Abstract
Cryogenic-electron microscopy single particle analysis has revolutionized the structural analysis of icosahedral viruses, including tailed bacteriophages. In recent years, localized (or focused) reconstruction has emerged as a powerful data analysis method to capture symmetry mismatches and resolve asymmetric features in icosahedral viruses. Here we describe the methods used to reconstruct the 2.65 MDa tail apparatus of the Shigella phage Sf6, a representative member of the Podoviridae superfamily.
Keywords: Cryo-EM, Single Particle Analysis, Localized Reconstruction, Bacteriophages, Electron density, Real-space refinement
1. Introduction
Classical crystallographic studies on icosahedral viruses have been largely supplanted by single-particle analysis of frozen-hydrated viruses, which provides a more rapid, cost-effective, and physiological-relevant way to study large molecular complexes [1]. Nonetheless, icosahedral symmetry averaging continues to be used in cryo-electron microscopy single particle analysis (cryo-EM SPA) to improve the signal-to-noise and the resolution of electron density maps. One of the problems of icosahedral averaging is the loss of information on components that do not obey icosahedral symmetry [2], which can be smeared or wiped off entirely from a density. In recent years, localized (or focused) reconstruction has emerged as a powerful data analysis method to capture symmetry mismatches and resolve asymmetric features in icosahedral viruses [3,4]. The tenet of this method is to extract smaller image areas (or sub-particles) corresponding to different symmetry-related subunits from particle images and process them as if they were individual particles. Compared to previously developed methods to handle symmetry mismatches, such as standard asymmetric refinement [5], symmetry relaxation [6], symmetry expansion combined with focused classification/refinement [7,8], and multibody refinement [9], localized reconstruction is less computationally intensive as the sub-particles extracted from the original particle are usually considerably smaller than the original particle. Another advantage of using localized reconstruction to visualize asymmetric features in icosahedral capsids is that the orientation of each sub-particle can be calculated using the orientation and symmetry of the original virion. In addition, if the sub-particle is symmetric (e.g., dodecameric portal protein, hexameric tail protein, etc.), it can be aligned to its own symmetry convention, and the final density improved by applying local averaging. Unsurprisingly, localized reconstruction has found valuable applications in the analysis of bacteriophage tails that vary significantly in size and complexity. Here, we describe the utilization of localized reconstruction to decipher the complete architecture of the Shigella-phage Sf6 tail apparatus [10]. The Sf6 tail is a ~2.65 MDa molecular machine consisting of 51 copies of five unique polypeptide chains. Using localized reconstruction, we identified a 3D class containing the phage tail that we subjected to 12- and 6-fold rotational averaging, yielding excellent maps at a maximum Fourier Shell Resolution (FSC) of 2.7 Å at 0.143 cut-off. These maps were used to de novo build 14,310 amino acids corresponding to 12 copies of the portal protein and the head-to-tail adaptor gp4, six copies of the tail adaptor gp10 and six trimeric copies of the tailspike N-termini. As the density for the tailspike body (amino acid residues 124-623) was poorly visible in the symmetrized localized reconstruction due to its flexibility, we computed a second focused reconstruction from the symmetry expansion of the C6 map. This new map allowed us to model 1,862 residues of the full-length Sf6 tailspike. Finally, the tail needle was modeled by fitting a previously determined crystallographic model [11,12] in a lower-resolution asymmetric (C1) map. Overall, all 51 polypeptide chains of the Sf6 tail apparatus were subjected to real-space refinement that yielded a final Correlation Coefficient (CC) of 0.84-0.9.
2. Materials
All steps of grid preparation, vitrification, clipping, and mounting require consumables and tools only available through Thermo Scientific™. Most computationally intensive steps of the cryo-EM structure determination workflow are carried out on NVIDIA GeForce RTX graphics processors (GPUs) using CUDA, a programming language developed by NVIDIA [13]. Please visit the RELION [14] website for more technical information about GPU computing.
