Abstract
A culture-independent molecular phylogenetic approach was used to survey constituents of microbial communities associated with an aquifer contaminated with hydrocarbons (mainly jet fuel) and chlorinated solvents undergoing intrinsic bioremediation. Samples were obtained from three redox zones: methanogenic, methanogenic-sulfate reducing, and iron or sulfate reducing. Small-subunit rRNA genes were amplified directly from aquifer material DNA by PCR with universally conserved or Bacteria- or Archaea-specific primers and were cloned. A total of 812 clones were screened by restriction fragment length polymorphisms (RFLP), approximately 50% of which were unique. All RFLP types that occurred more than once in the libraries, as well as many of the unique types, were sequenced. A total of 104 (94 bacterial and 10 archaeal) sequence types were determined. Of the 94 bacterial sequence types, 10 have no phylogenetic association with known taxonomic divisions and are phylogenetically grouped in six novel division level groups (candidate divisions WS1 to WS6); 21 belong to four recently described candidate divisions with no cultivated representatives (OP5, OP8, OP10, and OP11); and 63 are phylogenetically associated with 10 well-recognized divisions. The physiology of two particularly abundant sequence types obtained from the methanogenic zone could be inferred from their phylogenetic association with groups of microorganisms with a consistent phenotype. One of these sequence types is associated with the genus Syntrophus; Syntrophus spp. produce energy from the anaerobic oxidation of organic acids, with the production of acetate and hydrogen. The organism represented by the other sequence type is closely related to Methanosaeta spp., which are known to be capable of energy generation only through aceticlastic methanogenesis. We hypothesize, therefore, that the terminal step of hydrocarbon degradation in the methanogenic zone of the aquifer is aceticlastic methanogenesis and that the microorganisms represented by these two sequence types occur in syntrophic association.
The use of natural attenuation as a cleanup method for underground storage tank sites with petroleum-contaminated soil and groundwater has increased dramatically over the last few years. Since 1995, natural attenuation has been the most common treatment for contaminated groundwater and the second most common treatment for contaminated soil at these sites. About 17,000 contaminated groundwater sites (47% of active sites) and 29,000 contaminated soil sites (28% of active sites) are being remediated through natural attenuation (43). In order for a regulatory agency to approve the cleanup of a contaminated site via natural attenuation, the occurrence of natural attenuation must be demonstrated by various lines of evidence. One such line of evidence is historical groundwater and/or soil chemistry data that demonstrate decreasing contaminant mass and/or concentration over time at relevant sampling points (44). This decrease may be due to contaminant sorption, dilution, volatilization, and nonbiological and biological breakdown. The biological breakdown, or intrinsic bioremediation, of hydrocarbons is particularly beneficial, because it ultimately converts hydrocarbons to carbon dioxide, water, and methane, rather than simply repartitioning the hydrocarbons. Methods for demonstrating biological activity at a site include sampling for intermediates of biological hydrocarbon metabolism, demonstration of electron acceptor depletion, microcosm studies, and description of the microbial community at the site, specifically identifying organisms believed to be responsible for the metabolism of contaminants (5, 19, 44).
The identification of organisms has traditionally been achieved by cultivation techniques such as plate counting. However, because typically >99% of naturally occurring microorganisms are not cultivated by standard techniques (2), alternate methods must be used to describe community constituents. One such method is the identification of rRNA genes (rDNA) in DNA extracted directly from the environment, typically via PCR amplification, cloning, and sequencing (2, 31). A number of molecular microbial diversity studies of soil communities have been reported (6, 7, 20, 23, 24, 39), but there has been no similar analysis of a contaminated aquifer undergoing intrinsic bioremediation.
The subjects of the present study are the microbial communities associated with a contaminated aquifer located on the former Wurtsmith Air Force Base in Michigan. Waste fuels and chlorinated solvents were applied to site FT-2 (Fig. 1) from 1952 to 1986 for the purpose of fire-fighting training. Concentrations of benzene, toluene, ethylbenzene, and xylenes in the plume range from 20 to 1,000 μg/liter; concentrations of dichloroethane, chloroethane, and vinyl chloride range from 2 to 1,000 μg/liter (14, 45). Most of the contamination is associated with aquifer solids; total petroleum hydrocarbon analyses showed an average of 13,650 mg/kg between 4.5 and 5.7 m below the ground surface. The aquifer, composed of alternating eolian sands and glacial outwash material, averages 21 m thick and is highly permeable, with a hydraulic conductivity of approximately 30 m/day and a groundwater velocity of 0.1 to 0.3 m/day (15, 45). The water table at the sampling site is approximately 5.2 m below ground surface, fluctuating 0.3 to 1 m annually, and the groundwater temperature is nearly constant at 10 to 12°C. A number of organic acids have been detected at this site (3), providing evidence, along with stabilization of the plume and depletion of electron acceptors in the plume relative to the uncontaminated aquifer, for the occurrence of intrinsic bioremediation.
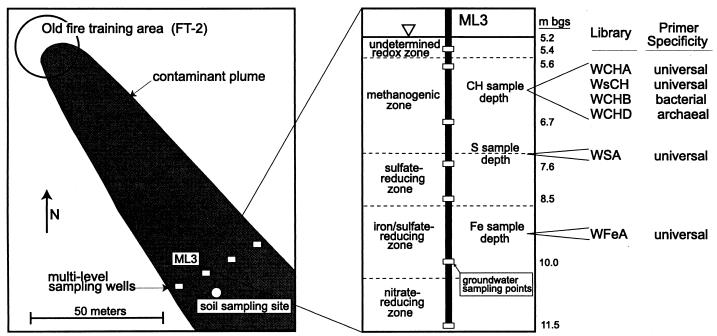
FIG. 1.
Diagram of sampling locations and profile of clone libraries. Approximate redox zone and soil sample depths are indicated along with groundwater sampling points from sampling well ML3. The water table (∇) is approximately 5.2 m below the ground surface (m bgs).
The general view of the course of intrinsic bioremediation of a hydrocarbon-contaminated aquifer is stepwise microbial utilization of electron acceptors, with sequential utilization of the electron acceptors that provide the most potential energy upon concomitant oxidation of the hydrocarbon. Thus, oxygen is utilized first, and then nitrate, bioavailable ferric iron, and sulfate are utilized. After sulfate is depleted, methanogenesis is thought to be the final mode of carbon cycling (5). Recent studies have shown, however, that sulfate reduction and methanogenesis may take place concurrently (32). Also, aquifer heterogeneity and influx of electron acceptors, such as nitrate and sulfate, from rainwater complicate the picture of redox zone distribution.
