Abstract
The escalating pandemic brought about by the novel SARS-CoV-2 virus is threatening global health, and thus, it is necessary to develop effective antiviral drugs. Usenamine A is a dibenzo-furan derivative separated from lichen Usnea diffracta showing broad-spectrum activity against different viruses. We evaluate that usenamine A has antiviral effects against novel SARS-CoV-2 Delta variant pseudotyped viruses (PVs) in A549 cells. In addition, usenamine A significantly suppresses SARS-CoV-2 PV-induced mitochondrial depolarization, elevated reactive oxygen species (ROS) levels, apoptosis, and inflammation. Usenamine A also causes the SARS-CoV-2 spike protein to become less stable. Thus, usenamine A shows potential as an antiviral drug that can provide protection against COVID-19.
Keywords: Natural products, SARS-CoV-2, Pseudotyped viruses, Antiviral activity, Reactive oxygen species
1. Introduction
The coronavirus (COVID-19) pandemic that resulted from the outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has seriously damaged public health and has brought about economic burden worldwide [1]. The SARS-CoV-2 spike protein is highly conservative properties, which is a key factor in its infection pathway [2,3]. The S1 domain is mainly responsible for binding the virus to receptors, and the S2 domain mainly contains HR domains that are closely related to viral fusion [4,5]. The SARS-CoV-2 spike protein binds to cells that it recognizes through the cell surface receptor ACE2. The virus enters the host cell and spike protein proteolysis activates the host to encode proteases, which remarkably affects replication and other coordinated processes [6,7]. Inhibiting the spike protein may mitigate the invasion of the virus into host cells, and this could serve as a protective therapeutic strategy against SARS-CoV-2 infections [8].
However, SARS-CoV-2 is a biosafety level-3 (BSL-3) pathogen, and experiments with actual pathogenic SARS-CoV-2 samples entail stricter requirements corresponding to biosafety level-3 (BSL-3) laboratory conditions. These restrictions make it difficult to perform antiviral evaluations [9]. Replication-restricted PVs that bear viral coat proteins can mimic some biological properties of actual viruses in vivo, and thus, they can serve as safe and useful alternatives for antiviral research under less stringent biosafety conditions [10]. The novelty of this research paper in the context of SARS-CoV-2 PVs lies in the utilization of liposomes. The PVs are primarily composed of liposomes containing the SARS-CoV-2 spike protein within their membranes, and their genetic composition closely mirrors that of a live SARS-CoV-2 virus but with a restricted capacity for replication within host cells. Additionally, these PVs encapsulate spike RNA from SARS-CoV-2, allowing for the monitoring of PV infection levels through PCR methods. Consequently, the PVs serve as a secure platform for emulating the properties of highly pathogenic viruses without necessitating high-level biosafety facilities. This article focuses on discussing the construction of a SARS-CoV-2 Delta variant pseudoviral production system and the potential use of such pseudoviruses for cell infection experiments.
According to the clinical data from studies on the novel coronavirus study, severe inflammation and lung damage can result from uncontrolled cytokine storms caused by the virus once patients are infected [11,12], and the intensity of such storms is linked to the severity of the disease. IL-1β is the main inflammatory mediator in the human body and can be inhibited to alleviate cytokine storms. Additionally, IL-6 is a multifunctional cytokine produced by immune cells to trigger inflammatory responses through multiple pathways [13]. TNF-α is the main pro-inflammatory cytokine in acute and chronic inflammation [14], the level of which may be significantly increased in the serum of patients with new coronary pneumonia [15,16]. Therefore, these three inflammatory factors were used in this study to assess the severity of cellular inflammation.
Despite the remarkable advancements in vaccine development for COVID-19, the search for effective therapeutics remains a crucial endeavor in controlling the pandemic [17]. Small molecule drugs have long been recognized as potent tools against viral infections due to their ability to target specific viral or host functions [18,19]. However, with the SARS-CoV-2 virus, many current small molecule drugs have exhibited limited efficacy or fewer bioavailability challenges in administration or resistance [20,21]. Natural products, with their rich history and variety, offer a reservoir of bioactive compounds that have the potential to address these challenges. Several natural products have demonstrated potent antiviral properties against various viral pathogens, suggesting their potential in the context of COVID-19 [22,23]. Furthermore, natural products as immunomodulatory agents in the context of aid in public health have garnered significant attention [24,25]. Immunomodulators hold promise in strengthening the body's immune response, mitigating inflammation, and enhancing antiviral immunity [26]. Considering this, the integration of insights from natural products into modern therapeutic development may offer an irreplaceable strategy in the fight against SARS-CoV-2.
Usnea sp. is a fruticose lichen renowned for its medicinal significance in various traditional practices. Several researchers have reported the antibacterial activity of the Usnea sp. even before the antibiotic era. Usnea sp are known to exhibit promising antibacterial effects on Gram-positive strains, including Staphylococcus aureus and Bacillus sp. For instance, U. florida and U. antarctic show antibacterial activity against S. aureus [27,28]. The diverse biomolecules in lichen extracts often correlate with its antioxidant activities [29]. The antioxidant potential of U. longissima methanolic extract showed potential DPPH scavenging activity [30]. Furthermore, extracts of Usnea sp have also exhibited anticancer properties. For example, the extract of U. barbata illustrated dose-dependent cytotoxicity in B16 (mouse melanoma cell line) and C6 (rat glioma cell line) yet remained non-toxic to the non-cancerous human keratinocyte HaCaT cell line [31]. Meanwhile, the methanol extract of U. filipendula induced apoptosis in human breast cancer cells (MCF-7 cell line and MDA-MB 231 cell line [32].
Small molecule drugs are excellent complements to vaccines as prophylactic and post-exposure therapeutic agents [18]. However, current small molecule drugs suffer from poor efficacy. As a key component of the COVID-19 treatment regimen, bioactive natural products [33] and traditional medicine may play an irreplaceable role in the treatment of SARS-CoV-2 infections [34]. Usnea diffracta, which belongs to the Usneaceae family, grows as a moss parasite in humid climates [35]. Like other varieties of Usneaceae, Usnea diffracta is widely known as “songluo” in China and Korea and is applied as a medicinal product to treat different ailments. In previous chemical research studies on Usnea diffracta, it has been suggested that the presence of phenyls, depsides, anthraquinones, dibenzofurans, and terpenoids could produce insecticidal, antitumor, antiviral, antibacterial, and anti-inflammatory effects [[36], [37], [38]]. In terms of antiviral efficacy, it is widely believed that Usnea diffracta affects the RNA replication of viruses [[39], [40], [41]].
This study presents a biological evaluation of usenamine A, a dibenzofuran compound extracted from the ethyl acetate part of Usnea diffracta, in SARS-CoV-2.
