Abstract
Background
Wohlfahrtiimonas chitiniclastica is an emerging fly-borne zoonotic pathogen, which causes infections in immunocompromised patients and some animals. Herein, we reported a W. chitiniclastica BM-Y from a dead zebra in China.
Methods
The complete genome sequencing of BM-Y showed that this isolate carried one chromosome and one novel type of blaVEB–1-carrying plasmid. Detailed genetic dissection was applied to this plasmid to display the genetic environment of blaVEB–1.
Results
Three novel insertion sequence (IS) elements, namely ISWoch1, ISWoch2, and ISWoch3, were found in this plasmid. aadB, aacA1, and gcuG were located downstream of blaVEB–1, composing a gene cassette array blaVEB–1–aadB–aacA1–gcuG bracketed by an intact ISWoch1 and a truncated one, which was named the blaVEB–1 region. The 5′-RACE experiments revealed that the transcription start site of the blaVEB–1 region was located in the intact ISWoch1 and this IS provided a strong promoter for the blaVEB–1 region.
Conclusion
The spread of the blaVEB–1-carrying plasmid might enhance the ability of W. chitiniclastica to survive under drug selection pressure and aggravate the difficulty in treating infections caused by blaVEB–1-carrying W. chitiniclastica. To the best of our knowledge, this is the first report of the genetic characterization of a novel blaVEB–1-carrying plasmid with new ISs from W. chitiniclastica.
Keywords: Wohlfahrtiimonas chitiniclastica, bla VEB–1 , plasmid, drug resistance, zoonotic bacteria, mobile genetic element
1. Introduction
Wohlfahrtiimonas chitiniclastica is an emerging fly-borne pathogen that causes bacteremia (Rebaudet et al., 2009), sepsis (Almuzara et al., 2011), cellulitis (de Dios et al., 2015), osteomyelitis (Suryalatha et al., 2015), wound infections (Nogi et al., 2016), and soft tissue and bone infections (Kõljalg et al., 2015) in humans and bacteremia (Thaiwong et al., 2014), endocarditis (Josue et al., 2015), and hoof fetlow (Qi et al., 2016) in animals. It was first identified from both the homogenate of the third-stage larvae of the parasitic fly Wohlfahrtia magnifica and the foregut of a third-stage maggot of W. magnifica collected from an infested vulval wound of a Romney sheep in Hungary in 2008 (Tóth et al., 2008). Since then, W. chitiniclastica has been associated with several obligate parasitic flies, including W. magnifica (Tóth et al., 2008), Chrysomya megacephala (Cao et al., 2013), Lucilia sericata (Campisi et al., 2015), Musca domestica (Gupta et al., 2012), and Lucilia illustris (Iancu et al., 2020). These flies can transmit W. chitiniclastica to humans and animals, which causes local or systemic infections originating from wounds infested with fly larvae. Diverse risk factors have been identified among infected patients and animals, including homelessness, alcoholism, tobacco abuse, poor hygiene, chronic open wounds, proximity to livestock, increasing age, low socioeconomic status, immunocompromised patients, chronic cardiovascular/circulatory disease, and neurological disease (Schröttner et al., 2017). To date, W. chitiniclastica has been identified in 10 countries in Europe (Tóth et al., 2008; Rebaudet et al., 2009; Campisi et al., 2015; Kõljalg et al., 2015; Schröttner et al., 2017; Pablo-Marcos et al., 2019; Dovjak et al., 2021; Hladík et al., 2021; De Smet et al., 2022; Karaca et al., 2022), four in Asia (Suryalatha et al., 2015; Zhou et al., 2016; Suraiya et al., 2017; Katanami et al., 2018), two in South America (Almuzara et al., 2011; Matos et al., 2016), one in North America (Thaiwong et al., 2014), one in Oceania (Connelly et al., 2019), and one in Africa (Hoffman et al., 2016).
It is speculated that W. chitiniclastica is intrinsically resistant to fosfomycin due to the fosfomycin efflux proteins, the gene homolog encoding for transferases, and the gene homologous to mdtG that related to fosfomycin resistance (Kopf et al., 2021). In addition, research has revealed that W. chitiniclastica shows resistance to amikacin (Kopf et al., 2021), tetracycline (Snyder et al., 2020), and tobramycin (Kopf et al., 2021), and it is intermediate to ampicillin (Qi et al., 2016). However, W. chitiniclastica is susceptible to the majority of the known antibiotics including β-lactams, quinolones, and trimethoprim/sulfamethoxazole (Kopf et al., 2022). Therefore, these three categories of antimicrobials may be the best options for treating W. chitiniclastica infections (Schröttner et al., 2017).
blaVEB–1 was initially identified from a clinical Escherichia coli isolated from Vietnam, and it encodes Vietnamese extended-spectrum β-lactamase (VEB-1) that displays high-level resistance to amoxicillin, piperacillin, cefotaxime, ceftazidime, and aztreonam (Poirel et al., 1999). Since then, blaVEB–1 has been found in IncA/C (Papagiannitsis et al., 2012), IncF (Paul et al., 2017), IncK (Paul et al., 2017), and IncP plasmids (Siebor et al., 2021) and in plasmids with undetermined Inc type from various Gram-negative species, including Enterobacteriaceae (Poirel et al., 1999), Morganellaceae (Oikonomou et al., 2016), Acinetobacter (e.g., Acinetobacter baumannii with the accession number LC107425), Aeromonas (e.g., Aeromonas caviae with the accession number KU886278), Shewanella (e.g., Shewanella algae with the accession number CP033574), Vibrio alginolyticus (Ye et al., 2016), Pseudomonas aeruginosa (Naas et al., 1999), Achromobacter xylosoxidans (with the accession number DQ393569), and Pasteurella multocida (with the accession number CP014157), but not in W. chitiniclastica. Generally, blaVEB–1 is carried by integrons, and aadB encoding an aminoglycoside adenyltransferase is located downstream of blaVEB–1 (Naas et al., 1999, 2000).
This study showed the complete genome sequences of a W. chitiniclastica from the pancreas of a dead zebra in China, which carried one chromosome and one novel type of blaVEB–1-carrying plasmid. Detailed genetic dissection was applied to this plasmid to display the genetic environment of blaVEB–1. The transcription start site and the promoter of the blaVEB–1 region were identified. The data presented here provided a deeper genomics and bioinformatics understanding of W. chitiniclastica for clinical treatment and pathogenesis research. To the best of our knowledge, this is the first report of the genetic characterization of a novel blaVEB–1-carrying plasmid from W. chitiniclastica.
2. Materials and methods
2.1. Bacterial isolation
The blood, heart, liver, spleen, lung, kidney, and pancreas specimens from a dead zebra were collected from Shenzhen Safari Park in 2013 and transferred to our laboratory in Changchun under sterile conditions for bacterial isolation. For each specimen, the tissue was plated onto brain heart infusion agar medium, 5% sheep blood agar medium, and MacConkey agar medium. The sample was then incubated at 37°C for 18 h under aerobic conditions.