To describe the microbial constituents of the Wurtsmith aquifer, we conducted a molecular phylogenetic survey of the main redox zones. The dominant redox processes in the three zones analyzed were proposed, on the basis of geochemical characterization, to be methanogenesis, methanogenesis-sulfate reduction, and iron or sulfate reduction. Small-subunit (16S or 18S) rDNAs were amplified by PCR from DNA extracts of core sections from these three zones, and clone libraries were prepared from the PCR products for sequencing. We identified metabolically important members of the community by phylogenetic comparison with cultivated organisms and developed the hypothesis that carbon flow in this environment occurs predominantly through aceticlastic methanogenesis.
MATERIALS AND METHODS
Sample collection.
An extensive geochemical characterization of the FT-2-associated plume can be found elsewhere (14, 45). Both soil sample collection and groundwater sample collection and analysis were conducted during the week of 15 to 21 October 1996. Groundwater chemistry was determined with water extracted from multilevel well ML3, approximately 100 m downgradient from the FT-2 source, with sampling ports at the depths indicated in Fig. 1. Soil samples were collected with a Geoprobe hydraulic punch 10 m downgradient from ML3. Cores were brought to the surface, capped, placed in a nitrogen-filled glove bag for subsampling, and frozen after subsampling. Although cores were processed as aseptically as possible, the exterior of the cores was presumed to be contaminated, and samples for analysis were taken from the interior of the cores.
The three cores analyzed in this study, CH, S, and Fe, were taken from approximately 6.1, 7.3, and 9.2 m below the ground surface, respectively (Fig. 1). The redox geochemistry of these cores was inferred from the chemistry of the groundwater from ML3. Core CH appeared to be methanogenic, core S appeared to be from the transition region between the sulfate-reducing and methanogenic zones, and core Fe appeared to be from a region in the aquifer where both sulfate reduction and iron reduction were occurring.
DNA extraction.
Community nucleic acids were extracted from sediment samples by use of a bead beating protocol based on that of Barns et al. (4). Briefly, 0.5 to 1.0 g of sediment was resuspended in modified 2× buffer A (200 mM Tris [pH 8.0], 50 mM EDTA, 200 mM NaCl, 2 mM sodium citrate, 10 mM CaCl2)–pyrophosphate (0.2%)–poly(A) (200 μg/ml)–lysozyme (3 mg/ml) in a 2-ml screw-cap tube and incubated for 40 min at 37°C. Proteinase K (to 1.2 mg/ml) and sodium dodecyl sulfate (to 0.3% [wt/vol]) were added, and the mixture was incubated for 30 min at 50°C. Samples were reciprocated on a Mini-Beadbeater (Biospec) at low speed for 2 min in the presence of 50% (vol/vol) phenol-chloroform-isoamyl alcohol (24:24:1), 5% (wt/vol) sodium dodecyl sulfate, and approximately 0.3 g of acid-washed zirconium-silica beads (0.1 mm diameter). Lysates were extracted with phenol-chloroform. Sodium acetate was added to 0.3 M, and nucleic acids were precipitated from the solution by the addition of 1 volume of isopropanol. Community nucleic acids were purified by passage through a ChromaSpin+TE-1000 column (Clontech Laboratories, Inc.).
PCR and cloning.
Community rDNAs were PCR amplified from 1 to 50 ng of bulk DNA in reaction mixtures containing (as final concentrations) 1× PCR buffer II (Perkin Elmer), 2.5 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, a 300 nM concentration of each forward and reverse primer, and 0.025 U of AmpliTaq Gold (Perkin Elmer) per μl. Reaction mixtures were incubated in a model PT-100 thermal cycler (MJ Research Inc.) at 94°C for 12 min (for initial denaturation and activation of AmpliTaq Gold), followed by 30 cycles at 94°C for 1 min, 50°C for 45 s, and 72°C for 2 min and then by a final extension period of 12 min at 72°C. Six clone libraries were prepared (Fig. 1). For all clone libraries, rDNAs were amplified with universal reverse oligonucleotide primer 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′). The universal forward oligonucleotide primer for clone libraries WCHA, WsCH, WSA, and WFeA was 533F (5′-GTG CCA GCM GCC GCG GTA A-3′). For clone library WCHB, the forward primer was 27F (specific for Bacteria) (5′-AGA GTT TGA TCC TGG CTC AG-3′) (21). For clone library WCHD, the forward primer was 25F (specific for Archaea) (5′-CYG GTT GAT CCT GCC RG-3′). PCR products were separated on a 0.7% agarose gel, excised, purified with an UltraClean kit (MoBio Laboratories, Inc.), and eluted in 10 mM Tris (pH 8.0). The purified PCR products were cloned with an Original TA Cloning Kit or a TOPO TA Cloning Kit in accordance with the manufacturer’s instructions (Invitrogen Corp.). Plasmid DNAs containing inserts were prepared for restriction fragment length polymorphism (RFLP) analysis and sequencing (see below) by use of a 96-well alkaline lysis procedure modified (18) from that of Ng et al. (27).
Screening of rDNA clones by RFLP.
rDNA inserts from recombinant clones were reamplified by PCR with reaction mixtures containing (as final concentrations) 1× Pfu reaction buffer (Stratagene), 2.5 mM MgSO4, a 100 μM concentration of each deoxynucleoside triphosphate, a 150 nM concentration of each vector-specific forward and reverse primer, ca. 0.01 U of Pfu DNA polymerase per μl, and 50 to 400 pg of purified plasmid per μl as a template. The cycle profile was the same as that for the initial amplification of the rDNA (see above). Aliquots (25 μl) of crude reamplified rDNA PCR products were digested with 1.5 U of each of the 4-base-specific restriction endonucleases HinPII and MspI in 1× NEB buffer 2 (New England Biolabs)–0.01% Triton X-100 in a final volume of 30 μl for 3 h at 37°C. Digested fragments were separated by agarose (2% MetaPhor; FMC BioProducts) gel electrophoresis and visualized by staining with ethidium bromide and UV illumination. RFLP patterns for each library were grouped visually, and representatives were selected (see Results) for sequencing.
Sequencing of rDNA clones.
