Abstract
Polycomb group response elements (PREs) mediate the mitotic inheritance of gene expression programs and thus maintain determined cell fates. By default, PREs silence associated genes via the targeting of Polycomb group (PcG) complexes. Upon an activating signal, however, PREs recruit counteracting trithorax group (trxG) proteins, which in turn maintain target genes in a transcriptionally active state. Using a transgenic reporter system, we show that the switch from the silenced to the activated state of a PRE requires noncoding transcription. Continuous transcription through the PRE induced by an actin promoter prevents the establishment of PcG-mediated silencing. The maintenance of epigenetic activation requires transcription through the PRE to proceed at least until embryogenesis is completed. At the homeotic bithorax complex of Drosophila, intergenic PRE transcripts can be detected not only during embryogenesis, but also at late larval stages, suggesting that transcription through endogenous PREs is required continuously as an anti-silencing mechanism to prevent the access of repressive PcG complexes to the chromatin. Furthermore, all other PREs outside the homeotic complex we tested were found to be transcribed in the same tissue as the mRNA of the corresponding target gene, suggesting that anti-silencing by transcription is a fundamental aspect of the cellular memory system.
Keywords: Polycomb, chromatin silencing, intergenic transcription, Drosophila
Chromosomal elements termed PREs (Polycomb group response elements) mediate the mitotic inheritance of transcriptional programs, thus ensuring the stable propagation of cell fates throughout development. In the repressed state, PREs act as silencers by recruiting Polycomb group (PcG) complexes to chromatin. When in the activated mode, however, PREs are controlled by the counteracting trithorax group (trxG) proteins, which in turn promote the formation of a transcriptionally competent chromatin structure (for review, see Ringrose and Paro 2004). Besides the well studied homeotic genes in the Drosophila bithorax complex (BX-C), PREs have been shown to be associated with a number of target genes (Kassis 1994; Maurange and Paro 2002; Bloyer et al. 2003; Ringrose et al. 2003). However, despite extensive analyses of the dynamics of PcG and trxG protein association with a PRE and requirements during the establishment of either the silenced or the activated state (Orlando et al. 1998; Poux et al. 2001; Mohd-Sarip et al. 2002; Déjardin and Cavalli 2004), it is not known how the decision between the recruitment of either repressive PcG or activating trxG complexes to a PRE is taken.
There is accumulating evidence that silencing represents the default state of these elements, whereas the conversion of a PRE into the active chromatin mode is triggered by the embryonic activation of the target gene promoter (Busturia and Bienz 1993; Chan et al. 1994; Cavalli and Paro 1998, 1999; Klymenko and Müller 2004; Sengupta et al. 2004). However, it is not clear how this transcriptional activity is communicated to a PRE and, as a consequence, which mechanism regulates the epigenetic switch of a PRE into the active mode.
Noncoding RNAs were identified in the BX-C, and it has been proposed that this regulated intergenic transcription may play a role in the opening of distinct chromatin domains (Lipshitz et al. 1987; Sanchez-Herrero and Akam 1989; Cumberledge et al. 1990). We and others found that a tight correlation exists between the relief of PcG-mediated silencing and the noncoding transcription through regulatory regions of the BX-C (Bender and Fitzgerald 2002; Hogga and Karch 2002; Rank et al. 2002). First, in a transgenic model system, we showed that the switch of silent PREs into the activated mode is accompanied by transcription through these elements themselves. The same PRE sequences at the endogenous BX-C locus are also transcribed in wild-type embryos. Importantly, the pattern of these transcripts reflects the expression pattern of the cognate target genes and thus the pattern in which these PREs are expected to be in the activated mode (Bae et al. 2002; Rank et al. 2002). In two parallel studies, ectopic transcription through intergenic regions of the BX-C originating from promoter insertions was shown to result in homeotic phenotypes reminiscent of PRE deregulation (Bender and Fitzgerald 2002; Hogga and Karch 2002). These results led to the proposition that the transcription through a repressed PRE or the noncoding RNA generated during this process may trigger the epigenetic activation of these elements (Rank et al. 2002).
Apart from the BX-C in Drosophila, a correlation of intergenic transcription with the opening of the chromatin structure was described for the β-globin locus in humans (Ashe et al. 1997; Gribnau et al. 2000) and the immunoglobulin heavy chain (Igh) locus in mice (Bolland et al. 2004). At the mouse Igh locus, antisense transcription through the VH region was found to correlate with the transition from DJH to VDJH recombination (Bolland et al. 2004). In the developmental regulation of globin gene expression at the human β-globin locus, regulated intergenic transcripts were shown to delineate differentially activated chromatin subdomains (Gribnau et al. 2000). Similar to the regulation of PREs in the Drosophila BX-C, it has been suggested that the transcription through intergenic regions of the mouse Igh and human β-globin loci is required for the remodeling of the chromatin structure resulting in a higher accessibility. However, the data so far do not allow us to exclude the possibility that the observed correlation of intergenic transcription with chromatin domain opening might reflect a consequence rather than be the cause for an open chromatin structure. To distinguish between these possibilities and to understand the mechanism regulating the epigenetic state of a PRE, it is essential to determine whether the noncoding transcription through these elements is of functional significance for their epigenetic activation.
Here we devised a transgenic reporter system allowing us to address this question and analyze the effects of transcription through the well characterized Fab-7 PRE in detail. We show that constitutive transcription through the PRE producing either sense or antisense Fab-7 RNA leads to the epigenetic activation of this element. We discuss a possible role of the transcription through PREs in the maintenance of the activated mode at later stages of development. We found PRE sequences outside the homeotic gene complexes to also be transcribed, raising the possibility that the epigenetic activation triggered by transcription may be widespread among the genes regulated by PREs.
Results
A transgenic reporter system to study the function of noncoding transcription through PREs
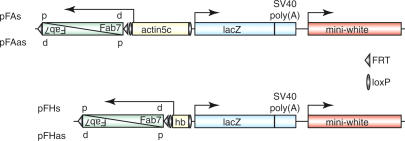
To determine whether transcription through PREs and the epigenetic activation of these elements are functionally related, we designed a transgenic reporter system in which transcription through the Fab-7 PRE is controlled by different promoters with defined activities (Fig. 1). In one set of transgenes, transcription through Fab-7 is controlled by the constitutively active actin5C promoter (Fig. 1; Vigoreaux and Tobin 1987), whereas in the second group, it is driven by the transiently active zygotic hunchback promoter (Fig. 1; Schröder et al. 1988). We cloned the Fab-7 PRE in both orientations on the transgene so that transcription from the promoters leads to the generation of either sense (pFAs and pFHs in Fig. 1) or antisense (pFAas and pFHas in Fig. 1) Fab-7 RNA with respect to the transcription of its endogenous target gene Abdominal-B (AbdB). The expression of the transformation marker miniwhite reflects the activity of the PRE, thus serving as a direct read-out of the epigenetic state of Fab-7. The LacZ reporter was not used in this study. An inherent problem in the analysis of transgenic PRE constructs is that the degree of silencing imposed by a PRE is strongly influenced by the chromosomal environment. To overcome this problem, we examined the Fab-7 PRE subjected to different transcription states at identical insertion sites of the transgenes. To achieve this, we cloned the promoters between loxP sites, whereas the Fab-7 PRE is flanked by FRT sites. This enabled us to separately excise the promoter or the PRE sequences from the transgene by Cre/loxP or Flp/FRT recombination, respectively (Chou and Perrimon 1996; Siegal and Hartl 1996).