2.1. Vitrification
1. Quantifoil grids 2/1 Cu 200-mesh or similar hole/spacing size; see specs on the Quantifoil website (https://www.quantifoil.com/).
2. Phage Sf6 at 1 × 1014 phages/ml in phosphate-buffered saline (PBS).
3. PELCO easiGlow™--Glow Discharge Cleaning System.
4. Thermo Scientific™ Vitrobot™ Mark IV.
5. Liquid nitrogen, holders, dewars.
2.2. Cryogenic electron microscopy
1. Clipping station, Thermo Scientific™.
2. Loading station, Thermo Scientific™.
3. Cryo-transmission electron microscope (cryo-TEM) equipped with an AutoLoader system, e.g., 200 kV Glacios, 300 kV Titan Krios, Krios G4 Cryo-TEM, Thermo Scientific™.
4. K3 detector, Gatan or Falcon4 detector, Thermo Scientific™.
3. Data collection software: EPU, Thermo Scientific™ or Latitude, Gatan.
4. Data management: 8 TB (or larger) hard drive.
2.3. Single particle analysis
1. Workstation requirements—Hardware, multi-core central processing unit (CPU) and Compute Unified Device Architecture (CUDA)-supported graphics processing unit (GPU), multiple hard drives, and solid-state drives; operating system, Ubuntu 18.04, 20.04 or latest version (or other Linux operating systems compatible with CUDA).
2. SPA Software—RELION [14]; utility software—Scipion 3.0 [15], UCSF Chimera [16].
2.4. Model building and analysis software
1. Model building software—UCSF Chimera [16], UCSF ChimeraX [17], PyMol [18].
2. Model refinement software—Phenix [19], Coot [20], UCSF Chimera [16].
3. Methods
Wear personal protective equipment, e.g., safety goggles, shields, and cryogenic gloves, when handling liquid nitrogen. Gloves-free is recommended to avoid electrostatics when handling grids.
3.1. Vitrification
1. Turn on the Thermo Scientific™ Vitrobot™ Mark IV. Fill the humidifier with deionized, ultrapure water. Set temperature to 4 degrees Celsius and humidity to 100%. Install filter paper on the blotting pads inside the Vitrobot chamber. Equilibrate the Vitrobot chamber for 30 minutes to reach desired conditions. To access the Thermo Scientific™ Vitrobot™ Mark IV user manual, please visit the Thermo Scientific™ website (https://www.thermofisher.com).
2. Equilibrate the coolant container with liquid nitrogen before pouring liquid ethane into the cold cryogen cup. For detailed assembly of the coolant container, please see the Vitrobot user manual.
3. Place Quantifoil grids of choice with the carbon side face upward on a grid holder block part of the PELCO easiGlow™ discharger or a parafilm-wrapped glass slide.
4. Glow discharge the grids at plasma current 15 mA, negative charge, hold 10 seconds, process 60 seconds, working vacuum 0.39 mbar. Glow-discharged grids should be used within 30 min.
5. Use the designated Vitrobot tweezer to hold one glow-discharged grid and assemble it onto the Vitrobot. Initiate sample application using foot paddle. Apply 2.5-3 μl of sample onto the grid inside the Vitrobot chamber. For a single blot, use 7-8 seconds for blot time, 0 seconds for drain time, using a blot force of 2-10, and a blot total of 1. After blotting, transfer the vitrified grid to a grid box pre-cooled with liquid nitrogen. Please see the user manual for detailed instructions.
3.2. Cryogenic electron microscopy
1. Use the clipping station to clip vitrified grids and mount them onto the AutoLoader. For detailed instructions for clipping and mounting tools, please see the Thermo Scientific™ electron microscopy AutoLoader manual.
2. Screen cryo-grids on a Thermo Scientific™ cryogenic electron microscope, 200 kV Glacios, or 300 kV Krios. Set up a data collection with the parameters in Table 1. Please note that the exact pixel size at a nominal magnification varies with the calibration of a specific microscope.
Table 1.