2. Materials and methods
2.1. Isolation of usenamine A
Ultrasonication was employed to extract dry Usnea diffracta (1.5 kg) three times with 100 % MeOH at room temperature overnight. We removed the solvent in vacuo to obtain a methanol extract (285.7g), which was suspended in distilled water and successively partitioned with EtOAc (2Lx3) or n-butanol (2Lx3) to generate crude extracts of EtOAc (125.4g) and n-butanol (80.1g), respectively. The EtOAc fraction was treated with silica gel CC and eluted with a gradient of CH2CL2-MeOH (100:1 to 1:100, v/v) as the mobile phase to produce eight fractions (E1-E8). Fraction E3 (35.1g) underwent treatment with RP C18-MPLC (10 %–100 % methanol) to obtain another 13 subfractions (M1-M13). Fraction M12 was again further separated with silica gel CC (hexane-ethyl acetate, 10:1 to 1:1, v/v = 100 % MeOH) to obtain five subfractions (M14-M18). Preparative HPLC was used to obtain usenamine A from subfraction M17 with 70 % acetonitrile.
The molecular formula of usenamine A was detected as C18H17NO6 from HR-ESI-MS data (m/z 344.0887 [M + H] +, calculated for C18H18NO6, 344.0891). 1H NMR spectrum showed the signals corresponding to four distinct methyl groups with shifts at δH 1.63, 1.96, 2.53, and 2.63. In addition, resonances for two hydroxy-group protons were noted at δH 13.40 and 12.30, and two amino protons at δH 11.54 and 9.83. An olefinic proton resonance was evident at δH 5.85 (H-4). 13C NMR revealed 18 carbon resonances were discerned, encompassing three ketocarbonyls, four oxygenated olefinic carbons, six olefinic carbons, four methyl carbons, and a sp3 quaternary carbon resonating at δC 56.5 (C-9b). By contrast with the compounds previously identified in U. diffracta [42,43], the dibenzofuran core structure was ascertained. Furthermore, the compound structure was further validated by referencing the spectral data of usenamine A has been previously reported [44].
2.2. Cell culture
The human lung cancer cell line A549 was obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea). HEK293T cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). The cells were placed in a culture dish containing Roswell Park Memorial Institute 1640 (RPMI-1640) medium or Dulbecco's Modified Eagle's Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) or supplemented with 10 % fetal bovine serum (FBS) and 1 % antibiotic-antimycotic (Thermo Fisher Scientific, Waltham, MA, USA) before being cultured at 5 % CO2 at 37 °C.
2.3. Microscopy
We seeded A549 cells in 6-well plates at 1 × 105 cells per well to perform microscopy analysis, and this was followed by a treatment process with the compound (10 and 20 μM). 48 h later, we examined the cell morphology, and an EVOS M5000 Imaging System (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to obtain cell images.
2.4. Cell viability assay
A549 cells underwent culturing in 96-well plates with 6 × 104 cells in each well before being exposed to various concentrations of usenamine A. After 48 h of incubation, 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyl-2 tetrazolium bromide (MTT) solution was added to each well, and the cells were then subjected to 2 h of incubation at 37 °C under 5 % CO2. A Synergy HTX Multi-Mode Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA) was used to evaluate the cell viability against the absorbance of the resulting solution at 490 nm.
2.5. Pseudovirus (PV) production
We co-transfected engineered pMD2.G (which encodes codon-optimized SARS-CoV-2 Delta spike proteins) and a psPAX2 plasmid into HEK293T cells with Metafectene. After 48 h of transfection, we collected the supernatant that contained the SARS-CoV-2 Delta pseudovirus and filtered the supernatant with cellulose acetate membranes (0.45 μm-pore). We replaced the medium with fresh DMEM with 10 % FBS for additional harvests 72 h post-transfection. To avoid the titer of PVs being decreased due to freezing or thawing, the supernatant was aliquoted to 1 mL and stored at −80 °C before use.
2.6. Real-Time PCR
We extracted total RNA from A549 cells that had been treated using usenamine A for 48 h with the RNeasy mini kit (Qiagen, Hilden, Germany) or the QIAamp viral RNA kit following the manufacturers’ protocols. With the same amount of RNA, cDNA was synthesized using the LunaScript RT SuperMix Kit (New England Biolabs, Ipswich, MA, USA). Nanodrop was used to measure RNA concentrations (Thermo Fisher Scientific, Waltham, MA, USA). PCR reactions were carried out with specific primers that had 10 μL of 2 × qPCR Master mix (SFC probes, Cheongju, Korea), 0.5 μL of PCR Forward Primer (10 μM), 0.5 μL of PCR Reverse Primer (10 μM), 1 μL of cDNA template, and 8 μL of PCR-grade water. The reaction underwent amplification via a flowing step at 95 °C by being denatured for 3 min followed by 40 cycles involving heating to 95 °C for 5 s and 60 °C for 30 s using a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific, Waltham, MA, USA). The primer sequences used in this study (5′ → 3′) were IL-6 (F: AGCCCTGAGAAAGGAGACAT, R: TGGAAGGTTCAGGTTGTTTT), IL-1β (F: CTGTCCTGCGTGTTGAAAGA, R: TTCTGCTTGAGAGGTGCTGA), TNF-α (F: AACCTCCTCTCTGCCATCAA, R: CCAAAGTAGACCTGCCCAGA), and Spike (F: TCTGCTTTACTAATGTCTATGC, R: GCTATAACGCAGCCTGTAAA, P: TCAGACAAATCGCTCCAGGGCA).
2.7. Apoptosis assay
The cells underwent culturing in 6-well plates and were exposed to different concentrations of usenamine A (10 μM). We seeded usenamine A-treated and untreated (control) A549 cells in 6-well plates and incubated them for 48 h at 37 °C. After 48 h of treatment, we collected the cells and washed them with phosphate-buffered saline (PBS). The cells were then resuspended in a binding buffer (1 × ). The cells underwent annexin V-PE and 7-AAD staining according to the instructions of the manufacturer. Then, we classified the cells into early (Annexin V+/7-AAD–) and late (Annexin V+/7-AAD+) apoptotic cells using a flow cytometer (BD FACSVerse, BD Biosciences, San Jose, CA, USA). The FlowJo software (Version 10, BD Biosciences) was also used to analyze cell percentages.
2.8. Tetramethylrhodamine methyl Ester Perchlorate (TMRM) assay
After 48 h of incubation, we collected the usenamine A-treated A549 cells. After being washed with PBS, the cells were incubated in 100 nM cell-permeable fluorescent indicator TMRM (ThermoFisher Scientific, Waltham, MA, USA). TMRM was prepared by diluting 10 mM stock in PBS for 30 min at 37 °C. To assess the mitochondrial membrane potential, we used PBS to wash the cells. We used a flow cytometer (BD FACSVerse, BD Biosciences, San Jose, CA, USA) for cell measurement, and the FlowJo v10 software (B Version 10, BD Biosciences) was used for cell analysis.
2.9. Measuring reactive oxygen species (ROS)
2,7-dichlorodihydrofluorescein diacetate (H2DCFDA; Thermo Fisher Scientific, Waltham, MA, USA) levels were assessed to analyze intracellular ROS levels. We gradually perfused cells with PBS buffer (37 °C) and used an inverted fluorescent microscope (Olympus) for imaging. The cells underwent half an hour of incubation in DCFDA (1 μM) at room temperature. Cold PBS was used to wash the cells, which were then resuspended in 0.5 mL of PBS with 1 % fetal bovine serum. A flow cytometer (BD Biosciences, USA) was used together with the FlowJo software for fluorescence analysis.
