Summary
Passion fruit, a valuable tropical fruit, faces climate-related growth challenges. Antioxidant enzymes are vital for both stress protection and growth regulation in plants. We first provided systemic analysis of enzymatic antioxidant gene families in passion fruit, identifying 90 members including 11 PeSODs, 45 PeAPXs, 8 PeCATs, 7 PeGPXs, 6 PeMDHARs, 8 PeDHARs, and 5 PeGRs. Gene members in each gene family with same subcellular localization showed closer phylogenetic relationship. Many antioxidant genes exhibited tissue- or developmental stage-specific expression patterns during floral and fruit development, with some widely expressed. Their co-expressed genes were linked to photosynthesis and energy metabolism, suggesting roles in protecting highly proliferating tissues from oxidative damage. Potential genes for enhancing temperature stress resistance were identified. The involvement of diverse regulatory factors including miRNAs, transcription factors, and CREs might contribute to the complex roles of antioxidant genes. This study informs future research on antioxidant genes and passion fruit breeding.
Subject areas: Plant physiology, Plant development, Genomics
Graphical abstract

Highlights
-
•
Found 11 SODs, 45 APXs, 8 CATs, 7 GPXs, 6 MDHARs, 8 DHARs, 5 GRs in passion fruit
-
•
Genes with same subcellular localization showed closer phylogenetic relationship
-
•
Highly expressed gene in growing flower/fruit may protect against oxidative damage
-
•
Identified candidates for enhancing passion fruit’s temperature stress resistance
Plant physiology; Plant development; Genomics
Introduction
Passion fruit (Passiflora edulis) is a popular fruit crop cultivated worldwide in tropical and subtropical areas as to its important edible, medicinal, and ornamental value.1 However, the growth and development of passion fruit, especially during the reproductive period, are greatly affected by climatic conditions including light, temperature, and water. Unfavorable climatic factors such as heat, cold, drought can result in restricted growth, few flower buds, low fruit setting rate, severe flower and fruit drop, et al. which seriously affect the economic value of passion fruit industry. Therefore, it is of great importance to investigate genes involved in regulation of plant growth and development coping with diverse stresses to develop strategies for plant molecular breading.
It’s well known that unfavorable environmental factors could induce the production of excessive reactive oxygen species (ROS) in plant cells, which would result in oxidative modification and cell damage, thus, leading to abnormal plant growth or death.2 However, emerging evidences suggest that ROS also play critical roles in maintaining normal plant growth/development, such as cellular proliferation and differentiation,3 cell wall formation and loosening,4 organogenesis5 and senescence.6 To minimize ROS-derived damages and keep the balance between ROS scavenging and generation, advanced antioxidant defense systems have been evolved in plants, including seven main classes of antioxidant enzymes, low-molecular antioxidants, and thioredoxin/glutaredoxin systems.
In plants, ROS exist in different types including superoxide anion radicals (O2−), hydrogen peroxide (H2O2), hydroxyl radicals (OH−), peroxyl radicals (HOO−) and singlet oxygens (1O2), and different ROS types with different oxidative capacity can affect different physiological and biochemical reactions which are regulated by different genes.7 Generally, superoxide is the precursor of diverse ROS due to its instability and strong oxidation/reducibility. SOD is a member of the metalloenzyme family and plays a pioneering role in scavenging ROS by catalyzing the dismutation reaction of neutralizing superoxide into H2O2. According to the classification of different metal ions bound to the catalytic center, SOD can be divided into three major categories including Cu/Zn-SOD (CSD), Fe-SOD (FSD), and Mn-SOD (MSD).8 Cu/Zn-SOD is mainly located in mitochondria, chloroplast, and cytosol, Fe-SOD is mainly located in mitochondria, chloroplast and peroxisome, and Mn-SOD is mainly located in mitochondria and peroxisome.9,10 Then, the formed hydrogen peroxide can be neutralized via three main ways catalyzed by CAT, GPX with reduced thioredoxins oxidized, and APX with the oxidation of ascorbate (AsA) to monodehydroascorbate (MDHA).11 CAT is a heme domain-containing oxidoreductase that is present in all aerobic species,12,13 while GPX belongs to the non-heme thiol peroxidase family.14 Differently, APX belongs to the heme peroxidase superfamily15,16 and is one of the major components the ascorbate-glutathione (AsA-GSH) cycle.8 In response to stresses, the AsA-GSH cycle plays vital roles in scavenging ROS, which also comprises of other three classes of antioxidant enzymes including monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), and glutathione reductase (GR).9 Unlike SOD, CAT, GPX, and APX, which mainly catalyze ROS decomposition, MDHAR, DHAR, and GR play roles in maintaining the level of reduced forms of antioxidants such as ascorbic acid and glutathione (GSH).11 The generated MDHA in the oxidation reaction catalyzed by APX may be regenerated to AsA by the reaction of MDHAR or spontaneously transformed into dehydroascorbate (DHA). Subsequently, DHA could be reduced to AsA under the reaction of DHAR with GSH oxidation to glutathione disulfide (GSSG). Finally, NADPH-dependent GR can reduce GSSG and plays a key role in controlling the levels of reduced glutathione.9
More and more investigations suggest that plant antioxidant enzymes do not only protect cells from stress induced oxidative damages but also can be implicated in the regulation of plant growth and development.7 In Arabidopsis, AtCAT2 is mainly expressed in leaves, is the major isoform in leaves,10 can be regulated by light, cold or circadian clock, is associated with plant photorespiration, leaf senescence,6 and stress responding including drought and cold17; Overexpression of the AtGPXL5 gene resulted in altered plant growth and redox status under salt stress18; GR2 mutants with excessive accumulation of oxidized glutathione results in root apical cells entering the oxidized state and eventually leads to abnormal growth.19 In rice, silencing of OsGPX1 affects normal plant development and photosynthesis under salinity conditions.20 Plants knocked out of OsGPX5 also showed increased sensitivity to salt stress, impaired seed germination, and plant growth and development.21 More importantly, antioxidant enzyme genes are precisely regulated by transcription factors, hormones, miRNAs or other factors, to keep the ROS levels well controlled and participate in the regulation of plant developmental process and stress responses. In rice, the transcription factor OsMADS3 regulates ROS homeostasis at later stages of another development, and abnormal expression of OsMADS3 results in the accumulation of superoxide and causes pollen sterility.22 In Arabidopsis, AtCSD1 and AtCSD2 are induced by downregulation of miR398, which favorably enhances plant tolerance to oxidative damage.23 In wheat, the knockdown of TaMDHARs by miRNA could result in improved resistance to Puccinia striiformis.24
In passion fruit, antioxidant enzymes have been reported to be closely related to various processes such as fruit development,25 postharvest storage,26 light response,27 and heat tolerance (our unpublished data), but there has been no systematic study of the characteristics and the expression patterns of the enzymatic antioxidant gene families in passion fruit. Our present work aims to provide a relatively complete profile of the passion fruit antioxidants enzyme gene families, which could lay a solid foundation for further research on the functional roles of the enzymatic antioxidant gene family in passion fruit, and also provide valuable gene resources for its resistance molecular breeding.
Results
Identification and phylogenetic analysis of enzymatic antioxidant genes
A total of 90 genes (11 PeSODs, 45 PeAPXs, 8 PeCATs, 7 PeGPXs, 6 PeMDHARs, 8 PeDHARs, and 5 PeGRs) encoding for antioxidant enzymes were identified in passion fruit genome, and the genes in each class were renamed according to their positional order on the chromosomes (Table S1). Physicochemical properties analysis results shown that, the proteins encoded by the 90 enzymatic antioxidant genes contained 105 (PeGPX5) to 842 (PeAPX31) amino acids with molecular weights (MWs) ranging from 13880.91 Da (PeGPX5) to 93602.86 Da (PeAPX31). Predicted protein isoelectric points (pI) ranged from 4.81 (PeAPX29) to 10.08 (PeGPX4). The results of instability index calculation showed that 60% of the enzymatic antioxidant proteins were stable in vitro, while the remaining 40% were unstable in vitro. Aliphatic amino acid index (A.I.) results indicate that the enzymatic antioxidant protein has a high aliphatic index with thermal stability between 55.25 (PeAPX32) and 100.14 (PeGR2). The grand average of hydropathicity (GRAVY) of almost all enzymatic antioxidant proteins was negative except for PeAPX39, indicating that they are predominantly hydrophilic.28 Subcellular localization prediction results showed that 90 enzymatic antioxidant genes were mainly localized in the chloroplast, cytoplasm, peroxisome, and mitochondrion, while few of them were localized in the vacuole (PeAPX10, PeAPX17, PeAPX25, PeAPX33, and PeAPX34) and nucleus (PeGPX3). The diversity of subcellular localization implies various biological functions within the members of passion fruit enzymatic antioxidant gene family.29
To investigate the evolutionary relationship between the enzymatic antioxidant gene family in passion fruit and Arabidopsis, the maximum likelihood (ML) phylogenetic trees were constructed separately for each gene family (Figure 1). For SOD gene family (Figure 1A), the phylogenetic tree showed that PeSOD and AtSOD proteins could be divided into three major groups: Mn-SOD, Fe-SOD, and Cu/Zn-SOD. The number of Fe-SOD and Mn-SOD in passion fruit was the same as the corresponding subfamily member number in Arabidopsis (3 PeFSDs and 2 PeMSDs, 3 AtFSDs and 2 AtMSDs), while the number of Cu/Zn-SOD was twice that in Arabidopsis (6 PeCSDs and 3 AtCSDs). Subcellular localization prediction results shown that, PeCSDs were mainly located in chloroplast except PeCSD4, which is located in cytoplasm and clustered as the outgroup of all Cu/Zn-SODs. PeFSD1/2 clustered in one branch was located in chloroplast whereas PeFSD3 clustered in another branch was located in mitochondrion. With regard to the APX gene family (Figure 1B), all of the 8 AtAPXs and 45 PeAPXs were clustered into six subgroups with one of them contains only passion fruit genes and named as APX subgroup. And the other five subgroups were named according to the phylogenetic relationships including APX-1/2 (peroxisome, PeAPX1/4, 2 AtAPXs), APX-3/5 (peroxisome, PeAPX16/24/44, 2 AtAPXs), APX-4 (chloroplast, PeAPX7/8, 1 AtAPXs), APX-6 (cytoplasm, PeAPX37/38, 1 AtAPXs), and stAPX (chloroplast/mitochondrion, PeAPX9/30/29/32, 2 AtAPXs), which was consistent with their subcellular localization. Whereas PeAPXs in the passion fruit specific APX subgroup were mainly located in cytoplasm (27 PeAPXs) and vacuole (5 PeAPXs), and the GO enrichment analysis shown these PeAPXs were mainly involved in biological processes including cellular oxidant detoxification, plant-type cell wall organization or biogenesis, cellular response to stimulus (Table S12). As to the CAT gene family (Figure 1C), PeCAT1/7/8 was closely clustered with AtCAT2, while other 5 PeCATs were closely clustered with AtCAT3, and all PeCATs were located in peroxisome. With regard to GPX gene family (Figure 1D), all the GPXs could be divided into four subgroups, and the branch containing PeGPX4/5 and AtGPX4/5 were clustered with highest bootstrap values, followed by the subgroup containing PeGPX3 and AtGPX2/3. Finally, for the MDHAR gene family (Figure 1E), every Arabidopsis AtMDHAR was closely clustered together with two passion fruit PeMDHARs, with two extra Arabidopsis AtMDHARs clustered as outgroups. In comparison to Arabidopsis, the number of PeDHARs was expanded, six PeDHARs were clustered in a single subgroup without Arabidopsis homologs, while the other two PeDHARs were clustered within three Arabidopsis genes (Figure 1F). For GR gene family (Figure 1G), PeGR1/5 were closely grouped with AtGR, and PeGR3/4 were clustered in a single subgroup, while AtGR1 and PeGR2 were served as outgroups. Most of the PeMDHARs were located in chloroplast except for PeMDHAR1 (cytoplasm), and most of PeDHARs were located in cytoplasm except for PeDHAR8 (chloroplast). The predicted subcellular localization of PeGPXs and PeGRs were diversified, and many of these genes were predicted to be localized in multiple subcellular compartments, such as PeGPX1/2/6/7 (Chloroplast/Mitochondrion) and PeGR1/5 (Chloroplast/Cytoplasm/Mitochondrion). Additionally, some proteins showed one-to-one homology, such as PeCSD5 and AtCSD2, PeFSD1 and AtFSD1, PeAPX1 and AtAPX2, PeDHAR8 and AtDHAR3, and most of these protein pairs shown similar subcellular localization.
