Abstract
The 16S rRNA sequences and selected phenotypic characteristics were determined for six recently isolated bacteria that can tolerate high levels of hydrolyzable and condensed tannins. Bacteria were isolated from the ruminal contents of animals in different geographic locations, including Sardinian sheep (Ovis aries), Honduran and Colombian goats (Capra hircus), white-tail deer (Odocoileus virginianus) from upstate New York, and Rocky Mountain elk (Cervus elaphus nelsoni) from Oregon. Nearly complete sequences of the small-subunit rRNA genes, which were obtained by PCR amplification, cloning, and sequencing, were used for phylogenetic characterization. Comparisons of the 16S rRNA of the six isolates showed that four of the isolates were members of the genus Streptococcus and were most closely related to ruminal strains of Streptococcus bovis and the recently described organism Streptococcus gallolyticus. One of the other isolates, a gram-positive rod, clustered with the clostridia in the low-G+C-content group of gram-positive bacteria. The sixth isolate, a gram-negative rod, was a member of the family Enterobacteriaceae in the gamma subdivision of the class Proteobacteria. None of the 16S rRNA sequences of the tannin-tolerant bacteria examined was identical to the sequence of any previously described microorganism or to the sequence of any of the other organisms examined in this study. Three phylogenetically distinct groups of ruminal bacteria were isolated from four species of ruminants in Europe, North America, and South America. The presence of tannin-tolerant bacteria is not restricted by climate, geography, or host animal, although attempts to isolate tannin-tolerant bacteria from cows on low-tannin diets failed.
The toxicity of phenolic compounds in the environment has fostered studies of bacteria that are able to tolerate and/or metabolize high levels of these compounds, particularly under anaerobic conditions (1, 4, 14, 21, 30, 36). Tannins are secondary polyphenolic compounds known primarily for their ability to bind to and precipitate proteins and other macromolecules. Tannins have been found in many habitats, including sewage sludge, forest litter, and the rumen (9, 14, 15, 28). Bacteria capable of degrading or tolerating tannins have been isolated from sewage sludge (14) and from the alimentary tracts of koalas (Phascolarctos cinereus) (33), goats (Capra hircus) (4, 30), and horses (Equus caballus) (31). Most of the isolates have been characterized phenotypically, and phylogenetic characterization has been limited to studies conducted in Australia (4, 34, 35) and Japan (31). Little is known about the geographic diversity and host species diversity of tannin-tolerant and tannin-degrading bacteria.
The objective of this study was to characterize six recently isolated tannin-tolerant bacteria by examining their phenotypic characteristics and molecular phylogeny. These bacteria were isolated from the ruminal contents of goats (C. hircus), sheep (Ovis aries), white-tail deer (Odocoileus virginianus), and Rocky Mountain elk (Cervus elaphus nelsoni), all of which had consumed forage containing tannins. Our goal was to genetically and biochemically characterize tannin-tolerant bacteria isolated from different host animals in various geographic locations.
MATERIALS AND METHODS
Chemicals.
Tannic acid, p-coumaric acid, ferulic acid, gallic acid, pyrogallol, and phloroglucinol were purchased from Sigma Chemical Company, St. Louis, Mo. Quebracho tannin is an extract of Schinopsis balansae and is commonly used as a standard compound in tannin assays (19). Spray-dried quebracho was purchased from Trask Chemical Company, Marietta, Ga. Desmodium (Desmodium ovalifolium) is a tropical forage legume which may contain more than 29% condensed tannin on a dry matter basis (17). Lyophilized desmodium was obtained from Honduras, and freeze-dried myrtle (Mirtus communis), a browse species from the Mediterranean region, was obtained from Sardinia. The reagents used, including acetic, propionic, and butyric acids, were analytical grade.
Condensed tannin purification.
Condensed tannins were purified with Sephadex LH-20 by using the method of Asquith and Butler (2), as modified by Hagerman and Butler (20).
Culture technique and media.
The anaerobic culture techniques of Hungate (22), with modifications (5), were used for all incubations. Rum 10 medium (26) was used for initial transfers of ruminal fluid from foreign animals before isolation of the tannin-tolerant bacteria. Ruminal fluid medium (6), which was used for all incubations except those in which the ability to utilize or tolerate different substrates was tested, contained 0.3% (wt/vol) glucose as the carbon source. Filter-sterilized carbohydrates were added after autoclaving. Ruminal fluid for the medium was obtained from a nonlactating cow fed medium-quality grass hay without tannins. The ruminal fluid was clarified aerobically by centrifugation at 20,000 × g for 20 min at 4°C.
For amino acid and casein utilization studies, we used modified Rum 10 medium (26) in which an amino acid mixture replaced the casein and a vitamin solution (8) replaced the yeast extract. The amino acid mixture consisted of 7.30% methionine, 18.25% cysteine, 14.60% threonine, 5.47% histidine, 8.76% arginine, 12.77% lysine, 14.60% tyrosine, 5.47% tryptophan, and 12.77% phenylalanine (37). The 0.3 g of casein in the original formulation was replaced by 0.3 g of the amino acid mixture, and the yeast extract was replaced by 1 ml of the vitamin solution. Defined medium (8) containing mineral salts, a vitamin solution, cysteine hydrochloride, and a volatile fatty acid solution was used for all other studies of carbohydrate, ammonia, and phenolic compound utilization and phenolic compound tolerance.
Medium was transferred in 9.7-ml portions to butyl rubber-stoppered Balch anaerobic culture tubes (Bellco Glass Inc., Vineland, N.J.) purged with oxygen-free CO2. The tubes were sterilized by autoclaving them at 120°C for 15 min. The pH of the medium after autoclaving was between 6.5 and 6.7. All incubations were performed anaerobically in batch culture at 39°C. When hydrolyzable tannins, condensed tannins, or phenolic monomers were included in the growth medium, they were added after autoclaving as prereduced, filter-sterilized solutions.