2.10. Western blot
A549 cells underwent 36 h of treatment with usenamine A (10 μM) in 6-well plates with 1 × 106 cells in each well. Total protein was extracted by using a 2x sampling buffer and then boiling for 15 min. The proteins separated by SDS-PAGE were transferred to a polyvinylidene difluoride (PVDF) membrane. Following 1.5 h of blocking in 5 % dry milk in TBST and 1 h of washing in 1 × TBST, the PVDF membrane was incubated with primary antibodies provided by Santa Cruz Biotechnology (Dallas, TX, USA) and Cell Signaling Technologies (Danvers, MA, USA) against green fluorescence protein (GFP) and β-actin. The membranes were first washed in 1x TBST and then underwent 2 h of incubation with anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies. Finally, the membranes were visualized with the assistance of enhanced chemiluminescent reagents purchased from Thermo Fisher Scientific. An Image Quant LAS mini (Fujifilm, Tokyo, Japan) was used to detect signals, and the relative protein level was normalized to β-actin.
2.11. Molecular docking
The structure of the delta spike protein (PDB ID: 7W92) was retrieved from the RCSB Protein Data Bank in PDB format [45,46]. Protein models were built by excluding heteroatoms and water molecules. The 3D structure of usenamine A and remdesivir were generated and their energy minimization was done using ChemOffice. Both receptor (delta spike protein) and ligands (usenamine A and remdesivir) were generated using AutoDock4 [47] and saved in PDBQT format. Active site residues including (LYS417, GLY446,TYR449, TRY453, LEU455, SER494, TYR495, GLY496, PHE497, GLN498, ASN501, and TYR505) were identified [48]. This active site was targeted through a grid box to target residues binding pocket was established with the following dimension: Å: (X, Y, Z) = (72 Å, 62 Å, 54 Å) and center (X, Y, Z) = (163.567 Å, −193.233 Å, 244.390 Å). A total of ten docking runs were implemented for each ligand. The most favorable binding poses of usenamine A and remdesivir were identified by determining the lowest binding energy (ΔG). Binding energies were optimized by applying the search genetic algorithm and the Lamarckian genetic algorithm with the assistance of AutoDock tools. The docked structures with high binding affinity were visualized using both PyMOL [49] and Discovery Studio Visualizer [50]. FDA-approved drug remdesivir was used as reference control.
2.12. ADMET profiling
The ADME attributes of usenamine A were computationally assessed using the SwissADME tool, developed by the Molecular Modeling Group at the Swiss Institute of Bioinformatics [51,52]. We used Lipinski's "rule-of-five" as a framework to gauge the biochemical properties of the compound that might impact its cellular permeation and absorption. To be considered drug-like, a molecule must meet at least three of Lipinski's stipulations: a molecular weight below 500 Da, a log P value under 5, less than 10 hydrogen-bond acceptors, and fewer than 5 hydrogen-bond donors. Additionally, the OSIRIS Property Explorer software was utilized to predict potential toxicities of usenamine A, specifically its mutagenic, tumorigenic, irritant, and reproductive effects [53,54].
2.13. Statistical analysis
GraphPad Prism (GraphPad Software, Inc., version 7, San Diego, CA, USA) was used for statistical analysis, and the results took the form of means ± SEM. The Student's t-test assisted in further analyzing the data. The resulting p-values indicated statistical significance (*p < 0.05, **p < 0.01, *** < 0.001, ****p ≤ 0.0001).
3. Results
3.1. Establishment of a SARS-CoV-2 delta pseudovirus system using A549 cells
SARS-CoV-2 delta PVs were produced via the co-transfection of human embryonic kidney (HEK) 293T cells with a pLVX-N1-Spike (Delta) lentiviral vector, an envelope protein expressing plasmid pMD2.G.Spike (Delta), and a packaging plasmid psPAX2 expressing Gag-pol. To obtain a high titer of PVs, the plasmid ratio was revised and optimized for transfection. Plasmid psPAX2 and modified pMD2.G were optimized to a 2:1 parameter to generate the pseudotyped virus capsid. Because of the high amount of plasmid needed to be transfected, Metafectene was used for its high transfection efficiency. HEK293T cells were also chosen due to their high infection efficiency. Thus, we constructed a system that produced PVs of the SARS-CoV-2 Delta variant for the following study.
3.2. Usenamine a has direct antiviral effects against SARS-CoV-2 infections in A549 cells
Fig. 1A displays the structure of usenamine A (Figs. S1–S7). Based on the MTT results (Fig. 1B), usenamine A possesses low cytotoxicity against A549 cells. To reveal the anti-infectivity of usenamine A for inhibiting SARS-CoV-2 PV infections in A549 cells in vitro, the concentration of usenamine A was set to 10 μM to quantitatively analyze the effect of viral RNA concentration in A549 cells. To better demonstrate the antiviral effect exerted by the drug on A549 cells, usenamine A was first added to the cells and cultured in an incubator for 4 h before transduction. Once the cells fully came into contact with usenamine A, the cells were infected with the PVs and then placed in an incubator for 36 h. As shown in Fig. 1C, the 10 μM and 20 μM usenamine A-treated groups exhibited significant and dose-dependent decreases in GFP signals (which indicates PV infection) compared to the no usenamine A treatment group. After the cells were infected with PVs, RNA was extracted from the supernatant and cells. Then, the viral copies of spike RNA were counted via quantitative PCR (qPCR), and the results showed a significant reduction in the amount of viral spike RNA in the cells treated with usenamine A of 20 μL concentration (Fig. 1D). This indicates that usenamine A can inhibit PV infections and that usenamine A exerts antiviral effects directly against SARS-CoV-2 PV infections in vitro. As a result, the PVs were barely able to enter the cells. Therefore, usenamine A shows potential as an effective agent against COVID-19.
Fig. 1.
Anti-infectivity effects of usenamine A against SARS-CoV-2 PVs in A549 cells. (A) Structure of usenamine A. (B) Cell viability under MTS assay measurements. A549 cells were exposed to different concentrations of usenamine A. (C) Fluorescence microscopy results to analyze the impact of usenamine A after pseudoviral transduction (scale bar: 200 μm). (D) RNA expression levels of SARS-CoV-2 PVs. Data were in the form of mean ± standard deviation (n = 3). **p < 0.01 and ***p < 0.001 relative to those in the control group.
3.3. Changes in mitochondrial membrane potential
Mitochondrial membrane potential (ΔΨm) can be used to effectively evaluate the function and state of mitochondria. To determine any connections between the protective effect of usenamine A and the mitochondrial membrane, mitochondrial depolarization was measured against the intensity of TMRM fluorescence through flow cytometry. As show in Fig. 2, the percentage of cells stripped of ΔΨm increased sharply in the group treated with PVs relative to the control group. That is to say, the pseudovirus caused ΔΨm to become unstable. However, usenamine A treatment reduced the percentage of cells stripped of ΔΨm, suggesting that usenamine A reversed the effect induced by PVs in the cells.
