Key Points
Complement activation is shaped by Ag density and Ab hinge length.
Unique IgG2 polymorphism shows less efficient complement activation.
Cellular context affects CDC independently of known complement regulators.
Abstract
Complement activation via the classical pathway is initiated when oligomeric Igs on target surfaces are recognized by C1 of the complement cascade. The strength of this interaction and activation of the complement system are influenced by structural variation of the Ab, including Ab isotype, subclass, and glycosylation profile. Polymorphic variants of IgG have also been described to influence Fc-dependent effector functions. Therefore, we assessed complement binding, deposition, and complement-dependent cytotoxicity (CDC) of 27 known IgG allotypes with anti-trinitrophenyl specificity. Differences between allotypes within subclasses were minor for IgG1, IgG3, and IgG4 allotypes, and more substantial for IgG2. Allelic variant IGHG2*06, containing a unique serine at position 378 in the CH3 domain, showed less efficient complement activation and CDC compared with other IgG2 polymorphisms. We also observed variable cell lysis between IgG1 and IgG3, with IgG3 being superior in lysis of human RBCs and Ramos cells, and IgG1 being superior in lysis of Raji and Wien133 cells, demonstrating that a long-standing conundrum in the literature depends on cellular context. Furthermore, we compared IgG1 and IgG3 under different circumstances, showing that Ag density and Ab hinge length, but not complement regulators, define the context dependency of Ab-mediated CDC activity. Our results point toward a variation in the capacity of IgG subclasses to activate complement due to single amino acid changes and hinge length differences of allotypes to activate complement, which might give new insights on susceptibility to infectious, alloimmune, or autoimmune diseases and aid the design of Ab-based therapeutics.
Introduction
The complement system is a fast and efficient first-line defense mechanism to fight invading pathogens. It consists of a series of soluble inactive precursor proteins in the blood that are sequentially activated on cell surfaces (1). This cascade of events is triggered by tick-over of the C3b component on foreign surfaces (alternative pathway), by lectin recognition (lectin pathway), or by Ab opsonization of cell-based Ags on pathogens or in autoimmune diseases (classical pathway) (1, 2). All pathways converge on the level of C3 cleavage into activated C3b, which can covalently bind the target surface via its reactive thioester. C3b-opsonized pathogens are either recognized and engulfed by phagocytes expressing the CR3 receptor or they are directly killed via the membrane attack complex, which is formed via the terminal pathway (1, 2). The expression of membrane complement regulators and recruitment of soluble complement regulators tightly regulate this process and prevent complement activation on host cells (3).
The classical pathway of the complement system is activated after Igs bind to cell-surface Ags and trigger the first component of the complement pathway (C1) (4) that is composed of three units, C1q, C1r, and C1s. The Fc domains of surface-bound Ig Abs are recognized by six globular head domains of C1q (5), which leads to activation of the classical complement pathway (6). The structural variation between Ig isotypes and IgG subclasses influences the interaction with C1q and therefore the capacity to activate the complement system (7–9). The pentameric or hexameric nature of IgM Abs is unique and facilitates a very efficient recognition by C1q (10–12). Other Ab isotypes and subclasses show great variation in the capacity to activate complement. IgG1 and IgG3 are the most potent activators of the classical complement pathway, with IgG2- and IgG4-induced complement activation being limited to high Ag and Ab concentrations (13–16). In contrast to activation through IgM, activation of the complement system requires interaction between multiple IgG molecules and C1q (5). The molecular assembly of an IgG hexamer on the pathogen surface through Fc–Fc interactions facilitates C1q binding and subsequent complement activation (4, 5, 17, 18). Numerous studies have compared complement activation by all IgG subclasses in various model systems in vitro with diverse outcomes depending on the target Ag, Ag density, and expression of complement regulators (13, 19–24). This context dependency of complement activation indicates that the requirements for efficient hexamer formation and complement activation are different for each subclass. Compared to IgG1 and IgG3, complement activation by IgG2 is less efficient, which is probably the result of the amino acid deletion at position 236 in the lower hinge (25, 26). Nevertheless, IgG2 Abs are relatively efficient in complement activation at high Ag density and are frequently raised to highly repetitive polysaccharide Ags on bacteria (27–31). In line with this, IgG2 Ab levels correlate with protection from several pathogens expressing polysaccharide Ags. Furthermore, IgG2 subclass deficiency leads to a higher susceptibility to bacterial infections, illustrating the relevance of IgG2-mediated clearance of bacterial pathogens (32).
Conflicting data have been reported on whether IgG1 or IgG3 is superior in activating the complement system. In general, IgG3 has been found to be most effective in binding complement components and triggering complement activation at low Ag densities, where a long and flexible hinge is thought to improve the formation of a hexameric complex (24, 28, 29, 33). At high Ag density, however, IgG1 is superior compared with IgG3 in activating the complement system (13, 20, 29, 34). The density dependency of IgG1 and IgG3 is not only explained by the difference in hinge length, as the insertion of an IgG1 hinge in an IgG3 Ab increased the capacity to activate the complement system even at low Ag density (22, 33, 34). It seems rather to be explained by the amino acid variation in the Fc domain between IgG1 and IgG3 (19). However, most of these studies show Ab concentration-dependent variation and do not demonstrate a density-dependent reversal in potency of IgG1 and IgG3 with different Ag levels on the same target cells. Different Ags and expression levels were only considered across different cell lines, which might impact subclass-specific superiority. This indicates that the requirements for efficient complement activation are extremely context-dependent and warrant further investigation.
Besides the functional differences between IgG subclasses, polymorphic variants of IgG are also likely to influence the Fc-dependent effector function of Abs (8, 9, 35). This interindividual diversity in IgG has been linked to predisposition to various diseases (36–40). For example, anti-malaria IgG3 Abs from mothers expressing IgG3-H435 are more efficiently transported across the placenta and protect the newborn child from malaria infection (36). It has been previously reported that determinants in the CH2 and hinge domain influenced FcγR binding (35) and Ab-dependent cellular cytotoxicity activity of IgG3 allotypes (41). This encouraged us to study the influence of IgG polymorphic variants on complement activation. In previous studies, the G3m(b*) IgG3 allotype was found to activate the complement system more efficiently than the G3m(g*) allotype (19, 20); no differences between the ability of allotypes G1m(za) and G1m(f) to cause cell lysis were reported (34), and G3m(s*) showed greater bactericidal activity than G3m(b*) and IgG1 (33). This indeed warrants a more extensive characterization of complement activation by all IgG allotypes. A recent study showed that the deposition of complement by six IgG3 allotypes showed no differences (35). However, to date, the full extent of the potential variability of the IgG3 allotypes, including their hinge length variation, remains unexplored regarding complement activity. Therefore, in this study, we characterized complement activation and complement-dependent cytotoxicity (CDC) by all known IgG allotypes (at the start of the study).
Materials and Methods
Human serum albumin–trinitrophenyl
IgG-depleted human serum albumin (HSA; 20%; Sanquin) at a concentration of 10 mg/ml was labeled with trinitrophenyl (TNP) in 0.15 M Na2HPO4 (Merck) with 0.33, 1, or 3 mM 2,4,6-trinitrobenzenesulfonic acid (TNBS; Sigma-Aldrich) at room temperature for 30 min. For further purification and to remove unbound TNBS, the solution was dialyzed in 2 l of PBS (Fresenius Kabi) using a dialysis cassette (Slide-A-Lyzer G2 cassette, 10-kDa molecular mass cutoff; Thermo Fisher Scientific) overnight at 4°C. After a second dialysis step in PBS, HSA-TNP aliquots were made and stored at −20°C. The concentration of HSA-TNP was determined based on the final volume compared with the starting volume.
Pooled normal human serum
Normal human serum (NHS) from at least five healthy volunteers was pooled and used across all experiments. All donors gave informed consent for this purpose.
RBCs
RBCs were isolated from whole human peripheral blood from healthy donors with blood type O rhesus D positive (RhD+), which were collected in sterile vacutainer tubes containing EDTA.
Cell lines
CD20+ Raji and Ramos (human Burkitt’s lymphoma) cell lines were cultured in B cell media, containing RPMI 1640 medium with l-glutamine, HEPES (Life Technologies, Thermo Fisher Scientific), 1% (v/v) penicillin (Sigma-Aldrich), 1% (v/v) streptomycin (Sigma-Aldrich), 0.1% (v/v) 2-ME (Sigma-Aldrich), 0.1% (v/v) IgG-depleted transferrin (Sigma-Aldrich), and 5% (v/v) FCS (Bodinco). CD20+ Wien133 (human Burkitt’s lymphoma) cells were cultured in IMDM with HEPES and l-glutamine (Lonza), supplemented with 10% (v/v) heat-inactivated donor bovine serum. Cell lines were maintained at 37°C in a 5% (v/v) CO2-humidified incubator.
