Abstract
Sarcomas are a diverse group of malignant neoplasms of mesenchymal origin. They develop rarely, but due to poor prognosis, they are a challenging and significant clinical problem. Currently, available therapeutic options have very limited activity. A better understating of sarcomas’ pathogenesis may help develop more effective therapies in the future. The Sonic hedgehog (Shh) signaling pathway is involved in both embryonic development and mature tissue repair and carcinogenesis. Shh pathway inhibitors are presently used in the treatment of basal cell carcinoma. Its increased activity has been demonstrated in many sarcomas, including osteosarcoma, Ewing sarcoma, chondrosarcoma, rhabdomyosarcoma, leiomyosarcoma, and malignant rhabdoid tumor. In vitro studies have demonstrated the effectiveness of inhibitors of the Hedgehog pathway in inhibiting proliferation in those sarcomas in which the components of the pathway are overexpressed. These results were confirmed by in vivo studies, which additionally proved the influence of Shh pathway inhibitors on limiting the metastatic potential of sarcoma cells. However, until now, the efficacy of sarcomas treatment with Shh pathway inhibitors has not been established in clinical trials. The reason for that may be the non-canonical activation of the pathway or interactions with other signaling pathways, such as Wnt or Notch. In this review, we present the Shh signaling pathway's role in the pathogenesis of sarcomas, including both canonical and non-canonical signaling. We also propose how this knowledge could be potentially translated into clinics.
Keywords: Sarcoma, Soft-tissue sarcoma, Bone sarcoma, Hedgehog pathway, Gli
Introduction
Signaling pathways, crucial in the physiological functions of cells and tissues, may, through dysregulation and ensuing dysfunctions, be significant factors in the process of tumorigenesis (Park et al. 2020). The Hedgehog (Hh) signaling pathway plays an important role in embryogenesis and in the upkeep of mature tissues and stem cells (Katoh and Katoh 2008). Mutations leading to dysregulation of the Hh pathway are consistently observed in the basal cell carcinoma (BCC) and medulloblastoma and sporadically in other cancers (Carpenter and Ray 2019). Currently, treatment with specific Shh pathway inhibitors has been approved in BCC by both European Medical Agency (EMA) and Food and Drug Administration (FDA) (Brancaccio et al. 2020), but new studies are being carried out in the hope of expanding these indications (Carballo et al. 2018).
Sarcomas develop from transformed mesenchymal cells and are usually divided into sarcomas arising from the soft tissues and the bones. They are further segregated into various subtypes, making them a very diverse group of tumors (Mehren et al. 2020; Gronchi et al. 2021; W.C.o.T.E. Board 2020). Though sarcomas are rare, accounting for just 1% of all adult malignant tumors, they are characterized by poor prognosis and unsatisfactory treatment options, which makes them a significant clinical challenge (Mastoraki et al. 2020).
In this article, we summarize the current data regarding the role played by the Shh pathway in the pathogenesis of sarcomas. Our goal is to emphasize that connection and start a discussion about the potential value of Shh targeted therapy in the treatment of sarcomas.
Sonic Hedgehog pathway activation and regulation
The Hedgehog (Hh) pathway is a ligand-dependent signaling pathway. In vertebrates, three Hh ligands have been described: Desert hedgehog (Dhh), Indian hedgehog (Ihh), and Sonic hedgehog (Shh), the last one being the primary ligand in humans (Lézot et al. 2020). Furthermore, the Hh signaling pathway consists of two 12-pass transmembrane receptors Ptch1 and Ptch2, Smoothened (Smo) receptor, and three transcription factors Gli1, Gli2, and Gli3 (Yao et al. 2018).
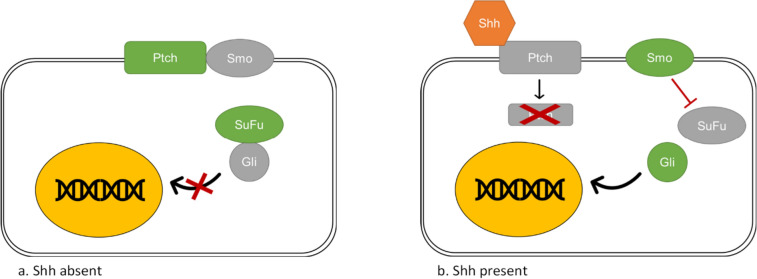
There are two pathways which activate the Shh cascade—a canonical and a non-canonical one (Lézot et al. 2020). Interaction between the Shh and the Ptch receptor is the base of the canonical cascade (Marigo et al. 1996). When the Hh ligand is absent, the Ptch receptor binds to a constitutively active Smo receptor, suppressing the Smo activity (Fig. 1) (Liu et al. 2017). The suppressor of fused (SuFu) is a negative regulator of the Shh pathway (Liao et al. 2020). In the absence of activated Smo, SuFu forms SuFu–Gli complexes and sequesters Gli proteins in the cytoplasm, restraining their activity (Zhang et al. 2017). However, recent data indicate that SuFu may also further increase Gli2 expression in cells with an already high Gli2 expression (Yin et al. 2019). The presence of the Hh ligand leads to the binding of the Hh ligand to the Ptch receptor, which results in endocytic degradation of Ptch in the lysosome (Incardona et al. 2000). Activated Smo induces the dissociation of SuFu–Gli complexes and translocation of Gli proteins into the nucleus, where it can regulate the expression of various genes (Ruel and Thérond 2009).
Fig. 1.
Shh pathway activation. The Shh pathway consists of the Sonic hedgehog ligand (Shh), Patched receptor (Ptch), Smoothened receptor (Smo), Suppressor of fused (SuFu) and transcription factors (Gli). The illustration shows the canonical activation of the Shh signaling pathway. a In the absence of the Shh ligand, Ptch binds to a constitutively active Smo, suppressing its activity. In the absence of activated Smo, SuFu forms SuFu–Gli complexes and restrains Gli activity. b In the presence of the Shh ligand, its binding with Ptch results in endocytic degradation of Ptch in the lysosome. Activated Smo induces the dissociation of SuFu–Gli complexes and translocation of Gli proteins into the nucleus
The non-canonical cascade refers to the activation of Gli transcription factors independently of Smo (Pietrobono et al. 2019). This can be caused by various signaling molecules and pathways, which can work separately or simultaneously (Pietrobono et al. 2019). Examples of these mechanisms will be discussed in more detail later. Non-canonical activation is involved in carcinogenesis connected with elevated Gli activity (Brechbiel et al. 2014) (see Fig. 2).
Fig. 2.
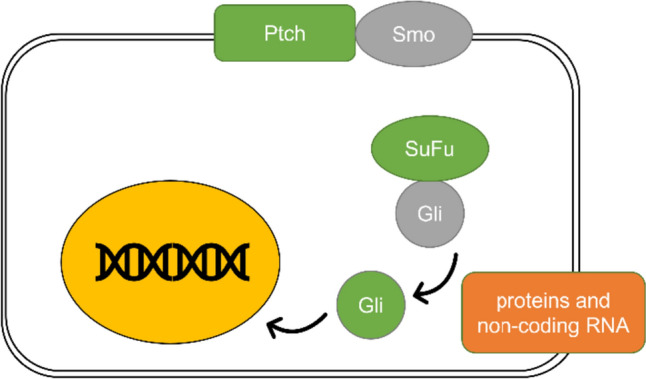
Non-canonical Shh pathway activation. The illustration shows an alternative way of Shh signaling pathway activation. The non-canonical pathway activation happens independently of Smo and directly through the activation of Gli by various proteins and non-coding RNA, some of which are described in more detail in the text
Various proteins and non-coding RNA regulate the Hh signaling pathway: microRNA (miRNA) and long non-coding RNA (lncRNA) (Yao et al. 2018). Notably, the regulators acting beyond Smo may participate in the non-canonical activation of the Shh pathway.
Kinases, transcription factors, glycoproteins, and pro- and anti-apoptotic factors have been connected to the activation or inhibition of the Hh pathway (Yao et al. 2018). Nek2A, an NIMA (never in mitosis gene a)-related kinase 2A, inhibits the Hh pathway and the transcriptional activity of Gli2 by stabilizing SuFu (Zhou et al. 2017). Rab23, a GTPase of the Rab family, also negatively regulates the Hh pathway through interaction with SuFu (Chi et al. 2012). Moreover, the methylation of Gli3, catalyzed by Set7, increases Gli3 stability and DNA-binding activity, which promotes the Shh pathway activation (Fu et al. 2016). Increased levels of Galectin-1 (Gal-1) activate the Hh signaling by increasing the transcription of Gli1 via a Smo-independent pathway (Chong et al. 2016). Another study demonstrated that silencing the B4GALT1 gene, which encodes Beta-1,4-galactosyl-transferase 1, in K562/ADR cells results in the inhibition of the Hh pathway (Zhou et al. 2012).
Proteins that interact directly with Smo also play a role in regulating the Hh pathway. RACK1, a receptor of activated kinase 1, initiates Gli1 transcription through interaction with activated Smo (Shi et al. 2012), while a GTPase Arl13b promotes Hh signaling by stabilizing Smo (Shao et al. 2017).
Gli1, Gli2, and Gli3 are also directly regulated by various proteins. Overexpression of NUSAP1 leads to upregulation of the Hh pathway’s target gene through induction of Gli1 translocation into the nucleus (Wu et al. 2017). Mastermind-like 1 (Maml1) binds to the Gli proteins and acts as a transcriptional coactivator, reinforcing the activation of Shh pathway target genes (Quaranta et al. 2017). Overexpression of beta1 integrin (ITGB1) results in the upregulation of Shh and Gli1 levels and the downregulation of SuFu, leading to the activation of Hh signaling (Song et al. 2015). FOXC1, a transcription factor, activates the Smo-independent Hh pathway by direct interaction with Gli2, increasing its DNA-binding and transcription-activating capacity (Han et al. 2015). Overexpression of Sloan–Kettering viral oncogene homolog (Ski), a protein which can function both as an oncoprotein and a tumor suppressor gene, leads to increased expression of Shh pathway components, such as Shh, Ptch-1, Smo, Gli1, and Gli2 (Song et al. 2016). Increased activity of mTORc2, one of the complexes formed by mTOR kinase (Fu and Hall 2020), has been shown to enhance the expression of Hh components (Gli1, Gli2, and Ptch1) and the target genes of the pathway (Cyclin D1, Cyclin D2, Cyclin E, Snail, Slug, and VEGF). Moreover, mTORc2 promotes stability and nuclear translocation of Gli2 (Maiti et al. 2017).