Figure 1.
Transgenic reporter constructs to study the function of noncoding transcription through PREs. The Fab-7 PRE is located downstream of either the constitutively active actin5c promoter (pFAs and pFAas) or the transiently active zygotic hunchback (hb) promoter (pFHs and pFHas). The orientation of the Fab-7 element is either proximal-distal (p-d) or distal-proximal (d-p) in relation to its orientation in the BX-C (Martin et al. 1995; see also Fig. 5). As such, either sense (d-p, pFAs and pFHs) or antisense (p-d, pFAas and pFHas) strand with respect to the Abdominal B (AbdB) mRNA direction is produced. The Fab-7 PRE is flanked by FRT sites (triangle), and the promoters are cloned between loxP sites (oval), allowing the separate excision of these sequences from the transgene by Flp/FRT and Cre/loxP recombination, respectively.
The Fab-7 PRE becomes activated upon constitutive transcription through the element
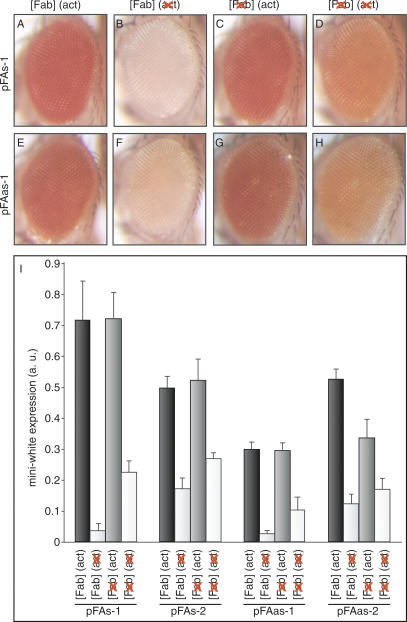
To test whether constitutive transcription through the Fab-7 element has any effect on the epigenetic state of this PRE, we used the actin5c promoter. We assessed the expression of the miniwhite marker in the pFAs (Fig. 2A-D) and pFAas (Fig. 2E-H) transgenic fly lines before and after all possible recombination steps. When the Fab-7 PRE on the transgene was transcribed from the actin5c promoter, the pFAs and pFAas adult flies hatched with a dark orange or red eye color (Fig. 2A,E). In all transgenic lines obtained (≥20 independent lines for each of the pFAs and pFAas constructs), we never observed silencing of the miniwhite gene under this condition, indicating that the Fab-7 PRE is switched into the activated mode. We verified that transcription through Fab-7 occurred in the expected pattern by RNA in situ hybridizations in embryos and third-instar larval tissues (Burn et al. 1989; data not shown). In contrast, after the excision of the actin5c promoter from the transgene, the miniwhite gene was almost completely repressed (Fig. 2B,F). Thus, in the absence of transcription, the Fab-7 PRE is in the repressed state, which results in the silencing of the reporter. When the Fab-7 element was removed from the transgene, the miniwhite gene was highly expressed in the presence of the actin5c promoter (Fig. 2C,G). The excision of the actin5c promoter from the transgene resulted in a moderate decrease in eye pigmentation (Fig. 2D,H), indicating that the vicinity of the promoter to the marker gene has an activating effect. The pFAs and pFAas transgenic lines behaved very similarly, demonstrating that the orientation of the Fab-7 PRE on the transgene is irrelevant for the observed effects.
Figure 2.
Fab-7 is activated upon constitutive transcription through the PRE. Shown are pictures of adult fly eyes of the pFAs and pFAas transgenic lines before and after all possible recombination steps. (A,E) Expression of the miniwhite marker, when the Fab-7 PRE is transcribed from the actin5c promoter. (B,F) Eye pigmentation in the presence of the Fab-7 PRE but absence of the actin5c promoter on the transgene. (C,G) miniwhite expression in the absence of Fab-7 when the actin5c promoter is in the vicinity of the marker gene. (D,H) Level of eye pigmentation when neither Fab-7 nor the actin5c promoter are on the transgene. (I) miniwhite expression level determined by photometric pigment measurements before and after Cre/loxP and/or Flp/FRT recombination in four independent transgenic lines. Shown are the mean values of three independent measurements.
To quantify the effect of transcription through the Fab-7 PRE on the miniwhite expression levels, we performed photometric pigment measurements in four independent transgenic lines (Fig. 2I). The increase in eye pigmentation upon transcription through Fab-7 from the actin5c promoter varied considerably, from 2.9- to 19.3-fold depending on the insertion site of the transgene. In contrast, the moderate elevation of miniwhite expression caused by the mere presence of the actin5c promoter alone remained relatively constant (1.9- to 3.2-fold). Comparison of the levels of eye pigmentation in flies bearing the activated Fab-7 element with those bearing the actin5c promoter but no PRE on the transgene, demonstrates that the activation of the Fab-7 PRE by transcription results in the complete derepression of the miniwhite marker. We conclude that constitutive transcription through the Fab-7 PRE results in the stable epigenetic activation of this element.
Epigenetic activation requires transcription to pass through the Fab-7 PRE
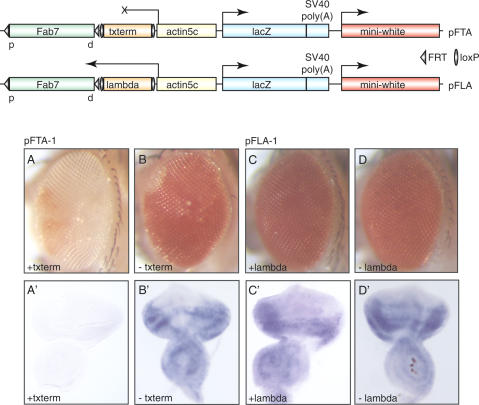
The actin5C promoter employed in the pFAs and pFAas transgenes is a strong promoter. Therefore, it is conceivable that the activation of the Fab-7 element might be achieved by the recruitment of transcriptional activators into its immediate vicinity. To examine whether the transcriptional machinery indeed has to pass through the PRE in order to trigger the epigenetic activation, we inserted a transcription termination signal between the Fab-7 element and the actin5C promoter (Fig. 3, pFTA). The termination signal we chose originates from the 3′UTR of the Drosophila hsp70 gene (Struhl and Basler 1993). In two of six transgenic pFTA lines obtained, the miniwhite gene was almost completely silenced, indicating that in this situation, the Fab-7 PRE is in the repressed state (Fig. 3A). In contrast, after excision of the terminator sequence from the transgene by Cre/loxP recombination, the Fab-7 PRE was again stably activated, as illustrated by the strong derepression of the miniwhite marker (Fig. 3B). RNA in situ hybridizations showed that in the pFTA-1 transgenic line, transcription through the Fab-7 PRE from the actin5C promoter was efficiently terminated, whereas after removal of the terminator sequence, Fab-7 RNA was again detectable (Fig. 3A′,B′). We tested two other pFTA transgenic lines in which the miniwhite gene was not repressed and detected Fab-7 transcripts even in the presence of the termination signal (data not shown), indicating that at these insertion sites of the transgene, transcription is not efficiently terminated. Moreover, the total number of recovered transgenic pFTA lines was surprisingly low, suggesting that genuine transgenic lines may have been missed due to a complete repression of the miniwhite transformation marker.
