Abstract
Two yeast isolates, a wine-making yeast first identified as a Mel+ strain (ex. S. uvarum) and a cider-making yeast, were characterized for their nuclear and mitochondrial genomes. Electrophoretic karyotyping analyses, restriction fragment length polymorphism maps of PCR-amplified MET2 gene fragments, and the sequence analysis of a part of the two MET2 gene alleles found support the notion that these two strains constitute hybrids between Saccharomyces cerevisiae and Saccharomyces bayanus. The two hybrid strains had completely different restriction patterns of mitochondrial DNA as well as different sequences of the OLI1 gene. The sequence of the OLI1 gene from the wine hybrid strain appeared to be the same as that of the S. cerevisiae gene, whereas the OLI1 gene of the cider hybrid strain is equally divergent from both putative parents, S. bayanus and S. cerevisiae. Some fermentative properties were also examined, and one phenotype was found to reflect the hybrid nature of these two strains. The origin and nature of such hybridization events are discussed.
The genus Saccharomyces can be divided into two major groups: sensu stricto and sensu lato (2). The sensu stricto yeasts, which include S. bayanus, S. cerevisiae, S. paradoxus, and S. pastorianus (syn. S. carlsbergensis), represent a closely related biological complex (14). S. cerevisiae is the major species found among wine yeasts, while S. bayanus represents a small part of them. The sensu stricto yeasts contain at least 16 distinctive nuclear chromosomes of small, medium, and large sizes, and each species appears to contain a unique karyotype (29). Their mitochondrial DNA (mtDNA) molecules range in size from 64 to 85 kb and contain a number of G+C clusters, among them three to nine ori-rep sequences (27). Molecular polymorphism is widespread among the sensu stricto yeasts, especially among yeast strains associated with the wine industry (5, 30), and almost every isolate has a characteristic karyotype and restriction pattern of digested mtDNA (27). However, among isolates belonging to the same species, similar karyotypes and restriction patterns are observed. In the laboratory, members of the sensu stricto group can be mated at low frequency and can generate viable offspring (19).
The lager brewing strain S. pastorianus (syn. S. carlsbergensis) is a partial amphitetraploid, which was generated upon an interspecific fusion-cross between two different yeasts (see, e.g., reference 11). One of the parental strains in this fusion-cross was S. cerevisiae and the second was a member of the S. bayanus species complex, possibly S. monacensis (8, 24, 27). In the characterized strains of S. pastorianus (syn. S. carlsbergensis), both sets of parental chromosomes are present (11), but the mtDNA molecule was inherited only from the non-S. cerevisiae parent (27). Initially, the hybrid zygote was possibly heteroplasmic regarding the mitochondrial genome, but apparently only one parental type was transmitted to the progeny.
In this report, two yeast isolates, a wine-making yeast first identified as Mel+ (ex. S. uvarum) and a cider-making yeast, are characterized for their nuclear and mitochondrial genome and are shown to be hybrids. In addition, some fermentation properties such as production of aroma compounds of these two yeasts are studied.
MATERIALS AND METHODS
Yeast strains and media.
The yeast strains used in this study are listed in Table 1. S. cerevisiae VKM Y-502 and S. bayanus VKM Y-1146 are monosporic cultures of reference strains (20). Except for strain S288C, which is a standard laboratory strain, all other strains of S. cerevisiae are industrial wine-making yeasts. S. bayanus CBS 380 and S. paradoxus CBS 432 are the type strains, and S. carlsbergensis Y385 used for the mtDNA restriction fragment length polymorphism (RFLP) experiment is a beer production strain. Other strains of S. bayanus are wine yeasts from the collection of the Faculté d’Oenologie de Bordeaux. CID1 is a cider yeast which was isolated from a mixed culture from the bottom of a home-fabricated apple cider from Brittany, France. S6U is a wine-producing yeast. Yeast were grown on YPG medium (20 g of l-glucose/liter, 10 g of BactoPeptone/liter, 10 g of yeast extract/liter) at 20, 25, or 30°C, depending on the species. Fermentations were carried out in grape juice of Vitis vinifera var. Sauvignon. The must was sterilized by filtration (turbidity, <5 NTU).
TABLE 1.