The hedgehog signaling pathway is crucial in embryonic processes: it controls cells differentiation, tissue polarity, and proliferation (Varjosalo and Taipale 2008). Timing aberrations of hedgehog signaling can generate embryological malformations (Ericson et al. 1996). To some degree, the Hh pathway remains active in mature organisms and participates in processes such as stem cell maintenance and tissue repair (Roma et al. 2012). Genes regulated by the Hh signaling pathway are crucial in cell proliferation and survival, cell cycle, cell invasion, and stem cell formation (Yao et al. 2018). However, in most tissues, the pathway remains inactive and is only activated when necessary, for example, in the regeneration of damaged tissues (Skoda et al. 2018).
The Hh pathway plays an essential role in osteogenesis, regulating endochondral and intramembranous ossification. It also promotes bone resorption through indirect activation of osteoclasts, making it a crucial factor in bone homeostasis and remodeling (Yang et al. 2015).
The role of Shh pathway in the pathogenesis of selected subtypes of sarcomas
Osteosarcoma
Osteosarcoma is a malignant bone tumor that produces osteoid and immature bone and comprises mesenchyme-derived cells (W.C.o.T.E. Board 2020; Biazzo and Paolis 2016). It is a high-grade sarcoma and appears mostly among children and young adults (Lo et al. 2014a). Osteosarcoma has a high metastatic potential; approximately one-fourth of patients with osteosarcoma have metastases at presentation (Tsukamoto et al. 2020). The clinical outcome for patients with lung metastases remains poor, since they are hard to control (Yao et al. 2018) and are resistant to standard chemotherapy (Saitoh et al. 2016).
Previous studies have shown that both the canonical and non-canonical Hh pathways may be involved in osteosarcoma tumorigenesis (Lo et al. 2014b). Osteosarcoma patients with higher levels of Gli1 are more likely to respond better to chemotherapy (Lézot et al. 2020; Lo et al. 2014a). Other evidence suggests a correlation between overexpression of Gli-2 and poor clinical outcomes (Yang et al. 2013). Gene expression analyses by real-time PCR revealed overexpression of Shh, Ihh, Ptch1, Smo, and Gli2 in osteosarcoma cell lines. In contrast, examination of osteosarcoma biopsy specimens showed overexpression of Smo, Ptch1, and Gli compared to normal bone tissue cells (Hirotsu et al. 2010) (Yang et al. 2013). Another group confirmed higher expression of Gli1 and Gli2 in canine osteosarcoma cell lines compared to normal canine osteoblasts. Moreover, there was a correlation between the Gli1 or Gli2 expression level and the expression of Ptch1 and PAX6 (Shahi et al. 2014).
Data report that Gli2 significantly promotes the proliferation, migration, and invasion of mesenchymal stem cells and osteosarcoma cells (Nagao-Kitamoto et al. 2015a). This has been confirmed by knockdown of Gli2, which promoted the arrest of osteosarcoma cells in the G1 phase of cell cycle and inhibited osteosarcoma growth, demonstrated in murine xenograft models (Nagao et al. 2011). One of the target genes of Gli2 is the RPS3 gene encoding the ribosomal protein S3, which is a component of the eukaryotic 40S ribosomal subunit and is involved in various processes, such as apoptosis, immune response, DNA repair, transcriptional regulation, and transformation (Yao et al. 2018; Gao and Hardwidge 2011). Overexpression of RPS3 increases the migration and invasion of osteosarcoma cells, playing a role in metastases formation (Nagao-Kitamoto et al. 2015a).
Exosomes, small vesicles secreted by various cells, contain proteins, lipids, and nucleic acids such as mRNA or miRNA. Exosomes acquired from mesenchymal stem cells derived from human bone marrow (hBMSC) have been shown to promote osteosarcoma cell growth by activating the Hh pathway. Hsp70 and CD63 expression has been detected in these exosomes (Qi et al. 2017).
The interaction of the Hh pathway with other signaling pathways can also be an essential factor in osteosarcoma progression, including metastasis formation (Yao et al. 2018). Some evidence suggests that aberrant Hh signaling leads to overexpression of the Yes-associated protein 1 (Yap-1), which acts as an oncogene. YAP1 is the effector of the Hippo pathway, which dysfunction can result in tumorigenesis and metastasis (Chan et al. 2014; Kovar et al. 2020). Moreover, both Wnt and Hh pathway components are significantly upregulated in metastatic cells compared to parental cell lines (Muff et al. 2015). Another study demonstrated the upregulation of target genes of both AKT/PI3K and Hh pathways in canine mammary osteosarcomas; this indicates that the interaction of these two pathways may be an important factor in osteosarcoma formation and proliferation (Pawlowski et al. 2011).
The Notch pathway has also been shown to influence the Hh pathway in the pathogenesis of osteosarcoma. DNMT3A, through methylation of miR-149, leads to its decreased activity, promoting overexpression of Notch components. This results in increased activity of the Hh pathway, which induces osteosarcoma's development and progression (Cheng and Wang 2022).
The role of the Hh pathway in the pathogenesis of osteosarcoma has also been shown in several studies assessing the preclinical efficacy of the Hh pathway inhibitors. Cyclopamine, a Smo inhibitor, promoted G1-phase arrest of the cell cycle, inhibited expression of cyclin D1, cyclin E1, SKP2, and pRb, and, as a result, restrained the growth of osteosarcoma in vitro (Hirotsu et al. 2010). In an animal model, it has been shown that cyclopamine decreased pulmonary osteosarcoma metastasis formation by 20% (Warzecha et al. 2012). Four acylguanidine and acylthiourea derivatives of cyclopamine, which have similar Smo-inhibiting properties, had been shown to have either cytotoxic or proliferation-inhibiting effects in osteosarcoma cell lines. The three drugs inhibiting proliferation decreased the expression of Gli1 and showed significant pro-apoptotic activity without causing severe side effects. Moreover, unlike the older generation of Smo inhibitors—cyclopamine, vismodegib, or sonidegib—they also effectively inhibit a chemoresistant form of Smo (Bernardini et al. 2018).
Examination of osteosarcoma cell viability revealed that treatment based on a combination of ATO and GANT61 (Gli inhibitors) or vismodegib (Smo inhibitor) decreased osteosarcoma cell migration. Moreover, inhibition of the osteosarcoma metastasis to the lung was observed during this combination treatment (Nagao-Kitamoto et al. 2015b). Decreased expressions of Gli1, Gli2, Ptch1, and PAX6 were observed after treatment of canine osteosarcoma cells using GANT61. Moreover, inhibition of cell growth was observed (Shahi et al. 2014). GANT61 has also been proven to inhibit the viability of certain osteosarcoma cell lines (Lo et al. 2014b) and to reduce the resistance of osteosarcoma cells to cisplatin in vivo (Chen et al. 2021). A Gli inhibitor ATO promotes apoptotic cell death in human osteosarcoma cells by accumulating DNA damage (Nakamura et al. 2013). Moreover, recent findings indicate that ATO inhibits the transcriptional activity of Gli2 and inhibits osteosarcoma cell invasion (Nagao-Kitamoto et al. 2015b).
Another study on patient-derived xenograft models of osteosarcoma evaluated the efficacy of treatment with saridegib, another Smo inhibitor. The drug effectively inhibited the canonical but not the non-canonical Hh pathway in the tumor and its microenvironment. The inhibition resulted in decreased expression of Ptch1 and Gli1, increased level of apoptosis, and decreased tumor weight and volume (Lo et al. 2014b).
Transcription of the genes associated with osteogenic differentiation is connected with the degree of the chromatin compaction (Montecino et al. 2020). There are different mechanisms that control chromatin organization in osteogenic cells, one of the major ones is a polycomb repressor complex 2 (PRC2) (Voigt et al. 2013; Shi et al. 2017).
PRC2 is involved in the modification of the chromatin during osteogenesis—it catalyzes the process of the trimethylation of histone H3 at lysine 27 (H3K27me3) and, as a result, induces chromatin condensation and transcriptional repression (Chamberlain et al. 2008). Enhancer of zeste homolog 2 (Ezh2) is one of the methylotransferases forming PRC2 (Shi et al. 2017). EZH2 is involved in the process of skeletal development, supporting self-renewal of mesenchymal stem cells and blocking osteogenic commitment of progenitor cells (Carrasco et al. 2023). It has been described that inactivation of Ezh2 leads to the activation of major osteogenic pathways and increased expression of bone formation-related genes and results in the pro-osteogenic effects (Dudakovic et al. 2020, 2015). A study revealed that the Ezh2 inhibitor-Tazometostat (EPZ6438), through the loss of H3K27me3 in the presence of osteogenic cues, enhanced osteogenic differentiation (Carrasco et al. 2023). Different results of Ezh2 inhibition have been described depending on the period of the Ezh2 inhibition (Carrasco et al. 2023). Short-term inactivation of Ezh2 (such as caused by Tazometostat-EPZ6438) activates the osteogenic process in the progenitor cells and stimulates bone formation in vivo, while persistent loss of Ezh may lead to a loss in the number of the osteoblasts (Dudakovic et al. 2020, 2016; Galvan et al. 2021).
Ewing sarcoma
Ewing sarcoma (ES) is the second most frequent bone sarcoma affecting children and adolescents (Balamuth and Womer 2010; Grünewald et al. 2018). It occurs predominantly in bones (long bones, pelvis, chest wall, and spine) and to a much lower extent in soft tissues (W.C.o.T.E. Board 2020; Lézot et al. 2020). Ewing sarcoma is associated with chromosomal translocation—usually t(11;22) (q12;q24), which leads to EWSR1–FLI1 genes fusion (Aurias et al. 1984). Nonetheless, in approximately 15–20% of Ewing sarcomas, EWSR1 is fused with members of the ETS family other than FLI1, most frequently ERG (Sorensen et al. 1994).
Both survival and tumorigenesis of the Ewing sarcoma family of tumors are keyed to the function of EWS-FLI1, and it was shown that Gli1 is a transcriptional target of EWS-FLI1 (Beauchamp et al. 2009). Data suggest that Gli1 upregulation by EWS-FLI1 is Smo-independent, which indicates an involvement of the non-canonical activation pathway (Zwerner et al. 2008; Joo et al. 2009).
