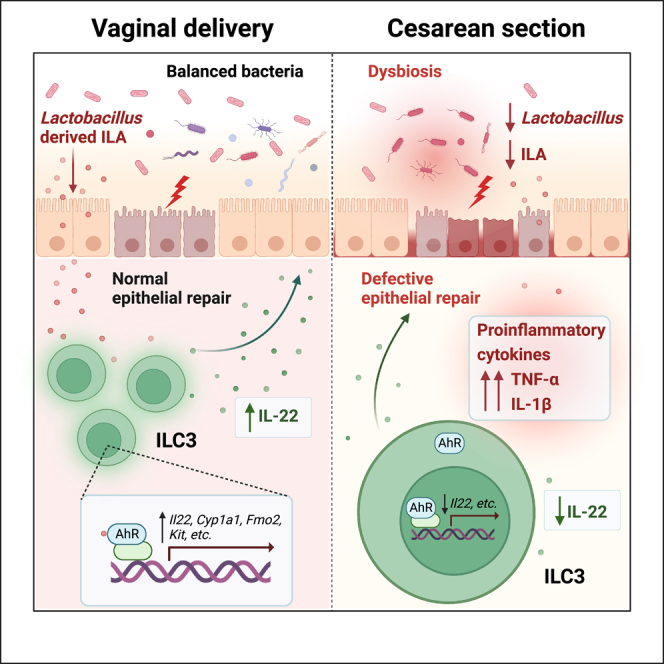
Summary
Cesarean section (CS) delivery is known to disrupt the transmission of maternal microbiota to offspring, leading to an increased risk of inflammatory bowel disease (IBD). However, the underlying mechanisms remain poorly characterized. Here, we demonstrate that CS birth renders mice susceptible to dextran sulfate sodium (DSS)-induced colitis and impairs group 3 innate lymphoid cell (ILC3) development. Additionally, CS induces a sustained decrease in Lactobacillus abundance, which subsequently contributes to the colitis progression and ILC3 deficiency. Supplementation with a probiotic strain, L. acidophilus, or its metabolite, indole-3-lactic acid (ILA), can attenuate intestinal inflammation and restore ILC3 frequency and interleukin (IL)-22 level in CS offspring. Mechanistically, we indicate that ILA activates ILC3 through the aryl hydrocarbon receptor (AhR) signaling. Overall, our findings uncover a detrimental role of CS-induced gut dysbiosis in the pathogenesis of colitis and suggest L. acidophilus and ILA as potential targets to re-establish intestinal homeostasis in CS offspring.
Subject areas: Biological sciences, Physiology, Immunology, Microbiology
Graphical abstract
Highlights
-
•
CS induces heightened susceptibility to DSS-induced colitis and gut dysbiosis
-
•
CS impairs ILC3 development both at steady state and during colitis
-
•
L. acidophilus-derived ILA ameliorates colitis and ILC3 deficiency in CS mice
-
•
ILA restores the level of IL-22 in ILC3 via AhR signaling pathway
Biological sciences; Physiology; Immunology; Microbiology
Introduction
Inflammatory bowel diseases (IBDs), including ulcerative colitis (UC) and Crohn’s disease (CD), are chronic and relapsing disorders of the gastrointestinal tract with increasing global incidence.1 Although the etiology of IBD is not fully understood, evidence suggests that both genetic predisposition and environmental factors contribute to the initiation and recurrence of these diseases.2 Recent studies have highlighted the role of intestinal microbiota in IBD pathophysiology.2,3,4 Germ-free mice experiments demonstrate the impact of gut microbiota on the early stage of intestinal immune system development, which potentially affects IBD susceptibility.5,6 For example, the colonization of neonatal germ-free mice with normal intestinal flora could alleviate oxazolone-induced colitis by abrogating the excessive accumulation of invariant natural killer T (iNKT) cells in the colon.7 Given the limitations of germ-free mice in recapitulating the complex host-microbiota interplay, it is imperative to explore whether gut microflora alterations caused by early-life disturbances in clinical interventions can have enduring effects on the development of intestinal immunity.
The colonization and expansion of gut microbiota are affected by various factors, among which cesarean section (CS) delivery plays a significant role.8,9,10 The incidence of CS has risen substantially in recent years, far exceeding the guidelines recommended by the World Health Organization.8,9 This increase in CS delivery is concerning due to its potential to deprive infants of the microbial exposure they would normally receive during a vaginal birth, which has been shown to be crucial for establishing a healthy gut microbiome.10,11,12 The majority of studies to date have indicated that CS-born infants are at a higher risk of immunologic and metabolic disorders, including allergy,13,14 IBDs,15,16 type 1 diabetes,17 and obesity.18,19 However, some of these associations remain controversial,20,21 and the underlying mechanisms are unclear. Numerous strategies have been tried out to restore a healthy gut microbiota composition in CS offspring, such as dietary probiotics, vaginal fluids seeding, and mother fecal transplantation,12,22,23 however, whether these interventions can reverse the long-term consequences related to CS remains poorly characterized. Therefore, it is crucial to evaluate the impact of CS delivery on gut microbiota and to assess the potential of intervention strategies in improving the associated health outcomes.
Lactobacillus, the most frequently isolated microorganism from the healthy human vagina,24 is important for promoting a healthy intestinal environment and restoring a balanced microbial population.25 Furthermore, several strains of Lactobacillus have exhibited immunomodulatory properties through the production of metabolites.26 Preclinical studies have shown its efficacy in alleviating colitis.27,28 However, infants born by CS display decreased relative abundance of Lactobacillus in the gut microbiome compared with their vaginal delivery (VD) counterparts.22,29 Whether this CS-induced reduction in Lactobacillus can affect gut immunity and IBD susceptibility remains to be further elucidated.
In this study, we investigated the effects of birth mode on gut microbiota and intestinal immunity in the dextran sulfate sodium (DSS)-induced colitis model. We observed that CS birth aggravated intestinal inflammation and impaired group 3 innate lymphoid cell (ILC3) activation both at steady state and during colitis. Furthermore, we detected significant alterations in gut microbiota composition induced by CS and identified the key roles of Lactobacillus acidophilus and its metabolite indole-3-lactic acid (ILA) in ameliorating colitis and restoring ILC3 function in CS offspring. Our findings provide additional insights into a microbiome-driven correlation between birth mode, gut immunity development, and colitis, which may have implications for the prevention and treatment of IBD.
Results
Cesarean section increases susceptibility to dextran sulfate sodium-induced colitis and alters the gut microbiome
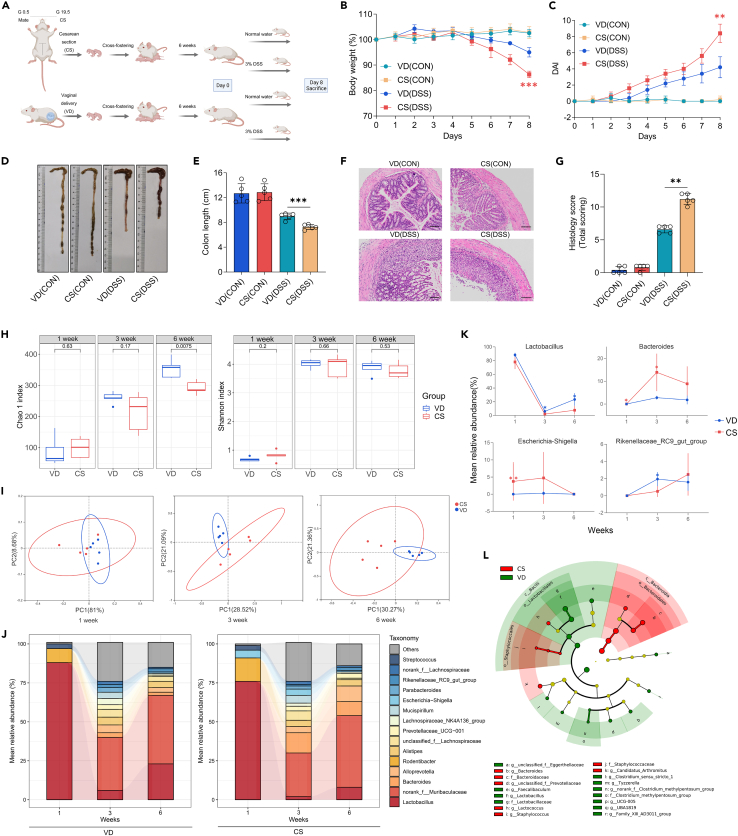
To explore the influence of birth mode on susceptibility to colitis later in life, ICR mice delivered via CS or vaginally were subjected to the continuous administration of 3% DSS or normal water for 8 days (Figure 1A). At steady state, no significant differences were observed comparing VD and CS mice. In the DSS-induced colitis group, CS-born mice exhibited aggravated intestinal inflammation, along with constant weight loss, higher disease activity index (DAI) score, shorter colon length, and increased disruption of epithelial crypt architecture compared with those born vaginally (Figures 1B–1G). Given the significant role of delivery mode in driving variation in the gut microbiota, we compared the composition of fecal flora in both VD and CS mice at different stages of their lives (early life: week 1, pre-weaning adolescence: week 3, and adulthood: week 6). Although the Shannon alpha diversity index was unchanged, the Chao1 index decreased significantly in CS mice at week 6 (p = 0.0015), indicating a lower microbial diversity in the feces of CS-born mice compared with VD mice in adulthood (Figure 1H). Principal coordinate analysis (PCoA) revealed that CS altered the beta diversity of the gut microbiota in mice, with the most pronounced changes occurring at the age of 6 weeks (Figure 1I). We analyzed the relative abundances of these taxa to identify the details of the bacterial alterations. At the genus level, the microbiota of VD and CS groups developed similarly, with a predominant abundance of Lactobacillus in early life and an expansion of norank_f_Muribaculaceae from 3 weeks onwards. However, the abundance of Lactobacillus was found to be decreased in CS mice compared to their VD counterparts, with particularly marked differences at 3 (p = 0.0253) and 6 (p = 0.0134) weeks of age. These findings are in line with prior clinical studies.22,29 By contrast, the Escherichia-Shigella genus, commonly associated with the environment, had a higher relative abundance level in CS mice compared with VD offspring at week 1 (p = 0.0079). From 3 weeks of age onwards, Bacteroides (p = 0.0166) was enriched while Rikenellaceae_RC9_gut_group (p = 0.0156) was significantly depleted in CS mice (Figures 1J and 1K). Differences in the abundance of other taxa between the two groups were minimal (Figure S1). The linear discriminant analysis effect size (LEfSe) results validated the finding that CS-born mice had a lower proportion of genus Lactobacillus (LDA = 5.367) in their intestines than the VD group in adulthood (Figure 1L).