Figure 3.
Epigenetic activation requires transcription to pass through the Fab-7 PRE. In the pFTA construct, the hsp70 3′UTR transcription termination signal (txterm) is inserted between the Fab-7 element and the actin5c promoter. In the pFLA transgene, the Fab-7 PRE is separated from the actin5c promoter by λ DNA of the same length as the hsp70 3′UTR lacking termination sequences. The termination signal and λ DNA are flanked by loxP sites, the Fab-7 PRE by FRT sequences. (A,B) miniwhite expression in the pFTA line before and after excision of the hsp70 3′UTR from the transgene, respectively. (A′,B′) In situ hybridizations detecting Fab-7 RNA to show that transcription from the actin5c promoter is efficiently terminated before the PRE is reached. (C,D) The level of eye pigmentation remains constant independent of the presence or absence of λ DNA on the transgene. C′,D′) RNA in situ hybridization to confirm that increasing the distance between the Fab-7 element and the actin5c promoter does not influence transcription through the PRE.
It is possible that the repression we observed in the pFTA lines is simply due to the increased distance between the PRE and the actin5C promoter. To test this, we generated control transgenic lines in which the Fab-7 element and the actin5C promoter are separated by λ DNA of the same length as the hsp70 3′UTR terminator lacking termination signals. In the 10 transgenic lines obtained, we never observed miniwhite repression (Fig. 3C). As expected, the excision of the λ DNA from the transgene by Cre/loxP recombination had no effect on the expression of the reporter (Fig. 3D). In situ hybridizations demonstrated that the Fab-7 PRE was transcribed in these flies (Fig. 3C′,D′), thus confirming that repression in the pFTA lines is caused specifically by the terminator sequence.
These results demonstrate that the epigenetic activation of the Fab-7 element indeed requires the transcriptional machinery to pass through the PRE, indicating that the recruitment of transcriptional activators into the vicinity of the PRE is not sufficient to overcome silencing. Moreover, these data demonstrate that the increase of miniwhite expression caused by the presence of the actin5C promoter on the transgene in the pFAs and pFAas lines (Fig. 2A,E) is the consequence of the activation of the Fab-7 PRE and is not due to a direct interaction of the actin5C promoter with the promoter of the miniwhite gene.
The maintenance of epigenetic activation requires transcription throughout embryogenesis
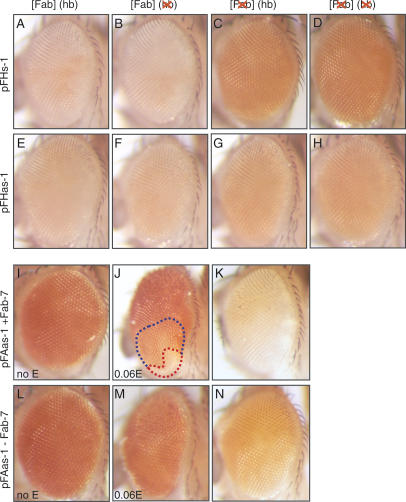
To better understand the mechanism underlying the epigenetic activation of PREs, we wished to test whether transcription only triggers the establishment, or if it is also required for the maintenance of the activated state throughout development. To this end, we generated transgenic flies using the constructs pFHs and pFHas depicted in Figure 1. In these flies, the Fab-7 PRE is transcribed from a 0.7-kb fragment of the zygotic hunchback (hb) promoter, which is active only during early embryogenesis (Schröder et al. 1988). Transcription from this promoter starts at cell division cycle 11-12, which is shortly before the endogenous transcripts through the PREs of the BX-C appear (Schröder et al. 1988; Sanchez-Herrero and Akam 1989). The activity of the minimal zygotic hb promoter ceases with the beginning of gastrulation. RNA in situ hybridization verified that the Fab-7 PRE on the transgenes was transcribed in the expected pattern (data not shown). In contrast to the actin5c lines pFAs and pFAas, which never showed repression of the miniwhite gene prior to recombination, the reporter was silenced in a pairing-sensitive manner in 50% (pFHs, n = 12) and 55% (pFHas, n = 11) of the transgenic lines. These are the typical rates of pairing-sensitive repression that are expected in transgenic reporter lines carrying a repressed PRE (Kassis 2002), suggesting that the Fab-7 element was not stably activated. Indeed, the comparison of eye pigmentation before and after excision of the zygotic hb promoter from the transgene showed that transcription through Fab-7 from this promoter was not sufficient to activate the PRE (Fig. 4, cf. A,B,E,F). When the Fab-7 PRE was removed from the transgene, the miniwhite gene was no longer silenced and was expressed to a level typical for the insertion site of the transgene (Fig. 4C,G). In the absence of the Fab-7 PRE, removal of the zygotic hb promoter also had no effect on the expression of the miniwhite reporter (Fig. 4, cf. C and D, G and H).
Figure 4.
The maintenance of epigenetic activation requires continuous transcription. (A-H) miniwhite expression in the pFHs and pFHas transgenic lines before and after all possible recombination steps. (A,E) Eye pigmentation when the Fab-7 PRE is transcribed from the zygotic hb promoter. (B,F) Level of miniwhite expression after removal of the zygotic hb promoter. (C,G) Excision of the Fab-7 element leads to derepression of the miniwhite marker. (D,H) Further excision of the zygotic hb promoter from the transgene does not change the activity of the miniwhite gene. (I-N) Change in the level of eye pigmentation by expression of the conditionally active Cre-EBD fusion protein. (I) In the absence of estrogen, the expression of the miniwhite gene was not changed. (J) Induction of Cre-EBD activity by transferring first-instar larvae to food containing 0.06 mg/mL estrogen resulted in clones with low eye pigmentation (red circle) in one out of 20 adult flies. (K) Expected level of eye pigmentation when the Cre-EBD and eyGAL4 transgenes are combined with the pFAas transgene. (L-N) Similar conditions as in I-K for a line lacking the Fab-7 PRE (no clones were observed in these cases.
We reasoned that this time frame of transcription may be too short to firmly establish the epigenetically activated state of the Fab-7 PRE. Consistent with this, endogenous noncoding PRE transcripts in the BX-C can also be detected in late embryogenesis (Rank et al. 2002). Additionally, in a transgenic assay we established previously, the Fab-7 PRE can be stably switched into the activated mode by the transient expression of a trans-activator in embryos, but the highest efficiency of switching was achieved when the trans-activator was expressed at later stages of embryogenesis (Cavalli and Paro 1998). Therefore, it is conceivable that the firm establishment of the epigenetically activated PRE state requires a longer time window of transcription, extending over a number of cell divisions.
