Abstract
Mosaicism represents a genuine real phenomenon, but its high prevalence and undisclosed clinical significance, stress the burden on genetic counseling and the management of PGT-A results. Even though the assumption of mosaicism from NGS intermediate chromosome copy number profiles may represent a reasonable interpretation, other potential technical reasons, including amplification bias, contamination, biopsy technique, or the analysis algorithms, may constitute alternative explanations. Thresholds confining mosaicism ranges are established according to models employing mixtures of normal and abnormal cells with steady conditions of quantity and quality which are unable to reflect the full extent of variability present in a trophectoderm (TE) biopsy specimen. When the concordance of TE with the ICM is considered, mosaic TE biopsies poorly correlate with the chromosomal status of the remaining embryo, displaying mostly ICM aneuploidy in cases of TE high-range mosaics diagnosis and euploidy when mosaicism grade in TE is less than 50% (low-mid range mosaicism), which implies an evident overestimation of mosaicism results. Indeed, a binary classification of NGS profiles that excludes mosaic ranges, including only euploid and aneuploid diagnosis, provides higher specificity and accuracy in identifying abnormal embryos and discarding them. As intermediate copy number profiles do not represent strong evidence of mosaicism but only an inaccurate and misleading assumption, and considering that no increased risk has been reported in the offspring, until diagnosis specificity is improved and its clinical implications are determined, laboratories should consider limiting predictions to euploid and aneuploid and stop reporting mosaicism.
Keywords: Mosaicism, Thresholds, PGT-A, NGS, Blastocyst, Trophectoderm, Inner cell mass
Introduction
Preimplantation genetic testing for aneuploidies (PGT-A) aims to improve IVF outcomes by avoiding the transfer of aneuploid embryos. Although mosaicism represents a genuine biological phenomenon, reported not only in PGT-A but also in PGT-M and PGT-SR, the prevalence of mosaic diagnosis has significantly grown with the use of next-generation sequencing (NGS) technologies based on chromosome copy numbers. Results of intermediate copy numbers for whole chromosomes or part of them (segmental) are assumed as evidence of a mixture of diploid and aneuploid cells and so diagnosed as diploid-aneuploid mosaicism. While aneuploid embryos are routinely discarded because they are mostly not viable, fail to implant, or are lost later in pregnancy [1–3], the clinical significance of mosaicism remains unclear. Scarce data are available on the outcomes of putative mosaic embryos, even though they suggest that a substantial number of mosaic embryos yield development potential and can lead to healthy pregnancies, but with increased implantation failure and miscarriage rates [4–7]. Overall, low-range mosaic embryos provide healthy outcomes comparable to those seen in the euploid group, whereas high-range mosaic embryos are more likely to result in poorer transfer outcomes and miscarriage.
A self-correcting mechanism to eliminate aneuploid cells through apoptosis and severe proliferative reduction has been demonstrated in the animal model [8], while in humans, although signs of embryonic plasticity are evident and similar models have been suggested [9–11], convincing evidence is still lacking [12]. However, it alone could not justify the poor correlation with the inner cell mass (ICM) found in most cases of mosaicism diagnosed from a trophectoderm (TE) biopsy [6, 13–15]. In this regard, the inaccuracy of the NGS-based copy number strategy to predict mosaicism has been recently highlighted by a review that found 57% of embryos diagnosed as mosaic to be truly euploid or fully aneuploid after rebiopsy [16]. In the lower range mosaics (< 50%), the presence of aneuploid cells in the ICM is extremely rare and seldom affects other areas of the embryo of mosaics [6, 14]. Contrarily, even though the concordance TE-ICM is once again significantly low in the high-range mosaics (> 50% aneuploid cells), this is because these putative mosaic embryos are uniformly aneuploid in the ICM [14, 17]. Thus, the prevalence of true mosaicism would be significantly lower than that previously reported by laboratories, indicating a clear overestimation [13].
The suboptimal validity to predict true mosaicism in the embryo calls into question the clinical value of reporting it, especially those mosaics in the low-medium range, mostly uniformly euploid but wrongly deselected for transfer, representing an evident negative impact on the clinical outcomes [13, 16]. It is worth noting that, the arbitrary severity of the criteria used to define the ranges for mosaicism (i.e., thresholds of 20–80% or 30–70%) can have a significant impact on the prevalence of mosaicism diagnosed; only by adjusting these cut-off values can result in different diagnoses using the same sample.
Assuming one embryo is mosaic based simply on an intermediate copy number result is clearly inaccurate and is leading to deselect a significant number of healthy embryos. Indeed, mosaicism constitutes merely one of the possible explanations, but these intermediate NGS profiles may also result from a variety of other technical factors, such as amplification bias, contamination, biopsy technique, or the analysis algorithms used to normalize data across the genome [13, 17, 18].