The W. chitiniclastica strain DSM 18708 (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Germany), isolated from artificial culture of W. magnifica in Hungary, was used as the reference strain in this study (Kõljalg et al., 2015).
2.2. Bacterial identification and phenotypic characteristics
Gram staining, BD Phoenix™-100 Automated Microbiology System (Becton, Dickinson and Company, USA) detection, PCR amplification of the 16S rRNA, rpoB, and gyrB genes followed by sequencing, and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, USA) were conducted for bacterial identification.
A transmission electron microscopy investigation was carried out. The motility of the bacteria was tested on motility agar (Tittsler and Sandholzer, 1936). To determine the bacterial growth curves, the optical density at 600 nm (OD600) was monitored for each culture with a 1-h interval during the first 12 h and a 2-h interval during the last 12 h using the NanoPhotometer N60 (Implen, Germany).
The minimum inhibitory concentrations (MICs) of piperacillin-tazobactam, ceftazidime, cefotaxime, cefepime, aztreonam, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, tetracycline, chloramphenicol, and trimethoprim-sulfamethoxazole were determined using the BD Phoenix™-100 Automated Microbiology System. The E. coli ATCC 25922 was used as an internal quality control.
The biochemical characteristics were tested using the BD Phoenix™-100 Automated Microbiology System, and the cellular fatty acid composition was detected using gas chromatography (Hewlett Packard 6890, USA) and identified using the Sherlock Microbial Identification System (MIDI, USA).
2.3. Sequencing and sequence assembly
The bacterial genomic DNA was isolated using the UltraClean microbial kit (Qiagen, Germany) and sequenced with a PacBio RS II sequencer (Pacific Biosciences, USA) (Fu et al., 2022; Li et al., 2022). The reads were assembled de novo using SMARTdenovo.1
2.4. Bacterial precise species identification
Bacterial precise species identification was conducted using pairwise average nucleotide identity (ANI) analysis between the bacterial genomic DNA sequenced in this study and the reference genome.2 A cutoff of ≥ 95% ANI was used to define a bacterial species (Richter and Rosselló-Móra, 2009).
2.5. Phylogenetic analysis
The core genes and specific genes of the bacterial genomes were analyzed using the CD-HIT rapid clustering of similar proteins software with a threshold of 50% pairwise identity and 0.7 length difference cutoff in amino acid (Li et al., 2001, 2002; Li and Godzik, 2006). The core amino acid sequences were extracted and aligned using MUSCLE (Edgar, 2004). Based on the core amino acid sequences, a maximum-likelihood phylogenetic tree was constructed using TreeBeST3 with a bootstrap iteration of 1,000.
2.6. Sequence annotation
RAST 2.0 (Brettin et al., 2015), blastp/blastn (Boratyn et al., 2013), and DANMEL (Wang et al., 2022) searches were used to predict open reading frames (ORFs). The online databases CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), and INTEGRALL (Moura et al., 2009) were used to find resistance genes and mobile elements. Inkscape 1.0 was used to draw gene organization diagrams.4
2.7. Identification of the transcription start site and the promoter of the blaVEB–1 region
The total RNA of the bacterial isolates was extracted using the RNeasy maxi kit (Qiagen, Germany). Rapid amplification of cDNA ends (RACE) was performed using 5′-RACE system version 2.0 (Invitrogen, USA) according to the manufacturer’s instructions. Three gene-specific primers GSP1 (5′-ATCCTTCTCATTGCTG-3′), GSP2 (5′-CTCCTATTCTGGCATTTTTTG-3′), and GSP3 primer (5′-AAGTTGTCAGTTTGAGCATTT-3′) were used. The final PCR products were cloned into the pMD18-T vector, and then, the positive clones were identified and sequenced. The 5′-RACE experiment was repeated three times. Five clones were selected randomly and sequenced each time. The transcription start site was determined according to the sequence comparison between the positive clones and the blaVEB–1 region. The online database BPROM (Cassiano and Silva-Rocha, 2020) was used for promoter prediction.
2.8. Conjugation and electroporation experiments
Conjugal transfer and electroporation were performed as described previously (Liang et al., 2018; Guan et al., 2022).
2.9. Data availability statement
The complete sequences of the chromosome of BM-Y and the plasmid pBM-Y were submitted to GenBank under the accession numbers CP115969 and CP115970, respectively.
3. Results
3.1. Identification of W. chitiniclastica BM-Y and its phenotypic characteristics
BM-Y was isolated from the pancreas specimens of a dead zebra. According to Gram staining and the sequences of the amplicon of the 16S rRNA, rpoB, and gyrB genes, BM-Y was identified as the Gram-negative bacteria W. chitiniclastica. The MALDI-TOF MS score of BM-Y was 2.418, confirming W. chitiniclastica. Scores above 2.300 represent a highly probable species identification. No other bacterial isolates were discovered in this study. BM-Y has a 97.19% ANI value with the reference genome DSM 18708 (accession number AQXD01000000). BM-Y contained one chromosome (2.0 Mb in length) and one plasmid pBM-Y (42.3 Kb in length), and they were fully sequenced herein. The chromosome carried no resistance genes. The plasmid pBM-Y harbored blaVEB–1, aadB, aacA1, and tetA(H).
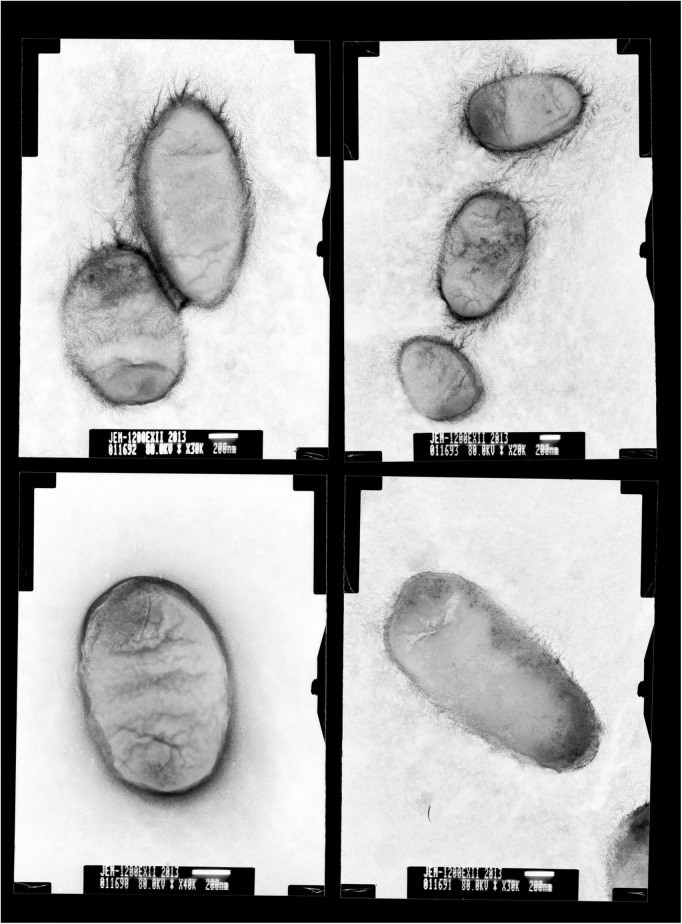
Colonies of BM-Y were small (colony diameter 0.8–1.0 mm), entire, convex, smooth, and glistening and were composed of short, straight, and non-motile rods (0.1–0.2 × 1.0–1.5 μm, Figure 1), which did not produce hemolysin on 5% sheep blood agar medium.