To precisely control the time of transcription through the Fab-7 element, we expressed a conditionally active Cre-EBD (estrogen-binding domain) fusion protein in all cells of the developing eye from a UASP-Cre-EBD304 transgene in our pFAas-1 transgenic line. The Cre-EBD fusion protein is active only in the presence of estrogen, which allowed us to control the time of excision of the actin5C promoter from the transgene (Heidmann and Lehner 2001). Thus, we limited the time window of Fab-7 transcription in the pFAas-1 line from the beginning of development until shortly after the completion of embryogenesis by transferring freshly hatched larvae onto estrogen-containing food. This represents a shorter time frame of transcription through Fab-7 than that driven by the actin5c promoter, which is active throughout development, but a longer time frame than that of the transiently active zygotic hb promoter.
We verified that recombination had occurred in the estrogen-treated larvae by genomic PCR on DNA recovered from the heads of the adult flies tested (data not shown). In the control pFAas group expressing the Cre-EBD fusion protein but not treated with estrogen, we detected background recombination in two out of five flies we analyzed. When first-instar larvae had been transferred onto estrogen-containing food, the number of flies showing recombination increased to four out of five. Although the PCR was performed under saturating conditions, the amount of PCR product in the cases where background recombination had occurred was considerably lower than that obtained in the estrogen-treated individuals, suggesting that the number of cells in which background recombination had occurred was very low. In the initial characterization of the UASP-Cre-EBD304 line, estrogen-independent recombination was not observed, whereas upon estrogen induction, recombination products were obtained in 100% of treated individuals (Heidmann and Lehner 2001). In that study, the clones in the adult eye in which recombination had occurred covered the majority of cells even after estrogen treatment for only 3 h during the first-instar larval stage. Therefore, we expect that recombination occurs in the majority of eye cells after transferring first-instar larvae onto estrogen-containing food.
We compared the eye colors of flies with and without exposure to estrogen, to determine the effect of excising the actin5c promoter at the end of embryogenesis. Control flies kept on food lacking estrogen showed the expected high expression of the miniwhite gene in the presence of both the actin5c promoter and the Fab-7 PRE on the transgene in combination with the eyGAL4 driver and the UASP-Cre-EBD304 construct (Fig. 4I). When first-instar larvae were transferred onto food containing 0.06 mg/mL estrogen, most of the adult flies had a similar eye color to the control flies (data not shown). In one of 10 to one of 20 cases, flies with small light-colored clones in the eyes were obtained, indicating that the miniwhite gene had become resilenced (Fig. 4J). Formally, we cannot rule out the possibility that this rare frequency of low miniwhite expression might be caused by some nonspecific recombination events. However, the degree of pigmentation in these clones closely matches that in pFAas-1 transgenic flies lacking the actin5C promoter combined with the UASP-Cre-EBD304 and eyGAL4 transgenes (Fig. 4K). We obtained similar results when larvae were transferred onto food containing 0.03 mg/mL or 0.09 mg/mL estrogen, which is consistent with the previous study of the UASP-Cre-EBD304 transgene, in which Cre recombinase activity was found to plateau at estrogen concentrations above 0.01 mg/mL (Heidmann and Lehner 2001).
In the control flies, which did not carry the Fab-7 PRE on the transgene, we did not observe flies with small light-colored clones in the eyes upon induction with estrogen, although large regions in the eyes showed an intermediate pigmentation (Fig. 4, cf. L and M). The comparison with control flies lacking the actin5c promoter suggests that this lighter pigmentation is probably due to the reduction of miniwhite expression by the removal of the actin5c promoter from the transgene (Fig. 4N). As stated above, in the presence of the Fab-7 PRE, the induction of Cre/loxP recombination by estrogen treatment resulted in flies with and without clones of low eye pigmentation. Genomic PCR indicated that recombination had occurred in both the flies with light-colored clones (Fig. 4J) and in those without clones (data not shown). In the latter, the presence of high uniform levels of miniwhite expression, concomitant with substantial recombination, indicates that despite the excision of the actin5c promoter in first-instar larvae, in most cases the Fab-7 PRE nevertheless remains active, and the miniwhite gene is thus strongly transcribed. These data suggest that generally, a time frame of transcription through the Fab-7 PRE on the transgene until the beginning of the first larval stage is sufficient for both the establishment and the maintenance of the activated PRE mode. In rare cases, however, we observed clones of low miniwhite expression, suggesting that in these cells, the epigenetic activation of the PRE is not stable enough and thus becomes re-silenced.
Endogenous BX-C PREs are transcribed at later stages of development
In a previous study, we showed that in wild-type Drosophila embryos, the characterized PREs of the BX-C are transcribed in a spatially and temporally regulated manner which reflects the pattern of activity of their associated genes (Rank et al. 2002). The analysis of our transgenic system indicates that transcription through the Fab-7 element until the beginning of the first-instar larval stage is sufficient for the stable activation of this PRE. As these data were obtained in a transgenic background, we next asked whether endogenous PRE sequences are transcribed only during early development or whether transcription persists until later stages.
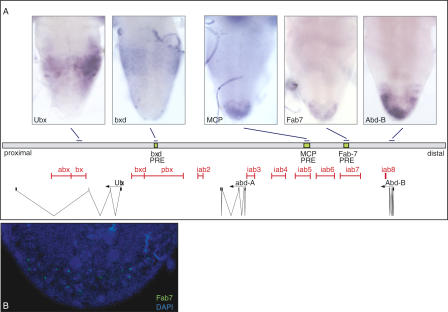
In the brain of third-instar larvae, the homeotic genes are expressed in a characteristic pattern along the anterior-posterior axis, following the principle of spatial colinearity (Duboule and Morata 1994). If transcription through PREs is required for the maintenance of epigenetic activation, we would expect the bxd, Mcp, and Fab-7 elements also to be transcribed in this tissue. By in situ hybridizations, we detected RNAs spanning the bxd, Mcp, and Fab-7 sequences in a pattern that reflects the expression of the target genes Ubx and AbdB, respectively (Fig. 5A). We detected PRE transcripts only in the sense direction with respect to the orientation of the coding genes. In contrast to the mRNAs, the noncoding RNAs showed a punctate staining in the brain tissue, suggesting that they are localized to the nuclei. Fluorescent in situ hybridizations in combination with DAPI staining showed the Fab-7 transcripts in the brain of third-instar larvae to appear as single or doublet spots within the cell nuclei (Fig. 5B). In line with previous studies, in which noncoding transcripts through infra-abdominal (iab) regions of the BX-C were found to be restricted to the nucleus in embryos (Drewell et al. 2002), we detected the Fab-7, Mcp, and bxd RNAs at discrete loci within the nucleus at this stage (data not shown). We expect that these signals represent nascent, noncoding transcripts, suggesting that they may be degraded following their synthesis. To determine the relative amounts of RNA accounting for these signals, we compared the expression levels of total and nascent AbdB mRNA with the level of noncoding Fab-7 RNA in SF4 tissue culture cells by real-time RT-PCR. The results from this analysis showed that the level of Fab-7 RNA is approximately sixfold less abundant than the amount of AbdB target mRNA (Supplementary Fig. 1).
Figure 5.
Noncoding transcripts of the BX-C were also transcribed at later stages of development. (A) In situ hybridizations using RNA probes which detect the Ubx and AbdB mRNAs as well as the noncoding bxd, Mcp, and Fab-7 PRE sense RNAs in the brain of third-instar larvae. Below the in situ hybridizations is a drawing showing the organization of the BX-C and the location of the sequences used for the preparation of RNA probes. (B) Fluorescent in situ hybridization showing the Fab-7 sense transcript in the posterior tip of the brain costained with DAPI to visualize the DNA.