Figure 1.
Phylogenetic trees of SOD
(A), APX (B), CAT(C), GPX (D), MDHAR (E), DHAR (F), and GR (G) proteins from passion fruit (Pe, Passiflora edulis) and Arabidopsis (At, Arabidopsis thaliana). The tree was constructed in MEGA 11 and IQ-TREE 2 software using the maximum likelihood (ML) option with 1000 bootstrap replicates. Blue and green stars indicate gene family members from passion fruit and Arabidopsis, respectively. Different colored branches represent different enzymatic antioxidant gene subfamilies, which were named according to previous studies.
Gene structure, conserved motifs, and CREs analysis of enzymatic antioxidant genes
The gene structure characteristics and conserved motif composition of seven different enzymatic antioxidant gene families of passion fruit were analyzed and shown according to their phylogenetic relationships (Figure 2). As to SODs, all the PeCSDs contained Sod_Cu domain and mainly contains motif SOD-Motif 1/2/6, while PeMSDs contained Sod_Fe_N/Sod_Fe_C domains and motif SOD-Motif 4/10/5/8/3/9, and PeFSDs contained Sod_Fe_N superfamily/Sod_Fe_C domains and motif SOD-Motif 4/7/5/8/3, indicating that higher sequence similarity was shared in PeFSDs and PeMSDs. With regard to PeAPXs, genes in different subfamilies shown diverse structure characteristics, and motifs APX-Motif 3/6/7/9/10 were only present in the passion fruit-specific APX subgroup genes. For PeCAT genes, genes closely clustered with Arabidopsis homologous genes including PeCAT1/7/8 contains all of the ten predicted motifs for CAT subfamilies, and showed more conserved gene structure and motif composition. Whereas other PeCAT genes in the same subfamily were different and lack certain motifs with higher gene structure and motif composition diversity. As to PeGPXs, GPX-Motif 6/3/1/4 were present in all PeGPXs, which constitute the most highly conserved domain. All PeMDHAR genes contained MDHAR-Motif 1–8 and 10 arranged with the same order, while MDHAR-Motif 9 was only present in PeMDHAR4/5. As to PeDHARs, closely clustered PeDHAR2/3/4 shown similar motif composition with PeDHAR5/6, while the subgroup PeDHAR1/7/8 shown diverse structures but all contains the specific DHAR-Motif 7. For PeGRs, genes in the same subgroups were also shown similar motif compositions, such as PeGR1/5, PeGR3/4. In order to gain an in-depth understanding of the gene structure diversity, we analyzed the exons and introns of the enzymatic antioxidant gene. The relative numbers of CDSs in the 90 enzymatic antioxidant genes ranged from 2 to 18, according to the results of the passion fruit GFF annotation file (Figure 2D). Among them, the number of PeMDHAR4/5 with 18 exons and CDSs are the largest, followed by PeGR3 with 15 exons and CDSs, suggesting that their alternatively spliced form may be the most complex.30 Among all of the 90 enzymatic antioxidant genes, closely clustered gene pairs shown similar motif compositions, conserved domain and exon-intron structures such as PeGPX6/7, PeMDHAR4/5, PeGR1/5, and PeAPX24/44, suggesting that phylogenetic relationships among gene family members were highly correlated with gene structure.
Figure 2.
The phylogenetic relationship, conserved motifs and gene structure of enzymatic antioxidant genes
(A) The maximum likelihood (ML) phylogenetic tree of enzymatic antioxidant proteins was constructed using full-length protein sequence with 1000 bootstrap replicates.
(B) Distribution of conserved motifs in enzymatic antioxidant proteins. Each gene family predicted 10 motifs, and the scale bar represents 30 to 200 aa range.
(C) Distribution of SOD, APX, CAT, GPX, MDHAR, DHAR, and GR domain of enzymatic antioxidant genes.
(D) The gene structures of the enzymatic antioxidant genes, include introns (black lines), exons (CDSs, orange rectangles), and untranslated regions (UTRs, blue rectangles).
The possible roles of enzymatic antioxidant genes in response to plant growth and development, phytohormones, and light and stress responses were further illustrated through the analysis of cis-regulatory elements (CREs) distributed in the putative gene promoter regions (2 kb upstream of gene transcriptional activation site). A total of 2043 CREs were predicted in the promoter regions of enzymatic antioxidant genes (Table S3), and Figure 3A shows 15 representative CREs. Among them, PeCSD5, PeDHAR6, and PeDHAR8 have the largest number of CREs with a total of 36 elements, while PeGPX6 has the least number of CREs with only 4 elements. All CREs can be divided into three broad categories including hormone responsiveness (812), growth and development (809), and stress responsiveness (422) (Figure 3B). Except for light responsive elements (706, 34.56%), other CREs involved in growth and development regulation were relatively insufficient and might be absent from many gene family such as CAT-box for meristem expression (45, 2.20%), GCN4_motif for endosperm expression (20, 0.98%), RY-element for seed-specific regulation (24, 0.73%), and circadian for circadian control (23, 1.13%). For CREs involved in hormone responsiveness, MeJA-responsiveness (CGGTA-motif and TGACG-motif, 349, 17.08%) and abscisic acid (ABA) responsiveness (ABRE, 256, 12.5%) are widely present in the promoter regions of enzymatic antioxidant genes in almost all gene families. Whereas CREs related with gibberellin (GA)-responsiveness (101, 4.94%) (61 in 90 enzymatic antioxidant genes) were relatively enriched in DHAR and MDHAR gene family, but absent from GPX and GR gene family. CREs associated with (IAA) auxin-responsiveness (58, 2.84%) and salicylic acid (SA) responsiveness (48, 2.35%) were irregularly present in some enzymatic antioxidant genes. Some enzymatic antioxidant genes such as PeAPX5/6/10/16/19/22, PeCAT1, PeFSD1, PeGPX2/4, and PeDHAR5/6 contain multiple identical hormone response elements in their promoter regions, suggesting that more rapid and robust responses to specific hormones may be possible. As to stress responsiveness related CREs, ARE for anaerobic induction (218, 10.67%) were abundant in all gene families, while other stress related CREs were relatively enriched in certain subfamilies. For example, (MYB) drought-inducibility (MBS, 106, 5.19%) was relatively abundant in APX and DHAR gene family relatively, but absent from MDHAR and GR gene family. Low-temperature responsiveness (52, 2.55%) and defense and stress responsiveness (36, 1.76%) related CREs were only present in few genes such as PeCSD3, PeDHAR7, PeGR1, and PeAPX4. In summary, these results showed that the composition and number of CREs in the promoter regions of different enzymatic antioxidant genes were highly diverse within and among subfamilies, suggesting that the expression of enzymatic antioxidant genes in passion fruit is regulated by diverse CREs.
Figure 3.
The CREs on the putative promoter of the enzymatic antioxidant genes
(A) Distribution of CREs identified in the 2000 bp upstream promoter region of enzymatic antioxidant genes.
(B) Venn diagram of various CREs.
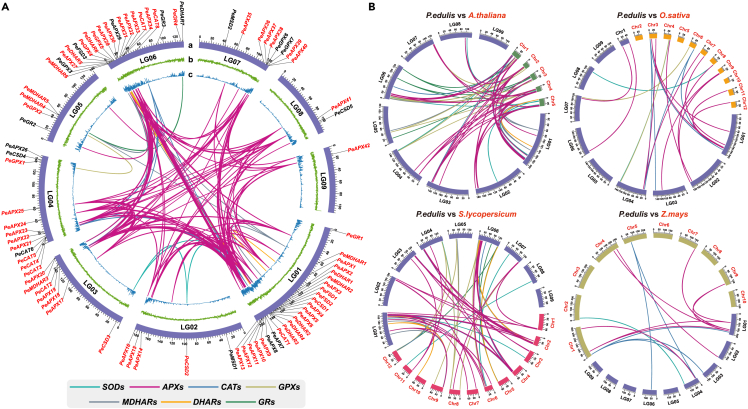
Chromosomal location and collinearity analysis of enzymatic antioxidant genes
Generally, the expansion of gene families is driven by different gene duplication patterns that are considered to be the driving force of species evolution.31 The 90 enzymatic antioxidant genes were unequally distributed on all nine passion fruit linkage groups (LG) (Figure 4A). The largest number of enzymatic antioxidant genes occurs on LG01 (24 genes), followed by LG08 (20 genes), LG04 (12 genes), LG07 (9 genes), LG03 (7 genes), LG02 (5 genes), LG05 (4 genes), LG08 (2 genes), and LG09 (1 gene). The MCScanX method was used to analyze the gene duplication events of passion fruit, and a total of 183 gene pairs with gene duplication events (177 segmental duplicated gene pairs and 6 tandem duplicated gene pairs) were identified. These results suggest that enzymatic antioxidant genes may arise through gene duplication, and segmental duplication events are the main driver of enzymatic antioxidant genes evolution. The substitution rate ratio Ka/Ks can be used to elucidate the evolutionary process and selection pressure of enzymatic antioxidant genes,32 and we calculated the Ka/Ks ratio for all 183 duplicated gene pairs (Table S4). Only one pair of segmental repeat genes (PeAPX30-PeAPX32) had a Ka/Ks value greater than 1, indicating that this pair of genes was strongly and positively selected during evolution and may have had an adaptive impact on the environment of passion fruit during evolution. The Ka/Ks ratio of remaining duplicated gene pairs were all less than 1, indicating that most of the enzymatic antioxidant gene pairs may have undergone purifying selection during evolution and played a key role in maintaining the conserved structure of the enzymatic antioxidant gene. Moreover, the divergence rate among duplicated enzymatic antioxidant genes was measured and the results showed that the divergence rate between duplicated enzymatic antioxidant genes was estimated to be between 0.0676 and 74.7395 million years ago (MYA).