FIGURE 1.
Transmission electron micrographs of Wohlfahrtiimonas chitiniclastica BM-Y. BM-Y was incubated on a brain heart infusion agar medium at 37°C for 18 h.
Both BM-Y and DSM 18708 reached the middle logarithmic phase (OD600 value of about 1.0) and the stationary stage at the fifth hour and the 12th hour, respectively (Figure 2).
FIGURE 2.
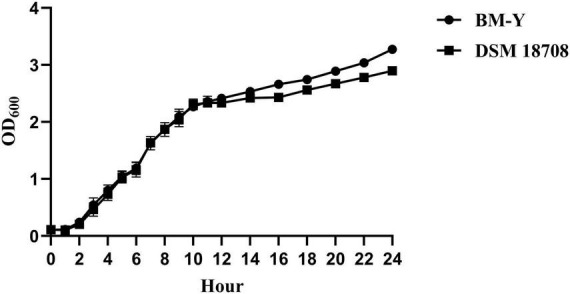
Bacterial growth curves of W. chitiniclastica BM-Y and DSM 18708.
The MICs of ceftazidime, cefotaxime, cefepime, aztreonam, gentamicin, and tetracycline of BM-Y were higher than that of DSM 18708. The MICs of 14 antimicrobial drugs are shown in Table 1.
TABLE 1.
Minimum inhibitory concentrations of 14 antimicrobial drugs.
| Antibiotics | Minimum inhibitory concentration (MIC, μg/mL) | ||
| BM-Y | DSM 18708 | ATCC 25922 | |
| Piperacillin-tazobactam | < 4/4 | > 64/4 | < 4/4 |
| Ceftazidime | > 16 | 16 | < 1 |
| Cefotaxime | 8 | < 1 | < 1 |
| Cefepime | > 16 | 8 | < 2 |
| Aztreonam | > 16 | 8 | < 2 |
| Imipenem | < 1 | < 1 | < 1 |
| Meropenem | < 1 | < 1 | < 1 |
| Amikacin | 16 | 16 | < 8 |
| Gentamicin | 8 | 4 | < 2 |
| Ciprofloxacin | < 0.5 | < 0.5 | < 0.5 |
| Levofloxacin | < 1 | < 1 | < 1 |
| Tetracycline | > 8 | < 2 | < 2 |
| Chloramphenicol | < 4 | < 4 | > 16 |
| Trimethoprim/ sulfamethoxazole |
< 0.5/9.5 | < 0.5/9.5 | < 0.5/9.5 |
The biochemical test results of BM-Y and DSM 18708 are shown in Table 2. A varied reaction between these two strains was observed for acetate, polymyxin B, and glycine. The cellular fatty acid profile (Table 3) of BM-Y revealed a proportionally high level of C18:1ω7c, C14:0, and C16:0, which was consistent with DSM 18708.
TABLE 2.
Biochemical characteristics of BM-Y and DSM 18708.
| Substrates | BM-Y | DSM 18708 |
| Arginine | − | − |
| Glutaryl-glycine-arginine | − | − |
| L-leucine | + | + |
| L-pyroglutamic acid | − | − |
| Acetate | − | + |
| Colistin | + | + |
| Malonate | − | − |
| 4MU-N-acetyl-BD-glucosaminide | − | − |
| PNP-BD-glucoside | − | − |
| β-gentiobiose | − | − |
| D-galactose | − | − |
| Sorbitol | − | − |
| L-arabinose | − | − |
| Maltulose | − | − |
| Ornithine | − | − |
| Glycine-proline | − | − |
| L-arginine | − | − |
| L-phenylalanine | + | + |
| L-tryptophan | − | − |
| Adonitol | − | − |
| D-mannitol | − | − |
| Polymyxin B | − | + |
| γ-glutamyl-NA | − | − |
| Bis (PNP) phosphate | + | + |
| Dextrose | − | − |
| D-gluconic acid | − | − |
| Sucrose | − | − |
| L-rhamnose | − | − |
| N-acetyl-galactosamine | − | − |
| Urea | − | − |
| Glycine | − | + |
| L-glutamic acid | − | − |
| L-proline | − | − |
| Lysine-alanine | − | − |
| Citrate | − | − |
| α-ketoglutaric acid | + | + |
| Tiglic acid | − | − |
| L-proline-NA | + | + |
| β-allose | − | − |
| D-fructose | − | − |
| D-melibiose | − | − |
| Galacturonic acid | − | − |
| Methyl-B-glucoside | − | − |
| N-acetyl-glucosamine | − | − |
| Esculin | − | − |
+, positive; −, negative.
TABLE 3.
Cellular fatty acid profiles of BM-Y and DSM 18708.
| Fatty acids | BM-Y (%) | DSM 18708 (%) | |
| Saturated acids | C12:0 | 5.68 | 5.10 |
| C14:0 | 17.70 | 14.40 | |
| C16:0 | 6.57 | 15.60 | |
| C18:0 | tr | 1.30 | |
| Unsaturated | C18:1ω7c | 50.03 | 47.30 |
| Branched acids | C12:0 3-OH | 3.23 | 1.00 |
| cyclo-C19:0ω8c | 5.68 | 7.60 | |
| Summed features | Summed feature 1 | 3.08 | 1.33 |
| Summed feature 2 | 5.64 | 2.86 | |
tr, trace amount (< 0.5%). Summed features represent two or three fatty acids that could not be separated by the Microbial Identification System. Summed feature 1 comprises C14:0 3-OH/iso-C16:1. Summed feature 2 comprises C16:1ω7c/iso-C15 2-OH.