Cryo-EM data collection parameters for Krios equipped with Gatan K3
| Microscope Imaging Mode | |
|---|---|
| Imaging Mode | Nanoprobe EFTEM |
| Camera Mode | Super Resolution |
| Cs | 2.7 mm |
| Camera type | Gatan K3 |
| Physical Pixel Size | 5 μm |
| Imaging Parameters | |
| Magnification | 81,000x |
| Nominal Dose | 50.0 e−/Å2 |
| Illuminated Area | 1.15 μm |
| Binning | 0.5 |
| Number of Frames | 40 |
| Defocus Range | −0.5 to −1.5 μm |
| Focus Interval | 12 μm |
| Filter Slit | 20 eV |
| Image Pixel Size | 0.561 Å |
| Spot Size | 7 |
| Dose rate | 13.13 e−/s/phys. pixel |
| Exposure Time | 4.8 sec |
| Settling Time | 2 sec |
| Defocus Step | 0.25 μm |
| Program | Latitude |
| Acquisition Method | Single Particle |
3. Adjust data collection time to collect ~50,000 phage particles.
3.3. Single particle analysis
The methods outlined below focus on the localized reconstruction of the phage Sf6 tail.
1. Software installation. Install the latest version of RELION, along with MotionCor2 [21] and CTFFIND4 [22]. Practice the RELION pipeline using RELION 3.1 tutorial (https://relion.readthedocs.io/en/release-3.1/SPA_tutorial/index.html) or RELION 4.0 tutorial (https://relion.readthedocs.io/en/release-4.0/STA_tutorial/Introduction.html). RELION tutorials provide users with detailed parameters and clear explanations.
2. Import micrograph movies and motion correction. Import movies collected on a K3 (or Falcon4) direct detector using parameters provided in Table 1. When doing motion correction, set bin to 2 to convert movies collected in super-resolution to un-binned micrographs. Make sure to apply gain reference with correct flipping according to which facility/detector the data were collected. Data collected on a Falcon4 will not have super-resolution, and the movies are post-gain referenced. Run RELION's implementation of motion correction using 1 MPI (Message Passing Interface) and as many threads available in the Central Processing Unit (CPU).
3. Contrast transfer function (CTF) estimation. Input the “corrected_micrographs.star” file from the completed “Motion correction” job and use CTFFIND-4.1 to estimate the contrast transfer function. Use 1 to 4 MPI to run this routine. For data curation, see note #1.
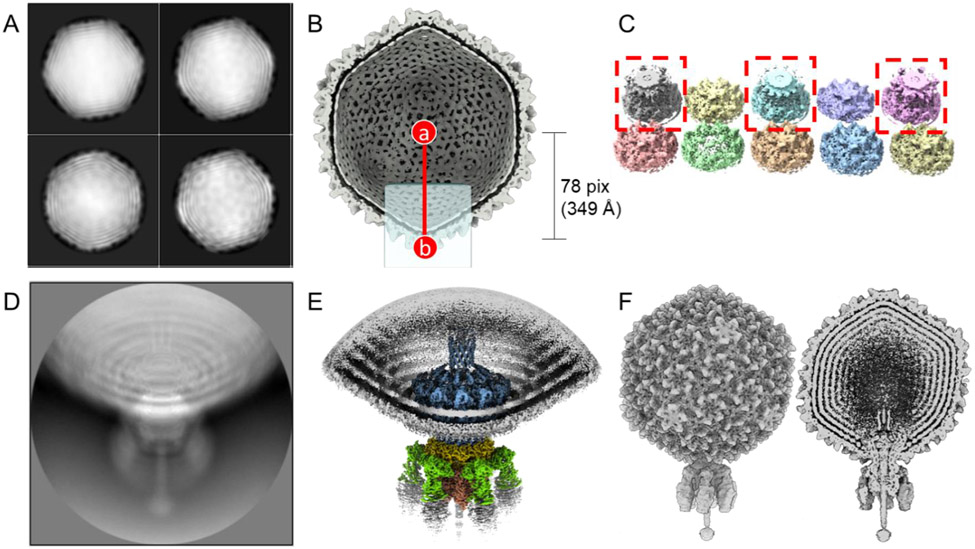
4. Particle picking and 2D classification. Use reference-free particle picking in RELION with a small set of micrographs (25 to 50 micrographs) followed by particle extraction to create a 2D reference for reference picking. For deep-learning particle picking, crYOLO [23] or TOPAZ [24] are excellent alternatives. For particle extraction of the Sf6 head that measures ~670 Å in diameter, use a box size of 180 pix (bin to 4, pixel size of 4.48 Å/pix). After referenced picking, extract binned particles from all micrographs. Carry out one round of default 2D classification (50 classes and 25 iterations) in RELION (Fig. 1A). See also note #2.