Plasmid templates from representative clones were sequenced with an ABI 373 Stretch DNA sequencer (Dye-Terminator Cycle Sequencing Ready Reaction FS Kit; PE Applied Biosystems) in accordance with the manufacturer’s instructions. Primers used for sequencing included vector primers and small-subunit (SSU) rDNA primers: 533F (see above), 356F (5′-ACT CCT ACG GGA GGC AGC A-3′), 907R (5′-CCG TCA ATT CCT TTR AGT TT-3′), and 1220R (5′-GTA GCR CGT GTG TMG CCC-3′).
Phylogenetic analyses and chimera detection.
Sequences were compared to available databases by use of the BLAST (Basic Local Alignment Search Tool) network service (1) to determine their approximate phylogenetic affiliations. Partial sequences were compiled in Sequence Navigator (PE Applied Biosystems); compiled sequences were aligned by use of the ARB database (40). Chimeric sequences were identified by use of the CHECK_CHIMERA program (25), by secondary-structure anomalies, and by branching-order discrepancies of independently inferred regions of the alignment as previously described (18). Sequence alignments used for phylogenetic inference were minimized by use of the Lane mask (22) for bacterial data sets (1,241 positions were used) or an archaeal mask designed by use of the ARB database (1,250 positions were used). All presented dendrograms were constructed by use of the ARB database with evolutionary distance (neighbor-joining algorithms with Olsen correction). The robustness of inferred topologies was tested by bootstrap resampling of trees calculated with evolutionary distance (test version 4.0d61 of PAUP*, written by David L. Swofford; neighbor-joining algorithm with either Kimura two-parameter correction or maximum-likelihood correction with an empirically determined gamma distribution model of site-to-site rate variation and empirically determined base frequencies), parsimony (test version 4.0d61 of PAUP*; heuristic search), and maximum likelihood (fastDNAml) (25). The data set used for the analysis is available at http://crab2.berkeley.edu/pacelab/179.htm.
Nucleotide sequence accession numbers.
The sequences of the rDNA clones have GenBank accession no. AF050526 to AF050629.
RESULTS
Analysis of libraries and compilation of sequence data.
To determine the microbial diversity associated with different redox zones in the contaminated aquifer at the former Wurtsmith Air Force Base, six rDNA clone libraries were prepared by PCR with bulk DNA extracted from aquifer samples and analyzed. Figure 1 shows a schematic diagram of the sampling location and the designations of clone libraries associated with the different sampling depths and proposed redox zones. Four libraries (two universal, one bacterial, and one archaeal) were prepared from the CH sample (methanogenic zone). One universal library was prepared from each of the S (sulfate-reducing–methanogenic zone) and Fe (iron- or sulfate-reducing zone) samples. One or two microtiter plates of clones (96 to 192 clones) were screened per library.
A total of 812 clones containing rDNA inserts from the six libraries were screened by analysis of RFLP patterns. Approximately 50% of the clones screened by RFLP type (with the exception of the archaeal library; see below) were unique, indicating that the microbial communities associated with the three redox zones studied are complex. Representative clones of all RFLP types that occurred more than once in a given library, as well as representatives of some of the unique RFLP types, were fully sequenced. Only four chimeric sequences were identified; all were clones with unique RFLP types in the WCHA library and were excluded from subsequent analyses. Groups of two or more highly related sequences (≥98% identical) were considered to belong to the same sequence type, resulting in the identification of 104 putatively nonchimeric sequence types from the six clone libraries. Table 1 summarizes the distribution of sequence types by divisions (main lines of descent within domains) and percent abundance in each library. Several sequences (WCHA2-01, WCHB1-60, and WCHB1-77) have unusual secondary structural features, such as large insertions or deletions relative to most 16S rDNAs, usually associated with variable regions of the molecule. Sequence WCHA2-01 has a 47-base insertion between nucleotides 597 and 598 (Escherichia coli numbering), transforming what is a stem-loop structure at this location in most SSU rRNAs into a proposed cruciform structure.
TABLE 1.
Sequence type distribution
| Sequence typea | % of type found in the following libraryb:
|
Putative division | Database match (≥95% identity) | |||||
|---|---|---|---|---|---|---|---|---|
| WCHA | WsCH | WCHB | WCHD | WSA | WFeA | |||
| WCHA2-13 | <1 | Acidobacteria | ||||||
| WCHB1-08, -18 | 1 | Actinobacteria | ||||||
| WCHB1-81 | <1 | 1 | Actinobacteria | |||||
| WFeA1-02 | 3 | Actinobacteria | ||||||
| WCHA1-01, -14, -2-47 | <1 | Cytophagales | ||||||
| WCHB1-29, -32, -53 | 1 | Cytophagales | ||||||