Isolation of bacteria.
Attempts were made to isolate bacteria from five species of ruminants in five different geographical locations, including Sardinian sheep, Honduran goats, Colombian goats, Rocky Mountain elk from Oregon, and domesticated cattle and white-tail deer from upstate New York. All of the animals except the cattle had consumed material containing tannins. The goats in Honduras and Colombia had been fed diets containing a tropical forage legume (Desmodium spp.), and the Sardinian sheep had been fed diets containing myrtle. White-tail deer in New York are known to consume oak (Quercus spp.), red maple (Acer rhubrum), and sugar maple (Acer saccharum) in the late fall. The Rocky Mountain elk in Oregon had been fed diets that contained fireweed (Epilobium angustifolium), red osier dogwood (Cornus stolonifera), Rocky Mountain maple (Acer glabrum), and alder (Alnus incana) (42). The cattle were fed predominantly grass hay and had never consumed feeds containing high levels of tannins.
Ruminal fluid was collected from sheep and goats orally with stomach tubes. Rocky Mountain elk ruminal contents were obtained with a stomach tube after the animals had been anesthetized, and samples of white-tail deer digesta were obtained from animals slaughtered during hunting season. Ruminal fluid from nonlactating Holstein cows was collected through fistulas.
Ruminal fluid from foreign animals was transferred twice in Rum 10 medium immediately after it was collected and prior to inoculation into ruminal fluid medium containing either tannic acid or quebracho at a concentration of 1 g/liter. The two successive transfers were necessary to meet importation requirements. Sardinian, Colombian, and Honduran samples were hand carried to Cornell University and arrived within 24 h of shipment. The elk ruminal fluid from Oregon was strained through cheesecloth and then inoculated directly into ruminal fluid medium containing either tannic acid or quebracho. Samples were shipped by overnight delivery and arrived within 24 h of collection. The elk samples were immediately frozen in a 20% glycerol solution and stored at −80°C. These samples were later thawed for culturing. The white-tail deer samples were hand carried in a thermos purged with oxygen-free CO2 immediately following slaughter of the animals. The samples were immediately strained through cheesecloth before being subcultured. Aliquots of the deer ruminal fluid were inoculated into enrichment medium within 6 h of collection.
Upon arrival at our laboratory, the ruminal bacteria from goats, sheep, and white-tail deer were subcultured three times with 2-day intervals between transfers in ruminal fluid medium containing either 1 g of tannic acid per liter or 1 g of quebracho per liter. The Rocky Mountain elk samples were thawed anaerobically before being subcultured. After the initial transfer, cultures were plated on ruminal fluid medium (6) containing 3% (wt/vol) agar and then were overlaid with 2 ml of a 10% (wt/vol) tannic acid solution or 2 ml of a 10% (wt/vol) purified quebracho solution. An anaerobic chamber (Coy, Ann Arbor, Mich.) with an atmosphere containing 94% CO2 and 6% H2 was used for plating experiments. Isolated colonies were selected and inoculated into ruminal fluid broth medium containing tannic acid or unpurified quebracho at a concentration of 1 g/liter. Weekly transfers were necessary for survival of the cultures. For long-term storage, cultures were frozen in 20% glycerol and stored at −80°C.
The cell wall types of the strains isolated were determined by the Gram stain method. Cultures for which the Gram stain results were variable were tested for monensin susceptibility as previously described (40). Cell size was measured during the mid-exponential phase of growth by phase-contrast microscopy by using a confocal scanning microscope.
Growth studies.
For all growth studies, 0.3-ml portions of mid-exponential-phase cultures grown in ruminal fluid medium were inoculated into 9.7-ml portions of the appropriate medium. The isolates were tested for the ability to utilize ammonium chloride, amino acids, and casein as nitrogen sources and the ability to use different carbohydrates as energy sources. Carbohydrate fermentation characteristics and enzymatic activities were determined for the gram-negative rod-shaped organism strain KN4 with an API 20E commercial identification kit (API System, Montalieu Vercieu, France).
The ability to utilize hydrolyzable and condensed tannins and phenolic monomers as sole energy sources and the ability to tolerate these compounds in the presence of supplemental carbohydrates were evaluated. Responses to oxygen also were tested under aerobic and microaerophilic conditions.
To study the tolerance of the bacteria to different phenolic compounds, cultures were inoculated into defined medium (8) lacking glucose (negative control), defined medium containing glucose (positive control), and defined medium containing glucose and the compound of interest (phenolic monomer, tannic acid, unpurified quebracho, or desmodium that had been purified with Sephadex LH-20 [2], as modified by Hagerman and Butler [20]). Tolerance was assessed by determining the maximum concentration of added phenolic compound at which there was growth, as measured by a change in absorbance at 600 nm (A600) when cultures were compared with the appropriate blank.
The phenolic monomers tested included p-coumaric acid, ferulic acid, gallic acid, pyrogallol, and phloroglucinol. After they were dissolved in phosphate buffer (pH 6.8) (18), the phenolic monomers were filter sterilized and added to the medium at final concentrations of 1, 5, 10, 20, 30, 40, and 50 mM. Tannic acid, unpurified quebracho, and purified desmodium were added at concentrations of 1.0, 2.0, 2.5, 3.0, 3.5, 4.0, 8.0, and 10.0 g/liter. The effect on the growth rate was monitored by measuring changes in the A600 caused by bacterial growth compared with the A600 of a blank containing medium and the appropriate concentration of the phenolic compound being tested. Values were obtained with a Spectronic 601 spectrophotometer (Milton Roy, Rochester, N.Y.).