Figure 1. The pipeline of localized reconstruction.
A. Representative 2D class averages of the S6 virion calculated using particles binned 4 times. B. Section view of the S6 icosahedral reconstruction with the 5-fold icosahedral axis aligned to the Z-axis (I3/I4 convention). Position (a) is the center of the icosahedral capsid, while (b) is the center of the re-extracted particle, re-centered from the icosahedral head. The light-blue square represents the position of the cylindrical mask, which covers the potential tail location. C. Ten 3D classes were obtained by non-sampling 3D classification of icosahedral expanded particles. 3D classes containing the Sf6-aligned tail are boxed in red. D. A representative 2D class average obtained from re-extracted tail particles. E. Localized reconstruction of aligned tail particles after applying C6 symmetry. F. Asymmetric reconstruction of the entire Sf6 virion (left panel) with both tail- and capsid-aligned particles. The right panel shows a section through the virion.
5. Icosahedral reconstruction. Run "Subset selection" to manually select particles from 2D classification in step 4. Make an initial map from selected particles using the "3D initial model" routine without symmetry applied (C1). Do a "3D classification" with icosahedral symmetry parameters I3 (or I4). Use the "Subset selection" routine to select one class with the best-aligned particles, higher estimated resolution, and larger number of particles. Use the "3D auto-refine" routine to finely align particles from selected 3D classes with the same icosahedral symmetry parameters. Detailed parameters for 2D classification and 3D refinement are in Table 2. See also note #3.
Table 2.
Initial parameters for 2D classification and 3D refinement
| 2D classification | 3D initial model | |
|---|---|---|
| Input particle and box pixel size | Input extracted particles from reference picking, 180 pix (bin 4) | Input selected 2D class particles, 180 pix (bin 4) |
| Reference map and symmetry | -- | Reference-free, C1 symmetry |
| Do CTF-correction | Yes | Yes |
| Number of classes | 50 | 1 |
| Regularization Parameter T | 2 | -- |
| Number of iterations | 25 | 25 (Initial); 100 (In-between); 25 (Final) |
| Mask diameter (Å) | 700 | -- |
| Sampling | Yes | -- |
| Running | 3 MPI 4-16 threads | 3 MPI 4-16 threads |
| Initial angular sampling (degrees) | -- | 15 |
| 3D classification | 3D auto-refine | |
| Input particle and box pixel size | Input particles from 3D initial model, 180 pix (bin 4) | Input selected 3D class particles, 180 pix (bin 4) |
| Reference map and symmetry | Input 3D initial model, low-pass filter to 60 Å and apply I3 or I4 symmetry | Input map from selected 3D class, low-pass filter to 60 Å and apply the same symmetry (I3 or I4) |
| Do CTF-correction | Yes | Yes |
| Number of classes | 4 | -- |
| Regularization Parameter T | 4 | -- |
| Number of iterations | 25 | -- |
| Mask diameter (Å) | 800 | 800 |
| Sampling | Yes | -- |
| Running | 3 MPI 4-16 threads | 3 MPI 4-16 threads |
| Initial angular sampling (degrees) | 7.5 | 7.5 (Initial), 1.8 (local search) |
6. Localized reconstruction: perform all steps described below and illustrated in Fig. 1:
(a) Particle expansion. Open a terminal window and type the command line "relion_particle_symmetry_expand --i run_data.star --o exp_data.star --sym I3" to expand the symmetry of aligned particles from the "3D auto-refine" job in step 5. Change I3 to I4 if I4 was used in 3D auto-refine. Refer to the RELION manual for command line details.
