Abstract
Although we have shown that steroid receptor coactivator-2 (SRC-2), a member of the p160/SRC family of transcriptional coregulators, is essential for decidualization of both human and murine endometrial stromal cells, SRC-2’s role in the earlier stages of the implantation process have not been adequately addressed. Using a conditional SRC-2 knockout mouse (SRC-2 d/d) in timed natural pregnancy studies, we show that endometrial SRC-2 is required for embryo attachment and adherence to the luminal epithelium. Implantation failure is associated with the persistent expression of Mucin 1 and E-cadherin on the apical surface and basolateral adherens junctions of the SRC-2 d/d luminal epithelium respectively. These findings indicate that the SRC-2 d/d luminal epithelium fails to exhibit a plasma membrane transformation (PMT) state known to be required for the development of uterine receptivity. Transcriptomics demonstrated that the expression of genes involved in steroid hormone control of uterine receptivity were significantly disrupted in the SRC-2 d/d endometrium as well as genes that control epithelial tight junctional biology and the emergence of the epithelial mesenchymal transition state, with the latter sharing similar biological properties with PMT. Collectively, these findings uncover a new role for endometrial SRC-2 in the induction of the luminal epithelial PMT state, which is a prerequisite for the development of uterine receptivity and early pregnancy establishment.
Keywords: Steroid receptor coactivator-2, mouse, endometrium, implantation, epithelium, transcriptome
1 |. INTRODUCTION
Steroid receptor coactivator-2 (SRC-2), also known as NCOA2 (or TIF2 in the human; GRIP1 in the mouse), is a member of the evolutionary conserved p160/SRC family of transcriptional coactivators, which includes SRC-1 and SRC-3 1, 2. Similar to other family members, SRC-2’s complex functional domain structure underpins its pleiotropic coregulator roles in both nuclear receptor (NR) and non-NR mediated transcriptional responses 2, 3. Such transcriptional responses are critical for a broad spectrum of physiological processes, from energy metabolism 4, 5, circadian rhythm homeostasis 6, fetal pulmonary development 7, 8 to reproduction 3. Concerning the latter, early studies showed that whole body SRC-2 knockout male mice display a severe subfertility phenotype, encompassing defects in spermiogenesis (teratozoospermia), incomplete adhesion of Sertoli cells to germ cells, and age-dependent testicular degeneration 9. These studies also demonstrated that SRC-2 knockout female mice exhibit a severe hypofertility phenotype due to placental hypoplasia arising from developmental impairments in maternal decidual cells that are juxtaposed to the placenta 9. These findings provided early support for conclusions drawn from observational studies on human endometrial tissue 10, 11 and the rat uterus 12 that endometrial SRC-2 is critical for ensuring both uterine function and health. In the case of the human endometrium, SRC-2 levels (along with SRC-3) do not markedly change with menstrual cycle phase 10; however, both SRCs exhibit significant increases in expression levels in epithelial and stromal cells of secretory-phase endometrial tissue when biopsied from patients diagnosed with polycystic ovarian syndrome (PCOS) 10. Apart from an increased susceptibility for endometrial cancer 10, 13, PCOS patients are predisposed to additional reproductive sequelae, which include low cycle fecundity and a high miscarriage rate that can reach 60–70% 14, 15. Collectively, these clinical studies indicate that perturbation in the homeostatic levels of SRC-2 may be causal for endometrial pathologies that lead to subfertility or infertility.
Using advanced conditional knockout technology, we revealed that postnatal abrogation of SRC-2 in a subset of cells expressing the progesterone receptor (PGR) results in infertility (rather than subfertility) in which the endometrium fails to enable embryo implantation, a necessary early step that leads to the establishment of the fetomaternal interface 16, 17. From human to mouse, the embryo implantation process advances through distinct interdependent stages: starting with embryo attachment to the luminal epithelium of the receptive endometrium, embryo invasion into the underlying endometrial stroma, and decidualization of the surrounding stromal fibroblasts into specialized epithelioid decidual cells 18. Encircling the conceptus, decidual cells control the development of the embryo and protect against cytotoxic stressors. While there is compelling support for the importance of SRC-2 in decidualization of human endometrial stromal cells and stromal cells of the murine endometrium 16, 19, 20, we have only a rudimentary understanding of endometrial SRC-2’s role during the earlier stages of the implantation process 16, particularly its influence on the endometrial epithelium. This knowledge insufficiency is significant since early implantation failure is not only causal for early pregnancy loss but undercuts the full potential of assisted reproductive technologies (ARTs) that rely on the transfer of healthy embryos into a receptive endometrium 21–24.
Using the SRC-2 conditional knockout mouse, the timed pregnancy model, and transcriptomic analysis, we demonstrate that endometrial SRC-2 is essential for the development of the receptive endometrium, and, in particular, the reprogramming of the luminal epithelium to enable attachment of the embryo and its subsequent invasion into the underlying stroma.
2 |. MATERIALS AND METHODS
2.1 |. Mouse models
The SRC-2 conditional knockout mouse (SRC-2 d/d) was previously described 16, 25. Briefly, the SRC-2 d/d bigenic was generated in a C57BL6J background by crossing our progesterone receptor cre knockin (Pgr cre/+) mouse 26 with a mouse carrying the SRC-2 floxed [L2 version] allele (SRC-2 f/f) 9. In an AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) accredited vivarium facility at Baylor College of Medicine, mice were housed in temperature-controlled rooms (22 ± 2oC) with a programmed 12-hour light/dark daily photocycle. Mice were fed an irradiated Formulab Diet (LabDiet/Lab Supply, Fort Worth, TX (#5008)) with access to fresh water ad libitum. Mice were treated humanely and surgical procedures were performed in accordance with the guidelines described in the Guide for the Care and Use of Laboratory Animals (“The Guide”, 8th edition, 2011), published by the National Research Council of the National Academies, Washington D.C. (www.nap.edu). All animal procedures were prospectively approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine.
2.2 |. Fertility studies
For timed pregnancy experiments, SRC-2 f/f and SRC-2 d/d females (8–10 weeks old) were housed overnight with fertility proven stud males. The following morning, coitus was visually confirmed in the female by the presence of a postcoital vaginal plug; the morning of detecting the vaginal plug was designated as the morning of gestation day 1 (GD1). Pregnant mice were individually housed before euthanasia on GD 4, 5, and 5.5 for the studies described below. Using uterine horns dissected from SRC-2 f/f and SRC-2 d/d females at GD4, embryo retrieval was performed by gently flushing the uterine lumen with 100 μl of sterile phosphate buffered saline (PBS). Retrieved embryos were digitally imaged within a petri dish using a color chilled AxioCam MRc5 digital camera with a Carl Zeiss AxioImager A1 upright microscope (Zeiss Inc., Jena, Germany). To visualize incipient implantation sites along both uterine horns of mice at GD5, Chicago sky blue dye (1% in PBS; 100μl per mouse) was injected into one lateral tail vein before mice were euthanized 2–5 minutes post-injection and their reproductive tract tissue dissected for macro-level digital imaging 27. The latest versions of the Photoshop and Illustrator programs in the Adobe Creative Suite software package (Adobe Systems Inc. San Jose CA) were used for general raw image processing, generation of image composites, and the insertion of annotations in the final figures. For serum isolation, whole blood was collected in microtainer tubes containing a serum separator microguard (Becton, Dickinson and Company, Franklin Lakes, NJ (#365967)) from mice at GD5 as previously described 27. Microtainer tubes containing drawn blood were kept at room temperature for 30 minutes to ensure complete blood coagulation before serum was separated by centrifugation at 2000 rpm for 10 minutes at 4oC; serum was stored at −80 oC until analysis. Serum 17β-estradiol and progesterone levels were measured using the ENZO ELISA kit (#ADI-900–174 and #ADI-900–011 respectively; R&D Systems Inc., Minneapolis, MN).
2.3 |. General histology and immunohistochemical analyses
Tissues were fixed in 4% paraformaldehyde, progressively dehydrated using increased concentrations of ethanol, then xylene cleared before fixed tissues were paraffin embedded. Paraffin blocks were sectioned at 5μm thickness and resultant sections stained with hematoxylin and eosin (H&E) 27. Immunohistochemical detection of SRC-2; mucin 1 (MUC 1); and Snail family zinc finger 1 (SNAI1) was conducted using the following primary rabbit polyclonal antibodies: NCOA2/SRC2 (Bethyl Laboratories, Waltham, MA #A300–345A, 1:500 dilution); MUC 1 (Abcam Inc., Waltham, MA #Ab15481, 1:100 dilution); and SNAI1 (ThermoFisher Scientific Inc., Carlsbad, CA #PA5–115940, 1:100 dilution). Following incubation with their respective primary antibody, tissue sections were incubated with a goat anti-rabbit secondary antibody conjugated with horseradish peroxidase (HRP; Vector Laboratories Inc., Burlingame, CA #P-1000, 1:200 dilution). Peroxidase activity was detected with the Vectastain Elite ABC-HRP kit (Vector Laboratories Inc. #PK-6100). Proliferating cells in S-phase of the cycle were visualized in tissue sections by immunohistochemical detection of 5-bromo-2’-deoxyuridine (BrdU) incorporation using the BrdU in situ detection kit (BD Biosciences, San Jose CA #551321). Following immunostaining, tissue sections were lightly counterstained with hematoxylin before applying Permount solution for placement of coverslips. To detect E-cadherin by immunofluorescence staining, the Alexa Fluor 488 conjugated donkey anti-rabbit secondary antibody (Invitrogen Inc., Carlsbad, CA #A-21206, 1:400 dilution) was used following incubation with a rabbit primary monoclonal antibody against mouse E-cadherin (Cell Signaling Technology Inc., Danvers, MA #3195, 1:200 dilution). In the case of claudin 7 (CLDN 7) immunofluorescence staining, the Alexa Fluor 594 donkey anti-rabbit secondary antibody (Invitrogen Inc., #A-21207, 1:400 dilution) was used following incubation with a rabbit polyclonal primary antibody against human CLDN 7 (Proteintech group Inc., Rosemont, IL, #10118–1-AP, 1:200 dilution). After incubation with the primary and secondary antibodies, slides were mounted with Vectashield Antifade mounting medium containing 4, 6-diamidino-2-phenylindole (DAPI; Vector Laboratories Inc. #H-1200–10). For certain immunohistochemical studies, the staining intensity of tissues was scored (H-score 28, 29) on a scale of 0= no staining, 1=faint staining, 2= moderate staining, and 3-strong staining. Tissue sections (n=5) per genotype group (n=3) were analyzed three times by two independent investigators, blinded to the identities of the tissue sections. The average values from the biological replicates are presented as histograms.