The finding that PRE transcripts can be detected in the brain of third-instar larvae suggests that in the context of the endogenous BX-C, continued transcription through PREs may be required for the stable maintenance of the epigenetically activated state.
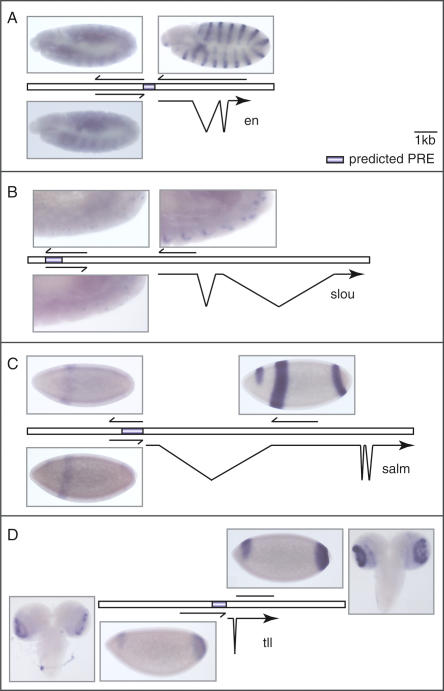
Apart from the homeotic genes, the PcG/trxG memory system is known to regulate many more targets (for review, see Ringrose and Paro 2004). We thus asked whether PcG-regulated gene transcription is accompanied by transcription through the associated PREs at loci other than the BX-C. To test this, we designed RNA probes directed against the genetically characterized engrailed (en) PRE (Kassis 1994), as well as against three predicted PREs that potentially regulate the genes slouch (slou; Dohrmann et al. 1990), spalt major (salm, Frei et al. 1988), and tailless (tll, Strecker et al. 1986; Daniel et al. 1999; Ringrose et al. 2003). The genetically characterized PRE of the en gene and the predicted PREs of the salm and tll genes are localized in the respective promoter regions, whereas the predicted PRE of the slou gene lies 3 kb upstream of the transcriptional start site (Kassis 1994; Ringrose et al. 2003). In situ hybridizations showed that the en PRE as well as the predicted PREs of the slou, salm, and tll genes are transcribed in embryos (Fig. 6). The transcription through these sequences shows the same spatial regulation as that of the mRNAs of the known (en) and predicted target genes (slou, salm, and tll). For the en, slou, and salm PREs, we detected transcripts of both sense and antisense strands (Fig. 6A-C), whereas transcripts of the tll PRE were only detected in the antisense orientation (Fig. 6D). In addition, the putative PRE transcripts display a nuclear localization similar to that of the bxd, Mcp, and Fab-7 RNAs. Three of the transcribed PRE sequences lie in the promoter regions of their associated genes. To exclude the possibility that upstream transcription is a general property of active promoters, we also examined the rosy (ry) gene, which is not regulated by a PRE (Ringrose et al. 2003). Using in situ hybridization, we did not detect transcription in the promoter of this gene, suggesting that upstream transcription is a PRE-specific phenomenon. Interestingly, the antisense strand of the predicted tll PRE is also transcribed in the optic lobes of the brain in third-instar larvae, where the tll mRNA is also expressed (Fig. 6D). In contrast, we did not detect transcription through the en and salm PREs at this stage, although the mRNAs for both genes are transcribed at high levels.
Figure 6.
Noncoding transcription through predicted PREs. Shown are in situ hybridizations using strand-specific probes to detect mRNAs as well as transcripts spanning predicted PRE sequences. (A) The en PRE was transcribed in sense and antisense orientation in a pattern that reflects the en mRNA distribution. (B) Both sense and antisense transcription through the predicted slou PRE were detected in the same cells that express the slou mRNA. (C) The predicted salm PRE was transcribed in both orientations in the same pattern as the respective mRNA. (D) Transcription through the predicted tll PRE was found only in the antisense direction, in a pattern that reflects the tll mRNA expression domain. For this PRE, transcripts were also detected in the optic lobes of third-instar larvae.
These results suggest that the activation of a gene does not result in spurious transcription through its promoter region, but rather that the transcription we observe in embryos is involved in triggering the epigenetic activation of the known and predicted PREs we tested. The fact that we detected transcripts only through the tll PRE and not through the en and salm PREs later in development suggests that the maintenance of the activated state may be regulated by different mechanisms in different tissues. Alternatively, the en and salm genes might be controlled by more than one PRE, which are differentially deployed in different tissues and would thus show a differential pattern of epigenetic activation and noncoding transcription.
Discussion
In the Drosophila genome, a particular set of a few hundred genes is stably and heritably inactivated by the PcG proteins. Developmental silencing imposed on these genes by the PcG proteins appears to represent a default state. For example, PREs on a transgene in most cases induce a global and tissue-independent repression of the adjacent reporter genes. Thus, in order to obtain a permanently active state of a PRE-regulated target gene, PcG-induced silencing needs to be counteracted. In this work, we present evidence that transcription through a PRE prevents silencing and switches the maintenance mode of the element to activity. Our results with the transgene system indicate that noncoding transcription is not a consequence of gene activation but is causally related to the prevention of silencing. The process of transcription appears to be central to this mechanism, while the RNAs produced seem to play a minor role, if they do have a function at all.
We and others have shown that intergenic transcription in the BX-C has a profound phenotypic effect on the expression of the Hox genes. Our results with transgenes suggest that the spatially and temporally regulated transcription of noncoding RNAs in the BX-C induces a remodeling of the chromatin, thereby preventing PcG-mediated silencing. The consequence of this is the segment-specific activation of the Hox genes. This is probably especially important in large gene complexes where PREs are located at long distances from the promoters they regulate. Déjardin and Cavalli (2004) showed that in a transgenic assay, the activation of a minimal 219-bp Fab-7 PRE is not accompanied by transcription through the element. However, in this case the minimal PRE was juxtaposed to the promoter, probably benefiting from the open chromatin environment induced by the bound transcription factors. Our own results (Rank et al. 2002), however, indicate that an 870-bp large Fab-7 PRE, under similar conditions but containing more PcG protein-binding sites, cannot be activated anymore. This suggests that over a certain threshold level of silencing, imposed by the stability of the silencing complexes, chromatin remodeling by transcription is required to remove PcG complexes in order to counteract their silencing activity in a mitotically heritable fashion.
Hox cluster regulation is only part of the entire PcG/trxG memory system, prompting us to analyze the transcription pattern of characterized en and predicted salm, slou, and tll target genes as well as their respective PRE sequences (Kassis 1994; Ringrose et al. 2003). RNA in situ hybridizations revealed that these sequences are also transcribed in a pattern that reflects the expression of the cognate target genes. This suggests that the mechanism of epigenetic activation of PREs initially described for the homeotic gene clusters may be required for the regulation of many more PcG target genes than previously thought. Transcription through these PREs can be either uni- (tll PRE) or bidirectional (en, slou, and salm PREs), further suggesting that the induction of epigenetic activation relies on the remodeling of chromatin induced by the transcriptional process, rather than by the noncoding RNAs.
