ABSTRACT
The genetic alterations contributing to migration proficiency, a phenotypic hallmark of metastatic cells required for colonizing distant organs, remain poorly defined. Here, we used single-cell magneto-optical capture (scMOCa) to isolate fast cells from heterogeneous human breast cancer cell populations, based on their migratory ability alone. We show that captured fast cell subpopulations retain higher migration speed and focal adhesion dynamics over many generations as a result of a motility-related transcriptomic profile. Upregulated genes in isolated fast cells encoded integrin subunits, proto-cadherins and numerous other genes associated with cell migration. Dysregulation of several of these genes correlates with poor survival outcomes in people with breast cancer, and primary tumors established from fast cells generated a higher number of circulating tumor cells and soft tissue metastases in pre-clinical mouse models. Subpopulations of cells selected for a highly migratory phenotype demonstrated an increased fitness for metastasis.
Keywords: Breast cancer, Metastasis, Applied optics, Microscopy, Photobleaching, Single-cell isolation, Biophysics, Cell migration
Summary: Single cells captured from a highly heterogeneous triple-negative breast cancer cell population exhibit higher migration speed and focal adhesion dynamics, which are retained by their progeny as a result of a motility-related transcriptomic profile.
INTRODUCTION
The inefficacy of current metastatic breast cancer treatments requires major improvements in our ability to probe the molecular biology of malignant cells (Riggi et al., 2018; Seyfried and Huysentruyt, 2013). Tumor heterogeneity and multiple mechanisms of therapeutic resistance (Polyak, 2011; Sun and Yu, 2015; Lawson et al., 2018) yield a myriad of signaling pathways as putative targets for novel therapeutic strategies, which partly explains the slow progress in preventing secondary tumors (Rashid et al., 2021). Many studies have investigated the molecular mechanisms enabling the colonization of distant organs by cancer cells, such as the ability to penetrate the peripheral bloodstream (intravasation), evade immune detection, extravasate and further proliferate in a new microenvironment (Fidler, 2003). Among all the cell phenotypes required for colonizing distant organs, migratory proficiency is widely accepted as a key characteristic of highly metastatic cells (Partin et al., 1989).
Several biophysical traits related to cell migration have been correlated with metastatic aggressivity. During migration, a cell applies dynamic traction forces on the surrounding matrix to drive directional movement (Mierke et al., 2008). This process requires deformation of the cell body and the establishment of new cell–matrix adhesion sites to enable displacement. Metastatic cells exert higher traction forces and are known to be more deformable and less rigid than their non-metastatic counterparts (Kraning-Rush et al., 2012), which provides the flexibility required to migrate through small pores and crawl more efficiently (Alibert et al., 2017; Guck et al., 2005; Cross et al., 2007; Gossett et al., 2012; Qi et al., 2015; Beri et al., 2018; Wisniewski et al., 2020; Liu et al., 2020). Cancer cells also rely on highly dynamic adhesions for efficient migration as stabilization of focal adhesions (FAs) dramatically reduces metastatic potential (Bijian et al., 2013). Conversely, reduced adhesiveness increases metastasis (Palmer et al., 2008; Beri et al., 2020). Furthermore, FA size is indicative of traction stress within the cell and predicts cell speed in a biphasic relationship (Case and Waterman, 2015). The formation of large mature adhesions can increase traction forces, which can reduce cell migration by anchoring the cell to the underlying substrate; however, dynamic adhesions can generate sufficient tractional forces but also dissociate quickly to ensure rapid cell migration (Kim and Wirtz, 2013).
The kinematic properties of cancer cells are often investigated by manipulating individual or several candidate genes, through functional inhibition or genetic knockout approaches, to study their impact on cell migration in vitro and metastatic potential in vivo. For example, knockdown of WAVE3 (also known as WASF3), an actin cytoskeleton-remodeling protein, reduces the migration, invasion and metastatic ability of MDA-MB-231 breast cancer cells (Sossey-Alaoui et al., 2007; Fernando et al., 2009). Knockdown of ShcA (also known as SHC1) or LPP, which localize to FAs and invadopodia, also reduces breast cancer cell migration, invasion and metastasis (Ngan et al., 2017; Kiepas et al., 2020b). Inhibition of SRC, a key player in the turnover of adherent junctions and focal FAs, reduces the metastatic burden of MDA-MB-231 cells and the overall lethality of osteoclast bone resorption, whereas its overexpression preferentially leads to the development of bone metastasis (Aleshin and Finn, 2010). miR34a-mediated knockdown of CD44, an adhesion molecule involved in colon and other cancers, markedly inhibits the metastasis of many cancer cell lines (Liu et al., 2011). Similarly, miR10b-mediated knockdown of RhoC, a member of the Rho small GTPases family involved in cell migration and matrix remodeling, impairs the development of metastasis (Ma et al., 2007). Overall, cell migration is a very complex and heterogeneous phenomenon regulated by many molecular mechanisms. Consequently, a candidate gene approach is not only labor intensive, but may also miss crucial interactions that occur during the metastatic cascade. Hence, physical isolation of cells based on biophysical parameters or migratory phenotypes coupled with comprehensive genetic analyses should facilitate the study of the underlying molecular mechanisms that promote metastatic fitness.
Few technologies enable the capture of cells based solely on their migratory characteristics. Recently, a specialized microfluidic device was developed to isolate cells capable of migrating through narrow channels (Yankaskas et al., 2019). Digital microfluidic microgels have also been employed to separate migrating cells by cutting layers of a hydrogel (Li et al., 2020). Transwell migration assays have also been used to isolate aggressive cells after multiple rounds of selection (Hapach et al., 2021). Here, we used single-cell magneto-optical capture (scMOCa) (Binan et al., 2019a) to capture migrating cells without any physical constraint based on their migratory velocity or distance travelled. We isolated and expanded intact cells to generate novel subpopulations from a highly heterogeneous triple-negative breast cancer cell (TNBC) population. We demonstrate that the progeny of these cells maintains a migratory phenotype, elevated adhesion dynamics and exhibits a strikingly different transcriptomic profile compared with the parental cell population. Importantly, in vivo analysis reveals that fast migratory cells have increased metastatic activity.
RESULTS
scMOCa reliably captures highly motile cells from a heterogeneous cell population
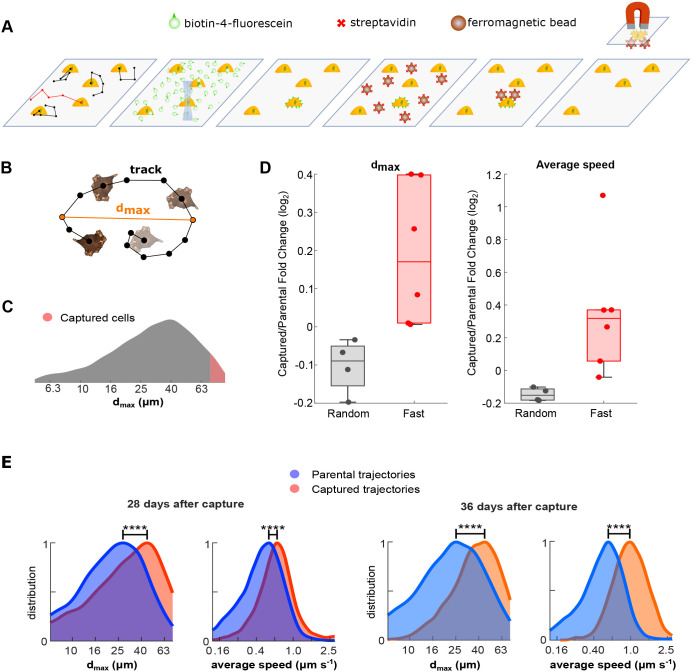
We sought to isolate cells displaying the fastest motility within heterogeneous cultures of MDA-MB-231 human TNBC cells (Fig. 1A). Briefly, scMOCa leverages confocal microscopy lasers to crosslink fluorescein-conjugated biotin molecules to the plasma membrane of individually selected cells. Target cells are identified in a field of view and illuminated to tether fluorescent biotin conjugates to cellular membranes via extracellular free radicals produced by photobleaching. Cells are then incubated with streptavidin-coated ferromagnetic beads and selectively captured with a simple magnet to isolate them from the population, with high purity. The procedure does not affect viability, proliferation rate or cell physiology (Binan et al., 2016, 2019a). Cells were plated at low density on optical-quality plastic dishes coated with fibronectin and imaged with darkfield illumination to acquire high-contrast, high-frequency time-lapse movies without inducing phototoxicity. (Antolović et al., 2014) Cells were tracked using a nearest neighbor algorithm (Jaqaman et al., 2008; Mazzaferri et al., 2015). We limited tracking time to a period of 2 h because extended imaging periods may allow cells to divide, increasing the complexity of real-time identification of fast cells. To verify that this short imaging time did not introduce biases, we analyzed cell migration over a 9-h period, divided the movies into shorter 2-h windows and compared the identification of the most motile cells across all movies according to seven track measures (Fig. S1A). The analysis showed that four out of seven motility parameters were robust to the choice of imaging time (Fig. S1A). Of these, the greatest distance between two arbitrary track points (dmax) was the most robust, with the fastest cells over a 9-h-long acquisition successfully identified with a probability of 80% from movies lasting only 2 h (Fig. S1B,C). Thus, for the rest of the study, cell migration was characterized in real time using dmax, and fast cells were defined as those in the 95th percentile of most motile cells (Fig. 1B,C).
Fig. 1.
Overview of cell migration analysis and scMOCa capture. (A) MDA-MB-231 cells were tracked using a nearest neighbor algorithm and the most motile cells were automatically illuminated with a low-powered laser to locally photobleach the fluorescein and biotinylate cell membranes. Streptavidin-coated ferromagnetic beads attached specifically to illuminated cells, which permitted magnetic capture and physical isolation of fast motile cells. (B) Cells were tracked and classified according to the greatest distance between any two arbitrary positions in their migration trajectories (dmax). (C) Only cells with dmax larger than the 95th percentile were captured, which is represented by the red region on the measured distribution in the original culture. (D) Fold change of dmax and average speed between capture replicates compared with their parental cells, 20 days after the capture. The captured cells come from fast (n=4) and random (n=6) selection. A dot represents an independent capture. The box plot displays the median, and the lower and upper quartiles. Whiskers shows the minimum and maximum values, excluding the outliers. Values were considered as outliers if they deviated from the upper or lower quartiles by 1.5 times the interquartile range. (E) Expanded cells were characterized at several time points after capture and exhibited more than 1.3 times larger dmax (****P<0.0001, 1500 bootstrap samples) compared with the parental cell population. Captured cells were more than two times faster (****P<0.0001, 1500 bootstrap samples) and maintained their phenotype after several cell divisions over a period greater than a month (approximately 20 cell divisions and 14 passages occurred in 36 days).