Generation of Raji and Ramos knockout cell lines
Knockout (KO) Raji and Ramos cell lines lacking the complement regulatory proteins CD46, CD55, and CD59 were generated either as single KOs or in combination. Briefly, guide RNA primers (Gene Universal) designed for high frameshift mutation (42) of the three target genes were annealed and ligated into lentiCRISPR v2 vector (plasmid 52961; Addgene) containing puromycin resistance. HEK293T cells (3.5 × 105) were seeded in a 12-well culture plate with IMDM++ (Lonza) supplemented with 10% FCS (v/v; Sigma-Aldrich), 100 U/ml penicillin (Invitrogen), and 100 µg/ml streptavidin (Invitrogen) as culture medium. The next day, HEK293T cells were transfected using 3 µg of polyethylenimine (Polysciences) with 0.9 µg of lentiviral vector for the desired target, and the packaging vectors pVSVg (0.18 µg, plasmid 8545; Addgene), psPAX2 (0.27 µg, plasmid 12260; Addgene), and pAdv (0.1 µg, plasmid E171A; Promega). Three and 4 d after transfection, the medium containing the virus was collected and frozen at −80°C until transduction.
Raji and Ramos cells were washed with fresh B cell media and plated into 96-well flat-bottom plates at 2000 cells/well. Additional wells for an empty vector (lentivirus) and nontransduced cells were included as controls. B cell media were removed via centrifugation and replaced with 60 µl of protamine sulfate medium (1:600 in B cell media) and 40 µl of each virus. The plate was wrapped in plastic foil, centrifuged (1.5 h, 2400 rpm at 37°C), and incubated at 37°C for 3 d.
To determine a suitable concentration of antibiotics, the concentration of antibiotics to kill parental cells was examined. Cells were washed with PBS and fresh B cell media. Then, cells were split to ∼2000 cells/well, and for each KO cell line, puromycin in B cell medium at concentrations of 0, 0.25, 0.5, 1, and 2 µg/ml puromycin was added. Ten days later, wells with the highest concentration of puromycin with living cells were selected. Cells were grown out in 12-well plates. Expression of CD46, CD55, and CD59 on KO Raji and Ramos cells was determined via flow cytometry using anti–CD46-FITC (1:229; in-house), anti–CD55-allophycocyanin (1:250, clone IA10; BD Biosciences), and anti–CD59-FITC (1:100, clone MEM-443; Invitrogen) Abs. Viable, negative for near-infrared fluorescent-reactive dye (1:1000; Invitrogen), and negative populations for individual complement regulators were cell sorted using a FACSAria III (BD Biosciences) and cultured in B cell media. The same procedure was used to generate double and triple KO cell lines, but instead of wild-type cell lines, single KO cells were used.
Ab cloning and production
All Ab constructs to produce anti-TNP Abs were expressed transiently in HEK293F cells under serum-free conditions and purified by protein A or G affinity chromatography as previously described (43). We have previously published on this anti-TNP allotype set and fully characterized these Abs for glycosylation profile, FcγR binding, and oligomerization (41). The sequences for anti-TNP mutants were ordered as gene fragments in pUC vector (IDT) and swapped with the anti-TNP VH sequences in the pcDNA3.1 vectors expressing IGHG1*03, IGHG1*03-A378S, IGHG2*01, and IGHG2*06 constant domains (European Union numbering conventions are used throughout). Anti-CD20 Ab H chain (HC) expression vectors were constructed by inserting de novo synthesized (GeneArt) codon-optimized HC coding regions into expression vector pcDNA3.3 (Invitrogen). The HC coding regions consisted of the VH regions of human mAbs 7D8 (human CD20-specific) (44) or b12 (HIV-1 gp120-specific) (45) genetically fused to the CH regions of human IGHG1*03, selected IgG3 allotypes (9), or one of the mutant variants (rch3 [reduced core-hinge consisting of three exons], rch1A, rch1B, h1 [G1 hinge], C219S). Likewise, separate L chain (LC) expression vectors were constructed by inserting the appropriate VL coding regions in frame with the LC coding regions of the human (J00241) κ L chain into expression vector pcDNA3.3 (Invitrogen). The variable domain sequence of the anti–wall teichoic acid (WTA) Abs was used as previously described (46). Batches of purified Ab were tested by high-performance size-exclusion chromatography for aggregates or degradation products and shown to be at least 95% monomeric. Their N-linked glycosylation profiles have been previously been published (41).
Complement ELISA
To determine complement deposition via ELISA, MaxiSorp plates (Nunc) were coated with 10 µg/ml HSA-TNP in PBS at room temperature overnight. Three different TNPylation levels were used (3, 1, and 0.33 mM). The plates were washed three times with PBS + 0.1% Tween 20 (wash buffer) either manually or by using an ELISA washer (405 LSRS washer; BioTek). In the following step, plates were incubated for 90 min with anti-TNP Abs, serially diluted in 0.1% poloxamer (plx)-PBS (BASF ChemTrade), and washed with wash buffer. Next, plates were incubated with 100 µl of 1:35 pooled serum with 0.1% plx in veronal buffer (VB)++ (1.8 mM sodium barbital and 3.1 mM barbituric acid [pH 7.3–7.4] + 1 mM CaCl2 + 0.5 mM MgCl2), shaking at 37°C for 60 min, and washed subsequently. C1q, C3, and C4 deposition was detected by incubating plates for 1 h with 1 µg/ml HRP-labeled anti–C1q-2 (47), anti–C3-19 (48), or anti–C4-10 (49) in 0.1% plx-PBS, respectively. Colorimetric detection was performed using tetramethylbenzidine mix. After 5 min, color development was stopped using 0.2 M sulfuric acid and absorbance was measured with an ELISA plate reader (Synergy 2; BioTek) at 450 nm, corrected for 540 nm.
Hemolytic assay
RBCs were TNPylated by incubating 40 × 106 RBCs with various concentrations (0.25, 0.5, 1, or 2 mM) of TNBS solution in 0.15 M Na2HPO4 buffer (pH 10) at room temperature for 10 min and then washed three times with PBS to remove unbound TNP. RBCs were resuspended in 3 ml of VB containing 0.05% gelatin (VBG; Sigma-Aldrich). The following was combined in a total volume of 100 µl per well in a round-bottom plate: a glass bead to agitate the RBCs (2 mm; Merck), 3 × 106 RBCs and anti-TNP Abs in VB++ containing 0.05% gelatin, and 10 or 50% NHS. The plate was incubated, shaking at 37°C for 60 min. Plates were centrifuged for 2 min at 1800 rpm and supernatants were transferred to a MaxiSorp plate. The ODs of the supernatants were measured using an ELISA plate reader (Synergy 2; BioTek) at 412 nm, corrected for 690 nm. Maximum hemoglobin release was determined by incubating RBCs with distilled water. The percentage CDC was determined by applying the following formula: % CDC = (ODsample/ODmaximal) × 100. The area under the curve (AUC) was determined to perform unpaired t test statistics.
Complement deposition via flow cytometry
To determine complement deposition on RBCs, a similar protocol was used as described above (hemolytic assay) with some adaptations. To prevent lysis of the RBCs and allow quantification of complement deposition, eculizumab (Amgen) was added to all wells (20 µg/ml). Furthermore, the hemolytic assay was optimized for FACS purposes by lowering the serum concentration (5% NHS) and choosing a low TNPylation level (0.5 mM TNBS). After an incubation at 37°C shaking for 1 h, plates were centrifuged and cells were washed three times with PBS to remove all serum proteins. Then, cells were incubated with 1 µg/ml anti–C3-19-FITC (48), anti–C4-10-allophycocyanin (49), or anti–IgG-PE (clone M1310G05; BioLegend) in 0.5% BSA-PBS. To study the expression of different complement regulators, conjugated Abs were tested at the following dilutions: mouse anti-human CD46-3–FITC (1:229; in-house mAb), mouse anti-human CD55-allophycocyanin (1:250; clone IA10; BD Biosciences), mouse anti-human CD59-FITC (1:100; clone MEM-443; Thermo Fisher Scientific), rabbit anti-human complement receptor 1 (CR1; 1:500; in-house mAb) in combination with goat anti–rabbit IgG Alexa Fluor 488 (1:500; clone A11034; Invitrogen) and mouse anti–human CR2-BV605 (1:100; clone BB-ly4; BD Biosciences). Isotype controls IgG1-FITC (PeliCluster; Sanquin), IgG1-allophycocyanin (eBioscience), and rabbit anti-human IgG (polyclonal M1023; Sanquin) were included to determine the background. The plate was incubated on ice for 30 min. After the incubation step, the cells were washed three times with 0.1% BSA-PBS. Complement deposition was measured on a FACSCanto II (BD Biosciences) flow cytometer. Results were corrected for cell size after background subtraction. However, a direct comparison between RBCs and the other cell types was not possible, because the expression on RBCs was not corrected for cell size.