(b) Mask creation and resampling. Create a cylindrical mask of radius = 125 pix and height = 500 pix using SPIDER [25] or Scipion 3.0 [15]. Open UCSF Chimera and load the cylindrical mask and the map reconstructed in the "3D auto-refine" routine in step 5. In Chimera, manually move the mask to the icosahedral five-fold facing toward you along the z-axis, and using the command line in Chimera, type "vop #0 resample onGrid #1", where #0 is the mask volume, and #1 is the capsid volume (Fig. 1B). Save the resampled mask as a new .mrc file to be used in the next step. For more details about mask making, see note #4.
(c) Masked non-sampling 3D classification. Run a "3D classification" routine with the following inputs: expanded I3/I4 particles.star file, and reference map from "3D auto-refine" job. Input the resampled cylindrical mask. Low-pass filter the density to 60 Å and set the symmetry to C1, the number of classes to 10, regularization T = 25, and iteration to 25. Set sampling to "No" image alignment; this is a crucial setting. Set MPI = 1 and 12-36 threads up to your CPU capacity. When the particle expansion search is completed, examine each class with UCSF Chimera and use the "Subset selection" routine to select the classes with tail densities (Fig. 1C). Also, use the "Subset selection" routine to remove duplicated particles.
(d) Particle re-extraction. Re-extract particles using the "Particle extraction" routine with the following settings: Input "micrograph STAR file" from CTF-estimated "micrographs.star", "re-extract refined particles" as "Yes", "refined particles STAR file" with duplicates removed "particles.star", "re-center refined coordinates" with "Yes", "re-center on – X, Y, Z" set to 0, 0, 78. Set box size of 512 pix without rescaling particles (unbinned). The above settings re-extract and re-center the particle along the Z-axis outward 78 pixels (bin 4) to the center of the portal-tail complex, ~349 Å away from the center of the phage head (Fig. 1B-(b)). See note #5 for details about particle extraction.
(e) 3D auto-refinement. After re-extraction, run a "2D classification" routine without sampling to verify the sub-particles (Fig. 1D). Use the command line "relion_reconstruct --i Extract/jobxxx/particles.star --sym C1" to create a reconstructed map directly from the "Particle extraction" job. Run "3D auto-refine" routine using the re-extracted particles and reconstructed map and apply C5 symmetry to finely align the capsid (Table 3). Perform symmetry expansion to C5, as detailed above. Run a non-sampling "3D classification" routine search for C6 (follow non-sampling 3D classification in Table 3). Now the particles are C5 (capsid)-C6 (tail) aligned. See note #6 for identification of map handedness.
Table 3.
Localized reconstruction parameters
| 3D classification (non-sampling) | 3D auto-refine (small angular) | |
|---|---|---|
| Input particle and box pixel size | Input expanded particles, 180 (bin 4) or 512 pix (bin 1) | Input re-extracted or aligned sub-particles, 512 pix (bin 1) |
| Mask | Resampled cylindrical mask | *** |
| Reference map and symmetry | Set C1 sym to I3/I4 expanded particles and map. Set C6 sym to C5 expanded particles and map. | Input the reconstructed map and set desired symmetry (C1, C3, C5, C6, C12, etc.) |
| Do CTF-correction | Yes | Yes |
| Number of classes | 10 | -- |
| Regularization Parameter T | 25 | -- |
| Number of iterations | 25 | -- |
| Mask diameter (Å) | −1 (do not apply a soft circular mask) | −1 (do not apply a soft circular mask) |
| Sampling | No | -- |
| Running | 1 MPI 12-36 threads | 3 MPI 4-16 threads |
| Initial angular sampling (degrees) | -- | 0.9 (Initial), 0.9 (local search) |
For the first small angular 3D auto-refine, the user can execute the routine without a mask. Next, use the “Mask Create” routine to create a mask from the reconstructed map and apply the tight mask in the second small angular 3D auto-refine.