Yeast strains used in this study
| Strain | Origin or sourcea | Species |
|---|---|---|
| VKM Y-502 | VKM | S. cerevisiae |
| CBS 1171 | CBS | S. cerevisiae (type strain) |
| CBS 380 | CBS | S. bayanus (type strain) |
| VKM Y-1146 | VKM | S. bayanus |
| EG8 | INRA Colmar | S. cerevisiae |
| VL3c | Faculté d’Oenologie de Bordeaux | S. cerevisiae |
| VL1 | Faculté d’Oenologie de Bordeaux | S. cerevisiae |
| SIHA3 | Darmstadt | S. cerevisiae |
| CID1 | Collection of J. Hansen | Species hybrid |
| S6U | Lallemand Inc. | Species hybrid |
| P3 | Faculté d’Oenologie de Bordeaux | S. bayanus |
| TB28 | Faculté d’Oenologie de Bordeaux | S. bayanus |
| CBS 432 | CBS | S. paradoxus (type strain) |
| Y385 | Collection of J. Piskur | S. carlsbergensis |
CBS, Centraal Bureau voor Schimmelcultures, Baarn, The Netherlands; VKM, National Collection of Microorganisms, Moscow, Russia; CLIB, Collection de Levures d’Intérêt Biotechnologique, Paris, France.
Contour-clamped homogeneous electric field gel electrophoresis.
Chromosomal DNA was prepared in agarose plugs (3) and separated on a 0.8% agarose gel (Agarose NA; Pharmacia) at 165 V and 10°C by using the following program (6): switch, 12.5 h, 40 to 90 s; switch, 16.5 h, 80 to 120 s.
MET2 PCR-RFLP.
The PCR amplification reaction was carried out on entire yeast cells after cultivation on solid YPG medium until the stationary phase (17). Amplification reactions were performed with a Perkin-Elmer DNA thermal Thermocycler 480, using synthetic oligonucleotide primers for MET2 amplification as described by Hansen and Kielland-Brandt (8). PCR products were precipitated, and aliquots were digested with EcoRI or PstI. The resulting DNA fragments were analyzed by electrophoresis on a 1.8% agarose gel (Agarose NA). A Boehringer Mannheim DNA molecular weight marker VIII was used.
Preparation and sequencing of MET2 gene fragments.
For preparative purposes, MET2 fragments from strains S6U and CID1 were amplified by PCR by using the primers 5′-CGGCTCTAGACGAAAACGCTCCAAGAGCTGG-3′ and 5′-CGGCTCTAGAGACCACGATATGCACCAGGCAG-3′, containing at their ends XbaI restriction sites and four arbitrary bases to allow for restriction endonuclease digestion. Genomic DNA was prepared from 10-ml liquid yeast cultures by the protocol of Hoffman and Winston (9). Ten microliters of a 100× dilution of each DNA preparation was used as template. The PCRs were performed on a Stratagene Robocycler 40 for 25 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C, followed by one cycle of 72°C for 10 min. Eight independent reactions for each DNA template were performed. Each series of identical reactions was pooled, and the amplified DNA was precipitated, washed, and redissolved in an appropriate volume of water and used for direct sequencing or cloning. Restriction digestions and ligation reactions were performed in accordance with the recommendations of the manufacturers. DNA fragments were isolated from agarose by using Bio-Rad Prep-A-Gene purification matrix. The sequencing reactions were performed on a Perkin-Elmer DNA Thermocycler 480, and sequences were run on an Applied Biosystems Sequenator 373A or 310 in accordance with the recommendations of the manufacturers. Sequencing primers for direct sequencing were identical to the ones used for the PCR amplification except that no restriction sites or additional arbitrary bases were included. Sequencing of the cloned fragments was performed employing the same primers or standard m13 primers. Both strands of the DNA were sequenced in all cases.
Cloning of MET2 DNA fragments.
PCR-amplified MET2 fragments from both strains were cloned into pUC19 as follows. Precipitated, redissolved DNA was restriction endonuclease digested with either PstI and XbaI or with EcoRI and XbaI. Uncut DNA fragments were purified as described above and ligated into pUC19 vector that was opened with XbaI and treated with calf intestine alkaline phosphatase. PCR-amplified DNA from several independent reactions were employed in these experiments. The resulting plasmids used for sequencing were pJH147 (strain S6U; EcoRI uncut), pJH150 (strain CID1; EcoRI uncut), pJH153 (strain S6U; PstI uncut) and pJH156 and pJH157 (strain CID1; PstI uncut).
