Summary
Base editing, a CRISPR-based genome engineering technique, enables precise single-nucleotide modifications while minimizing double-strand breaks. Here, we present a protocol for arrayed mutagenesis using base editors to identify regulatory elements within the gamma-globin locus. We describe steps for guide RNA (gRNA) cloning into lentiviral vectors, establishing stable cell lines with base editor expression, transducing gRNAs, and assessing editing efficiency. This protocol can be applied to diverse genomic regions and cell lines for arrayed screening, facilitating genetic research, and target discovery.
For complete details on the use and execution of this protocol, please refer to Ravi et al. (2022)1
Subject areas: Cell Culture, Genetics, Molecular Biology, CRISPR
Graphical abstract

Highlights
-
•
CRISPR-based base editing for precise single-nucleotide modification
-
•
Arrayed mutagenesis using base editors can identify regulatory elements
-
•
Protocol for genomic arrayed screening for cell lines at any locus
-
•
Instructions on gRNA design, stable preparation, and evaluating gene editing
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Base editing, a CRISPR-based genome engineering technique, enables precise single-nucleotide modifications while minimizing double-strand breaks. Here, we present a protocol for arrayed mutagenesis using base editors to identify regulatory elements within the gamma-globin locus. We describe steps for guide RNA (gRNA) cloning into lentiviral vectors, establishing stable cell lines with base editor expression, transducing gRNAs, and assessing editing efficiency. This protocol can be applied to diverse genomic regions and cell lines for arrayed screening, facilitating genetic research, and target discovery.
Before you begin
This protocol delineates the procedure for conducting arrayed mutagenesis within the specific genomic loci using adenine and cytosine base editing techniques. Specifically, we focused on screening a 300 bp region upstream of the transcription start site at the gamma globin promoter in human erythroid cell line. The objective was to identify targets that could enhance fetal hemoglobin expression which will offer potential therapeutic applications for beta hemoglobinopathies. Importantly, this protocol could be applied to explore other genomic regions of any size, enabling the identification of regulatory elements and facilitating the development of therapeutic gene editing strategies. It is worth noting that screening a large number of candidate guide RNAs (gRNAs) simultaneously by a single researcher may compromise the quality of the results. For the purpose of achieving consistent and reproducible outcomes, it is recommended to limit the number of gRNAs to approximately 50 per screening. Especially when conducting screening in multiple batches, it is essential to include appropriate internal controls and replicates during the screening process. The strict adherence to good laboratory practices is crucial to prevent microbial or cellular cross-contamination.
Prior to commencing the screening process, ensure the availability of the required lentiviral constructs and target gRNA oligos. In our study, we obtained the lentiviral constructs for the cloning of gRNAs from Addgene, and the respective identifiers are provided in the key resources table. Several web-based tools are available for gRNA design, many of which consider the specific base editor variant being used. Reliable options include BE-Designer,2 DeepBaseEditor,3 ACEofBASEs,4 and PnB designer.5 The links for all these tools are provided in the key resources table. Alternatively, gRNAs can be designed manually or using general gRNA design software such as Benchling or CHOPCHOP.6 In order to achieve the desired nucleotide conversions with higher efficiency, it is important to carefully select the gRNA with optimal editing windows for the specific base editor variant being used. It is recommended to include a gRNA targeting a safe harbor locus (e.g., AAVS1, CCR5) or a scramble control as a reference. The gRNAs should be synthesized with appropriate overhangs (Figure 1) to facilitate their cloning into the lentiviral vector, and desalted oligos are sufficient for this purpose. During the design of sequencing primers, careful consideration of the sequencing platform is essential, whether it is Next-Generation Sequencing (NGS) or Sanger sequencing. This critical assessment ensures that the primers are customized to meet the specific requirements of the chosen sequencing platform, thereby producing dependable and accurate sequencing data. The primers used in our study are provided in the key resources table.
Figure 1.