| WCHB1-69 | 2 | 1 | Cytophagales | |||||
| WsCH39 | 1 | Cytophagales | 95% Chryseobacterium indologenes (M58773) | |||||
| WCHA1-69 | 3 | 1 | 1 | Green nonsulfur bacteria | ||||
| WCHB1-05 | 5 | 1 | Green nonsulfur bacteria | |||||
| WCHB1-31 | 1 | 3 | 2 | Green nonsulfur bacteria | ||||
| WCHB1-43, -44, -50, -57, -80 | 1 | Green nonsulfur bacteria | ||||||
| WCHB1-62 | 1 | 1 | 2 | Green nonsulfur bacteria | ||||
| WCHB1-63 | 1 | Green nonsulfur bacteria | 98% Anaerobic digestor clone HB28 (U81745) | |||||
| WCHA1-94 | <1 | Green sulfur bacteria | ||||||
| WCHA1-17, -45, -53 | <1 | Low-G+C gram positive | ||||||
| WCHB1-20 | 1 | Low-G+C gram positive | ||||||
| WCHB1-21 | <1 | 6 | 2 | Low-G+C gram positive | ||||
| WCHB1-49, -54, -84 | 1 | Low-G+C gram positive | ||||||
| WCHB1-71 | 1 | 1 | Low-G+C gram positive | |||||
| WCHB1-77 | 1 | Low-G+C gram positive | 95% Desulfotomaculum orientis (Y11570) | |||||
| WCHB1-82 | 1 | Low-G+C gram positive | ||||||
| WCHB1-89 | 1 | Low-G+C gram positive | 95% Anaerobic digestor clone HB76 (U81754) | |||||
| WFeA1-16 | 4 | Low-G+C gram positive | 99% Exiguobacterium aurantiacum (X70316) | |||||
| WsCH1, 5, 8 | 1 | Low-G+C gram positive | ||||||
| WCHA1-79 | <1 | 2 | OP10 | |||||
| WsCH12 | 1 | OP10 | ||||||
| WCHA1-09 | 2 | 1 | OP11 | |||||
| WCHA1-11, -16, -20, -24; 2-01, -26 | <1 | OP11 | ||||||
| WCHB1-07 | 2 | OP11 | ||||||
| WCHB1-11 | <1 | 1 | OP11 | |||||
| WCHB1-26 | 13 | 18 | 2 | 6 | OP11 | |||
| WCHB1-56 | <1 | 2 | 1 | OP11 | ||||
| WCHB1-58 | 1 | OP11 | ||||||
| WCHB1-64 | <1 | 3 | OP11 | |||||
| WCHB1-02 | 1 | OP5 | ||||||
| WCHB1-03 | 3 | 2 | 8 | 3 | 8 | OP5 | ||
| WCHA1-39 | <1 | OP8 | ||||||
| WCHA1-83 | 1 | OP8 | ||||||
| WFeA1-35 | 1 | OP8 | ||||||
| WFeA1-59 | 1 | 7 | OP8 | |||||
| WCHA1-65 | <1 | Proteobacteria (α) | ||||||
| WCHB1-55 | 1 | Proteobacteria (α) | ||||||
| WCHB1-87 | 1 | Proteobacteria (α) | ||||||
| WCHA1-52 | <1 | Proteobacteria (β) | 98% Azoarcus sp. strain BS1-14 (AF011348) | |||||
| WCHA1-62 | 1 | 6 | 1 | Proteobacteria (β) | 98% Denitrifying Fe-oxidizing bacterium (U51101) | |||
| WFeA1-06c | 17 | Proteobacteria (β) | 97% Duganella zoogloeoides (X74914) | |||||
| WsCH66 | 1 | Proteobacteria (β) | ||||||
| WCHA1-76 | <1 | Proteobacteria (γ) | 95% Legionella birminghamensis (Z49717) | |||||
| WCHA1-85 | <1 | Proteobacteria (γ) | ||||||
| WCHA1-07 | <1 | Proteobacteria (δ) | 95% Syntrophus gentianae (X85132) | |||||
| WCHB1-12 | 7 | 16 | 7 | 4 | Proteobacteria (δ) | 95% Syntrophus gentianae (X85132) | ||
| WCHB1-27, -67 | 1 | Proteobacteria (δ) | ||||||
| WFeA1-10 | 4 | Proteobacteria (δ) | ||||||
| WsCH54 | 2 | Proteobacteria (δ) | ||||||
| WCHB1-30 | 4 | Spirochetes | ||||||
| WCHB1-40, -91 | 1 | Spirochetes | ||||||
| WCHA1-89 | <1 | Termite group I | ||||||
| WCHA1-33 | <1 | Verrucomicrobia | ||||||
| WCHA1-80 | 2 | 1 | Verrucomicrobia | |||||
| WCHB1-25 | 2 | Verrucomicrobia | ||||||
| WCHB1-41 | 1 | 1 | Verrucomicrobia | |||||
| WCHD3-88 | 1 | 1 | Verrucomicrobia | |||||
| WCHA1-48 | <1 | WS1 | ||||||
| WCHA1-78 | 2 | 2 | 1 | WS1 | ||||
| WCHA1-37 | <1 | WS2 | ||||||
| WCHA1-56 | <1 | WS3 | ||||||
| WsCH31 | 3 | WS4 | ||||||
| WCHB1-60 | 1 | WS5 | ||||||
| WCHA | WsCH | WCHB | WCHD | WSA | WFeA | |||
| WCHB1-01 | 13 | WS6 | ||||||
| WCHB1-06 | 7 | WS6 | ||||||
| WCHB1-15 | 4 | WS6 | ||||||
| WCHA1-38 | <1 | Crenarchaeota | 96% Marine clone C20 (U71114) | |||||
| WCHA1-57, 2-08 | <1 | Euryarchaeota | ||||||
| WCHD3-02 | 8 | Euryarchaeota | ||||||
| WCHD3-03 | 8 | 3 | 81 | 9 | Euryarchaeota | 97% Methanosaeta concilii (X16932) | ||
| WCHD3-07 | 3 | Euryarchaeota | 97% Methanospirillum sp. (L48407) | |||||
| WCHD3-16, -33 | 1 | Euryarchaeota | ||||||
| WCHD3-30 | <1 | 1 | Euryarchaeota | |||||
| WCHD3-34 | 2 | Euryarchaeota | ||||||
Groups of two or more highly related sequences (≥98% identical) are considered to belong to the same sequence type. Sequences listed on the same line represent different types belonging to the same division and detected only once within a library.
Based on direct sequence comparisons or inferred from RFLP patterns. Percentages do not add up to 100% because not all unique RFLPs were sequenced.
Potential contaminant (42).
Comparative analyses.
Comparative analyses of the Wurtsmith aquifer sequences to known 16S rDNA sequences revealed a broad spectrum of bacterial and, to a lesser extent, archaeal diversity (Table 1). Of the 104 sequence types identified, 94 were bacterial and 10 were archaeal. No eucaryal rDNAs were identified in any of the libraries. Primers specific for Bacteria and Archaea amplified rDNAs belonging to their respective domains, with the exception of bacterial clone WCHD3-88, which exhibited mispriming of the forward primer specific for Archaea around nucleotide 140 (E. coli numbering). Of the 104 sequence types, 42 were ≥90% identical to rDNA sequences available in GenBank as of March 1998 (only 7 of these were ≥97% identical). Of the 104 sequence types, 62 were <90% identical to known rDNA sequences.