To assess whether a fast-motility phenotype can be retained over time, highly motile cells were automatically detected, illuminated, captured and expanded. A total of 100-250 cells were effectively captured in each experiment. After a period of 20 days in culture, several motility features were retained (Fig. 1D). We thus analyzed parental and fast cells over a 1-month period (more than 12 passages), subjecting them to identical culture conditions and passage number. Fast cells maintained both significantly larger dmax (P<0.0001) and average speed (P<0.0001) (Fig. 1E), indicating that the phenotype is passed from mother to daughter cells. In particular, the distribution's median for dmax and average speed remained 62% [effect size (ES)=0.82)] and 77% (ES=1.19) greater, respectively, for the fast cells compared with the parental cells. Furthermore, this phenotype was maintained after freezing and thawing aliquots of this fast-cell population.
Selection and capture are based on complex cellular phenotypes rather than specific genotypes or protein expression profiles; isolated clones may differ between each experiment because fewer than 200 cells from approximately 4000 tracked cells were captured per experiment. To analyze the consistency of retained motility characteristics, we repeated the capture six times. Although new cell populations displayed a wide distribution of velocities, a median 30% increase (ES=0.51) in average speed remained consistent over 3 weeks in culture (Fig. 1D; Fig. S2A). Importantly, random cell selection was performed as a control (Fig. 1D; Fig. S2B). We did not observe differences between randomly selected cells and the parental population, further supporting the assumption that the procedure alone does not affect cellular motility (Binan et al., 2019a).
Highly motile cells engage transcriptional programs that enhance migration
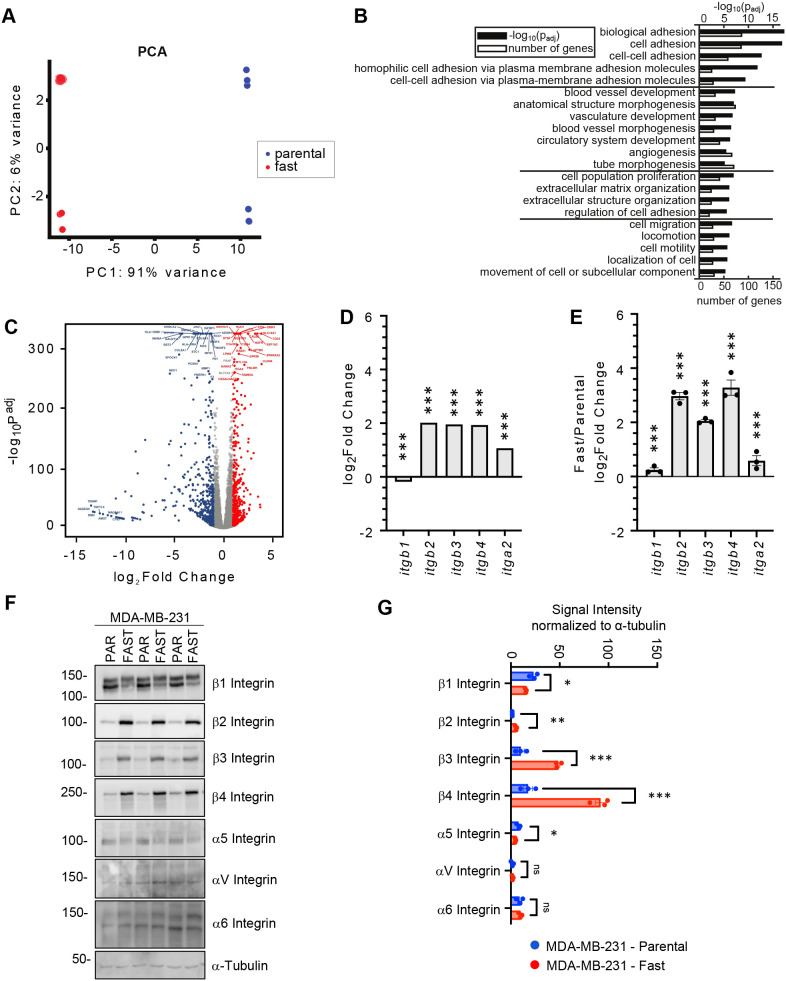
To investigate the molecular mechanisms driving the enhanced migratory phenotype of fast cells, we compared gene expression profiles in fast and parental MDA-MB-231 cell populations, at equal passage number, using RNA sequencing (RNA-seq) (Fig. 2A-C; Fig. S3). Principal component analysis revealed that fast and parental cells displayed markedly distinct global profiles, as 91% of the variance was explained by the first principal component (Fig. 2A). We observed a high degree of concordance between the RNA-seq data generated from two independent batches comprising three technical replicates of fast-cell populations, confirming that the phenotype obtained after laser capture is stable. Batch differences between two sets of experiments, separated by multiple rounds of thawing and expansion, were reflected in the second principal component, which explained only 6% of the variance. Differential expression analysis between fast and parental cells resulted in 932 genes with significant differences (adjusted P<0.05, log2 fold-change>1, average normalized expression>100) (Fig. 2B,C). Differentially expressed genes (Table S1) were highly reproducible in both batches, with 707 (75%) statistically significant genes shared between the two analyses (Fig. S3). Pathway analysis of these genes revealed that the most significant Gene Ontology (GO) biological process terms were in the areas of biological adhesion, followed by angiogenesis and blood vessel development, extracellular matrix organization, and migration and cell locomotion (Fig. 2B).
Fig. 2.
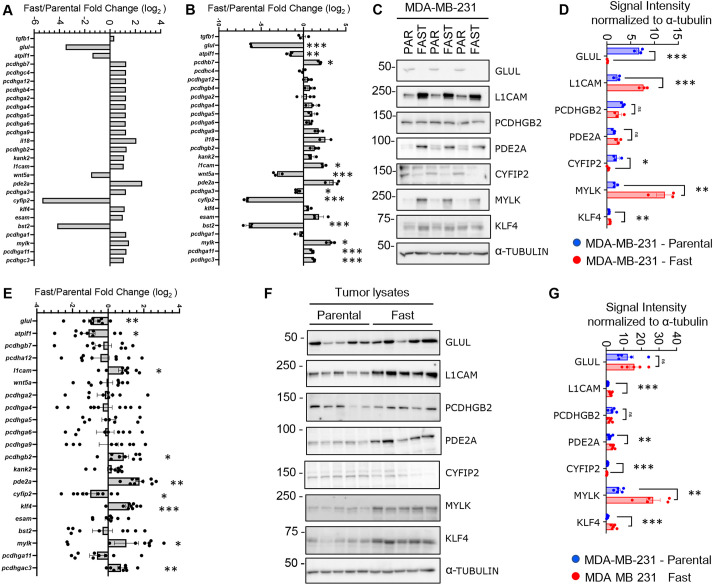
Genotypic characterization of captured MDA-MB-231 cells. (A) Principal component analysis (PCA) using the 1000 most variant genes as features. (B) Groups of the most important GO biological process terms from the analysis highlights terms related to adhesion. Groups were made with the k-means algorithm. (C) Differential expression analysis comparing parental and fast cells (n=3 biological replicates per group). Well-expressed genes (average normalized expression >100) are shown. Blue: upregulated genes in the parental population; red: upregulated genes in the fast population. (D) Normalized RNA expression for five integrins from RNA-seq data. Differences were validated with bootstrap statistics. (E) Validation of RNA expression of integrin subunits in fast and parental cells. Data are presented as fold change relative to parental MDA-MB-231 cells and normalized to β-actin (ACTB) and RPLP0. Differences were analyzed with multiple unpaired t-test. (F) Immunoblot of three independent cell lysate pairs from MDA-MB-231 cell populations. α-tubulin serves as a loading control. PAR, parental. (G) Quantification of the immunoblots presented in F. Data are presented as the signal intensity normalized to the loading control, α-tubulin. Differences were analyzed with multiple unpaired t-test. *P<0.05; **P<0.01; ***P<0.001. ns, not significant. Error bars represent s.e.m.
Integrin subunits are differentially expressed in fast relative to parental cells
Among the top upregulated genes in fast cells, belonging to all four GO term categories (Fig. 2B), were integrin subunits. The integrin family plays an essential role in cell adhesion, migration and invasion as well as tumor progression and metastasis (reviewed by Hamidi and Ivaska, 2018). The expression of several integrin subunits (ITGB2, ITGB3, ITGB4 and ITGA2) was significantly elevated in the fast-cell population compared with parental cells (Fig. 2D). We next validated the RNA expression of these integrin subunits in independent RNA preparations (Fig. 2E). Of the integrin subunits that were assessed, RNA transcript levels of ITGB2, ITGB3 and ITGB4 were the most highly differentially expressed between the fast and parental cells. Integrins function as heterodimeric proteins containing an α and β subunit. β1 integrin is the most promiscuous β subunit, associating with many α subunits (α1-11 and αV), whereas β2 integrin associates with αL, αM, αX or αD subunits. β3 integrin associates with αV and αIIB subunits, whereas β4 only associates with the α6 integrin subunit (reviewed by Hynes, 2002). Immunoblot analysis of cell lysates prepared from fast and parental cells confirmed the upregulation of β2, β3 and β4 integrin subunits, as well as a slight decrease in the expression of β1 integrin (Fig. 2F,G). Of the α subunits analyzed, only the α5 integrin subunit was downregulated in the fast-cell population relative to the parental cells (Fig. 2F,G).
Specific integrin pairs possess different substrate specificities. For example, β4 is almost exclusively involved in binding to laminin (Hynes, 2002; Cooper and Giancotti, 2019). Indeed, the α6β4 integrin receptor has been reported to preferentially recognize the laminin-332 (laminin-5) isoform (Nishiuchi et al., 2006). Laminin-332 is involved in processes linked to metastasis development (reviewed by Ramovs et al., 2017) and promotes cellular adhesion and migration more efficiently than other ECM molecules (Miyazaki, 2006; Xue and Hemmings, 2013). Whereas β4 was elevated in fast cells, the α6 integrin subunit was expressed at similar levels in both fast and parental MDA-MB-231 cells (Fig. 2F,G). The αVβ3 integrin receptor is also known to bind laminin as well as vitronectin; of note, β3, but not αV, was upregulated in fast cells. Interestingly, both α5 and β1 subunits were downregulated in fast cells. The α5β1 receptor binds fibronectin, which was the substrate used to select the fast-cell population. Accordingly, the fold change in fast cell speed relative to that of parental cells was 20% greater (ES=0.8, P<0.05) on laminin compared with collagen or fibronectin (Fig. S4A,B), suggesting that the β4 integrin plays a role in the captured phenotype. The β2 integrin subunit is typically expressed on leukocytes and is functionally involved with leukocyte trafficking. When heterodimerized with various α subunits, β2 integrin interacts with fibronectin, ICAM1 and VCAM1 (Guenther, 2022). Thus, differential expression of integrin subunits may affect FA dynamics and the metastatic ability of the fast-cell population.