In this paper, we explore several parameters with potential implications for the accuracy of the current NGS approaches for mosaicism detection. Some outstanding issues must be unraveled before the validation of new approaches to interpret the outburst of complex data and, so, the clinical significance of mosaicism. Therefore, the sensitivity and specificity of different diagnosis strategies are put in the spotlight to determine the appropriateness of reporting mosaicism, which ultimately defines the conditions that lead to the highest clinical outcomes, optimizing the implantation rate and reducing miscarriages and abnormal pregnancies. Undoubtably, strong implications for genetic counseling and the clinical management of PGT-A depend on the resolutions adopted.
Correlation TE-ICM
The value of a TE diagnosis depends on its ability to be representative of the rest of the embryo. Unfortunately, considering the mitotic origin of embryonic mosaicism during early development, an evenly distribution of abnormal cells through the embryo is unlikely [5]. Indeed, a TE biopsy remains an indirect estimation of the entire embryo’s status; it remains unknown to what extent a mosaic profile in the TE reflects the karyotype of the remaining blastocyst [19]. When mosaicism is present, mathematical modeling supports the limited clinical value of a single TE biopsy in diagnosing the genetic status of the remaining embryo [20]. Given their nature, euploidy and aneuploidy are prone to be uniformly distributed within the embryo, and the data from analyzing the distinct parts of the blastocyst confirm a high correlation between TE-ICM when a non-mosaic karyotype is found in the biopsied TE cells [9, 14, 16, 21, 22]. On the contrary, in cases of mosaicism, and even though data is still scarce, they mostly show a low correlation of TE with the remaining embryo, poorly reflecting the status of the ICM, which undoubtedly challenges the reliability of TE mosaic diagnosis to predict the embryo karyotype. A systematic review of studies including rebiopsy on embryos targeted after TE biopsy in the mosaic range revealed that most of them (57.4%) were unlikely to present mosaicism, displaying instead euploidy (29%) and 28.4% of fully aneuploidy [16]. Additional results came from new approaches using single nucleotide polymorphism (SNP) genotyping and karyomaping technologies to distinguish the cause of aneuploidies [17]; testing embryos previously identified by NGS-based PGT-A as mosaic, a significant overestimation and inaccuracy in the diagnosis of mosaicism was reported: Only 1 of 21 (4.8%) cases diagnosed as putative mosaic was confirmed; 42.8% were euploid, and 47.6% were displaying several types of aneuploidies, like meiotic trisomies, monosomies, and digynic triploidies [17].
Data from blastocyst disaggregation studies, where different samples from each embryo were analyzed, strongly supported this poor correlation in cases of TE mosaicism. The concordance TE-ICM was found to be likely dependent on the grade of mosaicism, so that it would be related to the moment when the segregation error had occurred [6]. Thus, when high-rate mosaicism (50–70%) was diagnosed in the diagnostic TE sample, 65% of blastocysts were uniformly aneuploid in the ICM and the rest of the analyzed pieces. On the contrary, in cases of low-medium mosaic (< 50% aneuploid cells in a single TE sample), the aneuploid cells were reported to rarely impact the ICM (limited to only 1% of cases), suggesting the presence of a single group of aneuploid cells highly confined to a restricted portion of the embryo [6]. These results were ratified by Wu et al. [14], who, after rebiopsying several regions of 101 putative mosaic blastocysts, found a concordance with the ICM of only 27.5%, displaying euploidy (63.7%) and aneuploidy (8.8%) instead. The higher rates of aneuploidy (37.5%) were reported when high-rate mosaicism was involved, corroborating the potential incidence of the grade of mosaicism on the concordance TE-ICM for mosaicism. Additionally, they identified different distributions related to the type of mosaicism, displaying a lower ICM concordance for segmental-chromosome mosaicism than when whole chromosomes are imbalanced [14]. More recently, results confirmed the low ability to predict the chromosomal status of the remaining embryo in cases of reported mosaicism in the original TE sample when very few cases showed the same karyotype in all 4 pieces analyzed, both for whole chromosome (2.29%) and segmental (2.15%), while the partial embryo concordance (diagnoses confirmed in at least 1 sample) were 39.08% and 41.94%, respectively [23].