Representative image of the gRNA oligos to be synthesized
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| DH10B competent cells | ECOS, Yeastern Biotech | CAT # FYE507-10VL |
| Chemicals, peptides, and recombinant proteins | ||
| GoTaq G2 Hot Start Master Mix | Promega | CAT # M7422 |
| T4 polynucleotide kinase | NEB | CAT # M0201 |
| T4 DNA ligase | NEB | CAT # M0202 |
| BsmB1 | NEB | CAT # R0580 |
| Polyethylene glycol | Sigma-Aldrich | CAT # 89510 |
| Lenti-X Concentrator | Takara | CAT # 631232 |
| SCF | Immuno Tools | CAT # 11343325 |
| EPO | Zydus Nephrosciences | Zyrop 4000 IU injection |
| Penstrep | Gibco | CAT # 15140122 |
| Dexamethasone | Alfa Aesar | CAS # 1177-87-3 |
| Doxycycline | Sigma-Aldrich | CAS # 24390-14-5 |
| Glutamine | Gibco | CAT # 25030081 |
| FBS | Gibco | CAT # 10270106 |
| PBS | HyClone | CAT # SH30256.02 |
| Polybrene | Sigma-Aldrich | CAS # 28728-55-4 |
| HEPES | Gibco | CAT # 15630080 |
| Puromycin | Gibco | CAT # A1113803 |
| StemSpan SFEM-II | STEMCELL Technologies | CAT # 09655 |
| DMEM | HyClone | CAT # SH30243.01 |
| Opti-MEM reduced serum media | Gibco | CAT # 31985062 |
| FuGENE HD | Promega | CAT # E2312 |
| LB agar | HiMedia | CAT # M1151 |
| LB broth | HiMedia | CAT # M1245 |
| Ampicillin sodium salt | SRL | CAT # 61314 |
| QuickExtract DNA extraction solution | Epicentre | CAT # QE09050 |
| HighPrep PCR | MagBio | REF # AC-60005 |
| HighPrep DTR | MagBio | REF # DT-70005 |
| Critical commercial assays | ||
| Zymoclean Gel DNA Recovery Kit | Zymo Research | CAT # D4001 |
| NucleoBond Xtra Midi | Macherey-Nagel | REF # 740410 |
| NucleoSpin Blood – DNA kit | Macherey-Nagel | REF # 740951 |
| NucleoSpin plasmid | Macherey-Nagel | REF # 740588 |
| BigDye Terminator v3.1 Cycle Sequencing Kit | Applied Biosystems | CAT # 4337458 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | |
| HUDEP-2 | Cell Engineering Division, RIKEN BioResource Center | |
| Oligonucleotides | ||
| gRNAs cloned in #57822 | Ravi et al.1 | Check Table S1 |
| Sequencing F: gagggcctatttcccatgat | Ravi et al.1 | Sanger sequencing primer for colony PCR |
| Sequencing R: tggatctctgctgtccctgt | Ravi et al.1 | |
| HBF 1 F: acaaaagaagtcctggtatc | Ravi et al.1 | |
| HBF 2 F: ttactgcgctgaaactgtgg | Ravi et al.1 | |
| HBF 1 R: cttcccagggtttctcctcc | Ravi et al.1 | |
| NGS 2 F: gctcttccgatct tgaatcggaacaaggcaaagg | Ravi et al.1 | |
| NGS 2 R: gctcttccgatct gtgaaatgacccatggcgtc | Ravi et al.1 | |
| NGS 3 F: gctcttccgatct cctggacctatgcctaaaaca | Ravi et al.1 | |
| NGS 3 R: gctcttccgatct agtttagccagggaccgttt | Ravi et al.1 | |
| Recombinant DNA | ||
| pLKO5.sgRNA.EFS.GFP | Addgene | #57822 |
| pLenti-FNLS-P2A-Puro | Addgene | #110841 |
| pLenti-ABERA-P2A-Puro | Addgene | #112675 |
| pMD2.G | Addgene | #12259 |
| psPAX2 | Addgene | #12260 |
| Software and algorithms | ||
| Synthego ICE | Synthego | |
| EditR | EditR: Edit Deconvolution by Inference of Traces in R (shinyapps.io) | |
| CRISPResso2 | CRISPResso2 (partners.org) | |
| Benchling | CRISPR gRNA Design Tool: Benchling | |
| SnapGene | SnapGene: software for everyday molecular biology | |
| BE-Designer | http://www.rgenome.net/be-designer | |
| DeepBaseEditor | http://deepcrispr.info/DeepBaseEditor | |
| ACEofBASEs | https://aceofbases.cos.uni-heidelberg.de | |
| PnB designer | https://fgcz-shiny.uzh.ch/PnBDesigner | |
| CHOPCHOP v3 | CHOPCHOP (uib.no) | |
Step-by-step method details
Designing gRNA and ligation
Timing: 2 days
Here, we outline the process of designing gRNA and performing ligation for CRISPR-based genome editing. This two-day procedure involves the preparation of a lentiviral vector backbone for gRNA cloning and the annealing of oligos, followed by ligation to create a functional construct. This detailed walkthrough will facilitate the precise design and assembly of gRNAs, a fundamental component of CRISPR-based experiments, allowing for targeted genomic modifications.
Day 1
-
1.Preparation of backbone for cloning gRNAs:
-
a.Digest the lentiviral vector (Addgene #57822)7 using the BsmB1 enzyme.
-
i.Assemble the reaction mixture as follows.
Reagent Volume Addgene # 57822 (500 ng) 8 μL NEB buffer 3.1 (10×) 5 μL BsmB1 enzyme (NEB) 10 U/μL 2 μL Nuclease free water Upto 50 μL Total 50 μL -
ii.Incubate at 55°C for 3 h to ensure complete digestion.