3.2. Phylogenetic analysis
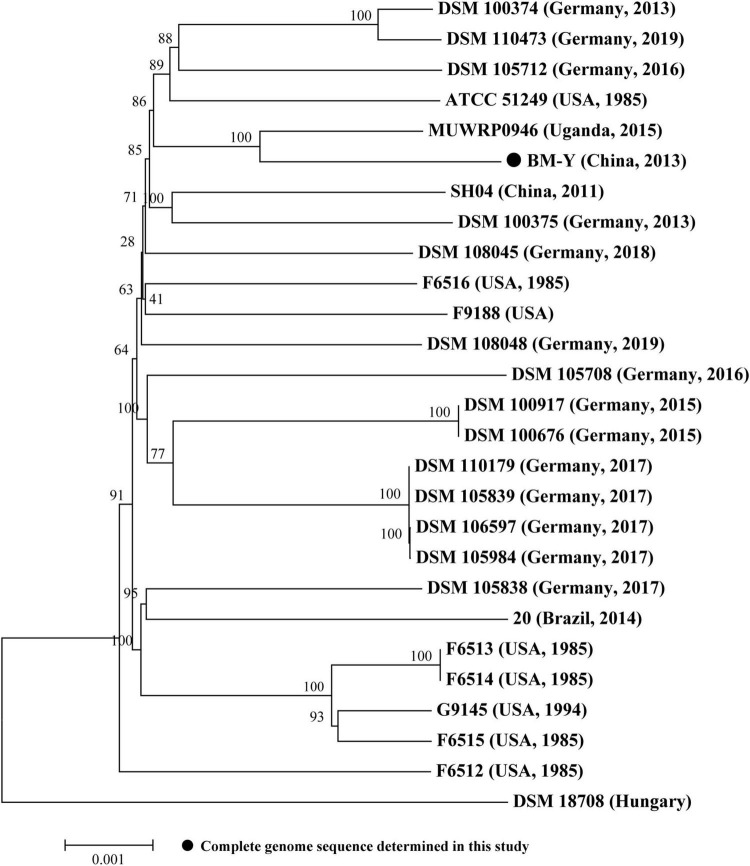
Based on the bacterial core/pan-genome analysis, we conducted the phylogenetic analysis of 27 sequenced W. chitiniclastica isolates (Supplementary Table 1), including one complete genome sequence from this study and 26 draft genome sequences from GenBank (last accessed March 17, 2023). Construction of a phylogenetic tree (Figure 3) based on core genes (1602/3597, 44.53%) of 27 W. chitiniclastica genomes revealed that all strains clustered in one subclade with the reference genome DSM 18708, and BM-Y shared the closest genetic relationship with MUWRP0946 from Uganda. This result indicated a surprisingly conserved core/pan genome without clear host or geographical clustering, suggesting a potential spread and transmission. This is in line with Kopf’s report (Kopf et al., 2022).
FIGURE 3.
Maximum-likelihood phylogenetic tree of 27 W. chitiniclastica isolates. The numbers above branches indicate bootstrap values of 1,000 times. The bar corresponds to a scale of sequence divergence.
3.3. Organization of the blaVEB–1-carrying plasmid pBM-Y
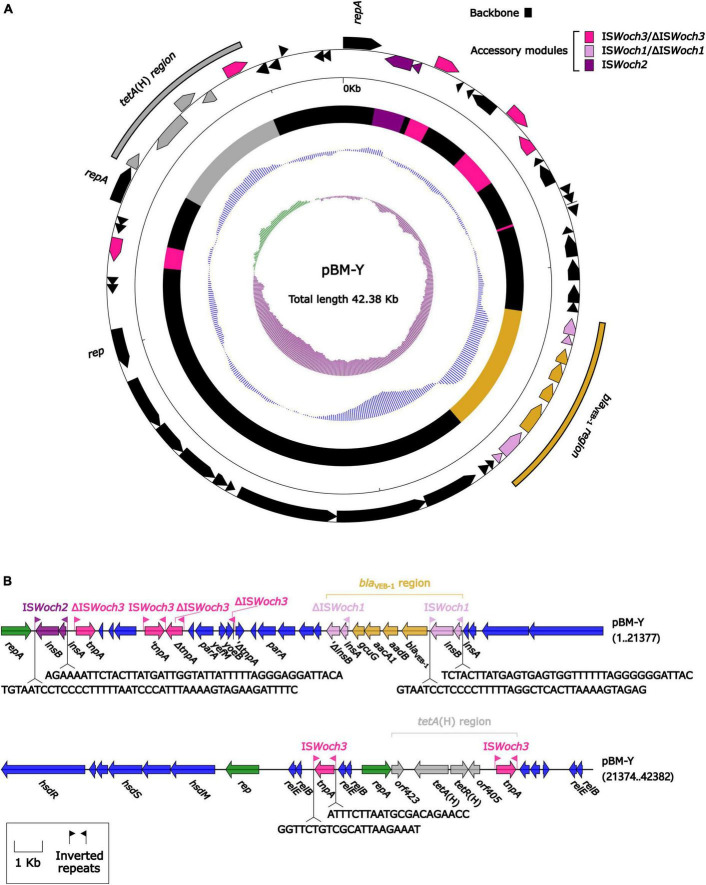
The modular structure of pBM-Y (Figure 4) was separated into the backbone and the accessory modules. The backbone contained three different rep (replication) with no similar DNA sequences (nucleotide identity < 95%) in GenBank, two parA (partition) with different sequences, three groups of relBE (a toxin–antitoxin system) with low nucleotide identities, one group of yefM/yoeB (a toxin–antitoxin system), and one group of hsdMSR (a type I restriction–modification system). No conjugal transfer genes were identified in the backbone. The accessory modules were composed of a blaVEB–1 region, a tetA(H) region, and intact or truncated insertion sequences (ISs) ISWoch2 and ISWoch3.
FIGURE 4.
Organization of the plasmid pBM-Y. (A) Schematic map of pBM-Y. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents the GC-skew [(G–C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents the GC content. (B) Linear structure of pBM-Y. Genes are denoted by arrows. The genes, accessory genetic elements (AGEs), and other features are colored based on their functional classification. The numbers in brackets indicate nucleotide positions within pBM-Y.
blaVEB–1 was originally found in Tn2000 in the plasmid pNLT1 from E. coli MG-1 (Poirel et al., 1999; Naas et al., 2001). Tn2000 was an IS26-composite transposon composed of ΔIn53 and was used as a reference herein. A detailed sequence comparison (Figure 5) was applied to the blaVEB–1-carrying ΔIn53 from Tn2000 and the blaVEB–1 region from pBM-Y. Both ΔIn53 and the blaVEB–1 region harbored the blaVEB–1 cassette (with a 133-bp attachment site of the cassette, attC), the aadB cassette (with a 60-bp attC), and the aacA1/gcuG fusion gene cassette (with a 118-bp attC) (Naas et al., 2001), but they showed three major modular variations: (i) an intact attC_aacA1/gcuG in ΔIn53, while an interrupted one in the blaVEB–1 region; (ii) a gene cassette array (GCA) qacL–aadB–aacA1–gcuG–blaVEB–1–aadB–arr-2–cmlA5–blaOXA–10–aadA1 in ΔIn53, while a GCA blaVEB–1–aadB–aacA1–gcuG in the blaVEB–1 region; and (iii) a truncated 5′-conserved segment (5′-CS) and a 3′-CS were upstream and downstream, respectively, of the GCA in ΔIn53, while a novel IS element ISWoch1 and a truncated one were upstream and downstream, respectively, of the GCA in the blaVEB–1 region. No direct repeats (DRs) were identified at the ends of the blaVEB–1 region from pBM-Y.