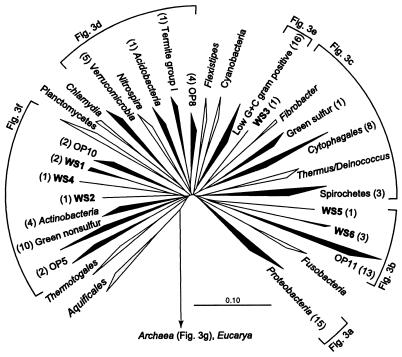
Figure 2 is an evolutionary distance tree of the bacterial domain showing the distribution of the 94 Wurtsmith bacterial sequence types among bacterial divisions, a rough description of the bacterial biodiversity at this site. The Wurtsmith sequences fell into 20 of the 35 to 40 known main phylogenetic divisions (16). The majority (63 of 94) of the bacterial sequence types were phylogenetically associated with 10 of the well-recognized divisions. Twenty-one of the novel bacterial sequence types were affiliated with four recently described candidate divisions that have no cultivated representatives (OP divisions; see Discussion). A candidate division is a monophyletic group of rDNA sequences with no specific association with known divisions. Ten of the 94 bacterial sequence types had no specific association with any of the known divisions or candidate divisions and were phylogenetically divided among six novel division level groups (candidate divisions WS1 to WS6). No consistent branching order of divisions in the tree shown in Fig. 2 could be established by bootstrap resampling, with the exception of the basal placement of the Aquificales division and the distant but specific relationships of the Chlamydia division to the Verrucomicrobia division and the green sulfur bacteria division to the Cytophagales division.
FIG. 2.
Diagrammatic radial representation of selected putative bacterial divisions. Filled wedges indicate that sequences representing these divisions were obtained in the present study. The numbers in parentheses next to the divisions indicate the number of sequences obtained from the respective divisions. Nonfilled wedges are reference divisions with no representation among the Wurtsmith clones. Wedges labeled WS indicate previously undetected candidate divisions.
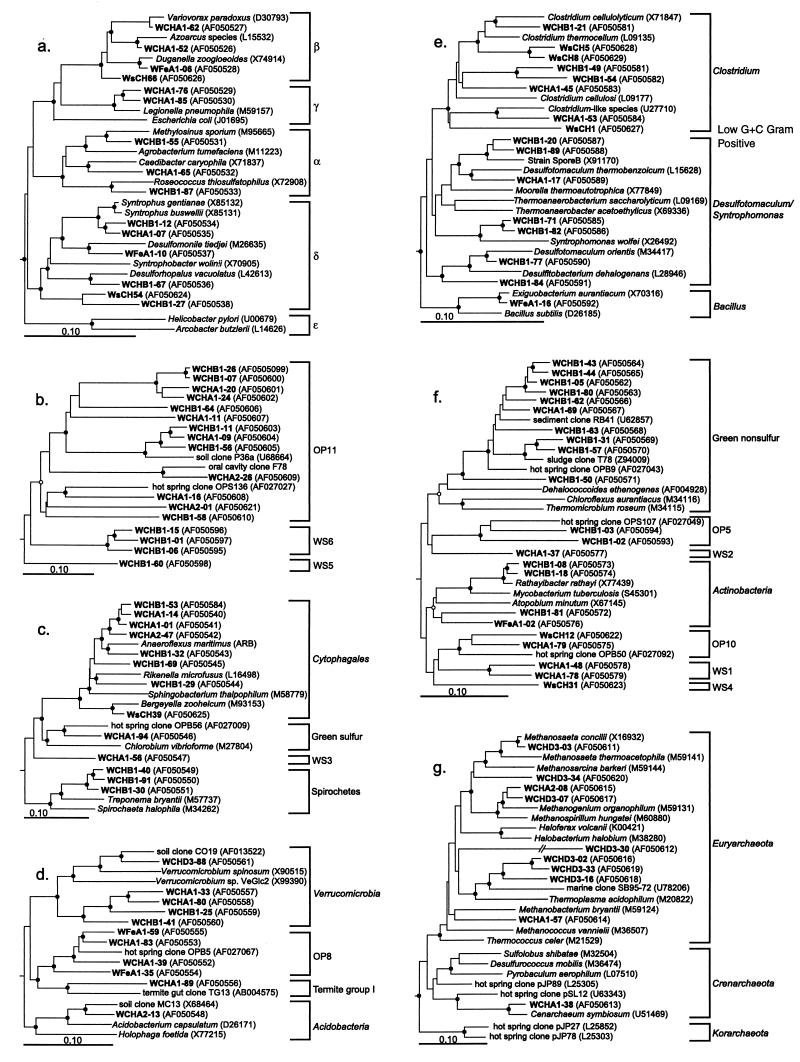
Figure 3 shows seven dendograms that comprise subsections of Fig. 2, expanded with more reference sequences. The grouping of divisions into dendrograms a to f in Fig. 3 is arbitrary due to the undefined division level branching order in the Bacteria (16, 18). Most Wurtsmith sequence types were placed with high bootstrap values into their respective divisions.
FIG. 3.
Evolutionary distance dendrograms of bacterial and archaeal 16S rDNA sequence types obtained from the Wurtsmith aquifer. Putative divisions are listed outside the brackets for panels b, c, d, f, and g; subdivisions are listed for panel a (Proteobacteria) and panel e (low-G+C gram-positive organisms). Six novel candidate divisions determined in this study are labeled WS1 to WS6. Reference sequences were chosen with the ARB parsimony insertion tool and database. Bacillus subtilis (D26185) and Synechococcus sp. strain PCC 6301 (X01296) were used as the outgroups for panels a, b, c, d, and f; Synechococcus sp. strain PCC 6301 and E. coli (J01695) were used as the outgroups for panel e; and Thermatoga maritima (M21774) and Aquifex pyrophilus (M83548) were used as the outgroups for panel g. Branch points supported (bootstrap values, >74%) by rate-corrected maximum likelihood, parsimony, and distance analyses are indicated by filled circles; open circles indicate branch points supported by some analyses but only marginally supported (bootstrap values, 50 to 74%) or not supported (bootstrap values, <50%) by others. Branch points without circles were not resolved (bootstrap values, <50%) as specific groups in different analyses.
To compare organisms that occupy different redox zones and to examine experimental reproducibility, four PCR libraries were prepared with universally conserved 16S rDNA primers: WCHA and WsCH with DNA from the methanogenic CH core, WSA with DNA from the methanogenic–sulfate-reducing S core, and WFeA with DNA from the iron- or sulfate-reducing Fe core. All libraries exhibited some sequence types that were numerically dominant (13 to 81% of the clones), as well as numerous sequence types that occurred only once or twice in a particular library. Libraries WCHA and WsCH were prepared from the same bulk DNA extracted from methanogenic soil core CH by independent but identical PCR and cloning steps to test the reproducibility of these steps. The two clone libraries were similar in their composition; most numerically dominant sequence types, such as WCHB1-26, WCHB1-12, and WCHD3-03, were dominant in both libraries (Table 1). The sequence types that differed between the two libraries were the relatively rare ones that, therefore, would have a good probability of not being resampled in these analyses. Library WSA was similar to libraries WCHA and WsCH; only the sequence types with RFLP patterns that appeared more than once in the WSA library were sequenced, and all were identical to numerically dominant sequence types in the WCHA and WsCH libraries. Library WFeA, from the iron- or sulfate-reducing zone, was markedly different from the other three libraries. Only the sequence types with RFLP patterns that appeared more than once in the WFeA library were sequenced; five of the seven types sequenced were not encountered in any of the other libraries.