Figure 1.
CS increases susceptibility to DSS-induced colitis and alters the gut microbiome
(A) Schematic representation of CS animal model and study design, created with BioRender.
(B–G) 3% DSS was administered to mice and disease assessments were carried out on day 8 by body weight loss (B), DAI scores (C), colon shortening (D and E), H&E staining with histopathological scores of the distal colon (F and G), scale bars, 100 μm (n = 5 per group).
(H) Boxplots of Chao1 and Shannon Diversity index calculated on fecal samples at week 1, week 3 and week 6 by unpaired t-test, compared between VD and CS groups (n = 5 per group).
(I) PCoA analysis of fecal microbiota structure in VD and CS mice at week 1 (ANOSIM; R = 0.328, p = 0.027), week 3 (ANOSIM; R = 0.532, p = 0.008), and week 6 (ANOSIM; R = 0.576, p = 0.004) based on Bray-Curtis distance, with ellipses indicating 95% confidence interval (CI) of datapoints (n = 5 per group).
(J) Changes in the mean relative abundances of bacterial genera in feces, sampled at week 1, week 3, and week 6 in both VD and CS groups. Only those genera with mean relative abundances of >2% across all samples are represented, while the remaining sequences are grouped as others (n = 5 per group).
(K) Several significantly changed microbes, including Lactobacillus, Bacteroides, Escherichia-Shigella and Rikenellaceae_RC9_gut_group are presented with their group means and standard errors (n = 5 per group).
(L) Cladogram based on LEfSe analysis from phylum to genus level with the value of the Kruskal–Wallis rank-sum test set to 0.05 on week 6 (n = 5 per group).
Each identified sequence was taxonomically categorized using the SILVA database (Release138 http://www.arb-silva.de) (J–L). Data shown as the mean ± SEM. One dot represents one mouse. ns, not significant, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. VD(CON): Mice born vaginally exposed to normal water; CS(CON): Mice born by CS exposed to normal water; VD(DSS): Mice born vaginally exposed to 3% DSS; CS(DSS): Mice born by CS exposed to 3% DSS.
Overall, these outcomes suggest that cesarean delivery in mice increases susceptibility to experimental colitis and causes significant alterations in the gut microbiome.
Gut microbiota plays a crucial role in the susceptibility of cesarean section mice to dextran sulfate sodium-induced colitis
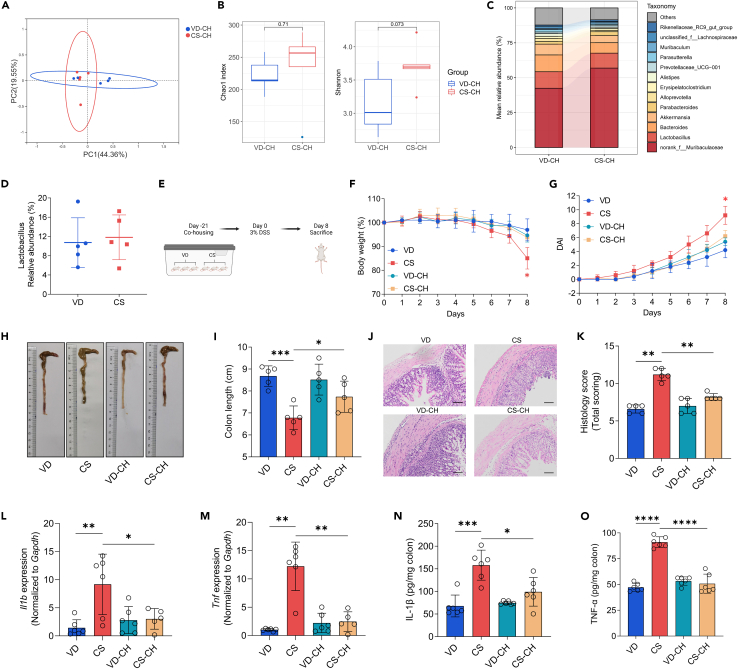
To examine the impacts of gut microbiota on experimental colitis susceptibility in CS mice, we conducted fecal microbiota transfer (FMT) via co-housing. To minimize litter effects, we randomly assigned littermates from multiple litters. The gut microbiota composition of co-housed VD (VD-CH) and co-housed CS (CS-CH) offspring was examined in adulthood. Co-housing reversed the lower alpha diversity indices in CS mice and there were no obvious differences in microbial community structure between the two groups based on PCoA analysis (Figures 2A and 2B). Most importantly, the reduction in genus Lactobacillus associated with CS was reversed by co-housing (Figures 2C and 2D). We administered 3% DSS in drinking water to VD, CS, VD-CH, and CS-CH mice (Figure 2E). Interestingly, compared with CS mice, CS-CH mice exhibited alleviated colitis symptoms, including reduced body weight loss, lower DAI and increased colon length (Figures 2F–2I). Histological analysis revealed that co-housing suppressed DSS-induced inflammatory cell infiltration and crypt destruction (Figures 2J and 2K). In addition, co-housing significantly reduced DSS-elevated levels of IL-1β and TNF-α in the colon of CS (Figures 2L–2O).
Figure 2.
Gut microbiota plays a crucial role in the susceptibility of CS mice to DSS-induced colitis
(A) PCoA analysis of fecal microbiota structure in co-housed VD and co-housed CS mice at steady state (n = 5 per group).
(B) Boxplots of Chao1 and Shannon Diversity index calculated on samples from co-housed VD and co-housed CS mice at steady state by unpaired t-test (n = 5 per group).
(C and D) Comparison of the mean relative abundance of fecal bacteria in co-housed VD and co-housed CS mice at the genus level (C) and the mean relative abundance of Lactobacillus in co-housed VD and co-housed CS mice at steady state (D). Each identified sequence was taxonomically categorized using the SILVA database (Release138 http://www.arb-silva.de) (n = 5 per group).
(E) Schematic representation of study design, created with BioRender.
(F–K) An 8-day course of 3% DSS was administrated to mice. Disease severity was assessed at day 8 by weight loss (F), DAI scores (G), colon shortening (H and I), H&E staining with histopathological scores of the distal colon (J and K), scale bars, 100 μm (n = 5 per group).
(L and M) qRT-PCR analysis of relative Il1b (L) and Tnf (M) expression in the colon tissue of mice from different groups treated with 3% DSS for 8 days (n = 5–6 per group).
(N and O) ELISA determined the protein levels of IL-1β (N) and TNF-α (O) in the colon tissue of mice from different groups treated with 3% DSS for 8 days (n = 6 per group).
Data shown as the mean ± SEM. One dot represents one mouse. ns, not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. VD-CH: VD co-housed; CS-CH: CS co-housed.
Our results collectively suggest that gut microbiota is partially responsible for the increased susceptibility to experimental colitis in CS mice.
Cesarean section impairs ILC3 development and IL-22 expression in the intestine
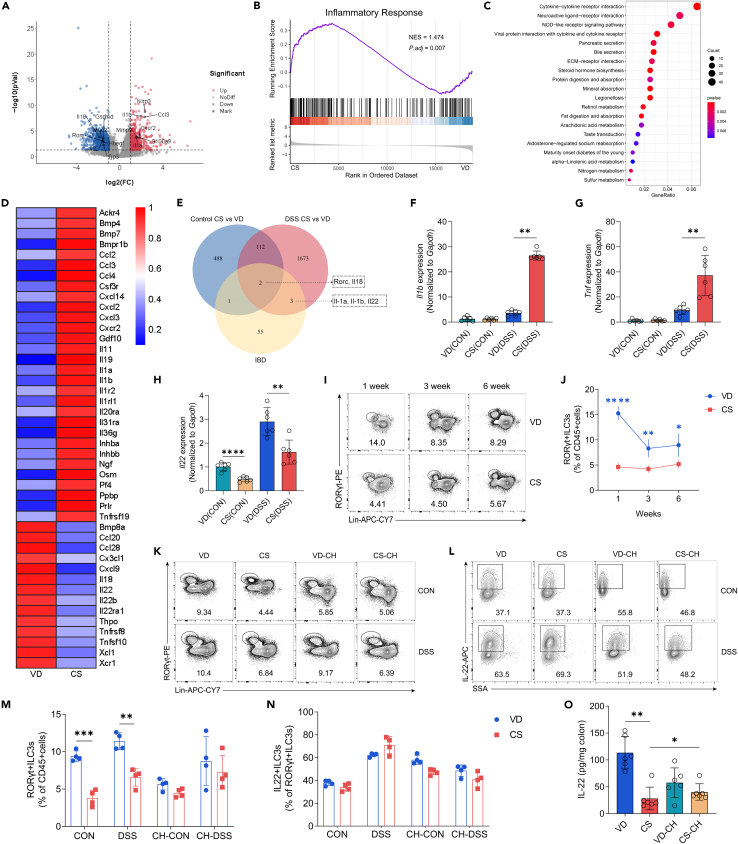
The impact of microbial colonization during early life on immune system development and host health is well-established.6,30 Therefore, it is imperative to investigate whether alterations in the intestinal microbiota resulting from CS would affect the colonic immune system. We conducted RNA-sequencing on colon tissues from VD and CS mice at steady state and after colitis induction. For comparison between CS and VD mice with DSS-induced colitis, we identified 1718 differentially expressed genes (DEGs). Specifically, transcripts associated with tissue repair (Il18, Il22, Hbegf), intestinal barrier (Muc2, Tjp3), and normal lymphoid organogenesis (Rorc) were down-regulated, and those linked to inflammatory responses (Il1a, Il1b, Nlrp3, S100a9) and immune cell recruitment (Ccl2, Ccl3, Cxcl2, Cxcl3, Cxcr2) were up-regulated in CS colitis mice (Figures 3A and 3C). Gene set enrichment analysis (GSEA) demonstrated an increase in inflammatory response gene set in CS colitis mice (Figure 3B). Furthermore, KEGG analysis of DEGs revealed the strongest enrichment of the cytokine-cytokine receptor pathway in CS colitis mice compared with VD colitis mice (Figures 3C and 3D). We also compared the colonic transcriptomes of CS and VD mice at steady state and identified 603 DEGs. Transcripts encoding calcium channels (Trpv1, Trpv3, Ephb6) were enriched in CS control mice (without DSS treatment), while those encoding proteins involved in gut homeostasis, including Ffar2, Rorc, Il18, Muc2, were down-regulated compared with VD control mice. Expression of Il22 was also decreased in CS control mice, but not to a significant degree. We further intersected the two gene sets (CS_CON versus VD_CON and CS_DSS versus VD_DSS) with genes in the "inflammatory bowel disease" list and identified 5 transcripts (Rorc, Il18, Il1a, Il1b, Il22) that were commonly dysregulated (Figure 3E). We then validated that colonic tissues from CS mice displayed lower Il22 and higher Il1b and Tnf expression than VD group by qRT-PCR (Figures 3F–3H). These data indicate that CS increases the expression of pro-inflammatory genes in colitis.