-
i.
-
b.Perform gel electrophoresis of the digested product on a 0.8% agarose gel.
-
i.Ensure that the samples are run for at least 90 min to achieve sufficient separation.
-
ii.To serve as a control for digestion, load the undigested plasmid in the adjacent well.
-
i.
-
c.Carefully excise the desired digested backbone (∼8000 bp) using a clean blade and purify it using the Zymo gel purification Kit as per the manufacturer’s protocol.
-
d.Elute the purified product in 25 μL of nuclease-free water and adjust the concentration to 50 ng/μL.Note: The digestion and gel purification of 4 μg of plasmid will yield approximately 2000–2500 ng, which is adequate for cloning 40–50 gRNAs. It is essential to digest an adequate amount of plasmid, considering the total number of gRNAs to be cloned.
-
a.
Day 2
-
2.Annealing of gRNA oligos:
-
a.Reconstitute the lyophilized forward and reverse oligos to 100 pm concentration.
-
b.Assemble the oligo annealing reaction as follow in a PCR tube:
Reagent Volume Forward Oligo (100 pm) 1 μL Reverse Oligo (100 pm) 1 μL NEB ligase buffer 10× 1 μL T4 polynucleotide kinase 10 U/μL (NEB) 0.5 μL Nuclease free water 6.5 μL Total 10 μL -
c.Set up the following reaction in a thermocycler.
Cycles 1 Cycle 1 Cycle 1 Cycle 1 Cycle Temperature (oC) 37°C 95°C 25°C 4°C Time (min) 45 min 5 min 1 min ∞ Ramp rate Ramp 100% Ramp 100% Ramp 3% -
d.Take 1 μL of annealed product and dilute it (1:200) using sterile water. Store at 4°C until use.Note: The steps described are for a single gRNA. Perform similar reactions for all gRNAs.
-
a.
-
3.Ligation:
-
a.Set up the reaction as follows.
Reagent Volume Vector backbone (50 ng/μL) 1 μL Oligo product (1:200 diluted in H2O) (Insert) 6 μL NEB ligase buffer 10× 2 μL NEB ligase 0.5 μL Nuclease free water 10.5 μL Total 20 μL -
b.Mix thoroughly and incubate at 16°C for 12–16 h, then store at 4°C until transformation.
-
a.
Cloning and confirmation of clones
Timing: 5 days
This protocol outlines the step-by-step process for cloning and validating the guide RNAs (gRNAs) for use in downstream editing experiments. The protocol spans five days and includes key procedures such as transformation, colony picking, glycerol stock preparation, colony PCR, Sanger sequencing, and plasmid isolation. These critical steps ensure the generation of plasmids containing accurately cloned gRNAs, crucial for successful editing.
Day 1
-
4.Transformation and plating:
-
a.Prepare LB Agar plate with ampicillin (100 μg/mL) as per the requirement.
-
b.Thaw 50 μL of DH10B competent cells on ice.
-
c.Add 6 μL of ligated product to the cells and mix by flicking the tubes.
-
d.As a control add 50 ng of digested backbone to another vial of competent cells.
-
e.Keep on ice for 30 min. Set up water bath at 42°C.
-
f.After 30 min, give heat shock for 45 s at 42°C.
-
g.Rapidly transfer to an ice bath and incubate for 5 min.
-
h.Plate the whole volume on LB Ampicillin plate and incubate at 37°C for 12–14 h.
-
a.
Day 2
-
5.Picking colonies and preparation of glycerol stock:
-
a.Check the control plate and ensure that there are no colonies.
-
b.Prepare LB broth (3 mL for each gRNA).
-
c.Arrange 2 mL tubes containing one mL of LB broth (with ampicillin), 3 tubes for each gRNA.
-
d.Pick and inoculate 3 isolated colonies into this LB broth from the plate.
-
e.The plates can be stored at 4°C for about 1 week as a backup.
-
f.Keep the inoculum in a shaker incubator for 6 h at 37°C.
-
g.The inoculum should be visibly turbid by then.
-
h.From one mL of the inoculum take 500 μL and prepare glycerol stock by adding 500 μL of 50% glycerol, and store immediately at −80°C.
-
i.Centrifuge the remaining inoculum at 2700 g for 5 min.
-
j.Remove the supernatant and add 500 μL of Sterile water.
-
k.Centrifuge again, discard the supernatant, and add 500 μL of sterile water.
-
l.Mix well and store at 4°C.
-
m.Use this as a template for the colony PCR as described below.
-
a.
Day 3
-
6.Colony PCR:
-
a.Set up the PCR reaction as follows.
Reagent Volume Forward Primer (10 pm) 1 μL Reverse Primer (10 pm) 1 μL Processed inoculum 2 μL Master mix 2× (Promega) 10 μL Nuclease free water 6 μL Total 20 μL -
b.Set up the following reaction in a thermocycler.