To determine the volatile fatty acid and lactate production profiles of isolated cultures, 1.5-ml aliquots of the culture medium were centrifuged at 4,000 × g for 5 min, and the supernatant was removed. A 360-μl aliquot of each sample was transferred to a microcentrifuge tube containing 40 μl of 50 mM H2SO4. After the tube contents were mixed and allowed to stand at room temperature for 10 min, each preparation was centrifuged again, and the supernatant was analyzed to determine the volatile fatty acid content by the high-pressure liquid chromatography method of Ehrlich et al. (11). A mixture of acetic, propionic, isobutyric, butyric, fumaric, and lactic acids was used as a calibration standard in all analyses.
To measure tannic acid degradation, cultures were grown in the presence of tannic acid at a concentration of 1 g/liter. At the end of each fermentation, 1-ml aliquots of culture medium were mixed with 50-mg portions of polyvinylpyrrolidone (PVP) to remove the residual tannic acid. Samples were incubated at room temperature for 60 min to permit PVP-tannin binding and then were centrifuged at 6,000 × g for 10 min. The affinity of phenolic monomers to PVP is low because of the limited number of hydroxyl groups available for binding (10). As a result, the breakdown products resulting from tannic acid degradation remained in the supernatants. One-milliliter aliquots of each supernatant were analyzed to determine the total phenolic compound content by using the Prussian blue method of Price and Butler (38), which quantified the concentration of phenolic hydroxyl groups in the sample but did not provide structural information on the phenolic compounds.
The presence of pyrogallol, phloroglucinol, and gallate was determined by gas chromatography-mass spectrometry. To detect pyrogallol, phloroglucinol, or gallate, trimethylsilyl derivatives were prepared and analyzed with a Hewlett-Packard model 5890 series 2 gas chromatograph. Isolated phenolic end products and nonvolatile components were prepared by extracting the supernatant from cells grown with the compounds in an equal volume of ethyl acetate and drying the extracts under CO2. Compounds were separated on an HP-5 column cross-linked with 5% phenyl methyl siloxane; the column was 30 cm long, had an internal diameter of 0.25 mm and a film thickness of 0.25 μm, and was obtained from Hewlett-Packard Co., Wilmington, Del. The temperature gradient used was 40 to 250°C. Chromatographic peaks were identified by comparison with a gas chromatography-mass spectrometry internal library or with authentic samples.
Degradation of hydrolyzable tannin-protein complexes was evaluated by a modification of the method of Osawa (32). The formation of zones of clearing when the bacteria were grown anaerobically on brain heart infusion agar plates (Difco Laboratories) which had been overlaid with 2 ml of a 10% (wt/vol) tannic acid solution was monitored. All of the isolates were tested simultaneously.
Extraction of DNA and amplification and cloning of the 16S rRNA genes.
Zirconium-silica beads (0.8 g of 0.1-mm-diameter beads) were added to 800-μl portions of cultures in the late-exponential phase of growth. The cells were disrupted with a Mini-BeadBeater-8 (Biospec Products Inc., Bartlesville, Okla.) for 3 min. Total DNA was extracted from each lysate with an equal volume of phenol-chloroform-isoamyl alcohol (50:49:1) (3). The DNA was resuspended in double-distilled water to the original volume (800 μl) and was used for the subsequent PCR.
The 16S rRNA gene was amplified by PCR (23) with a forward primer corresponding to nucleotide positions 8 to 27 of Escherichia coli 16S rRNA (24) (forward primer 8FPL; 5′-CGGATCCGCGCCGCTGCAGAGTTTGATCCTGGCTCAG-3′) and a primer corresponding to the reverse complement of positions 1510 to 1492 (reverse primer 1492RPL; 5′-GGCTCGAGCGGCCGCCCGGGTTACCTTGTTACGACTT-3′). Primer 8FPL is specific for members of the domain Bacteria, whereas 1492RPL is a universal primer and aligns with sequences conserved in all three phylogenetic domains (39). Each 50-μl reaction mixture contained each primer at a concentration of 0.1 μM, 0.2 mM dCTP, 0.2 mM dATP, 0.2 mM dGTP, 0.2 mM dTTP, 5 μl of a 10× Gibco PCR buffer (200 mM Tris-HCl [pH 8.4], 500 mM KCl) (Life Technologies, Grand Island, N.Y.), 2.5 U of Gibco Taq DNA polymerase, 3 mM MgCl2, and 5 μl of resuspended bacterial DNA, which corresponded to approximately 106 DNA molecules (23). Each amplification mixture was overlaid with mineral oil before it was incubated in an Amplitron II thermal cycler (Thermolyne, Dubuque, Iowa). The program consisted of 30 cycles consisting of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. There was a 5-min denaturation step at 94°C before the first cycle, and there was a 5-min extension step at 72°C after the 30th cycle. For each set of PCR, negative controls that contained no added template were included. Amplified DNA for cloning was electrophoresed and extracted from a 1.5% agarose gel by using a Qiaex II gel extraction kit (Qiagen, Hilden, Germany). The extracted DNA fragments were cloned into PCR 2.1 (Invitrogen, San Diego, Calif.). About 15 clones from each ligation transformation reaction were selected. Because each of the clones from the same ligation had the same insert size based on ligation product digests, one clone was selected for sequencing. Plasmid DNA was extracted by using the Wizard Minipreps DNA purification system (Promega). Extracts were prepared for sequencing by gel purification by using a BstEII digest of lambda DNA (US Biochemical Corporation, Cleveland, Ohio) as a molecular weight standard.
The purified plasmid preparations were sequenced directly with an ABI automated DNA sequencer by using a Prism dideoxy terminator cycle sequencing kit and the recommended sequencing protocol (Perkin-Elmer, Foster City, Calif.). Forward and reverse DNA-specific primers, as well as internal primers, were used for sequencing. The internal sequencing primers used to obtain the complete 16S ribosomal DNA sequences were P520 (5′-CAGCAGCCGCGGTAATAC; positions 520 to 537) and P1090 (5′-TTAAGTCCCGCAACGAGCG; positions 1090 to 1108).
Phylogenetic analyses.