Figure 3.
CS impairs ILC3 development and IL-22 expression in the intestine
(A) Volcano plot showing the significantly different up-regulated and down-regulated genes of colon tissues in CS (DSS) group compared with those in VD (DSS) group (n = 4 per group).
(B) GSEA of RNA-seq data revealed gene sets significantly enriched in CS (DSS) mice compared with VD (DSS) mice, particularly for gene sets involved in the inflammatory response signaling pathway (n = 4 per group).
(C) Bubble plot showing the top 20 enriched KEGG pathways for DEGs between VD (DSS) and CS (DSS) group (n = 4 per group).
(D) Heatmap showing expression levels of DEGs between VD (DSS) and CS (DSS) group in the list of “Cytokine-cytokine receptor pathway” term in the KEGG database (n = 4 per group).
(E) Venn diagram illustrating the number of shared DEGs among the 3 gene sets (n = 4 per group). Control CS vs. VD: DEGs between CS (CON) and VD (CON); DSS CS vs. VD: DEGs between CS (DSS) and VD (DSS); IBD: Genes in the list of “inflammatory bowel disease” term in the KEGG database.
(F–H) qRT-PCR analysis of relative Il1b (F), Tnf (G) and Il22 (H) expression in the colon tissue of VD and CS mice from untreated control group or 3% DSS-treated group (n = 6 per group).
(I and J) Representative flow cytometry plots and percentage of colonic ILC3s (liveCD45+Lin-RORγt+) from VD and CS mice at defined ages (n = 4 per group).
(K–N) Representative flow cytometry plots and percentage of colonic ILC3s (K and M) and IL-22+ILC3s (L and N) from adult VD, CS, co-housed VD and co-housed CS mice at steady state or after exposure to 3% DSS (n = 4 per group).
(O) ELISA determined the protein levels of IL-22 in the colon tissue of VD, CS, co-housed VD and co-housed CS mice treated with 3% DSS (n = 6 per group).
Data shown as the mean ± SEM. One dot represents one mouse. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
It is noteworthy that IL-22 plays a crucial role in early host defense, epithelial barrier function, and mucosal repair.31,32 Additionally, tissue-resident ILC3s, characterized by expressing the transcription factor RAR-related orphan receptor gamma t (RORγt), constitute the primary source of IL-22 after intestinal damage.33,34 Based on the above results, we hypothesized that CS mice exhibited impaired development of ILC3s. To test this hypothesis, we detected the colonic ILC3 frequency and IL-22 expression in VD and CS mice at week 1, week 3, and week 6 under steady state by flow cytometry (Figures 3I and 3J). Notably, the ILC3 populations exhibited a reduction in CS mice both at steady state and after colitis induction. Expression of IL-22 was unaltered when comparing ILC3 subsets between VD and CS groups, suggesting that the reduced IL-22 level is probably caused by the decreased ILC3 cell number in CS mice. Interestingly, cohousing could partially normalize the reduced ILC3s frequency and lower IL-22 level in CS-born mice (Figures 3K–3N). Similarly, we also examined the proportion of other immune cell populations in the lamina propria, including neutrophils, monocytes, macrophages, CD4+ T cells, and ILC2s. There were no significant alterations observed between the VD and the CS groups either before or after DSS-induced colitis (Figure S2). We confirmed a reduction in IL-22 level in the colonic tissue of CS-born mice following treatment with DSS via ELISA. Notably, co-housing was found to reverse this reduction (Figure 3O).
Together, these results suggest that CS delivery leads to long-term alterations in the colonic immune system, resulting in the impaired development of ILC3s and reduced level of IL-22, which are associated with gut dysbiosis.
L. acidophilus strain supplementation attenuates dextran sulfate sodium-induced colitis and restores group 3 innate lymphoid cell frequency and IL-22 level in cesarean section mice
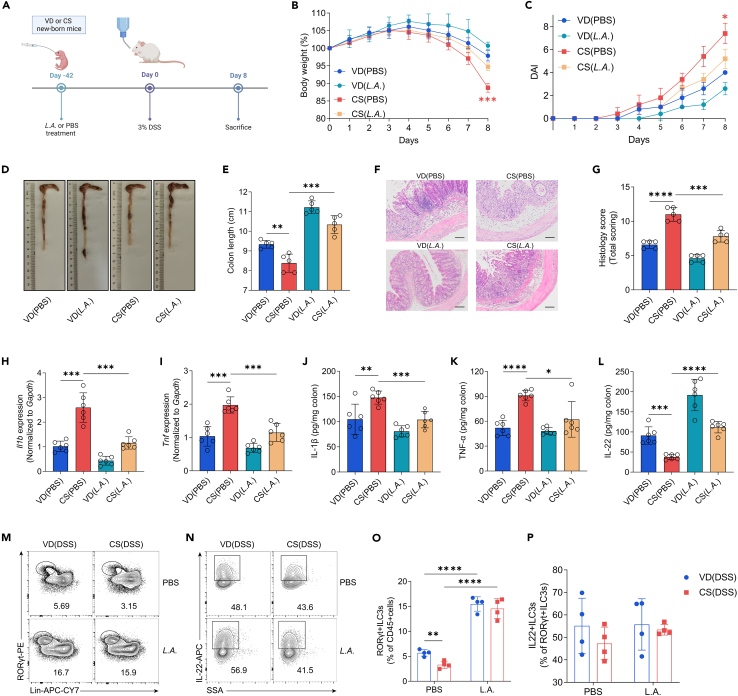
To investigate whether the reduced abundance of genus Lactobacillus contributed to the colitis severity and immune dysregulation in CS mice, we first determined the correlation between genus Lactobacillus relative abundance and ILC3 proportion in the intestine of VD and CS mice using Spearman correlation. Our results showed a significant positive correlation between genus Lactobacillus abundance and ILC3 frequency at week 3 and week 6 (Figure S3). Then, we treated mice with a probiotic strain, L. acidophilus, and an equal volume of PBS was used as a negative control (Figure 4A). We observed that probiotic-treated CS mice presented with more alleviated colitis symptoms including less weight loss, reduced DAI, and longer colon length as well as relatively milder histopathological outcomes than CS group treated with PBS. L. acidophilus treatment also reduced the colitis severity in VD-born mice but not to a significant degree (Figures 4B–4G). Moreover, CS colitis mice with probiotic-treatment showed lower levels of pro-inflammatory cytokines, including IL-1β and TNF-α compared with their PBS control counterparts (Figures 4H–4K). Although the IL-22 expression in ILC3s from L. acidophilus-treated mice was not significantly changed compared with that from PBS-treated CS mice, the proportion of ILC3s exhibited a significant increase in the colon of probiotic-treated CS mice (Figures 4M–4P). At steady state, the increase in the percentage of ILC3s in CS mice was also observed but less pronounced (Figure S4). ELISA confirmed that L. acidophilus administration alleviated the decreased expression of IL-22 in the colon of CS mice. (Figure 4L).
Figure 4.
L. acidophilus strain supplementation attenuates DSS-induced colitis and restores ILC3 frequency and IL-22 level in CS mice
(A) Schematic representation of L. acidophilus administration and experimental timeline, created with BioRender.
(B–G) Mice treated with L. acidophilus or PBS were given 3% DSS in drinking water for 8 days. Disease assessment was performed at day 8 by weight loss (B), DAI scores (C), colon shortening (D and E), H&E staining with histopathological scores of the distal colon (F and G), scale bars, 100 μm (n = 5 per group).
(H and I) qRT-PCR analysis of relative Il1b (H) and Tnf (I) expression in the colon tissue of mice from different groups treated with 3% DSS for 8 days (n = 6 per group).
(J–L) ELISA determined the protein levels of IL-1β (J), TNF-α (K), and IL-22 (L) in the colon tissue of mice from different groups treated with 3% DSS for 8 days (n = 6 per group).