CDC viability assay
Raji and Ramos cells, including KO cell lines, were washed in RPMI 1640 without supplements and counted. Then, 3 × 106 cells were incubated at 37°C for 15 min with 0.5 mM TNBS in a 0.2 M Na2HPO4 buffer (pH 10). Afterward, cells were washed once with B cell media and three times with PBS. TNPylated cells were counted and resuspended in B cell media at a concentration of 1 × 106 cells/ml. Unlabeled (control) or TNPylated cells (1 × 105 cells/well) were incubated with serially diluted IgG anti-TNP in B cell media for 15 min at room temperature. After this opsonization step, pooled NHS was added to the wells (20 or 50%) and incubated at 37°C shaking for 45 min. Afterward, cells were washed twice with 0.1% FCS-PBS to remove all serum proteins and subsequently stained with live/dead fixable near-infrared cell stain (1:1000; Invitrogen, Thermo Fisher Scientific) on ice in the dark for 30 min. After three washing steps with 0.1% FCS-PBS, FACS was performed on an LSRFortessa flow cytometer (BD Biosciences). Cells were gated based on forward and side scatter characteristics, and a forward scatter width versus forward scatter height plot was used for doublet exclusion. A total of 1 × 105 cells per well were analyzed. The percentage allophycocyanin-Cy7–positive cells (compared with total cells) was used to evaluate the degree of CDC.
The capacity of anti-CD20 Abs to induce CDC was assessed by preincubating with unlabeled Raji cells, Ramos cells, and Wien133 target cells. The following steps were the same as described above except for the testing of Wien133 cell viability, which was determined by staining with propidium iodide and detected using an iQue screener (IntelliCyt).
Complement binding and deposition on Staphylococcus aureus
GFP-labeled S. aureus (Newman spa/sbi-KO) (50) was incubated in a round-bottom 96-well plate with a concentration range of IgG plus 1% IgG- and IgM-depleted serum (51) as complement source at 37°C on a shaker (750 rpm) for 30 min. Samples were split, washed, and incubated with 1 µg/ml mouse anti-C1q or anti-C3c (both from Quidel) at 4°C on a shaker for 30 min. Bound anti-C1q and anti-C3c Abs were detected after incubation of washed bacteria with Alexa Fluor 647–labeled F(ab′)2 goat anti-mouse IgG (Jackson ImmunoResearch) for an additional 30 min. Then, samples were fixed with 1% paraformaldehyde. Complement binding and deposition were measured by flow cytometry by gating on GFP-positive bacteria.
Phagocytosis assay
Phagocytosis was measured using GFP-labeled S. aureus (Newman spa/sbi-KO and Wood-46). In a round-bottom 96-well plate, bacteria were mixed with a concentration range of IgG plus 1% IgG- and IgM-depleted serum as complement source at 37°C on a shaker (750 rpm) for 15 min. Subsequently, neutrophils were added (at a ratio of 10 bacteria per 1 neutrophil) and incubated for 15 min at 37°C on a shaker (750 rpm) in a final volume of 50 µl. The reaction was stopped with 1% ice-cold paraformaldehyde, and neutrophil-associated fluorescent bacteria were analyzed by flow cytometry after gating neutrophils based on forward and side scatter properties. Phagocytosis was defined by two parameters: the percentage of cells with a positive fluorescent signal (% positive cells), and the mean fluorescence intensity of all neutrophils representing the overall phagocytosis efficacy.
Results
Complement activation by IgG allotypes and subclasses
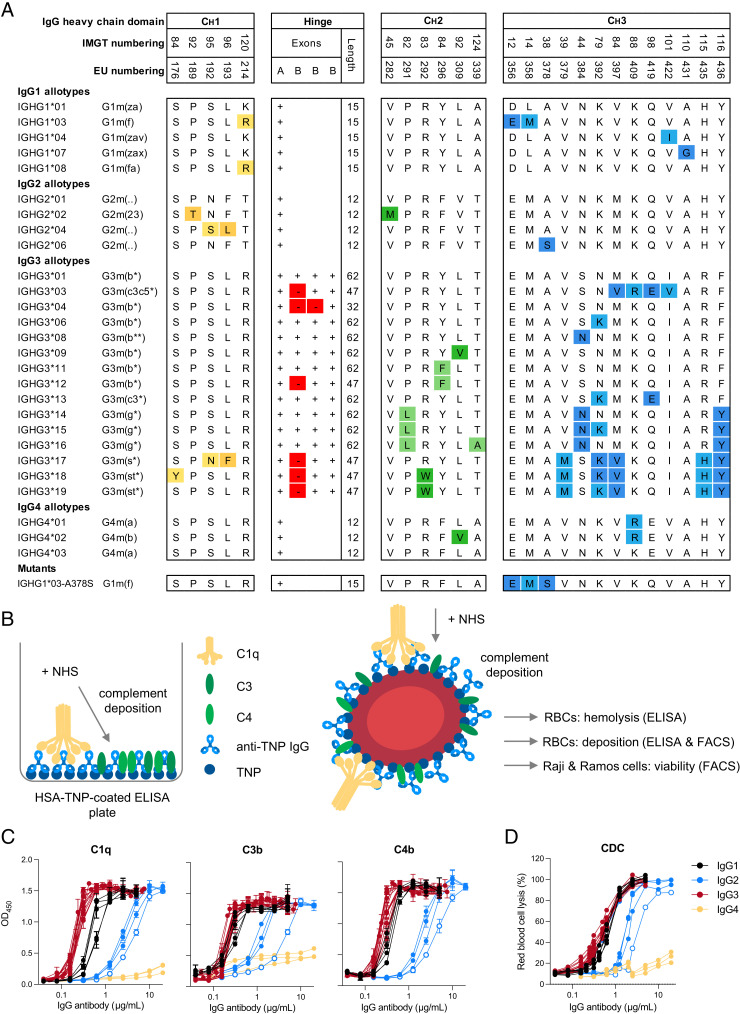
Previously, we produced a set of IgG allotypes with anti-TNP specificity and characterized glycosylation profile, FcγR binding, and Ab-dependent cellular cytotoxicity activity (41). The amino acid variation between polymorphic variants within each IgG subclass (27 allotypes in total) is illustrated in Fig. 1A. We used the same set of anti-TNP Abs to study complement activation by IgG allotypes. First, in an exploratory study, we determined complement binding (C1q) and deposition (C1q, C4b, and C3b) by all anti-TNP IgG allotypes on HSA-TNP (1 mM TNBS) in an ELISA setup as depicted in Fig. 1B. In line with previous studies, C1q binding was most efficient for IgG3 Abs followed by IgG1, IgG2, and IgG4 (Fig. 1C) (28, 29, 33). A similar subclass-specific pattern was observed at the level of C4b and C3b deposition, although IgG1 and IgG3 allotypes showed roughly similar levels of C3 deposition in contrast to the superior binding of C1q by IgG3 (Fig. 1C). Within the IgG1, IgG3, and IgG4 subclass we did not observe major allotype-specific differences in complement activation at the conditions tested. This is in contrast to previous studies, where G3m(b*) IgG3 allotype activated the complement system and induced RBC lysis more efficiently than did the G3m(g*) allotype (19, 20). Interestingly, one IgG2 allotype (IGHG2*06) showed less complement deposition compared with the other IgG2 allotypes, which was most profound at the level of C3 (Fig. 1C). To determine the functional relevance of these differences, we analyzed CDC of all IgG allotypes in a hemolytic assay with TNPylated RBCs (2 mM TNBS) as target cells (Fig. 1D). The CDC activity of all IgG allotypes showed a similar subclass pattern (IgG3 ≥ IgG1 > IgG2 > IgG4, from strongest to weakest CDC activity) as observed for deposition ELISAs (Fig. 1C), independent of their N-linked glycosylation profile at position 297 within each subclass (Supplemental Fig. 1). Furthermore, IgG2 allotype IGHG2*06 showed less efficient CDC of RBCs compared with other IgG2 allotypes (Fig. 1D). Thus, in addition to the variation in complement activation between IgG subclasses, polymorphic IgG2 variants also fluence the efficiency of complement activation.
FIGURE 1.
Complement activation by IgG subclasses and allotypes. (A) Overview of the amino acid variation (one-letter code) between different polymorphic variants (allotypes) for each individual IgG subclass. The presence or absence of a hinge exon in IgG3 allotypes is indicated with a + or – symbol, respectively (adapted from Ref. 41). (B) Schematic illustration of the complement deposition ELISA. Trinitrophenyl (TNP)ylated human serum albumin is coated on an ELISA plate, followed by incubation with anti-TNP IgG Abs and subsequently with pooled normal human serum (NHS). Schematic illustration of hemolytic assay and complement-dependent cytotoxicity (CDC) assay with RBCs, Raji cells, and Ramos cells as target cells. RBCs, Raji cells, and Ramos cells were opsonized with anti-TNP Abs and then incubated with NHS. Complement activation was either determined by flow cytometry (deposition of C4b and C3b), or hemoglobulin release from RBCs was measured via ELISA. (C) Complement activation by IgG1 (black), IgG2 (blue), IgG3 (red), and IgG4 allotypes (yellow) as measured by deposition of C1q, C4, and C3 in ELISA (1 mM TNPylated HSA) or (D) CDC activity in a hemolysis assay (2 mM TNPylated RBCs). IGHG2*06 is indicated with empty circles. All Abs were tested in a 2-fold serial dilution starting from 20 µg/ml for IgG2 and IgG4 allotypes and starting from 5 µg/ml for IgG1 and IgG3 allotypes.