2.4 |. Immunoblotting
Protein concentration was determined using the Bradford reagent (ThermoFisher Scientific Inc. #23225) before protein lysate preparations were resolved by a 10% polyacrylamide-SDS gel and transferred to a hydrophobic polyvinylidene difluoride (PVDF) membrane. After blocking PVDF membranes with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST), membranes were incubated with specific primary rabbit polyclonal antibodies, depending on the experiment. The following primary antibodies were used in these studies against the estrogen receptor (ESR1; Abcam Inc., Waltham MA #ab75635, 1:1000 dilution), the progesterone receptor (PGR; ThermoFisher Scientific Inc. #MA5–14505, 1:1000 dilution) or a mouse monoclonal primary antibody against β-actin (Novus Biologicals Inc., Centennial, CO #NB100–5874, 1:10,000 dilution) overnight at 4oC. The following morning, immunoblots were washed before incubation with an anti-rabbit HRP-conjugated secondary antibody (1:5000) or an anti-mouse HRP-conjugated secondary antibody for 1 hour at room temperature. Chemiluminescence signal detection was achieved using the SuperSignal West Pico PLUS Chemiluminescent substrate (ThermoFisher Scientific Inc. #34580). Immunoreactive protein bands were imaged using the Azure 600 Imaging Systems (Azure Biosystems, Sierra Court, Dublin CA).
2.5 |. Quantitative real-time PCR
Total RNA was prepared from tissues using the RNeasy Plus Mini kit (Qiagen Inc., Germantown, MD #74134). The NanoDrop 2000 UV/Visual spectrophotometer (ThermoFisher Scientific Inc.) was used for quantification of RNA prior to reverse transcription using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific Inc. #4368814) 30. Amplified cDNA was diluted to 10ng/μl before quantitative reverse transcription-PCR (qRT-PCR) was performed with the Fast TaqMan 2X Mastermix (Applied Biosystems/Life Technologies, Grand Island, NY #4352042); the TaqMan assays used in this study are listed in Table S1. All qRT-PCR experiments were performed using the 7500 Fast Real-Time PCR system (Applied Biosystems/ Life Technologies); delta-delta cycle threshold was used to normalize expression to the 18S reference.
2.6 |. Genome-wide RNA expression profiling
Genome scale RNA-sequencing (RNA-seq) and analysis were performed as previously described 31, 32. Briefly, total RNA purity and physical integrity were determined using the NanoDrop spectrophotometer (ThermoFisher Scientific Inc.) and the 2100 Bioanalyzer with RNA chips (Agilent Technologies Inc., Santa Clara CA) respectively. Total RNA preparations that scored a RNA integrity number (RIN) of 7 or greater were used in these experiments; RNA samples from three mice were used for each genotype. From 250 ng of RNA, sequencing libraries were prepared using the TruSeq Stranded mRNA kit (Illumina Inc., San Diego CA #20020594) and then PCR amplified. Quality analysis of resultant libraries was performed using the 4200 TapeStation with D1000 ScreenTape assays (Illumina Inc.). Adapter-ligated fragment concentration was estimated by the qRT-PCR assay with a KAPA Library Quantification kit (KAPA Biosystems, Wilmington MA #KR0405). After equimolar pooling, libraries were quantified with the 2100 Bioanalyzer (using the High Sensitivity DNA Kit with DNA chips) and the KAPA Library Quantification Kit. Sequencing of libraries was achieved using the Illumina NovaSeq 6000 sequencer. Raw paired-end 100 base pair (bp) sequencing reads in Illumina fastq file format were generated at mid-output and mapped to the mouse genome (Genome Reference Consortium Mouse Build 39; National Center for Biotechnology Information (NCBI)) through use of hierarchical indexing for spliced alignment of transcripts (HISAT2) software 33. The number of reads aligned to known genes was determined by the Python-based software package HTSeq 34 (http://www-huber.embl.de/users/anders/HTSeq). To reduce possible PCR bias, read duplicates were removed with Picard Tools (http://broad.institute.github.10/picard).
The Bioconductor package DESeq was applied to the gene expression dataset to detect differentially expressed genes (DEGs) between the two groups 35. The false discovery rate (FDR) for DEGs was estimated using the Benjamini and Hochberg method 36. Gene expression comparisons with an FDR ≤ 0.05 and an absolute fold change (IFCI) ≥ 1.5 or 2 (depending on the analysis) were considered significantly differentially expressed between the two groups. Genes with significantly altered expression were used to identify affected pathways 37. Fragments per kilobase of transcript per million (FPKM) values of transcripts were used for hierarchical clustering; the pheatmap package in R was used to generate the heatmap. Using raw gene count data, principal component analysis (PCA) was performed with the R function prcomp package (https://cran.r-project.org); raw data files were deposited in the Gene Expression Omnibus (GEO) repository at the NCBI (GSE237740; www.ncbi.nilm.go/geo). Gene ontology (GO) enrichment analysis was performed using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) functional annotation clustering tool (https://david.abcc.ncifcrff.gov) 38. Specific genes involved in major biological and signal transduction pathways were also determined using the Kyoto Encyclopedia of Genes and Genomes software (KEGG; http://www.genome.jp/kegg/). The Ingenuity Pathway Analysis (IPA) program (www.ingenuity.com) identified both upstream regulators and canonical pathways associated with genes of interest in the DEG dataset.
2.7 |. RNA-seq data correlations
Recently used by our group 39, the Illumina correlation engine (http://www.illumina.com/products/by-type/informatics-products/basespace-correlationengine.html (formerly NextBio)) is a web-based RNA sequencing and microarray database that has curated over 23,000 scientific studies to enable correlation analysis. The differentially expressed gene (DEG) dataset from the SRC-2 d/d vs SRC-2 f/f (GD5) RNA-seq study described here was uploaded to the correlation engine and compared with biosets in the database using the Running Fisher test 40. The biosets of interest represent curated DEG datasets from a number of published mouse models exhibiting suppressed progesterone responses and/or estrogen hypersensitivity within the murine endometrium 41–46. The Illumina correlation engine provides an assessment of the statistical significance of the correlation of the overlapping genes between a given bioset (Bioset 2) and the DEG dataset described here (Bioset 1), with a summary p-value as a major readout (a large –log (p-value) indicates a high degree of similarity).
2.8 |. Statistical analyses
Two-tailed unpaired Student t-tests were used to estimate the statistical significance of differences between the two groups, and one-way ANOVA between more than two groups. Unless otherwise stated, data were graphically presented as mean ± standard deviation (S.D.). Differences with p-values < 0.05 were considered statistically significant; asterisks represent the level of significance: * p<0.05; **p<0.001, ***p<0.001; and ****p<0.0001. Prism software version 9 (GraphPad Software Inc., San Diego CA) was used for the majority of the reported statistical analyses.
3 |. RESULTS
3.1 |. Development of the receptive endometrium requires SRC-2 for early pregnancy establishment
Our previous studies demonstrated that the SRC-2 d/d female mouse is infertile 16, 25, and that impairment in endometrial stromal decidualization (artificial decidual response) is at least one factor contributing to the SRC-2 d/d infertility phenotype 19, 20, 25. Because SRC-2 is expressed in both the epithelial and stromal compartments of the murine endometrium (Figure S1) and because the epithelial compartment along with the stroma plays a crucial role in the development of the receptive endometrium 47, we examined whether endometrial SRC-2 is required for the earlier stages of the implantation process using the timed natural pregnancy model. Figure 1A shows that the SRC-2 d/d female produces an equivalent number of pre-implantation blastocysts at GD4; however, embryo implantation fails to occur in the SRC-2 d/d female (Figure 1B, C). At GD5, we confirmed that normal ovarian function and associated 17β-estradiol and progesterone serum levels are detected in the SRC-2 d/d female (Figure S2A, B), confirming that the infertility phenotype is intrinsic to the SRC-2 d/d endometrium. We also found that the expression levels of the nuclear receptor for each steroid hormone are unchanged in the SRC-2 d/d uterus (Figure S2C).
FIGURE 1.
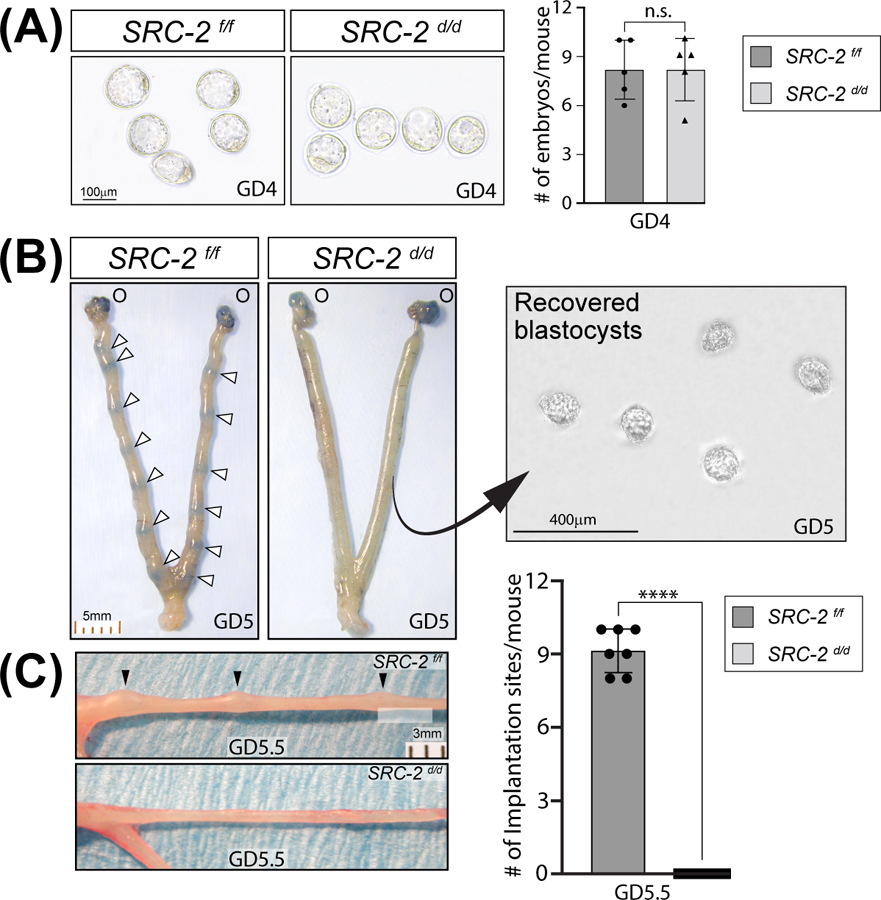
Embryo implantation is blocked in the SRC-2 d/d mouse. (A) An equivalent number of preimplantation blastocysts are produced by the SRC-2 f/f and SRC-2 d/d at GD4 (n = 5 mice per genotype); n. s. denotes not significant. (B) Tail vein injection of Chicago sky blue dye reveals the expected number of implantation sites in the SRC-2 f/f at GD5 (denoted by white arrowheads (O indicates the location of the ovaries)); the scale bar applies to both panels. The similarly treated SRC-2 d/d female does not show blue banding along the uterine horn as seen in the SRC-2 f/f control. The right panel shows recovered blastocysts from flushing one uterine horn of the SRC-2 d/d mouse at GD5; the result is representative of five mice. (C) By GD5.5, implantation sites can be visually detected (without the need for staining) along one of the two uterine horns of the SRC-2 f/f mouse at GD5.5 (top panel (black arrowheads)) whereas implantation sites are not present in the uterine horn of the SRC-2 d/d mouse (bottom panel); scale bar in the top panel applies to both panels. The histogram on the right graphically displays the number of implantation sites for each genotype at GD5.5 (n = 7 mice per genotype).