(M–P) Representative flow cytometry plots and percentage of colonic ILC3s (M and O) and IL-22+ILC3s (N and P) from DSS colitis mice treated with L. acidophilus or PBS (n = 4 per group).
Data shown as the mean ± SEM. One dot represents one mouse. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. VD(PBS): VD mice treated with PBS and exposed to 3% DSS; CS(PBS): CS mice treated with PBS and exposed to 3% DSS; VD(L.A.): VD mice treated with L. acidophilus and exposed to 3% DSS; CS(L.A.): CS mice treated with L. acidophilus and exposed to 3% DSS.
The findings suggest that Lactobacillus deficiency in the gut microbiota is a contributing factor to the increased susceptibility of CS mice to experimental colitis. Additionally, probiotic treatment with an L. acidophilus strain can alleviate colitis symptoms and rescue ILC3 deficiency.
Indole lactic acid administration mitigates colitis severity and counteracts the reduced group 3 innate lymphoid cell frequency and IL-22 expression in cesarean section mice
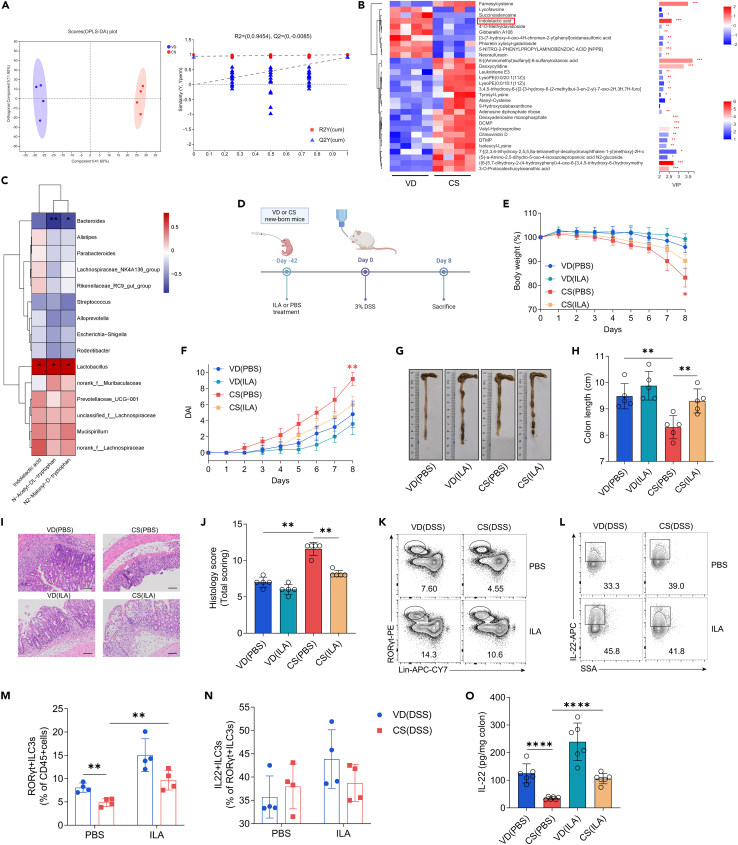
Since gut metabolites are essential bridges between the intestinal microflora and disease progression, we then investigated the changes in intestinal metabolic profiles in CS mice before colitis induction. Orthogonal partial least squares discriminant analysis (OPLS-DA) score plot revealed that metabolites of the two groups were distinctly separated (Figure 5A). Based on OPLS-DA coefficients (|FC| > 0.5) and VIP values (VIP >1), 30 significantly altered metabolites were selected. Among these, we focused on ILA, an important product involved in tryptophan metabolism,26,35 as it was significantly different between CS and VD groups and mainly produced by the genus Lactobacillus (Figure 5B). We observed a substantial positive correlation between three tryptophan metabolites and genus Lactobacillus abundance, but not other bacterial genera (Figure 5C). To investigate the role of ILA in the progression of experimental colitis in vivo, we administrated mice with ILA in the same way as L. acidophilus treatment (Figure 5D). Our results showed that CS mice treated with ILA had increased body weight, reduced DAI, less shortened colon length, and reduced histological destruction in colitis compared with CS control mice (Figures 5E–5J). Additionally, colonic ILC3 percentage and IL-22 expression in CS colitis mice with ILA treatment were higher than those in CS control offspring (Figures 5K–5N). ELISA showed that administering ILA effectively mitigated the reduction of IL-22 in the colon tissue of CS-born mice (Figure 5O).
Figure 5.
ILA administration mitigates colitis severity and counteracts the reduced ILC3 frequency and IL-22 expression in CS mice
(A) OPLS-DA score plots and permutation tests between adult VD and CS mice at steady state (n = 4 per group).
(B) Heatmap showing abundance of the top 30 metabolites based on VIP scores (VIP >1) of CS mice vs. VD controls (n = 4 per group).
(C) Heatmap illustrating Spearman’s rank correlation coefficients (two-sided tests) between the relative abundance of gut microbiota and concentrations of tryptophan catabolites in fecal samples from 6-week-old mice in both VD and CS groups. The genera of bacteria (with mean relative abundances of >2% across all samples) are derived from 16S rRNA gene sequencing. Each identified sequence was taxonomically categorized using the SILVA database (Release138 http://www.arb-silva.de).
(D) Schematic representation of ILA administration and experimental timeline, created with BioRender.
(E–J) Mice treated with ILA or PBS were given 3% DSS for 8 days. Disease assessment was conducted at day 8 by body weight loss (E), DAI scores (F), colon shortening (G and H), H&E staining with histopathological scores of the distal colon (I and J), scale bars, 100 μm (n = 5 per group).
(K–N) Representative flow cytometry plots and percentage of colonic ILC3s (K and M) and IL-22+ILC3s (L and N) from DSS colitis mice with different treatment (n = 4 per group).
(O) ELISA determined the protein levels of IL-22 in the colon tissue of mice from different groups treated with 3% DSS for 8 days (n = 6 per group).
Data shown as the mean ± SEM. One dot represents one mouse. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. VD(PBS): VD mice treated with PBS and exposed to 3% DSS; CS(PBS): CS mice treated with PBS and exposed to 3% DSS; VD(ILA): VD mice treated with ILA and exposed to 3% DSS; CS(ILA.): CS mice treated with ILA and exposed to 3% DSS.
These findings suggest that ILA supplementation alleviates the severity of experimental colitis and effectively mitigates the reduction of ILC3s and IL-22 in CS-born mice.
Indole-3-lactic acid affects group 3 innate lymphoid cell responses via the aryl hydrocarbon receptor signaling
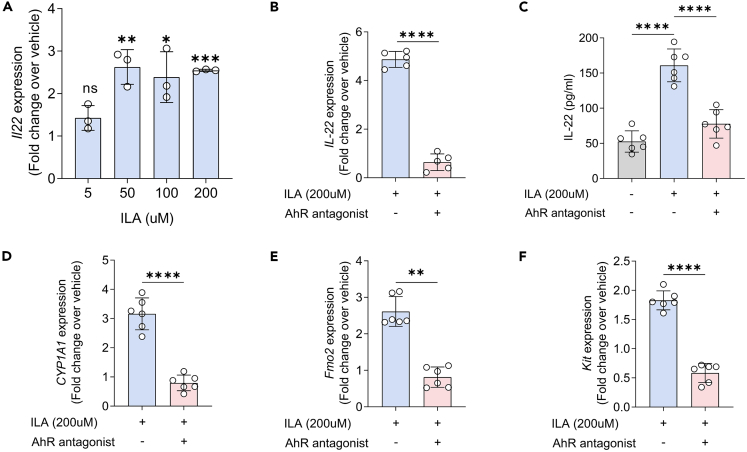
It has been demonstrated that ILA plays a crucial role in regulating intestinal and systemic homeostasis, particularly by binding to AhR.35 Considering that AhR is an essential transcription factor for ILC3 development and IL-22 secretion,36,37 we probed whether ILA affected ILC3 function through the AhR signaling. First, we isolated ILC3s from the lamina propria of the mice colon and treated these cells with ILA. We observed a dose-dependent IL-22 production by ILC3s following exposure to ILA, with the strongest effect at 200uM concentration (Figure 6A). Conversely, the ILA-induced IL-22 expression and secretion in ILC3s was inhibited with the addition of the AhR antagonist CH-223191 (Figures 6B and 6C). Furthermore, we also verified that the ILA-induced upregulation of AhR-targeted genes, such as Cyp1a1, Fmo2, and Kit37,38 were also blocked by the AhR antagonist in isolated ILC3s (Figures 6D–6F).
Figure 6.
ILA affects ILC3 responses via the AhR signaling
(A) Fold change in Il22 mRNA expression in isolated ILC3s stimulated with ILA at 5, 50, 100 and 200 μM as compared with vehicle (DMSO control) (n = 3 per group).
(B) Fold changes over vehicle (DMSO control) in Il22 mRNA expression in isolated ILC3s in the presence of 200 μM ILA with or without the AhR antagonist CH-223191 (n = 6 per group).
(C) ELISA determined the protein levels of IL-22 in isolated ILC3s in the presence of vehicle alone (DMSO) or 200 μM ILA with or without the AhR antagonist CH-223191 (n = 6 per group).
(D–F) Fold changes over vehicle (DMSO control) in Cyp1a1 (D), Fmo2 (E) and Kit (F) mRNA expression in isolated ILC3s in the presence of 200 μM ILA with or without the AhR antagonist CH-223191 (n = 6 per group).
Data shown as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Collectively, our results demonstrate that the influence of microbiota-derived ILA on IL-22 secretion by ILC3 subsets might be exerted through the AhR signaling pathway.