Isolation and sequencing of mtDNA.
mtDNA was isolated from various yeasts by using a modification of the bisbenzimide-CsCl gradient method (25, 27). RFLP was studied on purified mtDNA by using GC-clutters as enzymes, i.e., HaeIII and MspI. The sequence of the mitochondrial OLI1 gene was obtained by direct sequencing on purified mtDNA (7). The following two primers, with homology to the 5′ and 3′ ends of the OLI1 coding region, were used in direct sequencing: OLI1 YM-1 (forward primer), 5′-GCAATTAGTATTAGCAGCTAAATATATTGG-3′; and OLI1 YM-4 (reverse primer), 5′-AATAAGAATGAAACCATTAAACAGA-3′. The open reading frame of OLI1 is 228 bp long, and the sequence was determined on both strands except for the terminal 25 bases in the 5′ and 3′ ends, which were determined on only one strand.
Fermentation experiments.
The yeast inocula were obtained from overnight cultures grown on diluted must. The quantity of yeasts was measured by determining the optical density at 600 nm in order to inoculate the must at a level of 3 × 106 to 4 × 106 cells/ml. The fermentation test was carried out in 375-ml bottles containing grape juice. The turbidity of the juice was adjusted to 200 NTU with insoluble material of the must to improve the fermentation velocity (22). At the midpoint of the fermentation, control experiments were performed to ensure that the must had been inoculated with the correct yeast species and strains. The implantation of S. cerevisiae strains was verified by PCR amplification of delta sequences (17). For the S. bayanus strains, karyotyping by contour-clamped homogeneous electric field gel electrophoresis was used to confirm the inoculation. When the sugars were below 2 g/liter, bottles were placed at 10°C and SO2 was adjusted to 60 mg/liter and the wines were rapidly analyzed for content of higher alcohols and esters by gas chromatography coupled with a flame ionization detector (CARBOWAX 20M capillary column, type BP20; length, 50 m; internal diameter, 0.25 mm; film thickness, 0.50 μm; VARIAN 3400 gas chromatograph; Merck D-2500 chromatointegrator).
RESULTS
Electrophoretic karyotyping analyses.
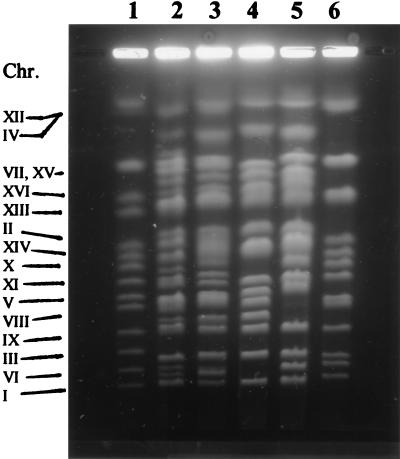
Wine S. cerevisiae strains are characterized by important variations in chromosomal length whereas wine yeasts, genetically identified as S. bayanus, do not exhibit large chromosomal polymorphism (20, 30). However, many authors in previous works have shown that electrophoretic karyotyping analyses can be used to differentiate S. cerevisiae and S. bayanus (4, 13, 20, 24). Chromosomal DNA patterns of strains S6U and CID1 displayed similar but specific band patterns of more than 20 bands. When these patterns were compared to the reference strains of S. cerevisiae and S. bayanus, a high proportion of bands corresponded to one or the other reference strain (Fig. 1). Actually, the karyotypes of S6U and CID1 contained an almost complete set of S. cerevisiae and S. bayanus chromosomes, indicating the hybrid nature of these two yeasts. According to our previous studies of various yeast isolates which were either S. cerevisiae- or S. bayanus-like, the existence of hybrids in nature is quite rare (16, 18). Karyotypes of different isolates belonging to the same species exhibit polymorphism, like the isolates belonging to the presented S. bayanus and S. cerevisiae strains. On the other hand, the two hybrid isolates displayed a similar karyotype. Thus, it is likely that the hybrids have a similar origin for the nuclear genome.
FIG. 1.
Electrophoretic karyotyping of standard S. cerevisiae YNN 295 (lane 1), species hybrids (lane 2, CID1; lane 3, S6U), two S. bayanus strains (lane 4, VKM Y-1146; lane 5, CBS 380) and a S. cerevisiae strain (lane 6, VKM Y-502).
