Model organisms (MOs), including yeast, worm (C. elegans), fruit fly (Drosophila), zebrafish, frog (Xenopus), mouse and rat, contribute greatly to our understanding of human development and disease. To be successful, MO research critically depends on many shared resources. Particularly important are MO stock centers and MO databases (MODs), without which most MO work would not be possible. This article focuses on MODs, which are mostly supported by grants from the National Institutes of Health (NIH), especially the National Human Genome Research Institute (NHGRI).
We are deeply concerned that the support for these vital databases is in jeopardy due to large cuts in their grant budgets. We fear these budget cuts will slow biomedical research worldwide and create increased waste of resources due to duplication of efforts. Indeed, the cuts threaten to erode access to reliable, expertly fact-checked data and cause an increase in mis-information due to the degraded organization of knowledge and information.
Why are MOs crucial to the modern biomedical research enterprise? Owing to the evolutionary conservation of genes and their functions, MOs provide insights into the molecular genetic basis of many fundamental biological phenomena, as well as mechanisms and treatments of human disease. This is best illustrated by the observation that, of the last 25 years of Nobel Prizes in Medicine or Physiology, 16 are based on research using one of the seven MO species listed in Table 1. This success of MO contributions to biomedical research reflects the investment made by the NIH to support research using these organisms.
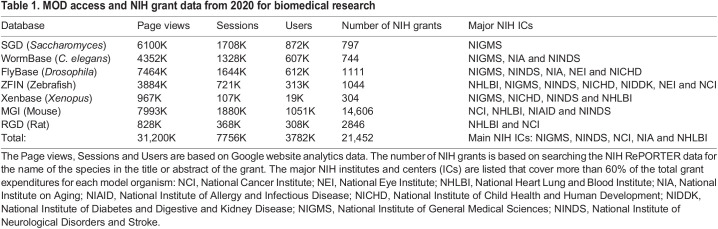
Table 1.
MOD access and NIH grant data from 2020 for biomedical research
MODs have been the primary means of cataloguing and organizing MO data for the past 25 years. They provide open access, searchable and well-curated information about the diverse biological properties of each organism, such as their respective genes, mutant and transgenic alleles, gene and protein expression, genetic and physical interactions, disease associations, mutant phenotypes, as well as other information relevant to MO researchers and others outside the field. MODs are also constantly innovating and collaborating with their respective communities to accelerate the research enterprise. The main MODs (listed in Table 1) are among the most extensively used databases in biomedical research based on Google search data. Indeed, more than 30 million page-views are reported each year by over 3.7 million users worldwide, demonstrating the extraordinary utility of these resources. In addition to researchers, users include high school and college students learning about biology and MOs, highlighting that MODs also represent an important educational resource. Science is reckoning with the existence of entrenched systems of unequal training and advancement opportunities for large segments of the population, and free access to MODs provides a powerful and equitable teaching tool.
The mission of MODs relies on software developers and expert biological curators to comb through the newly published literature, to properly tag and integrate the knowledge, and to convert it to a form amenable for searches and computation. MODs are collective resources across communities. They greatly facilitate an integrative as well as a comprehensive understanding of gene function. Because they integrate suites of genomic data repositories and computational biology analysis platforms, the impact of MODs on biomedical research has been transformational. Annotations generated by MODs, including gene ontology information, are integrated into hundreds of bioinformatics resources and have been foundational to data science innovations that rely on semantic reasoning to support predictive biology (Bult et al., 2018; Cherry et al., 2012; Gene Ontology Consortium et al., 2020; Harris et al., 2019; Karimi et al., 2018; Larkin et al., 2020; Ruzicka et al., 2018; Smith et al., 2019).
The financial support for MO research comes from different NIH institutes and centers (ICs), and varies from MO to MO (Table 1). At one extreme, yeast research is mostly supported by the National Institute of General Medicine (NIGMS), whereas work on zebrafish and mice relies on support from many different institutes. The number of grants supported by this funding in the year 2020 is also shown in Table 1. These numbers may be an underestimate or an overestimate, depending on the species, as they rely on grant applicants mentioning the species in the title, terms or abstract of the funded grant. We previously published a similar analysis (Wangler et al., 2015) and, based on additional analyses, the NIH estimated that, in the case of some MOs such as worms and flies, about 20-30% or more NIH R01 grants were supported than we had estimated (https://nexus.od.nih.gov/all/2016/07/14/a-look-at-trends-in-nihs-model-organism-research-support/). On the other hand, for mouse, the estimated number of grants may be slightly inflated as the number of grants supporting mouse research based on analyses from NIH in 2016 was closer to 12,500 (https://www.youtube.com/watch?v=f9FXNU1YWQo). The precise numbers, however, are not a major issue for our argument here. We estimate that a total of about 21-24,000 NIH grants supported research in one or more of the seven MOs in 2020, and numerous grants are also supporting investigations in other MOs, such as planarians, chick, fish species other than zebrafish, rabbits, guinea pigs, ferrets, axolotl and larger animals. This probably corresponds to $11-13 billion of the total NIH budget or approximately one quarter of the allocated budget of $42 billion. The main conclusion is that significant resources are allocated to MO research by the NIH, and that MO researchers in the USA and, more broadly, across the world, heavily rely on MODs.