Discussion
The relationship between birth mode and the onset of IBD has been debated for an extended period. Although some epidemiological studies have suggested a higher susceptibility to IBD in individuals born by CS,15,16 others have failed to identify such a relationship.21 In this study, we demonstrated that CS exacerbated the severity of DSS-induced colitis, which is consistent with a previous report.39 Another study showed that CS increased sensitivity to oxazolone-induced colitis.40 The oxazolone model is a Th2-mediated process,41,42 while the DSS-induced colitis tends to reveal more about the interaction between gut microbiota and innate immunity in colitis.43,44 Thus, it is possible that the biological mechanisms through which CS may impact the pathogenesis of IBD are complex and multifaceted.
The transition from the in utero to the postnatal environment has a profound influence on the development of the pioneer intestinal microbiota.45 Previous human studies have reported that in comparison with infants born vaginally, CS-born babies have lower levels of genus Lactobacillus, which is associated with the mother’s vaginal microbiota.22,29,46 In agreement, our findings showed that CS led to persistent structural changes in the offspring’s gut microbiome, predominantly characterized by a reduced relative abundance of genus Lactobacillus. However, there are also studies revealing a higher abundance of genus Prevotella47 or a depletion of genus Bifidobacterium48 as a perturbation signature caused by CS in C57BL/6 and NIH Swiss mice, respectively. The discrepancy between our findings and those of others may attribute to the differences in the genetic background of mice. Lactobacillus has long been advocated in clinical studies for the prevention and treatment of various gastrointestinal disorders, including enteric infection, antibiotic-associated diarrhea, necrotizing enterocolitis in preterm infants, IBD, and colorectal cancer.49 Furthermore, Lactobacillus is known to exert its functions in a strain-specific manner.50 For example, a previous study revealed that L. reuteri D8 could accelerate intestinal epithelial cell proliferation and thus ameliorate mucosal damage caused by TNF-α exposure.28 Another study suggested that L. johnsonii treatment improved symptoms of DSS-induced colitis via regulating the innate immune responses.51 Consistent with these studies, our findings demonstrated that supplementation with an L. acidophilus strain successfully mitigated the severity of experimental colitis in CS mice, indicating that the colitis phenotype linked to CS birth mode is at least partially mediated by the disrupted gut microflora. Although we observed a similar effect of FMT in reversing susceptibility to experimental colitis caused by CS, it should be acknowledged that the lack of a well-defined target and the potential for transferring pathogens limit its clinical use.52 Therefore, it will be of interest to develop targeted therapy acting on specific microbiota for reestablishing intestinal homeostasis.
Recent studies suggest that the establishment of the microbiome in early life occurs in tandem with immune system development, expansion, and education.7,30,53,54,55 Our data showed high levels of pro-inflammatory cytokines in CS mice, indicating that CS delivery affects the intestinal immune system in colitis. A previous study revealed that CS mice had a decreased proportion of FoxP3+ regulatory T cells in the mesenteric lymph node and spleen, as well as a higher frequency of iNKT cells in colonic intraepithelial lymphocytes.40 However, the influence of CS on other immune cell types remains unclear. Here, we found that the expression of ILC3-related genes, including Rorc and Il22, were significantly downregulated in the colon of CS mice both at steady state and after colitis induction. The decreased ILC3 frequency and IL-22 levels in CS offspring could be corrected by co-housing, indicating that gut microbiota is partially responsible for the immune disturbances caused by CS. Previous studies have demonstrated that signals from the microbiota critically influence ILC3 responses either directly or indirectly via regulating epithelial and myeloid responses.56,57,58 Lactobacillus has been shown to induce IL-22 production by ILC3s.26 In agreement, we found that administration with an L. acidophilus strain to CS mice restored ILC3 frequency and IL-22 levels in experimental colitis. However, the effects of L. acidophilus on CS-born mice were not prominent at steady state. These results indicate that inflammatory background may be a variable influencing the role of L. acidophilus in the regulation of ILC3s.
Previous studies have suggested the capability of Lactobacillus to regulate immune functions in part through their metabolic products, including ILA.26,28 In this study, we observed lower fecal ILA content in CS mice, and administration of ILA to CS offspring effectively reduced the susceptibility to experimental colitis and rescued the defective IL-22 production. These results may underscore a mechanism by which L. acidophilus reestablished intestinal immune homeostasis in CS offspring to protect them against colitis. Apart from Lactobacillus, other gut bacteria also have the ability to generate ILA. Several species of Clostridium and Bacteroides are capable of converting tryptophan into ILA.59,60 Furthermore, Bifidobacterium has been reported to be a major source of ILA in humans.30,35,61 Considering the intricate interplay between gut microbiota and the host, further research is necessary to explore whether ILA derived from other gut bacteria, such as Bifidobacteria, could offer similar or perhaps more potent benefits in mouse models or human subjects with increased susceptibility to colitis induced by CS.
ILA has been shown to activate AhR, which is a transcription factor that plays a pivotal role in the ILC3-regulated development of isolated lymphoid follicles and cryptopatches, as well as local IL-22 production.26,37,62,63 Deficiency in AhR ligand production can impair IL-22 level in ILC3s, which may disrupt epithelial barriers and immune defense.31,32 In line with these findings, we demonstrated that ILA could activate mouse ILC3s, upregulating the expression of genes associated with the AhR signaling pathway, which were suppressed by the AhR antagonist. These results indicate that the stimulatory effect of ILA on ILC3s is at least partially mediated by AhR signaling. Beyond affecting ILC3s, ILA has been demonstrated to affect other immune cells. For instance, ILA is known to modulate intestinal T cell differentiation to maintain the delicate balance between immunity and tolerance in the gut.64 It can also regulate pro-inflammatory cytokine production by monocytes35 or intestinal epithelial cells61,65 to avoid excessive inflammatory damage. Moreover, ILA has the capability to mediate immune suppression of macrophages.66 In addition to regulating immune responses, a more recent study has reported that ILA can also directly modulate the gut microbiota by promoting beneficial bacteria and reducing potentially harmful ones.67 In light of these multifaceted roles of ILA, it is conceivable that some of the protective effects we observed in our CS mice might be due to these alternative mechanisms beyond the rescue of ILC3s and IL-22. Therefore, further investigations, including the use of genetic engineering or other methods, are warranted to elucidate the specific mechanisms of ILA-mediated colitis amelioration in CS offspring.
In summary, our findings uncover the role of L. acidophilus and its metabolite ILA in mitigating the increased susceptibility to colitis in CS mice, as well as restoring the ILC3 population and IL-22 level. Our research provides an important rationale for future interventions directed at targeting gut microbiota and their metabolites to retain immune homeostasis of CS-born infants and prevent the potential long-term adverse outcomes associated with CS in their adulthood.
Limitations of the study
Our study is currently limited to animal models, and clinical samples are required to validate our findings. Furthermore, our findings support the potential use of L. acidophilus and ILA as targeted therapies for ameliorating colitis in individuals born by CS, however, additional research is required to determine the optimal dosage, duration, and safety of this intervention. Finally, while our results suggest a relationship between the gut microbiota, the ILC3-IL-22 axis, and the susceptibility to experimental colitis in CS offspring, the role of other immune-related alterations and non-immunological factors associated with delivery mode, such as physical stress and hormonal exposure, still needs further investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD4 Antibody | Biolegend | Cat# 100405; RRID: AB_312690 |
| APC/Cyanine7 anti-mouse CD19 Antibody | Biolegend | Cat# 152412; RRID: AB_2922473 |
| PE anti-mouse Ly-6C Antibody | Biolegend | Cat# 128007; RRID: AB_1186133 |
| Alexa Fluor® 700 anti-mouse CD45 Antibody | Biolegend | Cat# 103128; RRID: AB_493715 |
| APC/Cyanine7 anti-mouse CD11c Antibody | Biolegend | Cat# 117324; RRID: AB_830649 |
| APC/Cyanine7 anti-mouse CD3ε Antibody | Biolegend | Cat# 100330; RRID: AB_1877170 |
| APC/Cyanine7 anti-mouse CD11b Antibody | Biolegend | Cat# 101226; RRID: AB_830642 |
| PE/Cyanine7 anti-mouse CD11b Antibody | Biolegend | Cat# 101216; RRID: AB_312799 |
| FITC anti-mouse CD11b Antibody | Biolegend | Cat# 101205; RRID: AB_312788 |
| APC/Cyanine7 anti-mouse CD45R/B220 Antibody | Biolegend | Cat# 103224; RRID: AB_313007 |
| APC/Cyanine7 anti-mouse TER-119 Antibody | Biolegend | Cat# 116223; RRID: AB_2137788 |
| APC/Cyanine7 anti-mouse FcεRIα Antibody | Biolegend | Cat# 134326; RRID: AB_2572064 |
| APC/Cyanine7 anti-mouse CD5 Antibody | Biolegend | Cat# 100650; RRID: AB_2876396 |
| APC/Cyanine7 anti-mouse Ly-6G Antibody | Biolegend | Cat# 127624; RRID: AB_10640819 |
| FITC anti-mouse Ly-6G Antibody | Biolegend | Cat# 127605; RRID: AB_1236488 |
| APC/Cyanine7 anti-mouse CD16/32 Antibody | Biolegend | Cat# 101328; RRID: AB_2104158 |
| Brilliant Violet 421™ anti-mouse F4/80 Antibody | Biolegend | Cat# 123131; RRID: AB_10901171 |