Figure 4.
Intraspecies and interspecies synteny analysis in comparative genomics
(A) Distribution and collinearity of enzymatic antioxidant genes in passion fruit genome. Enzymatic antioxidant genes marked in red have collinearity, and those marked in black lack collinearity. (a): The 9 chromosomal pseudomolecules: units on the circumference are mega base values of pseudomolecules; (b): GC content; (c): gene density.
(B) Enzymatic antioxidant genes synteny analysis of passion fruit and 4 representative plants. Species names are prefixed with “A.thaliana”, “O.sativa”, “S.lycopersicum”, and “Z.mays”, denote Arabidopsis thaliana, Oryza sativa, Solanum lycopersicum, and Zea mays, respectively.
Collinear analysis of different species is a method to investigate their evolution and affinities.33 To further reveal the gene duplication timing of enzymatic antioxidant genes and infer their phylogenetic mechanism, we selected four representative species for comparative analysis of collinearity with passion fruit, including two dicots (Arabidopsis and tomato) and two monocots (rice and maize) (Figure 4B). A total of 55 enzymatic antioxidant genes were collinearly associated with the tomato genome, followed by Arabidopsis (47), rice (18), and maize (10) (Table S5). These results indicated that the collinearity between P. edulis and dicot genomes is greater than that between P. edulis and monocot genomes. Individual homologous genes exhibit one-to-many or many-to-one homozygosity. Among them, there are 6 enzymatic antioxidant genes (PeCSD4, PeAPX1, PeAPX16, PeAPX23, and PeCAT2) with collinear associations between all of these four selected species, suggesting that these genes in the enzymatic antioxidant gene family may play an important role during evolution.34
Secondary and tertiary structure analysis of enzymatic antioxidant genes
Structural analysis of proteins has important implications for understanding their functions.35 The results of secondary protein structure prediction (Table S6) showed that all enzymatic antioxidant proteins were mainly composed of α-helix (4.61%–58.30%), extended strand (9.04%–34.21%), β-turn (1.82%–11.82%), and random coil (25.23%–55.63%) composition. Based on the AlphaFold2 and SWISS-MODEL database, homology modeling of enzymatic antioxidant proteins was performed (Figure S1), and the structure with the highest GMQE and QMean (Table S6) was selected as the representative structure for each gene family (Figure 5). Detailly, PeMDHAR3 was selected to represent the MDHAR gene family. There is a non-covalent flavin-adenine dinucleotide (FAD) ligand in the structure of (5jci.1.A), and its binding domain adopts a typical α/β sheet, consisting of two anti-parallel β-sheets and one parallel β-sheet, surrounded by four α-helices. FAD binds to the gap on the FAD-binding domain of PeMDHAR3 via hydrogen bonding and van der Waals interactions.36 As to GR gene family, there are also two non-covalent FAD ligands in the structure of PeGR4 (1dxl.2.A), showing a symmetrical distribution. For APX gene family, there are two non-covalent PROTOPORPHYRIN IX CONTAINING FE (HEM) ligands in the structure of PeAPX1 (2y6b.1.A). With regard to the GPX gene family, the reduced form of PeGPX2 (2p5r.1.A) crystallizes as two dimers in an asymmetric unit and possess 5 non-covalent calcium ion (Ca+2) ligands in its structure. For DHAR gene family, there are a non-covalent glutathione (GSH) and calcium ion (Ca+2) ligands in the structure of PeDHAR1 (4pqi.1.A), respectively. As to CAT gene family, there are four acetate ions (ACT) of non-functional binders in the structure of PeCAT8 (4qol.1.A), and they show a similar mutual symmetrical distribution. Finally, for the SOD gene family, PeCSD1 (1xso.1.A) chelates two copper ions (Cu+2), while PeFSD1 (7bjk.1) and PeMSD2 (4c7u.1.A) chelates two zinc ions (Zn+2) and four manganese ions (Mn+2), none of these metal ions are covalently bonded to the peptide chain.37 Collectively, the homology models of these proteins provide a preliminary basis for further understanding the molecular functions of enzymatic antioxidant proteins.
Figure 5.
Structural analysis of the tertiary structure of enzymatic antioxidant protein and its small molecule ligand
Among them, for the per-residue confidence score (pLDDT) between 0 and 100. Some regions below 50 pLDDT may be unstructured in isolation. The confidence color schemes, residues are colored by their local quality value.
Transcription factor regulatory network analysis of enzymatic antioxidant genes
The potential TF regulatory network of all 90 enzymatic antioxidant genes in passion fruit was analyzed using the PTRM online database. The results showed that there were 2022 TFs from 33 different TF families were identified to be involved in the regulation of the 90 enzymatic antioxidant genes (Figure 6A). Lots of stress response related TFs including Dof, WRKY, HSF, bZIP were found in many enzymatic antioxidant genes. While diverse TFs related with plant growth and development including BBR-BPC, MIKC-MADS, LBD, bHLH, and AP2 were also identified in most of the enzymatic antioxidant genes (Figure 6B; Table S7). Additionally, phytohormone-related TFs were also predicted such as ERF and ARF. Among all the predicted TFs, the most abundant members belonged to the BBR-BPC family (428 members), followed by the ERF family (366 members), the Dof family (264 members), the MIKC-MADS family (176 members), and the AP2 family (122 members) (Figure 6C). Among all 90 enzymatic antioxidant genes, PeAPX15 was targeted by most TFs (136), followed by PeGR5 (94), PeCSD5 (75), PeAPX26 (74), and PeAPX25 (70). The complex regulatory network of transcription factors may enable these genes to be precisely regulated and involved in growth and development, as well as stress and hormone responsiveness.
Figure 6.
The putative TFs regulatory network analysis of enzymatic antioxidant genes
(A) Limegreen v nodes represent TFs; steelblue circular nodes represent enzymatic antioxidant genes.
(B) Wordcloud for TFs. The font size is positively correlated with the number of corresponding TFs.
(C) Statistical results of the number of TFs.
Prediction of putative miRNAs directing enzymatic antioxidant genes
MicroRNAs (miRNAs) are a class of single-stranded RNA molecules of around 22 nucleotides (nt) in length that regulate gene expression by binding to target gene transcripts to inhibit their translation or promote mRNA degradation.38,39 To better understand the regulatory mechanism of miRNAs in enzymatic antioxidant genes in passion fruit, a total of 25 miRNAs (ped-miRNAs) were identified, belonging to 17 different miRNA families (Table S8). The network interactions and schematic diagram of miRNA targeting sites in PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, and PeDHAR genes are presented in Figure 7. As shown in Figure 7A, the 17 identified miRNAs targeted 21 enzymatic antioxidant genes and results showed that ped-miR160c-5p, ped-miR171k-5p, and ped-miR828b targeted the most three enzymatic antioxidant genes, which are: (PeAPX27, PeAPX43, and PeMDHAR2), (PeCAT1, PeCAT7, and PeDHAR7), and (PeAPX29, PeAPX32, and PeAPX39), respectively. Meanwhile, ped-miR164b-5p targets two PeMDHARs (PeMDHAR4 and PeMDHAR5), ped-miR395a targets two PeAPXs (PeAPX5 and PeAPX6), while the other remaining miRNAs targeted only one enzymatic antioxidant gene. PeMDHAR2 was the one targeted by most miRNAs (Figure 7B), including ped-miR169a, ped-miR169b, and ped-miR160c-5p. Likewise, PeAPX2 and PeAPX26 were targeted by 2 miRNAs, including (ped-mir399e, ped-mir399d) and (ped-miR171j-3p, ped-mir171b-3p). In summary, the regulation of the enzymatic antioxidant gene family in passion fruit involves a complex regulatory network of TFs and miRNAs.
Figure 7.
Predicted miRNAs targeting enzymatic antioxidant genes
(A) Network illustration of predicted miRNA targeted enzymatic antioxidant genes. Green rectangle shapes represent the predicted miRNAs and steelblue circle shapes represent targeted enzymatic antioxidant genes.
(B) The schematic diagram indicates the enzymatic antioxidant genes targeted by miRNAs and the putative miRNAs sites are indicated by orange color, upper sequences are from the gene region and lower sequences are from the miRNAs.
Expression and GO/KEGG enrichment analysis of enzymatic antioxidant genes
To explore the possible functions of the enzymatic antioxidant genes, the expression profiles of the 90 enzymatic antioxidant genes in diverse passion fruit tissues of the floral organs and fruit at different developmental stages were characterized using transcriptomic data generated in our lab (Figure 8) (Tables S9 and S10). After filtering out the genes with low expression levels, the remaining 83 enzymatic antioxidant genes could be grouped into five blocks (block A–E) by the hierarchical clustering of expression profiles (Figure 8A). The enzymatic antioxidant genes in different blocks showed distinct temporal and spatial expression patterns: (A) Enzymatic antioxidant genes in block A showed a preferential expression mainly at the late development stage of bract, petal, and corona filament including PeAPX2/24/44/27/43, PeMDHAR2/6, PeGR4, PeCSD6, and PeGPX7. (B) The majority of block B genes showed preferential expression in some floral tissues, such as PeAPX7/8/20/14/11/12 preferentially highly expressed at the early developmental stage of stamen, other genes were mainly higher expressed in bract and sepal such as PeAPX5/13/35. Interestingly, this block only contained CAT and APX gene members, which were all involved in the scavenging H2O2; (C) Most of block C genes showed preferential expression at the early stage of certain tissues, such as PeMDHAR1, PeCAT3/4/5 were higher expressed at the early stages of bract, sepal, petal, corona filament, and ovules. Whereas some genes including PeAPX33/34/19/25, PeCSD2, PeGR3 were higher expressed during the whole development processes of ovules. (D) Three genes clustered in block D including PeMDHAR3 and PeAPX28/31, shown a relatively higher expression levels in ovules. (E) Almost a third of all the antioxidant enzyme genes were highly expressed at different stages of fruit development, which are grouped in block D. With fruit ripening, the expression of some antioxidant enzyme genes showed an up-regulated trend including PeGR1/2/5, PeAPX1, PeGPX1/6, PeCSD4. Whereas some genes showed a down-regulated trend, such as PeAPX15/29/30/32/4/17 and PeFSD1, PeMSD1/2. Besides, many genes shown a highest expression level at the second stage of fruit development including PeCSD3/5, PeAPX22/39, PeCAT1/7, PeGPX3, and PeDHAR1/8. In short, different enzymatic antioxidant genes showed diverse expression profiles within or between different subgroups. Many of these genes showed tissue- or developmental stage-specific expression patterns. For example, PeAPX20/14/11/12 in block B was explicitly and highly expressed only at the early developmental stage of the stamen. However, some genes also showed a relatively high expression level across all tissues or organs tested here, such as PeMSD1/2, PeFSD1, PeCSD3, PeGPX1/2, PeAPX16, PeMDHAR2, PeDHAR7, and PeGR2/4. Thus, most enzymatic antioxidant genes were highly expressed in certain tissues or at certain development stages, suggesting these genes might play some previously unknown roles in the regulation of passion fruit growth and development.