Highly motile cells exhibit increased adhesion dynamics on fibronectin and laminin
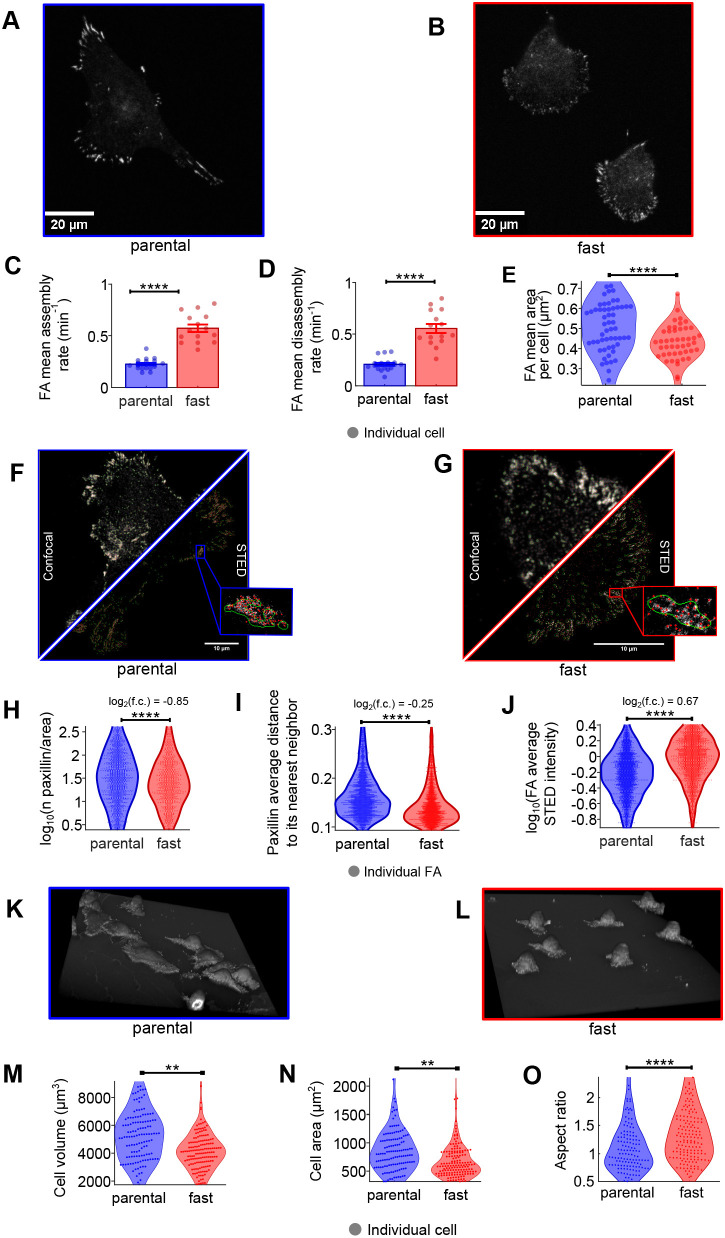
Cell migration on the extracellular matrix (ECM) is highly dependent on the formation and dissolution of cellular adhesions (De Martino et al., 2020). Smaller adhesions tend to exhibit more-rapid turnover rates, which enable faster locomotion (Kim and Wirtz, 2013). For example, nascent adhesions at the leading edge of migratory cells are smaller than 1 μm in diameter and have a lifespan shorter than 120 s (Choi et al., 2008). Therefore, to determine the assembly and disassembly rates of adhesions identified in each cell population, we performed continuous time-lapse imaging of mCherry-paxillin, an important component of all cell–matrix adhesion subtypes (Case and Waterman, 2015). Total internal reflection fluorescence (TIRF) microscopy (Fig. 3A,B) revealed that both the assembly and disassembly rates of adhesions in fast cells (∼0.57 and ∼0.55 min−1, respectively) were significantly faster (P<0.0001) than those of parental cells (∼0.23 and ∼0.20 min−1, respectively) when assessed on fibronectin-coated surfaces (Fig. 3C-E). When allowed to migrate on laminin-332, fast cells also displayed higher adhesion assembly and disassembly rates (∼0.54 and ∼0.45 min−1, respectively) compared with parental cells (∼0.33 and ∼0.26 min−1, respectively) (P=0.00004) (Fig. S4C-E). Although adhesion assembly rates in fast cells remained largely unchanged between fibronectin and laminin, adhesion disassembly rates were reduced in cells seeded on laminin compared with fibronectin.
Fig. 3.
Characterization of cellular adhesions in MDA-MB-231 fast and parental breast cancer cells. (A,B) Representative images showing paxillin-stained cellular adhesions imaged with TIRF microscopy. Parental cells (left) show large and well-defined focal adhesions (FA) compared with the fast cells (right). (C,D) Assembly and disassembly rates for cell populations. (E) FA size averaged per cell. Only one of the triplicates is shown here for simplicity; similar results were obtained for the others. Statistics for C-E were generated with two-sample t-tests. (F,G) Representative images of confocal and STED images showing paxillin-stained cellular adhesion. Green lines circle the FAs segmented on the confocal images; red points localize the centroid of paxillin clusters in STED images. (H) Number of paxillin clusters per FA area. (I) Distance of paxillin clusters to their nearest neighbor, averaged per FA. (J) Cluster intensity averaged over every FA. Statistics for H-J were accomplished with bootstrap statistics. (K,L) Representative 3D renderings of cell morphologies of the indicated cell populations. (M-O) Analysis of cell volume (M), cell area (N) and conical aspect ratio (height/average base diameter) (O) of the indicated cell populations are shown. Statistics for (M,N,O) were accomplished with bootstrap statistics. **P<0.01; ****P<0.0001. Error bars represent s.e.m.
We subsequently analyzed the size of cellular adhesions and paxillin density in the parental and fast MDA-MB-231 cell populations, which can indicate the FA maturation stage (Aziz et al., 2022). Immunofluorescence images of endogenous paxillin acquired with confocal microscopy revealed that fast cells possessed significantly smaller (P<0.0001, ES=0.40) adhesions than parental cells on fibronectin (∼0.42 µm2 compared with ∼0.50 µm2, respectively) and on laminin (P<0.0001, ES=0.52) (∼0.60 µm2 compared with ∼0.65 µm2, respectively). FAs were also significantly smaller on fibronectin than on laminin (P<0.0001, ES=0.55). To resolve the size and structure of small, nascent adhesions, super-resolution stimulated emission depletion (STED) microscopy was employed (Fig. 3F,G). STED imaging of endogenous paxillin revealed that fast cells possessed 1.8 times fewer (P<0.0001, ES=0.2) paxillin clusters per FA area (∼50 clusters µm2 for fast cells and ∼89 for parental cells) (Fig. 3H). Adhesion clusters in fast cells were on average 1.2 times closer to each other compared with parental (P<0.0001, ES=0.42), with average nearest-neighbor distances of 0.14 µm in fast cells compared with 0.18 µm for parental cells (Fig. 3I). Surprisingly, the average STED intensity over the FA area was significantly higher for the fast cells (ES=0.66), suggesting that higher concentrations of paxillin are localized within each individual adhesion (Fig. 3J). Finally, the difference in total intensity over all FAs was significantly higher for fast cells, but with a very small ES (P<0.0001, ES=0.06), suggesting that the total amount of paxillin is independent of the FA size and that its concentration decreases when the FA matures. Hence, the rapid adhesion turnover observed in fast cells does not allow complete FA maturation (Aziz et al., 2022).
Two major migration modes are found in 3D environments: mesenchymal and ameboid. Mesenchymal cell migration relies on cell–matrix adhesions to generate the necessary traction force for movement. Although this type of migration allows for efficient movement along ECM fibers, it is relatively slow compared with ameboid migration. Ameboid cell migration typically occurs in cells with low adhesion force or high actomyosin-mediated contractility. Importantly, cancer cells may undergo a mesenchymal-to-ameboid transition (MAT) (Wang et al., 2017; Yamada and Sixt, 2019; Clark and Vignjevic, 2015). Ameboid cells have small or diffusely organized adhesion sites and exhibit a more rounded morphology, much like what was observed for fast cells selected by scMOCa. We thus quantified the average cell size of the two populations by fluorescently staining the membranes of fixed cells and performing 3D confocal image reconstruction (Fig. 3K,L). Fast cells exhibited ∼24% smaller volume (median of 4077 µm3 compared with 5338 µm3, ES=0.18, P=7×10−3), 27% smaller contact area with the substrate (median of 593 compared with 809 µm2, ES=0.18, P=5×10−3), and 28% greater aspect ratio (height/area) (ES=0.55, P=9×10−5) (Fig. 3M-O), suggesting that fast cells are more contractile (Murrell et al., 2015). The dispersion of volume and area of the fast cells were ~50% of that of the parental cells, indicating that the fast-cell population was more homogeneous in terms of cell size and average FA area.
Highly motile cells generate more soft tissue metastases
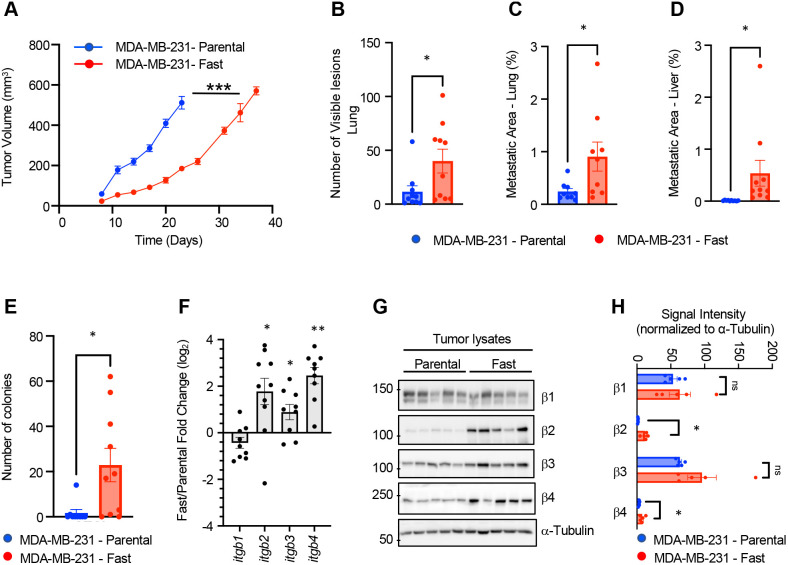
To evaluate tumor growth of fast and parental cells, we injected 1 million parental and fast-selected MDA-MB-231 cells into the mammary fat pad of immunocompromised mice and monitored tumor volume with regular caliper measurements until tumors reached a volume of 500 mm3 (Fig. 4A). The MDA-MB-231 fast-cell population exhibited impaired tumor growth compared with the parental population, despite no significant differences in proliferation (Ki67) and decreased apoptosis (cleaved caspase 3) in resected mammary tumors (Fig. S5A,B). Similarly, no significant difference in the doubling time was observed in vitro when cells were cultured on plastic (P=0.98) (Fig. S5C).