FIGURE 5.
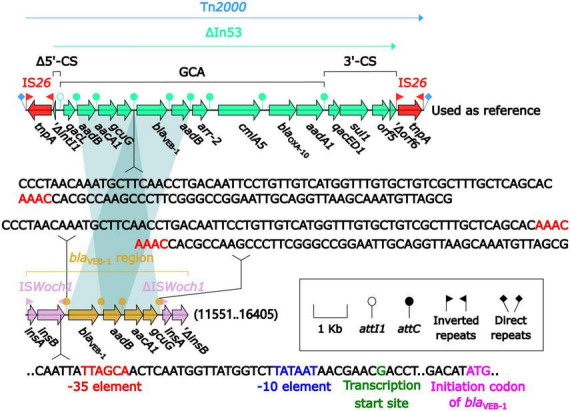
Comparison of the blaVEB–1 region from pBM-Y and blaVEB–1-carrying Tn2000. Genes are denoted by arrows. The genes, AGEs, and other features are colored based on their functional classification. The shading in light blue denotes regions of homology (nucleotide identity ≥ 95%). The 3′ fragment of attC_aacA1/gcuG is upstream of blaVEB–1, while the 5′ fragment is downstream of gcuG. The numbers in the brackets indicate the nucleotide positions within pBM-Y. Tn2000 (Naas et al., 2001) (accession number AF205943) was used as a reference.
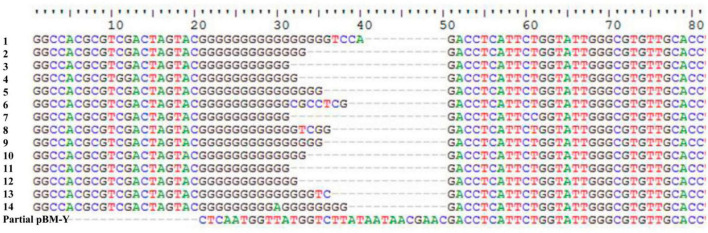
ISWoch1, ISWoch2, and ISWoch3 were named based on the first four letters of the species in which they were first discovered (Wohlfahrtiimonas chitiniclastica) and had IR of 37, 45, and 21 bp, respectively (Figure 4). ISWoch1 and ISWoch2 encoded two transposases, while ISWoch3 carried a single one. No DNA sequences similar (nucleotide identity < 95%) to these transposases were found using blastn analysis. The promoter of ISWoch1 was determined using 5′-RACE experiments. Fourteen of 15 positive clones were successfully sequenced. GACCT (Figure 6), the transcription start site of the blaVEB–1 region, was identified through multiple sequence alignments. This site was found in ISWoch1 and located 173 bp upstream of the initiation codon of blaVEB–1. A promoter with the −10/−35 region (TATAAT/TTAGCA) was located upstream of this site (Figure 5). The spacing between the −10/−35 region was 18 bp. Based on the results of the promoter prediction, ISWoch2 contained a −10/−35 region (TATCAT/ATACCA, with a 21-bp spacer), and ISWoch3 carried a −10/−35 region (TAAAAT/TTATCA, with a 19-bp spacer). ISWoch1–blaVEB–1–aadB–aacA1–gcuG–ΔISWoch1 was composed of the blaVEB–1 region (Figure 5). ISWoch2 was located upstream of the truncated ISWoch3 and downstream of the repA. Three intact ISWoch3 and three truncated ones were found in pBM-Y. ISWoch3–orf405–tetR(H)–tetA(H)–orf423 formed the tetA(H) region.
FIGURE 6.
Identification of the transcription start site of the blaVEB–1 region. Multiple sequence alignments of 14 positive clones and partial pBM-Y.
3.4. Conjugation and electroporation experiments
The blaVEB–1-carrying transconjugants could not be obtained, regardless of the number of times the conjugation experiments were performed. This result might be due to the absence of the essential conjugal transfer genes, including rlx (relaxase), oriT (origin of conjugative replication), pri (DNA primase), and cpl (coupling protein), and the type IV secretion system (T4SS) in pBM-Y.
The blaVEB–1-carrying electroporants were not obtained, no matter how many times the electroporation experiments were conducted. Because we failed to extract the plasmid pBM-Y from strain BM-Y.
4. Discussion
Wohlfahrtiimonas chitiniclastica is an emerging fly-borne zoonotic pathogen, which is often carried by different species of flies, and it causes infections in immunocompromised patients and some animals (Almuzara et al., 2011; Thaiwong et al., 2014), leading to bacteremia, sepsis, and other infections. β-lactams, quinolones, and trimethoprim/sulfamethoxazole are used to treat patients and animals infected by W. chitiniclastica (Schröttner et al., 2017).
Herein, BM-Y was isolated from the pancreas of a dead zebra in Shenzhen Safari Park in China in 2013. We speculated that W. chitiniclastica was possibly transferred to this park through either or both internal and international transport routes. Notably, W. chitiniclastica was first isolated and identified in China from C. megacephala captured from the Pudong International Airport in 2011 (Cao et al., 2013). This fly is one of the most common species found in South China (Liu et al., 2009), and it may have become the depositor of W. chitiniclastica in China. However, BM-Y shares the closest genetic relationship with MUWRP0946 from Uganda. W. chitiniclastica is more likely to be transferred from other countries to this park through international food (animal- and plant-based) trade or travel. We failed to isolate W. chitiniclastica from the flies collected in Shenzhen Safari Park, although we made several attempts.
We are not sure whether BM-Y was associated with the death of the zebra because we failed to find the maggots from the zebra. Generally, flies transmit W. chitiniclastica to the host by laying eggs that subsequently hatch into larvae inside an open wound (Schröttner et al., 2017). In this study, neither an open wound nor a maggot was found, but the flies might have carried W. chitiniclastica to deposit the eggs on the mucosal surfaces of the zebra (Schröttner et al., 2017). It is necessary to continuously monitor the spread of W. chitiniclastica, especially the ones that carry antimicrobial resistance genes, in the future.
pBM-Y is a very specific plasmid. It carries three different and novel rep genes. Currently, no other similar genes can be found in GenBank. pBM-Y encodes two toxin–antitoxin systems: RelBE and YefM/YoeB, which have been shown to increase plasmid maintenance (Gotfredsen and Gerdes, 1998). The HsdMSR type I restriction–modification system is also identified in pBM-Y. This system can stabilize plasmids by degrading the unmethylated incoming DNA (Oliveira et al., 2014). It is speculated herein that the loss of the toxin–antitoxin or restriction–modification system might lead to the instability of pBM-Y. No conjugative genes were identified; so, pBM-Y is putatively mobilized by the conjugative plasmids (Xu et al., 2021).