The efficacy of ATO, a direct Gli inhibitor, has been shown in both cell lines and xenograft models of the EWS-FLI1 Ewing sarcoma. Its effects included cell cytotoxicity and inhibition of cell migration and invasion (Beauchamp et al. 2011) (Zhang et al. 2012). In vitro studies experiments demonstrated that another Gli inhibitor, GANT61, reduces the growth of Ewing sarcoma cells, mainly by inducing caspase-3/7-dependent cell apoptosis. The SK-N-LO cell line, characterized by the presence of the EWS-FLI1 fusion, was the most sensitive line to GANT61 treatment (Mullard et al. 2020).
Chondrosarcoma
Chondrosarcoma is a malignant cartilage tumor, against which typically neither chemotherapy nor radiotherapy is effective (W.C.o.T.E. Board 2020; Fiorenza et al. 2002). Physiologically, Ihh and PTHrP (parathyroid hormone-related protein) regulate chondrocyte proliferation and differentiation. This signaling pathway is controlled through a negative feedback loop. A study showed that the Ihh–PTHrP pathway is dysregulated in chondrosarcoma cells, leading to constitutive ligand-dependent Hh signaling, as demonstrated by overexpression of Ptch1 and Gli1. However, no correlation was observed between the tumor grade and the level of Hh pathway components’ expression (Tiet et al. 2006). Another study demonstrated increased levels of Ihh mRNA compared to Shh mRNA and high levels of Ptch1, Smo, and Gli1 mRNA.
The efficacy of saridegib (Smo inhibitor) in the treatment of chondrosarcoma in primary xenografts was also evaluated. The observed effects included decreased volume and cellularity of the tumor, tumoral calcification, and decreased chondrocyte proliferation. The high efficacy of the treatment was likely due to the ligand-dependent nature of the Hh pathway present in chondrosarcoma (Campbell et al. 2014).
However, emerging evidence suggests that the non-canonical pathway activation might also play a role in its pathogenesis. It has been shown that Gli1 overexpression can be caused by the major vault protein (MVP) via mTOR/S6K1 signaling pathway (Wang et al. 2021). This might explain the unsatisfactory results of the clinical trials assessing the efficacy of Smo inhibitors—saridegib or vismodegib in patients with chondrosarcoma (Wagner et al. 2013; Italiano et al. 2013).
Rhabdomyosarcoma
Rhabdomyosarcoma is a high-grade tumor of skeletal myoblast-like cells and is the most common malignant soft-tissue sarcoma affecting children (W.C.o.T.E. Board 2020; Skapek et al. 2019; Dziuba et al. 2018). There are two major subtypes of RMS—embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS) (Yechieli et al. 2021). ARMS samples' analysis demonstrated that most cases are connected with chromosomal translocation (Gallego Melcón and Sánchez de Toledo Codina 2007; Davis et al. 1994). Such chromosomal aberrations can lead to gene fusions, which are observed in ARMS, where in the majority of samples PAX3/7–FOXO1 gene fusion is present (Kaleta et al. 2019). Chromosomal translocations have not been observed in ERMS cases; however, another study, which examined 12 embryonal rhabdomyosarcoma specimens from 10 patients, revealed gains and losses of some of the chromosomes or chromosomal regions. One of the most frequent ones was the loss of 9q22, a locus of Ptch (Bridge et al. 2000) (Roma et al. 2012). A cohort study revealed that in both ERMS and fusion-negative ARMS, Ptch1, Gli1, and Gli3 genes have higher expression levels than in fusion-positive ARMS. This study also demonstrated a correlation between high expression of Ptch1 and reduced overall survival in both ERMS and fusion-negative ARMS (Zibat et al. 2010). However, there is an ongoing discussion regarding whether the survival of ERMS patients is linked to Gli1 and Ptch1 expression level—microarray analysis of rhabdomyosarcoma samples showed that the survival rate, age, tumor stage, group, and the primary anatomic site did not correlate with the expression of Gli1 mRNA transcripts (Pressey et al. 2011). Furthermore, this study did not identify any correlation between Gli1 or Ptch1 expression and poor clinical outcomes in ERMS and gene-fusion-negative ARMS patients (Pressey et al. 2011). However, the authors of this study emphasize the necessity of carrying out more trials on larger cohorts of patients to understand the molecular background of ERMS pathogenesis properly.
Regarding the efficacy of Hh inhibitors, an in vivo study was carried out to assess the effectiveness of sonidegib (Smo inhibitor) treatment in the embryonal subtype of rhabdomyosarcoma (ERMS) with a mutation in Ptch. Sonidegib had a significant antitumor effect in murine models of ERMS, as evidenced by a reduction of tumor growth in monotherapy and combined with pictilisib, a PI3K inhibitor. This effect of sonidegib treatment correlated with a decreased expression of Gli1 in vitro. Another study with vismodegib showed a similar effect (Geyer et al. 2018). Moreover, ATO, a Gli inhibitor, was identified to reduce viability and clonal growth and induce apoptosis of both embryonal and alveolar rhabdomyosarcoma cell lines (Boehme et al. 2016).
Leiomyosarcoma
Leiomyosarcoma (LMS) is one of the most common soft-tissue sarcoma in adults, representing 10–20% of newly diagnosed cases. Uterine LMS is, in turn, the most common type of uterine sarcoma (George et al. 2018). Increased Smo, SuFu, and Gli1 expression was described in uterine LMS compared to normal myometrium (Garcia et al. 2016). Another study confirmed elevated levels of Smo and Gli1 and that the Hh pathway is deregulated in uterine LMS (Garcia et al. 2021). A study demonstrated that leiomyosarcoma cells showed decreased proliferation, migration, and invasion in response to treatment with Smo or Gli inhibitors (Garcia et al. 2020).
Interesting data come from analyses of different regulators of the Hh pathway in LMS. It has been shown that NKX6-1 plays an oncogenic role in LMS, and its overexpression modulates the Shh pathway, leading to the promotion of stem cell properties in tumor cells and poor prognosis. In vitro treatment of NKX6-1 overexpressing LMS cells with an inhibitor of the Shh pathway RU-SKI43 resulted in cell growth inhibition (Su et al. 2021). The effect of GANT61 (Gli inhibitor) on LMS was assessed using a leiomyosarcoma xenograft model. GANT61 caused significant regression of the leiomyosarcoma growth and decreased expression of Gli1 and its target genes: BMP4 and c-MYC (Garcia et al. 2022).
Malignant rhabdoid tumor
Malignant rhabdoid tumors (MRT) are a group of mainly soft-tissue cancers which most commonly develop in the kidney or the brain but can be found in any body part. Those located in the brain are referred to as atypical teratoid/rhabdoid tumors (ATRT). MRT mainly affect infants and is believed to develop during embryogenesis. They remain one of the most lethal pediatric cancers and have a particularly bad prognosis in case of metastases (W.C.o.T.E. Board 2020; Custers et al. 2021).
Most ATRTs are characterized by a biallelic mutation of the SMARCB1 gene and loss of encoded protein INI1/SNF5 (Frühwald et al. 2016). A recent meta-analysis has divided all MRT into three main subgroups based on their molecular and clinical features. One of them, ATRT-SHH, is characterized by an overexpression of Shh pathway components (such as Gli1 and Ptch1) and members of the Notch pathway (Ho et al. 2020). A study indicates that in MRTs, the Shh pathway is activated through the non-canonical pathway. The lack of SNF5 protein, typically involved in limiting Gli1 expression, leads to Gli1 overexpression and drives the growth of cancer cells. This theory was supported by the in vivo treatment of MRT with a small-molecule inhibitor of Gli, which led to the decrease of Gli1 levels and inhibition of tumor growth, whereas Smo inhibitors had no effect (Jagani et al. 2010) (see Table 1).
Table 1.
Hedgehog pathway components expression and effect of pathway inhibition in vitro and in vivo in various sarcomas
| Overexpression of Hh components in cell lines | Overexpression of Hh components in human tumors | Effect of Hh inhibition | Other pathways involved | |
|---|---|---|---|---|
| Osteosarcoma |
Shh Ihh Ptch1 Smo Gli |
Shh Ihh Ptch1 Smo Gli |
Cyclopamine (SMO inhibitor): Inhibition of growth and metastasis formation in preclinical models Vismodegib (SMO inhibitor): Decreased cell migration and metastasis formation (when used in combination treatment) Saridegib (SMO inhibitor): Effective inhibition of canonical pathway, noneffective inhibition of non-canonical pathway in xenograft model ATO (Gli inhibitor) Decreased cell migration and metastasis formation in preclinical models (when used in combination treatment) Increased apoptosis, reduced invasion GANT61 (Gli inhibitor) Reduction of osteosarcoma cells' resistance to cisplatin Inhibition of cell growth Inhibition of certain OS cell lines viability reduction of cell migration and metastasis formation in combination treatment in preclinical models Emodin Lowered radioresistance of osteosarcoma cells Degalactotigonin Suppression of osteosarcoma cells |
Wnt pathway Notch pathway Hippo pathway MAPK pathway PI3K/AKT/mTOR pathway |
| Ewing sarcoma | Gli | Gli |
GANT61 (Gli inhibitor) Reduced cell growth in preclinical models (particularly in those with the presence of the EWS-FLI1 Fusion gene) ATO (Gli inhibitor) Increased cytotoxicity, reduced cell migration and invasion in preclinical models of EWS-FLI1 Ewing-sarcoma |
EWS–FLI1 pathway IGF–1R pathway |
| Chondrosarcoma |
Ptch1 Gli1 |
Ptch1 Gli1 |
Saridegib (Smo inhibitor) Decreased volume and cellularity of the tumor Decreased tumoral calcification Decreased chondrocyte proliferation in xenograft models |
Ihh–PTHrP pathway PI3K/AKT/mTOR pathway SRC pathway |
| Rhabdomyosarcoma |
Ptch1 Gli1 Gli3 |
Ptch1 Gli1 Gli3 |
Vismodegib, Sonidegib (SMO inhibitors) Reduced tumor growth Reduced number of proliferating cells in ERMS with a mutation in Ptch in preclinical models ATO (Gli inhibitor) Increased cells apoptosis Reduced viability and clonal growth in preclinical models of both ERMS and ARMS |
Wnt pathway Notch pathway Hippo pathway p53 pathway |
| Leiomyosarcoma |
Smo Gli |
Smo SuFu Gli1 |
GANT61 (Gli inhibitor) Regression of leiomyosarcoma growth on xenograft model |
|
| Malignant rhabdoid tumor (ATRT-SHH subtype) | Gli1 | Gli1 |
small molecule inhibitor of Gli Inhibition of tumor growth in in vivo treatment Smo-inhibitors No effect on tumor growth inhibition |
Notch pathway |
Shh pathway inhibitors in the treatment of sarcomas
Smo inhibitors
Smo inhibitors include cyclopamine and its derivatives and analogues: vismodegib, saridegib (IPI-926), and sonidegib (erismodegib, LDE225). While cyclopamine is chemically unstable (Tremblay et al. 2009) and has significant side effects (Warzecha et al. 2012), the newer Smo inhibitors are characterized by a more favorable pharmacokinetic profile and, in some cases, a higher potency (Tremblay et al. 2009; Kumar and Fuchs 2015).