-
i.The primers used are mentioned in the key resources table.
Steps Temperature Time Cycles Initial Denaturation 95°C 10 min 1 Denaturation 95°C 20 s 35 cycles Annealing 55°C 20 s Extension 72°C 45 s Final extension 72°C 10 min 1 Hold 16°C Forever -
ii.Run 5 μL of the PCR product on 1% agarose gel.
-
iii.Positive colonies are expected to exhibit a band at ∼500 bp (based on the primers designed).
-
i.
-
a.
-
7.Sanger sequencing:
-
a.Proceed for Sanger sequencing with one positive colony from each gRNA group.
-
b.Use the 15 μL of PCR product that was remaining after the gel run for sequencing.
-
a.
Day 4
-
8.Analyzing Sanger sequencing data:
-
a.Using the Sanger sequencing data, confirm whether the gRNA sequence has been cloned correctly.
-
b.Make sure that the sequencing data obtained matches exactly with the gRNA sequence (Figure 2).
-
a.
Note: If any clone yields a negative result, proceed to the sequencing stage for the subsequent clones.
-
9.Inoculation of positive clones:
-
a.After confirming the clones, proceed to plasmid isolation by inoculating the positive clones.
-
b.For each gRNA, prepare 10 mL of LB broth (with ampicillin).
-
c.Using a sterile loop, transfer a small volume of the glycerol stock from the positive clone into the LB broth.
-
d.Keep in a shaker incubator for 16 h at 37°C.
-
a.
Note: ensuring that the glycerol stock does not thaw completely during the process.
Figure 2.
Representative Sanger sequencing data for successful cloning of gRNA into lentiviral backbone
Day 5
-
10.Isolation of plasmid DNA from the inoculum:
-
a.Isolate the plasmid using MN mini plasmid kit as per the manufacturer’s protocol.
-
b.Subsequently, dilute the isolated plasmids to achieve a final concentration of 500 ng/μL. Store the plasmid at −20°C until use.
-
a.
Lenti-virus production for gRNA
Timing: 4–5 days
This section outlines the steps involved in the production and collection of lentiviral particles packaged with gRNA. The focus here is to culture healthy and actively dividing HEK293T cells, followed by the transfection of gRNA plasmids along with lentiviral packaging vectors to generate lentivirus. The protocol spans 4–5 days and includes critical procedures such as cell seeding, transfection, verification of successful transfection through GFP expression, and virus collection. These steps are essential for obtaining high-quality viral particles.
Day 1
-
11.Culturing HEK293T cells:
-
a.Seed healthy and actively dividing HEK293T cells in one well of 6 well plate so that it would reach 70–80% confluency in 24 h.
-
b.Increase the number of wells based on the number of gRNAs.
-
a.
Day 2
-
12.Transfection:
-
a.Ensure that HEK293T cells are approximately 80% confluent and healthy.
-
b.Bring the transfection reagent (FuGENE HD) to 25–30°C.
-
c.Prepare the transfection mixture as given below:
Reagent Final concentration Volume OptiMEM 100 μL Addgene # 12259 (500 ng/μL) 500 ng 1 μL Addgene # 12260 (500 ng/μL) 700 ng 1.4 μL 57822 plasmid with gRNA (500 ng/μL) 800 ng 1.6 μL FuGENE HD 3:1 (reagent (μL): DNA (μg)) 6 μL Total 110 μL -
d.Incubate the mixture in 25–30°C for 15–20 min.
-
e.Remove the cell culture dish with HEK293T cells from the incubator and place it inside the biosafety cabinet.
-
f.Add the transfection reagent dropwise to the cells.
-
g.Gently swirl the dish to ensure even distribution of the mixture, then return the dishes and incubate for 48 h.
-
a.
Day 4
-
13.Confirmation of transfection:
-
a.Before proceeding with virus collection ensure that the transfection was successful and gRNA plasmids are expressing GFP by using fluorescence microscopy.
-
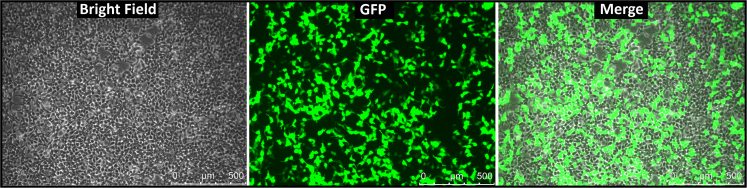
b.Successful transfection should result in over 75% of the HEK cells expressing GFP within 48 h (Figure 3).
-
a.
-
14.Virus collection:
-
a.Collect the supernatant from each well separately without disturbing the cells.
-
b.Clarify by spinning the supernatant at 400 g for 5 min at 25–30°C.