NGS based on chromosome copy number
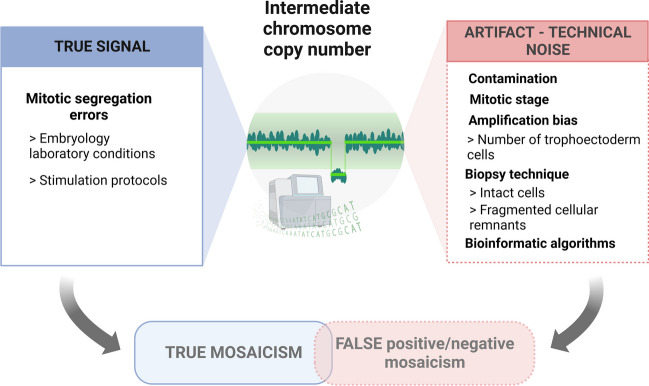
Current NGS technologies used in PGT-A to select euploid embryos from a simple trophectoderm sample are based on genome-wide chromosome copy number analysis [17]. NGS represents a leading technology with higher resolution and efficiency than comparative genome hybridization arrays (aCGH). Following the whole genome amplification (WGA), successive library construction, sequencing, and alignment of readings with the human reference genome are required to get the analysis result. Precisely, this necessary DNA extraction and amplification, known to be susceptible to errors, together with other methodological issues, might make it difficult to differentiate the true signal from the technical noise. The implementation of cut-off values for euploid assignment intends to discount this analytical disturbance, so deviations lower than the established threshold for 2 chromosome copy numbers are reported as euploidy. However, when a chromosome copy number deviation is higher and falls outside both euploidy and aneuploidy thresholds, it is considered to belong to the mosaic range. Thus , intermediate copy numbers outside these ranges are assumed to be consistent with the presence of both euploid and aneuploid cells among the biopsied trophectoderm and are, therefore, profiled as mosaics [17]. A mixture of euploid and aneuploid cells represents the most relevant type of mosaicism (diploid-aneuploid mosaic), and its frequency has markedly increased with the implementation of NGS as the prevalent technology for aneuploidy screening. Globally speaking, the incidence of mosaicism mostly ranges between 5 and 15% [6, 24, 25], even though high variability has been reported between clinics [16]. As inadequate stimulation protocols and suboptimal embryo culture methodologies may entail an increase in oxidative stress and promote segregation errors [26, 27], this could be consistent with the higher mosaic incidence reported in certain IVF units. Notwithstanding, other factors related to the genetic testing laboratory practices may entail different accuracy (sensitivity and specificity) and therefore have significant implications for the mosaicism rate reported (Fig. 1).
Fig. 1.
True chromosomal and technical artifacts leading to NGS intermediate copy number profiles
NGS technical limitations in estimating the karyotype
While NGS enabled higher resolution and dynamic range for PGT-A [4], attempts to design thresholds for detecting mosaicism are unable to reflect the full extent of variability in a current TE biopsy specimen. Most validation studies for mosaicism are based on models employing mixtures of normal and abnormal cell lines at different ratios, which intend to mirror the variation found in in-vivo samples. These cell mixtures have been mostly developed from Genomic DNA (gDNA) of cell lines with distinct known karyotypes [28] or alternatively by merging well-defined proportions of euploid and aneuploid cells [29]. However, a challenging limitation arises from any attempt to mimic true mosaicism samples since all models represent a highly stable scenario with no variation in the quantity or quality of cells analyzed. This condition certainly differs from an embryonic biopsy specimen, characterized by an uncertain number of intact cells together with fragmented cellular remnants derived from technical procedures. Thus, the extrapolation of mosaicism ranges from an unrealistic stable model clearly constrained the diagnosis efficacy in the event of biopsied highly variable cellular contents [15].
Interestingly, due to the high variability in TE samples, a suboptimal number of trophectoderm cells may lead to diagnosis errors by reporting ambiguous intermediate copy numbers. With the exception of polymerase chain reaction (PCR)-free methodologies, for most platforms, DNA amplification is required to generate sequencing-ready NGS libraries. At the time of quantification, the state of the PCR amplification plot may be critically affected by an insufficient or excessive number of cells, leading to an underestimation of the relative quantity of DNA [13]. Thus, the PCR amplification curve, consisting of well-described successive steps such as baseline, exponential, and plateau (saturation) phases, may be scrolled forward, accelerated, in cases with too many cells, reaching the saturation state of the reaction before quantification. Reaching the plateau stage, relative differences in copy number between the screened aneuploid cells and euploid controls would be wrongly diminished, displaying false intermediate copy numbers inferred as mosaicism. Similarly, the availability of too few cells would also impact the PCR amplification plot, but in this case, delaying the linear phase of amplification would lead to an underestimated DNA relative quantification and therefore, again, to an artifactually reported intermediate copy number result. Therefore, the number of cells available may potentially lead to inaccurate chromosome copy number profiles.