Immunohistochemical detection of BrdU incorporation revealed that while the SRC-2 f/f endometrium at GD4 display the typical cell type specific proliferative pattern for a receptive endometrium (i.e. an absence of luminal epithelial proliferation with a coincident increase in the number of proliferating cells within the subluminal stroma 47 (Figure 2A)), the SRC-2 d/d endometrium failed to generate this endometrial receptive response within the context of a natural pregnancy state (Figure 2B, D, and E). Cell counts of BrdU positive luminal epithelial cells revealed a nearly 10-fold increase in the average number of BrdU positive cells in the SRC-2 d/d luminal epithelium when compared with the SRC-2 f/f luminal epithelial compartment at GD4 (Figure 2E). Furthermore, while the SRC-2 f/f endometrium at GD5 exhibits extensive proliferation of stromal cells prior to their decidualization 24 hours later, the SRC-2 d/d endometrium displays a significantly lower number of proliferating stromal cells (Figure S3A-G). The results from these timed natural pregnancy experiments correlate with our previous findings using ovariectomized mice that were administered an established 17β-estradiol plus progesterone hormone treatment regimen to artificially elicit uterine receptivity 19, 48.
FIGURE 2.
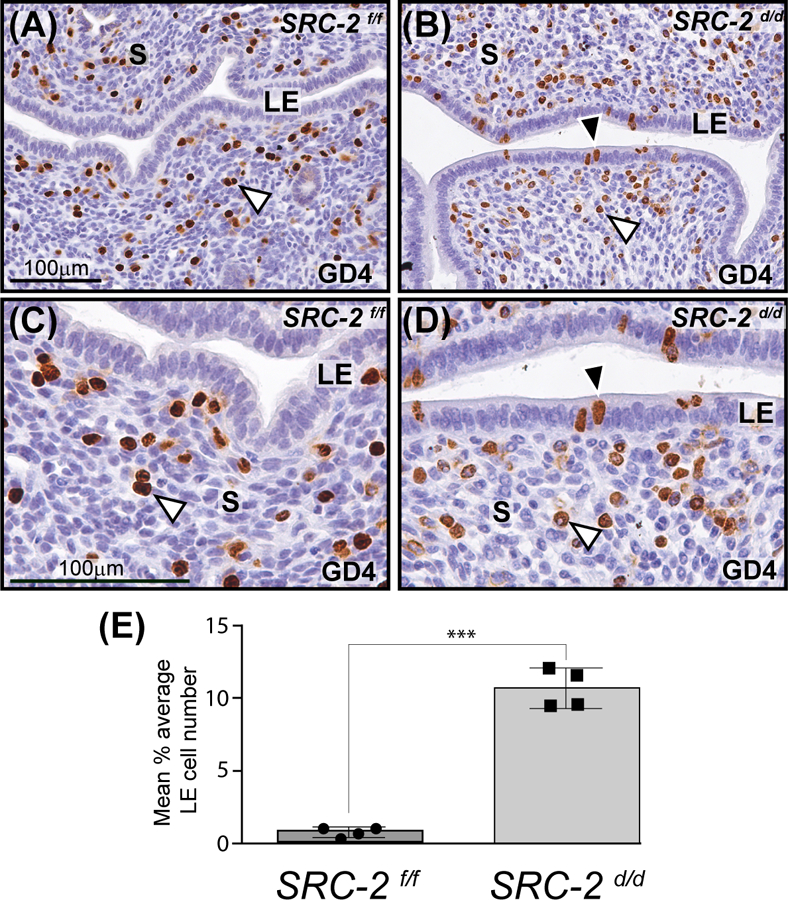
Retention of a subset of proliferating luminal epithelial cells in the SRC-2 d/d endometrium at GD4. (A) Immunohistochemical detection of cells that are positive for BrdU incorporation in the endometrium of the SRC-2 f/f at GD4. Note the presence of proliferating stromal cells in the endometrium of the SRC-2 f/f mouse at GD4 (white arrowhead). In contrast, the luminal epithelium (LE) of the SRC-2 f/f endometrium is negative for BrdU immunopositivity. Together, the epithelial and stromal cellular proliferative profiles are indicative of a receptive uterus at GD4 47. (B) A subset of luminal epithelial cells in the SRC-2 d/d endometrium at GD4 is consistently immunopositive for BrdU incorporation (black arrowhead); a subgroup of subluminal stromal cells is positive for BrdU incorporation (white arrowhead). Panels (C) and (D) are higher power magnifications of regions shown in panels (A) and (B) respectively. Again note the absence and presence of BrdU immunopositive luminal epithelial cells in the SRC-2 f/f and SRC-2 d/d endometrium respectively. Although qualitative, BrdU positive stromal cells in the SRC-2 f/f endometrium consistently register a stronger immunopositive signal than BrdU positive stromal cells in the SRC-2 d/d endometrium, compare the stromal (S) compartment in (C) with (D). Scale bar in (A) and (C) apply to (B) and (D) respectively. (E) Histogram graphically displays the average number of BrdU positive cells per 100 luminal epithelial cells counted from each of three separate tissue sections per mouse (4 mice per genotype).
The aberrant retention of a subset of proliferating cells in the luminal epithelial compartment of the SRC-2 d/d endometrium at GD4 (Figure 2B, D, and E) suggests that other cellular properties of this epithelial compartment, which are required for normal embryo implantation, may also be compromised. Immunohistochemical detection of mucin 1 (MUC 1 49, 50), the expression of which is attenuated in the endometrium of the normal murine endometrium at GD4 (Figure 3A 49, 50), remains significantly elevated in the SRC-2 d/d endometrium at GD4 and 5 (Figure 3A-F). Furthermore, immunofluorescence investigations confirmed that E-cadherin expression is absent in the basolateral junctional regions of luminal epithelial cells of the SRC-2 f/f endometrium that closely contact the trophectodermal cells of the implanting embryo (Figure 3G, I). The suppression of E-cadherin expression in these cellular regions is known as one cellular alteration that signals the emergence of a plasma membrane transformation (PMT) state 51, 52. Suppression of E-cadherin expression normally serves to loosen the tight intercellular adherens junctions within the epithelial compartment to facilitate embryo invasion into the underlying stroma 52–54. However, E-cadherin expression persists in the junctional zones of the SRC-2 d/d epithelium at GD5 (Figure 3H, J). Together, these results suggest that absence of SRC-2 in the SRC-2 d/d uterus results in disruption of epithelial reprogramming that underpins the development of the PMT state, which is a prerequisite for the generation of a receptive uterus for embryo implantation.
FIGURE 3.
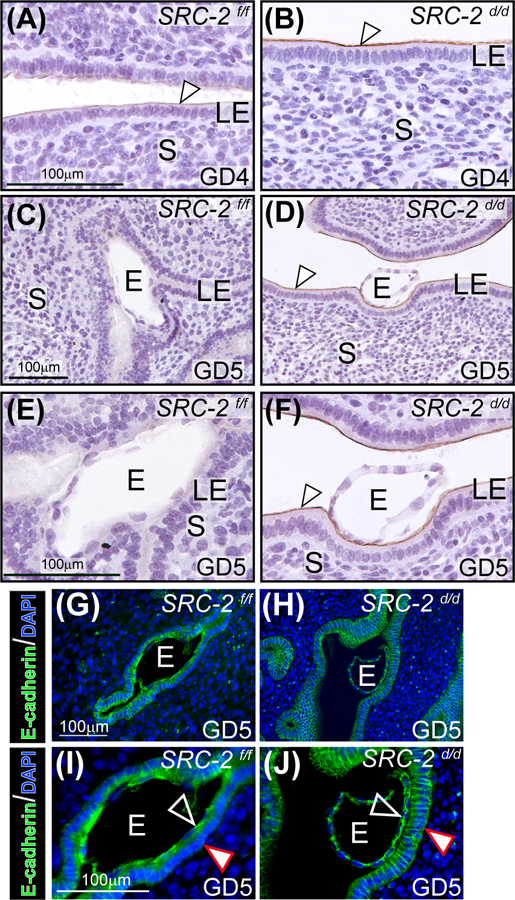
Luminal epithelial (LE) cell marker expression is altered in the SRC-2 d/d endometrium. (A) An endometrial tissue section obtained from a SRC-2 f/f mouse at GD4; section is immunohistochemically stained for MUC 1 expression. Note the low levels of MUC 1 expression on the apical surface of the LE (white arrowhead). (B) Expression of MUC 1 is noticeably stronger on the apical surface of the LE of the SRC-2 d/d endometrium at GD4 (white arrowhead). (C) By GD5, MUC 1 expression is absent in the endometrium of the SRC-2 f/f mouse; E indicates embryo. (D) At GD5, the SRC-2 d/d endometrium still retains strong MUC 1 immunopositivity on the apical surface of the LE (white arrowhead) despite the presence of an embryo. (E) and (F) show higher power magnification images shown in (C) and (D) respectively. Again note the absence of MUC 1 expression in the LE of the SRC-2 f/f endometrium at GD5 and the continued presence of MUC 1 expression on the apical surface of the LE compartment within the SRC-2 d/d endometrium (white arrowhead). (G) Immunofluorescence detection of E-cadherin in the epithelium of the SRC-2 f/f endometrium at GD5; E denotes embryo. Note E-cadherin expression is specific to the epithelial chamber of the SRC-2 f/f implantation site. (I) Higher power magnification image of region shown in (G). Note that E-cadherin expression is present in the trophectoderm of the embryo (black arrowhead) whereas there is significantly less E-cadherin expression in the basolateral regions of epithelial cells that are juxtaposed to the embryo (white arrowhead). (I) Immunofluorescence detection of the E-cadherin protein in the SRC-2 d/d endometrium at GD5. As in (G), E-cadherin immunopositivity is specifically localized to the epithelial compartment. (J) Higher power magnification image of a region shown in (H). Note the retention of E-cadherin expression in the apical and basolateral regions of the epithelial cells (white arrowhead); trophoblast cells of the embryo are positive for E-cadherin expression (black arrowhead). Scale bar in (G) and (I) apply to (H) and (J) respectively.