Fig. 4.
Characterization of tumor growth and metastasis of the fast-cell population. (A) Tumor growth curves of fast and parental cell populations in immunocompromised mice. (B) Number of lesions present on Hematoxylin & Eosin-stained sections of lung tissue isolated for tumor-bearing mice at endpoint. (C) Quantification of metastatic burden in the lung as assessed by vimentin-positive staining of lung tissue sections (percentage area of positive staining). (D) Metastatic area in the liver assessed by vimentin-positive staining of lung tissue sections (percentage area of positive staining). (E) Number of colonies identified from a culture of circulating tumor cells collected by cardiac puncture. (F) qPCR analyses of the expression of β-integrin subunits in tumors. Data are presented as the mean fold change difference between fast tumors relative to parental MDA-MB-231 tumors normalized to β-actin (ACTB) and RPLP0. (G) Immunoblot for the indicated β-integrin subunits from tumor lysates (n=5 for each tumor type). α-Tubulin is presented as a loading control. (H) Quantification of integrin expression normalized to α-tubulin. *P<0.05, **P<0.005, ***P<0.0005. ns, not significant. Unpaired t-test was used for statistics. Error bars represent s.e.m.
Despite the slower growth rate, mice bearing MDA-MB-231 fast tumors displayed significantly more visible lung-surface lesions (Fig. 4B) and greater metastatic burden in both the lung and liver at the time of tumor resection (500 mm3; parental, 23 days; fast, 7 days) (Fig. 4C,D). These data suggest that tumors derived from the fast population possessed greater metastatic potential compared with parental-derived mammary tumors. Indeed, when comparing mice with an equal primary tumor burden, significantly more circulating tumor cells were isolated from mice injected with the fast population relative to the parental MDA-MB-231 population (Fig. 4E). Representative images of lung and liver lesions are shown in Fig. S5D,E.
We previously demonstrated that specific β integrin subunits were upregulated in the fast-cell population relative to the parental MDA-MB-231 cells. Transcript levels for ITGB2, ITGB3 and ITGB4 remained elevated in fast cell tumors relative to parental tumors (Fig. 4F). However, at the protein level, only β2 and β4 integrin subunits remained elevated in fast cell mammary tumors relative to parental cell tumors (Fig. 4G,H).
Differentially expressed genes correlate with patient outcomes
To gain further insight into potential mechanisms driving the increased aggressive phenotype observed in the fast-cell population, we investigated our GO analysis more extensively. To identify genes that might be involved in fast cell aggressiveness, we selected genes that (1) were common in multiple sub-categories of each identified GO term, (2) displayed prognostic significance in the Breast Cancer TCGA data set (Fig. S6), and (3) had altered (up/down) expression levels in the fast-cell population relative to parental cells that were correlated with worse patient outcomes. These criteria resulted in a list of 27 differentially expressed genes between the fast and parental cell populations (Fig. 5A). Validation of these genes on independent RNA isolations from fast and parental cells revealed a subset of significantly modulated transcripts (Fig. 5B). We extended these analyses to assess a select number of candidates by immunoblot (Fig. 5C,D). Many of these significantly modulated proteins are known to promote tumor growth or enhance cell migration/invasion (GLUL, CYFIP2, MYLK, L1CAM and KLF4), suggesting additional targets that might promote the pro-metastatic phenotypes displayed by the fast-cell population. We next assessed the mRNA expression levels of these candidates from endpoint tumor material (Fig. 5E). Finally, we assessed the levels of protein expression for those genes that were significantly differentially expressed at the transcript level (Fig. 5F,G). Many of these targets retained the differential expression in tumors, suggesting they may play a functional role in modulating tumor growth and metastasis in vivo.
Fig. 5.
Validation of differentially expressed genes. (A) RNA-seq expression data of the 27 differentially expressed genes. (B) qPCR analysis of 27 prognostic targets identified from the RNA-seq data. Data are presented as the mean fold change difference between fast cells relative to parental MDA-MB-231 normalized to β-actin (ACTB) and RPLP0. Data from three independent RNA isolations are presented. (C) Immunoblot of selected proteins identified from the data shown in B. Three independent cell lysates are presented with a representative loading control, α-tubulin. (D) Quantification of the protein present on the immunoblot presented in C. Data are presented as signal normalized to α-tubulin on each individual immunoblot (data not shown). (E) qPCR analysis of 21 prognostic targets identified from the RNA-seq data and differential expression in B. Data are presented as the mean fold change difference between fast cells relative to parental MDA-MB-231 normalized to ACTB and RPLP0. Data from three independent RNA isolations are presented. (F) Immunoblot of selected proteins identified from the data shown in E. Five independent tumor lysates are presented with a representative loading control, α-tubulin. (G) Quantification of the protein present on the immunoblot presented in F. Data are presented as signal normalized to α-tubulin on each individual immunoblot (data not shown).*P<0.05, **P<0.005, ***P<0.0005. ns, not significant. Unpaired t-test was used for statistics. Error bars represent s.e.m.
Two targets revert the fast-migration phenotype
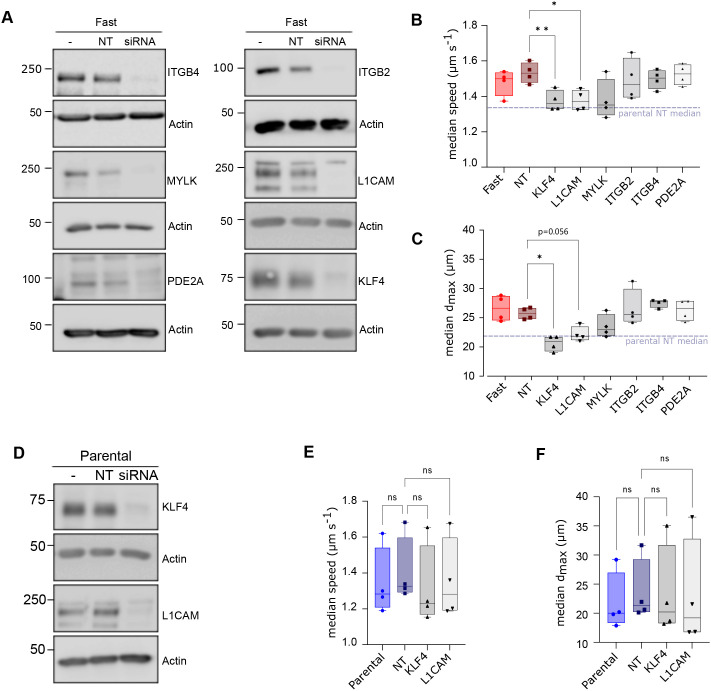
To determine whether the six differently expressed candidates that we identified in our in vivo analysis of the fast cell and parental populations (Figs 4 and 5) contributed to the migration phenotype, we transiently knocked down each target using small interfering RNA (siRNA) (Fig. 6A) and measured the effect on cellular migration in vitro. From four replicated experiments, L1CAM and KLF4 siRNAs significantly decreased the speed (ES=2.8 and 3.0, respectively) compared with the non-targeting siRNA (Fig. 6B). Although we measured a very large ES for both knockdowns on dmax (3.4 for L1CAM and 4.1 for KLF4) only KLF4 significantly decreased dmax compared with the non-targeting siRNA (Fig. 6C). Transient knockdown of either L1CAM or KLF4 in the parental cells (Fig. 6D) did not have a significant effect on motility, with a small ES on speed of 0.21 and 0.46 and on dmax 0.27 and 0.15 (Fig. 6E,F), respectively. Moreover, knockdown of KLF4 or L1CAM in the fast cells reduced the median speed and dmax to within 2-3% of the non-targeting knockdown parental control, suggesting that these two gene targets contribute to the fast cell phenotype.
Fig. 6.
Contribution of candidate target genes to the fast cell phenotype in vitro. (A) Immunoblots from fast-selected MDA-MB-231 breast cancer cells transfected with siRNAs against candidate gene targets (siRNA), or non-targeting (NT) controls, were performed 48-h post-transfection. Non-transfected fast MDA-MB-231 cells (-) served as an additional control for candidate protein expression. An immunoblot for actin served as a loading control. (B,C) Median speed and dmax of fast cells following transient knockdown of the indicated targets. Non-significant differences with the NT sample are omitted for simplicity. (D) Immunoblot analysis for KLF4 and L1CAM following transient knockdown in parental MDA-MB-231 cells. An immunoblot for actin served as a loading control. (E,F) Median speed and dmax of parental cells following knockdown of KLF4 and L1CAM. Differences are compared to the parental NT sample. Boxplots show minimum value, first quartile, median, third quartile and maximum value. *P<0.05; **P<0.005; ns, not significant. Statistics for the tracking experiments (B,C,E,F) were accomplished with paired one-way ANOVA corrected for multiple comparison with Geisser–Greenhouse correction. The immunoblots represent one of the replicates for tracking.
DISCUSSION
Isolation and expansion of single cells based on phenotypic rather than genotypic traits opens the door to uncovering molecular pathways underlying well-understood cellular behaviors. Here, we tracked single cells in a culture of metastatic TNBC cells and captured the most migratory cells from an analysis of their trajectories. We showed that this migratory phenotype is preserved after many cell divisions, indicating that this behavior is encoded in the genome or epigenome of the selected cells, and investigated the molecular underpinnings that could explain their enhanced migration kinetics.
Capture and expansion of single cells based on visual phenotypes may not necessarily render identical results in parallel experiments, as the underlying molecular pathways that determine similar behaviors can differ within a large heterogeneous cell population. Indeed, in our hands, a fast migratory phenotype that was preserved for longer than 6 weeks in culture was achieved for four out of six independent experiments. The fastest of these cell populations was studied thoroughly in vitro and in vivo. Despite the wide distribution of velocities that could be observed in each experiment, the differential in average velocity of both populations was statistically significant at all time points. The large width of all distributions is explained by the total short duration of movies, differences in cell cycle phase, as well as intrinsic variability. Furthermore, the fast cell lines we generated are not clonal, as several tens of cells are originally captured and expanded to increase the throughput of experiments.
The isolation and characterization of metastatic cancer cells remains an active area of research (Fidler, 2003; Entenberg et al., 2023; Fares et al., 2020). A fundamental characteristic of metastatic tumor cells is their ability to migrate (Partin et al., 1989). Here, we have isolated a fast-migrating population of tumor cells to determine whether an increased migratory ability correlates with metastatic fitness. To the best of our knowledge, very few attempts have been made to isolate metastatic cells based solely on migration-related phenotypes for transcriptomic analysis and in vivo experiments (Beri et al., 2020; Yankaskas et al., 2019; Li et al., 2020; Hapach et al., 2021). The microfluidic assay for quantification of cell invasion (MAqCI), (Yankaskas et al., 2019), cell invasion in digital microfluidic microgel systems (CIMMS), (Li et al., 2020) and the isolation of cells based on substrate adhesion strength by applying a shear stress in a custom flow chamber are analogous techniques that have addressed this goal (Beri et al., 2020). Cells isolated by MAqCI and flow chambers demonstrated enhanced velocity after isolation and displayed an enhanced propensity to form metastases, but their phenotypes returned to normal after 2 weeks (Yankaskas et al., 2019; Beri et al., 2020). Cells isolated by multiple rounds of trans-well migration kept their phenotype; however, this did not correlate with an increase in metastatic ability (Hapach et al., 2021).