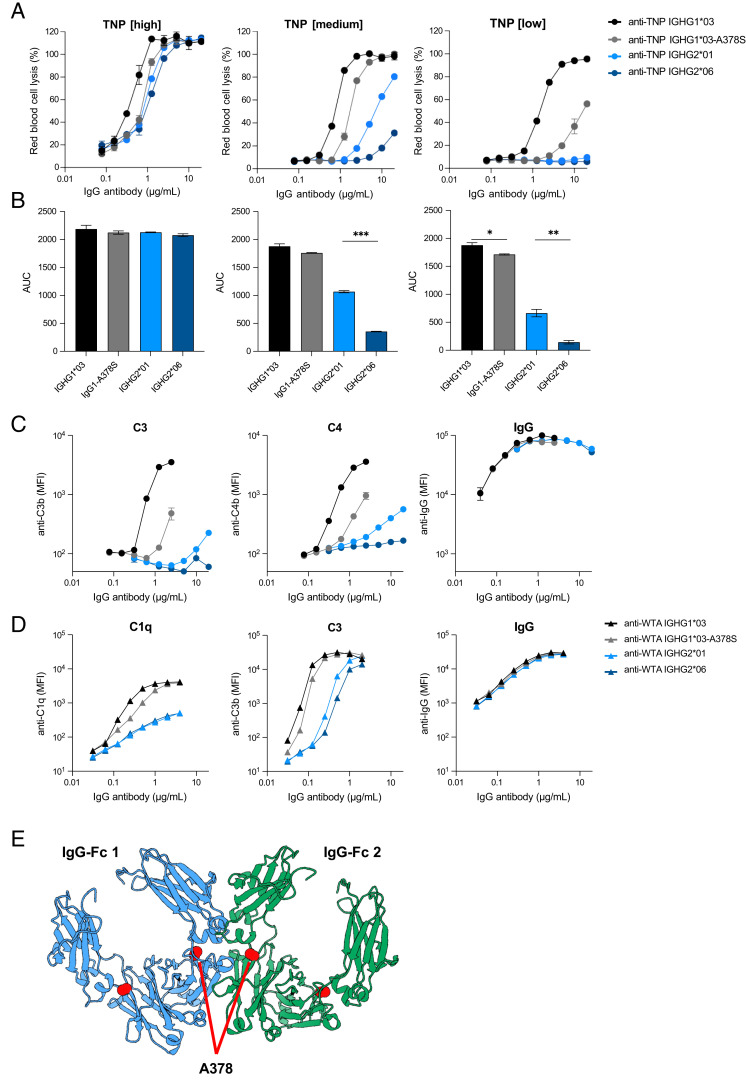
Serine 378 decreases complement activation of IgG1 and IgG2
IgG2 allotype IGHG2*06 harbors a unique serine at position 378 in the CH3 domain instead of an alanine at that position in IGHG2*01. To further investigate whether this variation is indeed influencing complement activation, we introduced this serine in an IgG1 background, creating an IGHG1*03-A378S mutant and compared it to IGHG1*03 and IgG2 allotype IGHG2*01 and IGHG2*06 in our complement assays. In ELISA, we observed a 2-fold reduction in C1q binding, C4b and C3b deposition for the IGHG1*03-A378S mutant compared with IGHG1*03, which was comparable to the 2-fold difference in complement deposition between IGHG2*01 (A378) and IGHG2*06 (S378) (Supplemental Fig. 2A). Next, we assessed the CDC activity of these variants in a hemolytic assay at different Ag densities and found that IGHG1*03-A378S mutant reduced CDC by 2- to 5-fold, depending on the Ag density (Fig. 2A, 2B). A stronger reduction of CDC activity (5-fold) was observed at lower Ag density (0.25 mM TNPylated RBCs), which is consistent with the data on complement deposition at lower Ag density in ELISA (Supplemental Fig. 2A). In line with reduced CDC activity, deposition of C3b and C4b on the RBCs was 4-fold lower for IGHG1*03-A378S compared with IGHG1*03 (Fig. 2C). The difference in complement activation between IGHG2*01 and IGHG2*06 was most profound at an Ag density of 0.5 mM TNP (Fig. 2A, Supplemental Fig. 2A). At the lowest Ag density (0.25 mM), both IgG2 allotypes failed to induce any CDC (Fig. 2A). However, the AUCs revealed significant differences for IGHG2*01 and IGHG2*06 at 0.5 mM TNBS (p = 0.0003) and IGHG1*03-A378S compared with IGHG1*03 (p = 0.0340) and IGHG2*01 compared with IGHG2*06 (p = 0.0098) at 0.25 mM TNBS (Fig. 2B).
FIGURE 2.
Serine 378 decreases complement activation of IgG1 and IgG2. (A) Complement-dependent cytotoxicity (CDC) activity of two IgG2 allotypes (IGHG2*01 in light blue and IGHG2*06 in dark blue), IGHG1*03 (black), and an IGHG1*03-378S mutant (gray) specific to trinitrophenyl (TNP) on RBCs at three different Ag densities (1 mM [high], 0.5 mM [medium], and 0.25 mM [low] TNBS concentration). Anti-TNP Abs were tested in duplicates in a 2-fold serial dilution starting at 20 µg/ml. (B) Area under the curve (AUC) of their CDC activity. (C) Complement deposition (C3b and C4b) and Ab binding (IgG) on RBCs (0.5 mM TNBS) was determined by flow cytometry for the same panel of anti-TNP allotypes. (D) Complement deposition (C1q and C3b) and IgG binding on S. aureus with the same allotypes, but with anti–wall teichoic acid (WTA) specificity was determined by flow cytometry. (E) Structural representation of two IgG1 Fc domains (blue and green) derived from the crystal structure matrix of IgG1 (PDB:1hzh). The alanine at position 378 in both Fc domains is depicted with red spheres and is positioned close to the interface between the Fc domains. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we cloned and produced the four IgG variants (IGHG1*03, IGHG1*03-A378S, IGHG2*01, and IGHG2*06) with a different specificity, namely S. aureus anti-WTA (46), assessed their quality (Supplemental Fig. 2B), and determined whether S378-containing Abs also reduce complement activation on a bacterial cell surface. In line with the deposition on RBCs, we observed that C1q binding and C3b deposition on S. aureus was less efficient with IgG1 IGHG1*03-A378S and IGHG2*06 compared with their A378-containing counterparts (Fig. 2D). Differences confirm the same trend as observed in the RBC assays. The small differences in complement deposition between anti-WTA variants seem to be functionally relevant, as we have also observed this trend in Ab-dependent cellular phagocytosis of bacteria by neutrophils for S378-containing Abs (Supplemental Fig. 2C). All Abs showed similar levels of opsonization in both model systems, indicating that the reduced complement activation of S378-containing Abs is not caused by a difference in Ag binding or opsonization efficiency (Fig. 2C, 2D). Instead, we hypothesize that the serine at position 378 interferes with efficient Fc–Fc interactions that are required for hexamer formation. Residue 378 is positioned near the interface between Fc domains and may influence hexamer formation (Fig. 2E) (17).
Complement activation via IgG1 or IgG3 is cell type–dependent
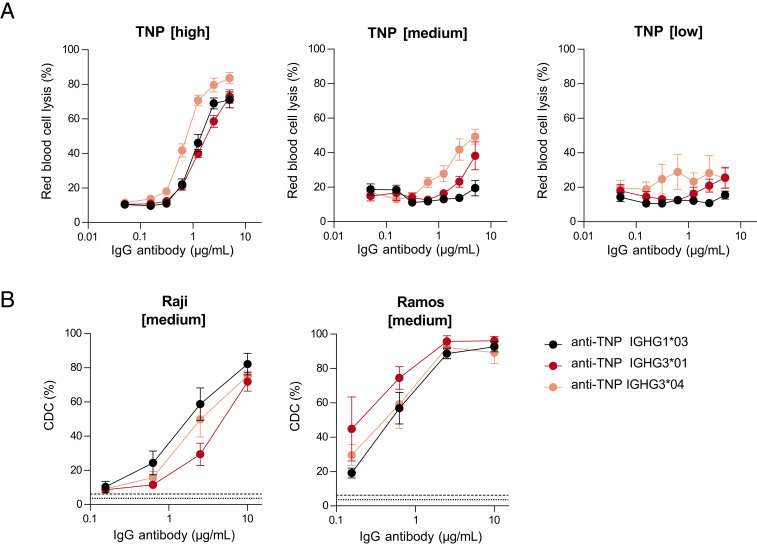
Depending on the model system used, IgG1 or IgG3 is most effective in activating the complement system (13, 20, 24, 28, 29, 33, 34, 52). We studied the effect of various parameters (nature of the Ag, Ag density, and target cell) on IgG1- versus IgG3-mediated complement activation. In the hemolytic CDC assay with RBCs as target cells (Fig. 1B), we found that IgG3 (IGHG3*04) has better CDC activity compared with IgG1 and IGHG3*01 in the presence of 20% NHS (Fig. 3A) and 50% NHS (low and medium TNP density, Supplemental Fig. 3A). We have included two different IgG3 allotypes to consider the variable IgG3 hinge length in our assays (IGHG3*01 [62 aa] and IGHG3*04 [32 aa]). At lower or complement suboptimal Ag density (0.25 and 0.5 mM TNBS), this superiority of both IgG3 variants over IgG1 was stronger than at higher Ag density. This illustrates that Ag density influences IgG1- and IgG3-mediated complement activation differently (Fig. 3A).