-
c.Transfer the 2 mL of clarified supernatant to another tube containing 700 μL of virus concentrator.
-
d.Mix well and incubate at 4°C for 1 h.
-
e.Centrifuge at 2700 g for 30 min at 4°C.
-
f.Discard the supernatant and resuspend the pellet in 100 μL of cold 1× PBS.
-
g.Store immediately at −80°C and do not thaw until use.
-
h.The HEK293T cells can be topped up with fresh media and the virus was collected again after 24 h.
-
i.This virus can be stored as a backup or used for trial transduction experiments if needed.
-
a.
Figure 3.
GFP expression in HEK293T cells 48 h after transfection
Preparation of base editor cell lines
Timing: 3 weeks
This protocol outlines steps essential for the production of lentiviral vectors and the establishment of stable cell lines harboring base editor plasmids. The protocol encompasses several key phases, starting with the preparation of endotoxin-free plasmids, followed by the production of lentivirus, and culminating in the creation of stable cell lines.
In this protocol, we have used codon optimized ABE and CBE max constructs with puromycin selectable marker from Lucas Dow (pLenti-FNLS-P2A-Puro (Addgene #110841-CBE) and pLenti-ABERA-P2A-Puro (Addgene #112675-ABE)).8 There are newer versions of base editors with increased processivity, modified PAMs, and editing windows, which can be obtained from plasmid repositories such as Addgene. To ensure consistent and reproducible results in downstream processes, we recommend using BE lentiviral constructs that incorporate antibiotic selection markers. These markers enable the isolation of pure transduced populations by conferring resistance to specific antibiotics, allowing the generation of stable cell lines with the desired characteristics. We have used second generation lentiviral packaging constructs in this study (Addgene #12259 and #12260), a gift from Didier Trono. Readers are directed to the review article by Anzalone et al. for a detailed comparison of various base editors available that would aid in choosing the BE for your application.9
Day 1
-
15.Endotoxin free plasmid preparation:
-
a.Inoculate the bacterial stock in LB broth with appropriate antibiotic (Ampicillin 100 μg/mL) and culture for 16 h.
-
b.Harvest the bacteria by centrifuging the inoculum at 2700 g for 10 min at 25–30°C in a 50 mL Falcon tube.
-
c.After centrifugation, carefully collect the pellet for plasmid isolation, and discard the supernatant.
-
d.Isolate the plasmid using NucleoBond Xtra Midi EF (Macherey-Nagel) kit.
-
a.
Note: Ensure that the final concentration of plasmid preparation is not below 500 ng/μL.
Day 2–5
-
16.Lentivirus production:Note: The steps described here are for preparing lentivirus with one of the above-mentioned constructs. Ensure the availability of reagents as per your need.
-
a.Day 2: Seeding cells.
-
i.Seed low passage HEK293T cells in a 10 cm cell culture dish that will result in 70–80% confluent in 24 h.
-
i.
-
b.Day 3: Transfection.
-
i.Make sure HEK293T cells are ∼80% confluent and are healthy.
-
ii.Bring the transfection reagent (FuGENE HD) to 25–30°C.
-
iii.Prepare the transfection mixture as given below in 1 mL Eppendorf tube:
Reagent Final concentration Volume OptiMEM 500 μL Addgene # 12259 (500 ng/μL) 2500 ng 5 μL Addgene # 12260 (500 ng/μL) 3500 ng 7 μL Addgene # 112675 (ABE7.10) (500 ng/μL) or Addgene # 110841 (BE 3) (500 ng/μL) 4000 ng 8 μL FuGENE HD 3:1 (reagent (μL): DNA (μg)) 30 μL Total 550 -
iv.Vortex well and incubate at 25–30°C for 15 min.
-
v.Take out the cell culture dish from the incubator and gently add the transfection reagent mixture dropwise to the cell culture medium inside the Bio Safety Cabinet (BSC).
-
vi.Swirl the dish to spread the mixture evenly and return the dishes to the incubator.
-
i.
-
c.Day 5: Harvesting the lentivirus.
-
i.48 h after transfection, collect the supernatant without disturbing the cells in a 15 mL falcon tube.
-
ii.Clarify by centrifugation at 400 g for 5 min to remove any unadhered cells and debris.
-
iii.Carefully transfer the supernatant to a falcon tube containing 4× Lenti X concentrator (Takara) or 4× PEG viral concentrator.
-
iv.The ratio of concentrator to media should be 1:3.
-
v.Mix well and incubate at 4°C for 1 h.Note: The HEK293T cells can be topped up with fresh media and the virus can be collected again after 24 h and will be processed as above. After collecting the virus bleach and discard the HEK293T cells.
-
vi.Composition for preparation of PEG virus concentrator.