In order to explore further the diversity of the methanogenic zone of the Wurtsmith aquifer and to examine the effects of different primers on the composition of clone libraries, libraries WCHB (bacterial primers) and WCHD (archaeal primers) were created with the same DNA as that used for the WCHA and WsCH libraries. The numerically dominant sequence type in the WCHD library (81% of RFLP patterns examined) was also the numerically dominant archaeal sequence type in the WCHA library (universal primers). The WCHB library, however, differed from the WCHA library; three of the most numerous sequence types in the WCHB library were not observed in the WCHA library (WCHB1-01, WCHB1-06, and WCHB1-15). These sequence types did not occur in the universal libraries because they contained four mismatches from the universal 515F primer (see Discussion).
DISCUSSION
Microbial diversity of the contaminated aquifer.
SSU rRNA-based molecular analyses have resulted in the description of over 30 major lineages (divisions) in the bacterial domain (16). The majority (70%) of the analyzed Wurtsmith rDNA sequence types were affiliated with recognized bacterial divisions and archaeal groups. Some main findings include a new subdivision of the Verrucomicrobia division defined exclusively by Wurtsmith sequence types (Fig. 3d) (16); significant phylogenetic expansion of the termite group I candidate division (Fig. 3d) (29); a great diversity of Wurtsmith sequence types in the green nonsulfur bacterium division (Fig. 3f) (16); a terrestrial close relative (WCHA1-38) of the marine crenarchaeote Cenarchaeum symbiosum (95% identity); and the first description of terrestrial representatives of a low-temperature, Thermoplasma-associated clade of Euryarchaeota (Fig. 3g), members of which previously had been detected only in marine environments (26).
Approximately 20% of the described Wurtsmith sequence types are affiliated reproducibly with four recently described candidate divisions from a Yellowstone hot spring (OP5, OP8, OP10, and OP11) (18). The association of the Wurtsmith sequence types with the OP candidate divisions expands the known diversity for these divisions and strengthens their division level status. Candidate division OP11 is represented by 13 Wurtsmith sequence types, including one type, WCHB1-26, that is abundant in the universal libraries of the methanogenic zone (ca. 15% of clones analyzed). Similarly, a representative of candidate division OP5, WCHB1-03, appears to be abundant in all redox zones of the contaminated aquifer (Table 1). The apparent abundance and diversity of OP sequence types suggest that organisms represented by these sequence types may play significant but unknown roles in the bioremediation process in the Wurtsmith aquifer, providing an impetus to attempt the cultivation of representatives of the OP11 and OP5 groups for further study.
The present study of the Wurtsmith aquifer suggests the existence of six novel candidate divisions (WS1 to WS6; Fig. 2 and 3), indicating that despite recent expansion, our knowledge of the extent of microbial diversity is still incomplete, even at the division level. Candidate divisions WS1 to WS6 are each represented by only one to three Wurtsmith sequence types but together comprise 10% of the total number of sequence types analyzed. Study of additional representatives of these groups will be required to verify their division level status.
Microbial distribution.
Four SSU rDNA libraries were created with universal PCR primers. Two universal libraries were created from a single extraction of DNA from the methanogenic zone (WCHA and WsCH), one originated from the interface between the methanogenic zone and sulfate-reducing zone (WSA), and one originated from the iron- or sulfate-reducing zone (WFeA) (Fig. 1). Libraries WCHA and WsCH were created to test the reproducibility of the PCR and cloning steps of the library construction protocol. The libraries had the same set of numerically dominant sequence types; sequence types that occurred infrequently differed between the libraries (Table 1). The composition of the WSA library was very similar to those of the WCHA and WsCH libraries, indicating that the two soil samples were possibly from geochemically similar environments. These results indicate that the classification of the two samples as originating from different redox zones may need reassessment. The WFeA library had a suite of sequence types different from those of the other libraries, consistent with the expectation that Fe3+ as the predominant electron acceptor results in a community physiologically distinct from the methanogenic community. Sequence type WFeA1-06, representing 17% of the WFeA library, is essentially identical to sequence types found in several environmental clonal studies performed on diverse habitats, by our laboratory and others, as well as in a negative control (no-sample) extraction, indicating that it may be a laboratory contaminant, possibly from widely used reagents (42). The amount of biomass in the WFeA sample, as approximated by the DNA extracted from samples, was much lower (<10-fold) than in the other samples. Since the Fe horizon is at the leading edge of hydrocarbon dispersal downward in the plume, there is less carbon source to drive communities than there is in the upper horizons. Laboratory contaminants presumably have a greater chance of being detected in libraries created from low-biomass samples (42). No sequences detected in parallel negative control extractions were encountered in the other libraries analyzed (data not shown).
Libraries WCHB (bacterial primers) and WCHD (archaeal primers) were created from the methanogenic CH sample. The composition of the WCHD library reinforces the results obtained with the universal methanogenic libraries, that WCHD3-03 is the dominant archaeal sequence type found in the methanogenic zone of the aquifer. Three of the dominant bacterial sequence types found in the WCHB library, comprising candidate division WS6 (Table 1), were not detected in the universal libraries, likely due to incomplete conservation of the universal 515 primer-binding site (see Fig. 4). This observation of variation in the putatively universal 515 primer-binding site indicates that different primer sets should be used in order to gauge the microbial diversity of an environment by the clonal library method or at least that universal primer 515F should not be used as the only forward primer.
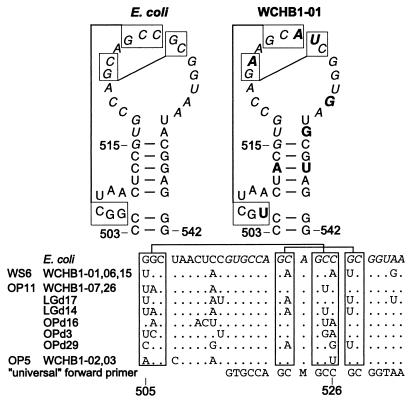
FIG. 4.