FIGURE 3.
Complement activation mediated by IgG1 and IgG3 is cell type–dependent. (A) Complement-dependent cytotoxicity (CDC) activity of IGHG1*03 (black), IGHG3*01 (long hinge, red), and IGHG3*04 (short hinge, pink) specific to trinitrophenyl (TNP) on RBCs at various Ag densities of 0.25 mM (low), 0.5 mM (medium), and 1 mM (high) TNBS in the presence of 20% serum. (B) CDC activity of same anti-TNP Abs was determined on Raji and Ramos B cells at an Ag density of 0.5 mM (medium) TNBS in the presence of 20% serum. All allotypes were tested in duplicates in a 4-fold serial dilution starting at 10 µg/ml. Negative controls are shown as dotted line for TNP unlabeled cells and dashed line represents TNPylated cells in the absence of Abs. Data represent mean ± SEM of n = 3.
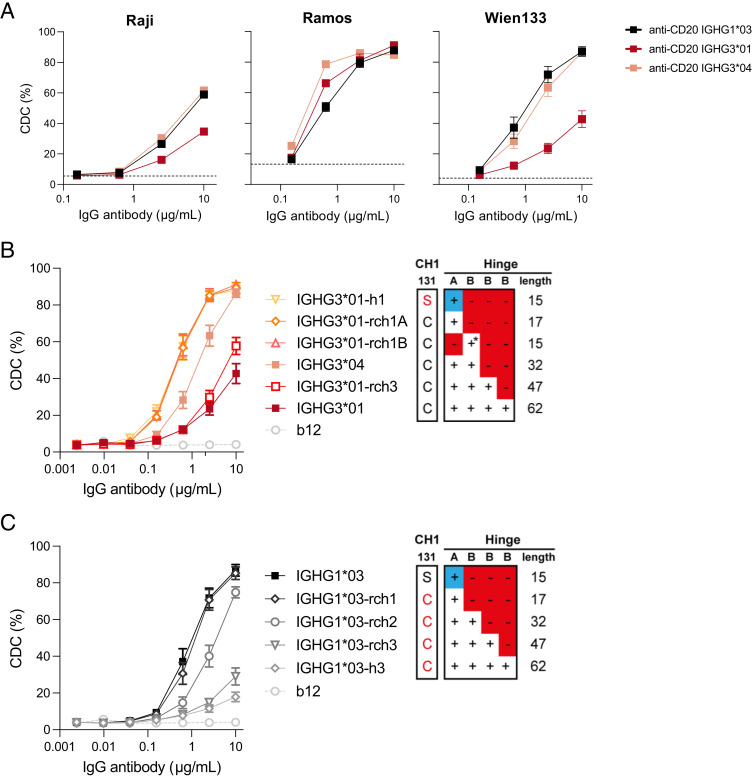
Then, in a different setup, where we used B cell lines (Raji and Ramos) as target cells, the CDC activity of anti-TNP Abs was investigated (Fig. 3B, Supplemental Fig. 3B). We found that IgG1 showed superior CDC activity compared with IgG3 for Raji cells, but not for Ramos cells (Fig. 3B). A similar pattern was observed when we compared CDC activity against Raji cells using anti-CD20 Abs, indicating that the superiority of IgG1 with Raji cells is not dependent on the Ag (Fig. 4A, Supplemental Fig. 3C), suggesting that cell characteristics are responsible. Next, we tested the anti-TNP and anti-CD20 Abs on Ramos cells and found slight superiority of IGHG3*01 and IGHG3*04 over IgG1 (Figs. 3B, 4A). This implies that specific characteristics of the Raji cells facilitate a more efficient complement activation by IgG1 compared with IgG3.
FIGURE 4.
Influence of the hinge length on CD20-targeted complement-dependent cytotoxicity. The capacity of CD20-specific IgG1*03 and IgG3*01 variants to induce complement-dependent cytotoxicity (CDC) was assessed in vitro. (A) CDC activity of anti-CD20 IGHG1*03 (black), IGHG3*01 (long hinge, red), and IGHG3*04 (short hinge, pink) of Raji, Ramos, and Wien133 cells. All allotypes were tested in duplicates or triplicates in a 4-fold serial dilution starting at 10 µg/ml in presence of 20% serum. The dashed gray line represents cells without any IgG Abs. (B) Matched set of natural (IGHG3*01 and IGHG3*04) and IGHG3*01 hinge mutants (containing deletions or substitutions of selected hinge exons) representing a range of hinge lengths (from yellow to dark red). Data represent mean ± SEM of n = 3. Red amino acid residues represent mutations to enable IgG1 hinge-like (top panel) or IgG3 hinge-like (bottom panel) HC-LC pairing. The blue + represents the IgG1 hinge exon; * indicates that C219S has been included to retain IgG3-like HC-LC pairing. (C) Matched set of natural (IGHG1*03) and IGHG1*03 hinge mutants (containing substitutions or insertions of selected hinge exons) representing a range of hinge lengths (from black to light gray). See (B) for details. Anti-HIV-1 gp120 clone b12 is used as negative control. Data represent mean ± SEM of n = 3.
These finding were corroborated using Wien133 cells, where the IGHG1*03 variant of the anti-CD20 Ab was again superior to the IGHG3*01 variant (Fig. 4A, Supplemental Fig. 3C). In the Wien133 model system, with a clear window between anti-CD20 IgG1- and IgG3-mediated CDC, we further examined the influence of hinge length on the CDC capacity. We found that selected IgG3 allotypes that contained identical CH2 sequences (and thus identical C1q binding sites) displayed a clear correlation with hinge length, with the shortest hinge, IGHG3*04 (two hinge exons), showing the highest CDC capacity (Fig. 4B, Supplemental Fig. 3D). This correlation was confirmed using a matched set of anti-CD20 IgG3 (and IgG1 hinge length variants), where the shortest hinges (single hinge exon) displayed the most potent CDC activity (Fig. 4B, 4C). Taken together, these data show that the hinge length can modulate the CDC activity of IgG3 allotypes.
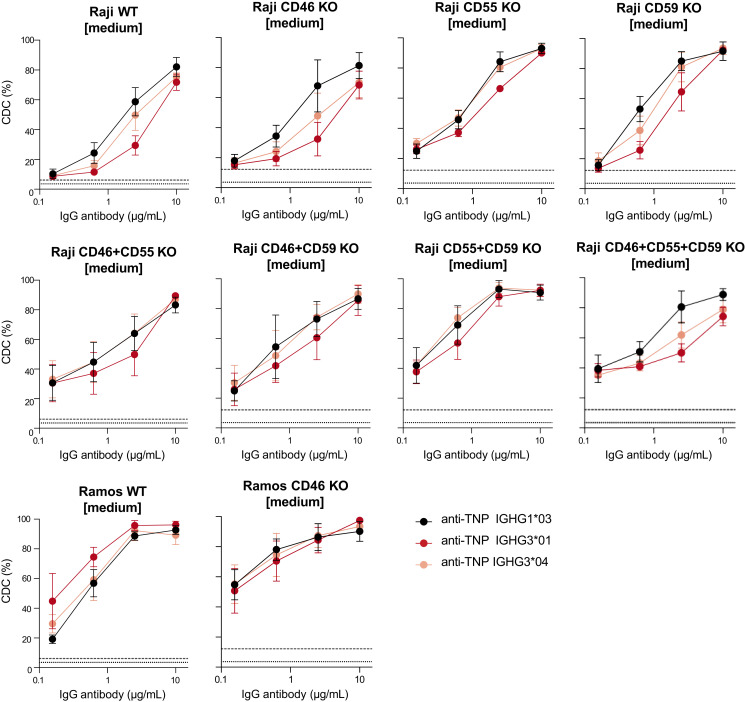
To investigate whether the differential expression of complement regulators (Supplemental Fig. 4) on the various target cells might influence IgG1- and IgG3-mediated complement activation, we generated Raji and Ramos KO cell lines that lack complement regulators. The KO cell lines included single, double, and triple KOs for CD46 (membrane cofactor protein, responsible for decay of convertases), CD55 (C3 convertase decay accelerating factor), and CD59 (a membrane attack complex inhibitor). No CR1 (removal of immune complexes and pathogens coated with C3b and C4b) KO cell lines were generated, because neither Raji nor Ramos cells express this receptor. As differential expression of CR2 on Raji and Ramos cells was identified, it warrants further investigation in future studies.