| Pacific Blue™ anti-mouse FOXP3 Antibody | Biolegend | Cat# 126410; RRID: AB_2105047 |
| BV421 Mouse Anti-GATA3 | BD Biosciences | Cat# 563349; RRID: AB_2738152 |
| ROR gamma (t) Antibody (AFKJS-9) PE | Thermo Fisher Scientific | Cat# 12-6988-82; RRID: AB_1834470 |
| IL-17A Antibody (eBio17B7) PE-eFluor™ 610 | Thermo Fisher Scientific | Cat# 61-7177-82; RRID: AB_2574656 |
| IL-22 Antibody (IL22JOP) APC | Thermo Fisher Scientific | Cat# 17-7222-82; RRID: AB_10597583 |
| T-bet Antibody (eBio4B10 (4B10)) PE | Thermo Fisher Scientific | Cat# 12-5825-82; RRID: AB_925761 |
| IFN gamma Antibody (XMG1.2) PE-Cyanine7 | Thermo Fisher Scientific | Cat# 25-7311-82; RRID: AB_469680 |
| Bacterial and virus strains | ||
| Lactobacillus acidophilus CGMCC 0460.2 | Grand Pharmaceutical Group Limited | N/A |
| Biological samples | ||
| Mice fecal samples | This study | N/A |
| Mice colon tissue | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Murine IL-2 | PeproTech | Cat# 212-12 |
| Recombinant Murine IL-7 | PeproTech | Cat# 217-17 |
| Recombinant Mouse IL-23 | Biolegend | Cat# 589002 |
| RPMI 1640 | Gbico | Cat# 11875119 |
| IMDM | Gbico | Cat# 12440053 |
| Fetal Bovine Serum (FBS) | Gbico | Cat# 10099141C |
| Penicillin/streptomycin | Gbico | Cat# 15070063 |
| HEPES | Gbico | Cat# 15630130 |
| EDTA | Invitrogen | Cat# AM9260G |
| Deoxyribonuclease I | Sigma | Cat# DN25-1G |
| Collagenase Type VIII | Sigma | Cat# C2139-5G |
| Percoll | Sigma | Cat# GE17-0891-09 |
| Indole-3-lactic acid | Sigma | Cat# I5508 |
| CH-223191 | Sigma | Cat# C8124 |
| Critical commercial assays | ||
| Mouse IL-1β ELISA Kit | Lianke Biotechnology | Cat# EK201B |
| Mouse TNF-a ELISA Kit | Lianke Biotechnology | Cat# EK282 |
| Mouse IL-22 ELISA Kit | Lianke Biotechnology | Cat# EK222 |
| Deposited data | ||
| 16S rRNA gene sequencing data | This paper | SRA: PRJNA958442 |
| RNA-seq dataset | This paper | GEO: GSE231473 |
| Experimental models: Organisms/strains | ||
| ICR Mouse | SPF(Beijing) Biotechnology Co., Ltd | N/A |
| Oligonucleotides | ||
| Primers for quantitative PCR, see Table S1 | This paper | N/A |
| Universal 16S rRNA V3-V4 region primers | Jin et al.55 | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.0.2 | GraphPad Software | N/A |
| FlowJo 10 software | FlowJo LLC | N/A |
| Gallios flow cytometer | Beckman Coulter | N/A |
| MoFlo Astrios EQ | Beckman Coulter | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Yanqing Li (liyanqing@sdu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
ICR mice were purchased from Beijing SPF Biotechnology Co., Ltd. ICR outbred female mice have greater maternal care than the frequently-used B6 mice, making them a good choice when choosing a mouse strain.68 Experiments were performed in sex-matched ICR mice aged 6–8 weeks, except for CS surgery. All mice were maintained in a specific pathogen-free (SPF) condition at Shandong University, under a 12:12-h dark-light cycle and stringent temperature and humidity (22 ± 2°C, 45%). Animal experiments were supported by the Ethical and Institutional Animal Care and Use Committee of Qilu Hospital of Shandong University (DWLL-2020-07).
Method details
CS surgery
Gestational day 0.5 (G0.5) was recorded by the discovery of a vaginal plug. On day 19.5 of gestation, pregnant female mice were euthanized and 70% ethanol was applied to sterilize the abdominal skin. An incision was made in the abdomen, then the uterus was dissected out and positioned on a sterile gauze with a heating pad underneath. The uterus was incised and gentle pressure was applied to remove the pups. Using sterile cotton swabs, pups were massaged until they were able to breathe spontaneously, which was followed by cutting the umbilical cord. The pups born by CS were fostered by a female giving birth on the same day. The litters of other pregnant females that delivered spontaneously were used as VD controls. The effects of foster care were controlled by cross-fostering in the vaginally born group (Figure 1A).
Co-housing procedure
The co-housing procedure was taken from Caruso et al.69 In brief, at 3 weeks old, VD and CS mice were weaned and co-housed together in a 1:1 ratio. A comparison was made between co-housed mice and their singly housed littermates.
Probiotic administration
The pups delivered by VD or CS received Lactobacillus acidophilus CGMCC 0460.2 at a dose of 1 × 109 CFU/kg body weight in sterile PBS or PBS alone as control once a day by oral gavage, starting from birth and throughout the experiment, which has been approved by the National Medical Products Administration in China. The probiotic was provided by Grand Pharmaceutical Group Limited.
DSS-induced colitis
DSS-induced colitis model was established as previously described.70 In brief, 6-week-old mice born by VD or CS were administered 3% DSS (MP Biologicals, USA) for 8 consecutive days. The control groups were provided with normal drinking water. A daily body weight measurement was conducted throughout the experiment. DAI scores were calculated as previously described.70 Mice were sacrificed on day 8 post-DSS. A colon length measurement was conducted, and colon tissues and fecal samples were collected for future analysis.
Histology
Distal colonic tissue was first fixed in 4% paraformaldehyde, followed by embedding in paraffin. Staining of 4-μm sections with hematoxylin and eosin (H&E) was performed following the manufacturer’s protocol. The histological scores were determined according to previously described methods.27
Quantitative real-time PCR (qRT-PCR)
Total RNA from colon tissues or isolated ILC3s was extracted using TRIzol (Invitrogen) and reversely transcribed into cDNA using ReverTra Ace® qPCR RT Kit (Toyobo). Then, qRT-PCR was done by using SYBR® Green Realtime PCR Master Mix (Toyobo). All the gene-specific primers were synthesized by Sangon Biotech (Sangon, Shanghai). Primer sequences are listed in Table S1. Gene expression was quantified using 2-ΔΔCt approach in relation to endogenous Gapdh control.
Enzyme-linked Immunosorbent Assay (ELISA)
For colonic tissues, weighted samples were homogenized, and supernatants were collected. The concentrations of IL-1β, TNF-α, and IL-22 were measured by corresponding Mouse ELISA Kits (Lianke Biotechnology), following the manufacturer’s instructions. For ILC3s, cell-free supernatants were harvested after treatment. The IL-22 concentrations were detected by Mouse IL-22 ELISA Kit (Lianke Biotechnology).
RNA sequencing (RNA-seq)
Total RNA from colon tissues was extracted and purified using TRIzol reagent under the manufacturer's instructions. A quantitative analysis of RNA abundance and purity was performed on each sample and then, RNA Integrity Number (RIN) was assessed. Transcriptome sequencing and bioinformatics analysis were performed by Lianchuan Biotechnology Company (Hangzhou, China). P-adjust threshold of 0.05 and an absolute log 2 (fold change) threshold of 1 were utilized as the screening criteria for differential genes. Data analysis was conducted using OmicStudio (http://www.omicshare.com/tools), an online platform.
Fecal 16S rRNA microbial analysis
Feces of the mice born by VD or CS at different time points (1 week, 3 weeks, and 6 weeks) were collected and stored in a −80°C freezer. DNA from fecal samples was extracted using FastDNA SPIN Kit (MP Biomedicals, Irvine, CA, USA). The 16S V3-V4 region was amplified by PCR using universal primers.55 The purified PCR products were sequenced by Majorbio (Shanghai, China). Each identified sequence was taxonomically categorized using the SILVA database (Release138 http://www.arb-silva.de). The statistical analysis was conducted on Majorbio I-Sanger Cloud Platform (https://cloud.majorbio.com/). To normalize the variations in sequencing depth, the estimates (after the exclusion of samples with <30,000 reads) were calculated by evenly sampling. Alpha diversities were analyzed using the Chao1 and the Shannon index calculated with the OTU. PCoA was conducted based on Bray-Curtis dissimilarity. The structural differences between VD and CS groups were investigated using analysis of similarity (ANOSIM). LEfSe was used to analyze differential abundances. Taxonomic cladograms, based on the significant taxa, were constructed on the Galaxy web application (http://huttenhower.org/galaxy/).
Untargeted fecal metabolomic analysis
Stool samples from VD and CS mice at 6 weeks were sent to Majorbio (Shanghai, China) for untargeted metabolomic analysis using LC-MS. Data analysis was carried out on Majorbio I-Sanger Cloud Platform (https://cloud.majorbio.com/).
Isolation of intestinal lamina propria lymphocytes (LPLs) and flow cytometry
Colonic LPLs were obtained following a previously established method.37 Briefly, colons were removed, opened longitudinally, sliced into 0.5 cm sections, and washed in pre-cold PBS. Intestinal epithelial cells were removed by incubation on a shaker in HBSS containing 10 mM HEPES (Gibco) and 2 mM EDTA (Invitrogen). The remaining pieces were then digested in RPMI-1640 medium (Gibco), addition with 5% fetal bovine serum (FBS; Gibco), 1 mg/mL collagenase VIII, and 0.1 mg/mL DNase I (Sigma-Aldrich) at 37°C for 1.5 h. After filtered through 70-μm cell strainers, LPLs were further enriched by a 40%/80% Percoll (Sigma-Aldrich) gradient centrifugation. For flow cytometry, single-cell suspensions were incubated with Aqua (Invitrogen) at ambient temperature for 25 minutes. Fc receptors were blocked by using unlabeled anti-CD16/32 (Invitrogen). The following antibodies were used for cell-surface staining: CD4 (GK1.5), CD19 (1D3/CD19), Ly-6C (HK1.4), CD11b (M1/70), CD45 (30-F11), CD11c (N418), F4/80 (BM8), and CD3ε (145-2C11). Lin comprised CD11c (N418), CD11b (M1/70), CD3ε (145-2C11), CD19 (1D3/CD19), CD45R/B220 (RA3-6B2), TER-119 (TER-119), FcεRIα (MAR-1), CD5 (53-7.3), Ly-6G (1A8), and CD16/32 (93). Antibodies used for intracellular staining included Foxp3 (MF-14), GATA3 (L50-823), RORγt (AFKJS-9), IFN-γ (XMG1.2), IL-17A (eBio17B7), IL-22 (IL22JOP) and T-bet (eBio4B10(4B10)). All these antibodies were purchased from Biolegend, Thermo Fisher Scientific, or BD Biosciences. A Gallios flow cytometer (Beckman Coulter) was used to detect samples, and analysis of flow cytometry data was carried out by FlowJo 10 software (FlowJo LLC).