Fig. 2.
Usenamine A stabilizing mitochondrial membrane potential changes induced by SARS-CoV-2 PVs. (A) Effects of 10 μM usenamine A for 48 h on mitochondrial membrane potential meas-ured by a TMRM reagent together with a flow cytometer. (B) Mitochondrial membrane potential quantification. Values indicate means ± SEM. (n = 3, ***p ≤ 0.001).
3.4. Changes in reactive oxygen species (ROS)
Mitochondrial membrane potential is capable of triggering ROS that can vary mitochondrial membrane permeability, and high ROS accumulation levels will result in cell damage and apoptosis by activating cell death signal pathways. We previously demonstrated that usenamine A affects mitochondrial depolarization, and thus, we explored how this affected the production of ROS. As shown in Fig. 3, ROS and mitochondrial ROS levels significantly increased to 60.9 % and 45.4 % in the PV treatment groups compared to the control group levels of 5.09 % and 7.94 %, respectively. The results also showed that PV-induced ROS generation was suppressed by usenamine A to 39.8 % and 26.4 % in the two staining groups.
Fig. 3.
Usenamine A suppressing ROS generation triggered by SARS-CoV-2 PVs in A549 cells. (A) (C) Intracellular ROS and mitochondrial ROS levels measured using a flow cytometer. (B) (D) Quantification of intracellular ROS and mitochondrial ROS levels. Values denote means ± SEM. (n = 3, ***p ≤ 0.001).
3.5. Usenamine a attenuates spike protein-induced apoptosis in A549 cells
To explore whether usenamine A could protect against spike protein-induced apoptosis, we carried out flow cytometric analysis on damaged cells with annexin V/propidium iodide (PI). According to the Annexin V-FITC/PI staining assay, late apoptotic cell numbers showed a significant increase in the PV-treated groups relative to the control group. When the cells were incubated with 10 μM usenamine A, this effectively alleviated the late apoptosis induced by the formation of spike protein–mediated syncytium, as shown in Fig. 4. In summary, usenamine A suppressed spike-induced late apoptosis in A549 cells, as confirmed by PI staining.
Fig. 4.
Usenamine A alleviating SARS-CoV-2 PV-triggered apoptosis in A549 cells. (A) Apoptosis analyzed by Annexin V-PI double staining together with flow cytometry. A549 cells were either treated or untreated with usenamine A for 48 h. (B) Early and late apoptotic cell quantification. Values are represented as means ± SEM. (n = 3, ***p ≤ 0.001).
3.6. Usenamine a alleviates PV-induced inflammation
To investigate the anti-inflammation effects exerted by usenamine A, we assessed the mRNA levels of IL-6, IL-1 beta, and TNF-alpha, which are common markers of inflammatory pathways. As shown in Fig. 5, the levels of the three aforementioned inflammatory factors were upregulated after PV transduction, which suggests the possibility of an inflammatory response similar to cytokine storms in cells. Conversely, the expression levels of the inflammatory factors were found to be significantly downregulated in the cells treated with usenamine A compared to the untreated group. That is to say, usenamine A is effective in suppressing the expression of inflammatory factors. Therefore, usenamine A could potentially be applied to reduce inflammatory responses and lung lesions in patients with new coronary pneumonia, thus reducing patient mortality.
Fig. 5.
Relative gene expression levels of the inflammatory markers IL-6, IL-1 beta, and TNF-alpha in A549 cells. Values were in the form of means ± SEM. (n = 3, **p ≤ 0.01, ***p ≤ 0.001).
3.7. Usenamine a decreases S-glycoprotein syncytium formation in 4 VOC
To reveal the antiviral mechanism of usenamine A, the effect of usenamine A on the spike protein was analyzed through western immunoblot assays. To increase spike protein synthesis, HEK293T cells were treated as hosts for the production of spike proteins. After transfecting the HEK293T cells with spike proteins of four types of VOC linked to GFP for 24 h, usenamine A of 10 μM concentration was added. Cell lysates were collected after 24 h of incubation and subsequently lysed. GFP levels were analyzed through western blotting to detect spike proteins. As suggested by the results in Fig. 6, the levels of the four VOC spike proteins declined significantly following usenamine A treatment in comparison with the DMSO-only group. This implies that usenamine A reduces the stability of the spike protein.
Fig. 6.
Expressions of four VOC spike proteins in HEK293T cells as obtained via Western blot analysis. Whole cell lysates were analyzed to confirm spike expressions after treating the cells with 10 μM usenamine A for 24 h.
3.8. Molecular docking
The binding efficiency of a molecule (usenamine A and remdesivir) to a target protein is evaluated through computer-assisted molecular docking by calculating the highest binding affinity and lowest binding energy. In our study, docking analysis revealed that usenamine A (−7.36 kcal/mol) exhibited a higher binding affinity than remdesivir (−6.23 kcal/mol). The potential binding pocket of the spike protein with remdesivir and usenamine A is shown in Fig. 7A and B. The usenamine A conformation was stabilized by the hydrogen bonds between the hydrogen atoms in amine, hydroxyl groups, and oxygen atoms in the aldehyde group. The 2D visualization of remdesivir and usenamine A interaction with delta spike protein complex demonstrated that the interaction occurred through extensive pi-alkyl bond interactions and pi-donor hydrogen bond interactions (Fig. 7C and D). The stabilization of the binding interactions between usenamine A and the spike protein were mediated by amino acid residue such as ARG403, TYR453, SER494, TYR495, GLY496, PHE497, ASN501, and TYR505. These results suggested that usenamine A and remdesivir exhibited similar docking pose and target active pocket. Therefore, the results of the usenamine A binds to spike proteins were agreement with the reference drug interactions thereby showing moderate binding affinity.
Fig. 7.
In silico binding analysis between remdesivir and the spike protein of COVID-19 and their interacting residues (TYR-453 and GLY 496) were labeled (A). In silico binding analysis between usenamine A and the spike protein, and their interacting residues (ARG 403 and GLY496) were labeled (B). 2D visualization of remdesivir and spike protein docking complex (C). 2D visualization of usenamine A and spike protein docking complex (D).