PCR-RFLP on the MET2 gene.
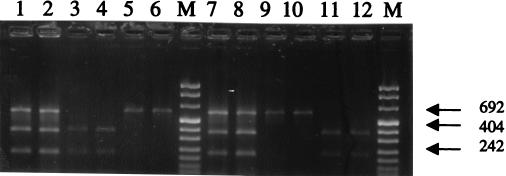
To substantiate the hypothesis that S6U and CID1 are hybrid yeasts containing genomic material related to that of S. cerevisiae and S. bayanus, we decided to employ RFLP on a 580-bp PCR-amplified fragment of a nuclear gene, MET2, as described previously (8, 18). The restriction endonuclease PstI cuts this MET2 sequence of S. bayanus, and there is no PstI site in the MET2 sequence of S. cerevisiae. On the contrary, EcoRI cuts the S. cerevisiae MET2 sequence but not the S. bayanus sequence (8, 18). The results obtained are shown in Fig. 2. S. cerevisiae VKM Y-502 and S. bayanus VKM Y-1146 and CBS 380 were used as reference strains. For the enzymes EcoRI and PstI, the restriction fragment profiles are characteristic of the two species S. cerevisiae and S. bayanus; EcoRI cleaves the S. cerevisiae MET2 fragment (two bands of 211 bp and 369 bp) but does not cleave the S. bayanus MET2 fragment. The behavior of PstI is different: for S6U and CID1, an EcoRI and a PstI restriction fragment pattern of three bands was obtained, with the same intensity and the same length as those obtained for S. cerevisiae with EcoRI and for S. bayanus with PstI (Fig. 2). These restriction fragment profiles appeared to consist of a mix of the profiles seen from S. cerevisiae and S. bayanus with a given enzyme and were identified for different subclones of S6U and CID1 from vegetative cells, which were isolated with a micromanipulator.
FIG. 2.
RFLP analysis of PCR-amplified MET2 gene fragment. Lanes 1 to 6, restriction analysis with EcoRI. Lanes 7 to 12, restriction analysis with PstI. Lanes 1 and 6, S6U; lanes 2 and 8, CID1; lanes 3 and 9, S. cerevisiae VKM Y-502; lanes 4 and 10, S. cerevisiae type strain 1171; lanes 5 and 11, S. bayanus VKM Y-1146; lanes 6 and 12, S. bayanus CBS 380. M, molecular weight marker (marker VIII from Boehringer Mannheim). Numbers and arrows are base pair markers.
Divergent MET2 sequences of the putative hybrid yeasts.
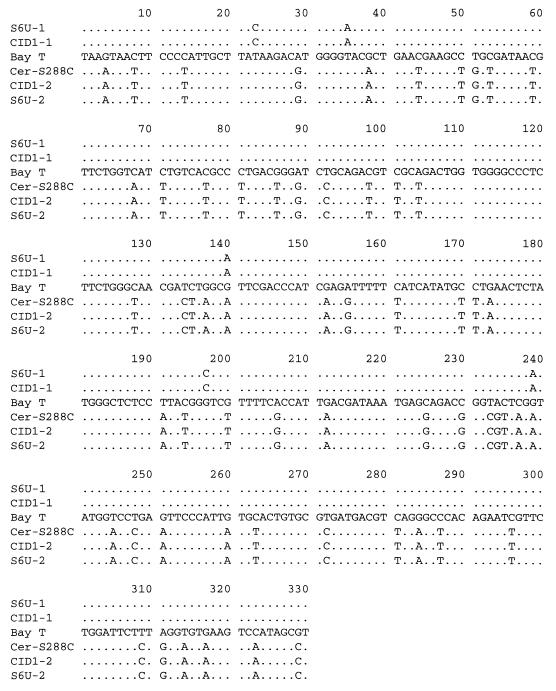
To obtain the nucleotide sequence of the central 330 bp of the amplified MET2 DNA fragments, we employed two techniques: direct sequencing of the PCR products, as described by Hansen and Kielland-Brandt (8) and sequencing of cloned PCR fragments. In the case of direct sequencing, the PCR fragments were thoroughly digested with either PstI or EcoRI. The DNA fragments in the restriction digests were separated on 2% agarose, and remaining uncut fragments were purified. In this manner, we were able to obtain unambiguous sequences from both fragments from strain S6U present after restriction digestion. Likewise, we obtained good sequences from the EcoRI-uncut MET2 fragment from strain CID1 and reasonably good sequences from the PstI-uncut MET2 fragment from the same strain. However, to resolve the identity of a few ambiguous nucleotides in the PstI-uncut MET2 from CID1, and in general to confirm the sequencing results, we decided to redo the sequencing on cloned MET2 PCR fragments. The cloning is described in the Materials and Methods section. The inserts of the plasmids pJH147, pJH150, pJH153, pJH156, and pJH157 were sequenced. The results from the direct sequencing of the S6U MET2 fragments were confirmed, and the sequences obtained from plasmids pJH156 and pJH157 were identical. As can be seen in Fig. 3, the 330 bp of the PstI-uncut MET2 alleles of both strains CID1 and S6U were completely identical to each other and to those of S. cerevisiae MET2 (15). The EcoRI-uncut MET2 alleles from both organisms were also identical, being 82% homologous to S. cerevisiae and 98.5% homologous to the MET2 allele of the S. bayanus type strain (8). We conclude that both strains are hybrid yeasts and that their genetic content may be regarded as derived, at least partially, from the genomes of S. cerevisiae and S. bayanus.