3.2 |. Abrogation of SRC-2 adversely alters the endometrial transcriptome required for embryo implantation
Because the embryo implantation process fails to advance in the SRC-2 d/d mouse at GD5 (Figure 1), coupled with the fact that SRC-2 is a coregulator of gene transcription 2, we used RNA-seq to identify genes for which normal transcription is dysregulated in the SRC-2 d/d uterus at GD5 (Figure 4). Therefore, the overall experimental design entailed bulk RNA-seq profiling of uterine tissue dissected from SRC-2 f/f control and SRC-2 d/d mutant mice at GD5, with triplicate tissue samples per genotype. The complete list of genes that are differentially expressed between the two genotypes is shown in the Excel table in the supplementary section (Supplementary Folder S1). The expression of a total number of 3,685 genes (1,452 downregulated and 2,233 upregulated) was identified as significantly changed (Figure 4A). The FKPM values for all 3,685 genes were analyzed by PCA (Supplementary Folder S1 (PCA tab)). The PCA confirmed that the SRC-2 f/f and SRC-2 d/d groups were significantly segregated with respect to their group triplicates. The total number of DEGs between the two genotypes, which reached the FDR (≤ 0.05) and FC (≥ 2) threshold parameters, was 1,685 genes (497 downregulated and 1188 upregulated) (Figure 4B). Applying the agglomeration and GO applications in DAVID, genes in the DEG list were grouped according to GO terms (Figure 4C). Note the enrichment for genes associated with cellular properties linked with the apical plasma membrane. In addition, the KEGG pathway annotation software revealed that protein families involved in tight junction biology received one of the highest scores in terms of the number of genes assigned to a given biological process between the two groups (Figure 4D). Ranking based on p-values and z-scores, IPA identified an overrepresented gene group in the DEG list that is significantly associated with EMT cellular processes (Table 1).
FIGURE 4.
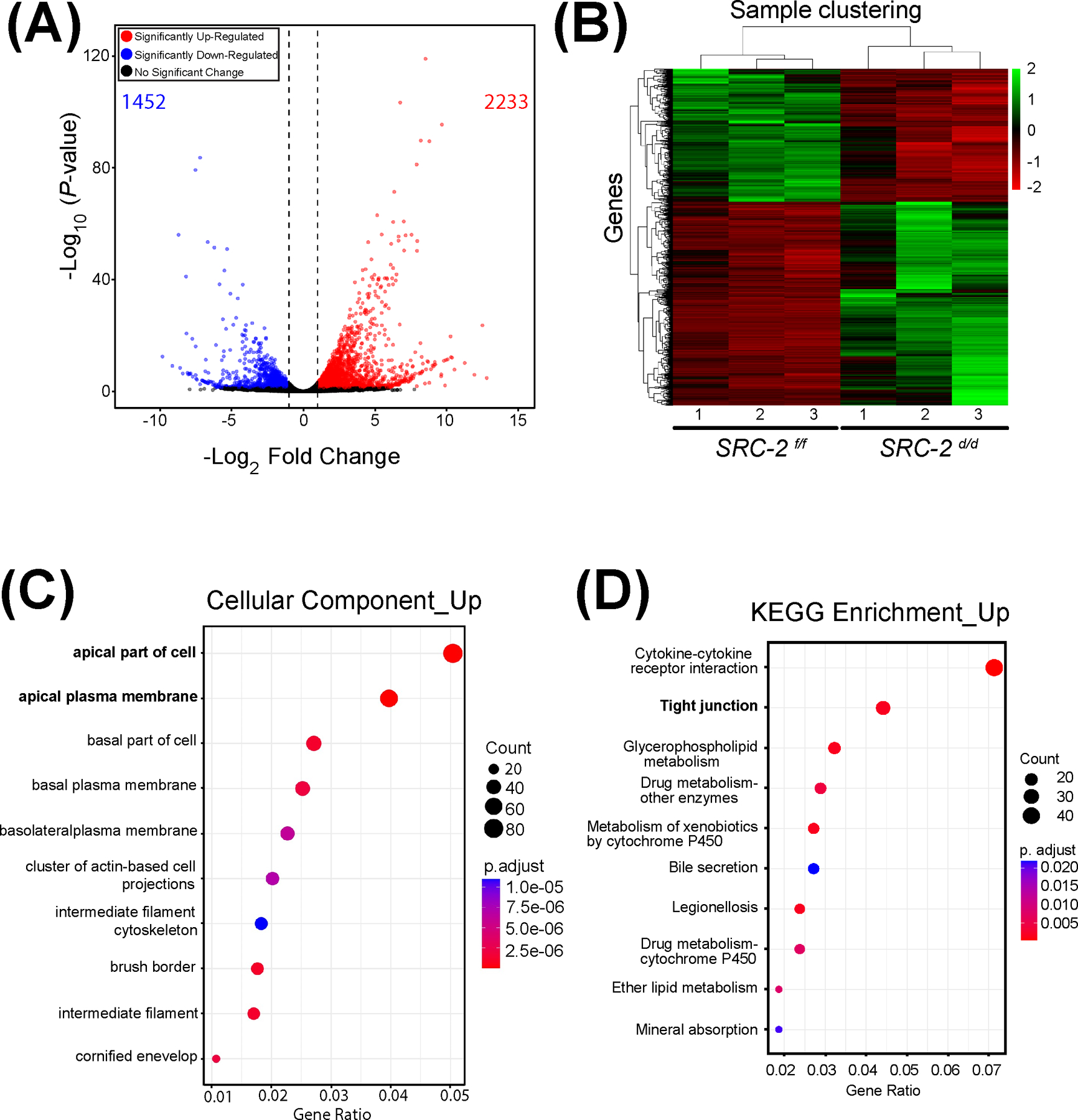
Genome-wide alteration of the transcriptome in the SRC-2 d/d uterus at GD5. (A) The volcano plot graphically displays the total number of genes differentially expressed between the SRC-2 f/f and SRC-2 d/d groups. Significance (p-value) versus fold change are plotted on the y- and x-axis respectively. The total number of genes differentially expressed between the two groups is 3,685 (1,452 and 2,233 downregulated and upregulated respectively). (B) The heatmap represents an unsupervised hierarchical clustergram of the total number of genes differentially expressed between the SRC-2 f/f and SRC-2 d/d groups that reached FDR and FC thresholds of ≤ 0.05 and ≥ 2 respectively. Each row represents a gene while each column denotes a sample replicate in the SRC-2 f/f or SRC-2 d/d group; the dendrogram on both axes show the arrangement of the clusters following analysis. The intensity of the color indicates the level of expression for each gene in the sample replicate. (C) Dot plot of enriched genes within the DEG dataset are stratified according to biological processes, the cellular component module is shown. Note apical and basolateral plasma membrane properties feature prominently in this analysis. (D) The KEGG analysis shows enrichment for tight junction biology (bold) for proteins encoded by genes for which expression is significantly changed between the SRC-2 f/f and SRC-2 d/d groups.
TABLE 1.