Figure 8.
Heatmap of transcription expression levels of enzymatic antioxidant genes
Hierarchical clustering of the expression profiles of the enzymatic antioxidant genes in (A) diverse tissues at different stages. br, bract; se, sepal; pe, petal; ca, corona filament; st, stamen; sg, stigma; ov, ovule; numbers represent developmental stages, 1 and 2 was early stage, 8 was late stage; (B) different tissues including leaf, petiole, bract, and bud under heat (T30, 30°C) and cold (T20, 20°C) stress conditions. The heatmap was created based on the log2(TPM +0.01) value of enzymatic antioxidant genes and normalized by row. The TPM value higher than 50 was shown as abundant genes and marked with “∗”. Differences in gene expression changes are shown in color as the scale, mediumvioletred for high expression and steelblue for low expression.
Enzymatic antioxidant genes play important roles in cell protection from stress induced oxidative damages, the expression profiles of passion fruit enzymatic antioxidant genes under heat and cold stress conditions were also explored using transcriptomic data (Figure 8B) (Table S9). The remaining 67 enzymatic antioxidant genes after filtering could also be clustered into five blocks (block I-V) by the hierarchical clustering of expression profiles. Genes in block II-IV showed a preferential expression in the green vegetative tissue leaf, bract, and petiole, respectively. Whereas genes in block I and V were preferentially higher expressed in buds. The expression of PeCSD2/3/4, PeAPX26/11/12/4, PeGPX3, and PeDHAR7 were induced immediately under heat stress treatment and shown highest expression levels in a short time. Some genes respond differently to heat and cold stress, such as the expression of PeMDHAR2 and PeGPX2 decreased in leaf, petiole, and bract in response to high temperature, but upregulated in bud induced by low temperature. While some genes were induced by both heat and cold stresses including PeGR2 and PeAPX30. Additionally, some genes including PeMSD1/2, PeFSD1, PeCSD2/3/5, PeCAT2, PeGPX1/2, PeAPX4/16/33/34, PeMDHAR2, PeDHAR1/8, and PeGR4 were abundant with a relatively high expression levels in all tissues before or after stress treatments.
GO and KEGG enrichment analyses of passion fruit were performed for 90 enzymic antioxidant genes and their co-expressed genes generated from WGCNA analysis using transcriptomic data (Tables S10 and S11). The passion fruit enzymic antioxidant genes were mainly significantly enriched in KEGG pathways such as biosynthesis of other secondary metabolites, peroxisome, glutathione metabolism, ascorbate and aldarate metabolism, phenylpropanoid biosynthesis, carbohydrate metabolism, indicating their important roles in diverse metabolic processes (Figure 9). Additionally, their co-expressed genes were significantly enriched in pathways such as photosynthesis, energy metabolism and carbon fixation, and other metabolism processes, suggesting that 90 enzymic antioxidant genes were involved in these processes. Similar results were also found in the GO enrichment analysis (Tables S10 and S11).
Figure 9.
Top 15 enriched KEGG pathway names for enzymic antioxidant family genes and co-expressed genes
The horizontal axis represents the rich factor, and the size of the black circle indicates the number of genes annotated with a given pathway name, and different colors represent p values.
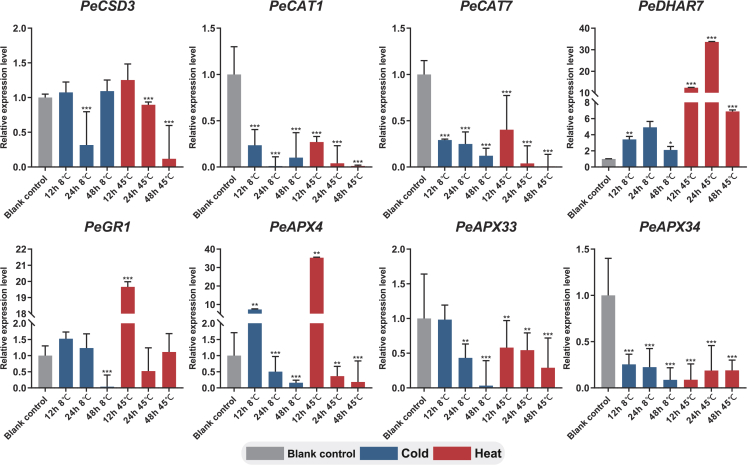
To further evaluate the expression profiles of the passion fruit enzymatic antioxidant genes under more pressurized heat and cold stress conditions, eight representative genes were selected for qRT-PCR analysis based on their significantly different expressions from transcriptomic results (Figure 10). Passion fruit seedlings were treated under heat (45°C) and cold (8°C) conditions, and leaf samples were collected for experimental analysis. In time series, many heat-responsive genes including PeDHAR7, PeGR1, and PeAPX4 were highly expressed in the short time. Most tested genes shown similar trends in response to heat and cold in leaves such as PeCAT1/7, PeDHAR7, and PeAPX4/33. Combined with transcriptomic results, PeGR1 was preferentially highly expressed in leaf and response to heat stress, while genes such as PeDHAR7 and PeAPX4 were induced with higher expression levels in diverse tissues including bud, petiole, and leaf under mild or severe heat stress.
Figure 10.
Expression profiles of 8 genes (PeCSD3, PeCAT1, PeCAT7, PeDHAR7, PeGR1, PeAPX4, PeAPX33, and PeAPX34) in response to cold and heat stress
Error bars represent standard deviations of the means of three independent biological replicates of qRT-PCR analysis. Asterisks indicate significant differences in transcript levels compared with those of blank control (0 h 0 mM). (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
To gain more insights into the potential roles of the passion fruit enzymatic antioxidant genes involved in phytohormone response, eight representative genes were selected for qRT-PCR analysis according to the predicted CREs and expression data from transcriptome (Figure 11). All of the eight test genes were induced with higher expression levels by GA, and seven genes but not PeAPX16 were induced with higher expression levels by MeJA. PeCAT1 and PeCSD3 were also induced with higher expression levels by ABA, while PeAPX19 and PeCAT1 were induced with higher expression levels by SA. Consistent with the CREs analysis results, these results suggested that the enzymatic antioxidant genes of passion fruit were related by diverse phytohormones and play important roles in various biological processes.
Figure 11.
Expression profiles of 8 genes (PeAPX16, PeAPX19, PeCAT1, PeCAT2, PeCSD3, PeFSD1, PeGPX2, and PeCSD6) in response to GA, SA, ABA, and MeJA treatments
Error bars represent standard deviations of the means of three independent biological replicates of qRT-PCR analysis. Asterisks indicate significant differences in transcript levels compared with those of blank control (0 h 0 mM). (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
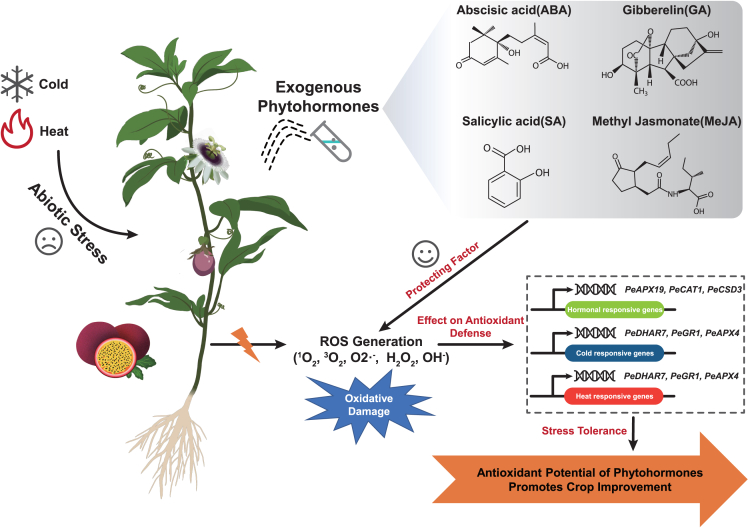
Throughout the life cycle of plants, ROS are dynamically produced or removed, which makes plants regulate their development to adapt to changing environments.7 Complex transcriptional and hormones regulation networks were also involved to keep the ROS concentration well-regulated.7 Here, candidate genes response to stresses and phytohormones were identified by qRT-PCR. By combining the bioinformatic analysis results with the experimental work, a schematic diagram was drawn in this research (Figure 12) to describe the potential roles of ROS and antioxidant genes in regulation of stress and hormone response in passion fruit. On the basis of these findings, the potential role of these enzymatic antioxidant genes can be predicted and shed lights for further investigations.
Figure 12.
Schematic diagram of enzymatic antioxidant genes abiotic stress resistance mechanism
Discussion
Passion fruit is a popular tropical fruit crop with important agricultural, economic, and ornamental value, but its growth and development are greatly affected by climatic conditions. Enzymatic antioxidant genes play key roles in regulating beneficial ROS accumulation for intracellular signals and impart plant tolerance to oxidative stress by scavenging excess of ROS.11 However, there has been no systematic investigations of enzymatic antioxidant genes in passion fruit yet.