Thresholds of mosaicism range
Cut-off values employed in NGS profiling are basically assumptions of technical noise that determine ranges for normal disomic chromosome values and for aneuploidy results [4, 30]. Intermediate copy number values falling outside these ranges of euploidy and aneuploidy are identified as putative mosaicism by testing laboratories in current practice. Clearly, the stringency of these thresholds has major implications for the incidence of mosaicism since, with different mosaic classification criteria, a single case may fall in or out of the mosaic range and therefore be differently diagnosed (Fig. 2). Estimating the presence of mosaicism from a single TE is dramatically affected by the criteria used [15]. While less stringent thresholds (20–80%) are implemented in most testing laboratories [31], others opt for more strict mosaicism cut-off values (30–70%), which imply a lower mosaicism incidence and the acceptance of higher analytical noise levels [28]. When a loose range (20–80%) for mosaicism is used, a significant prevalence of euploid and aneuploid embryos are incorrectly labeled as mosaic [6, 13–16]. Thus, any attempt at standardizing the prediction of mosaicism certainly requires a rigorous approach to increase the overall accuracy, improve the detection of true chromosomal imbalance, and reduce the technical noise.
Fig. 2.
Impact of thresholds on diagnosing mosaicism from NGS intermediate copy number profiles
In an effort to assess the accuracy of aneuploidy screening, the sensitivity and specificity of different mosaicism thresholds were assessed based on the same NGS copy number profiles in the largest blastocyst disaggregation study to date: 585 samples were analyzed (5 informative fractions per embryo) from 113 embryos [15]. Their goal was to define the most competent strategies, comparing not only 20–80% and 30–70% thresholds but also analyzing the accuracy of a binary classification approach (single cut-off at 50%) in which mosaics are not even considered, limiting results exclusively to euploidy or aneuploidy. As expected, the overall mosaicism rate reported in the TE biopsy using less stringent cut-offs for the mosaic range (20–80%) was higher (40.2%) than when 30–70% thresholds were performed (20.2%). However, the analysis of the subsequent biopsies of the remaining embryo revealed no net benefit; the increase in mosaics detected did not entail a higher specificity. Thus, the use of wider thresholds (20–80%) did not imply the detection of more true mosaic configurations but led to an increase in false-positive mosaicism (57.8 to 79.5%; P < 0.00001). By reducing the cut-off stringency from 30 to 20%, not even a single one of the additional mosaic diagnoses inferred could be truly confirmed in the rest of the embryo, which means that 100% of the additional mosaicism results were false positives. On the other hand, when the upper threshold (aneuploidy range) was raised from 70 to 80%, the concordance between aneuploid TE calls and the rest of the embryo segments (true constitutive aneuploidy) decreased from 95.7 to 87.2% (P = 0.00318), increasing again the misclassification of true aneuploid embryos in the high-range mosaicism group. Consequently, increasing the threshold stringency led to a lower error rate and a reduced inaccurate classification of embryos (either euploid or aneuploid) in the mosaic. Not only increasing the stringency from 20 to 30% entailed an overall higher accuracy, but tightening the thresholds up to a single cut-off at 50% (binary classification) represented the most accurate diagnostic approach, with a better specificity-sensitivity balance than those including mosaicism ranges. The accuracy, in terms of specificity, was reported to diminish from the higher results in the binary approach (98.2%) to the 30–70% range (81.9%) and the 20–80% range (54.5%), while the sensitivity just slightly increased (94%, 97.7%, 98.2%, respectively). According to their results, a dichotomous approach has proven to be a superior strategy in identifying and deselecting abnormal embryos (true aneuploid and true mosaic), which are likely to have adverse clinical outcomes [15].
Discussion
Even though embryo mosaicism is certainly a real phenomenon, its prevalence in PGT-A diagnosis has increased with NGS implementation as the current leading technology. However, the real impact of mosaicism diagnoses inferred from intermediate chromosome copy numbers on NGS profiles and its clinical relevance remains unclear, leading to a most challenging data interpretation and clinical management.
Currently, NGS intermediate chromosome copy number profiles are assumed as evidence of chromosomal mosaicism within a TE biopsy, although this assumption is clearly inaccurate and misleading. This misconception has resulted in a significant increase in false-positive mosaics and, consequently, an unacceptable depletion of healthy embryos. Far from an ideal situation in which both euploid and aneuploid cells from a TE biopsy would be analyzed and karyotyped individually, NGS intermediate results do not represent strong evidence of mosaicism but an interpretation of collective data that is inconsistent with the arbitrary standards for aneuploidy or euploidy. There are numerous other technical explanations for an intermediate copy number result apart from mosaicism, such as amplification bias, contamination, biopsy technique, or the analysis algorithms used to normalize data across the genome. Undoubtably, the poor correlation of mosaicism with the chromosomal status of the ICM and the rest of the embryo reinforces the need for further research to evaluate the impact of these factors on the NGS intermediate results. This putative TE mosaicism, far from being representative and distributed evenly through the whole embryo, mostly correlates with aneuploidy in cases of high-range mosaicism (> 50%) and with euploidy when they are allocated at the low-mid level (< 50%) [6, 13–15]. Considering that high-range mosaicism would frequently represent a technical variation for the aneuploidy range, and low-mid level mosaics would account for an artifact of the euploidy spectrum, is there any value in still reporting mosaicism, especially those allocated in the low-range, if doing so implies deselecting a significant number of viable embryos?