FIG. 3.
Partial nucleotide sequences of the MET2 genes from S. bayanus CBS 380 type strain (Bay T) (8), S. cerevisiae S288C (Cer-S288C) (15), the S. bayanus-like allele from S6U (S6U-1) and CID1 (CID1-1), and the S. cerevisiae-like allele from S6U (S6U-2) and CID1 (CID1-2). A dot denotes an identical nucleotide.
RFLP of mtDNA.
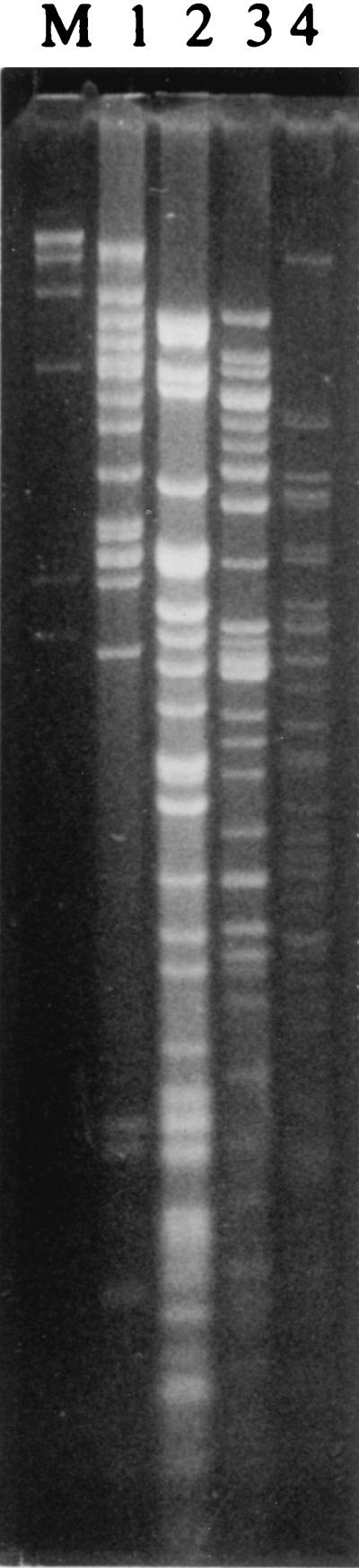
When a cross between two yeast cells occurs, the zygote contains the nuclear and mitochondrial genetic material from both parents. However, while the nuclear chromosomes are transmitted almost equally to the daughter cells, the mtDNA molecules segregate and even exhibit a bias in transmission (26). Therefore, the progeny initially contains a mixture of daughter cells which have the mitochondrial genome from one or another parent, or a novel recombinant mtDNA molecule. As mentioned before, in the case of S. carlsbergensis, only the non-S. cerevisiae mtDNA molecule was inherited. When mtDNAs from the two putative hybrid yeasts, S6U and CID1, were digested, they provided two completely different restriction patterns (Fig. 4), composed of more than 20 distinctive bands. The two patterns were also different from the pattern characteristic for mtDNA from S. paradoxus, S. carlsbergensis (Fig. 4), S. cerevisiae, and S. bayanus (27). Therefore, these restriction patterns can be used as fingerprints for identification of these yeasts. In addition, these data suggest that the mtDNA molecules from these two yeasts may not be very closely related to each other and could have a different origin. This possibility was more closely examined by sequencing of the mitochondrial OLI1 gene.