Cyclopamine inhibited growth and metastasis formation in preclinical models of osteosarcoma (Hirotsu et al. 2010; Warzecha et al. 2012). Vismodegib, used in combination treatment, was also successful in preclinical models, leading to a decrease in cell migration and metastasis formation in osteosarcoma (Nagao-Kitamoto et al. 2015b), and a reduction of tumor growth and the number of proliferating cells in the embryonal subtype of rhabdomyosarcoma with a mutation in Ptch (Geyer et al. 2018). Saridegib decreased tumor size and increased apoptosis in the preclinical models of osteosarcoma associated only with the canonical activation of the Hh pathway (Lo et al. 2014b) Sonidegib significantly reduced tumor growth and the number of proliferating cells in the preclinical models of the embryonal subtype of rhabdomyosarcoma with a mutation in Ptch (Geyer et al. 2018). Promising results have also been obtained in a study assessing the efficacy of four new cyclopamine derivatives in osteosarcoma cell lines. They significantly induced apoptosis and also effectively inhibited a chemoresistant form of Smo, unlike the older generations of Smo inhibitors (Bernardini et al. 2018).
While vismodegib and sonidegib have been approved by FDA as agents targeting the Hh pathway in the treatment of basal cell carcinoma, none of the Smo inhibitors has been approved for therapy of any sarcomas associated with Hh pathway dysfunction (Meiss et al. 2018; Casey et al. 2017) due to the lack of efficacy in clinical trials. The efficacy of vismodegib was assessed in a clinical trial conducted to determine if dual inhibition of the Notch and Hh pathways would result in a synergistic antitumor effect in advanced sarcomas (Gounder et al. 2022). Patients received a combination of vismodegib and a Notch inhibitor RO4929097 or Notch inhibitor alone. No patients had an objective response, and there was no difference in progression-free or overall survival between treatment arms. Paired tumor biopsies from a subset of patients demonstrated decreased expression of cleaved Notch and decreased phosphorylated Akt, suggesting successful inhibition of the gamma-secretase enzyme leading to downregulation of Notch signaling. Contrary, only two out of ten patients had a substantial decrease in Gli1 expression, implying that inhibition of the canonical Hh pathway was not very effective (Gounder et al. 2022).
It is important to note that the non-canonical pathway activation happens independently of Smo. Its inhibitors have thus no effect on the pathway in cases where this method of activation is predominant. This might explain the unsatisfactory results of the clinical trials assessing the efficacy of saridegib and vismodegib in patients with chondrosarcoma, where a significant role of the non-canonical activation is suspected (Wang et al. 2021; Wagner et al. 2013; Italiano et al. 2013).
Another possible explanation for the lack of efficacy of Smo inhibitors is the presence of specific mutations in the target protein. Several solutions have been proposed to overcome this issue: development of second-generation Smo inhibitors that would retain their inhibitory effect despite the presence of the mutation, for example, by targeting a different domain of the protein; targeting downstream molecules of Smo, such as Gli transcription factors; and finally genetic prescreening before initiating Smo inhibitor therapy (Nguyen and Cho 2022).
Another promising strategy could be the simultaneous inhibition of both the upstream and downstream levels of the Shh pathway. A study showed that a synthetic isoflavone, which targets Smo and Gli1 at the same time, had a significant anti-tumor effect both in in vitro and in vivo models of medulloblastoma. This form of therapy also has the potential to decrease the toxicity of individual Shh pathway inhibitors and should therefore be examined in sarcoma models (Lospinoso Severini et al. 2019).
Gli inhibitors
Gli inhibitors include GANT61 and arsenic trioxide (ATO). There is an ongoing discussion about their potential therapeutic value, particularly in the case of the non-canonical activation of the Shh pathway, where Smo inhibitor therapies have no effect (Beauchamp and Uren 2012).
GANT61 reduced cell growth in preclinical models of Ewing sarcoma, particularly the ones characterized by the EWS–FLI1 fusion gene (Mullard et al. 2020). It also reduced cell viability (Lo et al. 2014b; Shahi et al. 2014) as well as the resistance to cisplatin (Chen et al. 2021) in monotherapy and cell migration and metastasis formation in combination treatment (Nagao-Kitamoto et al. 2015b), in preclinical models of osteosarcoma. Finally, GANT61 caused a regression of growth in preclinical models of leiomyosarcoma (Garcia et al. 2022).
ATO in monotherapy in preclinical models of EWS–FLI1 Ewing sarcoma led to cell cytotoxicity, reduced both cell migration and invasion (Beauchamp et al. 2011; Zhang et al. 2012). Moreover, in preclinical osteosarcoma models, ATO has been observed to increase apoptosis and reduce invasion (Nagao-Kitamoto et al. 2015b; Nakamura et al. 2013). A study showed that targeting both embryonal and alveolar rhabdomyosarcoma with ATO resulted in increased apoptosis, reduced viability, and reduced clonal growth (Boehme et al. 2016). ATO combined with GANT61 or vismodegib reduced cell migration and metastasis formation in preclinical osteosarcoma models (Nagao-Kitamoto et al. 2015b).
These findings suggest that Gli inhibitors may have a potential therapeutic value in treating sarcomas associated with elevated Gli levels, particularly in the case of the non-canonical activation of the Shh pathway, where Smo inhibitors have no effect. However, while the FDA has approved ATO for the treatment of acute promyelocytic leukemia (Ferrara et al. 2022), neither drug has been registered as a treatment method for sarcomas. The significant cytotoxicity caused by Gli inhibitors might be a possible setback (Sigafoos et al. 2021).
Other drugs
Emodin (1, 3, 8-trihydroxy-6-methylanthraquinone) is a natural anthraquinone derivative which has been reported to have various desired pharmacological effects, such as an anti-neoplastic, antioxidant, anti-inflammatory, and anti-apoptotic potential (Semwal et al. 2021). Regarding emodin’s anticancer activity, its effects include induction of apoptosis and cell cycle arrest, anti-metastasis activity, and reversion of multidrug resistance (Dong et al. 2016). A study showed that emodin, through inhibition of the Shh pathway, can partially reverse the radioresistance of osteosarcoma cells. In this study, two cell lines have been used: human osteosarcoma cell line MG63 and, produced through 30-repeat low-dose X-ray irradiation cycles, cell line MG63R (radioresistant OS cells). The study revealed that emodin treatment before irradiation inhibited the nuclear translocation of Gli1 in MG63 cells and lowered the levels of Shh and BCL2 in MG63R cells (Qu et al. 2017). BCL2 is an antiapoptotic protein whose function has been linked to the mitochondrial pathway and has been described to be expressed at high levels in osteosarcoma cells (Chen et al. 2015). It has been described that BCL2 protein protects osteosarcoma cells from apoptosis, and on this account, silencing BCL-2 may have a positive outcome on the effectiveness of therapeutic strategies in osteosarcoma (Zhao et al. 2009).
Degalactotigonin (DGT), a substance extracted from the plant Solanum nigrum L., has been shown to inhibit the Hh pathway through GSK3 beta inactivation. This leads to the suppression of osteosarcoma proliferation and metastasis (Zhao et al. 2018). Glycogen synthase kinase-3β (GSK3β) is a serine/threonine protein kinase which levels have been described to have a direct impact on osteosarcoma cells (Tang et al. 2012). A study on patient-derived xenograft models has shown that inhibition of GSK-3β leads to inhibition of the NF- κB pathway and results in the apoptosis of osteosarcoma cells (Tang et al. 2012).
Conclusions and discussion
Dysregulation of the Hh pathway, leading to overexpression of its components, has been observed in some sarcomas, including osteosarcoma, Ewing-sarcoma, chondrosarcoma, rhabdomyosarcoma, leiomyosarcoma, and malignant rhabdoid tumor. This points to the critical role played by the Hh pathway in the tumorigenesis of sarcomas; however, Hh components do not seem to be the core mechanism in the pathogenesis of these tumors. Subsequently, the efficacy of sarcomas treatment with Hh inhibitors was assessed. Both in vitro and in vivo studies showed promising results, but they have not yet been confirmed in clinical trials. We believe that the observed lack of effect may result from two factors: the non-canonical pathway activation and interactions with other signaling pathways.
Due to its mechanism, the non-canonical pathway activation is less sensitive to Smo inhibition, making Gli inhibitors the only potentially effective Hh-targeted treatment in this group of sarcomas. Understanding which mechanism of Hh pathway activation is present in each sarcoma subtype (Table 2) is crucial to properly plan and execute preclinical experiments and clinical studies with Hh-targeted agents. Interactions between the Shh pathway and pathways, such as Wnt and Notch, create a complex network where activation of one pathway can increase the expression of the others’ components. Several connection points between Hh and Notch, Wnt, or TGF-β pathways show a reciprocal synergism contributing to tumorigenesis in various tumors but have not yet been thoroughly studied in sarcoma (Pelullo et al. 2019). This is thought to be one of the mechanisms of target-specific treatment evasion (Roma et al. 2012).
Table 2.