3.9. Drug-likeness and ADMET prediction of usenamine A
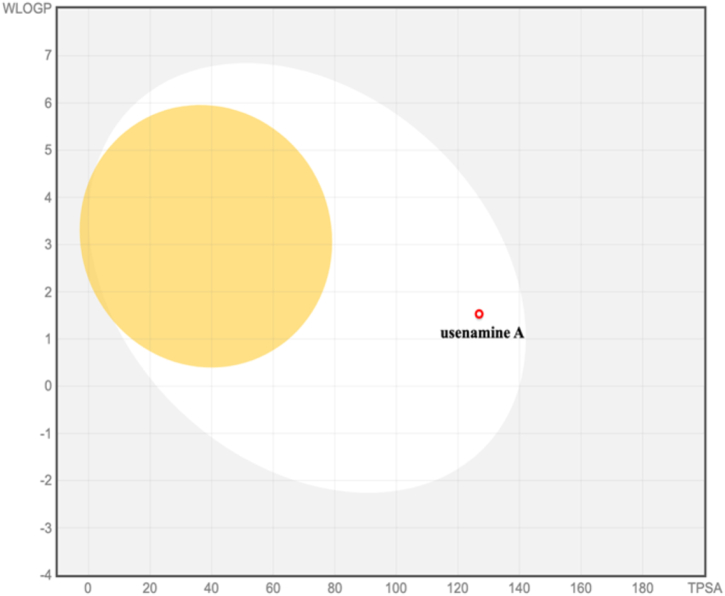
The pharmacokinetic profile of usenamine A was evaluated using in silico absorption, distribution, metabolism, and excretion (ADME) analyses. Lipinski's rule of five served as the guiding principle to anticipate the compound's druggability considering metrics such as molecular weight, lipophilicity (expressed as the octanol–water partition coefficient), and counts of hydrogen bond donors and acceptors. Notably, usenamine A met Lipinski's criteria, indicating promising bioavailability and drug-like properties (Table 1). The BOILED-Egg model, a visual representation correlating lipophilicity and polarity, was employed to forecast passive intestinal absorption and brain permeation potentials. Compounds situated in the yellow domain (yolk) are likely to penetrate the blood–brain barrier (BBB), whereas those in the white region are inclined towards passive absorption in the gastrointestinal (GI) tract (Fig. 8). Predictions highlighted usenamine A's aptitude for efficient GI absorption but limited BBB permeability. Toxicological predictions, encompassing mutagenicity, tumorigenicity, irritancy, and reproductive effects were carried out with the aid of the OSIRIS Property Explorer (Table 2). Encouragingly, usenamine A did not exhibit any discernible toxic tendencies.
Table 1.
Lipinski's rule of five for ADME analysis of usenamine A.
| Lipinski's Rule of Five |
||||||
|---|---|---|---|---|---|---|
| Compound |
Molecular weight (g/mol) |
Lipophilicity (MLogP) |
H-bond donors |
H-bond acceptors |
Rule violations |
Drug-likeness |
| <500 | <5 | <5 | <10 | <2 | ||
| Usenamine A | 343 g/mol | −0.52 | 3 | 6 | 0 | Yes |
Fig. 8.
Prediction of GI tract and brain permeation of usenamine A by brain or the intestinal estimated permeation predictive model (BOILED-Egg) method.
Table 2.
Predicted toxicity parameters of usenamine A.
| Compound | Mutagenicity | Tumorigenicity | Irritant Effect | Reproductive Toxicity |
|---|---|---|---|---|
| Usenamine A | None | None | None | None |
4. Discussion
As an emerging type of the severe acute respiratory infectious disease, COVID-19 has seriously threatened human health, the world economy, and social stability [55]. To date, it remains a challenge to find a cure for COVID-19 [56]. Anti-viral drugs that have been developed so far generally work in two ways [57]. One method involves reducing RdRP levels in the virus [58]. Another class of antiviral drugs aims to prevent viral infections by decreasing the levels of key proteases associated with viral replication [59]. The other method involves taking advantage of the lopinavir/ritonavir mechanism and has been clinically proven to be help improve the condition of COVID patients [60]. Besides these viral targets, researchers have also suggested using inflammatory cytokines generated in critically ill COVID-19 patients as therapeutic targets. This is because significant complications can arise due to molecular storms triggered by these cytokines [61,62]. For this reason, certain compounds that are capable of inhibiting inflammation (e.g., tocilizumab) may also be suitable for treating COVID-19 patients [63,64]. However, there is still no specific drug that can treat all related symptoms. Since naturally derived products have shown promising clinical effects when it comes to treating infectious diseases [65], it is necessary to explore more effective intervention strategies based on naturally derived products to prevent and treat COVID-19 cases.
Although our PV infection system is rapid and safe, it does not fully replicate the complexities of SARS-CoV-2 under in vivo conditions. However, we are working on establishing an appropriate in vivo model to study the in vivo antiviral activity of usenamine A. Furthermore, in in vivo studies, greater focus is placed on determining physiological factors, such as biodistribution, metabolism, and clearance on antiviral effects. These factors can potentially impact the efficacy and stability of the drug, necessitating comprehensive in vivo investigations to assess the drug's performance as well as its susceptibility to drug resistance stemming from the continuous evolution of SARS-CoV-2.
As new variants of the virus continue to emerge and evolve, the potential development of drug resistance to usenamine A must be taken into serious consideration. Viruses can adapt and mutate to develop resistance to most antiviral drugs [66,67]. A recent research paper discusses the emergence of mutations in SARS-CoV-2 to evade the effects of remdesivir [68], highlighting the risk of SARS-CoV-2 developing resistance to existing antiviral drugs. Therefore, we considered the potential of combining different antiviral drugs and immunomodulatory/anti-inflammatory drugs to enhance treatment efficacy against COVID-19 by mitigating the risk of drug-resistant virus variants being introduced. Furthermore, considering that the drug will be administered in tandem with active viral clearance by the immune system, including any potential resistant forms that may have emerged, it becomes advisable to focus surveillance efforts on immunocompromised individuals receiving usenamine A treatment.
As the medical community continues to grapple with the complexities of COVID-19, the introduction of new therapeutic agents like usenamine A is both promising and challenging. One of the potential complexities of usenamine A involves understanding and predicting its drug-drug interactions (DDIs) with existing therapeutic regimens. The first consideration is pharmacokinetics – how the body absorbs, distributes, metabolizes, and excretes the drug – as this can influence usenamine A's potential interactions with other drugs. If usenamine A is metabolized by the same liver enzymes as another anti-COVID-19 drug, there is potential for competitive inhibition where both drugs vie for the same metabolic pathway, leading to altered blood levels of one or both drugs [69,70]. Absorption dynamics can also play an important role. If usenamine A affects the gut's pH or interferes with transport proteins, it could influence the absorption rates of other orally administered drugs [71,72]. Given such considerations, in-depth studies are of paramount importance. Combination drug therapy trials – where usenamine A is administered alongside standard anti-COVID-19 drugs – can shed light on any synergistic or antagonistic effects. Additionally, computational models and databases can help predict potential interactions based on the known properties of usenamine A and other drugs [73]. While these models cannot replace clinical studies, they can guide researchers to potential areas of concern, making subsequent in vivo and in vitro investigations more targeted and efficient. As we look to integrate usenamine A into the arsenal against COVID-19, it is essential to understand not just its standalone effects, but also how it fits into the broader therapeutic landscape. Ensuring usenamine A's compatibility with existing drugs will be key to harnessing its full potential safely and effectively.
According to our research results, usenamine A, which is extracted from the ethyl acetate layer of Usnea diffracta, can exert inhibitory effects against SARS-CoV-2 PVs. These inhibitory effects may arise due to the cleavage of the coronavirus spike protein. Additionally, due to the similar crown structures of various spike proteins, we found that usenamine A also inhibited four other VOC mutant coronavirus infections. The data from our study proves the suitability of usenamine A for effective broad-spectrum anti-coronavirus therapies and suggest the potential of developing usenamine A as an antiviral agent that acts directly on spike proteins. Furthermore, we discovered that usenamine A exhibits anti-inflammatory effects and can suppress the synthesis of IL-6, IL-1β, and TNF-α, all of which have been reported to be associated with the severity of COVID-19.