Reagent Amount PEG-8000 (sigma) 200 g NaCl 35 g Milli Q water 200 mL 10x PBS (pH 7.4) HyClone 50 mL Dissolve by placing on a magnetic stirrer, with low heat if needed Makeup with MilliQ water 500 mL Vacuum filter using 500 mL 0.2 μ filter (Thermo) and store in 4°C -
vii.Centrifuge the incubated mixture at 2700 g for 30 min at 4°C.
-
viii.Remove the supernatant and resuspend the viral pellet in 200 μL of 1x PBS.
-
ix.Make two aliquots of 100 μL and store the virus at −80°C for the downstream process.Note: ensure that the lentivirus is transported and maintained on ice to prevent any drop in efficiency.
-
i.
-
a.
Day 6–26
-
17.Transduction of relevant cell line to prepare base editor stables:Note: This protocol describes the utilization of the HUDEP-2 cell line. However, it is important to note that the protocol can be readily customized and applied to different cell lines as needed.
-
a.Day 6: Revive approximately 0.5 million cells (1 million cells are needed on the day of transduction).
-
b.Day 8: Passage the cells and make sure they are healthy.
-
c.Day 10: Transduction.
-
i.Seed 0.5 million cells per well in 2 wells of a 6 well plate.
-
ii.To each well add 20 μL of 1 M HEPES buffer and 2 μL of polybrene (6 mg/mL).
-
iii.Carefully thaw the stored virus (100 μL) and add it into one well.
-
iv.The other well would serve as the non-transduced control.
-
v.Spinfection is carried out by centrifugation of the plate at 800 g for 30 min.
-
vi.Make sure the acceleration and brake are low (5 and 5).
-
vii.Keep the plates back in the incubator.Note: Use parafilm to seal the plate during centrifugation to avoid any accidental spillage as the culture contains lentivirus.
-
i.
-
d.Day 12 & 14: Selection of the stables.
-
i.Make sure that the transduced cells are viable.
-
ii.Reseed the cells in a fresh 6 well plate.
-
iii.Add 2 μL puromycin per mL of media (1 mg/mL) to both controls as well as the transduced samples.
-
iv.After two days check the cells under a microscope.
-
v.All cells in the control well must be dead and there would be ∼60%–80% cell death in the transduced well.
-
vi.Perform a media change and reseed the viable cells with the same concentration of puromycin.Note: If viable cells are persisting in the control well, increase the puromycin concentration until complete cell death is observed. Ensure that the same concentration is used for the transduced cells as well. Performing a literature search will provide insights into the suitable puromycin concentration for each specific cell line.
-
i.
-
e.Day 16–26: Stable cells maintenance and expansion.
-
i.Do media change on alternate days and maintain the cells in puromycin.
-
ii.Ensure the cells are healthy and expand enough.
-
iii.After 10 days of puromycin selection, the cells are ready for gRNA transduction.
-
iv.Freeze cells in multiple vials as backup and store them in liquid nitrogen (−196°C).Note: Transcriptome profiling of the stable cells can be performed and compared with wild type cells to ensure that during stable preparation the intrinsic properties of the cells are not altered significantly.
-
i.
-
a.
Transduction of gRNAs in base editor stables
Timing: 10 days
This comprehensive 10-day protocol focuses on transducing a base editing stable cell line with lentivirus carrying gRNA. The primary objective is to confirm the transduction and the editing obtained in the stable cells. The protocol encompasses crucial steps like maintaining optimal cell density, conducting lentiviral transduction, and assessing transduction efficiency through GFP expression via flow cytometry. Precise execution of these steps is imperative to achieve a transduction efficiency exceeding 95%, maximizing the potential for base substitutions across most cells. The protocol culminates in DNA isolation, target region amplification, and the assessment of base editing efficiency using data obtained from Sanger sequencing or NGS with appropriate software.
Day 1
-
18.Reviving stable cell line:
-
a.Revive and expand the cryopreserved base editor stable cells by culturing them in a growth medium supplemented with puromycin prior.
-
b.It is crucial to have approximately 0.2 million cells per guide RNA (gRNA) to facilitate simultaneous screening of the gRNAs.
-
c.Therefore, ensure an adequate quantity of cells to accommodate the screening of the gRNAs concurrently.
-
a.
-
19.Transduction:
-
a.Upon reaching the desired cell quantity, carefully distribute 0.2 million cells per well in a 12-well plate, utilizing 1 mL of media for each well.
-
b.To each well, add 10 μL of 1 M HEPES buffer and 1 μL of polybrene (6 mg/mL).
-
c.Thaw the stored lentivirus collected at 48 h and add it individually to each well.
-
d.Mix gently and perform spinfection by centrifuging at 800 g for 30 min with minimum acceleration and brake.
-
e.Incubate the cells at 37°C with 5% CO2 for 48 h.
-
a.