In the present study, a total of 90 genes (11 PeSODs, 45 PeAPXs, 8 PeCATs, 7 PeGPXs, 6 PeMDHARs, 8 PeDHARs, and 5 PeGRs) enzymatic antioxidant gene family members were identified in passion fruit genome. Each gene family could be classified into different subfamilies based on phylogenetic analysis with Arabidopsis homologous genes, respectively. For each gene family, gene members located in the same subcellular compartment seemed to be more related to each other than those genes from the same organism but with a different localization. This phenomenon was more obvious for the APX gene family, chloroplastic PeAPXs in subgroup APX-4 were clearly separated with other branches, peroxisomal PeAPXs in subgroup APX-3/5 and APX-1/2 were closely clustered together, while cytosolic PeAPXs in subgroup APX-6 were also closely clustered with the passion fruit specific subgroup APX, and most PeAPXs in APX subgroup were also located in cytoplasm (Figure 1). In addition, some proteins showed one-to-one homology, such as PeCSD5 and AtCSD2, PeFSD1 and AtFSD1, PeAPX1 and AtAPX2, PeDHAR8 and AtDHAR3, the subcellular localization analysis results shown that they were also share similar subcellular localization to their corresponded orthologous Arabidopsis proteins (Table S1). Similar with previous work,16 these results shown that a close relationship among enzymatic antioxidant proteins located in the same subcellular compartment, indicating these genes might be working together to keep ROS balance at the subcellular level. Motif and domain composition as well as gene exon/intron structure analysis showed that closely related gene members tend to show similar structural characteristics, as observed in other plants such as soybean40 and rice,16 which independently supports the phylogenetic analysis and classification results. Differently, genes in different subfamilies shown diverse structure characteristics. Additionally, homologous protein modeling results showed that the representative proteins of each gene family shown diverse 3D structure with unique characteristics related to their enzymatic functions, and the structural diversity among these genes might contribute to the functional diversity of enzymatic antioxidant gene families.11
Compared with Arabidopsis (8 AtSODs, 8 AtAPXs, 3 AtCATs, 7 AtGPXs, 5 AtMDHARs, 3 AtDHARs, and 2 AtGRs), passion fruit (11 PeSODs, 45 PeAPXs, 8 PeCATs, 7 PeGPXs, 6 PeMDHARs, 8 PeDHARs, and 5 PeGRs) has obviously higher gene number of CAT, DHAR, GR, and especially APX enzymes. According to collinearly analysis within passion fruit, a total of 183 gene pairs with gene duplication events (177 segmental duplicated gene pairs and 6 tandem duplicated gene pairs) were identified, and 152 of these gene pairs were PeAPXs related. These results suggest that enzymatic antioxidant genes may arise through gene duplication, and segmental duplication events are the main driver of enzymatic antioxidant genes evolution.41,42 Among all the duplicated gene pairs, only one pair of segmental repeat genes (PeAPX30-PeAPX32) had a Ka/Ks value greater than 1, indicating that this pair of genes was strongly and positively selected during evolution. Although both of these two genes were preferentially higher expressed during fruit development, only PeAPX30 could be induced under high and low temperature treatments. Although these two genes shown high sequence similarity and closely clustered in the same branch, the motif composition and exon-intron structure were differed a lot, which might contribute to their functional diversification. The structural analysis could help to provide more information about evolutionary duplication events. Some duplicated genes shown similar expression profiles and closely clustered together in the same subfamily, indicating they might have conserved function. For example, PeGR1 and PeGR5 were segmentally duplicated and all higher expressed at the late development stages of fruit and in leaf before and after stress treatment. These two genes share almost the same motif compositions and exon/intron structures. However, most of the duplicated genes evolved with differed structural characteristics and shown different expression patterns, such as PeMDHAR5 (block C and IV) and PeMDHAR6 (block A and II), PeGPX4 (block E and I) and PeGPX5 (block C and V), PeFSD1 (block E and II) and PeFSD2 (block C and I), indicating these genes might be suffered sub-functionalization or neo-functionalization after duplication. Compared with the model laboratory species Arabidopsis, passion fruit survived in a more complex environment, which needed to cope with more abundant abiotic stresses during its growth and development. The expansion of these enzymatic antioxidant genes and the diversification of their functions might provide more supports for plants to deal with diverse unfavorable conditions, thus, enhancing the ability of passion fruit to coping with changing environments.7
Lots of transcription factors have reported to be involved in the antioxidant regulation under diverse stresses43,44 or during growth/development.45 Here, various TFs targeting passion fruit enzymatic antioxidant genes were predicted and their regulatory network interactions with the targeted genes were constructed (Figure 6) (Table S7). Diverse TFs involved in stress response (Dof, WRKY, HSF, bZIP), plant growth/development (BBR-BPC, MIKC-MADS, LBD, bHLH, AP2) and phytohormone response (ERF and ARF) were found in many enzymatic antioxidant genes. In previous work, we identified an HSF transcription factor involved in the response of passion fruit to high temperature stress. VIGS silencing experiment showed that passion fruit plants were more sensitive to heat stress and the activities of antioxidant enzymes such as SOD, POD, and CAT decreased after silencing of this gene (unpublished data), indicating HSF TF was involved in the regulation of antioxidant enzymes. The complex transcriptional regulatory networks might contribute to the diversified functions of antioxidant enzymes in diverse biological processes.
Meanwhile, 17 putative ped-miRNAs targeting 21 passion fruit enzymatic antioxidant genes were identified (Figure 7), and some ped-miRNAs targeted different enzymatic antioxidant genes including ped-miR171k-5p (PeCAT1, PeCAT7 and PeDHAR7), ped-miR828b (PeAPX29, PeAPX32 and PeAPX39), ped-miR160c-5p (PeAPX27, PeAPX43 and PeMDHAR2), ped-miR164b-5p (PeMDHAR4 and PeMDHAR5), ped-miR395a (PeAPX5 and PeAPX6). We found that different genes targeted by same miRNA shown similar expression patterns with a few exceptions. For example, miR171 could regulate hormone crosstalk through GRAS TFs and involved in the fruit and floral development.46 PeCAT1, PeCAT7, and PeDHAR7 targeted by ped-miR171k-5p were all preferentially expressed during fruit development and also abundant during diverse floral organs development (Figure 8). Besides, miR171 has also reported to participate in the regulation of male fertility stability under high temperature stress response in soybean,47 drought response in potato.48 PeCAT1, PeCAT7, and PeDHAR7 were also induced with higher expression in flower bud under high temperature treatment (Figure 8). CREs prediction and qRT-PCR analysis results also shown that these genes were response to diverse hormones such as GA, SA, and MeJA (Figure 11). Thus, ped-miR171k-5p and its targets might also have multitasked roles as in passion fruit. Additionally, miR828 has been reported to be involved in anthocyanin and flavonol accumulation during fruit development in grapes,49 pineapple,50 and apple.51 All of the three genes PeAPX29/32/39 targeted by ped-miR828b were preferentially expressed during fruit development of passion fruit, indicating ped-miR828b might have conserved functions in the regulation of fruit development in passion fruit. These results suggested that the identified ped-miRNAs might play important roles in the regulation of growth/development or stress responding through modifying the transcriptional level of enzymatic antioxidant genes in passion fruit, which still needs further studies.
Although the individual enzymic antioxidant genes involved in stress response or growth/development is largely reported in previous work, the systematic investigations of the regulation of different enzymic antioxidant gene families in a single plant species remains limit. Furthermore, there are still few reports describing the developmental regulation of the enzymatic antioxidant genes. Gene expression analyses revealed the functional diversity of the enzymatic antioxidant genes during the growth and development of passion fruit, and many of these genes showed tissue- or developmental stage-specific expression patterns during floral and fruit development. For example, PeAPX20/14/11/12 in block B was explicitly and highly expressed only at the early developmental stage of the stamen, while PeAPX5/13/35 were preferentially expressed during the development of sepal. Besides, enzymatic antioxidant genes in block A showed a preferential expression mainly at the late development stage of bract, petal, and corona filament including PeAPX2/24/44/27/43, PeMDHAR2/6, PeGR4, PeCSD6, and PeGPX7, whereas genes in block C showed preferential expression at the early stage of certain tissues such as PeMDHAR1 and PeCAT3/4/5. Even in the same block E, some genes were highest expressed at the early stages such as PeAPX15/29/30/32/4/17, PeFSD1, and PeMSD1/2, while some genes shown a highest expression level at the middle stages including PeCSD3/5, PeAPX22/39, PeCAT1/7, PeGPX3, and PeDHAR1/8, and there were also some genes were highest expressed at the late development stages such as PeGR1/2/5, PeAPX1, PeGPX1/6, and PeCSD4. Previous investigations about APX genes in rice also found that, most of the OsAPX genes participating in ROS removal shown tissue/organ-specific expression profiles.52 Emerging evidences indicated that the important roles of ROS in the regulation of plant growth and development including the maintenance of plant vegetative apical meristems,3 the initiation of floral buds to maturation of reproductive organs,6 the development of different organs,53 fruit development, and senescence.54 Generally, low ROS levels are required for the progression of some basic biological processes, such as cellular proliferation and differentiation, but higher levels ROS could pose a significant threat to plants.7 For example, the primary accumulation of O2·- in the meristematic zone of the root tip is required for cell division, while the accumulation of H2O2 in the elongation zone confers cell differentiation.3 Similarly, active cell division and differentiation were taken place during floral organs and fruit development here in passion fruit, which are also very energy-intensive processes. The co-expressed genes with 90 passion fruit enzymic genes were also significantly enriched in energy metabolism related pathways (Tables S11 and S12). We noticed that PeMSDs (all located in mitochondrion, Table S1) were abundant across all test tissues or under stress treatments with high expression levels. Mn-SODs were reported to play critical roles in scavenging excess accumulation of O2·- and protecting mitochondria against oxidant damage.55 These widely abundantly expressed PeMSDs might play important roles in protecting mitochondria against oxidative damage caused by active energy consumption during floral and fruit development processes. Similar with PeMSDs, few other enzymic antioxidant genes including PeFSD1, PeGPX1/2, PeAPX16, PeMDHAR2, and PeGR4 were also abundant in all tested samples here, indicating they might also play roles in maintaining cell redox homeostasis in these processes. As we known, maintaining the balance of ROS levels is important for normal plant growth and development. For instance, GR2 mutants with excessive accumulation of oxidized glutathione results in root apical cells entering the oxidized state and eventually lead to abnormal growth in Arabidopsis.19 Here, the diversified expression profiles of different passion fruit enzymic antioxidant genes might also be associated with protecting the cells against the harmful effects of high ROS levels which are produced during highly proliferating floral organs and fruit.
The growth and development of passion fruit are highly sensitive to the climatic changes. Purple passion fruit is more susceptible to high ambient temperatures compared to yellow cultivars, and even mildly elevated temperatures (∼30°C) or decreased temperature (∼20°C) might affect its growth and development.56 Unfavorable conditions might cause stresses and immediately induce more generation of ROS, many genes including PeCSD2/3/4, PeAPX26/11/12/4, PeGPX3, and PeDHAR7 were induced immediately under heat stress treatment and shown highest expression levels in a short time, indicating their rapid response to stress conditions. Some genes respond differently to heat and cold stress and shown tissue-specific expression profiles, such as the expression of PeMDHAR2 and PeGPX2 decreased in leaf, petiole and bract under heat stress, but upregulated in bud induced by cold. Meanwhile, some genes were induced by both heat and cold stress treatments including PeGR2 and PeAPX30, while genes such as PeDHAR7 and PeAPX4 were induced with higher expression levels in diverse tissues including bud, petiole, and leaf under mild or severe heat stress. Engineering crop plants with improved antioxidative enzyme machinery provide a promising approach for better abiotic stress resistance. For example, overexpression of AtApx1 gene confers salinity stress tolerance by strengthening the antioxidative defense system in Brassica juncea.57 Over-expression of PaSOD and RaAPX in transgenic Arabidopsis improves cold stress tolerance through increase in vascular lignification.58 Thus, these candidate genes, which are not tissue specific or respond to diverse stresses, might have important application potential for the genetic improvement of passion fruit with better abiotic stress resistance. Besides, we found that many of these stress responsive enzymic antioxidant genes were preferentially expressed in certain tissues at specific developmental stages. For example, PeDHAR7 was induced under heat and cold stress, and also preferentially higher expressed during fruit development (Figure 9, clustered in block E and V). It contained diverse CREs associated with stress response (stress responsiveness, anaerobic induction), development (meristem expression element, light responsive element) and hormone response (ABA-responsiveness, GA-responsive elements) in its promoter region. The co-presence of different CREs in the promoter regions might be closely related to the diverse roles of enzymic antioxidant genes in the growth and development of passion fruit under different environmental changes. Additionally, multiple or diverse hormone response related CREs were found in the promoter region of many enzymic antioxidant genes, and qRT-PCR analysis of several representative genes under different hormone treatments showed that many enzymic antioxidant genes could respond to multiple hormones. Previous work shown that ROS could interact with hormones including ABA, MeJA, SA, and other signal molecules to regulate plant growth/development and stress response.7 Thus, the interaction between ROS and different hormone regulation networks might also contribute to the involvement of enzymic antioxidant genes in different biological processes.