Independent transcriptomic analyses revealed that fast and parental cell populations display highly distinct expression profiles, with the first component of principal component analysis accounting for 91% of the difference between parental and fast migrating MDA-MB-231 cells. We identified a list of significant GO terms that primarily highlight biological processes associated with adhesion complexes that are implicated in cell migration. Fast cells were isolated on fibronectin-coated substrates; however, increased motility was observed on laminin and collagen as well. This suggests that the molecular mechanisms driving rapid cell migration are – at least in part – upstream of interactions with the underlying substrate. However, it is notable that as cancer cells progress through the metastatic cascade, changes in ECM substrate composition are often co-incident with three-dimensional changes in pore size and tissue stiffness, which all impact metastases. How these factors affect the selected population of fast cells in comparison with the parental population remains to be determined.
Differences in cell migration phenotypes between captured and parental cells were associated with rapid turnover of smaller cell–matrix adhesions within the fast population. Our results show that fast migrating MDA-MB-231 cells exhibit a shift in their integrin profiles, with an upregulation of β4 relative to parental cells, which was also retained within mammary tumors. The a6β4 integrin receptor complex displays preferential binding to laminin-332 and has been implicated in the regulation of the actin cytoskeleton dynamics through the Rho family of small GTPases (Nishiuchi et al., 2006; Miyazaki, 2006). The greater aspect ratio (height/spread area) for the fast cells suggests that they are more contractile and result in shorter FA lifetimes (Murrell et al., 2015; Elosegui-Artola et al., 2018). In line with these findings, fast cells seeded on laminin-332 exhibited increased rates of adhesion assembly and disassembly relative to parental cells. However, the role of laminin-332 in cell motility remains understudied and controversial (Rousselle and Beck, 2013; Calaluce et al., 2004). Some studies argue that α3β1 primarily promotes keratinocyte motility on laminin-332, whereas α6β4 promotes stable ECM contacts owing to its role in promoting classical hemidesmosome (type I HD) formation. In contrast, other studies argue that α6β4 does regulate cell migration, as reduced expression of integrin β4 in human keratinocytes reduces cell motility on laminin-332.
Nevertheless, laminin-332 is aberrantly expressed at the invasive edge of numerous solid cancers, including breast cancer (Kwon et al., 2012; Carpenter et al., 2018; Kim et al., 2011). Our results indicate that MDA-MB-231 breast cancer cells produce laminin-332, which suggests that fast cells may increase their local migratory and invasive responses to this ECM constituent via increased α6β4 integrin expression. This phenomenon may explain the higher number of circulating tumor cells (CTCs) and increased metastatic burden observed with tumors derived from the fast-cell population. Finally, several ligand-independent functions of β4 integrin have been reported, including cooperation with receptor tyrosine kinase signaling to promote tumor cell invasion and metastasis and enhanced invasion of cancer cells through collagen I (Chao et al., 1996). Accordingly, we observed that mice bearing sized-matched mammary tumors derived from fast breast cancer cells exhibited increased CTCs and metastatic burden compared with mice bearing tumors from the parental MDA-MB-231 population.
Our broader transcriptomic analysis revealed several candidates that have known roles in tumor progression. The observation that L1CAM is highly expressed in the fast-cell population is consistent with literature linking L1CAM and cancer metastasis. L1CAM is a transmembrane glycoprotein that was initially identified for its role in neural development; however, its expression in breast cancer is associated with nodal involvement, high grade, and shorter disease-free period and overall survival (Schröder et al., 2009; Zhang et al., 2015). L1CAM expression promotes breast cancer cell migration and invasion (Zhang et al., 2015) as well as adhesion to activated endothelium (Dippel et al., 2013). In the brain, L1CAM expression on breast and lung cancer cells promotes the invasive growth of brain metastases (Valiente et al., 2014). In colorectal cancer, L1CAM expression promotes adhesion to ECM proteins of the vasculature and is associated with a stem cell-like metastasis phenotype (Ganesh et al., 2020).
Transcriptomic analyses also revealed upregulation of myosin light chain kinase (MYLK) in fast cells. MYLK phosphorylation of its substrate, myosin light chain, is required for the formation of contractile actomyosin filaments that play a crucial role in regulating cell morphology, contraction and motility (Kamm and Stull, 2001). In breast cancer, MYLK plays a role in promoting cell spreading (Betapudi et al., 2006), invadopodia formation and cell invasion (Sundararajan et al., 2015; Mierke et al., 2011). Consequently, inhibition of MYLK impairs the metastatic ability of breast cancer cells in vivo (Barkan et al., 2008). The consistent upregulation of MYLK in fast selected cells relative to parental MDA-MB-231 cells is consistent with the enhanced migratory and metastatic phenotypes exhibited by these cells.
The zinc finger-containing transcription factor KLF was also identified in our analysis. KLF has been shown to promote breast cancer cell migration, invasion and metastasis as well as maintenance of cancer stem cell phenotypes (Yu et al., 2011; Okuda et al., 2013). Stabilization of KLF4, via reduced KLF4 ubiquitylation, was associated with increased breast cancer metastasis in pre-clinical models and poor patient outcomes (Zou et al., 2019). However, the role of KLF4 in the context of breast cancer is controversial, with some studies suggesting the high levels of KLF4 are associated with good outcomes in people with breast cancer and can suppress metastasis in pre-clinical models (Zhu et al., 2022; Nagata et al., 2014; Yori et al., 2011).
Cytoplasmic FMR1 interacting protein 2 (CYFIP2) and phosphodiesterase 2A (PDE2A), two transcriptional targets identified in this analysis, have not been well studied in terms of their roles in breast cancer. CYFIP2 has well-documented roles in neural development and is a part of the WAVE regulatory complex, which controls actin polymerization and branching (Biembengut et al., 2021). CYFIP2 is also a p53-regulated, pro-apoptotic protein that, when downregulated in gastric cancer, promotes cell proliferation (Jiao et al., 2017). Similarly, loss of CYFIP2 promotes cell survival in colon cancer (Mongroo et al., 2011) and downregulation of CYFIP2 expression was also observed in people with clear cell renal cell carcinoma (Jiao et al., 2017). PDE2A is a cyclic nucleotide phosphodiesterase that hydrolyses cGMP and cAMP, thereby controlling the levels of these second messengers in cells. The role of PDE2A in the regulation of cancer cell migration is complex. In melanoma models, PDE2 has been shown to promote proliferation and invasion (Morita et al., 2013; Hiramoto et al., 2014); however, in osteosarcoma cell models PDE2 activity suppressed migration (Murata et al., 2019). The contribution of each of these genes in breast cancer requires further evaluation.
Transient knockdown of these six candidate genes individually revealed that L1CAM and KLF4 could, in part, be responsible for the fast-migration phenotype. Their knockdown significantly decreased the motility of the fast-cell population, with no significant effect on the parental cells, thus equalizing the migration profiles of both populations. Further in vivo studies would be required to determine the contribution of L1CAM, KLF4 and the remaining four candidates on the metastatic fitness of the fast-cell population.
Here, we have observed a correlation between increased migratory speed and enhanced metastatic ability; however, migration alone is not the only requirement for metastasis. Indeed, elegant intravital imaging studies have revealed interactions between tumor cells and macrophages. These cellular interactions modulated MENA (ENAH) transcripts within the tumor cells to increase their migratory capacity (Goswami et al., 2009), which resulted in increased metastasis. Cancer-associated fibroblasts have also been shown to enhance tumor cell migration through chemokine expression and microRNA expression (Elwakeel and Weigert, 2021). Our in vitro analysis identified two candidates that contribute to a fast-migration phenotype; further in vivo analysis may reveal insights to the role of our remaining four candidates in the metastatic process.
To our knowledge, scMOCa is the only technology that allows in vitro capture of single metastatic cells based on detailed analysis of motility without any genetic manipulations. The physical isolation of highly motile cells from a heterogeneous population demonstrates that migration characteristics alone can lead to an increased metastatic potential. Furthermore, migratory characteristics can be passed from generation to generation, allowing the underlying molecular mechanisms of highly motile cells to be probed based on transcriptomic traits. Other phenotypes associated with the metastatic potential of cancer cells could be investigated with this technology, as most cellular biomarkers that depend on time evolution, intracellular localization or contact are rarely accessible with standard cell separation methods because they require high-content imaging. This study illustrates the capability of probing the molecular underpinnings of rare cells detected visually that can drive major physiological changes in an organism and could be extended to other characteristics for which there are no available biochemical markers.
MATERIALS AND METHODS
Cell imaging and tracking
Cells were imaged with a custom-made device for darkfield illumination to obtain enhanced contrast and avoid toxicity caused by the photobleaching of fluorescent labels or the use of genetically modified cell lines. A phase ring IX-PH3 (Olympus, Japan) was inserted in the turret of a 0.55 NA brightfield condenser (Nikon), as previously described (Antolović et al., 2014). Cells were imaged with a CFI Plan Fluor 10× of 0.3 NA objective (Nikon) on an automated ECLIPSE Ti2 inverted microscope (Nikon). Images were acquired at 2-min intervals for 2 h on over 89 stage positions, covering the culture surface of the imaging dish. The dimension of a single field of view is 1769×1769 µm2. The cells covered about 5% of the observation area and we counted 30-50 cells per field of view. A MATLAB script performed live cell segmentation during acquisition based on intensity thresholding followed by cell tracking using a nearest neighbor algorithm previously described (Jaqaman et al., 2008; Mazzaferri et al., 2015). Cells were classified using dmax=max{[(xi−xj)2+(yi−yj)2]1/2}, where (xk,yk) is a point in a cell's trajectory, as illustrated in Fig. 1B. Because each tracking experiment quantified about 8000 cells per condition, we used bootstrapping to perform the statistical analysis, which consists of measuring the probability of the null hypothesis by numerically replicating the experiments 1500 times using resampled datasets from the original data. In other words, the two populations were compared thousands of times by randomly labeling cell trajectories to assess whether differences observed experimentally were significant. The ES between two populations was calculated by (M2−M1)/s, where Mx is the median value and s is the combined standard deviation.