Ingenuity Canonical Pathways identified by IPA analysis in differentially expressed genes between SRC-2f/f and SRC-2d/d mouse uteri at GD5
| Ingenuity Canonical Pathways | p- value | Z-Score | Molecules |
|---|---|---|---|
| Regulation Of The Epithelial Mesenchymal Transition In Development Pathway | 6.46E-04 | −2.529822128 | BCL9,CDH1,FZD1,FZD9,GLIS1,HNF1A,LEF1,mir-8,REL,SNAI1,WNT2,WNT6,WNT7B |
| Antioxidant Action of Vitamin C | 1.05E-04 | −2.496150883 | GSTO1,JAK2,MAPK11,MAPK13,NFKBIA,NOTUM,PLA2G10,PLA2G2D,PLA2G2E,PLA2G4F,PLA2G5,PLB1,PLCH1,PLCH2,REL,SLC23A3, STAT5A |
| Basal Cell Carcinoma Signaling | 1.38E-03 | −2.121320344 | BMP2,BMP5,BMP8A,FZD1,FZD9,GLIS1,HNF1A,LEF1,WNT2,WNT6,WNT7B |
| BEX2 Signaling Pathway | 1.23E-03 | −2.110579412 | HNF1A,ITGB6,KLK3,LEF1,MMP2,NFKBIA,PDGFC,PGF,PPP2R2B,PPP2R2C,REL,SPP1 |
| Pulmonary Healing Signaling Pathway | 1.48E-04 | −2.041241452 | BMPR1B,CDH1,DLK1,FGFR1,FRK,FZD1,FZD9,HNF1A,LEF1,MAPK11,MAPK13,MMP2,MMP3,NFKBIA,NOTCH4,PDGFC,PGF,PRKCG, RASD1,SMAD9, STAT5A,WNT2,WNT6,WNT7B |
| Colorectal Cancer Metastasis Signaling | 1.29E-03 | −1.963961012 | ADCY2,CDH1,DCC,FZD1,FZD9,GNG12,GNG4,HNF1A,JAK2,LEF1,MMP2,MMP3,NOS2,PDGFC,PGF,PIK3C2G,PIK3R3,RAC3,RASD1,REL, RHOBTB2, RHOV, TLR1,Tlr12,WNT2,WNT6,WNT7B |
| S100 Family Signaling Pathway | 1.29E-10 | −1.843908891 | ADCYAP1R1,ADGRA1,ADGRF1,ADGRG7,ADGRV1,ADORA1,ADORA2B,ADRA1A,ADRA1B,ADRA1D,ADRA2B,ANXA1,BDNF,BTC, CACNA1B, CACNA2D3,CDH1,CELSR1,CELSR2,CTSD,DRD4,EDNRB,EREG,ESR2,ETV4,F2R,F2RL3, FGFR1,FGFR3,FZD1,FZD9,GALR3, GPR160,GPRC5A,GPRC5C,HCAR2,HNF1A,HTR1D,HTR6,IFNK,Ighg2b,KLK3,LEF1,LPAR3,MAPK11,MAPK13,MMP2,MMP3,NAPSA,NMUR2, NOS2,NOTUM,NPY2R,OXTR,P2RY14, PDGFC,PGF,PIK3C2G, PIK3R3,PLA2G10,PLA2G2D,PLA2G2E,PLA2G4F,PLA2G5,PLCH1,PLCH2, PRKCG,PTGDR,PTGIR,RAC3,RXFP1,S100A14,S100A3, S100A6,S100A8,S1PR3,SMAD6,SMAD9,TACR1,TRPV5,VDR,VIPR2,WNT2,WNT6, WNT7B |
| Factors Promoting Cardiogenesis in Vertebrates | 1.29E-03 | −1.414213562 | BMP2,BMP5,BMP8A,BMPR1B,FZD1,FZD9,HNF1A,LEF1,NOG,NOTUM,NPPA,PLCH1,PLCH2,PRKCG,SMAD9,WNT2,WNT6,WNT7B |
| Pulmonary Fibrosis Idiopathic Signaling Pathway | 2.69E-05 | −1.333333333 | CDH1,COL13A1,COL22A1,COL25A1,COL26A1,Col6a4,COL6A5,COL6A6,EFNB2,F2R,FGF18,FGFR1,FGFR3,FZD1,FZD9,HNF1A,IL11, IL13RA1,ITGB6,JAK2, LEF1,MAPK11,MAPK13,MMP2,MMP3,NOTCH4,PDGFC,PIK3C2G,PIK3R3,PLAU, RASD1,REL,SNAI1,WNT2,WNT6, WNT7B |
| WNT/β-catenin Signaling | 5.50E-06 | −1.091089451 | BCL9,CDH1,CDH5,CDKN2A,DKK3,DKKL1,FRZB,FZD1,FZD9,GJA1,HNF1A,KREMEN2,LEF1,PPP2R2B,PPP2R2C,RARB,SFRP1,SFRP2, SFRP4,SFRP5,SOX7, SOX9,WNT2,WNT6,WNT7B |
3.3 |. Gene expression programs associated with murine uterine receptivity and epithelial cellular properties are significantly altered in the SRC-2 d/d mouse
Gene ontology enrichment scores along with focused biological, cellular, and molecular pathway analytics of DEGs between the SRC-2 f/f and SRC-2 d/d groups at GD5 revealed an overrepresentation of upregulated estrogen responsive genes in the SRC-2 d/d uterus (Figure 5A 55). In parallel, a downregulation of progesterone responsive genes is predicted by this analysis to occur in the SRC-2 d/d uterus at GD5 (Figure 5A 56). In addition, genes previously associated with uterine decidualization are predicted to be markedly downregulated in the SRC-2 d/d uterus at GD5 (Figure 5A 56). Interestingly, genes associated with epithelial cell tight junctions and EMT are predicted to be upregulated and downregulated respectively in the SRC-2 d/d uterus at GD5 (Figure 5A 52). In Figure 5B, results from quantitative real-time PCR analysis confirm the predicted expression changes for a number of the genes (shown in Figure 5A) in the SRC-2 d/d uterus at GD5. Interestingly, the expression of a subset of these genes are also altered in the SRC-2 d/d uterus at GD4 (Figure S4), supporting an important role for SRC-2 in the early expression changes of these genes during the development of the receptive uterus. Using the Illumina correlation engine (Figure 6 and Figure S5), we revealed that a significant number of genes in the 3111 DEG dataset (SRC-2 d/d vs SRC-2 f/f (GD5)) is present in DEGs reported for the PGR Activation Function 1 (AF1) domain mutant mouse (Pgr AF1/AF1, 453 genes) 45; Indian hedgehog (Ihh d/d, 361 genes) 42; SRY-box transcription factor 17 (Sox17 d/d, 978 genes) 46; Bone Morphogenetic protein 2 (Bmp2 d/d, 225 genes) 44; wingless-type MMTV integration site family, member 4 (Wnt4 d/d, 806 genes) 43; Sirtuin 1 (Sirt1 d/d, 1600 genes) 41; forkhead box A2 (Foxa2 d/d, 617 genes) 57; and the estradiol treated ovariectomized mouse (1317 genes (GSE 23241)). The correlative analysis revealed that the majority of overlapped genes followed a concordant (or positive) gene expression profile (both upregulated and downregulated): 92% with Pgr AF1/AF1; 81% with Ihh d/d; 63% with Sox17 d/d; 92% with Bmp2 d/d; 83% Wnt4 d/d; 94% Sirt1 d/d; 85% Foxa2 d/d; and 70% with the estradiol-treated ovariectomized mouse model. Together, our results indicate that SRC-2 is required for the progesterone induction of genes required to establish uterine receptivity 55, a subset of which are known to suppress estrogen responsive gene expression 58, a prerequisite for uterine receptivity. The results also support an important role for SRC-2 in endometrial epithelial reprogramming that normally manifests as a loosening of intercellular tight junctional connections and a triggering of a PMT cellular response 52, both of which are important for adherence between the apical surface of the luminal epithelium and the blastocyst and subsequent blastocyst invasion into the underlying stroma. As further support for this proposal, a number of genes—SNAI1 59, 60 and CLDN7 61, 62—associated with the EMT and PMT processes were shown to be deregulated in the SRC-2 d/d uterus at GD 5 (Figure S6 and S7).
FIGURE 5.
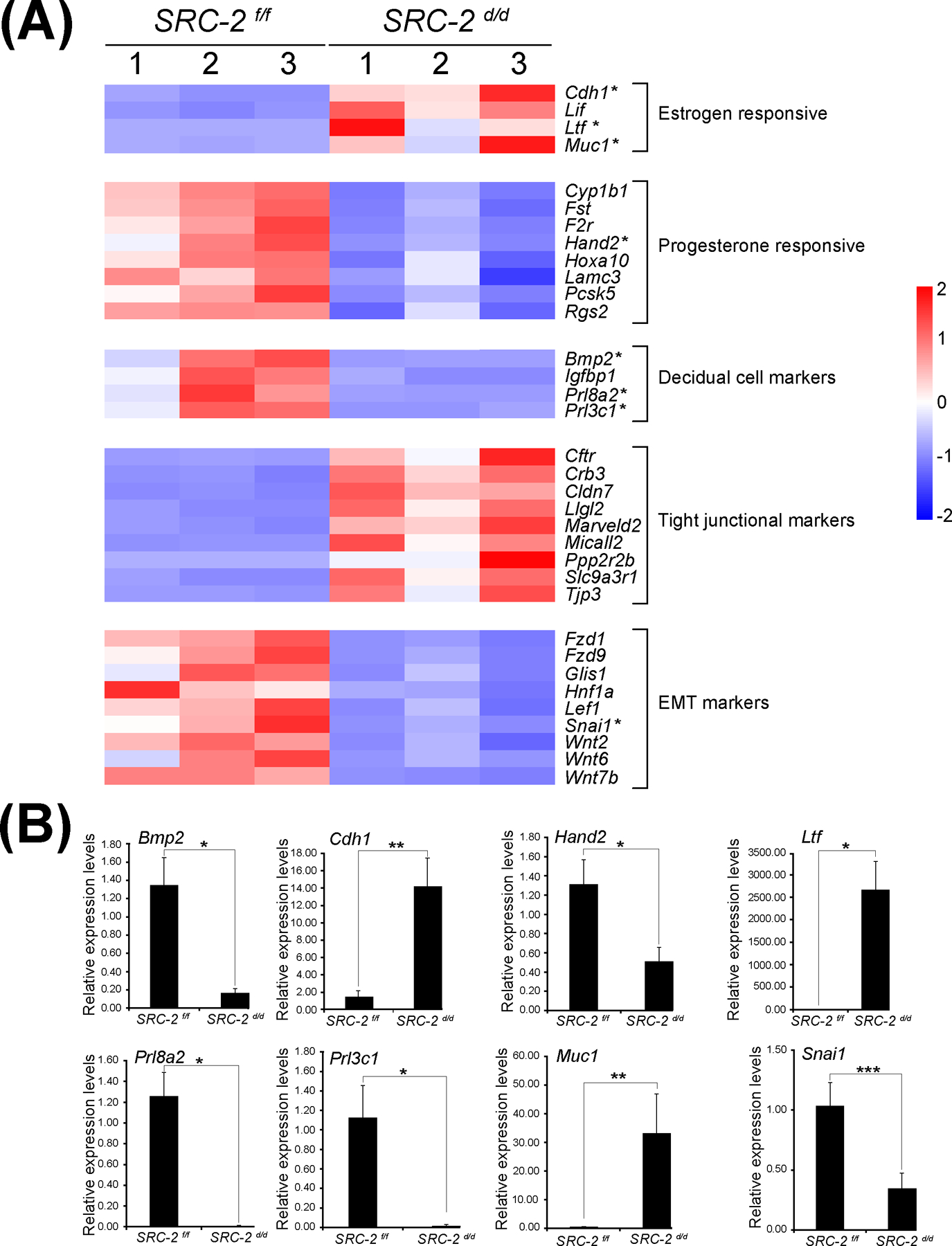
Aberrant expression of genes in the SRC-2 d/d uterus that are involved in uterine receptivity. (A) Heat map of gene expression changes between the SRC-2 f/f and SRC-2 d/d replicates predict an increase in the expression of estrogen responsive genes (Cadherin 1 (Cdh1); Leukemia inhibitory factor (Lif); Lactoferrin (Ltf); and Mucin 1 (Muc1)), a decrease in expression of progesterone responsive genes (Cytochrome P450 family 1 subfamily B member 1 (Cyp1b1); Follistatin (Fst); Coagulation F2 receptor (F2r); Heart and neural crest derivatives expressed 2 (Hand2); Homeobox A10 (Hoxa10); Laminin subunit gamma 3 (Lamc3); Proprotein convertase subtilisin kexin type 5 (Pcsk5); Regulator of G protein signaling 2 (Rgs2)), a decrease in expression of established decidual cell markers (Bone morphogenetic protein 2 (Bmp2); Insulin-like growth factor binding protein 1 (Igfbp1); Prolactin family 8, subfamily a, member 2 (Prl8a2); and Prolactin family 3, subfamily C, member 1 (Prl3c1)), an increase in expression of tight junctional markers (Cystic fibrosis transmembrane conductance regulator (Cftr); Crumbs cell polarity complex component 3 (Crb3); Claudin 7 (Cldn7); Lorelei-like-GPI-anchored protein 2 (Llg2); Marvel domain containing 2 (Marveld2); Molecule interacting with casl-like 2 (Micall2); Protein phosphatase 2, regulatory subunit B, beta (Ppp2r2b); Solute carrier family 9, member 3, regulator 1 (Slc9a3r1); and (Tight junction protein 3 (Tjp3)), and a decrease in expression of EMT markers (Frizzled class receptor 1 (Fzd1); Frizzled class receptor 9 (Fzd9); Glis family zinc finger 1 (Glis1); Hepatocyte nuclear factor 1 alpha (Hnf1a); Lymphoid enhancer binding factor 1 (Lef1); Snail family transcriptional repressor 1 (Snai1); Wnt family member 2 (Wnt2); Wnt family member 6 (Wnt6); and Wnt family member 7b (Wnt7b)). Asterisks denote genes chosen for further expression validation by qPCR shown below. (B) Quantitative real time PCR results for a selection of genes with asterisks listed in the heat maps shown in panel (A). Important to note: the RNA samples used in these quantitative real time PCR experiments were separate from the RNA samples used for the RNA-seq study; n= 3 per genotype.