(f) Applying local symmetry. Run "3D auto-refine" with small initial and local auto-sampling to both 0.9 degrees and apply C1, C6, or C12 to generate maps with the expected symmetry features (Fig. 1E, F).
(g) Map refinement and polishing. The aligned particles can be further refined to higher resolution using "Mask creation", "Post-processing", "CTF refinement", and "Bayesian polishing" routines. Follow the default procedures described in the RELION tutorial to set up these routines. As the particles are aligned locally, small initial and local angular sampling should always be used for 3D auto-refine (Table 3).
3.4. Model building
1. Map sharpening. Use the phenix.auto_sharpen [26] routine to automatically identify optimal map-sharpening/blurring parameters without reference to a model. The routine will write out an optimized version of a cryo-EM map in ccp4 format. See also note #7.
2. Prediction of 3D models. Use AlphaFold [27] for monomeric proteins and AlphaFold2 [28] to predict the structure of oligomeric models. If the oligomerization stoichiometry is unknown, predict different oligomers with symmetry close to the capsid 5-fold or tail 6-fold rotational symmetry.
3. Placing initial models into the sharpened density. Open sharpened maps in Chimera [16] (or ChimeraX [17]), read the initial model obtained from homologous proteins or predicted by AlphaFold, and manually drag the model into the density. Use Chimera's 'Fit in Map' command to optimize the fit of atomic coordinates to the density map locally. Write out the repositioned model.
4. De novo model building. Open the sharpened cryo-EM density map and the initial model in Coot. To identify the amino acid register, focus on major secondary structure elements, typically α-helices. Use 'Real Space Refine Zone' to fit atoms (either built de novo or from a prediction/homology model) to the density. Click "Real Space Refine Zone" in the vertical modeling toolbar and select the atoms to specify a residue range. Because real space refinement ignores geometry, use appropriate restraints depending on the region to refine (e.g., helical restraints for residues in a helix, etc.). Leave residues that have poor side chain density as alanine. Write out the initial model.
5. Positional refinement. Use phenix.real_space_refinement [29] to refine the initial model against the cryo-EM density. It is best to use the original (unsharpened) density map generated by RELION. Also include an initial round of rigid body and group ADP (B-factor) refinement that restrains one isotropic B-factor per residue. See also note #8.
4. Notes
1. Contrast transfer function (CTF) estimation. It is possible, and often convenient, to curate micrographs according to the metadata generated from CTF estimation using the “Subset selection” routine, in the “Subsets” tab, under “Select based on metadata values”. Set “Yes” and “Metadata label for subset selection” to “rlnCtfMaxResolution”. For instance, the user can set “Maximum metadata value” to “6” to cut off micrographs with an estimated resolution worse than 6 Å.
2. Particle picking and sorting. For particle extraction of the Sf6 icosahedral head (~670 Å in diameter), using unbinned particles will slow initial particle picking and 2D/3D classification. Four times binning (or box size smaller than 256 pixels) will facilitate computing from particle picking to localized reconstruction. Limit the box size to 512 pixels (or a maximum of 640 pixels) to avoid slowing down computation. Suitable box sizes can be found on the EMAN2 website (https://blake.bcm.edu/emanwiki/EMAN2/BoxSize).
3. Icosahedral reconstruction. RELION employs five types of icosahedral symmetries, named I1, I2, I3, I4, and I5; they differ primarily in how the icosahedron is oriented on each cartesian plane axes, with I3 symmetry being the original convention used in SPIDER [25], whereas I4 is RELION's icosahedral symmetry implementation. Both I3 and I4 place the 5-fold icosahedral axis on the cartesian Z-axis. It is recommended to run both I3 and I4 symmetries and compare resolutions and map features.