Cell capture and culture
Prior to tracking, MDA-MB-231 cells (ATCC, tested for contamination with IMPACT testing) were plated in DMEM (Wisent) supplemented with 10% fetal bovine serum (Wisent) and 1% penicillin-streptomycin-neomycin (Gibco, Thermo Fisher Scientific) with 40 mg ml−1 biotin-4-fluorescein (Sigma-Aldrich) and plated at very low density on a plastic-bottom 35 mm µ-dish (ibidi) coated with 5 µg cm−2 of human plasma fibronectin (Sigma-Aldrich). Cells covered about 3-5% of the 3.5 cm2 culture area of the imaging dish. During the 2-h acquisition, cells were kept in a stage-top incubator (ibidi). After live segmentation and tracking, the 5% most motile cells were identified and illuminated automatically with a 473 nm laser at 6 mW for 0.1 s focused with the same objective used for imaging. The laser's waist measured 9 µm at e−2 of the peak intensity, thus restricting illumination to single cell bodies. After five washes with PBS, we swept streptavidin-coated ferromagnetic beads of 2.8 µm diameter (Thermo Fisher Scientific) on the bottom of the culture dish by manually scanning a magnetized nail under the dish. After washing away unbound beads, only illuminated cells became decorated with the ferromagnetic beads. Cells were detached with 0.25% trypsin (Thermo Fisher Scientific) and placed in a vertical magnetic field to attract labeled cells alone, against gravity. The collection chamber was placed 6 mm above the imaging dish surface and kept in place by a custom-made holder. Captured cells were expanded for 2 weeks before further characterization. A more detailed protocol is available elsewhere (Binan et al., 2019a,b).
RNA extraction, sequencing and qPCR validation
Fast and parental cultures were subjected to the same number of passages. Three technical replicates (plated the same day) and two biological replicates (plated after two weeks of passages, frozen and thawed) were performed. RNA was extracted with RNeasy Micro Kit (QIAGEN) from three cultures (triplicate) per condition containing ∼40,000 cells. Dishes were coated with 5 µg cm−2 of human plasma fibronectin. RNA was then sequenced on Illumina Novaseq 6000 for 25 M paired ended, 100 nt reads with a library prepared with the NEBNext Ultra kit (New England BioLabs).
Raw reads were trimmed using Trimmomatic v0.32. First, adaptors and other Illumina-specific sequences from each read were removed using palindrome mode. Then, a four-nucleotide sliding window removed the bases once the average quality within the window fell below 30. Next, the first four bases at the start of each read were removed. Finally, reads shorter than 30 base pairs were filtered out. Reads were aligned to the human reference genome build hg19 using STAR v2.3.0e with default settings (Dobin et al., 2013). Reads mapping to more than ten locations in the genome (MAPQ<1) were discarded. Gene expression levels were estimated by quantifying primary alignments mapping to at most two locations (MAPQ≥3) to exonic regions (the maximal genomic locus of each gene and its known isoforms) using feature Counts v1.4.4 and the hg19 ensGene annotation set from Ensembl. Normalization (mean of ratios), variance-stabilized transformation of the data and differential gene expression analysis were performed using DESeq2 v1.14.1. Multiple control metrics were obtained using FastQC v0.11.2, samtools v0.1.20, BEDtools v2.17.0 and custom scripts. GO analysis of differentially expressed genes was performed with g:Profiler with an ordered query. To reduce the number of terms for visualization purposes, GO terms were subsequently grouped using a k-means algorithm based on similarity (overlap) in the gene list of each term.
Isolated RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer's directions. Quantitative PCR was performed using fast-start universal (Sigma-Aldrich), run on a Bio-Rad CFX instrument and analyzed by standard methods (Livak and Schmittgen, 2001) with normalization to reference genes.
In vitro characterization at the microscope
Adhesion dynamics assay
MDA-MB-231 cells were infected with a pMSCV-blast vector containing mCherry paxillin. Cells were seeded onto 35 mm glass-bottom dishes (ibidi) coated with 5 µg cm−2 human plasma fibronectin. Cells were imaged on a TIRF-Spinning Disk Spectral Diskovery System (Spectral Applied Research) coupled to a DMI6000B Leica microscope using with a Plan-Apochromat 63×1.47 NA. Each cell was illuminated continuously with a 561 nm laser for 15 min. TIRF illumination was used to limit fluorescence excitation to a depth of ∼100 nm. Pixel binning (2×2) was used to enhance signal. Assembly and disassembly rate were than analyzed using a custom MATLAB algorithm described previously (Kiepas et al., 2020a,b).
FA characterization
Cells were seeded onto 35 mm glass-bottom dishes (ibidi) coated with 5 µg cm−2 human plasma fibronectin. Cells were than fixed with 3% paraformaldehyde and 0.1% Triton X-100 in PBS and rinsed with PBS with 3.8 mg ml−1 of glycine. Cells were incubated overnight at 4°C with 1:200 purified mouse anti-paxillin (BD Transduction Laboratories) in 90% PBS, 10% fetal bovine serum, 0.05% Triton X-100 and 10 mg ml−1 bovine serum albumin. The cells were then incubated for 1 h with in 1:250 Alexa Fluor 647 goat anti-mouse (Thermo Fisher Scientific) in the same solution. The z-stacks of the cells were acquired over 1 µm above the glass surface at an interval of 0.2 µm using a IX71 inverted microscope (Olympus) equipped with a confocal microscopy upgrade (Thorlabs) and a 100×1.45 NA objective. The mean z-projection was then used to quantify the size of the FAs with a custom-made MATLAB script.
FA super-resolution imaging
Cells were stained as for FA characterization. For each condition (fast and parental), 11 cells were imaged on a Stellaris 8 STED microscope (Leica) with a 100×1.40 NA objective. Every horizontal line of the scan – fast axis – was acquired twice, one time with the confocal modality and one time with the STED modality, before moving to the next line of the scan. The depletion laser for the STED was set at 20% of the excitation laser intensity.
Cell morphology
Cells were fixed in 4% paraformaldehyde in PBS at room temperature and incubated with 1:200 wheat germ agglutinin conjugated with Alexa Fluor 488 (Thermo Fisher Scientific) in HBSS medium (Thermo Fisher Scientific) to label plasma membranes. z-stacks of the cells were acquired using a IX71 inverted microscope (Olympus) equipped with a confocal microscopy upgrade (Thorlabs) and a 100×1.45 NA objective. A custom-made MATLAB program was written to analyze the z-stacks.
Transient knockdown of gene targets using siRNA
For each condition, 400,000 cells were seeded into a 60 mm dish. After 24 h, cells were transfected with 20 nmols of SMART-pool siRNA (Horizon Discovery Biosciences Ltd) using 5 µl Lipofectamine RNAiMAX (Thermo Fisher Scientific) in 5 ml of media. The following day, cells were seeded onto fibronectin-coated, plastic-bottom 8-well plates for imaging and 60 mm dishes to confirm efficient target knockdown by immunoblotting. The next day (48 h post-transfection), the cells were imaged and tracked to determine the effect of reduced target expression on cell migration.
In vivo experiments
One million MDA-MB-231 parental or Fast cells were injected into the mammary fad pad of 6-week-old NOD-SCID Gamma female mice (The Jackson Laboratory). Tumors were measured weekly using caliper measurements and tumor volumes calculated using the formula 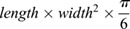 . CTCs and primary mammary tumor tissues when tumors reached ∼500 mm3 and lung/liver tissues were collected at endpoint. Lung and liver sections were stained for vimentin using a Ventana autostainer (Roche), digitally scanned, and metastatic area scored using ImageScope (Leica Biosystems). A more detailed protocol for CTC isolation has been previously described (Ngan et al., 2017).
. CTCs and primary mammary tumor tissues when tumors reached ∼500 mm3 and lung/liver tissues were collected at endpoint. Lung and liver sections were stained for vimentin using a Ventana autostainer (Roche), digitally scanned, and metastatic area scored using ImageScope (Leica Biosystems). A more detailed protocol for CTC isolation has been previously described (Ngan et al., 2017).
A minimum of eight samples are required to provide statistical power to these experiments; we injected ten mice per arm of the experiment to account for attrition that may occur in these types of experiments. Student's t-test was used to analyze the in vivo data. All animal studies were approved by the Animal Resource Centre at McGill University and comply with guidelines set by the Canadian Council of Animal Care.
Immunoblotting
Membranes were prepared and processed as previously described (Annis et al., 2018). Immunoblots were developed on a ChemiDoc Imager (Bio-Rad) and analyzed using Image Studio Lite software (LI-COR). See Table S2 for the antibodies used in this study.
Supplementary Material
Acknowledgements
We acknowledge the Goodman Cancer Institute histology core facility (McGill University) for routine histological services and the Advanced Bioimaging Facility (ABIF) for TIRF microscopy access.
Footnotes
Author contributions
Conceptualization: N.D.-L., M.G.A., P.M.S., S.C.; Methodology: N.D.-L., M.G.A., A.N., P.M.S., S.C., S.H.; Software: N.D.-L., S.C., S.H.; Formal analysis: N.D.-L., M.G.A., A.N., A.K., C.L.K., P.M.S., S.C.; Investigation: N.D.-L., M.G.A., A.N., L.B., J.R., G.M., S.H., C.L.K., P.M.S., S.C.; Resources: P.M.S., S.C.; Data curation: S.C., P.M.S.; Writing - original draft: N.D.-L.; Writing - review & editing: M.G.A., C.L.K., P.M.S., S.C.; Visualization: S.C.; Supervision: P.M.S., S.C.; Project administration: P.M.S., S.C.; Funding acquisition: S.C., P.M.S.
Funding
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (to S.C.), the Cancer Research Society of Canada (to S.C.), Fonds de Recherche du Québec – Nature et Technologie (to S.C.), and a Canadian Cancer Society i2I grant (to P.M.S.). S.C. holds a salary award from the Fonds de Recherche du Québec – Santé and P.M.S. is a McGill University William Dawson Scholar. N.D.-L. and G.M. holds scholarships from the Fonds de Recherche en Ophtalmologie de l'Université de Montréal. N.D.-L. and A.K. hold a scholarship from the Natural Sciences and Engineering Research Council of Canada. A.K. holds a scholarship from Fonds de Recherche du Québec – Santé. A.N. holds a scholarship from the Natural Sciences and Engineering Research Council of Canada. A.N. holds a scholarship from Fonds de Recherche du Québec – Nature et Technologie.
Data availability
The gene expression raw data and processed files are available on NCBI Gene Expression Omnibus (GEO) data repository (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE188224.
Contributor Information
Peter M. Siegel, Email: peter.siegel@mcgill.ca.