Limitations of the study
In the present study, we first provided a systemic analysis of enzymatic antioxidant genes in passion fruit. As a result, a total of 90 genes (11 PeSODs, 45 PeAPXs, 8 PeCATs, 7 PeGPXs, 6 PeMDHARs, 8 PeDHARs, and 5 PeGRs) encoding for antioxidant enzymes were identified. Each gene family could be classified into different subfamilies based on phylogenetic analysis with Arabidopsis homologous genes, respectively, and closely related gene members tend to show similar structural characteristics. Besides, gene members in each gene family located in the same subcellular compartment shown a closer phylogenetic relationship. Compared with Arabidopsis, the gene number of PeCATs, PeDHARs, PeGRs, and especially PeAPXs were obviously expanded. Collinear analysis shown that the expansion of enzymatic antioxidant gene family might be carried out mainly through segmental duplication, and the structural diversity of duplicated genes may contribute to their functional diversity. Gene expression analyses revealed many enzymatic antioxidant genes showed tissue- or developmental stage-specific expression patterns during floral and fruit development in passion fruit, the widely abundantly expressed enzymic antioxidant genes such as PeMSDs, PeFSD1, PeGPX1/2, PeAPX16 might play important roles in protecting cells against oxidative damage caused by active energy consumption in the highly proliferating floral and fruit tissues. Candidate genes such as PeDHAR7 and PeAPX4 were induced with higher expression levels in diverse tissues under mild or severe heat stress, might have important application potential for the genetic improvement of passion fruit with better abiotic stress resistance. Different CREs co-exist in the promoter region of enzymatic antioxidant genes and the complex interactions of diverse transcription factors, miRNAs and hormones regulation might be closely related to a variety of regulatory effects of enzymatic antioxidant genes in passion fruit. This genome-wide diversity analysis of enzymatic antioxidant genes in passion fruit provides new insights for future functional characterization of the regulation of important horticultural traits such as development, fruit ripening, and multiple hormone treatment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Floral and fruit samples of P.edulis | Sampled from the orchard of the Institute of Horticulture, Guangxi Academy of Agricultural Sciences | N/A |
| Tissues under stress treatment | Sampled from the intelligent greenhouse at Pingtan Technology Research Institute | N/A |
| Critical commercial assays | ||
| HiScript II Q RT SuperMix for qPCR (+gDNA wiper) | Vazyme | R223-01 |
| Taq Pro Universal SYBR qPCR Master Mix | Vazyme | Q712-02 |
| Deposited data | ||
| Genome data | the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/) database | GWHAZTM00000000 |
| Raw data for RNA-seq | the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/) database | CNP0002747 |
| Oligonucleotides | ||
| qPCR primers sequences | This paper | Table S13 |
| Software and algorithms | ||
| MEGA 11 | Kumar et al.59 | https://www.megasoftware.net/ |
| IQ-TREE 2 | Minh et al.60 | http://www.iqtree.org/ |
| Evolview v3 | Subramanian et al.61 | https://www.evolgenius.info/evolview |
| iTOL | Letunic et al.62 | https://itol.embl.de/ |
| SMART | Letunic et al.63 | http://smart.embl-heidelberg.de/ |
| ProtParam | Gasteiger et al.64 | https://web.expasy.org/protparam/ |
| Cell-PLoc 2.0 | Chou et al.65 | http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/ |
| MEME v5.5.1 | Bailey et al.66 | http://meme-suite.org/ |
| PlantCARE | Lescot et al.67 | http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ |
| Circos | Krzywinski et al.68 | http://circos.ca/software/download/circos/ |
| MCScanX | Wang et al.69 | https://github.com/wyp1125/MCScanX |
| TBtools | Chen et al.70 | https://github.com/CJ-Chen/TBtools/releases |
| KaKs_Calculator 3.0 | Zhang et al.71 | https://ngdc.cncb.ac.cn/biocode/tools/BT000001/releases/3.0 |
| SWISS-MODEL | Marco et al.72 | https://swissmodel.expasy.org/ |
| PyMOL | Schrodinger Sales Center | https://pymol.org/2/ |
| PTRM | Tian et al.73 | http://plantregmap.gao-lab.org/ |
| Cytoscape | Shannon et al.74 | https://cytoscape.org/ |
| psRNATarget | Dai et al.75 | http://plantgrn.noble.org/psRNATarget/ |
| eggNOG | Cantalapiedra et al.76 | http://eggnog-mapper.embl.de/ |
| Prism 9.0 | GraphPad | https://www.graphpad.com/ |
| R 4.3.1 | The R Foundation | https://www.r-project.org/ |
| ggplot2 | Wickham, Hadley77 | https://ggplot2.tidyverse.org/ |
| clusterProfiler | Wu et al.78 | https://guangchuangyu.github.io/software/clusterProfiler/ |
| pheatmap | Kolde et al.79 | https://github.com/raivokolde/pheatmap |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Ping Zheng (zhengping13@mails.ucas.ac.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The data presented in this study are available in the article, Supplementary Materials and online repositories. Genome data and RNA-seq data used in this work were deposited in the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/) database: GWHAZTM00000000 and NGDC: CNP0002747, respectively.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Plant materials and abiotic stress treatment
The P. edulis purple passion fruit used in this study was planted in the orchard of the Institute of Horticulture, Guangxi Academy of Agricultural Sciences. Diverse floral tissues at different developmental stages were sampled as previously described.80 Abiotic stress and hormone treatments were applied to fully grown (with well-developed roots and shoots) healthy and disease-free passion fruit plants with three biological repeats. As to cold stress treatment, healthy plants were placed in a growth chamber with the temperature set at 8°C; as to heat stress treatment, plants were kept in a growth chamber with the temperature set at 45°C; as to hormones treatment, gibberellin (GA, 100 μM), salicylic acid (SA, 100 μM), abscisic acid (ABA, 100 μM) and methyl jasmonate (MeJA, 100 μM) were applied to healthy plants respectively. The samples under diverse treatments were collected at 24 h and 48 h time intervals, respectively, with normal plants that were not treated used as the control. All the collected samples were immediately stored in liquid nitrogen prior to total RNA extraction.
Method details
Identification of enzymatic antioxidant genes in passion fruit
The whole genome data of passion fruit was downloaded from the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/) database with the accession number GWHAZTM00000000. Whole genome information for Arabidopsis, rice, tomato, maize and grapes genomic data were downloaded from the Ensembl Plants database (http://plants.ensembl.org/index.html). Amino acid sequences of the SOD gene family (8 genes), APX gene family (8 genes), CAT gene family (3 genes), GPX gene family (7 genes), MDHAR gene family (6 genes), DHAR gene family (3 genes), and GR gene family (2 genes) in Arabidopsis (Table S2) were downloaded from TAIR Arabidopsis genome database (http://www.arabidopsis.org/index.jsp),81 which were used to identify the members of the enzymatic antioxidant genes in passion fruit. Furthermore, the accuracy of deduced protein sequences was confirmed by searching for homologous sequences deposited in GenBank database (non-redundant) using the BLASTp tool (Basic Local Alignment Search Tool for proteins). The candidate protein sequences were obtained and identified using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/) (E-value of 10-5, other parameters set to defaults)82 and SMART databases.63 The confirmed enzymatic antioxidant genes were renamed according to their positions on the passion fruit chromosomes. The physic-chemical properties of molecular weight (MW), isoelectric point (pI) and grand average of hydropathicity (GRAVY) were analyzed by the ExPASy ProtParam tool.64 Finally, the subcellular localization of the protein was determined by the Cell-PLoc 2.0 server.65
Phylogenetic and evolutionary analysis
To observe the evolutionary relationship of the enzymatic antioxidant gene family, we constructed a phylogenetic tree about passion fruit and Arabidopsis protein sequences. Multiple sequence alignment of the protein sequences was conducted by MUSCLE83 implemented in MEGA 11 (Molecular Evolutionary Genetics Analyses) software.59 The maximum likelihood (ML) method phylogenetic tree based on the protein sequence alignment was then constructed according to the Whelan and Goldman substitution model (WAG+Gamma)84 by MEGA 11 and IQ-TREE 2 software.60 Relative branch support was evaluated by 1000 bootstrap replicates, the branch lengths were calculated by pairwise comparisons of genetic distances, and missing data were treated by pairwise deletions of gaps. For better visualization, the phylogenetic tree was visualized using the online tool Evolview v361 and iTOL (Interactive Tree of Life).62
Gene structure, conserved motif and cis-regulatory elements analysis
The intron-exon distributions of the enzymatic antioxidant genes were obtained using GFF annotation files from the passion fruit genome. Protein sequence analysis of Multiple Em for Motif Elicitation (MEME v5.5.1) online program66 is used to identify the identified conservative motif in PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, PeDHAR, and PeGR proteins. The optimization parameters are as follows: the number of repetitions is arbitrary; the maximum number of motifs is 10. The classification of DNA-binding motifs of enzymatic antioxidant genes was determined according to the basic regions of SOD, APX, CAT, GPX, MDHAR, DHAR, and GR domains described in previous studies. The upstream 2000 bp sequence of each PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, PeDHAR, and PeGR gene was retrieved by TBtools (v1.125) software70 according to the genomic full-length DNA sequences of passion fruit. Then, the PlantCARE database67 was used to predict putative promoter regions of enzymatic antioxidant genes in passion fruit cis-regulatory elements. Finally, the results were visualized using TBtools.
Chromosomal distribution and gene duplication analysis
All enzymatic antioxidant genes were anchored to their corresponding chromosomes using Circos68 against physical location information from the passion fruit genome database. To demonstrate the synteny of orthologous SOD, APX, CAT, GPX, MDHAR, DHAR, and GR genes obtained from passion fruit and other selected species, we analyzed gene duplication events by using the Multicollinearity Scanning Toolkit (MCScanX),69 setting default parameters, and using Circos and Dual Synteny Plot visualization results in TBtools software.70 The non-synonymous substitution (Ka) and synonymous substitution (Ks) rates as well as the Ka/Ks values of all duplicated gene pairs were calculated using the NG model of the KaKs_Calculator 3.0,71 and Ka/Ks ratio between homologous gene pairs. Its parameters are set as: NG as the Method (-m) and Standard Code as the Genetic code table (-c). The time of divergence (MYA: million years ago) of enzymatic antioxidant genes was measured using the following reference formula: T = Ks/2λ (where λ = 6.38 × 10−9).85
Secondary and tertiary structure analysis
The protein secondary structures of PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, PeDHAR, and PeGR proteins were predicted using SOPMA server.86 The tertiary structure was obtained from AlphaFold Protein Structure Database and the SWISS-MODEL with default parameters.72 Furthermore, tertiary structures of these proteins were examined by PyMOL.87
Transcription factors (TFs) regulatory network analysis
A regulation prediction tool, the Plant Transcriptional Regulatory Map (PTRM) (http://plantregmap.gao-lab.org/)73 was used to infer potential regulatory interactions of TFs in the upstream (2000-bp) regions of PeSODs, PeAPXs, PeCATs, PeGPXs, PeMDHARs, PeDHARs, and PeGRs with threshold (P-value≤1e−7). The Arabidopsis was the selected plant species. The predicted TFs were visualized into a network using the Cytoscape software (v3.10).74 Furthermore, the wordcloud is generated by the ggplot277 package88 in R.