Proposed mechanism of Hedgehog pathway activation in various types of sarcomas
| Type of sarcoma | Proposed hedgehog pathway activation (canonical, non-canonical, both) | Explanation |
|---|---|---|
| Osteosarcoma | Both |
Increased Smo expression and susceptibility to Smo inhibition Other cell lines: lack of susceptibility to Smo inhibition but susceptibility to direct Gli inhibition |
| Ewing sarcoma | Non-canonical | Gli1 upregulation by the EWS–FLI1 fusion gene, independent of Smo |
| Chondrosarcoma | Both |
Dysregulation of the Ihh–PTHrP pathway leads to constitutive ligand-dependent Hh signaling Gli1 overexpression due to MVP activity via mTOR/S6K1 signaling |
| Rhabdomyosarcoma (ERMS and fusion gene-negative ARMS) | Canonical |
Mutation in Ptch1, leading to increased Ptch1, Gli1 and Gli3 expression Susceptibility to Smo inhibition |
| Leiomyosarcoma | Canonical |
Increased Smo expression Susceptibility to Smo inhibition |
| Malignant rhabdoid tumor (ATRT-SHH subtype) | Non-canonical | Lower expression of the SNF5 protein leads to Gli1 overexpression |
Another aspect of the Shh pathway’s role in the treatment of sarcomas is its possible influence on susceptibility to immunotherapy. Reduced expression of the Hedgehog signaling pathway and the presence of CD8 + T cells have been shown to correlate with the best clinical outcome in patients with sarcoma undergoing immunotherapy (D’Angelo et al. 2022). Similar observations were also made in other tumors, such as gastric, breast, and basal cell carcinoma. It has been shown that the inhibition of Hh signaling results in decreased PD-L1 expression and tumor cell proliferation in mouse-derived gastric cancer organoids (Chakrabarti et al. 2018). Shh pathway inhibitors used in basal cell carcinoma have led to tumor regression accompanied by beneficial changes in the tumour’s microenvironment, such as upregulation of MHC class I expression, alteration of the local cytokine network, and infiltration of CD8 + T cells (Otsuka et al. 2015). Changes in the tumor microenvironment connected with Hh pathway inhibition have also been investigated in immunocompetent breast cancer murine models. It has been shown that Hh inhibition resulted in the reduction of immune-suppressive cells and an increased number of cytotoxic immune cells (Hanna et al. 2019). These effects could be potentially beneficial for treating sarcomas, which are generally considered “cold” tumors with a low number of infiltrating lymphocytes and rather immunosuppressive microenvironment (Petitprez et al. 2020).
A better understanding of the role played by the Shh pathway in the pathogenesis of sarcomas is necessary. We propose that further research should primarily focus on the role of the non-canonical pathway and the development of its inhibitors as well as potential combination therapies, which would simultaneously target not only the Shh pathway but also other signaling pathways. Such studies could be used to develop an effective therapy for sarcomas connected with Shh pathway dysregulations and thus help solve one of oncology’s most prominent problems (see Table 2).
Author contributions
All authors contributed to the study's conception and design. PS had the primary idea for this review. Material preparation, data collection, and the first draft of the manuscript were written by NB, DK, and KK. PS and PR revised the first draft, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Conflict of interest
Pawel Sobczuk has received travel grants from MSD, BMS, Roche, Novartis, and Pierre Fabre; honoraria for lectures from Swixx BioPharma, BMS, Gilead and Sandoz; honoraria for Advisory Boards from Sandoz; is a stock owner of Celon Pharma; Non-financial interests: European Society of Medical Oncology—Officer; Polish Society of Clinical Oncology—Member of Board of Directors. Katarzyna Kozak has received honoraria for lectures and travel grants from BMS, MSD, Novartis, Pfizer, and Pierre Fabre. Piotr Rutkowski has received honoraria for lectures from BMS, Merck, MSD, Novartis, Pierre Fabre, Sanofi; honoraria for Advisory Boards from Blueprint Medicines, BMS, Merck, MSD, Novartis, Pierre Fabre, Sanofi; Institutional funding support from BMS and Pfizer; Non-financial interests: ASCO—Officer, Polish Society of Surgical Oncology—Member of Board of Directors, Polish Oncological Society—President. Natalia Banaszek and Dominika Kurpiewska declare no conflict of interest.
Ethical approval
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Natalia Banaszek, Dominika Kurpiewska have contributed equally to this work.
References
- Aurias A, Rimbaut C, Buffe D, Zucker JM, Mazabraud A. Translocation involving chromosome 22 in Ewing's sarcoma. A cytogenetic study of four fresh tumors. Cancer Genet Cytogenet. 1984;12:21–25. doi: 10.1016/0165-4608(84)90003-7. [DOI] [PubMed] [Google Scholar]
- Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/s1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- Beauchamp EM, Uren A. A new era for an ancient drug: arsenic trioxide and Hedgehog signaling. Vitam Horm. 2012;88:333–354. doi: 10.1016/b978-0-12-394622-5.00015-8. [DOI] [PubMed] [Google Scholar]
- Beauchamp E, Bulut G, Abaan O, Chen K, Merchant A, Matsui W, Endo Y, Rubin JS, Toretsky J, Uren A. GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein. J Biol Chem. 2009;284:9074–9082. doi: 10.1074/jbc.M806233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC, Peaceman D, Ozdemirli M, Rodriguez O, Macdonald TJ, Albanese C, Toretsky JA, Uren A. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J Clin Invest. 2011;121:148–160. doi: 10.1172/jci42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini G, Geminiani M, Gambassi S, Orlandini M, Petricci E, Marzocchi B, Laschi M, Taddei M, Manetti F, Santucci A. Novel smoothened antagonists as anti-neoplastic agents for the treatment of osteosarcoma. J Cell Physiol. 2018;233:4961–4971. doi: 10.1002/jcp.26330. [DOI] [PubMed] [Google Scholar]
- Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82:690–698. [PubMed] [Google Scholar]
- Boehme KA, Zaborski JJ, Riester R, Schweiss SK, Hopp U, Traub F, Kluba T, Handgretinger R, Schleicher SB. Targeting hedgehog signalling by arsenic trioxide reduces cell growth and induces apoptosis in rhabdomyosarcoma. Int J Oncol. 2016;48:801–812. doi: 10.3892/ijo.2015.3293. [DOI] [PubMed] [Google Scholar]
- Brancaccio G, Pea F, Moscarella E, Argenziano G. Sonidegib for the treatment of advanced basal cell carcinoma. Front Oncol. 2020;10:582866. doi: 10.3389/fonc.2020.582866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechbiel J, Miller-Moslin K, Adjei AA. Crosstalk between hedgehog and other signaling pathways as a basis for combination therapies in cancer. Cancer Treat Rev. 2014;40:750–759. doi: 10.1016/j.ctrv.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, Qualman S, Barr F, Sorensen P, Triche T, Suijkerbuijk R. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes Chromosomes Cancer. 2000;27:337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Campbell VT, Nadesan P, Ali SA, Wang CY, Whetstone H, Poon R, Wei Q, Keilty J, Proctor J, Wang LW, Apte SS, McGovern K, Alman BA, Wunder JS. Hedgehog pathway inhibition in chondrosarcoma using the smoothened inhibitor IPI-926 directly inhibits sarcoma cell growth. Mol Cancer Ther. 2014;13:1259–1269. doi: 10.1158/1535-7163.MCT-13-0731. [DOI] [PubMed] [Google Scholar]
- Carballo GB, Honorato JR, de Lopes GPF, Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter RL, Ray H. Safety and tolerability of sonic hedgehog pathway inhibitors in cancer. Drug Saf. 2019;42:263–279. doi: 10.1007/s40264-018-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco ME, Thaler R, Nardocci G, Dudakovic A, van Wijnen AJ. Inhibition of Ezh2 redistributes bivalent domains within transcriptional regulators associated with WNT and Hedgehog pathways in osteoblasts. J Biol Chem. 2023 doi: 10.1016/j.jbc.2023.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey D, Demko S, Shord S, Zhao H, Chen H, He K, Putman A, Helms W, Keegan P, Pazdur R. FDA approval summary: sonidegib for locally advanced basal cell carcinoma. Clin Cancer Res. 2017;23:2377–2381. doi: 10.1158/1078-0432.Ccr-16-2051. [DOI] [PubMed] [Google Scholar]
- Chakrabarti J, Holokai L, Syu L, Steele NG, Chang J, Wang J, Ahmed S, Dlugosz A, Zavros Y. Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget. 2018;9:37439–37457. doi: 10.18632/oncotarget.26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–4866. doi: 10.1038/onc.2013.433. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhou J, Chen X, Yang B, Wang D, Yang P, He X, Li H. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol. 2015;36:8221–8229. doi: 10.1007/s13277-015-3568-y. [DOI] [PubMed] [Google Scholar]