Santiago Costantino, Email: santiago.costantino@umontreal.ca.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/lookup/doi/10.1242/jcs.260835.reviewer-comments.pdf
References
- Aleshin, A. and Finn, R. S. (2010). SRC: a century of science brought to the clinic. Neoplasia 12, 599-607. 10.1593/neo.10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibert, C., Goud, B. and Manneville, J.-B. (2017). Are cancer cells really softer than normal cells? Biol. Cell 109, 167-189. 10.1111/boc.201600078 [DOI] [PubMed] [Google Scholar]
- Annis, M. G., Ouellet, V., Rennhack, J. P., L'Esperance, S., Rancourt, C., Mes-Masson, A.-M., Andrechek, E. R. and Siegel, P. M. (2018). Integrin-uPAR signaling leads to FRA-1 phosphorylation and enhanced breast cancer invasion. Breast Cancer Res. 20, 9. 10.1186/s13058-018-0936-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolović, V., Marinović, M., Filić, V. and Weber, I. (2014). A simple optical configuration for cell tracking by dark-field microscopy. J. Microbiol. Methods 104, 9-11. 10.1016/j.mimet.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Aziz, A. U. R., Deng, S., Jin, Y., Li, N., Zhang, Z., Yu, X. and Liu, B. (2022). The explorations of dynamic interactions of paxillin at the focal adhesions. Biochim. Biophys. Acta Proteins Proteomics 1870, 140825. 10.1016/j.bbapap.2022.140825 [DOI] [PubMed] [Google Scholar]
- Barkan, D., Kleinman, H., Simmons, J. L., Asmussen, H., Kamaraju, A. K., Hoenorhoff, M. J., Liu, Z.-Y., Costes, S. V., Cho, E. H. and Lockett, S. (2008). Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 68, 6241-6250. 10.1158/0008-5472.CAN-07-6849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beri, P., Matte, B. F., Fattet, L., Kim, D., Yang, J. and Engler, A. J. (2018). Biomaterials to model and measure epithelial cancers. Nat. Rev. Materials 3, 418-430. 10.1038/s41578-018-0051-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beri, P., Popravko, A., Yeoman, B., Kumar, A., Chen, K., Hodzic, E., Chiang, A., Banisadr, A., Placone, J. K. and Carter, H. (2020). Cell adhesiveness serves as a biophysical marker for metastatic potential. Cancer Res. 80, 901-911. 10.1158/0008-5472.CAN-19-1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betapudi, V., Licate, L. S. and Egelhoff, T. T. (2006). Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 66, 4725-4733. 10.1158/0008-5472.CAN-05-4236 [DOI] [PubMed] [Google Scholar]
- Biembengut, Í. V., Silva, I. L. Z., de Souza, T. D. A. C. B. and Shigunov, P. (2021). Cytoplasmic FMR1 interacting protein (CYFIP) family members and their function in neural development and disorders. Mol. Biol. Rep. 48, 6131-6143. 10.1007/s11033-021-06585-6 [DOI] [PubMed] [Google Scholar]
- Bijian, K., Lougheed, C., Su, J., Xu, B., Yu, H., Wu, J. H., Riccio, K. and Alaoui-Jamali, M. (2013). Targeting focal adhesion turnover in invasive breast cancer cells by the purine derivative reversine. Br. J. Cancer 109, 2810-2818. 10.1038/bjc.2013.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binan, L., Mazzaferri, J., Choquet, K., Lorenzo, L.-E., Wang, Y. C., Affar, E. B., De Koninck, Y., Ragoussis, J., Kleinman, C. L. and Costantino, S. (2016). Live single-cell laser tag. Nat. Commun. 7, 11636. 10.1038/ncomms11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binan, L., Belanger, F., Uriarte, M., Lemay, J. F., de Koninck, J. C. P., Roy, J., Affar, E. B., Drobetsky, E., Wurtele, H. and Costantino, S. (2019a). Opto-magnetic capture of individual cells based on visual phenotypes. eLife 8, e45239. 10.7554/eLife.45239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binan, L., Roy, J. and Costantino, S. (2019b). Opto-magnetic selection and isolation of single cells. Bio-protocol 9, e3428. 10.21769/BioProtoc.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calaluce, R., Bearss, D. J., Barrera, J., Zhao, Y., Han, H., Beck, S. K., Mcdaniel, K. and Nagle, R. B. (2004). Laminin-5 β3A expression in LNCaP human prostate carcinoma cells increases cell migration and tumorigenicity. Neoplasia 6, 468-479. 10.1593/neo.03499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, P. M., Ziogas, A., Markham, E. M., Cantillep, A. S., Yan, R. and Anton-Culver, H. (2018). Laminin 332 expression and prognosis in breast cancer. Hum. Pathol. 82, 289-296. 10.1016/j.humpath.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, L. B. and Waterman, C. M. (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955-963. 10.1038/ncb3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, C., Lotz, M. M., Clarke, A. C. and Mercurio, A. M. (1996). A function for the integrin α6β4 in the invasive properties of colorectal carcinoma cells. Cancer Res. 56, 4811-4819. [PubMed] [Google Scholar]
- Choi, C. K., Vicente-Manzanares, M., Zareno, J., Whitmore, L. A., Mogilner, A. and Horwitz, A. R. (2008). Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039-1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G. and Vignjevic, D. M. (2015). Modes of cancer cell invasion and the role of the microenvironment. Curr. Opin. Cell Biol. 36, 13-22. 10.1016/j.ceb.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Cooper, J. and Giancotti, F. G. (2019). Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347-367. 10.1016/j.ccell.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, S. E., Jin, Y.-S., Rao, J. and Gimzewski, J. K. (2007). Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2, 780-783. 10.1038/nnano.2007.388 [DOI] [PubMed] [Google Scholar]
- De Martino, M., Daviaud, C., Diamond, J. M., Kraynak, J., Alard, A., Formenti, S. C., Miller, L. D., Demaria, S. and Vanpouille-Box, C. (2020). Activin A promotes regulatory T-cell-mediated immunosuppression in irradiated breast cancer. Cancer Immunol. Res. 9, 89-102. 10.1158/2326-6066.CIR-19-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippel, V., Milde-Langosch, K., Wicklein, D., Schumacher, U., Altevogt, P., Oliveira-Ferrer, L., Jänicke, F. and Schröder, C. (2013). Influence of L1-CAM expression of breast cancer cells on adhesion to endothelial cells. J. Cancer Res. Clin. Oncol. 139, 107-121. 10.1007/s00432-012-1306-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosegui-Artola, A., Trepat, X. and Roca-Cusachs, P. (2018). Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356-367. 10.1016/j.tcb.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Elwakeel, E. and Weigert, A. (2021). Breast cancer CAFs: spectrum of phenotypes and promising targeting avenues. Int. J. Mol. Sci. 22, 11636. 10.3390/ijms222111636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenberg, D., Oktay, M. H. and Condeelis, J. S. (2023). Intravital imaging to study cancer progression and metastasis. Nat. Rev. Cancer 23, 25-42. 10.1038/s41568-022-00527-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares, J., Fares, M. Y., Khachfe, H. H., Salhab, H. A. and Fares, Y. (2020). Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target. Ther. 5, 28. 10.1038/s41392-020-0134-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando, H. S., Kynaston, H. G. and Jiang, W. G. (2009). WASP and WAVE proteins: vital intrinsic regulators of cell motility and their role in cancer. Int. J. Mol. Med. 23, 141-148. [PubMed] [Google Scholar]
- Fidler, I. J. (2003). The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer 3, 453-458. 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Ganesh, K., Basnet, H., Kaygusuz, Y., Laughney, A. M., He, L., Sharma, R., O'Rourke, K. P., Reuter, V. P., Huang, Y.-H., Turkekul, M.et al. (2020). L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat. Cancer 1, 28-45. 10.1038/s43018-019-0006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett, D. R., Tse, H. T. K., Lee, S. A., Ying, Y., Lindgren, A. G., Yang, O. O., Rao, J., Clark, A. T. and Di Carlo, D. (2012). Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc. Natl Acad. Sci. USA 109, 7630-7635. 10.1073/pnas.1200107109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, S., Philippar, U., Sun, D., Patsialou, A., Avraham, J., Wang, W., Di Modugno, F., Nistico, P., Gertler, F. B. and Condeelis, J. S. (2009). Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin. Exp. Metastasis 26, 153-159. 10.1007/s10585-008-9225-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guck, J., Schinkinger, S., Lincoln, B., Wottawah, F., Ebert, S., Romeyke, M., Lenz, D., Erickson, H. M., Ananthakrishnan, R., Mitchell, D.et al. (2005). Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 88, 3689-3698. 10.1529/biophysj.104.045476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther, C. (2022). β2-integrins–regulatory and executive bridges in the signaling network controlling leukocyte trafficking and migration. Front. Immunol. 13, 809590. 10.3389/fimmu.2022.809590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi, H. and Ivaska, J. (2018). Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533-548. 10.1038/s41568-018-0038-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapach, L. A., Carey, S. P., Schwager, S. C., Taufalele, P. V., Wang, W., Mosier, J. A., Ortiz-Otero, N., Mcardle, T. J., Goldblatt, Z. E., Lampi, M. C.et al. (2021). Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res. 81, 3649-3663. 10.1158/0008-5472.CAN-20-1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto, K., Murata, T., Shimizu, K., Morita, H., Inui, M., Manganiello, V. C., Tagawa, T. and Arai, N. (2014). Role of phosphodiesterase 2 in growth and invasion of human malignant melanoma cells. Cell. Signal. 26, 1807-1817. 10.1016/j.cellsig.2014.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R. O. (2002). Integrins: bidirectional, allosteric signaling machines. Cell 110, 673-687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Jaqaman, K., Loerke, D., Mettlen, M., Kuwata, H., Grinstein, S., Schmid, S. L. and Danuser, G. (2008). Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695-702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, S., Li, N., Cai, S., Guo, H. and Wen, Y. (2017). Inhibition of CYFIP2 promotes gastric cancer cell proliferation and chemoresistance to 5-fluorouracil through activation of the Akt signaling pathway. Oncol. Lett. 13, 2133-2140. 10.3892/ol.2017.5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm, K. E. and Stull, J. T. (2001). Dedicated myosin light chain kinases with diverse cellular functions. J. Biol. Chem. 276, 4527-4530. 10.1074/jbc.R000028200 [DOI] [PubMed] [Google Scholar]
- Kiepas, A., Voorand, E., Mubaid, F., Siegel, P. M. and Brown, C. M. (2020a). Optimizing live-cell fluorescence imaging conditions to minimize phototoxicity. J. Cell Sci. 133, jcs242834. 10.1242/jcs.242834 [DOI] [PubMed] [Google Scholar]