FIGURE 6.
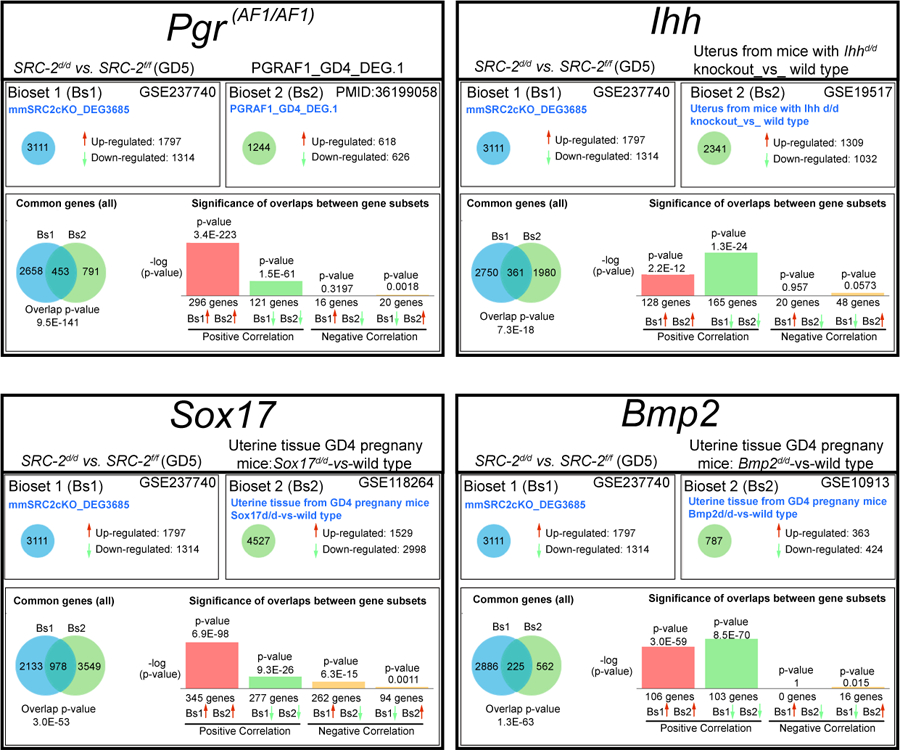
Significant correlation in the expression of a gene subset in the SRC-2 d/d versus SRC-2 f/f DEG dataset and in uterine DEG datasets from mutant mouse exhibiting progesterone suppression and concomitant estrogen hypersensitivity. The Illumina correlation engine was used to determine the significance of positive correlation in gene expression changes (up (red histogram) and down (green histogram)) between gene subsets in the SRC-2 d/d vs SRC-2 f/f DEG dataset and in DEG datasets reported for uterine tissue from four mutant mouse models that exhibit a progesterone suppression and estrogen hypersensitivity phenotype 42, 44–46.
DISCUSSION
In both the human and mouse, pregnancy establishment relies on the implantation of a high quality blastocyst into a receptive endometrium in which the latter’s luminal epithelium is the first contact site for the blastocyst 17, 63. To facilitate blastocyst attachment and adhesion to the luminal epithelium, the epithelium undergoes a set of cellular and molecular changes, many of which promote a PMT state that is crucial for embryo implantation 51, 52. For uterine receptivity to manifest, apical alterations of the luminal epithelium occur in which its extracellular glycoprotein composition is changed and cell-surface charge reduced to enable embryo attachment and adherence 49, 64. In parallel, apical, basolateral, and planar polarity of luminal epithelial cells is lost, resulting in a weakening of lateral epithelial cell surface interactions as well as a disruption of focal adhesions between the epithelium and basal lamina. These intercellular contact alterations are essential for the penetration of embryonic trophectodermal cells through the luminal epithelium into the underlying stroma 65–67. In the case of the mouse, one study has shown that luminal epithelial cells are lost at the contact site by entosis soon after blastocyst attachment 68. Notwithstanding these advances in our understanding, the identification of the key regulatory signals that control the emergence of endometrial receptivity in general and in the luminal epithelial PMT state in particular remains incomplete.
Applying the timed pregnancy model to the SRC-2 d/d mouse, we report that SRC-2 is required for the development of endometrial receptivity. Examination of established markers of uterine receptivity revealed that signals associated with the PMT state are significantly derailed in the SRC-2 d/d endometrium at GD5. For example, downregulation of MUC 1 expression at the apical surface of the luminal epithelium did not occur in the SRC-2 d/d endometrium at GD5. A prominent constituent of the glycocalyx layer on the apical surface of the luminal epithelium of the human endometrium 69, MUC 1 is known to exhibit anti-adhesive properties on the apical surface of the luminal epithelium 64. In vitro studies have also shown that MUC 1 expression is suppressed in primary endometrial epithelial cells at the site of human blastocyst attachment 70 while clinical investigations have revealed a link between abnormal endometrial MUC 1 levels and implantation failure 71. With parallels to the human, MUC 1 expression was shown to be markedly reduced in the luminal epithelium as the murine endometrium reaches the receptive state 49, 50. The downregulation of E-cadherin at adherens junctions of luminal epithelial cells, another PMT signal, is also compromised in the SRC-2 d/d endometrium at GD5. As a result of blocking E-cadherin downregulation, embryo penetration to the stroma is prevented due to a persistently polarized SRC-2 d/d luminal epithelium with intact tight intercellular junctions.
Transcriptomics uncovered a broader spectrum of genes associated with steroid hormone control of uterine receptivity, decidualization, and tight junctional and EMT biology. In keeping with SRC-2’s role in PGR-mediated signaling 16, 19, 25, 72–76, the induction in expression of a significant number of progesterone responsive genes is attenuated in the SRC-2 d/d endometrium at GD5. A number of these genes (i.e. Hand 2) have been shown to act as paracrine signals from the subepithelial stroma to the luminal epithelium to facilitate progesterone suppression of estrogen-induced signaling in the luminal epithelium 58. The switch in luminal epithelial proliferative status accompanies the change in the structural and functional properties of this cellular compartment, which leads to PMT and uterine receptivity. The persistent expression of a subset of estrogen responsive genes in the SRC-2 d/d luminal epithelium indicates an unwarranted heightened estrogen response, which is known to be linked to implantation failure in the mouse 77–79 and indirectly in the human 80, 81.
In addition to the stroma, SRC-2 is expressed in the epithelia of the human 10 and murine endometrium (Figure S1), suggesting that SRC-2 directly or indirectly regulates EMT processes during the periimplantation period; our transcriptomics analysis showed EMT is a biological process significantly altered in the SRC-2 d/d uterus at GD5. Interestingly, EMT shares similar properties with PMT such as loss of apical-basolateral polarity, a breakdown of cell-cell junctions, and an increase in extracellular matrix degradation 52. Together, these findings provide compelling support for epithelial-derived SRC-2 in directly controlling the PMT state that determines progression of the epithelium to a receptive phenotype for embryo attachment and adherence. In the case of the mouse, this proposal will be addressed in the future through selective SRC-2 ablation in the endometrial epithelial compartment using cre/lox methods 82.
Our previous studies have established stromal-derived SRC-2 as a critical coregulator in progesterone-dependent decidualization and pregnancy establishment 19, 20, 25. Using the timed natural pregnancy model, we reveal here that SRC-2 is pivotal for the PMT changes that must occur in the luminal epithelium to enable embryo attachment and adherence within the context of a receptive uterus. Given the cell-type specific roles of SRC-2 in endometrial biology and pathobiology 10, 19, 20, 25, 83, 84 and the knowledge-gap associated with early pregnancy establishment, our findings provide a foundation for further investigations into the specific involvement of this coregulator in the endometrial epithelium in ensuring normal endometrial function and health.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Yan Ying and Rong Zhao for their invaluable technical expertise. The technical assistance of the Gene and RNA Profiling (GARP) core at Baylor College of Medicine and CD Genomics, Shirley, New York USA is acknowledged.
FUNDING INFORMATION
The research was funded in part by the Intramural Research Program of the National Institute of Environmental Health Sciences: Project Z1AE8103311-01 (FJD) and National Institutes of Health (NIH)/ National Institute of Child Health and Human Development (NICHD) grants: R01 HD-07857/HD-08818, R01HD104813, and R01 HD-042311 to BWO, RK, and JPL respectively.