- Chen D, Kang X, Li Z, Chen L, Ma Q, Fan P. Hedgehog/GLI1 signaling pathway regulates the resistance to cisplatin in human osteosarcoma. J Cancer. 2021;12:6676–6684. doi: 10.7150/jca.61591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Wang W. DNMT3A regulates miR-149 DNA methylation to activate NOTCH1/hedgehog pathway to promote the development of junctional osteosarcoma. Biomed Res Int. 2022;2022:3261213. doi: 10.1155/2022/3261213. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chi S, Xie G, Liu H, Chen K, Zhang X, Li C, Xie J. Rab23 negatively regulates Gli1 transcriptional factor in a Su(Fu)-dependent manner. Cell Signal. 2012;24:1222–1228. doi: 10.1016/j.cellsig.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y, Tang D, Gao J, Jiang X, Xu C, Xiong Q, Huang Y, Wang J, Zhou H, Shi Y, Wang D. Galectin-1 induces invasion and the epithelial-mesenchymal transition in human gastric cancer cells via non-canonical activation of the hedgehog signaling pathway. Oncotarget. 2016;7:83611–83626. doi: 10.18632/oncotarget.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custers L, Khabirova E, Coorens THH, Oliver TRW, Calandrini C, Young MD, Vieira Braga FA, Ellis P, Mamanova L, Segers H, Maat A, Kool M, Hoving EW, van den Heuvel-Eibrink MM, Nicholson J, Straathof K, Hook L, de Krijger RR, Trayers C, Allinson K, Behjati S, Drost J. Somatic mutations and single-cell transcriptomes reveal the root of malignant rhabdoid tumours. Nat Commun. 2021;12:1407. doi: 10.1038/s41467-021-21675-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo SP, Richards AL, Conley AP, Woo HJ, Dickson MA, Gounder M, Kelly C, Keohan ML, Movva S, Thornton K, Rosenbaum E, Chi P, Nacev B, Chan JE, Slotkin EK, Kiesler H, Adamson T, Ling L, Rao P, Patel S, Livingston JA, Singer S, Agaram NP, Antonescu CR, Koff A, Erinjeri JP, Hwang S, Qin L-X, Donoghue MTA, Tap WD. Pilot study of bempegaldesleukin in combination with nivolumab in patients with metastatic sarcoma. Nat Commun. 2022;13:3477. doi: 10.1038/s41467-022-30874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Dong X, Fu J, Yin X, Cao S, Li X, Lin L, Ni J. Emodin: a review of its pharmacology, toxicity and pharmacokinetics. Phytother Res. 2016;30:1207–1218. doi: 10.1002/ptr.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, van Wijnen AJ. Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J Biol Chem. 2015;290:27604–27617. doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Camilleri ET, Riester SM, Paradise CR, Gluscevic M, O'Toole TM, Thaler R, Evans JM, Yan H, Subramaniam M, Hawse JR, Stein GS, Montecino MA, McGee-Lawrence ME, Westendorf JJ, van Wijnen AJ. Enhancer of zeste homolog 2 inhibition stimulates bone formation and mitigates bone loss caused by ovariectomy in skeletally mature mice. J Biol Chem. 2016;291:24594–24606. doi: 10.1074/jbc.M116.740571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakovic A, Samsonraj RM, Paradise CR, Galeano-Garces C, Mol MO, Galeano-Garces D, Zan P, Galvan ML, Hevesi M, Pichurin O, Thaler R, Begun DL, Kloen P, Karperien M, Larson AN, Westendorf JJ, Cool SM, van Wijnen AJ. Inhibition of the epigenetic suppressor EZH2 primes osteogenic differentiation mediated by BMP2. J Biol Chem. 2020;295:7877–7893. doi: 10.1074/jbc.RA119.011685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziuba I, Kurzawa P, Dopierała M, Larque AB, Januszkiewicz-Lewandowska D. Rhabdomyosarcoma in children - current pathologic and molecular classification. Pol J Pathol. 2018;69:20–32. doi: 10.5114/pjp.2018.75333. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Ferrara F, Molica M, Bernardi M. Drug treatment options for acute promyelocytic leukemia. Expert Opin Pharmacother. 2022;23:117–127. doi: 10.1080/14656566.2021.1961744. [DOI] [PubMed] [Google Scholar]
- Fiorenza F, Abudu A, Grimer RJ, Carter SR, Tillman RM, Ayoub K, Mangham DC, Davies AM. Risk factors for survival and local control in chondrosarcoma of bone. J Bone Joint Surg Br. 2002;84:93–99. doi: 10.1302/0301-620x.84b1.11942. [DOI] [PubMed] [Google Scholar]
- Frühwald MC, Biegel JA, Bourdeaut F, Roberts CW, Chi SN. Atypical teratoid/rhabdoid tumors-current concepts, advances in biology, and potential future therapies. Neuro Oncol. 2016;18:764–778. doi: 10.1093/neuonc/nov264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Hall MN. Regulation of mTORC2 signaling. Genes (Basel) 2020 doi: 10.3390/genes11091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Wu H, Cheng SY, Gao D, Zhang L, Zhao Y. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. Elife. 2016 doi: 10.7554/eLife.15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GallegoMelcón S, Sánchez de Toledo Codina J. Molecular biology of rhabdomyosarcoma. Clin Transl Oncol. 2007;9:415–419. doi: 10.1007/s12094-007-0079-3. [DOI] [PubMed] [Google Scholar]
- Galvan ML, Paradise CR, Kubrova E, Jerez S, Khani F, Thaler R, Dudakovic A, van Wijnen AJ. Multiple pharmacological inhibitors targeting the epigenetic suppressor enhancer of zeste homolog 2 (Ezh2) accelerate osteoblast differentiation. Bone. 2021;150:115993. doi: 10.1016/j.bone.2021.115993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Hardwidge PR. Ribosomal protein s3: a multifunctional target of attaching/effacing bacterial pathogens. Front Microbiol. 2011;2:137. doi: 10.3389/fmicb.2011.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia N, Bozzini N, Baiocchi G, da Cunha IW, Maciel GA, Soares Junior JM, Soares FA, Baracat EC, Carvalho KC. May Sonic Hedgehog proteins be markers for malignancy in uterine smooth muscle tumors? Hum Pathol. 2016;50:43–50. doi: 10.1016/j.humpath.2015.08.026. [DOI] [PubMed] [Google Scholar]
- Garcia N, Al-Hendy A, Baracat EC, Carvalho KC, Yang Q. targeting hedgehog pathway and DNA methyltransferases in uterine leiomyosarcoma cells. Cells. 2020 doi: 10.3390/cells10010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia N, Ulin M, Ali M, Al-Hendy A, Carvalho KC, Yang Q. Evaluation of hedgehog pathway inhibitors as a therapeutic option for uterine leiomyosarcoma using the xenograft model. Reprod Sci. 2022;29:781–790. doi: 10.1007/s43032-021-00731-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia N, Ulin M, Al-Hendy A, Yang Q. The role of hedgehog pathway in uterine leiomyosarcoma. J Cell Sci Ther. 2021;12(Suppl 5):314. [PMC free article] [PubMed] [Google Scholar]
- George S, Serrano C, Hensley ML, Ray-Coquard I. Soft tissue and uterine leiomyosarcoma. J Clin Oncol. 2018;36:144–150. doi: 10.1200/JCO.2017.75.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer N, Ridzewski R, Bauer J, Kuzyakova M, Dittmann K, Dullin C, Rosenberger A, Schildhaus HU, Uhmann A, Fulda S, Hahn H. Different response of Ptch mutant and Ptch wildtype rhabdomyosarcoma toward SMO and PI3K inhibitors. Front Oncol. 2018;8:396. doi: 10.3389/fonc.2018.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounder MM, Rosenbaum E, Wu N, Dickson MA, Sheikh TN, D'Angelo SP, Chi P, Keohan ML, Erinjeri JP, Antonescu CR, Agaram N, Hameed MR, Martindale M, Lefkowitz RA, Crago AM, Singer S, Tap WD, Takebe N, Qin L-X, Schwartz GK. A phase Ib/II randomized study of RO4929097, a gamma secretase or notch inhibitor with or without vismodegib, a hedgehog inhibitor, in advanced sarcoma. Clin Cancer Res. 2022 doi: 10.1158/1078-0432.Ccr-21-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronchi A, Miah AB, Dei Tos AP, Abecassis N, Bajpai J, Bauer S, Biagini R, Bielack S, Blay JY, Bolle S, Bonvalot S, Boukovinas I, Bovee JVMG, Boye K, Brennan B, Brodowicz T, Buonadonna A, De Álava E, Del Muro XG, Dufresne A, Eriksson M, Fagioli F, Fedenko A, Ferraresi V, Ferrari A, Frezza AM, Gasperoni S, Gelderblom H, Gouin F, Grignani G, Haas R, Hassan AB, Hecker-Nolting S, Hindi N, Hohenberger P, Joensuu H, Jones RL, Jungels C, Jutte P, Kager L, Kasper B, Kawai A, Kopeckova K, Krákorová DA, Le Cesne A, Le Grange F, Legius E, Leithner A, Lopez-Pousa A, Martin-Broto J, Merimsky O, Messiou C, Mir O, Montemurro M, Morland B, Morosi C, Palmerini E, Pantaleo MA, Piana R, Piperno-Neumann S, Reichardt P, Rutkowski P, Safwat AA, Sangalli C, Sbaraglia M, Scheipl S, Schöffski P, Sleijfer S, Strauss D, Strauss S, Sundby Hall K, Trama A, Unk M, van de Sande MAJ, van der Graaf WTA, van Houdt WJ, Frebourg T, Casali PG, Stacchiotti S. Soft tissue and visceral sarcomas: ESMO–EURACAN–GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up<sup>☆</sup>. Ann Oncol. 2021;32:1348–1365. doi: 10.1016/j.annonc.2021.07.006. [DOI] [PubMed] [Google Scholar]
- Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, Sorensen PH, Delattre O, Dirksen U. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. doi: 10.1038/s41572-018-0003-x. [DOI] [PubMed] [Google Scholar]
- Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, Zhang X, Li N, Bose S, Wang Q, Sakkiah S, Abrol R, Jensen TW, Berman BP, Tanaka H, Johnson J, Gao B, Hao J, Liu Z, Buttyan R, Ray PS, Hung MC, Giuliano AE, Cui X. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-like Breast Cancer. Cell Rep. 2015;13:1046–1058. doi: 10.1016/j.celrep.2015.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna A, Metge BJ, Bailey SK, Chen D, Chandrashekar DS, Varambally S, Samant RS, Shevde LA. Inhibition of Hedgehog signaling reprograms the dysfunctional immune microenvironment in breast cancer. Oncoimmunology. 2019;8:1548241. doi: 10.1080/2162402x.2018.1548241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu M, Setoguchi T, Sasaki H, Matsunoshita Y, Gao H, Nagao H, Kunigou O, Komiya S. Smoothened as a new therapeutic target for human osteosarcoma. Mol Cancer. 2010;9:5. doi: 10.1186/1476-4598-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Johann PD, Grabovska Y, De Dieu Andrianteranagna MJ, Yao F, Frühwald M, Hasselblatt M, Bourdeaut F, Williamson D, Huang A, Kool M. Molecular subgrouping of atypical teratoid/rhabdoid tumors-a reinvestigation and current consensus. Neuro Oncol. 2020;22:613–624. doi: 10.1093/neuonc/noz235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc Natl Acad Sci U S A. 2000;97:12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Italiano A, Le Cesne A, Bellera C, Piperno-Neumann S, Duffaud F, Penel N, Cassier P, Domont J, Takebe N, Kind M, Coindre JM, Blay JY, Bui B. GDC-0449 in patients with advanced chondrosarcomas: a French Sarcoma Group/US and French National Cancer Institute Single-Arm Phase II Collaborative Study. Ann Oncol. 2013;24:2922–2926. doi: 10.1093/annonc/mdt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, Klekota J, Tamayo P, Nguyen PT, Tolstorukov M, Park PJ, Cho YJ, Hsiao K, Buonamici S, Pomeroy SL, Mesirov JP, Ruffner H, Bouwmeester T, Luchansky SJ, Murtie J, Kelleher JF, Warmuth M, Sellers WR, Roberts CW, Dorsch M. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med. 2010;16:1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J, Christensen L, Warner K, States L, Kang HG, Vo K, Lawlor ER, May WA. GLI1 is a central mediator of EWS/FLI1 signaling in Ewing tumors. PLoS ONE. 2009;4:e7608. doi: 10.1371/journal.pone.0007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaleta M, Wakulińska A, Karkucińska-Więckowska A, Dembowska-Bagińska B, Grajkowska W, Pronicki M, Łastowska M. OLIG2 is a novel immunohistochemical marker associated with the presence of PAX3/7-FOXO1 translocation in rhabdomyosarcomas. Diagn Pathol. 2019;14:103. doi: 10.1186/s13000-019-0883-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review) Int J Mol Med. 2008;22:271–275. [PubMed] [Google Scholar]