FIG. 4.
mtDNA isolated from S. paradoxus (lane 1), S. carlsbergensis (lane 2), CID1 (lane 3), and S6U (lane 4) was digested with HaeIII, and the resulting fragments were separated on a 1% agarose gel. Lambda DNA cut with HindIII was used as a marker (M).
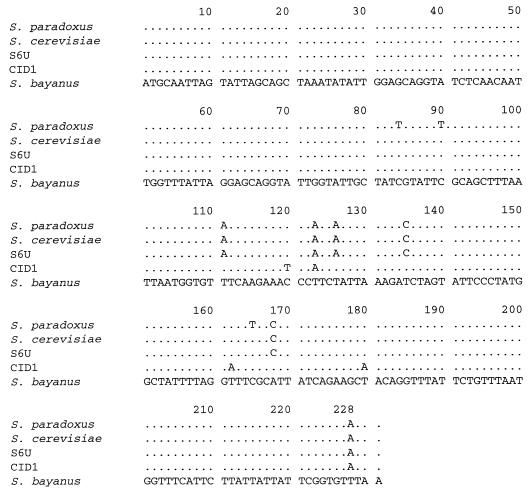
Sequence analysis of the mitochondrial OLI1 gene.
The OLI1 gene is one of the shortest and most conserved mitochondrial genes. In S. cerevisiae and S. paradoxus-S. douglasii, the open reading frame consists of 228 bp, which corresponds to 76 amino acids, and only three “silent” substitutions were found between these two species (21, 23). The open reading frames of the OLI1 gene originating from the two hybrid species, S6U and CID1, as well as from S. bayanus, were also found to be 228 bp long (Fig. 5). The amino acid sequence was identical in all cases, but several silent substitutions were observed among the analyzed species (Fig. 5). It is likely that these differences represent neutral mutations and can be directly used in reconstruction of the origin of the two hybrid yeasts. Nucleotide divergence within OLI1 among the tested species is shown in Table 2. Apparently, S. cerevisiae and S. douglasii-S. paradoxus are more closely related to each other than to S. bayanus. This observation fits well with the previous analysis of the mtDNA molecules from these three species (7, 27). The hybrid strains, S6U and CID1, show a divergence of 2.2%, which is almost as high as that between S. cerevisiae and S. bayanus, 2.6%. While the sequence of the OLI1 gene from S6U appears to be the same as for the S. cerevisiae gene, the CID1 OLI1 gene is equally divergent, 2.2%, from both putative parents, S. bayanus and S. cerevisiae (Table 2). These results unambiguously show that the mtDNA molecules of the two hybrid strains are different and are likely to have a different origin.
FIG. 5.
The open reading frame of the mitochondrial OLI1 gene in various yeast species. The sequences begin with the start codon, ATG, and finish with the stop codon, TAA. A dot denotes an identical nucleotide. The type strain of S. bayanus was used in this study. The accession numbers of the sequences are Y16964 (Saccharomyces sp. OLI1 gene, strain CID1), Y16965 (S. bayanus OLI1 gene), and Y16966 (Saccharomyces sp. OLI1 gene, strain S6U).
TABLE 2.
Nucleotide divergence among the sequences of the mitochondrial OLI1 gene from various yeast speciesa
| Species or hybrid strain | Nucleotide divergence
|
|||
|---|---|---|---|---|
| S. paradoxus | S. cerevisiae | S6U | CID1 | |
| S. paradoxus | ||||
| S. cerevisiae | 3 (1.3%) | |||
| S6U | 3 (1.3%) | 0 (0%) | ||
| CID1 | 6 (2.6%) | 5 (2.2%) | 5 (2.2%) | |
| S. bayanus | 9 (3.9%) | 6 (2.6%) | 6 (2.6%) | 5 (2.2%) |
Phenotypic divergence: production of esters.