However, we found no differences in IgG1- and IgG3-mediated complement activation independent of complement regulators for the KO cell lines (Fig. 5). This indicates that other target cell–specific factors may also impact the regulation of complement activation despite complement regulatory proteins.
FIGURE 5.
Complement activation of IgG1 and IgG3 is independent of complement regulators. Complement-dependent cytotoxicity (CDC) activity of IGHG1*03 (black), IGHG3*01 (long hinge, red), and IGHG3*04 (short hinge, pink) specific to trinitrophenyl (TNP) on Raji and Ramos B cells including CD46, CD55, and CD59 knockout cell lines at an Ag density of 0.5 mM (medium) TNBS. Negative controls are shown as dotted line for TNP unlabeled cells, and a dashed line represents TNPylated cells in the absence of Abs. Data represent mean ± SEM of n = 3.
Discussion
In this study, we investigated the functional relevance of polymorphic variants within IgG subclasses on complement activation via the classical pathway. Complement activation by an IgG2 allotype (IGHG2*06) that uniquely contains a serine at position 378 was markedly lower compared with other IgG2 allotypes, especially at lower Ag densities. By generating an IgG1-A378S mutant, we confirmed that S378-containing Abs display less efficient complement activation and CDC at low Ag density. Furthermore, we found that complement activation potency of IgG1 or IgG3 is differentially influenced by parameters, such as Ag density and target cells.
The presence of a serine at position 378 in the CH3 domain is the only amino acid variation that discriminates IGHG2*01 from IGHG2*06. This variation is responsible for a reduction of complement activation at the level of C1q binding. There are two possible explanations for the reduced C1q binding of S378-containing variants. The serine alters the CH2–CH3 interactions in the Fc domain, which affects the conformation of the CH2 domain or the angle to which it approaches the CH3 domain. These structural changes might have an allosteric effect on the C1q binding epitope and reduce the affinity for C1q (53). Alternatively, the serine at position 378 interferes with the Fc–Fc interactions that are required for IgG hexamer formation (5, 18). Residue 378 is positioned close to the CH2–CH3 elbow residues that are involved in these Fc–Fc interactions. In fact, the A378S mutation was previously found to reduce CDC activity of IgG1 anti-CD20 as part of a Fc domain mutant library to identify residues involved in hexamer formation (17). Based on the crystal structure of IgG1 Fc, we speculate that the serine interferes with the hydrogen bond formed between K248 in the CH2 domain and E380 in the CH3 domain. This might change the overall conformation of the CH2–CH3 elbow regions.
We found that the IgG2 polymorphic variant IGHG2*06 is less efficient in initiating complement activation at low Ag density. When targeting a highly abundant cell wall glycopolymer (WTA) that comprises 40% of the staphylococcal cell (54), complement activation via IGHG2*06 was not substantially lower, suggesting that this polymorphic variation does not have a large functional impact. Thus, the situation in which complement activation via IgG2 is involved in pathogen clearance (high Ag density) is minimally influenced by the IgG2 polymorphic variant IGHG2*06 (55). However, this small reduction in complement activation might prevent an overactivation of the immune system and sepsis-related problems by the induction of a cytokine storm (56). Future studies are necessary to determine whether people bearing the IGHG2*06 allotype are more sensitive to bacterial infections. The prevalence of IGHG2*06 is relatively high in African ethnic groups (prevalence of 25–40%) (57), encouraging further research on susceptibility to bacterial infections, in particular situations in which complement activation via IgG2 is involved in pathogen clearance (58).
IgG1 and IgG3 are generally described as potent Abs in terms of complement activation. Whether IgG1 or IgG3 is most effective in activating the complement system is still under debate and has previously been described to depend on Ag density and epitope patchiness (29, 33). In line with this, we found that IgG3 was more efficient in complement activation at low Ag (TNP) density. In contrast, IgG3-mediated CDC was also slightly stronger than IgG1-mediated CDC at high Ag density. In addition to Ag density, complement activation of IgG subclasses has previously been described to be dependent on the nature of the Ag, Ag density, serum source, and complement regulators (13, 30, 33, 59, 60). Similarly, relative potency for complement activation for IgM versus IgG depends critically on Ag density (11). We found that IgG1 was only superior compared with IgG3 in CDC activity when Raji or Wien133 cells were used as target cell, independently of the Ag (CD20 or TNP). This is in contrast to what has previously been described that Ag density rather than cell line–specific factors appeared to be responsible for differences between IgG1 and IgG3 (24). The superior CDC activity on Wien133 cells was also shown to be dependent on the hinge length. A recent study also observed that hinge length influenced complement deposition for both IgG1 and IgG3 (21). In this study, an increase in hinge length exhibited an increase in complement deposition; however, even longer hinges showed compromised activity, possibly due to reduced likelihood of efficient Fc multimerization (21). In contrast to previous studies (19, 20, 33), we did not observe any major differences between IgG3 allotypes at either different densities or Ab concentrations for either complement deposition or CDC.
The membrane composition and/or membrane-bound proteins such as complement regulators may also differentially affect complement activation via IgG1 or IgG3. We speculated that the difference in hinge length between IgG1 and IgG3 is affecting the distance of C1q from the membrane. Depending on this distance, the deposition of C4b and formation of the C3 convertase (C4bC2a) might be at a different location on the cell membrane, influencing the regulation of these complement components by complement regulators such as CD46, CD55, CD59, and CR1. In comparison with RBCs and Ramos cells, Raji cells express CD55 at high levels and RBCs and Ramos cells do not express CD46 and CD59, respectively. However, we could not detect any differences in CDC across the Raji KO cell lines. The difference in hinge length between IgG1 and IgG3 and the resulting distance of C1q from the membrane may instead lead to suboptimal deposition of C4b on the cell membrane, influencing the subsequent formation of the C3 convertase. Other cell target–specific features, including the expression of other inhibitory IgG receptors, such as CD32 or Fc receptor–like proteins, other complement regulators (CR2), or the composition on the cell membrane, should therefore be considered in future explorations. Opposite to previous studies, we could not detect any differences between IgG3 allotypes at either different Ag densities or Ab concentrations for either complement deposition or CDC (19, 20, 33).
The discovery of novel IgG polymorphisms continues, and the tested IgG allotypes included all variants known to the time point when the Abs were expressed. New IgG polymorphisms should be examined regarding their capacity to activate complement. Furthermore, a conserved glycan attached to asparagine at position 297 in the CH2 domain (61) can have an impact on the ability of Abs to engage with complement, which consequently modulates CDC (62). Especially, galactosylation of the Fc domain impacts the formation of IgG hexamers and C1q binding (63). Our anti-TNP allotypes were fully characterized for N-linked Fc glycosylation, and the glycan profile across all variants in each subclass was the same, except for some IgG3 allotypes (41). However, we could not observe any differences between IgG3 allotypes in C1q binding based on their galactosylation profiles. In addition, unique to one IgG2 (IGHG2*15; not tested in the present study) and several IgG3 allotypes a conserved glycan attached to asparagine 392 (N392) in the CH2 domain (64) and O-linked glycosylation sites in the hinge region of IgG3 (65) may also have an effect on the conformation of the Ab and consequently on complement activation.
In summary, the findings of this study on the functional differences of IgG subclasses and allotypes in complement activation highlight the context dependency of this Fc-mediated effector function. Understanding the key determinants that shape Ab-mediated functions is crucial in designing more effective Ab-based therapeutics. Therefore, exploring the associations between IgG allotypes and effector functions could open new strategies for Ab-based therapeutics and might link polymorphic variants with a predisposition to bacterial infectious diseases.
Supplementary Material
Acknowledgments
We thank Peter Ligthart from the Erythrocyte Serology Department at Sanquin for providing RhD+ RBCs for the hemolysis assays. We also thank Thijs van Osch, Nienke Oskam, Richard Pouw, and Janita Oosterhoff for technical guidance in the complement assays and cell culture, respectively. Furthermore, we acknowledge the Flow Cytometry Core Unit at Sanquin for assisting in cell sorting.
Footnotes
This work was supported by Genmab (to T.D. and S.W.d.T.).
The online version of this article contains supplemental material.
- AUC
- area under the curve
- CDC
- complement-dependent cytotoxicity
- CR1
- complement receptor 1
- HC
- H chain
- HSA
- human serum albumin
- KO
- knockout
- LC
- L chain
- NHS
- normal human serum
- plx
- poloxamer
- TNBS
- 2, 4, 6-trinitrobenzenesulfonic acid
- TNP
- trinitrophenyl
- VB
- veronal buffer
- WTA
- wall teichoic acid
Disclosures
A.F.L., M.M.M. and J.S. are former employees of Genmab and own stock and/or warrants. T.D. and S.W.d.T. were awarded Genmab-sponsored research funding. The other authors have no financial conflicts of interest.