Proposed secondary and tertiary interactions in the region from positions 503 to 542 (E. coli numbering) of 16S rRNA sequences. Sequences in italic type represent the binding site for the universal 515F primer. Connected boxes indicate proposed tertiary interactions. Nucleotides in bold type indicate differences between the WCHB1-01 and E. coli rRNAs.
Covariation analysis.
Sequences obtained from the environment can be valuable resources for covariation analysis of RNA structure (8). Some of the sequences detected in this study support the existence of base pairs in 16S rRNA that previously were indicated only by covariation in a limited number of sequences. Figure 4 shows the helix at positions 500 to 545 for E. coli and representatives of the WS6, OP11, and OP5 candidate divisions (16–18). Two proposed tertiary interactions (13) are indicated in Fig. 4. The only previous evidence for these tertiary interactions was the occurrence of covariations in rRNA sequences from some organelles and a few Mycoplasma spp. As shown in Fig. 4, the environmental sequences significantly expand support for the proposed interactions. Although most of the new sequences support the proposed tertiary interactions, some do not show canonical covariation in the pair at bp 504 and 526 (U-C in WCHB1-07 and WCHB1-26 and A-G in OPd16). As mentioned above, these changes in this otherwise universal sequence compromise positions 515 to 533 as a universal primer target for obtaining rRNA genes from the environment.
Physiological hypotheses.
Environmental rDNA sequences, in principle, can be used to infer some properties of the organisms that they represent. If a sequence is related to those of a group of cultivated organisms with common properties, then the environmental organism represented only by the sequence also is expected to have those properties. One barrier to deriving physiological hypotheses from this study is that many of the sequences are not specifically related to cultivated organisms. Indeed, the numerically dominant sequence types in the bacterial and universal libraries from methanogenic soil in this study are members of newly recognized candidate divisions, OP11 and OP5, for which no cultivated representatives are known. Nothing is known about the physiology of these bacteria, since they are defined only by sequences obtained from the environment. Any physiological interpretations must therefore rely heavily on the environmental conditions of a sample from which a sequence is isolated. Based on environmental chemistry, Hugenholtz et al. (18) speculated that organisms in a Yellowstone hot spring represented by OP11 sequences may engage in sulfate reduction. Of course, many organisms that are closely related by 16S rRNA genes have different physiologies; energy metabolism, for instance, can be highly variable in evolution (30).
Some of the sequences encountered at the Wurtsmith site, however, are related sufficiently closely to studied organisms that reasonable physiological hypotheses can be formulated. Two of the most abundant sequence types in this study represent members of phylogenetic groups with consistent modes of energy metabolism. Sequence WCHD3-03, representing 8% of the universal library and 81% of the archaeal library created from methanogenic zone soil, is 97% identical to the archaeon Methanosaeta concilii and monophyletic with the two known Methanosaeta spp. (Fig. 3g). The only recognized form of energy production in this genus is aceticlastic methanogenesis (47), indicating that the archaeon represented by the WCHD3-03 sequence also engages in aceticlastic methanogenesis. Sequence WCHB1-12, representing 8% of the universal library created from methanogenic zone soil, is 96% identical to the SSU rDNA of Syntrophus gentianae and monophyletic with Syntrophus spp., to the exclusion of all other δ-Proteobacteria (Fig. 3a). Syntrophus spp. obtain energy from the anaerobic oxidation of organic acids to acetate and hydrogen (46), suggesting a similar metabolic capability for the environmental bacteria represented by sequence WCHB1-12. S. gentianae and M. concilii recently were shown to interact syntrophically in a triculture with Methanospirillum hungatei or Desulfovibrio desulfuricans (for hydrogen removal). The utilization of benzoate by S. gentianae was greatly increased by the presence of M. concilii and the consequent removal of acetate, as was thermodynamically predicted (35). A similar syntrophic association between the organisms represented by sequences WCHD3-03 and WCHB1-12 seems possible (see below).
Although the relatively high levels of these Syntrophus-related and Methanosaeta-related sequences in the methanogenic zone libraries are consistent with the significant involvement of the corresponding organisms in methanogenic hydrocarbon degradation, potential quantitative biases inherent in the clonal library approach must be recognized. These biases include potential variability in rDNA copy number, cell lysis, DNA extraction, PCR, and cloning (2, 10, 33, 41). Quantitative assessments of microbial distribution are better made directly, for instance, by use of whole-cell in situ rDNA probes or quantitative hybridization to extracted RNA (2). In the few studies that have compared PCR-based clone libraries with other methods, however, the correspondence is good (11, 12, 38). Consequently, we believe that the determination of abundances of sequence types in a clonal diversity survey is a useful first step in predicting abundances of microorganisms in the environment.
In a methanogenic ecosystem, polymers such as polysaccharides, proteins, lipids, and nucleic acids typically are broken down by the action of extracellular hydrolytic enzymes into oligomers and monomers such as amino acids, sugars, purines, pyrimidines, glycerol, and fatty acids (34, 48). Fatty acids, in turn, are fermented to simpler acids (including acetate), hydrogen, and carbon dioxide by syntrophic bacteria that rely on the removal of acetate and hydrogen by other microorganisms to render the energetics of this fermentation favorable. The fate of acetate, hydrogen, and carbon dioxide depends on the organisms involved: hydrogen-oxidizing methanogens can produce methane from hydrogen and carbon dioxide, homoacetogens can produce acetate from hydrogen and carbon dioxide, and aceticlastic methanogens can produce methane and carbon dioxide from acetate. Environmental conditions dictate the most favorable processes. Recent studies on lake sediments and other cold, freshwater environments have shown that, at temperatures lower than 20°C, methane production occurs predominantly by aceticlastic methanogenesis and that hydrogen is consumed preferentially by homoacetogens (9, 28, 36, 37). This temperature effect also has been rationalized thermodynamically by Schink (34). The low temperature (10 to 12°C) of the Wurtsmith aquifer and the dominance in the methanogenic zone of probable aceticlastic methanogens indicate that aceticlastic methanogenesis is primarily important in hydrocarbon degradation in this environment as well. The low abundance of sequences that correspond to organisms known for H2-CO2 methanogenesis is consistent with this notion and suggests that hydrogen utilization may be dominated by acetogenic bacteria that are not yet identifiable by rDNA sequences.
