C. A determinative scheme for the fluorescent plant pathogenic pseudomonads. J Appl Bacteriol. 1966;29:470–489. doi: 10.1111/j.1365-2672.1966.tb03499.x. [DOI] [PubMed] [Google Scholar]
- 18.Little E L, Gilbertson R L. Phenotypic and genotypic characters support placement of Pseudomonas syringae strains from tomato, celery, and cauliflower into distinct pathovars. In: Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, von Kietzell J, editors. Pseudomonas syringae pathovars and related pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 542–547. [Google Scholar]
- 19.Louws F J, Fulbright D W, Stephens C T, de Bruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa J M, English H. Diseases of temperate zone tree fruit and nut crops. Publication 3345. Oakland: University of California Division of Agriculture and Natural Resources; 1991. [Google Scholar]
- 21.Opgenorth D C, Smart C D, Louws F J, de Bruijn F J, Kirkpatrick B C. Identification of Xanthomonas fragariae field isolates by rep-PCR genomic fingerprinting. Plant Dis. 1996;80:868–873. [Google Scholar]
- 22.Otta J D, English H. Serology and pathology of Pseudomonas syringae. Phytopathology. 1971;61:443–452. [Google Scholar]
- 23.Quigley N B, Gross D C. Syringomycin production among strains of Pseudomonas syringae pv. syringae: conservation of the syrB and syrD genes and activation of phytotoxin production by plant signal molecules. Mol Plant-Microbe Interact. 1994;7:78–90. doi: 10.1094/mpmi-7-0078. [DOI] [PubMed] [Google Scholar]
- 24.Roos I M M, Hattingh M J. Weeds in orchards as potential source of inoculum for bacterial canker of stone fruit. Phytophylactica. 1986;18:5–6. [Google Scholar]
- 25.Roos I M M, Hattingh M J. Pathogenicity and numerical analysis of phenotypic features of Pseudomonas syringae strains isolated from deciduous fruit trees. Phytopathology. 1987;77:900–908. [Google Scholar]
- 26.Rudolph K. Bacterial brown spot disease of bush bean (Phaseolus vulgaris L.) in Germany, incited by Pseudomonas syringae van Hall s. s. pathovar phaseoli. Z Pflanzenkr Pflanzenschutz. 1979;86:75–85. [Google Scholar]
- 27.Saad S M, Hagedorn D J. Relationship of isolate source to virulence of Pseudomonas syringae on Phaseolus vulgaris. Phytopathology. 1972;62:678–680. [Google Scholar]
- 28.Schaad N W, Azad H, Peet R C, Panopoulos N J. Identification of Pseudomonas syringae pv. phaseolicola by a DNA hybridization probe. Phytopathology. 1989;79:903–907. [Google Scholar]
- 29.Seemüller E, Arnold M. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria. 1978. Pathogenicity, syringomycin production and other characteristics of pseudomonad strains isolated from deciduous fruit trees; pp. 703–710. [Google Scholar]
- 30.Sokal R R, Sneath P H A. Principles of numerical taxonomy. San Francisco, Calif: W. H. Freeman & Co.; 1963. pp. 169–210. [Google Scholar]
- 31.Sundin G W, Demezas D H, Bender C L. Genetic and plasmid diversity within natural populations of Pseudomonas syringae with various exposures to copper and streptomycin bactericides. Appl Environ Microbiol. 1994;60:4421–4431. doi: 10.1128/aem.60.12.4421-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weingart H, Völksch B. Genetic fingerprinting of Pseudomonas syringae pathovars using ERIC-, REP-, and IS50-PCR. J Phytopathol. 1997;145:339–345. [Google Scholar]
- 34.Young J M. Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. syringae van Hall 1902. Ann Appl Biol. 1991;118:283–298. [Google Scholar]