Phylogenetic analyses were performed by using the PHYLIP phylogeny inference package, version 3.5c (12, 13). In the initial analysis, the sequences obtained were compared with the 16S rRNA sequences of organisms in the domain Bacteria obtained from the Ribosomal Database Project (RDP) (27) by using the SUGGEST TREE function. Aligned sequences of the resulting closest relatives and other sequences known to belong to the same phylogenetic group were retrieved from the RDP library. Our 16S rRNA sequences were manually aligned with these retrieved sequences.
Pairwise evolutionary distances were computed from the aligned sequences by using the PHYLIP program (12), DNADIST, and a Kimura model for nucleotide substitution, and trees were generated by using NEIGHBOR. The stability associated with treeing orders was evaluated by using the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE within the PHYLIP program. One hundred bootstrap trees were generated for each dataset.
Nucleotide sequence accession numbers.
The designations and GenBank accession numbers of the bacteria used in the phylogenetic analyses are listed below. The accession numbers of the bacteria used in the analysis of the streptococci (Fig. 1) were ATCC 43077, ATCC 27823, ATCC 27335, ATCC 33317, NCDO 597, ACM 3969, ATCC 19642, ATCC 33748, ATCC 9812, X94337, ATCC 35911, NCTC 3165, ATCC 25175, ATCC 19615, ATCC 19645, ATCC 15300, ATCC 33478, ATCC 27958, and NCDO 2156.
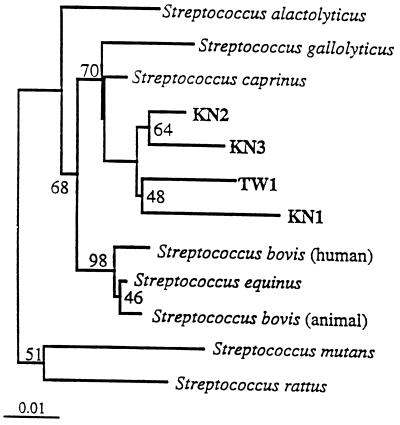
FIG. 1.
Phylogenetic tree based on the 16S rRNA sequences (>1,460 bp) of four recently isolated tannin-tolerant streptococci (KN1, KN2, KN3, and TW1), the sequences of some of their close relatives, and sequences obtained from the RDP (27). Lactococcus garviaea was used as the outgroup in this analysis. Branching distances were computed by using DNADIST and FITCH in PHYLIP (12). Scale bar = 1 inferred nucleotide substitution per 100 nucleotides.
The bacteria and accession numbers used in the analysis of the low-G+C-content gram-positive bacterium were X75788, ATCC 824, L04165, M23927, ATCC 25763, ATCC 25783, ATCC 35319, ATCC 25537, X73450, ATCC 43204, ATCC 19401, L11305, X73447, ATCC 29065, ATCC 25620, ATCC 17861, ATCC 13124, ATCC 49002, ATCC 33906, ATCC 33455, ATCC 25775, ATCC 19403, ATCC 25781, L04167, ATCC 25774, X66000, X72868, DSM 1237, L09176, ATCC 7956, NCTC 279, ATCC 49623, X62176, M23728, M99574, ATCC 43171, ATCC 43058, DSM4000, ATCC 29066, ATCC 35991, M59120, DSM 3985, D14150, D14149, D14143, ATCC 49914, and R12B1 (a cellulolytic thermophile).
The accession numbers or names of the bacteria used in the analysis of the enteric bacterium (Fig. 2) were Y13249, M25588, AE000452, E05133, E05134, ATCC 7001, Z96978, X80682, Z96978, X80682, U90316, X70906, Escherichia coli 5 (= c600), Salmonella dublin, Salmonella typhi, ATCC 9610, Z75320, ATCC 19428, and Z21939.
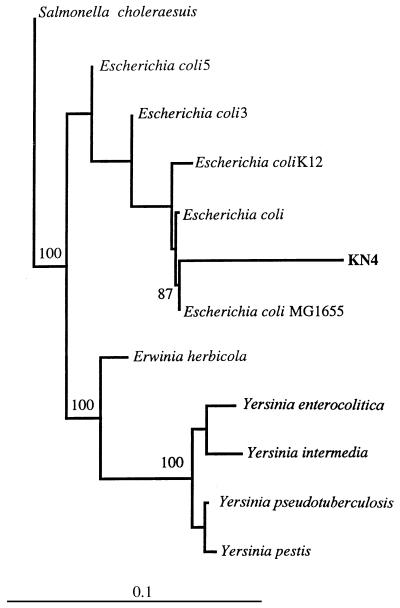
FIG. 2.
Phylogenetic tree based on the 16S rRNA sequences (>1,460 bp) of a recently isolated tannin-protein complex-degrading bacterium (KN4) isolated from the rumen of a white-tail deer and some of its close relatives. Clostridium sardiensis was used as the outgroup in this analysis. Branching distances were computed by using DNADIST and FITCH in PHYLIP (12). Scale bar = 10 inferred nucleotide substitutions per 100 nucleotides.
The nucleotide sequences of strains KN1, KN2, KN3, KN4, TW1, and TW2 have been deposited in the GenBank database under accession no. AF084832, AF084833, AF084834, AF084835, AF084836, and AF084837, respectively.
RESULTS
Physiological characteristics.
When ruminal contents were plated onto agar overlaid with tannic acid, we were able to isolate colonies from digesta of all of the animals except the cows at Cornell University. Despite repeated attempts at enrichment and isolation, we were unable to isolate tannin-tolerant bacteria from the cows, which had no history of tannin consumption. Six tannin-tolerant colonies, two each from goats and elk and one each from sheep and deer, were selected for further characterization. None of these bacteria was able to grow in medium containing condensed tannins as the sole carbon source. Based on phylogenetic and phenotypic characterization, the bacteria fell into three groups, which are discussed below.