Indeed, getting a truly comprehensive analysis of different types of aneuploidies (meiotic or mitotic) and being able to identify true mosaicism requires the use of SNP genotyping/karyomapping in conjunction with the NGS copy number analysis. NGS-based copy number alone cannot distinguish the mitotic or meiotic origin of aneuploidies, so additional approaches are needed to increase the specificity of mosaicism predictions, including meiopapping of polar bodies (PB) and genome-wide single nucleotide polymorphism (SNP) genotyping/karyomapping of PB and trophectoderm samples. In combination with NGS-based copy number analysis of TE cells, this approach can target different haplotypes, providing a second analysis that can identify maternal or paternal backgrounds of trisomies and monosomies and even detect polyploidy [17]. The higher cost and the need for genotyping both parents are the main disadvantages of this technology. Nevertheless, it represents a strong strategy to increasing accuracy and minimizing false mosaic positives/negatives profiles, which, in our opinion, must be implemented in order to determine the real clinical outcomes of true mosaic embryos. It becomes evident that we cannot unravel the clinical significance of mosaic embryos if mosaicism cannot be properly identified.
It is worth highlighting that the primary goal of PGT-A is to improve IVF success by optimizing the implantation rate and reducing pregnancy loss and abnormal pregnancies derived from aneuploidy. Recent data discourages the identification, reporting, and deselection of mosaic embryos based exclusively on NGS copy number profiles [15]; the use of mosaic ranges does not increase sensitivity, especially with 20–80% thresholds, but rather reduces diagnosis specificity, resulting in more false-positive mosaics. More importantly, their results suggest that avoiding the mosaicism classification represents the most accurate diagnostic approach for PGT-A, identifying abnormal embryos more efficiently and discarding them for transfer. That would be consistent with the good success rates globally reported for the transfer of lower mosaic embryos and the poor outcomes reported with mosaics in the high range and therefore a reflection of the outcomes of transferring euploid and aneuploid ones [13, 16]. Indeed, excluding putative mosaic embryos from transfer was previously suggested to be beneficial to achieving a live delivery and that, in order to preserve a net benefit with negligible harm, only aneuploid embryos should be eliminated from transfer [32].
To date, any attempt at classifying types of mosaicism and assigning them a clinical outcome has been based on inaccurate and misleading assumptions that classify embryos as low or high mosaic when, in fact, they are mostly euploid or aneuploid. Mosaicism is known to be a real phenomenon, but current NGS approaches are clearly overestimating it, and consequently any attempt to link the distinct types of mosaics to implantation rates is, at the very least, questionable because is mainly based on incorrect diagnoses. Logically, this does not exclude the possibility that a few embryos diagnosed as mosaic could be truly mosaics, but they are certainly much less prevalent than real euploid or aneuploid embryos misclassified as putative mosaics. Therefore, even excluding mosaicism represents a more accurate strategy to reach the PGT-A goal of selecting true euploid embryos with high implantation potential and discarding aneuploid embryos that will fail to implant, this approach logically assumes the chance of transferring a small percentage of real mosaic embryos. Due to their unclear reproductive potential, putative mosaic embryos are frequently chosen as a last resort and are given a lower transfer priority; however, when they are considered for transfer, their implications should be thoroughly discussed with the patient while taking into account the most recent professional practice recommendations.
The main concern with mosaicism is elucidating whether mosaic embryo transfer would lead to a higher frequency of congenital abnormalities in newborns. Except for two cases where mosaicism was confirmed in advanced prenatal stages, the rate of mosaicism verified in pregnancies is exceedingly low [33, 34], and the results to date appear encouraging until further investigations are released. Indeed, there is no evidence that mosaicism is more prevalent in pregnancies resulting from IVF than in natural conceptions, even when IVF embryos from no PGT-A cycles, which include euploids, aneuploids, and mosaics, are “blindly” transferred [35]. Therefore, concerns about the health of pregnancies appear to be overstated, and the potential transfer of a few real mosaic embryos would represent a minimal risk [36]. The low correlation between embryonic and extra-embryonic cells is in line with the prenatal diagnosis results of chorionic villus sampling (CVS) at 10–11 weeks gestation, when a small fraction (13.1%) of CVS mosaics were confirmed after amniotic fluid analysis, dramatically reducing the possibility of abnormalities appearing in fetal tissues [37]. However, we should consider that, even though congenital abnormalities seem not to be increasing, more studies certainly must be conducted to examine the longer-term outcomes (i.e., cognitive development or behavior) before the safety of putative mosaic embryo transfers can be determined.