ILC3s sorting and culture
The gating strategy used to analyze ILC3s in the large intestine was described previously.37 The cell isolation was conducted in a MoFlo Astrios EQ (Beckman Coulter). Sorted ILC3s were cultured in Iscove's Modified Dulbecco's Medium (IMDM; Gibco) supplemented with 10% FBS, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin (all from Gibco) in the presence of IL-2 (10μg/mL, Peprotech), IL-7 (10μg/mL, Peprotech), and IL-23 (20μg/mL, Biolegend). Cells were treated with ILA (Sigma I5508) dissolved in 0.1% DMSO at the final concentrations of 5, 50, 100, or 200 μM. 0.1% DMSO alone was used as vehicle control. Before adding the above compounds, ILC3s were pretreated for 1 hour with the AhR antagonist CH-223191 (Sigma C8124) at a concentration of 10 μM. Stimulated cells and supernatant were harvested after 24h treatment and stored at –80°C for subsequent use.
Quantification and statistical analysis
Statistical analysis was carried out using GraphPad Prism 8.0.2 software. Data were expressed as mean ± SD. Normality was verified by Shapiro-Wilk test. Statistical significance was determined by ANOVA or unpaired two-tailed Student’s t-tests. Mann–Whitney U tests or Wilcoxon-signed rank tests were performed as necessary. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Jie Yan for her valuable advice during the article drafting. This study was supported by the National Natural Science Foundation of China (82070552 and 81873550), Key Research and Development Program of Shandong Province (2021CXGC010506). This study is also supported by the Taishan Scholars Program of Shandong Province.
Author contributions
Y.-N.X. and Y.-Q.L. conceived the project. Y.-N.X. performed the research and analyzed the data. C.L. helped with experiments and data analyses. Y.-N.X. and C.L. wrote the article. R.-J.L. contributed to mouse models. M.-Q.Z., B.-C.F., J.-H.G., and X.L. provided scientific advice and technical support. L.-X.L. and S.-Y.L. revised the article. X.-L.Z. and Y.-Q.L. supervised the project and provided financial support. All authors contributed to the article and approved it for publication.
Declaration of interests
The authors declare no competing interests.
Published: October 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108279.
Contributor Information
Xiuli Zuo, Email: zuoxiuli@sdu.edu.cn.
Yanqing Li, Email: liyanqing@sdu.edu.cn.
Supplemental information
Data and code availability
-
•
Data: The 16S rRNA gene amplicon sequencing data and RNA-seq data have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database and Gene Expression Omnibus (GEO) database, respectively, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Other items: No other new unique reagent was generated. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Molodecky N.A., Soon I.S., Rabi D.M., Ghali W.A., Ferris M., Chernoff G., Benchimol E.I., Panaccione R., Ghosh S., Barkema H.W., Kaplan G.G. Increasing Incidence and Prevalence of the Inflammatory Bowel Diseases With Time, Based on Systematic Review. Gastroenterology. 2012;142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 2.de Souza H.S.P., Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 3.Abraham C., Cho J.H. Inflammatory Bowel Disease. N. Engl. J. Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lees C.W., Barrett J.C., Parkes M., Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 5.Torres J., Hu J., Seki A., Eisele C., Nair N., Huang R., Tarassishin L., Jharap B., Cote-Daigneault J., Mao Q., et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut. 2020;69:42–51. doi: 10.1136/gutjnl-2018-317855. [DOI] [PubMed] [Google Scholar]
- 6.Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olszak T., An D., Zeissig S., Vera M.P., Richter J., Franke A., Glickman J.N., Siebert R., Baron R.M., Kasper D.L., Blumberg R.S. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumbiganon P., Laopaiboon M., Gülmezoglu A.M., Souza J.P., Taneepanichskul S., Ruyan P., Attygalle D.E., Shrestha N., Mori R., Nguyen D.H., et al. Method of delivery and pregnancy outcomes in Asia: the WHO global survey on maternal and perinatal health 2007–08. Lancet. 2010;375:490–499. doi: 10.1016/S0140-6736(09)61870-5. [DOI] [PubMed] [Google Scholar]
- 9.Betran A., Torloni M., Zhang J., Gülmezoglu A., The WHO Working Group on Caesarean Section. Aleem H., Althabe F., Bergholt T., Bernis L., Carroli G., et al. WHO Statement on Caesarean Section Rates. BJOG: Int J Obstet Gy. 2016;123:667–670. doi: 10.1111/1471-0528.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsson H.E., Abrahamsson T.R., Jenmalm M.C., Harris K., Quince C., Jernberg C., Björkstén B., Engstrand L., Andersson A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 11.Shao Y., Forster S.C., Tsaliki E., Vervier K., Strang A., Simpson N., Kumar N., Stares M.D., Rodger A., Brocklehurst P., et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korpela K., Helve O., Kolho K.-L., Saisto T., Skogberg K., Dikareva E., Stefanovic V., Salonen A., Andersson S., de Vos W.M. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell. 2020;183:324–334.e5. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 13.Bager P., Wohlfahrt J., Westergaard T. Caesarean delivery and risk of atopy and allergic disesase: meta-analyses. Clin. Exp. Allergy. 2008;38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 14.Eggesbø M., Botten G., Stigum H., Nafstad P., Magnus P. Is delivery by cesarean section a risk factor for food allergy? J. Allergy Clin. Immunol. 2003;112:420–426. doi: 10.1067/mai.2003.1610. [DOI] [PubMed] [Google Scholar]
- 15.Andersen V., Möller S., Jensen P.B., Møller F.T., Green A. Caesarean Delivery and Risk of Chronic Inflammatory Diseases (Inflammatory Bowel Disease, Rheumatoid Arthritis, Coeliac Disease, and Diabetes Mellitus): A Population Based Registry Study of 2,699,479 Births in Denmark During 1973–2016. CLEP. 2020;12:287–293. doi: 10.2147/CLEP.S229056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bager P., Simonsen J., Nielsen N.M., Frisch M. Cesarean section and offspringʼs risk of inflammatory bowel disease: A national cohort study. Inflamm. Bowel Dis. 2012;18:857–862. doi: 10.1002/ibd.21805. [DOI] [PubMed] [Google Scholar]
- 17.Cardwell C.R., Stene L.C., Joner G., Cinek O., Svensson J., Goldacre M.J., Parslow R.C., Pozzilli P., Brigis G., Stoyanov D., et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008;51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 18.Kuhle S., Tong O.S., Woolcott C.G. Association between caesarean section and childhood obesity: a systematic review and meta-analysis: Caesarean section and childhood obesity. Obes. Rev. 2015;16:295–303. doi: 10.1111/obr.12267. [DOI] [PubMed] [Google Scholar]
- 19.Huh S.Y., Rifas-Shiman S.L., Zera C.A., Edwards J.W.R., Oken E., Weiss S.T., Gillman M.W. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch. Dis. Child. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuelsson U., Lindell N., Bladh M., Åkesson K., Carlsson A., Josefsson A. Caesarean section per se does not increase the risk of offspring developing type 1 diabetes: a Swedish population-based study. Diabetologia. 2015;58:2517–2524. doi: 10.1007/s00125-015-3716-3. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein C.N., Banerjee A., Targownik L.E., Singh H., Ghia J.E., Burchill C., Chateau D., Roos L.L. Cesarean Section Delivery Is Not a Risk Factor for Development of Inflammatory Bowel Disease: A Population-based Analysis. Clin. Gastroenterol. Hepatol. 2016;14:50–57. doi: 10.1016/j.cgh.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez-Bello M.G., De Jesus-Laboy K.M., Shen N., Cox L.M., Amir A., Gonzalez A., Bokulich N.A., Song S.J., Hoashi M., Rivera-Vinas J.I., et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat. Med. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korpela K., Salonen A., Vepsäläinen O., Suomalainen M., Kolmeder C., Varjosalo M., Miettinen S., Kukkonen K., Savilahti E., Kuitunen M., de Vos W.M. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 2018;6:182. doi: 10.1186/s40168-018-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S.K., McCulle S.L., Karlebach S., Gorle R., Russell J., Tacket C.O., et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein E.J.C., Tyrrell K.L., Citron D.M. Lactobacillus Species: Taxonomic Complexity and Controversial Susceptibilities. Clin. Infect. Dis. 2015;60:S98–S107. doi: 10.1093/cid/civ072. [DOI] [PubMed] [Google Scholar]
- 26.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Jang Y.J., Kim W.-K., Han D.H., Lee K., Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microb. 2019;10:696–711. doi: 10.1080/19490976.2019.1589281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou Q., Ye L., Liu H., Huang L., Yang Q., Turner J.R., Yu Q. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018;25:1657–1670. doi: 10.1038/s41418-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrick B.M., Rodriguez L., Lakshmikanth T., Pou C., Henckel E., Arzoomand A., Olin A., Wang J., Mikes J., Tan Z., et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. S0092867421006607. [DOI] [PubMed] [Google Scholar]