5. Conclusion and perspectives
Herein, we proposed that usenamine A could effectively help in inhibiting SARS-CoV-2 delta PV infections and provide cells with protection against cell damage via mitochondrial depolarization, elevated ROS levels, and cell death. Moreover, usenamine A was found to be effective in reducing molecular storms caused by the virus and in suppressing the expression of inflammatory factors. Usenamine A is also capable of destabilizing other coronavirus spike proteins, which means that it could potentially be applicable for a variety of antiviral drugs. This study aimed to assess the anti-SARS-CoV-2 activity of usenamine A by establishing pseudovirus infection models through human lung epithelial cells, namely, A549 cells. According to the research results, usenamine A suppressed PV infections, and thus, we conclude it has the potential to produce anti-SARS-CoV-2 effects.
Funding
This research was supported by the “Establishment of measurement standards for Chemistry and Radiation” (grant number KRISS-2023-GP2023-0006) funded by the Korea Research Institute of Standards and Science. In addition, this work was supported by a grant from the National Research Foundation (NRF) of Korea funded by the Korean government (MSIT) (grant number: NRF-2022R1C1C1009626).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Zijun Li: performed experiments and data analysis and prepared the Figs. Joo-Eun Lee: performed experiments and data analysis and prepared the Figs. Namki Cho: conceived and designed the study and wrote the manuscript. And all authors approved the final version of the manuscript. Hee Min Yoo: conceived and designed the study and wrote the manuscript. And all authors approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Seil Kim for sharing the viral gene material.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21742.
Contributor Information
Namki Cho, Email: cnamki@chonnam.ac.kr.
Hee Min Yoo, Email: hmy@kriss.re.kr.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Nalbandian A., Sehgal K., Gupta A., V Madhavan M., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., Ahluwalia N., Bikdeli B., Dietz D., Der-Nigoghossian C., Liyanage-Don N., Rosner G.F., Bernstein E.J., Mohan S., Beckley A.A., Seres D.S., Choueiri T.K., Uriel N., Ausiello J.C., Accili D., Freedberg D.E., Baldwin M., Schwartz A., Brodie D., Garcia C.K., V Elkind M.S., Connors J.M., Bilezikian J.P., Landry D.W., Wan E.Y. Post-acute COVID-19 syndrome. Nat Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., Schoehn G., Bernardi A., Delgado R., Fieschi F. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. PLoS Pathog. 2021;17 doi: 10.1371/JOURNAL.PPAT.1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hennrich A.A., Sawatsky B., Santos-Mandujano R., Banda D.H., Oberhuber M., Schopf A., Pfaffinger V., Wittwer K., Riedel C., Pfaller C.K., Conzelmann K.K. Safe and effective two-in-one replicon-and-VLP minispike vaccine for COVID-19: protection of mice after a single immunization. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand P., Puranik A., Aravamudan M., Venkatakrishnan A.J., Soundararajan V. SARS-CoV-2 strategically mimics proteolytic activation of human ENaC. Elife. 2020;9 doi: 10.7554/ELIFE.58603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/S41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 8.Berger I., Schaffitzel C. The SARS-CoV-2 spike protein: balancing stability and infectivity. Cell Res. 2020;30:1059–1060. doi: 10.1038/s41422-020-00430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaufer A.M., Theis T., Lau K.A., Gray J.L., Rawlinson W.D. Laboratory biosafety measures involving SARS-CoV-2 and the classification as a Risk Group 3 biological agent. Pathology. 2020;52:790–795. doi: 10.1016/j.pathol.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M., Zhang X.E. Construction and applications of SARS-CoV-2 pseudoviruses: a mini review. Int. J. Biol. Sci. 2021;17:1574–1580. doi: 10.7150/ijbs.59184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohamed Khosroshahi L., Rezaei N. Dysregulation of the immune response in coronavirus disease 2019. Cell Biol. Int. 2021;45:702–707. doi: 10.1002/cbin.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z.H., Yang D.L. A meta-analysis of the impact of COVID-19 on liver dysfunction. Eur. J. Med. Res. 2020;25:54. doi: 10.1186/s40001-020-00454-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollo K., Ducza L., Hegyi Z., Docs K., Hegedus K., Bakk E., Papp I., Kis G., Meszar Z., Bardoczi Z., Antal M. Interleukin-1 receptor type 1 is overexpressed in neurons but not in glial cells within the rat superficial spinal dorsal horn in complete Freund adjuvant-induced inflammatory pain. J. Neuroinflammation. 2017;14:125. doi: 10.1186/s12974-017-0902-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.V Fernandez M., Miller E., Krammer F., Gopal R., Greenbaum B.D., Bhardwaj N. Ion efflux and influenza infection trigger NLRP3 inflammasome signaling in human dendritic cells. J. Leukoc. Biol. 2016;99:723–734. doi: 10.1189/jlb.3A0614-313RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda K., Baba Y., Nagai Y., Miyazaki K., Malykhin A., Nakamura K., Kincade P.W., Sakaguchi N., Coggeshall K.M. IL-6 blocks a discrete early step in lymphopoiesis. Blood. 2005;106:879–885. doi: 10.1182/BLOOD-2005-02-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao B., Zhang Z., Viennois E., Kang Y., Zhang M., Han M.K., Chen J., Merlin D. Combination therapy for ulcerative colitis: orally targeted nanoparticles prevent mucosal damage and relieve inflammation. Theranostics. 2016;6:2250–2266. doi: 10.7150/thno.15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumari M., Lu R.M., Li M.C., Huang J.L., Hsu F.F., Ko S.H., Ke F.Y., Su S.C., Liang K.H., Yuan J.P.Y., Chiang H.L., Sun C.P., Lee I.J., Li W.S., Hsieh H.P., Tao M.H., Wu H.C. A critical overview of current progress for COVID-19: development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci. 2022;29 doi: 10.1186/S12929-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L., Wang Z. Bench-to-bedside: innovation of small molecule anti-SARS-CoV-2 drugs in China. Eur. J. Med. Chem. 2023;257 doi: 10.1016/J.EJMECH.2023.115503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kronenberger T., Laufer S.A., Pillaiyar T. COVID-19 therapeutics: small-molecule drug development targeting SARS-CoV-2 main protease. Drug Discov. Today. 