Note: The lentiviral transduction protocol mentioned above was optimized specifically for the HUDEP-2 cell line. For other cell lines, the viral titer and polybrene concentration may need to be adjusted accordingly to achieve a transduction efficiency exceeding 95%.
Note: Polybrene, HEPES buffer, and spinfection enhance lentivirus transduction efficiency by facilitating better viral entry, maintaining stable conditions, and increasing contact between viral particles and target cells, respectively.
Note: Measuring MOI values for each gRNA in the screening would be time consuming and incur additional cost. This is the reason why we recommend optimizing the transduction protocol to achieve maximum transduction efficiency.
Day 2
-
20.Evaluation of transduction efficiency:
-
a.Utilize approximately 150 μL of the cell culture for analysis.
-
b.Prior to analysis, wash the cells with 1x PBS.
-
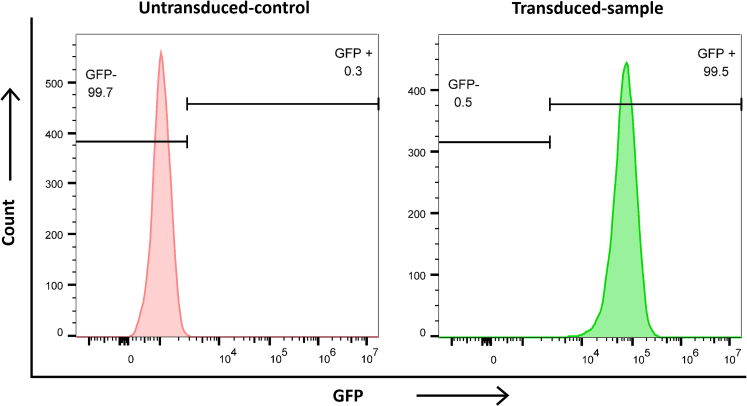
c.Evaluate transduction efficiency using flow cytometry by measuring the percentage of GFP expression (Figure 4).
-
d.Include a control group consisting of un-transduced cells for comparison.
-
e.Under ideal conditions and with careful execution of all protocol steps, it is anticipated that more than 95% of the cells will exhibit GFP expression.
-
f.If all cells are transduced efficiently with respective gRNA, transfer the cells to fresh media in a 6 well plate.
-
a.
Figure 4.
Representative flow cytometry data for evaluating transduction efficiency using intracellular GFP
Day 4
-
21.Maintenance and cryopreservation:
-
a.Perform media change by centrifuging the culture at 200 g and resuspending the cell pellet in fresh media.
-
b.Cryopreserve adequate cells in appropriate freezing media for backup storage prior to media change.
-
a.
Day 6–8
-
22.Maintenance:
-
a.Perform media change every 2 days to maintain the cells healthy and discard any excess cells.
-
a.
Note: If your functional assay requires more number of cells, expand the cells rather than discarding them.
Day 10
-
23.Evaluation of base editing in the stable cell line:
-
a.Isolate DNA from 0.5 million cells using NucleoSpin Blood – DNA kit (MN) as per the manufacturer’s protocol.
-
b.You can freeze the remaining cells or continue to culture them depending on the functional assays that need to be performed.Note: The protocol for evaluation of HbF expression can be found in Ravi et al.1 The control edited sample can be used as a base line to evaluate the HbF elevation induced by the gRNA.
-
c.Amplify the target region and perform Sanger sequencing/ targeted NGS using the primer mentioned in the key resources table.
- d.
-
a.
Figure 5.
Sequencing output from Sanger sequencing and NGS for Base editor
(A and B) Sanger sequencing data was analyzed for samples edited with ABE(A) and CBE(B)using EditR (the red color boxes indicate the desired converted bases and their substitution percentage). The chromatogram represents a substitution in A>G in ABE and C>T in CBE.
(C and D) Targeted deep sequencing (by NGS) of samples edited with ABE(C) and CBE(D) analyzed by CRISPResso-2. All possible outcomes in base editing are represented in the processed data from NGS.
Expected outcomes
The successful completion of this protocol will enable arrayed mutagenesis in specific genomic loci using adenine and cytosine base editing techniques. Our study focuses on screening a 300 bp region upstream of the gamma globin promoter in a human erythroid cell line to identify targets for enhancing fetal hemoglobin expression, potentially benefiting beta hemoglobinopathies and identifying new transcription regulators. By utilizing this approach, base substitutions can be introduced at any desired genomic sites, enabling the investigation of regulatory elements and the development of therapeutic gene editing strategies. This method serves both basic science research and translational applications. However, it is essential to ensure that the target region complies with the specific rules governing base editors, such as the availability of PAM sequences and the presence of target bases within the editing window region. The protocol outlines steps including gRNA design and cloning, lentivirus production, stable cell line generation, and assessment of editing efficiency using Sanger sequencing or NGS. The editing efficiency of gRNAs at a genomic locus varies depending on factors such as the target region, the position of the base within the target region, and the specific variants of base editors utilized. Consequently, different gRNAs may exhibit diverse editing efficiencies and functional outcomes. To obtain a more comprehensive insight into the editing and functional outcomes, we recommend referring to Ravi et al.1
Limitations
This protocol is specifically applicable for screening a constrained set of guide RNA (gRNA) targets. To accomplish saturation mutagenesis for more extensive DNA regions, alternative approaches such as pooled lentiviral screening should be employed. Arrayed screening, while providing greater reliability, entails higher costs and demands more labor-intensive procedures.