- 31.Lindemans C.A., Calafiore M., Mertelsmann A.M., O’Connor M.H., Dudakov J.A., Jenq R.R., Velardi E., Young L.F., Smith O.M., Lawrence G., et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W., Sonnenberg G.F. Activation and Suppression of Group 3 Innate Lymphoid Cells in the Gut. Trends Immunol. 2020;41:721–733. doi: 10.1016/j.it.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 35.Laursen M.F., Sakanaka M., von Burg N., Mörbe U., Andersen D., Moll J.M., Pekmez C.T., Rivollier A., Michaelsen K.F., Mølgaard C., et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021;6:1367–1382. doi: 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S., Bostick J.W., Ye J., Qiu J., Zhang B., Urban J.F., Avram D., Zhou L. Aryl Hydrocarbon Receptor Signaling Cell Intrinsically Inhibits Intestinal Group 2 Innate Lymphoid Cell Function. Immunity. 2018;49:915–928.e5. doi: 10.1016/j.immuni.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Heller J.J., Bostick J.W., Lee A., Schjerven H., Kastner P., Chan S., Chen Z.E., Zhou L. Ikaros Inhibits Group 3 Innate Lymphoid Cell Development and Function by Suppressing the Aryl Hydrocarbon Receptor Pathway. Immunity. 2016;45:185–197. doi: 10.1016/j.immuni.2016.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes T., Briercheck E.L., Freud A.G., Trotta R., McClory S., Scoville S.D., Keller K., Deng Y., Cole J., Harrison N., et al. The Transcription Factor AHR Prevents the Differentiation of a Stage 3 Innate Lymphoid Cell Subset to Natural Killer Cells. Cell Rep. 2014;8:150–162. doi: 10.1016/j.celrep.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Zhang J., Feng L. Disrupted metabolic signatures in amniotic fluid associated with increased risk of intestinal inflammation in cesarean section offspring. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1067602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachariassen L.F., Hansen A.K., Krych L., Nielsen D.S., Holm T.L., Tougaard P., Hansen C.H.F. Cesarean section increases sensitivity to oxazolone-induced colitis in C57BL/6 mice. Mucosal Immunol. 2019;12:1348–1357. doi: 10.1038/s41385-019-0207-8. [DOI] [PubMed] [Google Scholar]
- 41.Boirivant M., Fuss I.J., Chu A., Strober W. Oxazolone Colitis: A Murine Model of T Helper Cell Type 2 Colitis Treatable with Antibodies to Interleukin 4. J. Exp. Med. 1998;188:1929–1939. doi: 10.1084/jem.188.10.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heller F., Fuss I.J., Nieuwenhuis E.E., Blumberg R.S., Strober W. Oxazolone Colitis, a Th2 Colitis Model Resembling Ulcerative Colitis, Is Mediated by IL-13-Producing NK-T Cells. Immunity. 2002;17:629–638. doi: 10.1016/S1074-7613(02)00453-3. [DOI] [PubMed] [Google Scholar]
- 43.Katsandegwaza B., Horsnell W., Smith K. Inflammatory Bowel Disease: A Review of Pre-Clinical Murine Models of Human Disease. IJMS. 2022;23:9344. doi: 10.3390/ijms23169344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elson O., Sartor R.B., Tennyson G.S., Riddellii R.H. Experimental models of inflammatory bowel disease. Gastroenterology. 1995;109:1344–1367. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 45.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., Belzer C., Delgado Palacio S., Arboleya Montes S., Mancabelli L., et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017;81:e000366-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou L., Qiu W., Wang J., Zhao A., Zhou C., Sun T., Xiong Z., Cao P., Shen W., Chen J., et al. Effects of vaginal microbiota transfer on the neurodevelopment and microbiome of cesarean-born infants: A blinded randomized controlled trial. Cell Host Microbe. 2023;31:1232–1247.e5. doi: 10.1016/j.chom.2023.05.022. S1931312823002159. [DOI] [PubMed] [Google Scholar]
- 47.Zachariassen L.F., Krych L., Rasmussen S.H., Nielsen D.S., Kot W., Holm T.L., Hansen A.K., Hansen C.H.F. Cesarean Section Induces Microbiota-Regulated Immune Disturbances in C57BL/6 Mice. J. Immunol. 2019;202:142–150. doi: 10.4049/jimmunol.1800666. [DOI] [PubMed] [Google Scholar]
- 48.Morais L.H., Golubeva A.V., Moloney G.M., Moya-Pérez A., Ventura-Silva A.P., Arboleya S., Bastiaanssen T.F.S., O’Sullivan O., Rea K., Borre Y., et al. Enduring Behavioral Effects Induced by Birth by Caesarean Section in the Mouse. Curr. Biol. 2020;30:3761–3774.e6. doi: 10.1016/j.cub.2020.07.044. [DOI] [PubMed] [Google Scholar]
- 49.Lebeer S., Vanderleyden J., De Keersmaecker S.C.J. Genes and Molecules of Lactobacilli Supporting Probiotic Action. Microbiol. Mol. Biol. Rev. 2008;72:728–764. doi: 10.1128/MMBR.00017-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Baarlen P., Troost F., van der Meer C., Hooiveld G., Boekschoten M., Brummer R.J.M., Kleerebezem M. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. USA. 2011;108:4562–4569. doi: 10.1073/pnas.1000079107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia D.-J.-C., Wang Q.-W., Hu Y.-Y., He J.-M., Ge Q.-W., Qi Y.-D., Chen L.-Y., Zhang Y., Fan L.-N., Lin Y.-F., et al. Lactobacillus johnsonii alleviates colitis by TLR1/2-STAT3 mediated CD206 + macrophages IL-10 activation. Gut Microb. 2022;14 doi: 10.1080/19490976.2022.2145843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao T., Hsu R., Hacein-Bey C., Zhang W., Gao L., Kurth M.J., Zhao H., Shuai Z., Leung P.S.C. The Evolving Landscape of Fecal Microbial Transplantation. Clin. Rev. Allergy. 2023;65:101–120. doi: 10.1007/s12016-023-08958-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cahenzli J., Köller Y., Wyss M., Geuking M.B., McCoy K.D. Intestinal Microbial Diversity during Early-Life Colonization Shapes Long-Term IgE Levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zegarra-Ruiz D.F., Kim D.V., Norwood K., Kim M., Wu W.-J.H., Saldana-Morales F.B., Hill A.A., Majumdar S., Orozco S., Bell R., et al. Thymic development of gut-microbiota-specific T cells. Nature. 2021;594:413–417. doi: 10.1038/s41586-021-03531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin B.-Y., Li Z., Xia Y.-N., Li L.-X., Zhao Z.-X., Li X.-Y., Li Y., Li B., Zhou R.-C., Fu S.-C., et al. Probiotic Interventions Alleviate Food Allergy Symptoms Correlated With Cesarean Section: A Murine Model. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.741371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satoh-Takayama N., Vosshenrich C.A.J., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.-J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. Microbial Flora Drives Interleukin 22 Production in Intestinal NKp46+ Cells that Provide Innate Mucosal Immune Defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat. Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mortha A., Chudnovskiy A., Hashimoto D., Bogunovic M., Spencer S.P., Belkaid Y., Merad M. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dodd D., Spitzer M.H., Van Treuren W., Merrill B.D., Hryckowian A.J., Higginbottom S.K., Le A., Cowan T.M., Nolan G.P., Fischbach M.A., Sonnenburg J.L. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehrlich A.M., Pacheco A.R., Henrick B.M., Taft D., Xu G., Huda M.N., Mishchuk D., Goodson M.L., Slupsky C., Barile D., et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020;20:357. doi: 10.1186/s12866-020-02023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu J., Heller J.J., Guo X., Chen Z.m.E., Fish K., Fu Y.-X., Zhou L. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y.Y., Wang X.J., Su Y.L., Wang Q., Huang S.W., Pan Z.F., Chen Y.P., Liang J.J., Zhang M.L., Xie X.Q., et al. Baicalein ameliorates ulcerative colitis by improving intestinal epithelial barrier via AhR/IL-22 pathway in ILC3s. Acta Pharmacol. Sin. 2022;43:1495–1507. doi: 10.1038/s41401-021-00781-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., et al. Lactobacillus reuteri induces gut intraepithelial CD4 + CD8αα + T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meng D., Sommella E., Salviati E., Campiglia P., Ganguli K., Djebali K., Zhu W., Walker W.A. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr. Res. 2020;88:209–217. doi: 10.1038/s41390-019-0740-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hezaveh K., Shinde R.S., Klötgen A., Halaby M.J., Lamorte S., Ciudad M.T., Quevedo R., Neufeld L., Liu Z.Q., Jin R., et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. 2022;55:324–340.e8. doi: 10.1016/j.immuni.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Q., Zhao Q., Li T., Lu L., Wang F., Zhang H., Liu Z., Ma H., Zhu Q., Wang J., et al. Lactobacillus Plantarum-Derived Indole-3-Lactic Acid Ameliorates Colorectal Tumorigenesis via Epigenetic Regulation of CD8+ T Cell Immunity. Cell Metab. 2023;35:943–960.e9. doi: 10.1016/j.cmet.2023.04.015. [DOI] [PubMed] [Google Scholar]
- 68.Champagne F.A., Curley J.P., Keverne E.B., Bateson P.P.G. Natural variations in postpartum maternal care in inbred and outbred mice. Physiol. Behav. 2007;91:325–334. doi: 10.1016/j.physbeh.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 69.Caruso R., Ono M., Bunker M.E., Núñez G., Inohara N. Dynamic and Asymmetric Changes of the Microbial Communities after Cohousing in Laboratory Mice. Cell Rep. 2019;27:3401–3412.e3. doi: 10.1016/j.celrep.2019.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wirtz S., Popp V., Kindermann M., Gerlach K., Weigmann B., Fichtner-Feigl S., Neurath M.F. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: The 16S rRNA gene amplicon sequencing data and RNA-seq data have been deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database and Gene Expression Omnibus (GEO) database, respectively, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Code: This paper does not report original code.
-
•
Other items: No other new unique reagent was generated. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.