Abbreviations:
- AAALAC
Association for assessment and accreditation of laboratory animal care
- ANOVA
Analysis of variance
- ARTs
Assisted reproductive technologies
- ATCC
American type culture collection
- BrdU
5-bromo-2’-deoxyuridine
- CL
Corpus luteum
- DAPI
4, 6-diamidino-2-phenylindole
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DEGs
Differentially expressed genes
- DPBS
Dulbecco’s phosphate buffered saline
- EMT
Epithelial mesenchymal transition
- FBS
Fetal bovine serum
- FC
Fold change
- FDR
False discovery rate
- FPKM
Fragments per kilobase of transcript per million
- GD
Gestation day
- GRIP1
Glucocorticoid receptor interacting protein 1
- GEO
Gene expression omnibus
- GO
Gene ontology
- GE
Glandular epithelium
- H&E
Hematoxylin and eosin
- Hand2
Heart and neural crest derivatives expressed 2
- HISAT2
hierarchical indexing for spliced alignment of transcripts 2
- HRP
Horse radish peroxidase
- IPA
Ingenuity pathway analysis
- LE
Luminal epithelium
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MUC 1
Mucin 1
- NCBI
National center for biotechnology information
- NCOA
Nuclear receptor coactivator
- NR
Nuclear receptor
- NT
Non-targeting
- PCA
Principal component analysis
- PGR
Progesterone receptor
- PVDF
Polyvinylidene difluoride
- PMT
Plasma membrane transformation
- qRT-PCT
quantitative real-time PCR
- RIN
RNA integrity number
- RNA-seq
RNA sequencing
- RPM
Revolutions per minute
- S
Stroma
- SD
Standard deviation
- SDS
Sodium dodecyl sulfate
- SNAI1
Snail family zinc finger 1
- SRC
Steroid receptor coactivator
- TBST
Tris-buffered saline with Tween
- TIF2
Transcriptional intermediary factor 2
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPLEMENTARY MATERIAL
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
All data reported in the manuscript and support information is publically available. The RNA-seq data described in this manuscript has been deposited into GEO (accession number: GSE237740).
REFERENCES
- 1.Xu J, Wu RC, and O’Malley BW (2009) Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leo C, and Chen JD (2000) The SRC family of nuclear receptor coactivators. Gene 245, 1–11 [DOI] [PubMed] [Google Scholar]
- 3.Szwarc MM, Kommagani R, Lessey BA, and Lydon JP (2014) The p160/steroid receptor coactivator family: potent arbiters of uterine physiology and dysfunction. Biol Reprod 91, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Malley BW (2020) SRC-2 Coactivator: a role in human metabolic evolution and disease. Mol Med 26, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, He Y, Liu H, Zhou W, Wang C, Xu P, Cai X, Liu H, Yu K, Pei Z, Hyseni I, Fukuda M, Tong Q, Xu J, Sun Z, O’Malley BW, and Xu Y (2021) Hypothalamic steroid receptor coactivator-2 regulates adaptations to fasting and overnutrition. Cell Rep 37, 110075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Reineke LC, Dasgupta S, Dean A, Stevenson CR, Sivasubramanian N, Sreekumar A, Demayo F, York B, Fu L, and O’Malley BW (2014) SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell Rep 6, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao L, Rabbitt EH, Condon JC, Renthal NE, Johnston JM, Mitsche MA, Chambon P, Xu J, O’Malley BW, and Mendelson CR (2015) Steroid receptor coactivators 1 and 2 mediate fetal-to-maternal signaling that initiates parturition. J Clin Invest 125, 2808–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendelson CR (2009) Minireview: fetal-maternal hormonal signaling in pregnancy and labor. Mol Endocrinol 23, 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, and Chambon P (2002) The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol 22, 5923–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory CW, Wilson EM, Apparao KB, Lininger RA, Meyer WR, Kowalik A, Fritz MA, and Lessey BA (2002) Steroid receptor coactivator expression throughout the menstrual cycle in normal and abnormal endometrium. J Clin Endocrinol Metab 87, 2960–2966 [DOI] [PubMed] [Google Scholar]
- 11.Kershah SM, Desouki MM, Koterba KL, and Rowan BG (2004) Expression of estrogen receptor coregulators in normal and malignant human endometrium. Gynecol Oncol 92, 304–313 [DOI] [PubMed] [Google Scholar]
- 12.Konno T, Graham AR, Rempel LA, Ho-Chen JK, Alam SM, Bu P, Rumi MA, and Soares MJ (2010) Subfertility linked to combined luteal insufficiency and uterine progesterone resistance. Endocrinology 151, 4537–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu L, Luo J, Deng J, Huang C, and Li C (2023) Polycystic ovary syndrome is associated with a higher risk of premalignant and malignant endometrial polyps in premenopausal women: a retrospective study in a tertiary teaching hospital. BMC Womens Health 23, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dor J, Itzkowic DJ, Mashiach S, Lunenfeld B, and Serr DM (1980) Cumulative conception rates following gonadotropin therapy. Am J Obstet Gynecol 136, 102–105 [DOI] [PubMed] [Google Scholar]
- 15.Sagle M, Bishop K, Ridley N, Alexander FM, Michel M, Bonney RC, Beard RW, and Franks S (1988) Recurrent early miscarriage and polycystic ovaries. BMJ 297, 1027–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee A, Soyal SM, Fernandez-Valdivia R, Gehin M, Chambon P, Demayo FJ, Lydon JP, and O’Malley BW (2006) Steroid receptor coactivator 2 is critical for progesterone-dependent uterine function and mammary morphogenesis in the mouse. Mol Cell Biol 26, 6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurya VK, DeMayo FJ, and Lydon JP (2021) Illuminating the “Black Box” of Progesterone-Dependent Embryo Implantation Using Engineered Mice. Front Cell Dev Biol 9, 640907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gellersen B, and Brosens JJ (2014) Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev 35, 851–905 [DOI] [PubMed] [Google Scholar]
- 19.Kommagani R, Szwarc MM, Kovanci E, Gibbons WE, Putluri N, Maity S, Creighton CJ, Sreekumar A, DeMayo FJ, Lydon JP, and O’Malley BW (2013) Acceleration of the glycolytic flux by steroid receptor coactivator-2 is essential for endometrial decidualization. PLoS Genet 9, e1003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szwarc MM, Hai L, Gibbons WE, White LD, Mo Q, Kommagani R, Lanz RB, DeMayo FJ, O’Malley BW, and Lydon JP (2018) Retinoid signaling controlled by SRC-2 in decidualization revealed by transcriptomics. Reproduction 156, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellver J, and Simon C (2018) Implantation failure of endometrial origin: what is new? Curr Opin Obstet Gynecol 30, 229–236 [DOI] [PubMed] [Google Scholar]
- 22.Chard T (1991) Frequency of implantation and early pregnancy loss in natural cycles. Baillieres Clin Obstet Gynaecol 5, 179–189 [DOI] [PubMed] [Google Scholar]
- 23.Dimitriadis E, Menkhorst E, Saito S, Kutteh WH, and Brosens JJ (2020) Recurrent pregnancy loss. Nat Rev Dis Primers 6, 98. [DOI] [PubMed] [Google Scholar]
- 24.Macklon NS, Geraedts JP, and Fauser BC (2002) Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum Reprod Update 8, 333–343 [DOI] [PubMed] [Google Scholar]
- 25.Kommagani R, Szwarc MM, Kovanci E, Creighton CJ, O’Malley BW, Demayo FJ, and Lydon JP (2014) A murine uterine transcriptome, responsive to steroid receptor coactivator-2, reveals transcription factor 23 as essential for decidualization of human endometrial stromal cells. Biol Reprod 90, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, and Lydon JP (2005) Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 41, 58–66 [DOI] [PubMed] [Google Scholar]
- 27.Hai L, Szwarc MM, He B, Lonard DM, Kommagani R, DeMayo FJ, and Lydon JP (2018) Uterine function in the mouse requires speckle-type poz protein. Biol Reprod 98, 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CLL, Bolon B, Rudmann DG, Gianani R, Koegler SR, Krueger J, and Young GD (2017) The Gold Standard Paradox in Digital Image Analysis: Manual Versus Automated Scoring as Ground Truth. Arch Pathol Lab Med 141, 1267–1275 [DOI] [PubMed] [Google Scholar]
- 29.Meyerholz DK, and Beck AP (2018) Principles and approaches for reproducible scoring of tissue stains in research. Lab Invest 98, 844–855 [DOI] [PubMed] [Google Scholar]
- 30.Maurya VK, Szwarc MM, Lonard DM, Gibbons WE, Wu SP, O’Malley BW, DeMayo FJ, and Lydon JP (2022) Decidualization of human endometrial stromal cells requires steroid receptor coactivator-3. Front Reprod Health 4, 1033581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szwarc MM, Hai L, Gibbons WE, Mo Q, Lanz RB, DeMayo FJ, and Lydon JP (2019) Early growth response 1 transcriptionally primes the human endometrial stromal cell for decidualization. J Steroid Biochem Mol Biol 189, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurya VK, Szwarc MM, Fernandez-Valdivia R, Lonard DM, Yong S, Joshi N, Fazleabas AT, and Lydon JP (2022) Early growth response 1 transcription factor is essential for the pathogenic properties of human endometriotic epithelial cells. Reproduction 164, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Langmead B, and Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, and Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, McCarthy DJ, and Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y, and Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 57, 289–300 [Google Scholar]