The MICs of ceftazidime, cefotaxime, cefepime, aztreonam, gentamicin, and tetracycline of BM-Y were higher than that of DSM 18708, which was related to the resistance genes blaVEB–1, aadB, aacA1, and tetA(H). These genes were identified in pBM-Y using the CARD and ResFinder online databases. This indicates that these genes can express effectively and are involved in the increase in MICs. The blaVEB–1 gene cassette, the aadB gene cassette, and the aacA1/gcuG fusion cassette are individual mobile units and are usually found inserted into an integron (Partridge et al., 2018). In this study, they formed a GCA (blaVEB–1–aadB–aacA1–gcuG) different from that (qacL–aadB–aacA1–gcuG–blaVEB–1–aadB–arr-2–cmlA5–blaOXA–10–aadA1) in ΔIn53 from Tn2000 (Naas et al., 2001). This demonstrates that these gene cassettes might undergo reassembly via homologous recombination. An intact and a truncated ISWoch1 were at the 5′ and 3′ ends of blaVEB–1–aadB–aacA1–gcuG in pBM-Y, respectively, which indicated that this GCA might be captured by ISWoch1 to form a translocatable unit (TU, one copy of ISWoch1 and an adjacent region) (Partridge et al., 2018). Then, this TU inserts into a recipient that lacks ISWoch1 (intermolecular replicative transposition) or targets an existing copy of ISWoch1 (intermolecular conservative transposition) (Partridge et al., 2018). However, one copy of ISWoch1 has been truncated during transposition.
Two copies of ISWoch1 participated in the movement of blaVEB–1–aadB–aacA1–gcuG. A single IS can also move an adjacent region that includes one or more resistance genes by forming a TU (Partridge et al., 2018). Herein, ISWoch3 was located upstream of orf405–tetR(H)–tetA(H)–orf423, indicating that it might be involved in the mobilization of tetA(H), and this tetA(H) region might preferentially insert next to an existing copy of ISWoch3 in the recipient molecule, generating a structure of ISWoch3–orf405–tetR(H)–tetA(H)–orf423–ISWoch3 (Partridge et al., 2018). It should also be noted that six intact or truncated copies of ISWoch3 were presented in pBM-Y, demonstrating that they participated in complex homologous recombination events and promoted the assembly of complex structures as observed in pBM-Y. A promoter was found or predicted in each of ISWoch1, ISWoch2, and ISWoch3, meaning these three ISs might provide a promoter for captured genes. Taken together, ISWoch1, ISWoch2, and ISWoch3 might promote the dissemination of drug resistance genes and affect the expression of these genes to regulate antimicrobial resistance (Noel et al., 2022). Surveillance studies for these three ISs are necessary.
The promoter region (containing the −10 and −35 elements) of the blaVEB–1 region was located in ISWoch1. The optimal spacing between the −10 and 35 elements is 16–18 bp (Aoyama et al., 1983), and in this study, it was 18 bp. This indicates that ISWoch1 provides a strong promoter for the blaVEB–1 region. Therefore, BM-Y can express β-lactamase and show resistance to ceftazidime, cefepime, and aztreonam. This means that some β-lactams are not suitable for treating infections caused by blaVEB–1-carrying W. chitiniclastica, while quinolones and trimethoprim/sulfamethoxazole can still be the first choices.
To the best of our knowledge, this is the first report of the genetic characterization of a novel blaVEB–1-carrying plasmid with three new ISs from W. chitiniclastica. The acquisition of this plasmid can give bacteria the fitness advantage for adapting to mammals and enable bacteria to acquire new antimicrobial resistance genes. The resistance of W. chitiniclastica to ceftazidime, cefepime, aztreonam, and tetracycline might enhance its ability to survive under drug selection pressure and aggravate the difficulty in treating infections caused by W. chitiniclastica. It is necessary to continuously monitor the spread of the blaVEB–1-carrying W. chitiniclastica and the possibility of acquiring other drug resistance genes in W. chitiniclastica.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
Ethics statement
The animal study was approved by the Laboratory Animal Welfare and Ethics Committee of the Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JiaG: Formal analysis, Investigation, Writing – original draft. WZ: Formal analysis, Writing – original draft. JinG: Formal analysis, Writing – original draft. LZhe: Investigation, Writing – original draft. GL: Formal analysis, Writing – original draft. FH: Resources, Writing – original draft. ML: Formal analysis, Writing – original draft. XJ: Investigation, Writing – original draft. YS: Investigation, Writing – original draft. LZhu: Conceptualization, Writing – review and editing. XG: Conceptualization, Writing – review and editing.
Acknowledgments
We are grateful to Professor Dongsheng Zhou from the State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology for the assistance with the drawing of the gene organization diagrams.
Funding Statement
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (31872486).