- Kovar H, Bierbaumer L, Radic-Sarikas B. The YAP/TAZ pathway in osteogenesis and bone sarcoma pathogenesis. Cells. 2020 doi: 10.3390/cells9040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RM, Fuchs B. Hedgehog signaling inhibitors as anti-cancer agents in osteosarcoma. Cancers (basel) 2015;7:784–794. doi: 10.3390/cancers7020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lézot F, Corre I, Morice S, Rédini F, Verrecchia F. SHH signaling pathway drives pediatric bone sarcoma progression. Cells. 2020 doi: 10.3390/cells9030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Cai J, Liu C, Shen L, Pu X, Yao Y, Han B, Yu T, Cheng SY, Yue S. Protein phosphatase 4 promotes Hedgehog signaling through dephosphorylation of Suppressor of fused. Cell Death Dis. 2020;11:686. doi: 10.1038/s41419-020-02843-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Huang W, Wang J, Liu X, Yang J, Zhang Y, Geng Y, Tan W, Zhang A. Discovery of novel macrocyclic hedgehog pathway inhibitors acting by suppressing the Gli-mediated transcription. J Med Chem. 2017;60:8218–8245. doi: 10.1021/acs.jmedchem.7b01185. [DOI] [PubMed] [Google Scholar]
- Lo WW, Pinnaduwage D, Gokgoz N, Wunder JS, Andrulis IL. Aberrant hedgehog signaling and clinical outcome in osteosarcoma. Sarcoma. 2014 doi: 10.1155/2014/261804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WW, Wunder JS, Dickson BC, Campbell V, McGovern K, Alman BA, Andrulis IL. Involvement and targeted intervention of dysregulated Hedgehog signaling in osteosarcoma. Cancer. 2014;120:537–547. doi: 10.1002/cncr.28439. [DOI] [PubMed] [Google Scholar]
- LospinosoSeverini L, Quaglio D, Basili I, Ghirga F, Bufalieri F, Caimano M, Balducci S, Moretti M, Romeo I, Loricchio E, Maroder M, Botta B, Mori M, Infante P, Di Marcotullio L. A Smo/Gli multitarget hedgehog pathway inhibitor impairs tumor growth. Cancers (Basel) 2019 doi: 10.3390/cancers11101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Mondal S, Satyavarapu EM, Mandal C. mTORC2 regulates hedgehog pathway activity by promoting stability to Gli2 protein and its nuclear translocation. Cell Death Dis. 2017;8:e2926. doi: 10.1038/cddis.2017.296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- Mastoraki A, Schizas D, Vlachou P, Melissaridou NM, Charalampakis N, Fioretzaki R, Kole C, Savvidou O, Vassiliu P, Pikoulis E. Assessment of synergistic contribution of histone deacetylases in prognosis and therapeutic management of sarcoma. Mol Diagn Ther. 2020;24:557–569. doi: 10.1007/s40291-020-00487-2. [DOI] [PubMed] [Google Scholar]
- Meiss F, Andrlová H, Zeiser R. Vismodegib. Recent Results Cancer Res. 2018;211:125–139. doi: 10.1007/978-3-319-91442-8_9. [DOI] [PubMed] [Google Scholar]
- Montecino M, Carrasco ME, Nardocci G. Epigenetic control of osteogenic lineage commitment. Front Cell Dev Biol. 2020;8:611197. doi: 10.3389/fcell.2020.611197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff R, Rath P, Ram Kumar RM, Husmann K, Born W, Baudis M, Fuchs B. Genomic instability of osteosarcoma cell lines in culture: impact on the prediction of metastasis relevant genes. PLoS ONE. 2015;10:e0125611. doi: 10.1371/journal.pone.0125611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard M, Cadé M, Morice S, Dupuy M, Danieau G, Amiaud J, Renault S, Lézot F, Brion R, Thepault RA, Ory B, Lamoureux F, Corre I, Brounais-LeRoyer B, Rédini F, Verrecchia F. Sonic hedgehog signature in pediatric primary bone tumors: effects of the GLI Antagonist GANT61 on Ewing's sarcoma tumor growth. Cancers (Basel) 2020 doi: 10.3390/cancers12113438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao H, Ijiri K, Hirotsu M, Ishidou Y, Yamamoto T, Nagano S, Takizawa T, Nakashima K, Komiya S, Setoguchi T. Role of GLI2 in the growth of human osteosarcoma. J Pathol. 2011;224:169–179. doi: 10.1002/path.2880. [DOI] [PubMed] [Google Scholar]
- Nagao-Kitamoto H, Setoguchi T, Kitamoto S, Nakamura S, Tsuru A, Nagata M, Nagano S, Ishidou Y, Yokouchi M, Kitajima S, Yoshioka T, Maeda S, Yonezawa S, Komiya S. Ribosomal protein S3 regulates GLI2-mediated osteosarcoma invasion. Cancer Lett. 2015;356:855–861. doi: 10.1016/j.canlet.2014.10.042. [DOI] [PubMed] [Google Scholar]
- Nagao-Kitamoto H, Nagata M, Nagano S, Kitamoto S, Ishidou Y, Yamamoto T, Nakamura S, Tsuru A, Abematsu M, Fujimoto Y, Yokouchi M, Kitajima S, Yoshioka T, Maeda S, Yonezawa S, Komiya S, Setoguchi T. GLI2 is a novel therapeutic target for metastasis of osteosarcoma. Int J Cancer. 2015;136:1276–1284. doi: 10.1002/ijc.29107. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Nagano S, Nagao H, Ishidou Y, Yokouchi M, Abematsu M, Yamamoto T, Komiya S, Setoguchi T. Arsenic trioxide prevents osteosarcoma growth by inhibition of GLI transcription via DNA damage accumulation. PLoS ONE. 2013;8:e69466. doi: 10.1371/journal.pone.0069466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NM, Cho J. hedgehog pathway inhibitors as targeted cancer therapy and strategies to overcome drug resistance. Int J Mol Sci. 2022 doi: 10.3390/ijms23031733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A, Dreier J, Cheng PF, Nägeli M, Lehmann H, Felderer L, Frew IJ, Matsushita S, Levesque MP, Dummer R. Hedgehog pathway inhibitors promote adaptive immune responses in basal cell carcinoma. Clin Cancer Res. 2015;21:1289–1297. doi: 10.1158/1078-0432.Ccr-14-2110. [DOI] [PubMed] [Google Scholar]
- Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype signaling and therapeutic targets. Cells. 2020 doi: 10.3390/cells9102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski KM, Majewska A, Szyszko K, Dolka I, Motyl T, Krol M. Gene expression pattern in canine mammary osteosarcoma. Pol J Vet Sci. 2011;14:11–20. doi: 10.2478/v10181-011-0002-2. [DOI] [PubMed] [Google Scholar]
- Pelullo M, Zema S, Nardozza F, Checquolo S, Screpanti I, Bellavia D. Wnt, notch, and TGF-β pathways impinge on hedgehog signaling complexity: an open window on cancer. Front Genet. 2019;10:711. doi: 10.3389/fgene.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitprez F, de Reyniès A, Keung EZ, Chen TW, Sun CM, Calderaro J, Jeng YM, Hsiao LP, Lacroix L, Bougoüin A, Moreira M, Lacroix G, Natario I, Adam J, Lucchesi C, Laizet YH, Toulmonde M, Burgess MA, Bolejack V, Reinke D, Wani KM, Wang WL, Lazar AJ, Roland CL, Wargo JA, Italiano A, Sautès-Fridman C, Tawbi HA, Fridman WH. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- Pietrobono S, Gagliardi S, Stecca B. Non-canonical hedgehog signaling pathway in cancer: activation of GLI transcription factors beyond smoothened. Front Genet. 2019;10:556. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressey JG, Anderson JR, Crossman DK, Lynch JC, Barr FG. Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer. 2011;57:930–938. doi: 10.1002/pbc.23174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Zhou Y, Jiao Z, Wang X, Zhao Y, Li Y, Chen H, Yang L, Zhu H, Li Y. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth through hedgehog signaling pathway. Cell Physiol Biochem. 2017;42:2242–2254. doi: 10.1159/000479998. [DOI] [PubMed] [Google Scholar]
- Qu W, Wang Y, Wu Q, Hao D, Li D. Emodin impairs radioresistance of human osteosarcoma cells by suppressing sonic hedgehog signaling. Med Sci Monit. 2017;23:5767–5773. doi: 10.12659/MSM.907453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta R, Pelullo M, Zema S, Nardozza F, Checquolo S, Lauer DM, Bufalieri F, Palermo R, Felli MP, Vacca A, Talora C, Di Marcotullio L, Screpanti I, Bellavia D. Maml1 acts cooperatively with Gli proteins to regulate sonic hedgehog signaling pathway. Cell Death Dis. 2017;8:e2942. doi: 10.1038/cddis.2017.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma J, Almazán-Moga A, Sánchez de Toledo J, Gallego S. Notch, wnt, and hedgehog pathways in rhabdomyosarcoma: from single pathways to an integrated network. Sarcoma. 2012 doi: 10.1155/2012/695603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Thérond PP. Variations in Hedgehog signaling: divergence and perpetuation in Sufu regulation of Gli. Genes Dev. 2009;23:1843–1848. doi: 10.1101/gad.1838109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y, Setoguchi T, Nagata M, Tsuru A, Nakamura S, Nagano S, Ishidou Y, Nagao-Kitamoto H, Yokouchi M, Maeda S, Tanimoto A, Furukawa T, Komiya S. Combination of Hedgehog inhibitors and standard anticancer agents synergistically prevent osteosarcoma growth. Int J Oncol. 2016;48:235–242. doi: 10.3892/ijo.2015.3236. [DOI] [PubMed] [Google Scholar]