4. Choice of Mask. A suitable mask would result in a map with enhanced features and better resolution. The user can create a mask based on the entire 3D models via RELION-Mask creation. Adjust the parameters of “Initial binarisation threshold”, “Extend binary map this pixels”, “add a soft-edge of this many pixels” to obtain the best mask. To reconstruct localized areas with unknown structures, it is often helpful to use a generic cylindrical mask, attainable using Scipion-create 3D mask-geometry mask. Generally, a mask with radius = 125 px and height = 500 px will suffice in finding the unique 5-fold vertex containing the tail, at least for short-tailed Podoviridae like Sf6 or P22.
5. Particle extraction. This step is crucial in determining how much of a non-icosahedral object (e.g., a phage tail) one wants to reconstruct in the final map. When re-extracting the particles, the position where one moves the Z-axis determines the center of the subsequent reconstruction, and the box size determines the extent of the tail that will be observed. For longer tails, the user must find a balance between box size and the new center location. In designing a mask for localized reconstruction in Scipion 3, the user should have a way to estimate the most accurate mask size, as the mask should not cut out any part of the tail density. Also, to rescale (adjust pixel size) or re-box a cylindrical mask, use the resample feature in Chimera with the icosahedral map, as this will resample the mask with the same pixel size and box size as the icosahedral map. With the command line, use vop resample #0 OnGrid #1. Do not use RELION's relion_image_handler tools (e.g., new_box, _rescale_angpix) on masks, as this leads to off-centering of the resulting mask, which is problematic for “3D refinement” later.
6. Inspection of map handedness. Even though the same particle pool is used to generate the density map, the stochastic gradient descent (SGD) algorithm in the “3D initial model” routine has a 50% chance of being in the different handedness (see details in the RELION tutorial). Therefore, it is vital to check the handedness of a cryo-EM map by visual inspection. This can be done by inspecting the density for an α-helix, which should be right-handed; otherwise, invert the handedness by typing the command: relion_image_handler --i map.mrc --o map_invert.mrc --invert_hand.
7. Density Sharpening. To resolve areas of weak density possibly due to high mobility at that region, it is helpful to first extract the weak density from the map by docking a protein model into the density in Chimera and then running a phenix.map_box job in PHENIX. The cut-out density can then be sharpened by running phenix.auto_sharpen. If an initial model is available (from AlphaFold or a homologous protein), it is also helpful to rerun phenix.auto_sharpen using an initial model that covers the electron density.
8. Real space refinement. For densities better than 3.5 Å resolution, disable 'NCS constraints' in phenix.real_space_refinement, which is enabled by default, and instead select “NCS restraints”. For monomeric atomic models, check the “rigid body” option to avoid the models moving out of their density.
Acknowledgment
This work was supported by the National Institutes of Health grants R01 GM100888, R35 GM140733, and S10 OD030457 to G.C. This research was partly supported by the National Cancer Institute's National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E.