References
- 1. Merle, N. S., Church S. E., Fremeaux-Bacchi V., Roumenina L. T.. 2015. Complement system part I—molecular mechanisms of activation and regulation. Front. Immunol. 6: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ricklin, D., Reis E. S., Mastellos D. C., Gros P., Lambris J. D.. 2016. Complement component C3—the “Swiss army knife” of innate immunity and host defense. Immunol. Rev. 274: 33–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmidt, C. Q., Lambris J. D., Ricklin D.. 2016. Protection of host cells by complement regulators. Immunol. Rev. 274: 152–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Strasser, J., de Jong R. N., Beurskens F. J., Wang G., Heck A. J. R., Schuurman J., Parren P. W. H. I., Hinterdorfer P., Preiner J.. 2019. Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Lett. 19: 4787–4796. [DOI] [PubMed] [Google Scholar]
- 5. Diebolder, C. A., Beurskens F. J., De Jong R. N., Koning R. I., Strumane K., Lindorfer M. A., Voorhorst M., Ugurlar D., Rosati S., Heck A. J. R.. 2014. Complement is activated by IgG hexamers assembled at the cell surface. Science 343: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ugurlar, D., Howes S. C., de Kreuk B.-J., Koning R. I., de Jong R. N., Beurskens F. J., Schuurman J., Koster A. J., Sharp T. H., Parren P. W. H. I.. 2018. Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 359: 794–797. [DOI] [PubMed] [Google Scholar]
- 7. Damelang, T., Rogerson S. J., Kent S. J., Chung A. W.. 2019. Role of IgG3 in infectious diseases. Trends Immunol. 40: 197–211. [DOI] [PubMed] [Google Scholar]
- 8. de Taeye, S. W., Rispens T., Vidarsson G.. 2019. The ligands for human IgG and their effector functions. Antibodies (Basel) 8: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vidarsson, G., Dekkers G., Rispens T.. 2014. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 5: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiramoto, E., Tsutsumi A., Suzuki R., Matsuoka S., Arai S., Kikkawa M., Miyazaki T.. 2018. The IgM pentamer is an asymmetric pentagon with an open groove that binds the AIM protein. Sci. Adv. 4: eaau1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oskam, N., Ooijevaar-de Heer P., Derksen N. I. L., Kruithof S., de Taeye S. W., Vidarsson G., Reijm S., Kissel T., Toes R. E. M., Rispens T.. 2022. At critically low antigen densities, IgM hexamers outcompete both IgM pentamers and IgG1 for human complement deposition and complement-dependent cytotoxicity. J. Immunol. 209: 16–25. [DOI] [PubMed] [Google Scholar]
- 12. Sharp, T. H., Boyle A. L., Diebolder C. A., Kros A., Koster A. J., Gros P.. 2019. Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl. Acad. Sci. USA 116: 11900–11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giuntini, S., Reason D. C., Granoff D. M.. 2012. Combined roles of human IgG subclass, alternative complement pathway activation, and epitope density in the bactericidal activity of antibodies to meningococcal factor H binding protein. Infect. Immun. 80: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lighaam, L. C., Rispens T.. 2016. The immunobiology of immunoglobulin G4. Semin. Liver Dis. 36: 200–215. [DOI] [PubMed] [Google Scholar]
- 15. Zwarthoff, S. A., Widmer K., Kuipers A., Strasser J., Ruyken M., Aerts P. C., de Haas C. J. C., Ugurlar D., den Boer M. A., Vidarsson G., et al. 2021. C1q binding to surface-bound IgG is stabilized by C1r2s2 proteases. Proc. Natl. Acad. Sci. USA 118: e2102787118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oskam, N., Damelang T., Streutker M., Ooijevaar-de Heer P., Nouta J., Koeleman C., Van Coillie J., Wuhrer M., Vidarsson G., Rispens T.. 2023. Factors affecting IgG4-mediated complement activation. Front. Immunol. 14: 1087532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Jong, R. N., Beurskens F. J., Verploegen S., Strumane K., van Kampen M. D., Voorhorst M., Horstman W., Engelberts P. J., Oostindie S. C., Wang G., et al. 2016. A novel platform for the potentiation of therapeutic antibodies based on antigen-dependent formation of IgG hexamers at the cell surface. PLoS Biol. 14: e1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang, G., de Jong R. N., van den Bremer E. T. J., Beurskens F. J., Labrijn A. F., Ugurlar D., Gros P., Schuurman J., Parren P. W. H. I., Heck A. J. R.. 2016. Molecular basis of assembly and activation of complement component C1 in complex with immunoglobulin G1 and antigen. Mol. Cell 63: 135–145. [DOI] [PubMed] [Google Scholar]
- 19. Bindon, C. I., Hale G., Brüggemann M., Waldmann H.. 1988. Human monoclonal IgG isotypes differ in complement activating function at the level of C4 as well as C1q. J. Exp. Med. 168: 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brüggemann, M., Williams G. T., Bindon C. I., Clark M. R., Walker M. R., Jefferis R., Waldmann H., Neuberger M. S.. 1987. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 166: 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chu, T. H., Crowley A. R., Backes I., Chang C., Tay M., Broge T., Tuyishime M., Ferrari G., Seaman M. S., Richardson S. I., et al. 2020. Hinge length contributes to the phagocytic activity of HIV-specific IgG1 and IgG3 antibodies. PLoS Pathog. 16: e1008083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Natsume, A., In M., Takamura H., Nakagawa T., Shimizu Y., Kitajima K., Wakitani M., Ohta S., Satoh M., Shitara K., Niwa R.. 2008. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 68: 3863–3872. [DOI] [PubMed] [Google Scholar]
- 23. Vidarsson, G., van Der Pol W.-L., van Den Elsen J. M. H., Vilé H., Jansen M., Duijs J., Morton H. C., Boel E., Daha M. R., Corthésy B., van De Winkel J. G. J.. 2001. Activity of human IgG and IgA subclasses in immune defense against Neisseria meningitidis serogroup B. J. Immunol. 166: 6250–6256. [DOI] [PubMed] [Google Scholar]
- 24. Rösner, T., Derer S., Kellner C., Dechant M., Lohse S., Vidarsson G., Peipp M., Valerius T.. 2013. An IgG3 switch variant of rituximab mediates enhanced complement-dependent cytotoxicity against tumour cells with low CD20 expression levels. Br. J. Haematol. 161: 282–286. [DOI] [PubMed] [Google Scholar]
- 25. Brekke, O. H., Michaelsen T. E., Sandlie I.. 1995. The structural requirements for complement activation by IgG: does it hinge on the hinge? Immunol. Today 16: 85–90. [DOI] [PubMed] [Google Scholar]
- 26. Morgan, A., Jones N. D., Nesbitt A. M., Chaplin L., Bodmer M. W., Emtage J. S.. 1995. The N-terminal end of the CH2 domain of chimeric human IgG1 anti-HLA-DR is necessary for C1q, Fc gamma RI and Fc gamma RIII binding. Immunology 86: 319–324. [PMC free article] [PubMed] [Google Scholar]
- 27. Barrett, D. J., Ayoub E. M.. 1986. IgG2 subclass restriction of antibody to pneumococcal polysaccharides. Clin. Exp. Immunol. 63: 127–134. [PMC free article] [PubMed] [Google Scholar]
- 28. Garred, P., Michaelsen T. E., Aase A.. 1989. The IgG subclass pattern of complement activation depends on epitope density and antibody and complement concentration. Scand. J. Immunol. 30: 379–382. [DOI] [PubMed] [Google Scholar]
- 29. Michaelsen, T. E., Garred P., Aase A.. 1991. Human IgG subclass pattern of inducing complement-mediated cytolysis depends on antigen concentration and to a lesser extent on epitope patchiness, antibody affinity and complement concentration. Eur. J. Immunol. 21: 11–16. [DOI] [PubMed] [Google Scholar]
- 30. Riechmann, L., Clark M., Waldmann H., Winter G.. 1988. Reshaping human antibodies for therapy. Nature 332: 323–327. [DOI] [PubMed] [Google Scholar]
- 31. Siber, G. R., Schur P. H., Aisenberg A. C., Weitzman S. A., Schiffman G.. 1980. Correlation between serum IgG-2 concentrations and the antibody response to bacterial polysaccharide antigens. N. Engl. J. Med. 303: 178–182. [DOI] [PubMed] [Google Scholar]
- 32. Kuijpers, T. W., Weening R. S., Out T. A.. 1992. IgG subclass deficiencies and recurrent pyogenic infections, unresponsiveness against bacterial polysaccharide antigens. Allergol. Immunopathol. (Madr.) 20: 28–34. [PubMed] [Google Scholar]