- Kiepas, A., Voorand, E., Senecal, J., Ahn, R., Annis, M. G., Jacquet, K., Tali, G., Bisson, N., Ursini-Siegel, J., Siegel, P. M.et al. (2020b). The SHCA adapter protein cooperates with lipoma-preferred partner in the regulation of adhesion dynamics and invadopodia formation. J. Biol. Chem. 295, 10535-10559. 10.1074/jbc.RA119.011903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. H. and Wirtz, D. (2013). Focal adhesion size uniquely predicts cell migration. FASEB J. 27, 1351-1361. 10.1096/fj.12-220160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B. G., An, H. J., Kang, S., Choi, Y. P., Gao, M.-Q., Park, H. and Cho, N. H. (2011). Laminin-332-rich tumor microenvironment for tumor invasion in the interface zone of breast cancer. Am. J. Pathol. 178, 373-381. 10.1016/j.ajpath.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraning-Rush, C. M., Califano, J. P. and Reinhart-King, C. A. (2012). Cellular traction stresses increase with increasing metastatic potential. PLoS ONE 7, e32572. 10.1371/journal.pone.0032572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S.-Y., Chae, S. W., Wilczynski, S. P., Arain, A. and Carpenter, P. M. (2012). Laminin 332 expression in breast carcinoma. Appl. Immunohistochem. Mol. Morphol. 20, 159. 10.1097/PAI.0b013e3182329e8f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. and Werb, Z. (2018). Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20, 1349-1360. 10.1038/s41556-018-0236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. B., Scott, E. Y., Chamberlain, M. D., Duong, B. T. V., Zhang, S., Done, S. J. and Wheeler, A. R. (2020). Cell invasion in digital microfluidic microgel systems. Sci. Adv. 6, eaba9589. 10.1126/sciadv.aba9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., Patrawala, L., Yan, H., Jeter, C., Honorio, S.et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211-215. 10.1038/nm.2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Lee, S. J., Park, S., Konstantopoulos, K., Glunde, K., Chen, Y. and Barman, I. (2020). Cancer cells display increased migration and deformability in pace with metastatic progression. FASEB J. 34, 9307-9315. 10.1096/fj.202000101RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402-408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma, L., Teruya-Feldstein, J. and Weinberg, R. A. (2007). Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682-688. 10.1038/nature06174 [DOI] [PubMed] [Google Scholar]
- Mazzaferri, J., Roy, J., Lefrancois, S. and Costantino, S. (2015). Adaptive settings for the nearest-neighbor particle tracking algorithm. Bioinformatics 31, 1279-1285. 10.1093/bioinformatics/btu793 [DOI] [PubMed] [Google Scholar]
- Mierke, C. T., Rösel, D., Fabry, B. and Brábek, J. (2008). Contractile forces in tumor cell migration. Eur. J. Cell Biol. 87, 669-676. 10.1016/j.ejcb.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke, C. T., Frey, B., Fellner, M., Herrmann, M. and Fabry, B. (2011). Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J. Cell Sci. 124, 369-383. 10.1242/jcs.071985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, K. (2006). Laminin–5 (laminin–332): unique biological activity and role in tumor growth and invasion. Cancer Sci. 97, 91-98. 10.1111/j.1349-7006.2006.00150.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongroo, P. S., Noubissi, F. K., Cuatrecasas, M., Kalabis, J., King, C. E., Johnstone, C. N., Bowser, M. J., Castells, A., Spiegelman, V. S. and Rustgi, A. K. (2011). IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 71, 2172-2182. 10.1158/0008-5472.CAN-10-3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, H., Murata, T., Shimizu, K., Okumura, K., Inui, M. and Tagawa, T. (2013). Characterization of phosphodiesterase 2A in human malignant melanoma PMP cells. Oncol. Rep. 29, 1275-1284. 10.3892/or.2013.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, T., Shimizu, K., Kurohara, K., Tomeoku, A., Koizumi, G. and Arai, N. (2019). Role of Phosphodiesterase2A in proliferation and migration of human osteosarcoma cells. Anticancer Res. 39, 6057-6062. 10.21873/anticanres.13812 [DOI] [PubMed] [Google Scholar]
- Murrell, M., Oakes, P. W., Lenz, M. and Gardel, M. L. (2015). Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 16, 486-498. 10.1038/nrm4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Shimada, Y., Sekine, S., Hori, R., Matsui, K., Okumura, T., Sawada, S., Fukuoka, J. and Tsukada, K. (2014). Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer 21, 96-101. 10.1007/s12282-012-0357-y [DOI] [PubMed] [Google Scholar]
- Ngan, E., Stoletov, K., Smith, H. W., Common, J., Muller, W. J., Lewis, J. D. and Siegel, P. M. (2017). LPP is a Src substrate required for invadopodia formation and efficient breast cancer lung metastasis. Nat. Commun. 8, 15059. 10.1038/ncomms15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi, R., Takagi, J., Hayashi, M., Ido, H., Yagi, Y., Sanzen, N., Tsuji, T., Yamada, M. and Sekiguchi, K. (2006). Ligand-binding specificities of laminin-binding integrins: A comprehensive survey of laminin–integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 25, 189-197. 10.1016/j.matbio.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Okuda, H., Xing, F., Pandey, P. R., Sharma, S., Watabe, M., Pai, S. K., Mo, Y.-Y., Iiizumi-Gairani, M., Hirota, S., Liu, Y.et al. (2013). miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 73, 1434-1444. 10.1158/0008-5472.CAN-12-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, C. P., Mycielska, M. E., Burcu, H., Osman, K., Collins, T., Beckerman, R., Perrett, R., Johnson, H., Aydar, E. and Djamgoz, M. B. A. (2008). Single cell adhesion measuring apparatus (SCAMA): application to cancer cell lines of different metastatic potential and voltage-gated Na+ channel expression. Eur. Biophys. J. 37, 359-368. 10.1007/s00249-007-0219-2 [DOI] [PubMed] [Google Scholar]
- Partin, A. W., Schoeniger, J. S., Mohler, J. L. and Coffey, D. S. (1989). Fourier analysis of cell motility: correlation of motility with metastatic potential. Proc. Natl Acad. Sci. USA 86, 1254-1258. 10.1073/pnas.86.4.1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak, K. (2011). Heterogeneity in breast cancer. J. Clin. Invest. 121, 3786-3788. 10.1172/JCI60534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, D., Gill, N. K., Santiskulvong, C., Sifuentes, J., Dorigo, O., Rao, J., Taylor-Harding, B., Wiedemeyer, W. R. and Rowat, A. C. (2015). Screening cell mechanotype by parallel microfiltration. Sci. Rep. 5, 17595. 10.1038/srep17595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramovs, V., Te Molder, L. and Sonnenberg, A. (2017). The opposing roles of laminin-binding integrins in cancer. Matrix Biol. 57-58, 213-243. 10.1016/j.matbio.2016.08.007 [DOI] [PubMed] [Google Scholar]
- Rashid, N. S., Grible, J. M., Clevenger, C. V. and Harrell, J. C. (2021). Breast cancer liver metastasis: current and future treatment approaches. Clin. Exp. Metastasis 38, 263-277. 10.1007/s10585-021-10080-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi, N., Aguet, M. and Stamenkovic, I. (2018). Cancer metastasis: a reappraisal of its underlying mechanisms and their relevance to treatment. Annu. Rev. Pathol. Mech. Dis. 13, 117-140. 10.1146/annurev-pathol-020117-044127 [DOI] [PubMed] [Google Scholar]
- Rousselle, P. and Beck, K. (2013). Laminin 332 processing impacts cellular behavior. Cell Adhes. Migr. 7, 122-134. 10.4161/cam.23132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, C., Schumacher, U., Fogel, M., Feuerhake, F., Müller, V., Wirtz, R. M., Altevogt, P., Krenkel, S., Jänicke, F. and Milde-Langosch, K. (2009). Expression and prognostic value of L1-CAM in breast cancer. Oncol. Rep. 22, 1109-1117. 10.3892/or_00000543 [DOI] [PubMed] [Google Scholar]
- Seyfried, T. N. and Huysentruyt, L. C. (2013). On the origin of cancer metastasis. Crit. Rev. Oncog. 18, 43-73. 10.1615/CritRevOncog.v18.i1-2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui, K., Safina, A., Li, X., Vaughan, M. M., Hicks, D. G., Bakin, A. V. and Cowell, J. K. (2007). Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am. J. Pathol. 170, 2112-2121. 10.2353/ajpath.2007.060975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.-X. and Yu, Q. (2015). Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol. Sin. 36, 1219-1227. 10.1038/aps.2015.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan, V., Gengenbacher, N., Stemmler, M. P., Kleemann, J. A., Brabletz, T. and Brabletz, S. (2015). The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget 6, 27083. 10.18632/oncotarget.4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente, M., Obenauf, A. C., Jin, X., Chen, Q., Zhang, X. H.-F., Lee, D. J., Chaft, J. E., Kris, M. G., Huse, J. T., Brogi, E.et al. (2014). Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002-1016. 10.1016/j.cell.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, Q., Lu, D., Zhou, F., Wang, D., Feng, R., Wang, K., Molday, R., Xie, J. and Wen, T. (2017). A biosystems approach to identify the molecular signaling mechanisms of TMEM30A during tumor migration. PLoS ONE 12, e0179900. 10.1371/journal.pone.0179900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski, E. O., Mistriotis, P., Bera, K., Law, R. A., Zhang, J., Nikolic, M., Weiger, M., Parlani, M., Tuntithavornwat, S., Afthinos, A.et al. (2020). Dorsoventral polarity directs cell responses to migration track geometries. Sci. Adv. 6, eaba6505. 10.1126/sciadv.aba6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, G. and Hemmings, B. A. (2013). PKB/Akt–dependent regulation of cell motility. J. Natl. Cancer Inst. 105, 393-404. 10.1093/jnci/djs648 [DOI] [PubMed] [Google Scholar]
- Yamada, K. M. and Sixt, M. (2019). Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20, 738-752. 10.1038/s41580-019-0172-9 [DOI] [PubMed] [Google Scholar]
- Yankaskas, C. L., Thompson, K. N., Paul, C. D., Vitolo, M. I., Mistriotis, P., Mahendra, A., Bajpai, V. K., Shea, D. J., Manto, K. M., Chai, A. C.et al. (2019). A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3, 452-465. 10.1038/s41551-019-0400-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yori, J. L., Seachrist, D. D., Johnson, E., Lozada, K. L., Abdul-Karim, F. W., Chodosh, L. A., Schiemann, W. P. and Keri, R. A. (2011). Krüppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer. Neoplasia 13, I601-IN5. 10.1593/neo.11260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Li, J., Chen, H., Fu, J., Ray, S., Huang, S., Zheng, H. and Ai, W. (2011). Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 30, 2161-2172. 10.1038/onc.2010.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Yang, F., Ding, Y., Zhen, L., Han, X., Jiao, F. and Tang, J. (2015). Overexpression of L1 cell adhesion molecule correlates with aggressive tumor progression of patients with breast cancer and promotes motility of breast cancer cells. Int. J. Clin. Exp. Pathol. 8, 9240. [PMC free article] [PubMed] [Google Scholar]
- Zhu, K.-Y., Tian, Y., Li, Y.-X., Meng, Q.-X., Ge, J., Cao, X.-C., Zhang, T. and Yu, Y. (2022). The functions and prognostic value of Krüppel-like factors in breast cancer. Cancer Cell Int. 22, 1-12. 10.1186/s12935-022-02449-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H., Chen, H., Zhou, Z., Wan, Y. and Liu, Z. (2019). ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4. Cancer Lett. 467, 19-28. 10.1016/j.canlet.2019.09.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.