Troubleshooting
Problem 1
No GFP is expressed after transfection with one or more gRNA(s).
Potential solution
The lack of GFP expression might have occurred due to the mutation of GFP gene in the plasmid. It is advisable to select an alternate clone, perform sequence confirmation to ensure integrity, and subsequently utilize the plasmid isolated from the selected clone for conducting the experiments.
Problem 2
GFP expression is low in HEK cells 48 h after transfection.
Potential solution
Check the quality of the transfection reagent and optimize the ratio of reagent to DNA, if needed.
Problem 3
GFP expression is low after transduction.
Potential solution
Check the integrity of lentiviral packaging plasmids and perform restriction digestion for all the plasmids used for lentivirus production. Also, ensure that the lentivirus is stored and used properly.
Problem 4
Cells are dying after transduction.
Potential solution
Adjust the lentiviral titer to minimize cell death.
Problem 5
No editing with any gRNAs even after achieving >90% transduction efficiency.
Potential solution
Ensure puromycin selection is complete for cells expressing the base editors. Increase the concentration of puromycin until all the control cells are dead.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mohankumar KM (mohankumarkm@cmcvellore.ac.in)
Materials availability
This study did not generate new unique reagents. All reagents listed here can be found in the main article Ravi et al. (2022).
Data and code availability
This study did not generate any new data.
Acknowledgments
We thank all the laboratory members for their help in establishing the protocols and useful discussions. We thank Lokesh Panigrahi for critical reading of the manuscript. The research reported in this work was supported by NAHD grant BT/PR17316/MED/31/326/2015 (Department of Biotechnology, New Delhi, India), EMR grant EMR/2017/004363 (Science and Engineering Research Board [SERB], New Delhi, India), Indo-US GETin Fellowship_2018_066 (Indo-US Science and Technology Forum [IUSSTF]), and DBT grant BT/PR38392/GET/119/301/2020. We sincerely acknowledge CSCR (a unit of inStem, CMC Campus, Vellore, India) for providing the startup funds. N.S.R. and A.G. are supported by a Senior Research Fellowship from Council of Scientific & Industrial Research, India. We would like to acknowledge the CSCR core facility for supporting us with all the required instrumentation.
Author contributions
Conceptualization, N.S.R. and K.M.M.; investigation, N.S.R. and A.G.; writing – original draft, N.S.R. and A.G.; writing – review & editing, N.S.R., A.G., and K.M.M.; funding acquisition, K.M.M.; supervision, K.M.M.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Nithin Sam Ravi, Email: nithinsam91@gmail.com.
Kumarasamypet M. Mohankumar, Email: mohankumarkm@cmcvellore.ac.in.
References
- 1.Ravi N.S., Wienert B., Wyman S.K., Bell H.W., George A., Mahalingam G., Vu J.T., Prasad K., Bandlamudi B.P., Devaraju N., et al. Identification of novel HPFH-like mutations by CRISPR base editing that elevate the expression of fetal hemoglobin. Elife. 2022;11 doi: 10.7554/ELIFE.65421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018;19:542–547. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquart K.F., Allam A., Janjuha S., Sintsova A., Villiger L., Frey N., Krauthammer M., Schwank G. Predicting base editing outcomes with an attention-based deep learning algorithm trained on high-throughput target library screens. Nat. Commun. 2021;12:5114. doi: 10.1038/s41467-021-25375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornean A., Gierten J., Welz B., Mateo J.L., Thumberger T., Wittbrodt J. Precise in vivo functional analysis of DNA variants with base editing using ACEofBASEs target prediction. Elife. 2022;11:e72124–e72134. doi: 10.7554/eLife.72124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegner S.M., Karasu M.E., Schröder M.S., Kontarakis Z., Corn J.E. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinf. 2021;22:1–12. doi: 10.1186/s12859-021-04034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/NAR/GKW398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heckl D., Kowalczyk M.S., Yudovich D., Belizaire R., Puram R.V., McConkey M.E., Thielke A., Aster J.C., Regev A., Ebert B.L. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat. Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951.Generation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zafra M.P., Schatoff E.M., Katti A., Foronda M., Breinig M., Schweitzer A.Y., Simon A., Han T., Goswami S., Montgomery E., et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018;36:888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any new data.