2023;28 doi: 10.1016/J.DRUDIS.2023.103579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galan L.E.B., dos Santos N.M., Asato M.S., Araújo J.V., de Lima Moreira A., Araújo A.M.M., Paiva A.D.P., Portella D.G.S., Marques F.S.S., Silva G.M.A., de Sousa Resende J., Tizolim M.R., Santos P.L., Buttenbender S.F., de Andrade S.B., Carbonell R.C.C., Da Rocha J.G., de Souza R.G.S., da Fonseca A.J. Phase 2 randomized study on chloroquine, hydroxychloroquine or ivermectin in hospitalized patients with severe manifestations of SARS-CoV-2 infection. Pathog. Glob. Health. 2021;115:235–242. doi: 10.1080/20477724.2021.1890887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vallejos J., Zoni R., Bangher M., Villamandos S., Bobadilla A., Plano F., Campias C., Chaparro Campias E., Medina M.F., Achinelli F., Guglielmone H.A., Ojeda J., Farizano Salazar D., Andino G., Kawerin P., Dellamea S., Aquino A.C., Flores V., Martemucci C.N., Martinez S.M., Segovia J.E., Reynoso P.I., Sosa N.C., Robledo M.E., Guarrochena J.M., Vernengo M.M., Ruiz Diaz N., Meza E., Aguirre M.G. Ivermectin to prevent hospitalizations in patients with COVID-19 (IVERCOR-COVID19) a randomized, double-blind, placebo-controlled trial. BMC Infect. Dis. 2021;21 doi: 10.1186/S12879-021-06348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Shabat S., Yarmolinsky L., Porat D., Dahan A. Antiviral effect of phytochemicals from medicinal plants: applications and drug delivery strategies. Drug Deliv. Transl. Res. 2020;10:354–367. doi: 10.1007/S13346-019-00691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashaolu T.J., Zarei M., Agrawal H., Kharazmi M.S., Jafari S.M. A critical review on immunomodulatory peptides from plant sources; action mechanisms and recent advances. Crit. Rev. Food Sci. Nutr. 2023 doi: 10.1080/10408398.2023.2183380. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.H., Kim D.H., Jo S., Cho M.J., Cho Y.R., Lee Y.J., Byun S. Immunomodulatory functional foods and their molecular mechanisms. Exp. Mol. Med. 2022;54 doi: 10.1038/S12276-022-00724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notarte K.I.R., Quimque M.T.J., Macaranas I.T., Khan A., Pastrana A.M., Villaflores O.B., Arturo H.C.P., Pilapil IV D.Y.H., Tan S.M.M., Wei D.Q., Wenzel-Storjohann A., Tasdemir D., Yen C.H., Ji S.Y., Kim G.Y., Choi Y.H., Macabeo A.P.G. Attenuation of lipopolysaccharide-induced inflammatory responses through inhibition of the NF-κB pathway and the increased NRF2 level by a flavonol-enriched n-butanol fraction from uvaria alba. ACS Omega. 2023;8:5377–5392. doi: 10.1021/ACSOMEGA.2C06451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmeda-Hirschmann G., Tapia A., Lima B., Pertino M., Sortino M., Zacchino S., De Arias A.R., Feresin G.E. A new antifungal and antiprotozoal depside from the Andean lichen Protousnea poeppigii. Phyther. Res. 2008;22:349–355. doi: 10.1002/PTR.2321. [DOI] [PubMed] [Google Scholar]
- 28.Shrestha G., Raphael J., Leavitt S.D., St L.L. Clair, in vitro evaluation of the antibacterial activity of extracts from 34 species of North American lichens. Pharm. Biol. 2014;52:1262–1266. doi: 10.3109/13880209.2014.889175. [DOI] [PubMed] [Google Scholar]
- 29.Poulsen-Silva E., Gordillo-Fuenzalida F., Atala C., Moreno A.A., Otero M.C. Bioactive lichen secondary metabolites and their presence in species from Chile. Metabolites. 2023;13:805. doi: 10.3390/METABO13070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim M.-S., Cho H.-B. Melanogenesis inhibitory effects of methanolic extracts of Umbilicaria esculenta and Usnea longissima. J. Microbiol. 2007;45:578–582. https://pubmed.ncbi.nlm.nih.gov/18176544/ [PubMed] [Google Scholar]
- 31.Zugic A., Jeremic I., Isakovic A., Arsic I., Savic S., Tadic V. Evaluation of anticancer and antioxidant activity of a commercially available CO2 supercritical extract of old man's beard (Usnea barbata) PLoS One. 2016;11 doi: 10.1371/JOURNAL.PONE.0146342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Çeli̇kler Kasimoğullari Seyhan Oran Ferda Ari S., Authors SERAP ÇELİKLER KASIMOĞULLARI A., Oran S., Ari F., Ulukaya E., Aztopal N., Sarimahmut M., Öztürk Ş., Biol T.J., Çeli̇kler Kasimoğullari S., Ulukaya E., Aztopal N. Genotoxic, cytotoxic, and apoptotic effects of crude extract of Usnea filipendula Stirt. in vitro. Turkish J. Biol. 2014;38:940–947. doi: 10.3906/biy-1405-23. [DOI] [Google Scholar]
- 33.Wang Z., Wang N., Yang L., Song X.Q. Bioactive natural products in COVID-19 therapy. Front. Pharmacol. 2022;13 doi: 10.3389/FPHAR.2022.926507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z., Yang L. Chinese herbal medicine: fighting SARS-CoV-2 infection on all fronts. J. Ethnopharmacol. 2021;270 doi: 10.1016/J.JEP.2021.113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J., Lee J., Kim G.J., Yang I., Wang W., Nam J.W., Choi H., Nam S.J., Kang H. Mycousfurans A and B, antibacterial usnic acid congeners from the fungus mycosphaerella sp., isolated from a marine sediment. Mar. Drugs. 2019;17 doi: 10.3390/md17070422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovici V., Bucur L., Popescu A., Schroder V., Costache T., Rambu D., Cucolea I.E., Gird C.E., Caraiane A., Gherghel D., Vochita G., Badea V. Antioxidant and cytotoxic activities of Usnea barbata (L.) F.H. Wigg. Dry extracts in different solvents. Plants. 2021;10 doi: 10.3390/plants10050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rankovic B., Kosanic M., Stanojkovic T., Vasiljevic P., Manojlovic N. Biological activities of Toninia candida and Usnea barbata together with their norstictic acid and usnic acid constituents. Int. J. Mol. Sci. 2012;13:14707–14722. doi: 10.3390/ijms131114707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salgado F., Albornoz L., Cortez C., Stashenko E., Urrea-Vallejo K., Nagles E., Galicia-Virviescas C., Cornejo A., Ardiles A., Simirgiotis M., Garcia-Beltran O., Areche C. Secondary metabolite profiling of species of the genus Usnea by UHPLC-ESI-OT-MS-MS. Molecules. 2017;23 doi: 10.3390/molecules23010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L., Shi Q., Fang J.L., Mei N., Ali A.A., Lewis S.M., Leakey J.E.A., Frankos V.H. Review of usnic acid and Usnea barbata toxicity. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2008;26:317–338. doi: 10.1080/10590500802533392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Araujo H.D.A., Silva H., Silva Junior J.G.D., Albuquerque M., Coelho L., Aires A.L. The natural compound hydrophobic usnic acid and hydrophilic potassium usnate derivative: applications and comparisons. Molecules. 2021;26 doi: 10.3390/molecules26195995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingólfsdóttr K. Usnic acid, Phytochemistry. 2002;61:729–736. doi: 10.1016/S0031-9422(02)00383-7. [DOI] [PubMed] [Google Scholar]