Abbreviations
VEB, Vietnamese extended-spectrum β -lactamase; OD, optical density; MIC, minimum inhibitory concentration; ANI, average nucleotide identity; ORF, open reading frame; RACE, rapid amplification of cDNA end; GSP, gene-specific primer; rep, replication; par, partition; IS, insertion sequence; GCA, gene cassette array; CS, conserved segment; DR, direct repeat; rlx, relaxase; oriT, origin of conjugative replication; pri, primase; cpl, coupling protein; T4SS, type IV secretion system; TU, translocatable unit.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1276314/full#supplementary-material
References
- Almuzara M. N., Palombarani S., Tuduri A., Figueroa S., Gianecini A., Sabater L., et al. (2011). First case of fulminant sepsis due to Wohlfahrtiimonas chitiniclastica. J. Clin. Microbiol. 49 2333–2335. 10.1128/JCM.00001-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Takanami M., Ohtsuka E., Taniyama Y., Marumoto R., Sato H., et al. (1983). Essential structure of E. coli promoter: Effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 11 5855–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., et al. (2013). BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 41 W29–W33. 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L., Mahobia N., Clayton J. J. (2015). Wohlfahrtiimonas chitiniclastica bacteremia associated with myiasis, United Kingdom. Emer. Infect. Dis. 21 1068–1069. 10.3201/eid2106.140007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Chen T., Xu L., Yao L., Qi J., Zhang X., et al. (2013). Complete genome sequence of Wohlfahrtiimonas chitiniclastica strain SH04, isolated from Chrysomya megacephala collected from Pudong International Airport in China. Genome Announc. 1:e0011913. 10.1128/genomeA.00119-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiano M. H. A., Silva-Rocha R. (2020). Benchmarking bacterial promoter prediction tools: Potentialities and limitations. MSystems 5 e439–e420. 10.1128/mSystems.00439-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly K. L., Freeman E., Smibert O. C., Lin B. (2019). Wohlfahrtiimonas chitiniclastica bloodstream infection due to a maggot-infested wound in a 54-year-old male. J. Glob. Infect. Dis. 11 125–126. 10.4103/jgid.jgid_58_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dios A., Jacob S., Tayal A., Fisher M. A., Dingle T. C., Hamula C. L. (2015). First report of Wohlfahrtiimonas chitiniclastica isolation from a patient with cellulitis in the United States. J. Clin. Microbiol. 53 3942–3944. 10.1128/JCM.01534-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet D., Goegebuer T., Ho E., Vandenbroucke M., Lemmens A. (2022). First case of Wohlfahrtiimonas chitiniclastica isolation from a patient with a foot ulcer infection in Belgium. Acta Clin. Belg. 78 245–247. 10.1080/17843286.2022.2090770 [DOI] [PubMed] [Google Scholar]
- Dovjak P., Kroißenbrunner M., Iglseder B. (2021). Myiasis absent Wohlfahrtiimonas chitiniclastica bacteremia in a lung cancer patient: A case report. Eur. J. Med. Res. 26:101. 10.1186/s40001-021-00576-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Zhang J., Yang L., Ding N., Yue L., Zhang X., et al. (2022). Precision methylome and in vivo methylation kinetics characterization of Klebsiella pneumoniae. Genomics Proteomics Bioinform. 20 418–434. 10.1016/j.gpb.2021.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen M., Gerdes K. (1998). The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29 1065–1076. [DOI] [PubMed] [Google Scholar]
- Guan J., Li L., Zheng L., Lu G., Wang Y., Lakoh S., et al. (2022). First report of the colistin resistance gene mcr-10.1 carried by IncpA1763–KPC plasmid pSL12517-mcr10.1 in Enterobacter cloacae in Sierra Leone. Microbiol. Spect. 10:e0112722. 10.1128/spectrum.01127-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. K., Nayduch D., Verma P., Shah B., Ghate H. V., Patole M. S., et al. (2012). Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol. Ecol. 79 581–593. 10.1111/j.1574-6941.2011.01248.x [DOI] [PubMed] [Google Scholar]
- Hladík M., Lipovy B., Kaloudova Y., Hanslianova M., Vitkova I., Deissova T., et al. (2021). Human infections by Wohlfahrtiimonas chitiniclastica: A mini-review and the first report of a burn wound infection after accidental myiasis in Central Europe. Microorganisms 9:1934. 10.3390/microorganisms9091934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman R., Fortuin F., Newton-Foot M., Singh S. (2016). First report of Wohlfahrtiimonas chitiniclastica bacteraemia in South Africa. S. Afr. Med. J. 106:1062. 10.7196/SAMJ.2016.v106i11.11449 [DOI] [Google Scholar]
- Iancu L., Necula-Petrareanu G., Purcarea C. (2020). Potential bacterial biomarkers for insect colonization in forensic cases: Preliminary quantitative data on Wohlfahrtiimonas chitiniclastica and Ignatzschineria indica dynamics. Sci. Rep. 10:8497. 10.1038/s41598-020-65471-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45 D566–D573. 10.1093/nar/gkw1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josue D.-D., Eva S., Isabel V. A., Lucas D., Marisa A., Manuel A., et al. (2015). Endocarditis associated with Wohlfahrtiimonas chitiniclastica in a short-beaked common dolphin (Delphinus delphis). J. Wildlife Dis. 51 283–286. 10.7589/2014-03-072 [DOI] [PubMed] [Google Scholar]
- Karaca M. O., Gürler M., Afacan M., Terzi M. M., Evren E., Çınar Aydın G., et al. (2022). Wohlfahrtiimonas chitiniclastica-related soft-tissue infection and osteomyelitis: A rare case report. Ulus. Travma Acil Cerrahi Derg. Turk. J. Trauma Emerg. Surg. 28 1038–1041. 10.14744/tjtes.2022.01409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanami Y., Kutsuna S., Nagashima M., Takaya S., Yamamoto K., Takeshita N., et al. (2018). Wohlfahrtiimonas chitiniclastica bacteremia hospitalized homeless man with squamous cell carcinoma. Emerg. Infect. Dis. 24 1746–1748. 10.3201/eid2409.170080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg S., Telling K., Huik K., Murruste M., Saarevet V., Pauskar M., et al. (2015). First report of Wohlfahrtiimonas chitiniclastica from soft tissue and bone infection at an unusually high northern latitude. Folia Microbiol. 60 155–158. 10.1007/s12223-014-0355-x [DOI] [PubMed] [Google Scholar]
- Kopf A., Bunk B., Coldewey S. M., Gunzer F., Riedel T., Schröttner P. (2021). Identification and antibiotic profiling of Wohlfahrtiimonas chitiniclastica, an underestimated human pathogen. Front. Microbiol. 12:712775. 10.3389/fmicb.2021.712775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf A., Bunk B., Coldewey S. M., Gunzer F., Riedel T., Schröttner P. (2022). Comparative genomic analysis of the human pathogen Wohlfahrtiimonas Chitiniclastica provides insight into the identification of antimicrobial resistance genotypes and potential virulence traits. Front. Cell. Infect. Microbiol. 12:912427. 10.3389/fcimb.2022.912427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Jiang X., Yang T., Ju Y., Yin Z., Yue L., et al. (2022). Genomic epidemiology of carbapenemase-producing Klebsiella pneumoniae in China. Genomics Proteomics Bioinform. 20 1154–1167. 10.1016/j.gpb.2022.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics (Oxford, England) 22 1658–1659. [DOI] [PubMed] [Google Scholar]
- Li W., Jaroszewski L., Godzik A. (2001). Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics (Oxford, England) 17 282–283. [DOI] [PubMed] [Google Scholar]