Strains KN1, KN2, KN3, and TW1 were gram-positive cocci that were isolated from the ruminal contents of a goat in Colombia, a goat in Honduras, a sheep in Sardinia, and a Rocky Mountain elk in the United States, respectively. The isolate from the Colombian goat, KN1, was described elsewhere recently (30). A gram-negative rod-shaped organism, strain KN4, was isolated from the ruminal contents of a white-tail deer from upstate New York, and strain TW2 is a gram-positive rod-shaped organism that was isolated from Rocky Mountain elk ruminal contents in Oregon (Table 1). Some of the physiological and biochemical characteristics of the isolates are shown in Tables 1 and 2. All of the isolates were able to utilize ammonia, amino acids, or casein as a sole N source and grew on glucose, lactose, galactose, or cellobiose as a carbon source.
TABLE 1.
Phenotypic characteristics of six ruminal tannin-tolerant isolates from animals from different geographical locationsa
| Strain | Geographic origin | Animal originb | Morphology | Gram stain reaction | Color of coloniesc | Cell size (μm) | Aerotoleranced | End products | Utilization of C sources
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Starch | Arabinose | Sucrose | Fructose | Mannitol | Cellulose | |||||||||
| KN1 | Colombia | Goat | Cocci | + | White | 0.82 | + | Lactate, fumarate | + | + | + | + | − | − |
| KN2 | Sardinia | Sheep | Cocci, chains | + | White | 0.75 | + | Lactate, fumarate | + | + | + | + | + | − |
| KN3 | Honduras | Goat | Single cocci | + | White | 0.83 | + | Lactate, fumarate | + | − | + | + | + | − |
| TW1 | United States | Elk | Cocci, chains | + | White | 0.85 | + | Lactate, acetate | + | + | + | + | + | − |
| TW2 | United States | Elk | Rods | + | Clear | 3.2 × 1.0 | − | Lactate, acetate, butyrate | − | + | + | − | − | + |
| KN4 | United States | Deer | Rods | − | Yellow | 1.9 × 1.0 | + | Lactate, succinate, fumarate | − | + | − | + | + | − |
The bacteria were grown in defined medium (8) without tannins unless indicated otherwise.
The bacteria were obtained from animal rumens.
Cells were grown on rumen fluid medium plates containing 3% (wt/vol) agar.
Ability of the cells to grow in the presence of oxygen and in aerobic medium.
TABLE 2.
Highest concentrations of phenolic monomers and tannins at which six tannin-tolerant ruminal bacteria and S. bovis JB1 grewa
| Strain | Highest concn at which bacteria grew
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Pyrogallol (mM) | Phloroglucinol (mM) | p-Coumaric acid (mM) | Ferulic acid (mM) | Gallic acid (mM) | Tannic acid (g/liter) | Unpurified quebracho (g/liter) | Purified desmodium (g/liter) | |
| KN1 | 40 | 30 | 50 | 50 | 50 | 30 | 8 | 0.8 |
| KN2 | 30 | 30 | 30 | 30 | 30 | 2 | 4 | 0.7 |
| KN3 | 10 | 5 | 20 | 10 | 30 | 2 | 2 | 0.6 |
| KN4 | 5 | 5 | 5 | 5 | 10 | 2 | 8 | 0.4 |
| S. bovis JB1 | 5 | 5 | 5 | 5 | 5 | 0.2 | 1 | 0.002 |
| TW1 | 30 | 10 | 30 | 30 | 20 | 8 | 8 | 0.6 |
| TW2 | 20 | 20 | 20 | 20 | 20 | 2 | 8 | 0.5 |
Growth was defined as a change in A600 when inoculated medium was compared with a blank containing medium and the phenolic compound being studied. All tests were performed in defined medium (8).
The four gram-positive cocci (KN1, KN2, KN3, and TW1) had physiological characteristics which suggested that they were very similar to each other. Three of these isolates, KN2, KN3, and TW1, fermented mannitol (Table 1), a characteristic which is typical of human Streptococcus bovis isolates (29). Two of the four gram-positive cocci, KN1 and TW1, could degrade tannic acid with gallic acid and pyrogallol as end products. Zones of clearing were evident on plates of brain heart infusion medium overlaid with tannic acid with two of the gram-positive cocci, KN1 and TW1, while KN1 was the bacterium that was best able to tolerate phenolic monomers and hydrolyzable and condensed tannins. All of the recently isolated gram-positive cocci examined were able to tolerate between 300 and 400 times as much purified desmodium and between 10 and 150 times as much tannic acid as another gram-positive coccus, S. bovis JB1, which turned out to be a close relative of these organisms (see below). Differences in the ability to tolerate phenolic monomers also were evident.
All of the gram-positive cocci were facultative anaerobes, could utilize ammonia in the growth medium as a sole nitrogen source, and produced lactic acid as an end product. Three of the newly isolated gram-positive cocci (KN1, KN2, and KN3) produced lactate and formate, while TW1 produced lactate and acetate.
The gram-negative rod-shaped organism KN4 was the only isolate that produced zones of clearing on tannin-protein agar plates. This isolate tolerated far lower concentrations of phenolic monomers and tannins than the other isolates. KN4 grew on a variety of sugars but was not able to utilize sucrose, starch, or cellulose. Lactate, succinate, and formate were the only detectable fermentation products. It grew well aerobically. The API 20E results indicated that KN4 had ornithine decarboxylase, β-galactosidase, arginine dihydrolase, and lysine decarboxylase activities but was not able to hydrolyze urea, did not produce hydrogen sulfide, and did not have tryptophan deaminase or oxidase activity. It could utilize sorbitol, rhamnose, and melibiose but not citrate, gelatin, inositol, saccharose, or amygdalin.
The gram-positive, rod-shaped organism TW2 was a strict anaerobe. Endospores were not detected microscopically, and no growth occurred after incubation at 80°C for 10 min. This bacterium utilized a variety of sugars and was capable of degrading cellulose. This isolate produced lactate, acetate, and butyrate as fermentation products.