- 42.Hao Y.M., Yan Y.C., Zhang Q., Liu B.Q., Wu C.S., Wang L.N. Phytochemical composition, antimicrobial activities, and cholinesterase inhibitory properties of the lichen Usnea diffracta Vain. Front. Chem. 2023;10 doi: 10.3389/FCHEM.2022.1063645/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng J., Yang X. New dibenzofuran and anthraquinone from Usnea longissima. Zhongguo Zhongyao Zazhi. 2009;34:852–853. https://pubmed.ncbi.nlm.nih.gov/19623979/ [PubMed] [Google Scholar]
- 44.Yu X., Guo Q., Su G., Yang A., Hu Z., Qu C., Wan Z., Li R., Tu P., Chai X. Usnic acid derivatives with cytotoxic and antifungal activities from the lichen Usnea longissima. J. Nat. Prod. 2016;79:1373–1380. doi: 10.1021/ACS.JNATPROD.6B00109. [DOI] [PubMed] [Google Scholar]
- 45.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/NAR/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., Feng Z., Ghosh S., Goodsell D.S., Green R.K., Guranović V., Guzenko D., Hudson B.P., Kalro T., Liang Y., Lowe R., Namkoong H., Peisach E., Periskova I., Prlić A., Randle C., Rose A., Rose P., Sala R., Sekharan M., Shao C., Tan L., Tao Y.P., Valasatava Y., Voigt M., Westbrook J., Woo J., Yang H., Young J., Zhuravleva M., Zardecki C. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/NAR/GKY1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris G.M., Ruth H., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/JCC.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong F.C., Ong J.H., Chai T.T. SARS-CoV-2 spike protein-, main protease- and papain-like-protease-targeting peptides from seed proteins following gastrointestinal digestion: an in silico study. Phytomedicine. 2021;1 doi: 10.1016/J.PHYPLU.2020.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.PyMOL | www.pymol.org, (n.d.). https://www.pymol.org/pymol.html? (accessed September 11, 2022).
- 50.Life Sciences and Material Sciences Biovia – dassault systèmes. https://www.3ds.com/products-services/biovia/ (n.d.)
- 51.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/SREP42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daina A., Zoete V. Application of the SwissDrugDesign online resources in virtual screening. Int. J. Mol. Sci. 2019;20:4612–4620. doi: 10.3390/IJMS20184612. 2019. 4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brogi S., Quimque M.T., Notarte K.I., Africa J.G., Hernandez J.B., Tan S.M., Calderone V., Macabeo A.P. Virtual combinatorial library screening of quinadoline B derivatives against SARS-CoV-2 RNA-dependent RNA polymerase. Computation. 2022;10:7. doi: 10.3390/COMPUTATION10010007/S1. [DOI] [Google Scholar]
- 54.Quimque M.T., Notarte K.I., Letada A., Fernandez R.A., Pilapil D.Y., Pueblos K.R., Agbay J.C., Dahse H.M., Wenzel-Storjohann A., Tasdemir D., Khan A., Wei D.Q., Gose Macabeo A.P. Potential cancer- and alzheimer's disease-targeting phosphodiesterase inhibitors from uvaria alba: insights from in vitro and consensus virtual screening. ACS Omega. 2021;6:8403–8417. doi: 10.1021/ACSOMEGA.1C00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spicer A.J., Jalkanen S. Why haven't we found an effective treatment for COVID-19? Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.644850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jukič M., Kores K., Janežič D., Bren U. Repurposing of drugs for SARS-CoV-2 using inverse docking fingerprints. Front. Chem. 2021;9 doi: 10.3389/FCHEM.2021.757826/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen R., Lan Z., Ye J., Pang L., Liu Y., Wu W., Qin X., Guo Y., Zhang P. Cytokine storm: the primary determinant for the pathophysiological evolution of COVID-19 deterioration. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.589095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama C., Han J., Yau L., Reiss W.G., Kramer B., Neidhart J.D., Criner G.J., Kaplan-Lewis E., Baden R., Pandit L., Cameron M.L., Garcia-Diaz J., Chávez V., Mekebeb-Reuter M., Lima de Menezes F., Shah R., González-Lara M.F., Assman B., Freedman J., Mohan S.V. Tocilizumab in patients hospitalized with covid-19 pneumonia. N. Engl. J. Med. 2021;384:20–30. doi: 10.1056/NEJMOA2030340/SUPPL_FILE/NEJMOA2030340_DATA-SHARING.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afra K., Chen L.Y.C., Sweet D. Tocilizumab for hospitalized patients with COVID-19. CMAJ (Can. Med. Assoc. J.) 2021;193:E521. doi: 10.1503/cmaj.210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K.J., Liu X., Komabayashi T., Jeong S.I., Selli S. Natural products for infectious diseases. Evid Based Complement Altern. Med. 2016;2016 doi: 10.1155/2016/9459047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Irwin K.K., Renzette N., Kowalik T.F., Jensen J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016;2 doi: 10.1093/VE/VEW014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vitiello A. Sars-Cov-2 and risk of antiviral drug resistance, Ir. J. Med. Sci. 2022;191:2367. doi: 10.1007/S11845-021-02820-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szemiel A.M., Merits A., Orton R.J., MacLean O.A., Pinto R.M., Wickenhagen A., Lieber G., Turnbull M.L., Wang S., Furnon W., Suarez N.M., Mair D., Da Silva Filipe A., Willett B.J., Wilson S.J., Patel A.H., Thomson E.C., Palmarini M., Kohl A., Stewart M.E. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17 doi: 10.1371/JOURNAL.PPAT.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saha N. Clinical pharmacokinetics and drug interactions. Pharm. Med. Transl. Clin. Res. 2018:81–106. doi: 10.1016/B978-0-12-802103-3.00006-7. [DOI] [Google Scholar]
- 70.Palleria C., Di Paolo A., Giofrè C., Caglioti C., Leuzzi G., Siniscalchi A., De Sarro G., Luca G. Pharmacokinetic drug-drug interaction and their implication in clinical management. J. Res. Med. Sci. 2013;18:601–610. https://pubmed.ncbi.nlm.nih.gov/24516494/ [PMC free article] [PubMed] [Google Scholar]
- 71.Niu J., Straubinger R.M., Mager D.E. Pharmacodynamic drug-drug interactions. Clin. Pharmacol. Ther. 2019;105:1395–1406. doi: 10.1002/CPT.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corrie K., Hardman J.G. Mechanisms of drug interactions: pharmacodynamics and pharmacokinetics. Anaesth. Intensive Care Med. 2017;18:331–334. doi: 10.1016/J.MPAIC.2017.04.006. [DOI] [Google Scholar]
- 73.Ferdousi R., Safdari R., Omidi Y. Computational prediction of drug-drug interactions based on drugs functional similarities. J. Biomed. Inf. 2017;70:54–64. doi: 10.1016/J.JBI.2017.04.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.