Medicine has the moral obligation and the Hippocratic imperative to provide net medical benefit to patients with minimal harm [38]. In theory, mosaicism reporting could be considered a safer and more beneficial strategy in terms of reducing the risk of transferring embryos with a real gain or loss of chromosomal material, pregnancy loss, and congenital abnormalities. However, the more strict approach (20%–80%) results in a slight increase in terms of sensitivity (4.2%), but on the other hand, it comes with a high reduction of specificity (43.7%) which implies a significant boost of false-positive results and deselection of healthy embryos, in comparison with a strategy based on a single cut-off at 50% (binary classification). Thus, far from being considered beneficial for the patient, considering the better specificity-sensitivity balance showed and the extremely rare cases of congenital abnormalities, mosaicism diagnosis clearly represents a suboptimal embryo selection criterion, a significant embryo wastage and reduction of clinical outcomes. Reductio ad absurdum: in a test where hypothetically every embryo is removed, the risk of miscarriage will be eliminated, but since there will be no live births, the procedure cannot be logically viewed as beneficial. Therefore, decision-making should be based on evidence and clinical utility, and it has become evident that copy number criteria cannot accurately predict mosaicism in preimplantation embryos or their clinical outcomes. Overreporting mosaicism is leading to an unnecessary burden on the clinical management of these embryos. It seems logical that professionals should refrain from counseling patients based on misinterpreted simple copy number profiles that do not accurately reflect their true nature and jeopardize the chance of pregnancy.
Predictions of mosaicism by intermediate copy number thresholds should not be reported only because they are observed but because of their evident clinical significance. It could be argued that patients are entitled to be informed; however, sharing data that has little clinical value simply because it is available is creating confusion among professionals, struggling with their management, and leading to the discarding of healthy embryos as well as other unfavorable effects on patients, such as anxiety from making a decision after additional genetic counseling or the risk of an unnecessary prenatal test.
Certainly, NGS has demonstrated its clinical utility in diagnosing euploidy and aneuploidy, but when it comes to mosaicism and predicting its outcomes, copy number thresholds have proven to be inaccurate and unreliable. New validated strategies are needed to increase the accuracy of diagnosis and reduce false positives, better strategies like the combination of NGS with other diagnostic techniques like SNP genotyping and karyomapping of PB/trophectoderm samples. Properly designed, large-scale, randomized clinical trials should be conducted to provide sufficient high-level evidence about the validity of PGT-A for identifying mosaicism and unraveling its reproductive potential. Until then, the current practice of diagnosing mosaicism by intermediate copy number thresholds should be restricted to research settings until IVF treatment’s benefits are unequivocally demonstrated. At present, laboratories should be cautious and consider limiting predictions to aneuploid and euploid, which will avoid deselecting healthy embryos and ultimately lead to the highest clinical outcomes.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy B, Sigurjonsson S, Pettersen B, et al. Genomic imbalance in products of conception: single-nucleotide polymorphism chromosomal microarray analysis. Obstet Gynecol. 2014;124:202–209. doi: 10.1097/AOG.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 3.Tiegs AW, Tao X, Zhan Y, Whitehead C, Kim J, Hanson B, et al. A multicenter, prospective, blinded, nonselection study evaluating the predictive value of an aneuploid diagnosis using a targeted next-generation sequencing-based preimplantation genetic testing for aneuploidy assay and impact of biopsy. Fertil Steril. 2021;115:627–637. doi: 10.1016/j.fertnstert.2020.07.052. [DOI] [PubMed] [Google Scholar]