With our transgenic reporter system, we show that the constitutive transcription through the Fab-7 PRE from the actin5c promoter results in the stable epigenetic activation of this element. This raises the question of how this could be achieved mechanistically. As has been proposed for the developmental regulation of globin gene expression (Ashe et al. 1997; Gribnau et al. 2000) and the regulation of VDJH-recombination in mice (Bolland et al. 2004), the opening of the chromatin structure at a transcribed PRE may be induced by the passing of the transcriptional machinery through the regulatory sequence. It has been shown that the elongating RNA polymerase II complex is associated with the SWI/SNF remodeling complex and a histone acetyl transferase activity (Wilson et al. 1996; Cho et al. 1998; Wittschieben et al. 1999). Such enzymatic activities linked with the transcription machinery may catalyze the epigenetic modification of chromatin.
In this respect, it is interesting to note that most of the trxG mutants were initially uncovered as suppressors of the Pc phenotype (Kennison and Tamkun 1988), and that the combination of PcG with trxG mutations can restore the typical phenotype of the single mutations to wild type (Ingham 1985). The molecular mechanism behind this antagonism is not clear. Using clonal analysis, Klymenko and Müller (2004) recently reported that the trxG proteins Ash1 and Trx do not function as coactivators of Hox gene expression, but that they are required as anti-repressors to prevent PcG-induced silencing. In contrast, the Brahma complex containing the trxG proteins Brm, Osa, and Mor was shown to act as a coactivator of transcription (Kal et al. 2000), and a subset of trxG genes encode components of the Mediator complex (Kennison and Tamkun 1988; Boube et al. 2000; Treisman 2001). Our finding that transcription through Fab-7 induces the epigenetic activation of this PRE may explain how trxG complexes involved in more general transcriptional processes antagonize the establishment of PcG silencing.
Interestingly, in budding yeast, intergenic transcription through a promoter was recently shown to prevent the binding of a transcriptional activator to its target sequences (Martens et al. 2004). It is possible that, in a similar fashion, the transcription through PREs may lead to the displacement of repressive PcG complexes from the chromatin and/or the prevention of PcG recruitment to the PRE in the first place. Enzymatic activities carried by the RNA polymerase II complex may subsequently or in addition modify histones at PREs with positive epigenetic marks like acetyl or methyl moieties (Cavalli and Paro 1999; Beisel et al. 2002; Klymenko and Müller 2004). Interestingly, it was recently shown that the sequential induction of HoxB gene expression in mouse embryonic stem (ES) cells by retinoic acid correlates with the orchestrated looping out of this locus from chromosome territories (Chambeyron and Bickmore 2004). This indicates that, in addition to locus-wide changes in the chromatin structure such as histone modifications, the transcriptional activity of genes may be regulated by an additional order of complexity. It is possible that, in addition to inducing “small-scale” changes in the chromatin structure, transcription through PREs may lead to a subnuclear relocation of the target gene locus from a repressive into a transcriptionally permissive environment.
Removal or inhibition of binding of PcG silencing complexes to PREs by tissue-specific transcription appears to be an attractive mechanism to counteract the constant pressure of the repressive system acting by default. However, with such a solution, the problem of epigenetic maintenance is simply moved to another level, as we are now faced with the question of what prevents the intergenic transcription from being silenced by the PcG. The simple answer that promoters of noncoding transcripts are not sensitive to PcG/PRE silencing is probably not valid. Noncoding transcripts of the BX-C are activated by the same set of early segmentation genes as the Hox protein encoding mRNAs (Casares and Sanchez-Herrero 1995). As such, their subsequent regulation might be subjected to the same regiment of factors as the protein-encoding transcripts. However, the problem of transcriptional memory might be reduced to the problem of how to inhibit PcG silencing in particular cells/tissues, while the rest will be down-regulated by default.
With the processive transcription as the central issue, the cell cycle might become an important factor for the maintenance of active transcription. In Drosophila, PcG proteins dissociate from the chromosomes at mitosis (Buchenau et al. 1998). Thus, if after mitosis intergenic transcription started before PcG proteins rebind to the PREs, the chromatin would be turned into an active mode, and would thus be protected from PcG-mediated silencing until the next round of cell division. At this point, it remains an open question whether continuous transcription of noncoding RNAs is required throughout the cell cycle to prevent the PcG complexes from rebinding, or whether the initial setting of positive epigenetic marks by the early transcription process is sufficient to prevent silencing during interphase. This proposed mechanism further reduces the problem of how transcriptional activity is maintained to the problem of how only a positive epigenetic mark is maintained during DNA replication and mitosis. Here, recent advances in studies of histone variants propose some attractive candidate marks. In particular, the histone variant H3.3 associated with the transcription of active genes could be envisaged as a possible positive signal that is locally maintained and propagated at cell division (Ahmad and Henikoff 2002a; Tagami et al. 2004). As has been suggested before, targeted deposition of the H3.3 variant at sites of active transcription may serve to remove repressive epigenetic marks such as methylation (Ahmad and Henikoff 2002b). It was shown that the establishment of stable PcG silencing complexes not only requires a sequence component, but is also accompanied by the methylation of K9 and K27 of histone H3 (Czermin et al. 2002; Müller et al. 2002; Fischle et al. 2003; Ringrose et al. 2004). In contrast, positive marks, which have been shown to be mainly associated with H3.3 compared to H3 (McKittrick et al. 2004), would be specifically enriched at a transcribed PRE and transmitted through mitosis. After cell division, these epigenetic marks may then in turn provide a platform for noncoding transcription through the PRE early in the cell cycle, which itself may re-establish the full active chromatin environment, unsuitable for PcG protein binding and silencing. Additionally, the reported result that an activated PRE is still maintained over a certain period after transcription has ceased (by removal of the promoter by the inducible Cre recombinase, Fig. 4) suggests that this positive epigenetic signal is quite stable and is only diluted out by multiple cell divisions.
In summary, we propose that transcriptional maintenance during development by the PcG/trxG system is primarily a process of preventing PcG silencing to occur at those target genes that need to be maintained active in a defined cell lineage. The advantage of this mode of action is that a positive epigenetic mark, surviving DNA replication and mitosis, is sufficient to ensure stable and heritable maintenance of gene expression patterns, as the silenced mode is created by default. As such, transcription of intergenic sequences would serve as an anti-silencing mechanism that would continually counteract the initiation of this PcG-mediated silencing. In the future, it will be important to pursue the involvement of the various trxG components in the establishment and maintenance of regulatory transcription mechanisms and to analyze the link to cell cycle control and the identity and propagation of the positive epigenetic marks required to sustain active transcription.