References
- 1.McMullan G, Faruqi AR, Henderson R (2016) Direct Electron Detectors. Methods Enzymol 579:1–17. doi: 10.1016/bs.mie.2016.05.056 [DOI] [PubMed] [Google Scholar]
- 2.Parent KN, Schrad JR, Cingolani G (2018) Breaking Symmetry in Viral Icosahedral Capsids as Seen through the Lenses of X-ray Crystallography and Cryo-Electron Microscopy. Viruses 10 (2). doi:v10020067 [pii] 10.3390/v10020067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrishami V, Ilca SL, Gomez-Blanco J, Rissanen I, de la Rosa-Trevin JM, Reddy VS, Carazo JM, Huiskonen JT (2021) Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog Biophys Mol Biol 160:43–52. doi: 10.1016/j.pbiomolbio.2020.05.004 [DOI] [PubMed] [Google Scholar]
- 4.Ilca SL, Kotecha A, Sun X, Poranen MM, Stuart DI, Huiskonen JT (2015) Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat Commun 6:8843. doi: 10.1038/ncomms9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo F, Liu Z, Vago F, Ren Y, Wu W, Wright ET, Serwer P, Jiang W (2013) Visualization of uncorrelated, tandem symmetry mismatches in the internal genome packaging apparatus of bacteriophage T7. Proc Natl Acad Sci U S A 110 (17):6811–6816. doi: 10.1073/pnas.1215563110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morais mC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG (2001) Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J Struct Biol 135 (1):38–46. doi: 10.1006/jsbi.2001.4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheres SH (2016) Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol 579:125–157. doi: 10.1016/bs.mie.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 8.Grigorieff N (2016) Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Methods Enzymol 579:191–226. doi: 10.1016/bs.mie.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakane T, Kimanius D, Lindahl E, Scheres SH (2018) Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife 7. doi: 10.7554/eLife.36861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Hou CD, Yang R, Whitehead R 3rd, Teschke CM, Cingolani G (2022) High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine. Sci Adv 8 (49):eadc9641. doi: 10.1126/sciadv.adc9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhardwaj A, Molineux IJ, Casjens SR, Cingolani G (2011) Atomic structure of bacteriophage Sf6 tail needle knob. J Biol Chem 286 (35):30867–30877. doi:M111.260877 [pii] 10.1074/jbc.M111.260877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olia AS, Casjens S, Cingolani G (2007) Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol 14 (12):1221–1226 [DOI] [PubMed] [Google Scholar]
- 13.Kimanius D, Forsberg BO, Scheres SH, Lindahl E (2016) Accelerated cryo-EM structure determination with parallelisation using GPUs in RELiOn-2. Elife 5. doi: 10.7554/eLife.18722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zivanov J, Oton J, Ke Z, von Kugelgen A, Pyle E, Qu K, Morado D, Castano-Diez D, Zanetti G, Bharat TAM, Briggs JAG, Scheres SHW (2022) A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Elife 11. doi: 10.7554/eLife.83724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez-Moreno A, Del Cano L, Martinez M, Ramirez-Aportela E, Cuervo A, Melero R, Sanchez-Garcia R, Strelak D, Fernandez-Gimenez E, de Isidro-Gomez FP, Herreros D, Conesa P, Fonseca Y, Maluenda D, Jimenez de la Morena J, Macias JR, Losana P, Marabini R, Carazo JM, Sorzano COS (2021) Cryo-EM and Single-Particle Analysis with Scipion. J Vis Exp (171). doi: 10.3791/62261 [DOI] [PubMed] [Google Scholar]
- 16.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25 (13):1605–1612 [DOI] [PubMed] [Google Scholar]
- 17.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30 (1):70–82. doi: 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janson G, Paiardini A (2021) PyMod 3: a complete suite for structural bioinformatics in PyMOL. Bioinformatics 37 (10):1471–1472. doi: 10.1093/bioinformatics/btaa849 [DOI] [PubMed] [Google Scholar]
- 19.Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75 (Pt 10):861–877. doi: 10.1107/s2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallographica Section D 66 (4):486–501. doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14 (4):331–332. doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohou A, Grigorieff N (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192 (2):216–221. doi: 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill P, Gatsogiannis C, Raunser S (2019) SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2:218. doi: 10.1038/s42003-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bepler T, Morin A, Rapp M, Brasch J, Shapiro L, Noble AJ, Berger B (2019) Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 16 (11):1153–1160. doi: 10.1038/s41592-019-0575-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116 (1):190–199 [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC, Sobolev OV, Afonine PV, Adams PD (2018) Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D Struct Biol 74 (Pt 6):545–559. doi: 10.1107/S2059798318004655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skolnick J, Gao M, Zhou H, Singh S (2021) AlphaFold 2: Why It Works and Its Implications for Understanding the Relationships of Protein Sequence, Structure, and Function. J Chem Inf Model 61 (10):4827–4831. doi: 10.1021/acs.jcim.1c01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD (2018) Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74 (Pt 6):531–544. doi:S2059798318006551 [pii] 10.1107/S2059798318006551 [DOI] [PMC free article] [PubMed] [Google Scholar]