- 4.Munné S, Blazek J, Large M, Martinez-Ortiz PA, Nisson H, Liu E, Tarozzi N, Borini A, Becker A, Zhang J, et al. Detailed investigation into the cytogenetic constitution and pregnancy outcome of replacing mosaic blastocysts detected with the use of high-resolution next-generation sequencing. Fertil Steril. 2017;108:62–71. doi: 10.1016/j.fertnstert.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Lin PY, Lee CI, Cheng EH, Huang CC, Lee TH, Shih HH, Pai YP, Chen YC, Lee MS. Clinical outcomes of single mosaic embryo transfer: high-level or low-level mosaic embryo, does it matter? J Clin Med. 2020;9:1695. doi: 10.3390/jcm9061695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capalbo A, et al. Mosaic human preimplantation embryos and their developmental potential in a prospective, non-selection clinical trial. Am J Hum Genet. 2021;108:2238–2247. doi: 10.1016/j.ajhg.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viotti M, Victor AR, Barnes FL, Zouves CG, Besser AG, et al. Using outcome data from one thousand mosaic embryo transfers to formulate an embryo ranking system for clinical use. Fertil Steril. 2021;115:1212–1224. doi: 10.1016/j.fertnstert.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 8.Bolton H, Graham SJL, Van der Aa N, Kumar P, Theunis K, Gallardo EF, Voet T, Zernicka-Goetz M. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploidy cells and normal developmental potential. Nat Commun. 2016;7:665–666. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victor AR, Griffin DK, Brake AJ, Tyndall JC, Murphy AE, LepkowskyLal LTA, Zouves CG, Barnes FL, McCoy RC, et al. Assessment of aneuploidy concordance between clinical trophectoderm biopsy and blastocyst. Hum Reprod. 2019;34:181–192. doi: 10.1093/humrep/dey327. [DOI] [PubMed] [Google Scholar]
- 10.Starostik MR, Sosina OA, McCoy RC. Single-cell analysis of human embryos reveals diverse patterns of aneuploidy and mosaicism. Genome Res. 2020;30:814–825. doi: 10.1101/gr.262774.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang M, Rito T, Metzger J, Naftaly J, Soman R, Hu J, et al. Depletion of aneuploid cells in human embryos and gastruloids. Nat Cell Biol. 2021;23:314–321. doi: 10.1038/s41556-021-00660-7. [DOI] [PubMed] [Google Scholar]
- 12.Coticchio G, Barrie A, Lagalla C, Borini A, Fishel S, Griffin D, Campbell A. Plasticity of the human preimplantation embryo: developmental dogmas, variations on themes and self-correction. Hum Reprod Update. 2021;27(5):848–865. doi: 10.1093/humupd/dmab016. [DOI] [PubMed] [Google Scholar]
- 13.Treff NR, Marin D. The, “mosaic” embryo: misconceptions and misinterpretations in preimplantation genetic testing for aneuploidy. Fertil Steril. 2021;116:1205–1211. doi: 10.1016/j.fertnstert.2021.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Wu L, Jin L, Chen W, Liu JM, Hu J, Yu Q, Ren XL, Huang B, He H. The true incidence of chromosomal mosaicism after preimplantation genetic testing is much lower than that indicated by trophectoderm biopsy. Hum Reprod. 2021;36:1691–1701. doi: 10.1093/humrep/deab064. [DOI] [PubMed] [Google Scholar]
- 15.Girardi L, Figliuzzi M, Poli M, Serdarogullari M, Patassini C, Caroselli S, Pergher I, Cogo F, Coban O, Boynukalin FK, Bahceci M, Navarro R, Rubio R, Findikli N, Simón C, Capalbo A. The use of copy number loads to designate mosaicism in blastocyst stage PGT-A cycles: fewer is better. Hum Reprod. 2023;38:982–991. doi: 10.1093/humrep/dead049. [DOI] [PubMed] [Google Scholar]
- 16.Marin D, Xu J, Treff NR. Preimplantation genetic testing for aneuploidy: a review of published blastocyst reanalysis concordance data. Prenat Diagn. 2021;41:545–553. doi: 10.1002/pd.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handyside AH, McCollin A, Summers MC, Ottolini CS. Copy number analysis of meiotic and postzygotic mitotic aneuploidies in trophectoderm cells biopsied at the blastocyst stage and arrested embryos. Prenat Diagn. 2021;41:525–535. doi: 10.1002/pd.5816. [DOI] [PubMed] [Google Scholar]
- 18.Xiong S, Liu W, Wang J, Liu J, Gao Y, Wu L, Zhu J, Hao X, Li J, Liu D, Han W, Huang G. Trophectodrm biopsy protocols may impact the rate of mosaic blastocysts in cycles with preimplantation genetic testing for aneuploidy. J Assist Reprod Genet. 2021;38:1153–1162. doi: 10.1007/s10815-021-02137-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulson RJ, Treff NR. Isn’t it time to stop calling preimplantation embryos mosaic? F S Rep. 2020;1:164–165. doi: 10.1016/j.xfre.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gleicher N, Orvieto R. Is the hypothesis of preimplantation genetic screening (PGS) still supportable? A review J Ovarian Res. 2017;10:21. doi: 10.1186/s13048-017-0318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachdev NM, McCulloh DH, Kramer Y, Keefe D, Grifo JA. The reproducibility of trophectoderm biopsies in euploid, aneuploid, and mosaic embryos using independently verified next-generation sequencing (NGS): a pilot study. J Assist Reprod Genet. 