- Semwal RB, Semwal DK, Combrinck S, Viljoen A. Emodin - A natural anthraquinone derivative with diverse pharmacological activities. Phytochemistry. 2021;190:112854. doi: 10.1016/j.phytochem.2021.112854. [DOI] [PubMed] [Google Scholar]
- Shahi MH, Holt R, Rebhun RB. Blocking signaling at the level of GLI regulates downstream gene expression and inhibits proliferation of canine osteosarcoma cells. PLoS ONE. 2014;9:e96593. doi: 10.1371/journal.pone.0096593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Xu L, Chen L, Lu Q, Xie X, Shi W, Xiong H, Shi C, Huang X, Mei J, Rao H, Lu H, Lu N, Luo S. Arl13b promotes gastric tumorigenesis by regulating smo trafficking and activation of the hedgehog signaling pathway. Cancer Res. 2017;77:4000–4013. doi: 10.1158/0008-5472.Can-16-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Deng YZ, Zhao JS, Ji XD, Shi J, Feng YX, Li G, Li JJ, Zhu D, Koeffler HP, Zhao Y, Xie D. RACK1 promotes non-small-cell lung cancer tumorigenicity through activating sonic hedgehog signaling pathway. J Biol Chem. 2012;287:7845–7858. doi: 10.1074/jbc.M111.315416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang XX, Zhuang YW, Jiang Y, Melcher K, Xu HE. Structure of the PRC2 complex and application to drug discovery. Acta Pharmacol Sin. 2017;38:963–976. doi: 10.1038/aps.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigafoos AN, Paradise BD, Fernandez-Zapico ME. Hedgehog/GLI signaling pathway: transduction, regulation, and implications for disease. Cancers (Basel) 2021 doi: 10.3390/cancers13143410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skapek SX, Ferrari A, Gupta AA, Lupo PJ, Butler E, Shipley J, Barr FG, Hawkins DS. Rhabdomyosarcoma. Nat Rev Dis Primers. 2019;5:1. doi: 10.1038/s41572-018-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: a comprehensive review. Bosn J Basic Med Sci. 2018;18:8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang J, Wang J, Wang J, Guo X, Dong W. β1 integrin mediates colorectal cancer cell proliferation and migration through regulation of the Hedgehog pathway. Tumour Biol. 2015;36:2013–2021. doi: 10.1007/s13277-014-2808-x. [DOI] [PubMed] [Google Scholar]
- Song L, Chen X, Gao S, Zhang C, Qu C, Wang P, Liu L. Ski modulate the characteristics of pancreatic cancer stem cells via regulating sonic hedgehog signaling pathway. Tumour Biol. 2016 doi: 10.1007/s13277-016-5461-8. [DOI] [PubMed] [Google Scholar]
- Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing's sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor. ERG, Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- Su PH, Huang RL, Lai HC, Chen LY, Weng YC, Wang CC, Wu CC. NKX6-1 mediates cancer stem-like properties and regulates sonic hedgehog signaling in leiomyosarcoma. J Biomed Sci. 2021;28:32. doi: 10.1186/s12929-021-00726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang QL, Xie XB, Wang J, Chen Q, Han AJ, Zou CY, Yin JQ, Liu DW, Liang Y, Zhao ZQ, Yong BC, Zhang RH, Feng QS, Deng WG, Zhu XF, Zhou BP, Zeng YX, Shen JN, Kang T. Glycogen synthase kinase-3β, NF-κB signaling, and tumorigenesis of human osteosarcoma. J Natl Cancer Inst. 2012;104:749–763. doi: 10.1093/jnci/djs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiet TD, Hopyan S, Nadesan P, Gokgoz N, Poon R, Lin AC, Yan T, Andrulis IL, Alman BA, Wunder JS. Constitutive hedgehog signaling in chondrosarcoma up-regulates tumor cell proliferation. Am J Pathol. 2006;168:321–330. doi: 10.2353/ajpath.2006.050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, Yu LC, Behnke ML, Nair SJ, Hagel M, White K, Conley J, Manna JD, Alvarez-Diez TM, Hoyt J, Woodward CN, Sydor JR, Pink M, MacDougall J, Campbell MJ, Cushing J, Ferguson J, Curtis MS, McGovern K, Read MA, Palombella VJ, Adams J, Castro AC. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926) J Med Chem. 2009;52:4400–4418. doi: 10.1021/jm900305z. [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Errani C, Angelini A, Mavrogenis AF. Current treatment considerations for osteosarcoma metastatic at presentation. Orthopedics. 2020;43:e345–e358. doi: 10.3928/01477447-20200721-05. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mehren M, Kane JM, Bui MM, Choy E, Connelly M, Dry S, Ganjoo KN, George S, Gonzalez RJ, Heslin MJ, Homsi J, Keedy V, Kelly CM, Kim E, Liebner D, McCarter M, McGarry SV, Meyer C, Pappo AS, Parkes AM, Paz IB, Petersen IA, Poppe M, Riedel RF, Rubin B, Schuetze S, Shabason J, Sicklick JK, Spraker MB, Zimel M, Bergman MA, George GV. NCCN guidelines insights: soft tissue sarcoma, Version 1.2021: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2020;18:1604–1612. doi: 10.6004/jnccn.2020.0058. [DOI] [PubMed] [Google Scholar]
- W.C.o.T.E. Board (2020) WHO classification of soft tissue and bone tumours, 5th edn
- Wagner A, Hohenberger P, Okuno S, Eriksson M, Patel S, Ferrari S, Casali P, Chawla S, Woehr M, Ross R (2013) In: New York, New York, USA: Presented at the Connective Tissue Oncology Society Annual Meeting, Results from a phase 2 randomized, placebo-controlled, double blind study of the hedgehog pathway antagonist IPI-926 in patients with advanced chondrosarcoma
- Wang W, Yan T, Guo W, Niu J, Zhao Z, Sun K, Zhang H, Yu Y, Ren T. Constitutive GLI1 expression in chondrosarcoma is regulated by major vault protein via mTOR/S6K1 signaling cascade. Cell Death Differ. 2021;28:2221–2237. doi: 10.1038/s41418-021-00749-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha J, Dinges D, Kaszap B, Henrich D, Marzi I, Seebach C. Effect of the Hedgehog-inhibitor cyclopamine on mice with osteosarcoma pulmonary metastases. Int J Mol Med. 2012;29:423–427. doi: 10.3892/ijmm.2011.851. [DOI] [PubMed] [Google Scholar]
- Wu X, Xu B, Yang C, Wang W, Zhong D, Zhao Z, He L, Hu Y, Jiang L, Li J, Song L, Zhang W. Nucleolar and spindle associated protein 1 promotes the aggressiveness of astrocytoma by activating the Hedgehog signaling pathway. J Exp Clin Cancer Res. 2017;36:127. doi: 10.1186/s13046-017-0597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Liu X, Choy E, Mankin H, Hornicek FJ, Duan Z. Targeting hedgehog-GLI-2 pathway in osteosarcoma. J Orthop Res. 2013;31:502–509. doi: 10.1002/jor.22230. [DOI] [PubMed] [Google Scholar]
- Yang J, Andre P, Ye L, Yang YZ. The Hedgehog signalling pathway in bone formation. Int J Oral Sci. 2015;7:73–79. doi: 10.1038/ijos.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Han L, Chen Y, He F, Sun B, Kamar S, Zhang Y, Yang Y, Wang C, Yang Z. Hedgehog signalling in the tumourigenesis and metastasis of osteosarcoma, and its potential value in the clinical therapy of osteosarcoma. Cell Death Dis. 2018;9:701. doi: 10.1038/s41419-018-0647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechieli RL, Mandeville HC, Hiniker SM, Bernier-Chastagner V, McGovern S, Scarzello G, Wolden S, Cameron A, Breneman J, Fajardo RD, Donaldson SS. Rhabdomyosarcoma. Pediatr Blood Cancer. 2021;68(Suppl 2):e28254. doi: 10.1002/pbc.28254. [DOI] [PubMed] [Google Scholar]
- Yin WC, Satkunendran T, Mo R, Morrissy S, Zhang X, Huang ES, Uusküla-Reimand L, Hou H, Son JE, Liu W, Liu YC, Zhang J, Parker J, Wang X, Farooq H, Selvadurai H, Chen X, Ngan ES, Cheng SY, Dirks PB, Angers S, Wilson MD, Taylor MD, Hui CC. Dual regulatory functions of SUFU and targetome of GLI2 in SHH subgroup medulloblastoma. Dev Cell. 2019;48:167–183.e165. doi: 10.1016/j.devcel.2018.11.015. [DOI] [PubMed] [Google Scholar]
- Zhang S, Guo W, Ren TT, Lu XC, Tang GQ, Zhao FL. Arsenic trioxide inhibits Ewing's sarcoma cell invasiveness by targeting p38(MAPK) and c-Jun N-terminal kinase. Anticancer Drugs. 2012;23:108–118. doi: 10.1097/CAD.0b013e32834bfd68. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shen L, Law K, Zhang Z, Liu X, Hua H, Li S, Huang H, Yue S, Hui CC, Cheng SY. Suppressor of fused chaperones Gli proteins to generate transcriptional responses to sonic hedgehog signaling. Mol Cell Biol. 2017 doi: 10.1128/mcb.00421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang CL, Zeng BF, Wu XS, Gao TT, Oda Y. Enhanced chemosensitivity of drug-resistant osteosarcoma cells by lentivirus-mediated Bcl-2 silencing. Biochem Biophys Res Commun. 2009;390:642–647. doi: 10.1016/j.bbrc.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Jia Q, Wu MS, Xie X, Wang Y, Song G, Zou CY, Tang Q, Lu J, Huang G, Wang J, Lin DC, Koeffler HP, Yin JQ, Shen J. Degalactotigonin, a natural compound from Solanum nigrum L., inhibits growth and metastasis of osteosarcoma through GSK3beta inactivation-mediated repression of the hedgehog/Gli1 pathway. Clin Cancer Res. 2018;24:130–144. doi: 10.1158/1078-0432.CCR-17-0692. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zhang Z, Liu C, Jin C, Zhang J, Miao X, Jia L. B4GALT1 gene knockdown inhibits the hedgehog pathway and reverses multidrug resistance in the human leukemia K562/adriamycin-resistant cell line. IUBMB Life. 2012;64:889–900. doi: 10.1002/iub.1080. [DOI] [PubMed] [Google Scholar]
- Zhou F, Huang D, Li Y, Hu G, Rao H, Lu Q, Luo S, Wang Y. Nek2A/SuFu feedback loop regulates Gli-mediated Hedgehog signaling pathway. Int J Oncol. 2017;50:373–380. doi: 10.3892/ijo.2016.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibat A, Missiaglia E, Rosenberger A, Pritchard-Jones K, Shipley J, Hahn H, Fulda S. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29:6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
- Zwerner JP, Joo J, Warner KL, Christensen L, Hu-Lieskovan S, Triche TJ, May WA. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27:3282–3291. doi: 10.1038/sj.onc.1210991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.