- 33. Giuntini, S., Granoff D. M., Beernink P. T., Ihle O., Bratlie D., Michaelsen T. E.. 2016. Human IgG1, IgG3, and IgG3 hinge-truncated mutants show different protection capabilities against meningococci depending on the target antigen and epitope specificity. Clin. Vaccine Immunol. 23: 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Redpath, S., Michaelsen T., Sandlie I., Clark M. R.. 1998. Activation of complement by human IgG1 and human IgG3 antibodies against the human leucocyte antigen CD52. Immunology 93: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crowley, A. R., Richardson S. I., Tuyishime M., Jennewein M., Bailey M. J., Lee J., Alter G., Ferrari G., Morris L., Ackerman M. E.. 2023. Functional consequences of allotypic polymorphisms in human immunoglobulin G subclasses. Immunogenetics 75: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dechavanne, C., Dechavanne S., Sadissou I., Lokossou A. G., Alvarado F., Dambrun M., Moutairou K., Courtin D., Nuel G., Garcia A., et al. 2017. Associations between an IgG3 polymorphism in the binding domain for FcRn, transplacental transfer of malaria-specific IgG3, and protection against Plasmodium falciparum malaria during infancy: a birth cohort study in Benin. PLoS Med. 14: e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O’Hanlon, T. P., Rider L. G., Schiffenbauer A., Targoff I. N., Malley K., Pandey J. P., Miller F. W.. 2008. Immunoglobulin gene polymorphisms are susceptibility factors in clinical and autoantibody subgroups of the idiopathic inflammatory myopathies. Arthritis Rheum. 58: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oxelius, V. A., Pandey J. P.. 2013. Human immunoglobulin constant heavy G chain (IGHG) (Fcγ) (GM) genes, defining innate variants of IgG molecules and B cells, have impact on disease and therapy. Clin. Immunol. 149: 475–486. [DOI] [PubMed] [Google Scholar]
- 39. Pandey, J. P., Namboodiri A. M., Mohan S., Nietert P. J., Peterson L.. 2017. Genetic markers of immunoglobulin G and immunity to cytomegalovirus in patients with breast cancer. Cell. Immunol. 312: 67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tomescu-Baciu, A., Vartdal F., Holmøy T., Vedeler C. A., Lossius A.. 2018. G1m1 predominance of intrathecal virus-specific antibodies in multiple sclerosis. [Published erratum appears in 2019 Ann. Clin. Transl. Neurol. 6: 2608.] Ann. Clin. Transl. Neurol. 5: 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Taeye, S. W., Bentlage A. E. H., Mebius M. M., Meesters J. I., Lissenberg-Thunnissen S., Falck D., Sénard T., Salehi N., Wuhrer M., Schuurman J., et al. 2020. FcγR binding and ADCC activity of human IgG allotypes. Front. Immunol. 11: 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shen, M. W., Arbab M., Hsu J. Y., Worstell D., Culbertson S. J., Krabbe O., Cassa C. A., Liu D. R., Gifford D. K., Sherwood R. I.. 2018. Predictable and precise template-free CRISPR editing of pathogenic variants. [Published erratum appears in 2019 Nature 567: E1–E2.] Nature 563: 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dekkers, G., Treffers L., Plomp R., Bentlage A. E. H., de Boer M., Koeleman C. A. M., Lissenberg-Thunnissen S. N., Visser R., Brouwer M., Mok J. Y., et al. 2017. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front. Immunol. 8: 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teeling, J. L., French R. R., Cragg M. S., van den Brakel J., Pluyter M., Huang H., Chan C., Parren P. W. H. I., Hack C. E., Dechant M., et al. 2004. Characterization of new human CD20 monoclonal antibodies with potent cytolytic activity against non-Hodgkin lymphomas. Blood 104: 1793–1800. [DOI] [PubMed] [Google Scholar]
- 45. Roben, P., Moore J. P., Thali M., Sodroski J., Barbas C. F. III, Burton D. R.. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68: 4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lehar, S. M., Pillow T., Xu M., Staben L., Kajihara K. K., Vandlen R., DePalatis L., Raab H., Hazenbos W. L., Hiroshi Morisaki J., Kim J., et al. 2015. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 527: 323–328. [DOI] [PubMed] [Google Scholar]
- 47. McGrath, F. D. G., Brouwer M. C., Arlaud G. J., Daha M. R., Hack C. E., Roos A.. 2006. Evidence that complement protein C1q interacts with C-reactive protein through its globular head region. J. Immunol. 176: 2950–2957. [DOI] [PubMed] [Google Scholar]
- 48. Hack, C. E., Paardekooper J., Smeenk R. J., Abbink J., Eerenberg A. J., Nuijens J. H.. 1988. Disruption of the internal thioester bond in the third component of complement (C3) results in the exposure of neodeterminants also present on activation products of C3. An analysis with monoclonal antibodies. J. Immunol. 141: 1602–1609. [PubMed] [Google Scholar]
- 49. Leito, J. T. D., Ligtenberg A. J. M., van Houdt M., van den Berg T. K., Wouters D.. 2011. The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol. Immunol. 49: 185–190. [DOI] [PubMed] [Google Scholar]
- 50. Cruz, A. R., Boer M. A. D., Strasser J., Zwarthoff S. A., Beurskens F. J., de Haas C. J. C., Aerts P. C., Wang G., de Jong R. N., Bagnoli F., et al. 2021. Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc. Natl. Acad. Sci. USA 118: e2016772118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zwarthoff, S. A., Magnoni S., Aerts P. C., van Kessel K. P. M., Rooijakkers S. H. M.. 2021. Method for depletion of IgG and IgM from human serum as naive complement source. Methods Mol. Biol. 2227: 21–32. [DOI] [PubMed] [Google Scholar]
- 52. Rösner, T., Lohse S., Peipp M., Valerius T., Derer S.. 2014. Epidermal growth factor receptor targeting IgG3 triggers complement-mediated lysis of decay-accelerating factor expressing tumor cells through the alternative pathway amplification loop. J. Immunol. 193: 1485–1495. [DOI] [PubMed] [Google Scholar]
- 53. Orlandi, C., Deredge D., Ray K., Gohain N., Tolbert W., DeVico A. L., Wintrode P., Pazgier M., Lewis G. K.. 2020. Antigen-induced allosteric changes in a human IgG1 Fc increase low-affinity Fcγ receptor binding. Structure 28: 516–527.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brown, S., Santa Maria J. P., Walker S.. 2013. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67: 313–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saeland, E., Vidarsson G., Leusen J. H. W., Van Garderen E., Nahm M. H., Vile-Weekhout H., Walraven V., Stemerding A. M., Verbeek J. S., Rijkers G. T., et al. 2003. Central role of complement in passive protection by human IgG1 and IgG2 anti-pneumococcal antibodies in mice. J. Immunol. 170: 6158–6164. [DOI] [PubMed] [Google Scholar]
- 56. Ricklin, D., Reis E. S., Lambris J. D.. 2016. Complement in disease: a defence system turning offensive. Nat. Rev. Nephrol. 12: 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bashirova, A. A., Zheng W., Akdag M., Augusto D. G., Vince N., Dong K. L., O’hUigin C., Carrington M.. 2021. Population-specific diversity of the immunoglobulin constant heavy G chain (IGHG) genes. Genes Immun. 22: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Auton, A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., Korbel J. O., Marchini J. L., McCarthy S., McVean G. A., Abecasis G. R., et al. 1000 Genomes Project Consortium . 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Michaelsen, T. E., Sandlie I., Bratlie D. B., Sandin R. H., Ihle O.. 2009. Structural difference in the complement activation site of human IgG1 and IgG3. Scand. J. Immunol. 70: 553–564. [DOI] [PubMed] [Google Scholar]
- 60. Olafsen, T., Munthe Lund C. K., Bruland Ø. S., Sandlie I., Michaelsen T. E.. 1999. Complement-mediated lysis of cultured osteosarcoma cell lines using chimeric mouse/human TP-1 IgG1 and IgG3 antibodies. Cancer Immunol. Immunother. 48: 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rispens, T., Davies A. M., Ooijevaar-de Heer P., Absalah S., Bende O., Sutton B. J., Vidarsson G., Aalberse R. C.. 2014. Dynamics of inter-heavy chain interactions in human immunoglobulin G (IgG) subclasses studied by kinetic Fab arm exchange. J. Biol. Chem. 289: 6098–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee, C.-H., Romain G., Yan W., Watanabe M., Charab W., Todorova B., Lee J., Triplett K., Donkor M., Lungu O. I., et al. 2017. IgG Fc domains that bind C1q but not effector Fcγ receptors delineate the importance of complement-mediated effector functions. [Published erratum appears in 2017 Nat. Immunol. 18: 1173.] Nat. Immunol. 18: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Osch, T. L. J., Nouta J., Derksen N. I. L., van Mierlo G., van der Schoot C. E., Wuhrer M., Rispens T., Vidarsson G.. 2021. Fc galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. J. Immunol. 207: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Haan, N., Falck D., Wuhrer M.. 2020. Monitoring of immunoglobulin N- and O-glycosylation in health and disease. Glycobiology 30: 226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Plomp, R., Dekkers G., Rombouts Y., Visser R., Koeleman C. A. M., Kammeijer G. S. M., Jansen B. C., Rispens T., Hensbergen P. J., Vidarsson G., Wuhrer M.. 2015. Hinge-region O-glycosylation of human immunoglobulin G3 (IgG3). Mol. Cell. Proteomics 14: 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.