- 37.Huang da W, Sherman BT, and Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, and Lempicki RA (2007) DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh Y, Quiroz E, Wang T, Medina-Laver Y, Redecke SM, Dominguez F, Lydon JP, DeMayo FJ, and Wu SP (2023) The NR2F2-HAND2 signaling axis regulates progesterone actions in the uterus at early pregnancy. Front Endocrinol (Lausanne) 14, 1229033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, Shekar M, Wang H, Park J, Cui W, Wall GD, Wisotzkey R, Alag S, Akhtari S, and Ronaghi M (2010) Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummings MJ, Yu H, Paudel S, Hu G, Li X, Hemberger M, and Wang X (2022) Uterine-specific SIRT1 deficiency confers premature uterine aging and impairs invasion and spacing of blastocyst, and stromal cell decidualization, in mice. Mol Hum Reprod 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco HL, Lee KY, Broaddus RR, White LD, Lanske B, Lydon JP, Jeong JW, and DeMayo FJ (2010) Ablation of Indian hedgehog in the murine uterus results in decreased cell cycle progression, aberrant epidermal growth factor signaling, and increased estrogen signaling. Biol Reprod 82, 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Large MJ, Wetendorf M, Lanz RB, Hartig SM, Creighton CJ, Mancini MA, Kovanci E, Lee KF, Threadgill DW, Lydon JP, Jeong JW, and DeMayo FJ (2014) The epidermal growth factor receptor critically regulates endometrial function during early pregnancy. PLoS Genet 10, e1004451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee KY, Jeong JW, Wang J, Ma L, Martin JF, Tsai SY, Lydon JP, and DeMayo FJ (2007) Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol 27, 5468–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SH, Lim CL, Shen W, Tan SMX, Woo ARE, Yap YHY, Sian CAS, Goh WWB, Yu WP, Li L, and Lin VCL (2022) Activation function 1 of progesterone receptor is required for progesterone antagonism of oestrogen action in the uterus. BMC Biol 20, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Li X, Wang T, Wu SP, Jeong JW, Kim TH, Young SL, Lessey BA, Lanz RB, Lydon JP, and DeMayo FJ (2018) SOX17 regulates uterine epithelial-stromal cross-talk acting via a distal enhancer upstream of Ihh. Nat Commun 9, 4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cha J, Sun X, and Dey SK (2012) Mechanisms of implantation: strategies for successful pregnancy. Nat Med 18, 1754–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong W, and Pollard JW (1999) Progesterone inhibits estrogen-induced cyclin D1 and cdk4 nuclear translocation, cyclin E- and cyclin A-cdk2 kinase activation, and cell proliferation in uterine epithelial cells in mice. Mol Cell Biol 19, 2251–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeSouza MM, Mani SK, Julian J, and Carson DD (1998) Reduction of mucin-1 expression during the receptive phase in the rat uterus. Biol Reprod 58, 1503–1507 [DOI] [PubMed] [Google Scholar]
- 50.Surveyor GA, Gendler SJ, Pemberton L, Das SK, Chakraborty I, Julian J, Pimental RA, Wegner CC, Dey SK, and Carson DD (1995) Expression and steroid hormonal control of Muc-1 in the mouse uterus. Endocrinology 136, 3639–3647 [DOI] [PubMed] [Google Scholar]
- 51.Dudley JS, Murphy CR, Thompson MB, and McAllan BM (2017) Epithelial cadherin disassociates from the lateral plasma membrane of uterine epithelial cells throughout pregnancy in a marsupial. J Anat 231, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitby S, Zhou W, and Dimitriadis E (2020) Alterations in Epithelial Cell Polarity During Endometrial Receptivity: A Systematic Review. Front Endocrinol (Lausanne) 11, 596324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh H, and Aplin JD (2009) Adhesion molecules in endometrial epithelium: tissue integrity and embryo implantation. J Anat 215, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiwari A, Ashary N, Singh N, Sharma S, and Modi D (2021) Modulation of E-Cadherin and N-Cadherin by ovarian steroids and embryonic stimuli. Tissue Cell 73, 101670. [DOI] [PubMed] [Google Scholar]
- 55.Pawar S, Hantak AM, Bagchi IC, and Bagchi MK (2014) Minireview: Steroid-regulated paracrine mechanisms controlling implantation. Mol Endocrinol 28, 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Large MJ, and DeMayo FJ (2012) The regulation of embryo implantation and endometrial decidualization by progesterone receptor signaling. Mol Cell Endocrinol 358, 155–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Filant J, Lydon JP, and Spencer TE (2014) Integrated chromatin immunoprecipitation sequencing and microarray analysis identifies FOXA2 target genes in the glands of the mouse uterus. FASEB J 28, 230–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Kannan A, DeMayo FJ, Lydon JP, Cooke PS, Yamagishi H, Srivastava D, Bagchi MK, and Bagchi IC (2011) The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 331, 912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma XH, Hu SJ, Yu H, Xu LB, and Yang ZM (2006) Differential expression of transcriptional repressor snail gene at implantation site in mouse uterus. Mol Reprod Dev 73, 133–141 [DOI] [PubMed] [Google Scholar]
- 60.Baulida J, and Garcia de Herreros A (2015) Snail1-driven plasticity of epithelial and mesenchymal cells sustains cancer malignancy. Biochim Biophys Acta 1856, 55–61 [DOI] [PubMed] [Google Scholar]
- 61.Schumann S, Buck VU, Classen-Linke I, Wennemuth G, and Grummer R (2015) Claudin-3, claudin-7, and claudin-10 show different distribution patterns during decidualization and trophoblast invasion in mouse and human. Histochem Cell Biol 144, 571–585 [DOI] [PubMed] [Google Scholar]
- 62.Poon CE, Madawala RJ, Day ML, and Murphy CR (2013) Claudin 7 is reduced in uterine epithelial cells during early pregnancy in the rat. Histochem Cell Biol 139, 583–593 [DOI] [PubMed] [Google Scholar]
- 63.Muter J, Lynch VJ, McCoy RC, and Brosens JJ (2023) Human embryo implantation. Development 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Achache H, and Revel A (2006) Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update 12, 731–746 [DOI] [PubMed] [Google Scholar]
- 65.Kaneko Y, Day ML, and Murphy CR (2013) Uterine epithelial cells: Serving two masters. Int J Biochem Cell Biol 45, 359–363 [DOI] [PubMed] [Google Scholar]
- 66.Murphy CR (2000) Junctional barrier complexes undergo major alterations during the plasma membrane transformation of uterine epithelial cells. Hum Reprod 15 Suppl 3, 182–188 [DOI] [PubMed] [Google Scholar]
- 67.Murphy CR (2004) Uterine receptivity and the plasma membrane transformation. Cell Res 14, 259–267 [DOI] [PubMed] [Google Scholar]
- 68.Li Y, Sun X, and Dey SK (2015) Entosis allows timely elimination of the luminal epithelial barrier for embryo implantation. Cell Rep 11, 358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gipson IK, Ho SB, Spurr-Michaud SJ, Tisdale AS, Zhan Q, Torlakovic E, Pudney J, Anderson DJ, Toribara NW, and Hill JA 3rd. (1997) Mucin genes expressed by human female reproductive tract epithelia. Biol Reprod 56, 999–1011 [DOI] [PubMed] [Google Scholar]
- 70.Meseguer M, Aplin JD, Caballero-Campo P, O’Connor JE, Martin JC, Remohi J, Pellicer A, and Simon C (2001) Human endometrial mucin MUC1 is up-regulated by progesterone and down-regulated in vitro by the human blastocyst. Biol Reprod 64, 590–601 [DOI] [PubMed] [Google Scholar]
- 71.Salamonsen LA, Nie G, Hannan NJ, and Dimitriadis E (2009) Society for Reproductive Biology Founders’ Lecture 2009. Preparing fertile soil: the importance of endometrial receptivity. Reprod Fertil Dev 21, 923–934 [DOI] [PubMed] [Google Scholar]
- 72.Mukherjee A, Amato P, Allred DC, Fernandez-Valdivia R, Nguyen J, O’Malley BW, DeMayo FJ, and Lydon JP (2006) Steroid receptor coactivator 2 is essential for progesterone-dependent uterine function and mammary morphogenesis: insights from the mouse--implications for the human. J Steroid Biochem Mol Biol 102, 22–31 [DOI] [PubMed] [Google Scholar]
- 73.Hofman K, Swinnen JV, Verhoeven G, and Heyns W (2002) Coactivation of an endogenous progesterone receptor by TIF2 in COS-7 cells. Biochem Biophys Res Commun 295, 469–474 [DOI] [PubMed] [Google Scholar]
- 74.Jeong JW, Lee KY, Han SJ, Aronow BJ, Lydon JP, O’Malley BW, and DeMayo FJ (2007) The p160 steroid receptor coactivator 2, SRC-2, regulates murine endometrial function and regulates progesterone-independent and -dependent gene expression. Endocrinology 148, 4238–4250 [DOI] [PubMed] [Google Scholar]
- 75.Fernandez-Valdivia R, Mukherjee A, Amato P, Allred DC, Nguyen J, DeMayo FJ, and Lydon JP (2007) Progesterone-action in the murine uterus and mammary gland requires steroid receptor coactivator 2: relevance to the human. Front Biosci 12, 3640–3647 [DOI] [PubMed] [Google Scholar]
- 76.Yu X, Yi P, Panigrahi AK, Lumahan LEV, Lydon JP, Lonard DM, Lutdke SJ, Wang Z, and O’Malley BW (2022) Spatial definition of the human progesterone receptor-B transcriptional complex. iScience 25, 105321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma WG, Song H, Das SK, Paria BC, and Dey SK (2003) Estrogen is a critical determinant that specifies the duration of the window of uterine receptivity for implantation. Proc Natl Acad Sci U S A 100, 2963–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun X, Bartos A, Whitsett JA, and Dey SK (2013) Uterine deletion of Gp130 or Stat3 shows implantation failure with increased estrogenic responses. Mol Endocrinol 27, 1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kurihara I, Lee DK, Petit FG, Jeong J, Lee K, Lydon JP, DeMayo FJ, Tsai MJ, and Tsai SY (2007) COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet 3, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pellicer A, Valbuena D, Cano F, Remohi J, and Simon C (1996) Lower implantation rates in high responders: evidence for an altered endocrine milieu during the preimplantation period. Fertil Steril 65, 1190–1195 [DOI] [PubMed] [Google Scholar]
- 81.Simon C, Garcia Velasco JJ, Valbuena D, Peinado JA, Moreno C, Remohi J, and Pellicer A (1998) Increasing uterine receptivity by decreasing estradiol levels during the preimplantation period in high responders with the use of a follicle-stimulating hormone step-down regimen. Fertil Steril 70, 234–239 [DOI] [PubMed] [Google Scholar]
- 82.Daikoku T, Ogawa Y, Terakawa J, Ogawa A, DeFalco T, and Dey SK (2014) Lactoferrin-iCre: a new mouse line to study uterine epithelial gene function. Endocrinology 155, 2718–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szwarc MM, Kommagani R, Jeong JW, Wu SP, Tsai SY, Tsai MJ, O’Malley BW, DeMayo FJ, and Lydon JP (2014) Perturbing the cellular levels of steroid receptor coactivator-2 impairs murine endometrial function. PLoS One 9, e98664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szwarc MM, Kommagani R, Putluri V, Dubrulle J, Stossi F, Mancini MA, Coarfa C, Lanz RB, Putluri N, DeMayo FJ, Lydon JP, and O’Malley BW (2018) Steroid Receptor Coactivator-2 Controls the Pentose Phosphate Pathway through RPIA in Human Endometrial Cancer Cells. Sci Rep 8, 13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in the manuscript and support information is publically available. The RNA-seq data described in this manuscript has been deposited into GEO (accession number: GSE237740).