Phylogenetic analysis of the 16S rRNA sequences.
The use of primers which corresponded to more than one conserved region allowed us to generate nearly complete 16S rRNA sequences for the six isolates (24). The sequences corresponded to positions 28 to 1491 of the E. coli 16S rRNA sequence. When the four gram-positive cocci were compared to representatives of the Bacteria included in the RDP database library, they formed a distinct cluster within the genus Streptococcus of the lactic acid bacteria and were most closely related to the recent ruminal isolates Streptococcus caprinus (4) and Streptococcus gallolyticus (34). The intercluster identity values for the 16S rRNA sequences of these four tannin-tolerant cocci ranged from 94 to 97%. The phylogenetic gene tree which resulted from our multiple-sequence analysis is shown in Fig. 1.
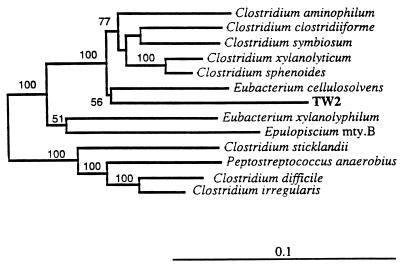
The gram-positive Rocky Mountain elk isolate (TW2) is a previously undescribed member of the genus Eubacterium. Figure 3 is a phylogenetic tree showing the evolutionary distances between this isolate and some other low-G+C-content gram-positive bacteria. Based on the phylogenetic classification of Collins et al. (7), TW2 is a member of subcluster XIVa, the subcluster that contains its closest relative, Eubacterium cellulosolvens, which also is cellulolytic.
FIG. 3.
Phylogenetic tree based on the 16S rRNA sequences (>1,460 bp) of a recently isolated anaerobic, tannin-tolerant bacterium (TW2) and some of its close relatives. Syntrophobacter wolinii was used as the outgroup in this analysis. Branching distances were computed by using DNADIST and FITCH in PHYLIP (12). Scale bar = 10 inferred nucleotide substitutions per 100 nucleotides.
Gram-negative isolate KN4, which was obtained from the ruminal contents of white-tail deer, is an enteric bacterium. Figure 2 is a phylogenetic tree which shows the positions of this isolate and some its relatives. Strain KN4 clustered with the gram-negative enteric bacteria in the γ subdivision of the class Proteobacteria in the domain Bacteria. Of the organisms in the RDP database, it was most closely related to, but was not identical to, several strains of E. coli.
Similarities to other tannin-tolerant and tannin-protein complex-degrading bacteria.
None of the six isolates exhibited 100% rRNA sequence similarity with any available 16S rRNA gene sequence. The highest levels of similarity were between the gram-positive bacteria and their closest relatives, S. caprinus and S. gallolyticus. S. caprinus is a tannin-tolerant bacterium that was isolated from the ruminal contents of a feral goat (4). Recent comparisons have revealed that S. caprinus 2.3 is the same organism as S. gallolyticus, which was isolated from several species of animals, including koalas, pigs, horses, and dogs (41). All of the S. gallolyticus isolates that have been characterized have gallate decarboxylase activity. As S. gallolyticus was isolated before S. caprinus, the name S. gallolyticus has nomenclatural priority (41). We included both 16S rRNA sequences and names in our phylogenetic analyses. The four recent tannin-tolerant isolates obtained in our study clustered near S. gallolyticus and S. caprinus.
DISCUSSION
Sequencing of the 16S rDNA enabled us to compare the recently isolated tannin-tolerant bacteria with available bacterial sequences in the EMBL and RDP (27) databases. Four of the six isolates which were examined formed a tight cluster within the genus Streptococcus. The studies of S. gallolyticus and S. caprinus of Osawa et al. (34) and Brooker et al. (4) indicated that there is a group of streptococci that are able to tolerate hydrolyzable and condensed tannins, as well as phenolic monomers. Some members of this group which had diverse origins also have gallate decarboxylase and/or tannase activities.
Osawa et al. (34) and Brooker et al. (4) identified bacteria isolated in Australia and Japan. In the present study we examined ruminal contents from three continents, Europe, North America, and South America, and found that the ability of ruminal bacteria to tolerate and/or degrade tannins is widespread. The ruminal contents of domestic temperate cattle from Cornell University did not yield tannin-tolerant bacteria. This was most likely because the animals had not previously consumed material containing tannins. S. bovis JB1, a well-studied ruminal strain, was quite intolerant of tannins and other phenolic compounds (Table 2).
In addition to having similar phylogenetic positions, three of our four ruminal streptococci could ferment mannitol, as could 90% of the S. gallolyticus strains isolated by Osawa et al. (34). S. bovis JB1 was not able to ferment mannitol. Our streptococci, however, could tolerate much higher levels of tannins and other phenolic compounds than S. bovis JB1 but could not cleave tannin-protein complexes, as determined by the zone-clearing method described by Osawa et al. (34) (Table 2). Some of the other important differences between our isolates and the strains isolated in Australia were the inability of S. caprinus or S. gallolyticus to use ammonia as a sole source of nitrogen and the production of different fermentation products. Our isolates produced lactate, fumarate, and acetate predominantly, while S. caprinus and S. gallolyticus produced small amounts of acetate and ethanol in addition to lactate.
Based on the results of biochemical characterization with an API test kit, gram-negative isolate KN4 is a member of a subgroup of E. coli (subgroup 7144552). This subgroup is very heterogeneous, and its members are uncommon strains of E. coli. The phylogenetic analyses demonstrated that KN4 is nested among E. coli strains and therefore most likely is an E. coli strain. An isolate from the rumen of a white-tail deer, KN4 is phenotypically similar to tannin-protein complex-degrading enterobacteria isolated from the alimentary tracts of koalas and described by Nemoto et al. (31) and Osawa (33). Osawa’s isolates were variable in size and included a mixture of short rods and long filamentous rods. These rods produced zones of clearing on tannin-protein complex plates but were not able to use ammonia as a sole nitrogen source. They also could not ferment mannitol. These isolates have not been characterized yet at the genotypic level, so our comparisons were limited to biochemical data.