Overall, then, we propose that the degradation of petroleum hydrocarbons in the methanogenic zone of the Wurtsmith aquifer proceeds by initial hydrolytic attack on hydrocarbons; fermentation of the resulting organic acids by syntrophic bacteria, including fermentation of the simple acids by microorganisms represented by the Syntrophus-related sequence type WCHB1-12; and aceticlastic methanogenesis by microorganisms represented by the Methanosaeta-related sequence type WCHD3-03. These two types of organisms may be involved in a syntrophic relationship in the Wurtsmith aquifer and may be useful indicator organisms for the evaluation of methanogenic intrinsic bioremediation in this and other aquifers. It remains to be seen, however, if these or similar types of microorganisms are present in other contaminated aquifers undergoing methanogenic intrinsic bioremediation.
ACKNOWLEDGMENTS
We thank Brett Goebel for insightful discussions during the study and other members of the Pace Lab for advice.
This research was supported by grants to N.R.P. from NIH and DOE. Funding for collection of the samples and data on aquifer geochemistry was provided in part by the U.S. Geological Survey Toxic Substances Hydrology Program; the National Center for Integrated Bioremediation Research and Development at the University of Michigan, through the Department of Defense Strategic Environmental Research and Development Program under cooperative agreement CR822922 with the U.S. Environmental Protection Agency; and by grant BIR 9120006 to the Center for Microbial Ecology at Michigan State University.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcelona M J, Lu J, Tomczak D M. Organic acid derivatization techniques applied to petroleum hydrocarbon transformation in surface environments. Ground Water Monit Remed. 1995;15:114–124. [Google Scholar]
- 4.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borden R C. Natural bioremediation of hydrocarbon-contaminated ground water. In: Norris R D, et al., editors. Handbook of bioremediation. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 177–199. [Google Scholar]
- 6.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown J W, Nolan J M, Haas E S, Rubio M A T, Major F, Pace N R. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc Natl Acad Sci USA. 1995;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conrad R, Bak F, Seitz H J, Thebrath B, Mayer H P, Schultz H. Hydrogen turnover by psychrotrophic homoacetogenic and mesophilic methanogenic bacteria in anoxic paddy soil and lake sediments. FEMS Microbiol Ecol. 1989;62:285–294. [Google Scholar]
- 10.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni S J, Rappe M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutell R R, Larsen N, Woese C R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haack, S. K., P. Adriaens, F. H. Chapelle, L. J. Forney, M. A. Henry, and D. T. Long. 1998. Unpublished data.
- 15.Huffman G C, Cummings T R, Gillespie J L, Brannen J R. Wurtsmith Air Force Base, Michigan. Investigation of soil and ground water contamination at selected sites. Installation Restoration Program Phase II—Confirmation/Quantification Stage 2. U.S. Lansing, Mich: Geological Survey; 1995. [Google Scholar]
- 16.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hugenholtz P, Hershberger K L, Flanagan J L, Kimmel B, Pace N R. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Widespread distribution of a novel phylum-depth bacterial lineage in nature, abstr. N-23; p. 385. [Google Scholar]
- 18.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampbell D H, Widemeier T H, Hanses J E. Intrinsic bioremediation of fuel contamination in ground water at a field site. J Hazard Mater. 1996;49:197–204. [Google Scholar]
- 20.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 22.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng W-L, Schummer M, Cirisano F D, Baldwin R L, Karlan B Y, Hood L. High-throughput plasmid mini preparations facilitated by micro-mixing. Nucleic Acids Res. 1996;24:5045–5047. doi: 10.1093/nar/24.24.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nozhevnikova A N, Kotsyurbenko O R, Simankova M V. Acetogenesis at low temperature. In: Drake H L, editor. Acetogenesis. London, England: Chapman & Hall, Ltd.; 1994. pp. 416–431. [Google Scholar]
- 29.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 31.Pace N R, Stahl D A, Lane D J, Olsen G J. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 32.Raskin L, Rittman B E, Stahl D A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schocke L, Schink B. Energetics of methanogenic benzoate degradation by Syntrophus gentianae in syntrophic coculture. Microbiology. 1997;143:2345–2351. doi: 10.1099/00221287-143-7-2345. [DOI] [PubMed] [Google Scholar]
- 36.Schulz S, Conrad R. Influence of temperature on pathways to methane production in the permanently cold profundal sediment of Lake Constance. FEMS Microb Ecol. 1996;20:1–14. [Google Scholar]
- 37.Schulz S, Matsuyama H, Conrad R. Temperature dependence of methane production from different precursors in a profundal sediment (Lake Constance) FEMS Microbiol Ecol. 1997;22:207–213. [Google Scholar]
- 38.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K-H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 40.Strunk, O., O. Gross, B. Reichel, M. May, S. Hermann, N. Struckmann, B. Nonhoff, M. Lenke, A. Vilbig, T. Ludwig, A. Bode, K. H. Schleifer, and W. Ludwig. ARB: a software environment for sequence data. Submitted for publication.
- 41.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner M, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tulis D S. Issues associated with natural attenuation. Washington, D.C: U.S. Environmental Protection Agency; 1997. http://www.epa.gov/OUST/rbdm/issues.htm . [Google Scholar]
- 44.U.S. Environmental Protection Agency. Use of monitored natural attenuation at superfund, RCRA corrective action, and underground storage tank sites. OSWER directive 9200.4-17. Washington, D.C: U.S. Environmental Protection Agency; 1997. http://www.epa.gov/OUST/directiv/9200_417 .htm. [Google Scholar]
- 45.U.S. Geological Survey. Wurtsmith Air Force Base, Michigan: Investigations of ground-water and soil contamination at selected sites. U.S. Lansing, Mich: Geological Survey; 1991. [Google Scholar]
- 46.Wallrabenstein C, Gorny N, Springer N, Ludwig W, Schink B. Pure culture of Syntrophus buswellii, definition of its phylogenetic status, and description of Syntrophus gentianae sp. nov. Syst Appl Microbiol. 1995;18:62–66. [Google Scholar]
- 47.Whitman W B, Bowen T L, Boone D R. The methanogenic bacteria. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 739–745. [Google Scholar]
- 48.Zehnder A J B. Ecology of methane formation. In: Mitchell R, editor. Water pollution microbiology. London, England: John Wiley & Sons; 1978. pp. 349–376. [Google Scholar]