Materials and methods
DNA vectors and cloning strategy
The pUZ vector (Lyko et al. 1997) was modified by ET-cloning (Zhang et al. 1998; Muyrers et al. 2000) to remove the UAS site and introduce BglII and AscI restriction sites (pUZmod; for all primer sequences see Supplemental Material). Briefly, primers containing homology regions to pUZ (ET upper and ET lower) were used to amplify the CAT-gene from the 705Cre vector (Zhang et al. 1998). The resulting PCR fragment was cloned by recombination into the pUZ vector and the CAT gene used for selection was removed by AscI digest. The minimal 0.7-kb zygotic hb promoter was amplified from genomic DNA, and the actin5c promoter was excised from the pMK26/SV40BS vector (kindly provided by M. Koelle, Yale University). These promoters were then subcloned into the pSV-paX1 vector (Buchholz et al. 1996) via HindIII/blunt end and BamHI/HindIII restriction sites. The resulting loxP-actin5c-loxP and loxP-hb-loxP fragments were PCR-amplified and cloned into the pUZmod vector via AscI and BglII/AscI restriction sites, respectively, yielding the pUZmod-act5c and pUZmod-hb vectors. The 3.6-kb Fab-7 HindIII fragment (Zink and Paro 1995) was subcloned into the pSV-paZ11 vector (Buchholz et al. 1996), and the resulting FRT-Fab-7-FRT fragment was PCR-amplified and cloned via NotI restriction sites into the pUZmod-act5c and pUZmod-hb vectors, yielding the pFAs, pFAas, pFHs, and pFHas constructs. The hsp70 3′UTR poly(A) signal excluding the scs' sequence from the pBS669 vector (Struhl and Basler 1993) and the 2-kb λ control DNA (DNA marker II, Roche) were PCR-amplified and subcloned into the pSV-paX1 vector (Buchholz et al. 1996) via BamHI/HindIII restriction sites. The resulting loxP-txterm-loxP and loxP-λ-loxP fragments were PCR-amplified and cloned via AscI sites into a pFAs vector from which loxP sites had been removed and the actin5c promoter reintroduced by blunt-end ligation, yielding the pFTA and pFLA constructs. All constructs were sequenced.
Fly strains and handling
The transgenic lines used here were p[FAs]-1 (chromosome 2), P[pFAs]-2 (chr. 3), P[pFAas]-1 (chr. 3), P[pFAas]-2 (chr. 3), P[pFHs]-1 (chr. 2), P[pFHas]-1 (chr. 3), P[pFTA]-1 (chr. 3), and P[pFLA]-1 (chr. 3). Quantitative pigment measurements were performed as described and analyzed using 24-h-old adult females (Reuter and Wolff 1981). Germline Cre/loxP and Flp/FRT recombination was done using the P[w+, cre] (Bloomington stock no. 1092) and P[ry+, hsFLP] (Bloomington stock no. 1929). The estrogen-inducible Cre/loxP recombination using the UASP-Cre-EBD304 II.6 transgenic line was essentially done as described (Heidmann and Lehner 2001). The eyGAL4 driver line was kindly provided by Stefan Schönfelder (University of Heidelberg; unpubl.). Recombination was verified by single-fly genomic PCR (Gloor et al. 1993) using whole flies or the heads of adult flies (for estrogen-induced recombination) as templates.
RNA in situ hybridizations
RNA in situ hybridization on embryos was done as described previously (Rank et al. 2002) with the following modifications. Hybridization solution contained 50% formamide, 5× SSC, 50 μg/mL heparin, 100 μg/mL herring sperm DNA (Invitrogen, cat. no. 15634-017), 0.1% Tween-20 at pH 5. Hybridization was performed overnight at 60°C. RNA in situ hybridization on larval tissues was essentially performed as described (Wolff 2000). For the fluorescent detection of RNAs, we used a monoclonal mouse anti-DIG antibody (1:400, Roche cat. no. 1 333 062) and goat anti-mouse Alexa 488-coupled secondary antibody (1:200, Molecular Probes cat. no. A-11029). Cell nuclei were visualized by DAPI (0.5 μg/mL, Sigma cat. no. D 9542) staining. All antibody incubations were done in PBS/Tween-20 (0.1%) containing 5% goat serum.
Acknowledgments
We are grateful to K. Basler for the pBS669 vector, P. Becker (Ludwig-Maximilians-Universität, Munich) for the SF4 cell line, C. Lehner for the UASP-Cre-EBD304II.6 transgenic line, and S. Schönfelder for providing the eyGAL4 driver line. We thank A. Löwer and L. Ringrose for helpful discussions and critical reading of the manuscript and S. Schönfelder and M. Hild for discussions. S.S. was supported by a Boehringer Ingelheim Fonds Fellowship and R.P. was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB/Transregio 5).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.326205.
References
- Ahmad K. and Henikoff, S. 2002a. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191-1200. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. 99: 16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe H.L., Monks, J., Wijgerde, M., Fraser, P., and Proudfoot, N.J. 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes & Dev. 11: 2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E., Calhoun, V.C., Levine, M., Lewis, E.B., and Drewell, R.A. 2002. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99: 16849-16852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857-862. [DOI] [PubMed] [Google Scholar]
- Bender W. and Fitzgerald, D.P. 2002. Transcription activates repressed domains in the Drosophila bithorax complex. Development 129: 4923-4930. [DOI] [PubMed] [Google Scholar]
- Bloyer S., Cavalli, G., Brock, H.W., and Dura, J.M. 2003. Identification and characterization of polyhomeotic PREs and TREs. Dev. Biol. 261: 426-442. [DOI] [PubMed] [Google Scholar]
- Bolland D.J., Wood, A.L., Johnston, C.M., Bunting, S.F., Morgan, G., Chakalova, L., Fraser, P.J., and Corcoran, A.E. 2004. Antisense intergenic transcription in V(D)J. recombination. Nat. Immunol. 5: 630-637. [DOI] [PubMed] [Google Scholar]
- Boube M., Faucher, C., Joulia, L., Cribbs, D.L., and Bourbon, H.-M. 2000. Drosophila homologues of transcriptional mediator complex subunits are required for adult and segment identity specification. Genes & Dev. 14: 2906-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchenau P., Hodgson, J., Strutt, H., and Arndt-Jovin, D.J. 1998. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos: Impact on models for silencing. J. Cell. Biol. 141: 469-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F., Angrand, P.O., and Stewart, A.F. 1996. A simple assay to determine the functionality of Cre or Flp recombination targets in genomic manipulation constructs. Nucleic Acids Res. 24: 3118-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn T.C., Vigoreaux, J.O., and Tobin, S.L. 1989. Alternative 5C actin transcripts are localized in different patterns during Drosophila embryogenesis. Dev. Biol. 131: 345-355. [DOI] [PubMed] [Google Scholar]
- Busturia A. and Bienz, M. 1993. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 12: 1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G. and Paro, R. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93: 505-518. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286: 955-958. [DOI] [PubMed] [Google Scholar]
- Casares F. and Sanchez-Herrero, E. 1995. Regulation of the infra-abdominal regions of the bithorax complex of Drosophila by gap genes. Development 121: 1855-1866. [DOI] [PubMed] [Google Scholar]
- Chambeyron S. and Bickmore, W.A. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & Dev. 18: 1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Rastelli, L., and Pirrotta, V. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13: 2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Orphanides, G., Sun, X., Yang, X.J., Ogryzko, V., Lees, E., Nakatani, Y., and Reinberg, D. 1998. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 18: 5355-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. and Perrimon, N. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S., Zaratzian, A., and Sakonju, S. 1990. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 87: 3259-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., and Pirrotta, V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185-196. [DOI] [PubMed] [Google Scholar]
- Daniel A., Dumstrei, K., Lengyel, J.A., and Hartenstein, V. 1999. The control of cell fate in the embryonic visual system by atonal, tailless and EGFR signalling. Development 126: 2945-2954. [DOI] [PubMed] [Google Scholar]