Despite the significant ongoing and successful contributions of MO research to biomedical research, the NIH budget to maintain and innovate the MODs has actually been decreasing since 2016. Particularly alarming are the recent budget reductions that will lead to a 50% cut in support for the MODs compared with the 2016 support levels. Interestingly, even though numerous NIH institutes support MO research, a single institute and one that is not listed in Table 1, the National Human Genome Research Institute or NHGRI, has been supporting 90% of the funding of the MODs. Only Xenbase is supported by another institute, the National Institute of Child Health and Human Development (NICHD). Unfortunately, very little to no support is provided to any of the MODs by international grants, yet international users represent more than 50% of users for each database. The vast majority of these users are located in Europe, China and Japan, and, with few exceptions, no or very little support has been obtained from these countries, despite efforts from NIH and some principal investigators of MODs.
This lack of MOD support from diverse sources is now more important than ever before in the face of the recent severe NIH funding cuts. These cuts significantly reduce the capability of expert curation in all MODs, which is crucial to the support mission of these databases. Moreover, papers published more recently typically incorporate far more diverse data than papers published even a few years ago. This information is spread throughout the literature, and is almost impossible to access and mine systematically if it is not collated in a MOD. Recently implemented technologies based on transcriptomics (including single cell RNA sequencing), metabolomics, protein mass spectroscopy and microCT/histotomography provide a plethora of data that need to be integrated within and between MODs (https://orip.nih.gov/about-orip/workshop-reports). Furthermore, integration of genotypic and molecular data together with phenotypic data is required. Phenotyping was once descriptive and idiosyncratic, making it poorly suited to computational mechanisms of discovery and searching. However, new technologies produce quantitative, organism-wide, digital morphological, physiological and cell biological information that can be anchored to other phenotypic and genetic information. The MODs are also well suited to flag each gene and related features using controlled vocabulary to allow computational biologists to take advantage of these rich resources. It has been argued that the annotations and curation should rely on artificial intelligence (AI). Whereas AI has been implemented for some data curation and this is a laudable goal, it has so far failed to fully capture the complexity of datasets and to generate reliable information for users. Given the nature of the scientific literature, the well-trained human curators of the MODs are still irreplaceable and are likely to remain so.
One reason that funding for MODs has been decreased is a NHGRI strategy to try to capture some of the complexities associated with MODs by developing and expanding the Alliance of Genome Resources (AGR) (Alliance of Genome Resources Consortium et al., 2019). This project aims to develop and maintain sustainable genome information resources that capture information from MODs, and the GO Consortium (Gene Ontology Consortium et al., 2020), and provide a platform that is similar across MOs to allow inter-MO data sharing. This strategy is welcomed and should facilitate the use of diverse MOs in understanding the genetic and genomic basis of human biology, health and disease. Nevertheless, the AGR crucially relies on the curation of data from MODs and, consequently, the usefulness of the AGR is being compromised by these funding cuts. Thus, the funding cuts threaten the success of MO research by weakening both the MODs as well as the AGR. Moreover, numerous additional resources rely on MODs, including projects such as the Monarch Initiative (Shefchek et al., 2019) and MARRVEL (Wang et al., 2017), which are used by scientists and clinicians world-wide to probe information related to human disease genes. Therefore, the decrease in funding for MODs will impact many scientists and clinicians who do not work directly with MOs, and will have a lasting impact on the biomedical research enterprise.
We propose several strategies that should be explored to mitigate the problem and restore MOD funding to a workable level to sustain research. The first is based on the simple observation that many NIH institutes support MO research that crucially depends on the MODs (Table 1; which is not a comprehensive list). If these institutes were to join forces with the NHGRI and agree on an NIH-wide stable support system for the MODs, each MOD could maintain its required curation and continue to innovate. These institutes are a driving force of scientific innovation and the MODs dramatically enhance the power of the grants that are being supported by each institute. Therefore, it seems logical that they all contribute to the support of the MODs. The second strategy is to tap discretionary funds of the Director of the NIH that are meant to prioritize important research issues. The third strategy is to charge each NIH grant using a MO fee that is proportional to the allocated funds. This fee could vary with a maximum of $1000 per year, and could be adapted over time to the needs of each MOD. The fourth strategy is to directly charge users of MODs. This strategy has been considered by some of the MODs. However, in addition to creating administrative hurdles, it is cumbersome and would unfairly affect users with limited funds. Fifth, the MODs could rely on voluntary contributions of users. This strategy is being implemented at FlyBase, but the funds that have been collected cover only a minor proportion of the support needed (Norbert Perrimon, personal communication). Last, an agreement with foreign national research institutes based on use could be implemented. Given that MO researchers in Europe, China and Japan constitute a large user MOD group (nearly 50%), seeking support from the European Research Council, the National Natural Science Foundation of China, and the Ministries of Education, Health, Labor and Welfare or the Agency for Medical Research and Development in Japan is a possibility.