Prediction of putative miRNA analysis of enzymatic antioxidant genes
The prediction of putative miRNA sites in the PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, PeDHAR, and PeGR gene was achieved by downloading the published passion fruit mature miRNAs,89 and then the CDS sequences were submitted to the online psRNATarget database75 with default parameters. The network interaction among the equivalent PeSOD, PeAPX, PeCAT, PeGPX, PeMDHAR, PeDHAR, and PeGR target genes and putative predicted miRNAs was constructed and displayed by Cytoscape software.74
GO and KEGG annotation analysis
The GO and KEGG annotation analyses were performed by submitting all the 90 enzymatic antioxidant gene protein sequences to the online database eggNOG,76 and enrichment analysis was performed using R package clusterProfiler.78 Co-expressed genes with 90 enzymic antioxidant genes were extracted from the analysis results of WGCNA (weighted gene co-expression network analysis) using RNA-seq data.
Expression profiles analysis based on RNA-seq data
Diverse floral tissues at an early stage when the archesporial cell had formed and the late stages when ovules had fully differentiated were used for the RNA-seq (Table S9). Fruit samples were collected at the fruit juice formation stage (53 days after pollination/DAP), fruit juice color transformation stage (60 DAP), peel color transformation stage (100 DAP), and fruit ripening stage (128 DAP). Additionally, four different tissues including leaves, petioles, bracts and buds were sampled at 1h, 4h, 12h, and 24h after high temperature (30°C) and low temperature (20°C) treatments. Detailed sampling information was also listed in Table S10.
RNA extraction and Illumina sequencing were performed as previously described,90 with 1 μg RNA per sample and three independent biological replicates for each tissue. The cDNA libraries were constructed using the NEBNext Ultra™ RNA Library Prep Kit for Illumina (NEB, Beverly city, MA, USA), following standard protocols. The transcript abundance of PeSODs, PeAPXs, PeCATs, PeGPXs, PeMDHARs, PeDHARs, and PeGRs was calculated as TPM (Transcripts Per Kilobase Million). The heatmap was generated by pheatmap packages in R based the log2(TPM + 1).
qRT-PCR analysis of key enzymatic antioxidant genes
The qRT-PCR analysis was used to explore the expression of representative enzymic antioxidant genes response to hormones and abiotic stresses. The Trizol method (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA, and the ThermoScript RT-PCR kit (Thermo Fisher Scientific, Carlsbad, CA, USA) was used to conduct the reverse-transcribed experiment. Real-time PCR was performed by using the SYBR Premix Ex Taq II system (TaKaRa Perfect Real Time) in the Bio-Rad Real-time PCR system (Foster City, CA, USA), and the primers used are listed in Table S13. The qRT-PCR program was: 95°C for 30 s; 40 cycles of 95°C for 5 s; 60°C for 34 s; 95°C for 15 s. The passion fruit EF1a was used as an internal control to normalize the mRNA levels.91 For each analysis, three technical replicates from three biological replicates were performed, the fold change of genes was calculated using the 2-ΔΔCT method.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad software version 9.0. Specifications of tests exploited and sample size for each experiment are mentioned in the figure legends. Data are presented as the standard error of the mean (SEM). Differences were analyzed by the student’s t-test and one-way ANOVA as indicated in the figure legends. Asterisk (∗) indicates P < 0.05 against each control group respectively.
Additional resources
Additional resources are provided in the supplemental information.
Acknowledgments
This work was supported by Science and Technology Innovation Project of Pingtan Science and Technology Research Institute (PT2021003, PT2021007), General Project of Fujian Natural Science Foundation (2020J01594). We thank Zhenjiang Zheng from Fujian Lianmi Ecological Agriculture Development Co., LTD for his assistance during sample collection.
Author contributions
Investigation, formal analysis, visualization, methodology, J.L.; writing - original draft, J.L., L.L., and P.Z.; validation, L.L. investigation, resources, W.Z. and X.W.; data curation, M.S., M.C., C.A., and S.C.; funding acquisition, X.W. and Y.Q.; conceptualization, writing - review & editing, R.L. and P.Z.; conceptualization, supervision, project administration, Y.Q. and P.Z. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108329.
Contributor Information
Yuan Qin, Email: yuanqin@fafu.edu.cn.
Ping Zheng, Email: zhengping13@mails.ucas.ac.cn.
Supplemental information
References
- 1.López-Vargas J.H., Fernández-López J., Pérez-Álvarez J.A., Viuda-Martos M. Chemical, physico-chemical, technological, antibacterial and antioxidant properties of dietary fiber powder obtained from yellow passion fruit (Passiflora edulis var. flavicarpa) co-products. Food Res. Int. 2013;51:756–763. doi: 10.1016/j.foodres.2013.01.055. [DOI] [Google Scholar]
- 2.Hasanuzzaman M., Hossain M.A., da Silva J.A.T., Fujita M. In: Crop stress and its management: perspectives and strategies. Venkateswarlu B., Shanker A.K., Shanker C., Maheswari M., editors. Springer; 2012. Plant response and tolerance to abiotic oxidative stress: antioxidant defense is a key factor; pp. 261–315. [DOI] [Google Scholar]
- 3.Tsukagoshi H., Busch W., Benfey P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Kärkönen A., Kuchitsu K. Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry. 2015;112:22–32. doi: 10.1016/j.phytochem.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Steffens B., Kovalev A., Gorb S.N., Sauter M. Emerging Roots Alter Epidermal Cell Fate through Mechanical and Reactive Oxygen Species Signaling. Plant Cell. 2012;24:3296–3306. doi: 10.1105/tpc.112.101790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann P., Heinlein C., Orendi G., Zentgraf U. Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 2006;29:1049–1060. doi: 10.1111/j.1365-3040.2005.01459.x. [DOI] [PubMed] [Google Scholar]
- 7.Huang H., Ullah F., Zhou D.X., Yi M., Zhao Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019;10:800. doi: 10.3389/fpls.2019.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazzarotto F., Teixeira F.K., Rosa S.B., Dunand C., Fernandes C.L., de Vasconcelos Fontenele A., Silveira J.A.G., Verli H., Margis R., Margis-Pinheiro M. Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 2011;191:234–250. doi: 10.1111/j.1469-8137.2011.03659.x. [DOI] [PubMed] [Google Scholar]
- 9.Pang C.-H., Wang B.-S. In: Ascorbate-Glutathione Pathway and Stress Tolerance in Plants. Anjum N.A., Chan M.-T., Umar S., editors. Springer Netherlands; 2010. Role of Ascorbate Peroxidase and Glutathione Reductase in Ascorbate–Glutathione Cycle and Stress Tolerance in Plants; pp. 91–113. [DOI] [Google Scholar]
- 10.Queval G., Issakidis-Bourguet E., Hoeberichts F.A., Vandorpe M., Gakière B., Vanacker H., Miginiac-Maslow M., Van Breusegem F., Noctor G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007;52:640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- 11.Bobrovskikh A., Zubairova U., Kolodkin A., Doroshkov A. Subcellular compartmentalization of the plant antioxidant system: an integrated overview. PeerJ. 2020;8:27. doi: 10.7717/peerj.9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nandi A., Yan L.J., Jana C.K., Das N. Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/9613090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frugoli J.A., Zhong H.H., Nuccio M.L., McCourt P., McPeek M.A., Thomas T.L., McClung C.R. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;112:327–336. doi: 10.1104/pp.112.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bela K., Horváth E., Gallé Á., Szabados L., Tari I., Csiszár J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015;176:192–201. doi: 10.1016/j.jplph.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Landi M. Commentary to: "Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds" by Hodges et al., Planta (1999) 207:604-611. Planta. 2017;245:1067. doi: 10.1007/s00425-017-2699-3. [DOI] [PubMed] [Google Scholar]
- 16.Teixeira F.K., Menezes-Benavente L., Margis R., Margis-Pinheiro M. Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: Inferences from the rice genome. J. Mol. Evol. 2004;59:761–770. doi: 10.1007/s00239-004-2666-z. [DOI] [PubMed] [Google Scholar]
- 17.McClung C.R. Regulation of catalases in Arabidopsis. Free Radic. Biol. Med. 1997;23:489–496. doi: 10.1016/s0891-5849(97)00109-3. [DOI] [PubMed] [Google Scholar]
- 18.Riyazuddin R., Bela K., Horváth E., Rigó G., Gallé Á., Szabados L., Fehér A., Csiszár J. Overexpression of the Arabidopsis glutathione peroxidase-like 5 gene (AtGPXL5) resulted in altered plant development and redox status. Environ. Exp. Bot. 2019;167 doi: 10.1016/j.envexpbot.2019.103849. [DOI] [Google Scholar]
- 19.Yu X., Pasternak T., Eiblmeier M., Ditengou F., Kochersperger P., Sun J., Wang H., Rennenberg H., Teale W., Paponov I., et al. Plastid-Localized Glutathione Reductase2-Regulated Glutathione Redox Status Is Essential for Arabidopsis Root Apical Meristem Maintenance. Plant Cell. 2013;25:4451–4468. doi: 10.1105/tpc.113.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima-Melo Y., Carvalho F.E.L., Martins M.O., Passaia G., Sousa R.H.V., Neto M.C.L., Margis-Pinheiro M., Silveira J.A.G. Mitochondrial GPX1 silencing triggers differential photosynthesis impairment in response to salinity in rice plants. J. Integr. Plant Biol. 2016;58:737–748. doi: 10.1111/jipb.12464. [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Fang G., Yang J., Li Y. A Thioredoxin-Dependent Glutathione Peroxidase (OsGPX5) Is Required for Rice Normal Development and Salt Stress Tolerance. Plant Mol. Biol. Rep. 2017;35:333–342. doi: 10.1007/s11105-017-1026-2. [DOI] [Google Scholar]
- 22.Hu L., Liang W., Yin C., Cui X., Zong J., Wang X., Hu J., Zhang D. Rice MADS3 Regulates ROS Homeostasis during Late Anther Development. Plant Cell. 2011;23:515–533. doi: 10.1105/tpc.110.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunkar R., Kapoor A., Zhu J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng H., Wang X., Zhang Q., Fu Y., Feng C., Wang B., Huang L., Kang Z. Monodehydroascorbate reductase gene, regulated by the wheat PN-2013 miRNA, contributes to adult wheat plant resistance to stripe rust through ROS metabolism. Biochim. Biophys. Acta. 2014;1839:1–12. doi: 10.1016/j.bbagrm.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 25.de Oliveira A.B., de Almeida Lopes M.M., Moura C.F.H., de Siqueira Oliveira L., de Souza K.O., Filho E.G., Urban L., de Miranda M.R.A. Effects of organic vs. conventional farming systems on quality and antioxidant metabolism of passion fruit during maturation. Sci. Hortic. 2017;222:84–89. doi: 10.1016/j.scienta.2017.05.021. [DOI] [Google Scholar]