- Déjardin J. and Cavalli, G. 2004. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 23: 857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann C., Azpiazu, N., and Frasch, M. 1990. A new Drosophila homeobox gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes & Dev. 4: 2098-2111. [DOI] [PubMed] [Google Scholar]
- Drewell R.A., Bae, E., Burr, J., and Lewis, E.B. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99: 16853-16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. and Morata, G. 1994. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 10: 358-364. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang, Y., Jacobs, S.A., Kim, Y., Allis, C.D., and Khorasanizadeh, S. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 17: 1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E., Schuh, R., Baumgartner, S., Burri, M., Noll, M., Jürgens, G., Seifert, E., Nauber, U., and Jäckle, H. 1988. Molecular characterization of spalt, a homeotic gene required for head and tail development in the Drosophila embryo. EMBO J. 7: 197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G.B., Preston, C.R., Johnson-Schlitz, D.M., Nassif, N.A., Phillis, R.W., Robertson, H.M., and Engels, W.R. 1993. Type I repressors of P element mobility. Genetics 135: 81-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J., Diderich, K., Pruzina, S., Calzolari, R., and Fraser, P. 2000. Intergenic transcription and developmental remodelling of chromatin subdomains in the human β-globin locus. Mol. Cell 5: 377-386. [DOI] [PubMed] [Google Scholar]
- Heidmann D. and Lehner, C.F. 2001. Reduction of Cre recombinase toxicity in proliferating Drosophila cells by estrogen-dependent activity regulation. Dev. Genes Evol. 211: 458-465. [DOI] [PubMed] [Google Scholar]
- Hogga I. and Karch, F. 2002. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129: 4915-4922. [DOI] [PubMed] [Google Scholar]
- Ingham P.W. 1985. A clonal analysis of the requirement for the trithorax gene in diversification of segments in Drosophila. J. Embryol. Exp. Morphol. 89: 349-365. [PubMed] [Google Scholar]
- Kal A.J., Mahmoudi, T., Zak, N.B., and Verrijzer, P. 2000. The Drosophila Brahma complex is an essential coactivator for the trithorax group protein Zeste. Genes & Dev. 14: 1058-1071. [PMC free article] [PubMed] [Google Scholar]
- Kassis J.A. 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6kb region. Genetics 136: 1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46: 421-438. [DOI] [PubMed] [Google Scholar]
- Kennison J.A. and Tamkun, J.W. 1988. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. 85: 8136-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T. and Müller, J. 2004. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 5: 373-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshitz H.D., Peattie, D.A., and Hogness, D.S. 1987. Novel transcripts from the Ultrabithorax domain in the bithorax complex. Genes & Dev. 1: 307-322. [DOI] [PubMed] [Google Scholar]
- Lyko F., Brenton, J.D., Surani, M.A., and Paro, R. 1997. An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nat. Genet. 16: 171-173. [DOI] [PubMed] [Google Scholar]
- Martens J.A., Laprade, L., and Winston, F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571-574. [DOI] [PubMed] [Google Scholar]
- Martin C.H., Mayeda, C.A., Davis, C.A., Ericsson, C.L., Knafels, J.D., Mathog, D.R., Celniker, S.E., Lewis, E.B., and Palazzolo, M.J. 1995. Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. 92: 8398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C. and Paro, R. 2002. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes & Dev. 16: 2672-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E., Grafken, P.R., Ahmad, K., and Henikoff, S. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 101: 1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Venturini, F., Chalkley, G.E., and Verrijzer, C.P. 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22: 7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Hart, C.M., Francis, N.J., Vargas, M.L., Sengupta, A., Wild, B., Miller, E.L., O'Connor, M.B., Kingston, R.E., and Simon, J.A. 2002. Histone methyltransferase activitiy of a Drosophila Polycomb group repressor complex. Cell 111: 197-208. [DOI] [PubMed] [Google Scholar]
- Muyrers J.P., Zhang, Y., Buchholz, F., and Stewart, A.F. 2000. RecE/RecT and Reda/Redb initiate double-stranded break repair by specifically interacting with their respective partners. Genes & Dev. 14: 1971-1982. [PMC free article] [PubMed] [Google Scholar]
- Orlando V., Jane, E.P., Chinwalla, V., Harte, P.J., and Paro, R. 1998. Binding of trithorax and Polycomb proteins to the bithorax complex: Dynamic changes during early Drosophila embryogenesis. EMBO J. 17: 5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S., Melfi, R., and Pirrotta, V. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes & Dev. 15: 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22: 8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G. and Wolff, I. 1981. Isolation of dominant suppressor mutations for position-effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 182: 516-519. [DOI] [PubMed] [Google Scholar]
- Ringrose L., and Paro, R. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38: 413-443. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Rehmsmeier, M., Dura, J.M., and Paro, R. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melonogaster. Dev. Cell 5: 759-771. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Ehret, H., and Paro, R. 2004. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of Polycomb complexes. Mol. Cell. 16: 641-653. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E. and Akam, M. 1989. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107: 321-329. [DOI] [PubMed] [Google Scholar]
- Schröder C., Tautz, D., Seifert, E., and Jäckle, H. 1988. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 7: 2881-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A.K., Kuhrs, A., and Müller, J. 2004. General transcriptional silencing by a Polycomb response element in Drosophila. Development 131: 1959-1965. [DOI] [PubMed] [Google Scholar]
- Siegal M.L. and Hartl, D.L. 1996. Transgene coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144: 715-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker T.R., Kongsuwan, K., Lengyel, J.A., and Merriam, J.R. 1986. The zygotic mutant tailless affects the anterior and posterior ectodermal regions of the Drosophila embryo. Dev. Biol. 113: 64-76. [DOI] [PubMed] [Google Scholar]
- Struhl G. and Basler, K. 1993. Organizing activity of wingless protein in Drosophila. Cell 72: 527-540. [DOI] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51-61. [DOI] [PubMed] [Google Scholar]
- Treisman J.E. 2001. Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128: 603-615. [DOI] [PubMed] [Google Scholar]
- Vigoreaux J.O. and Tobin, S.L. 1987. Stage-specific selection of alternative transcriptional initiation sites from the 5C actin gene of Drosophila melanogaster. Genes & Dev. 1: 1161-1171. [DOI] [PubMed] [Google Scholar]
- Wilson C.J., Chao, D.M., Imbalzano, A.N., Schnitzler, G.R., Kingston, R.E., and Young, R.A. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodelling. Cell 84: 235-244. [DOI] [PubMed] [Google Scholar]
- Wittschieben B.O., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C.D., Tempst, P., and Svejstrup, J.Q. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell. 1: 123-128. [DOI] [PubMed] [Google Scholar]
- Wolff T. 2000. Histological techniques for the Drosophila eye. Part I: Larva and pupa. In Drosophila protocols (eds. W. Sullivan, M. Ashburner, and W. Hawley), pp. 216-220. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Zhang Y., Buchholz, F., Muyrers, J.P., and Stewart, A.F. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20: 123-128. [DOI] [PubMed] [Google Scholar]
- Zink D. and Paro, R. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14: 5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]