It is difficult to overstate how devastating the loss or crippling of these resources would be for the continued success of basic research. The MODs and GO consortia have become indispensable resources; they save tremendous amounts of time and effort as they centrally collate information and reduce wasteful duplication of effort. By documenting the existence of animal, genetic and molecular reagents that have been deposited elsewhere in MO stock centers, the MODs increase research rigor and reproducibility, while minimizing generation of duplicate animal lines and other reagents. MODs thus accelerate research and, importantly, avoid waste. The curation of information also serves another NIH mission as it makes information paid for by US taxpayers publicly accessible and thus makes biomedical research more transparent.
The NIH has put out a request for information (RFI) on user experience with scientific data sources and tools (https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-182.html). We would appreciate it if you would let the NIH know how much you value MODs and how reduced funding for MODs would impact your productivity by completing the survey link on the RFI page.
In summary, the lack of adequate support for MODs will have a large, long-term and negative impact on scientific research and biomedical discovery. We urge the NIH to endorse their past, current and future investments in successful MO research by stabilizing funding for MODs. This funding needs to be at a level that will maintain both the quality of MODs and their capacity for continued innovation in disseminating the valuable data generated by MO researchers.
References
- Alliance of Genome Resources Consortium, Agapite, J., Albou, L.-P., Aleksander, S., Argasinska, J., Arnaboldi, V., Attrill, H., Bello, S. M., Blake, J. A., Blodgett, O.et al. (2019). Alliance of Genome Resources Portal: unified model organism research platform. Nucleic Acids Res. 48, D650-D658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult, C. J., Blake, J. A., Smith, C. L., Kadin, J. A., Richardson, J. E., Anagnostopoulos, A., Asabor, R., Baldarelli, R. M., Beal, J. S., Bello, S. M.et al. (2018). Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 47, gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. M., Hong, E. L., Amundsen, C., Balakrishnan, R., Binkley, G., Chan, E. T., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S. R.et al. (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700-D705. 10.1093/nar/gkr1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium, Carbon, S., Douglass, E., Good, B. M., Unni, D. R., Harris, N. L., Mungall, C. J., Basu, S., Chisholm, R. L., Dodson, R. J.et al. (2020). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T. W., Arnaboldi, V., Cain, S., Chan, J., Chen, W. J., Cho, J., Davis, P., Gao, S., Grove, C. A., Kishore, R.et al. (2019). WormBase: a modern Model Organism Information Resource. Nucleic Acids Res. 48, D762-D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, K., Fortriede, J. D., Lotay, V. S., Burns, K. A., Wang, D. Z., Fisher, M. E., Pells, T. J., James-Zorn, C., Wang, Y., Ponferrada, V. G.et al. (2018). Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 46, D861-D868. 10.1093/nar/gkx936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, A., Marygold, S. J., Antonazzo, G., Attrill, H., dos Santos, G., Garapati, P. V., Goodman, J. L., Gramates, L. S., Millburn, G., Strelets, V. B.et al. (2020). FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899-D907. 10.1093/nar/gkaa1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka, L., Howe, D. G., Ramachandran, S., Toro, S., Van Slyke, C. E., Bradford, Y. M., Eagle, A., Fashena, D., Frazer, K., Kalita, P.et al. (2018). The Zebrafish Information Network: new support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. 47, gky1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefchek, K. A., Harris, N. L., Gargano, M., Matentzoglu, N., Unni, D., Brush, M., Keith, D., Conlin, T., Vasilevsky, N., Zhang, X. A.et al. (2019). The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 48, D704-D715. 10.1093/nar/gkz997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. R., Hayman, G. T., Wang, S.-J., Laulederkind, S. J. F., Hoffman, M. J., Kaldunski, M. L., Tutaj, M., Thota, J., Nalabolu, H. S., Ellanki, S. L. R.et al. (2019). The Year of the Rat: The Rat Genome Database at 20: a multi-species knowledgebase and analysis platform. Nucleic Acids Res. 48, D731-D742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Al-Ouran, R., Hu, Y., Kim, S.-Y., Wan, Y.-W., Wangler, M. F., Yamamoto, S., Chao, H.-T., Comjean, A., Mohr, S. E.et al. (2017). MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am J Hum Genetics 100, 843-853. 10.1016/j.ajhg.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangler, M. F., Yamamoto, S. and Bellen, H. J. (2015). Fruit Flies in Biomedical Research. Genetics 199, 639-653. 10.1534/genetics.114.171785 [DOI] [PMC free article] [PubMed] [Google Scholar]