- Li W., Jaroszewski L., Godzik A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics (Oxford, England) 18 77–82. [DOI] [PubMed] [Google Scholar]
- Liang Q., Jiang X., Hu L., Yin Z., Gao B., Zhao Y., et al. (2018). Sequencing and genomic diversity analysis of IncHI5 plasmids. Front. Microbiol. 9:3318. 10.3389/fmicb.2018.03318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Shi Y., Wang H., Zhang R. (2009). Determination of malathion levels and its effect on the development of Chrysomya megacephala (Fabricius) in South China. Forensic Sci. Int. 192 14–18. 10.1016/j.forsciint.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Matos J., Queiroga A. P., de Oliveira Pedroza Bindi dos Reis C. C., Ribeiro R. L., Teixeira L. M., Albano R. M., et al. (2016). First report of the emerging zoonotic agent Wohlfahrtiimonas chitiniclastica isolated from a retail frozen chicken in Rio de Janeiro, Brazil. Antonie Van Leeuwenhoek 109 729–734. 10.1007/s10482-016-0673-x [DOI] [PubMed] [Google Scholar]
- Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. (2009). INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics (Oxford, England) 25 1096–1098. 10.1093/bioinformatics/btp105 [DOI] [PubMed] [Google Scholar]
- Naas T., Benaoudia F., Massuard S., Nordmann P. (2000). Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J. Antimicrob. Chemother. 46 703–711. [DOI] [PubMed] [Google Scholar]
- Naas T., Mikami Y., Imai T., Poirel L., Nordmann P. (2001). Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T., Poirel L., Karim A., Nordmann P. (1999). Molecular characterization of In50, a class 1 integron encoding the gene for the extended-spectrum β-lactamase VEB-1 in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 176 411–419. [DOI] [PubMed] [Google Scholar]
- Noel H. R., Petrey J. R., Palmer L. D. (2022). Mobile genetic elements in Acinetobacter antibiotic-resistance acquisition and dissemination. Ann. N. Y. Acad. Sci. 1518 166–182. 10.1111/nyas.14918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi M., Bankowski M. J., Pien F. D. (2016). Wohlfahrtiimonas chitiniclastica infections in 2 elderly patients, Hawaii. Emerg. Infect. Dis. 22 567–568. 10.3201/eid2203.151701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou O., Liakopoulos A., Phee L. M., Betts J., Mevius D., Wareham D. W. (2016). Providencia stuartii isolates from Greece: Co-carriage of cephalosporin (blaSHV–5, blaVEB–1), carbapenem (blaVIM–1), and aminoglycoside (rmtB) resistance determinants by a multidrug-resistant outbreak clone. Microb. Drug Resistance (Larchmont, N.Y.) 22 379–386. 10.1089/mdr.2015.0215 [DOI] [PubMed] [Google Scholar]
- Oliveira P. H., Touchon M., Rocha E. P. C. (2014). The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 42 10618–10631. 10.1093/nar/gku734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pablo-Marcos D., Siller-Ruiz M., Fernandez-Ayala M., Agüero J., Calvo J. (2019). First case of infection by Wohlfahrtiimonas chitiniclastica described in Spain. Rev. Esp. Quimioter. 32 558–560. [PMC free article] [PubMed] [Google Scholar]
- Papagiannitsis C. C., Miriagou V., Kotsakis S. D., Tzelepi E., Vatopoulos A. C., Petinaki E., et al. (2012). Characterization of a transmissible plasmid encoding VEB-1 and VIM-1 in Proteus mirabilis. Antimicrob. Agents Chemother. 56 4024–4025. 10.1128/AAC.00470-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S. R., Kwong S. M., Firth N., Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 e88–e17. 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Ingti B., Bhattacharjee D., Maurya A. P., Dhar D., Chakravarty A., et al. (2017). An unusual occurrence of plasmid-mediated blaOXA–23 carbapenemase in clinical isolates of Escherichia coli from India. Int. J. Antimicrob. Agents 49 642–645. 10.1016/j.ijantimicag.2017.01.012 [DOI] [PubMed] [Google Scholar]
- Poirel L., Naas T., Guibert M., Chaibi E. B., Labia R., Nordmann P. (1999). Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Gao Y., Wang G., Li L., Li L., Zhao X., et al. (2016). Identification of Wohlfahrtiimonas chitiniclastica isolated from an infected cow with hoof fetlow, China. Infect. Genetics Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 41 174–176. 10.1016/j.meegid.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Rebaudet S., Genot S., Renvoise A., Fournier P. E., Stein A. (2009). Wohlfahrtiimonas chitiniclastica bacteremia in homeless woman. Emer. Infect. Dis. 15 985–987. 10.3201/eid1506.080232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröttner P., Rudolph W. W., Damme U., Lotz C., Jacobs E., Gunzer F. (2017). Wohlfahrtiimonas chitiniclastica: Current insights into an emerging human pathogen. Epidemiol. Infect. 145 1292–1303. 10.1017/S0950268816003411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebor E., de Curraize C., Varin V., Magallon A., Neuwirth C. (2021). Mobilisation of plasmid-mediated blaVEB–1 gene cassette into distinct genomic islands of Proteus mirabilis after ceftazidime exposure. J. Glob. Antimicrob. Resist. 27 26–30. 10.1016/j.jgar.2021.07.011 [DOI] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder S., Singh P., Goldman J. (2020). Emerging pathogens: A case of Wohlfahrtiimonas chitiniclastica and Ignatzschineria indica bacteremia. IDCases 19:e00723. 10.1016/j.idcr.2020.e00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraiya S., Zuraina N., Ahmad F., Rahman Z. A. (2017). Fatal Wohlfahrtiimonas chitiniclastica bacteremia in an immunocompromised patient. Clin. Microbiol. Newslett. 39 172–173. 10.1016/j.clinmicnews.2017.07.003 [DOI] [Google Scholar]
- Suryalatha K., John J., Thomas S. (2015). Wohlfahrtiimonas chitiniclastica-associated osteomyelitis: A rare case report. Fut. Microbiol. 10 1107–1109. 10.2217/fmb.15.44 [DOI] [PubMed] [Google Scholar]
- Thaiwong T., Kettler N. M., Lim A., Dirkse H., Kiupel M. (2014). First report of emerging zoonotic pathogen Wohlfahrtiimonas chitiniclastica in the United States. J. Clin. Microbiol. 52 2245–2247. 10.1128/JCM.00382-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittsler R. P., Sandholzer L. A. (1936). The use of semi-solid agar for the detection of bacterial motility. J. Bacteriol. 31 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth E. M., Schumann P., Borsodi A. K., Kéki Z., Kovács A. L., Márialigeti K. (2008). Wohlfahrtiimonas chitiniclastica gen. nov., sp. nov., a new gammaproteobacterium isolated from Wohlfahrtia magnifica (Diptera: Sarcophagidae). Int. J. Systematic Evol. Microbiol. 58(Pt 4) 976–981. 10.1099/ijs.0.65324-0 [DOI] [PubMed] [Google Scholar]
- Wang P., Jiang X., Mu K., Jing Y., Yin Z., Cui Y., et al. (2022). DANMEL: A manually curated reference database for analyzing mobile genetic elements associated with bacterial drug resistance. mLife 1 460–464. 10.1002/mlf2.12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang J., Wang M., Liu M., Liu G., Qu H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13:119. 10.1186/s13073-021-00936-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Li R., Lin D., Zhou Y., Fu A., Ding Q., et al. (2016). Characterization of an IncA/C multidrug resistance plasmid in Vibrio alginolyticus. Antimicrob. Agents Chemother. 60 3232–3235. 10.1128/AAC.00300-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Li M., Zhu L., Hua F., Ji X., Sun Y., et al. (2016). Complete genome sequence of Wohlfahrtiimonas chitiniclastica strain BM-Y, isolated from the pancreas of a zebra in China. Genome Announc. 4 e643–e616. 10.1128/genomeA.00643-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.