2020;37:559–571. doi: 10.1007/s10815-020-01720-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capalbo A, Poli M, Jalas C, Forman EJ, Treff NR. On the reproductive capabilities of aneuploid human preimplantation embryos. Am J Hum Genet. 2022;109:1572–1581. doi: 10.1016/j.ajhg.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Tao X, Cheng M, Steward A, Guo V, Zhan Y, Scott RT, Jr, Jalas C. The concordance rates of an initial trophectoderm biopsy with the rest of the embryo using PGTseq, a targeted next-generation sequencing platform for preimplantation genetic testing-aneuploidy. Fertil Steril. 2022;117:315–323. doi: 10.1016/j.fertnstert.2021.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Munne S, Grifo J, Wells D. Mosaicism: “survival of the fittest” versus “no embryo left behind”. Fertil Steril. 2016;105:1146–1149. doi: 10.1016/j.fertnstert.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigo L, Clemente-Ciscar M, CamposGalindo I, Peinado V, Simon C, Rubio C. Characteristics of the IVF cycle that contribute to the incidence of mosaicism. Genes (Basel) 2020;11:1151. doi: 10.3390/genes11101151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Ha S, Li Z, Li J, Xiao W. CHK1-CENP B/MAD2 is associated with mild oxidative damage-induced sex chromosome aneuploidy of male mouse embryos during in vitro fertilization. Free Radic Biol Med. 2019;137:181–193. doi: 10.1016/j.freeradbiomed.2019.04.037. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Ha S, Li Z, Huang Y, Lin E, Xiao W. Aurora B prevents aneuploidy via MAD2 during the first mitotic cleavage in oxidatively damaged embryos. Cell Prolif. 2019;52:e12657. doi: 10.1111/cpr.12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcıa-Pascual CM, Navarro-Sanchez L, Navarro R, Martınez L, Jimenez J, Rodrigo L, Simon C, Rubio C. Optimized NGS approach for detection of aneuploidies and mosaicism in PGT-A and imbalances in PGT-SR. Genes (Basel) 2020;11:724. doi: 10.3390/genes11070724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodrich D, Xing T, Tao X, Lonczak A, Zhan Y, Landis J, Zimmerman R, Scott RT, Jr, Treff NR. Evaluation of comprehensive chromosome screening platforms for the detection of mosaic segmental aneuploidy. J Assist Reprod Genet. 2017;34:975–981. doi: 10.1007/s10815-017-0924-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinella F, Fiorentino F, Biricik A, Bono S, Ruberti A, Cotroneo E, Baldi M, Cursio E, Minasi MG, Greco E. Extent of chromosomal mosaicism influences the clinical outcome of in vitro fertilization treatments. Fertil Steril. 2018;109:77–83. doi: 10.1016/j.fertnstert.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 31.Leigh D, Cram DS, Rechitsky S, Handyside A, Wells D, Munne S, Kahraman S, Grifo J, Katz-Jaffe M, Rubio C, et al. PGDIS position statement on the transfer of mosaic embryos 2021. Reprod Biomed Online. 2022;45:19–25. doi: 10.1016/j.rbmo.2022.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Scriven PN. Elucidating the PGT-A paradox: marginalising the detriment relegates the benefit. J Assist Reprod Genet. 2022;39:2475–2481. doi: 10.1007/s10815-022-02640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahraman S, Cetinkaya M, Yuksel B, Yesil M, Pirkevi CC. The birth of a baby with mosaicism resulting from a known mosaic embryo transfer: a case report. Hum Reprod. 2020;35:727–733. doi: 10.1093/humrep/dez309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlade-Bartusia K, Strong E, Zhu O, Mackie J, Salema D, Volodarsky M, et al. Mosaic embryo transfer—first report of a live born with non-mosaic partial aneuploidy and uniparental disomy 15. FS Rep. 2022;3:192–198. doi: 10.1016/j.xfre.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang A, Adusumalli J, Patel S, Liem J, Williams J, 3rd, Pisarska MD. Prevalence of chromosomal mosaicism in pregnancies from couples with infertility. Fertil Steril. 2009;91:2355–2360. doi: 10.1016/j.fertnstert.2008.03.044. [DOI] [PubMed] [Google Scholar]
- 36.Gleicher N, Patrizio P, Mochizuki L, Barad DH. Previously reported and here added cases demonstrate euploid pregnancies followed by PGT-A as “mosaic” as well as “aneuploid” designated embryos. Reprod Biol Endocrinol. 2023;21:25. doi: 10.1186/s12958-023-01077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stetten G, Escallon CS, South ST, McMichael JL, Saul DO, Blakemore KJ. Reevaluating confned placental mosaicism. Am J Med Genet A. 2004;131:232–239. doi: 10.1002/ajmg.a.30363. [DOI] [PubMed] [Google Scholar]
- 38.Gillon R. Medical ethics: four principles plus attention to scope. BMJ. 1994;309(6948):184–188. doi: 10.1136/bmj.309.6948.184. [DOI] [PMC free article] [PubMed] [Google Scholar]