- 26.Li C., Xin M., Li L., He X., Liu G., Li J., Sheng J., Sun J. Transcriptome profiling helps to elucidate the mechanisms of ripening and epidermal senescence in passion fruit (Passiflora edulia Sims) PLoS One. 2020;15:e0236535. doi: 10.1371/journal.pone.0236535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang D., Yousef A.F., Wei X., Ali M.M., Yu W., Yang L., Oelmüller R., Chen F. Increasing the performance of Passion fruit (Passiflora edulis) seedlings by LED light regimes. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong H., Chen Q., Dai Y., Hu W., Zhang S., Huang X. Genome-wide identification of PbrbHLH family genes, and expression analysis in response to drought and cold stresses in pear (Pyrus bretschneideri) BMC Plant Biol. 2021;21:86. doi: 10.1186/s12870-021-02862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A., Sharma M., Gahlaut V., Nagaraju M., Chaudhary S., Kumar A., Tyagi P., Gajula M.N.V.P., Singh K.P. Genome-wide identification, characterization, and expression profiling of SPX gene family in wheat. Int. J. Biol. Macromol. 2019;140:17–32. doi: 10.1016/j.ijbiomac.2019.08.105. [DOI] [PubMed] [Google Scholar]
- 30.Wang R., Li Y., Gao M., Han M., Liu H. Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L. BMC Genom. 2022;23:548. doi: 10.1186/s12864-022-08782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch M., Conery J.S. The Evolutionary Fate and Consequences of Duplicate Genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 32.Ding A., Ding A., Li P., Wang J., Cheng T., Bao F., Zhang Q. Genome-Wide Identification and Low-Temperature Expression Analysis of bHLH Genes in Prunus mume. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.762135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J., Xie Q., Li C., Dong Y., Zhu S., Chen J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta. 2020;251:81. doi: 10.1007/s00425-020-03364-8. [DOI] [PubMed] [Google Scholar]
- 34.Yuan T., Liang J., Dai J., Zhou X.R., Liao W., Guo M., Aslam M., Li S., Cao G., Cao S. Genome-Wide Identification of Eucalyptus Heat Shock Transcription Factor Family and Their Transcriptional Analysis under Salt and Temperature Stresses. Int. J. Mol. Sci. 2022;23:8044. doi: 10.3390/ijms23148044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han L.M., Hua W.P., Cao X.Y., Yan J.A., Chen C., Wang Z.Z. Genome-wide identification and expression analysis of the superoxide dismutase (SOD) gene family in Salvia miltiorrhiza (vol 742C, 144603, 2020) Gene. 2020;742 doi: 10.1016/j.gene.2020.144639. [DOI] [PubMed] [Google Scholar]
- 36.Park A.K., Kim I.S., Do H., Jeon B.W., Lee C.W., Roh S.J., Shin S.C., Park H., Kim Y.S., Kim Y.H., et al. Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica. Sci. Rep. 2016;6 doi: 10.1038/srep33903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.del Río L.A., Sevilla F., Sandalio L.M., Palma J.M. Nutritional effect and expression of SODs: induction and gene expression; diagnostics; prospective protection against oxygen toxicity. Free Radic. Res. Commun. 1991;12–13:819–827. doi: 10.3109/10715769109145863. [DOI] [PubMed] [Google Scholar]
- 38.Huo C., He L., Yu T., Ji X., Li R., Zhu S., Zhang F., Xie H., Liu W. The Superoxide Dismutase Gene Family in Nicotiana tabacum: Genome-Wide Identification, Characterization, Expression Profiling and Functional Analysis in Response to Heavy Metal Stress. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidner C.A., Martienssen R.A. The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBarry J.D., Kissinger J.C. Jumbled Genomes: Missing Apicomplexan Synteny. Mol. Biol. Evol. 2011;28:2855–2871. doi: 10.1093/molbev/msr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeling M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu. Rev. Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z., Wang F., Ma Y., Dang H., Hu X. Transcription Factor SlAREB1 Is Involved in the Antioxidant Regulation under Saline-Alkaline Stress in Tomato. Antioxidants. 2022;11:1673. doi: 10.3390/antiox11091673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T., Li W., Hu X., Guo J., Liu A., Zhang B. A Cotton MYB Transcription Factor, GbMYB5, is Positively Involved in Plant Adaptive Response to Drought Stress. Plant Cell Physiol. 2015;56:917–929. doi: 10.1093/pcp/pcv019. [DOI] [PubMed] [Google Scholar]
- 45.Xu N., Chu Y., Chen H., Li X., Wu Q., Jin L., Wang G., Huang J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018;14:e1007662. doi: 10.1371/journal.pgen.1007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cedillo-Jimenez C.A., Feregrino-Perez A.A., Guevara-Gonzalez R.G., Cruz-Hernandez A. MicroRNA regulation during the tomato fruit development and ripening: A review. Sci. Hortic. 2020;270:8. doi: 10.1016/j.scienta.2020.109435. [DOI] [Google Scholar]
- 47.Ding X., Guo J., Zhang Q., Yu L., Zhao T., Yang S. Heat-Responsive miRNAs Participate in the Regulation of Male Fertility Stability in Soybean CMS-Based F-1 under High Temperature Stress. Int. J. Mol. Sci. 2021;22:2446. doi: 10.3390/ijms22052446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hwang E.W., Shin S.J., Yu B.K., Byun M.O., Kwon H.B. miR171 Family Members are Involved in Drought Response in Solanum tuberosum. J. Plant Biol. 2011;54:43–48. doi: 10.1007/s12374-010-9141-8. [DOI] [Google Scholar]
- 49.Tirumalai V., Swetha C., Nair A., Pandit A., Shivaprasad P.V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 2019;70:4775–4792. doi: 10.1093/jxb/erz264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rock C.D. A role for MIR828 in pineapple fruit development. F1000Res. 2020;9:16. doi: 10.12688/f1000research.21779.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B., Yang H.J., Yang Y.Z., Zhu Z.Z., Li Y.N., Qu D., Zhao Z.Y. mdm-miR828 Participates in the Feedback Loop to Regulate Anthocyanin Accumulation in Apple Peel. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teixeira F.K., Menezes-Benavente L., Galvão V.C., Margis R., Margis-Pinheiro M. Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 2006;224:300–314. doi: 10.1007/s00425-005-0214-8. [DOI] [PubMed] [Google Scholar]
- 53.Zeng J., Dong Z., Wu H., Tian Z., Zhao Z. Redox regulation of plant stem cell fate. EMBO J. 2017;36:2844–2855. doi: 10.15252/embj.201695955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huan C., Jiang L., An X., Yu M., Xu Y., Ma R., Yu Z. Potential role of reactive oxygen species and antioxidant genes in the regulation of peach fruit development and ripening. Plant Physiol. Biochem. 2016;104:294–303. doi: 10.1016/j.plaphy.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Ying Y., Chen J., Wang X. Transgenic Arabidopsis overexpressing Mn-SOD enhanced salt-tolerance. Plant Sci. 2004;167:671–677. doi: 10.1016/j.plantsci.2004.03.032. [DOI] [Google Scholar]
- 56.Fischer G., Miranda D. Review on the ecophysiology of important Andean fruits: Passiflora L. Rev. Fac. Nac. Agron. Medellin. 2021;74:9471–9481. doi: 10.15446/rfnam.v74n2.91828. [DOI] [Google Scholar]
- 57.Saxena S.C., Salvi P., Kamble N.U., Joshi P.K., Majee M., Arora S. Ectopic overexpression of cytosolic ascorbate peroxidase gene (Apx1) improves salinity stress tolerance in Brassica juncea by strengthening antioxidative defense mechanism. Acta Physiol. Plant. 2020;42:45. doi: 10.1007/s11738-020-3032-5. [DOI] [Google Scholar]
- 58.Shafi A., Dogra V., Gill T., Ahuja P.S., Sreenivasulu Y. Simultaneous Over-Expression of PaSOD and RaAPX in Transgenic Arabidopsis thaliana Confers Cold Stress Tolerance through Increase in Vascular Lignifications. PLoS One. 2014;9:e110302. doi: 10.1371/journal.pone.0110302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian B., Gao S., Lercher M.J., Hu S., Chen W.H. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letunic I., Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chou K.C., Shen H.B. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 66.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouzé P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:14. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Z. KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Dev. Reprod. Biol. 2022;20:536–540. doi: 10.1016/j.gpb.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tian F., Yang D.C., Meng Y.Q., Jin J., Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48:D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kohl M., Wiese S., Warscheid B. In: Data Mining in Proteomics: From Standards to Applications. Hamacher M., Eisenacher M., Stephan C., editors. Humana Press Inc, 999 Riverview Dr, Ste 208; 2011. Cytoscape: Software for Visualization and Analysis of Biological Networks; pp. 291–303. [DOI] [Google Scholar]
- 75.Dai X., Zhao P.X. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cantalapiedra C.P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021;38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wickham H. ggplot2. WIREs Computational Stats. 2011;3:180–185. [Google Scholar]
- 78.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kolde R., Kolde M.R. Package ‘pheatmap’. R package. 2015;1:790. [Google Scholar]
- 80.Liang J., Hou Z., Liao J., Qin Y., Wang L., Wang X., Su W., Cai Z., Fang Y., Aslam M., et al. Genome-Wide Identification and Expression Analysis of LBD Transcription Factor Genes in Passion Fruit (Passiflora edulis) Int. J. Mol. Sci. 2022;23:4700. doi: 10.3390/ijms23094700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113–119. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 85.Ma D., Dong S., Zhang S., Wei X., Xie Q., Ding Q., Xia R., Zhang X. Chromosome-level reference genome assembly provides insights into aroma biosynthesis in passion fruit (Passiflora edulis) Mol. Ecol. Resour. 2021;21:955–968. doi: 10.1111/1755-0998.13310. [DOI] [PubMed] [Google Scholar]
- 86.Geourjon C., Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 87.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ginestet C. ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A-Stat. Soc. 2011;174:245–246. doi: 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
- 89.Paul S., de la Fuente-Jiménez J.L., Manriquez C.G., Sharma A. Identification, characterization and expression analysis of passion fruit (Passiflora edulis) microRNAs. 3 Biotech. 2020;10:25. doi: 10.1007/s13205-019-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen P., Li Y., Zhao L., Hou Z., Yan M., Hu B., Liu Y., Azam S.M., Zhang Z., Rahman Z.U., et al. Genome-Wide Identification and Expression Profiling of ATP-Binding Cassette (ABC) Transporter Gene Family in Pineapple (Ananas comosus (L.) Merr.) Reveal the Role of AcABCG38 in Pollen Development. Front. Plant Sci. 2017;8:2150. doi: 10.3389/fpls.2017.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu Y., Tian Q., Huang W., Liu J., Xia X., Yang X., Mou H. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Passiflora edulis under stem rot condition. Mol. Biol. Rep. 2020;47:2951–2962. doi: 10.1007/s11033-020-05385-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data presented in this study are available in the article, Supplementary Materials and online repositories. Genome data and RNA-seq data used in this work were deposited in the National Genome Data Center (NGDC) (https://ngdc.cncb.ac.cn/) database: GWHAZTM00000000 and NGDC: CNP0002747, respectively.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.