Osawa (33) suggested that his enterobacteria were related to Enterobacter agglomerans (reclassified as Pantoea agglomerans [16]), which is a human pathogen isolated from plants and animals. The 16S rRNA sequence of P. agglomerans was not available so it could not be included in our phylogenetic comparisons, but phenotypic and genetic characterization of P. agglomerans (16) showed that it consists of several biogroups. In addition, there is an Erwinia-P. agglomerans complex with at least 10 relatedness groups, all of which have taxonomic problems. Although KN4 may constitute another relatedness group within the Erwinia-P. agglomerans complex, it is different from the type strain of P. agglomerans, which is negative for ornithine decarboxylase, arginine dihydrolase, and lysine decarboxylase and can grow on sucrose.
Gram-positive rod-shaped strain TW2, which was isolated from the ruminal contents of a Rocky Mountain elk, was closely related to E. cellulosolvens, a gram-positive cellulolytic rod (Fig. 3). E. cellulosolvens has been isolated from the ruminal contents of sheep and cows and the intestinal tract of a hog (25). Based on the phylogenetic reclassification proposed by Collins et al. (7), isolate TW2 is a member of the phenotypically diverse taxon subcluster XIVa, and, not surprisingly, its closest neighbor is cellulolytic.
In this study, we identified three phylogenetically distinct groups of ruminal bacteria which tolerate tannins and other phenolic compounds. Not only do these bacteria differ phylogenetically, but the tannin-tolerant enterobacteria and streptococci are the first groups of tannin-tolerant bacteria known to have members that occur in markedly different geographical locations. All of the tannin-tolerant bacteria described here were able to use both ammonia and amino acids as N sources, suggesting that tannin tolerance is not dependent on the form of N utilized. Climate, geography, and host animal do not seem to affect the ability of the bacteria to tolerate tannins. This was demonstrated by the streptococcal isolates described in this paper. These isolates are similar in both biochemical characteristics and 16S rRNA sequences, yet they were isolated from four different species of animals living in temperate and tropical climates on three continents. Our phylogenetic analysis suggested that our streptococci (KN1, KN2, KN3, and TW1, as well as S. caprinus and S. gallolyticus) and the ancestor of the S. bovis complex have a common ancestor (Fig. 1).
Examination of the genomes of tannin-tolerant isolates for the genes which confer tannin tolerance, complete evaluation of the structure of the proteins associated with tannin tolerance, and ecological studies which allow monitoring of these novel bacteria in their natural setting are the next logical steps in interpreting interactions between tannins and bacteria.
ACKNOWLEDGMENTS
We thank Yueh-Tyng Chien, Grace Lee, and Dave Hinman for advice on experiments. John Cook, a wildlife research biologist at the National Council of the Paper Industry for Air and Stream Improvement, LaGrande, Oreg., generously let us use his elk and provided useful advice which enabled us to collect ruminal samples from the elk. We are grateful to Miguel Vélez and Antonio Flores (Pan American School for Agriculture, Tegucigalpa, Honduras), Antonello Cannas (University of Sassari, Sassari, Sardinia, Italy), E. John Pollak (Cornell University), and Carlos Lascano (C.I.A.T, Cali, Colombia) for providing ruminal fluid. The technical assistance and recommendations of James Van Ee and Carol Bayles at the Sequencing Facility of the New York Center for Advanced Technology at Cornell University were greatly appreciated.
REFERENCES
- 1.Allison M J, Hammond A C, Jones R J. Detection of ruminal bacteria that degrade toxic dihydroxypyridine compounds produced from mimosine. Appl Environ Microbiol. 1990;56:590–594. doi: 10.1128/aem.56.3.590-594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asquith T N, Butler L G. Use of dye-labeled protein as spectrophotometric assay for protein precipitants such as tannin. J Chem Ecol. 1985;11:1535–1544. doi: 10.1007/BF01012199. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: Wiley and Sons; 1995. [Google Scholar]
- 4.Brooker J D, O’Donovan L A, Skene I, Clarke K, Blackall L, Muslera P. Streptococcus caprinus sp. nov., a tannin-resistant ruminal bacterium from feral goats. Lett Appl Microbiol. 1994;18:313–318. [Google Scholar]
- 5.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 6.Bryant M P, Burkey L A. Cultural methods and some characteristics of some of the more numerous groups of bacteria in the bovine rumen. J Dairy Sci. 1953;36:205–217. [Google Scholar]
- 7.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 8.Cotta M A, Russell J B. Effect of peptides and amino acids on efficiency of rumen bacterial protein synthesis in continuous culture. J Dairy Sci. 1982;65:226–234. [Google Scholar]
- 9.Deschamps A M. Nutritional capacities of bark and wood decaying bacteria with particular emphasis on condensed tannin degrading strains. Eur J For Pathol. 1982;12:252–257. [Google Scholar]
- 10.Doner L W, Bécard G, Irwin P L. Binding of flavonoids by polyvinylpolypyrrolidone. J Agric Food Chem. 1993;41:753–757. [Google Scholar]
- 11.Ehrlich G G, Goerlitz D F, Bourell J H, Eisen G V, Godsy E M. Liquid chromatographic procedure for fermentation product analysis in the identification of anaerobic bacteria. Appl Environ Microbiol. 1981;42:878–885. doi: 10.1128/aem.42.5.878-885.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 14.Field J A, Lettinga G. The methanogenic toxicity and anaerobic degradability of a hydrolyzable tannin. Water Res. 1987;21:367–374. [Google Scholar]
- 15.Field J A, Leyendeckers M J H, Alvarez R S, Lettinga G, Habets L H A. The methanogenic toxicity of bark tannins and the anaerobic biodegradability of water soluble bark matter. Water Sci Technol. 1988;20:219–240. [Google Scholar]
- 16.Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, DeLey J. Transfer of Enterobacter agglomerans Beijerinck 1888 Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersas. Int J Syst Bacteriol. 1989;39:337–345. [Google Scholar]
- 17.Giner-Chavez B, Van Soest P J, Robertson J B, Pell A N, Lascano C E, Reed J D. A method for isolating condensed tannins from crude plant extracts with trivalent ytterbium. J Sci Food Agric. 1997;74:359–368. [Google Scholar]
- 18.Goering H K, Van Soest P J. Forage fiber analysis (apparatus, reagents, procedures, and some applications) US Dep Agric Agric Handb. 1970;379:1–20. [Google Scholar]
- 19.Hagerman A E, Butler L G. Choosing appropriate methods and standards for assaying tannin. J Chem Ecol. 1989;15:1795–1810. doi: 10.1007/BF01012267. [DOI] [PubMed] [Google Scholar]
- 20.Hagerman A E, Butler L G. Assay of condensed tannins or flavonoid oligomers and related flavonoids in plants. Methods Enzymol. 1994;234:429–437. doi: 10.1016/0076-6879(94)34113-3. [DOI] [PubMed] [Google Scholar]
- 21.Huang L, Forsberg C W, Lam J S, Cheng K J. Antigenic nature of the chloride-stimulated cellobiosidase and other cellulases of Fibrobacter succinogenes subsp. succinogenes S85 and related fresh isolates. Appl Environ Microbiol. 1990;56:1229–1234. doi: 10.1128/aem.56.5.1229-1234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hungate R E. The anaerobic mesophilic cellulolytic bacteria. Bacteriol Rev. 1950;14:1–49. doi: 10.1128/br.14.1.1-49.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. New York, N.Y: Academic Press; 1990. pp. 3–12. [Google Scholar]
- 24.Johnson J L. Similarity analysis of rRNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 683–700. [Google Scholar]
- 25.Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Baltimore, Md: The Williams and Wilkins Co.; 1984. [Google Scholar]
- 26.Latham M J, Brooker B E, Pettipher G L, Harris P J. Ruminococcus flavefaciens cell coat and adhesion to cotton cellulose and to cell walls in leaves of perennial ryegrass (Lolium perenne) Appl Environ Microbiol. 1978;35:156–165. doi: 10.1128/aem.35.1.156-165.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;24:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller-Harvey I, McAllan A B, Theodorou M K, Beever D E. Phenolics in fibrous crop residues and their effects on the digestion and utilisation of carbohydrates and proteins in ruminants. In: Reed J D, Capper B S, Neate P J H, editors. Plant breeding and the nutritive value of crop residues. Addis Ababa, Ethiopia: ILCA; 1988. pp. 97–132. [Google Scholar]
- 29.Nelms L F, Odelson D A, Whitehead T R, Hespell R B. Differentiation of ruminal and human Streptococcus bovis strains by DNA homology and 16S rRNA probes. Curr Microbiol. 1995;31:294–300. doi: 10.1007/BF00314583. [DOI] [PubMed] [Google Scholar]
- 30.Nelson K E, Pell A N, Schofield P, Zinder S. Isolation and characterization of an anaerobic hydrolyzable tannin-degrading bacterium. Appl Environ Microbiol. 1995;61:3293–3298. doi: 10.1128/aem.61.9.3293-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemoto K, Osawa R, Hirota K, Ono T, Miyake Y. An investigation of gram-negative tannin-protein complex degrading bacteria in fecal flora of various mammals. J Vet Med Sci. 1995;57:921–926. doi: 10.1292/jvms.57.921. [DOI] [PubMed] [Google Scholar]
- 32.Osawa R. Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Appl Environ Microbiol. 1990;56:829–831. doi: 10.1128/aem.56.3.829-831.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osawa R. Tannin-protein complex-degrading enterobacteria isolated from the alimentary tracts of koalas and a selective medium for their enumeration. Appl Environ Microbiol. 1992;58:1754–1759. doi: 10.1128/aem.58.5.1754-1759.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osawa R, Fujisawa T, Sly L I. Streptococcus gallolyticus sp. nov., gallate degrading organisms formerly assigned to Streptococcus bovis. Syst Appl Microbiol. 1995;18:74–78. [Google Scholar]
- 35.Osawa R, Rainey F A, Fujisawa T, Lang E, Busse H J, Walsh T P, Stackebrandt E. Lonepinella koalarum gen. nov., sp. nov., a new tannin-protein complex degrading bacterium. Syst Appl Microbiol. 1995;18:368–373. [Google Scholar]
- 36.Osawa R, Walsh T P, Cork S J. Metabolism of tannin-protein complex by facultatively anaerobic bacteria isolated from koala feces. Biodegradation. 1993;4:91–99. [Google Scholar]
- 37.Pichard G R. Forage nutritive value. Continuous and batch in vitro rumen fermentations and nitrogen solubility. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1977. [Google Scholar]
- 38.Price M L, Butler L G. Rapid visual estimation and spectrophotometric determination of tannin content of sorghum grain. J Agric Food Chem. 1977;25:1268–1277. [Google Scholar]
- 39.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell J B. A proposed mechanism of monensin action in inhibiting ruminal bacterial growth: effects on ion flux and protonmotive force. J Anim Sci. 1987;64:1519–1525. doi: 10.2527/jas1987.6451519x. [DOI] [PubMed] [Google Scholar]
- 41.Sly L I, Cahill M M, Osawa R, Fujisawa T. The tannin-degrading species Streptococcus gallolyticus and Streptococcus caprinus are subjective synonyms. Int J Syst Bacteriol. 1997;47:893–894. doi: 10.1099/00207713-47-3-893. [DOI] [PubMed] [Google Scholar]
- 42.Woolston, T. K. 1995. Personal communication.