Some strains of S. bayanus have been reported to have specific and somewhat unusual fermentation properties. Some are cryophilic, having a higher growth rate and a better fermentability at low temperatures as compared to S. cerevisiae strains (13), and these produce wines with higher than usual amounts of flavor-active esters, especially β-phenylethyl alcohol and β-phenylethyl acetate (12, 28). Moreover S. cerevisiae-S. bayanus hybrids obtained by hybridization in the laboratory exhibited such fermentation characteristics at intermediate values (12, 28). We set out to investigate whether the hybrid nature of the genomes of S6U and CID1 was in any way phenotypically reflected in the production of these aroma compounds. The same Sauvignon blanc grape must was inoculated with four industrial S. cerevisiae wine yeast strains (EG8, VL3c, VL1, and SIHA3), two indigenous S. bayanus wine yeast strains (P3 and TB28), and the strains S6U and CID1. At the midpoint of the alcoholic fermentation, the strain implantation was verified by PCR associated with the delta sequence (S. cerevisiae strains and the strains S6U and CID1) (16) (data not shown) or electrophoretic karyotyping (S. bayanus strains). The amounts of β-phenylethyl alcohol and its acetate ester obtained for each strain are reported in Table 3. According to previous reports, the values obtained for the S. bayanus strains were 7.5 to 10 times higher for β-phenylethyl alcohol and 3 to 13 times higher for β-phenylethyl acetate compared to S. cerevisiae wine yeast (12). The wines produced by S6U and CID1 contain intermediate amounts of the two compounds, thus indicating that the genetic hybrid nature of these yeasts seems to be somewhat reflected in at least one phenotype of importance to the wine industry.
TABLE 3.
Production of β-phenylethyl alcohol and β-phenylethyl acetate by different strains of S. cerevisiae and S. bayanus and by hybrid strains
| Species or type | Yeast strain | β-Phenylethyl alcohol (mg/liter) | β-Phenylethyl acetate (mg/liter) |
|---|---|---|---|
| S. cerevisiae | EG8 | 15.6 | 0.63 |
| S. cerevisiae | VL3c | 22.8 | 0.68 |
| S. cerevisiae | VL1 | 16.5 | 0.45 |
| S. cerevisiae | SIHA3 | 18.6 | 0.91 |
| S. bayanus | P3 | 256 | 12.3 |
| S. bayanus | TB28 | 118 | 1.31 |
| Hybrid | S6U | 47.6 | 2.17 |
| Hybrid | CID1 | 48.3 | 1.61 |
DISCUSSION
When the karyotypes of the two wine and cider yeasts, S6U and CID1, were compared to the karyotypes of some known yeast species, it was apparent that their nuclear genomes contain S. cerevisiae-like and S. bayanus-like chromosomes. Therefore, it seemed likely that the two yeast strains are hybrids between two species, S. cerevisiae and S. bayanus. This theory was substantiated by the analysis of the nuclear MET2 gene, the sequence of which differs characteristically between S. cerevisiae and S. bayanus (8, 18). RFLP maps of a PCR-amplified MET2 gene fragment appeared as mixes of the RFLP maps of S. cerevisiae and S. bayanus, thus supporting the notion that S6U and CID1 constitute hybrids between S. cerevisiae and S. bayanus. Verification of this theory was obtained by the sequence analysis of a part of the two MET2 gene alleles, supposedly present in both yeasts: in S6U and CID1, there are indeed two alleles of the gene, one identical to S. cerevisiae MET2, and one almost identical to S. bayanus MET2. It is furthermore interesting that both copies of this gene were almost identical in both hybrid strains. Therefore, it is likely that the nuclear genomes of both hybrids have a similar origin. The origin, a cross between S. cerevisiae and a S. bayanus-like yeast, is reminiscent of the situation found in S. carlsbergensis lager brewing yeast. However, while S. carlsbergensis inherited the non-S. cerevisiae-like mtDNA molecule (27), the mitochondrial inheritance pattern is different in S6U and CID1.
While in yeast crosses, nuclear DNA is inherited from both parents, mtDNA exhibits a non-Mendelian pattern of inheritance (26). In the progeny, only one or the other parental mtDNA molecule, or a recombinant one, is found. The S6U and CID1 hybrids contained two different mtDNA molecules. The mtDNA molecule in S6U appears to originate from the S. cerevisiae-like parent, whereas the CID1 mtDNA molecule differs from that of S. cerevisiae as well as that from S. bayanus. Phylogenetically, the latter mtDNA molecule could be positioned between the S. cerevisiae and S. bayanus mtDNA molecules. Therefore, it is likely that the two hybrid strains do not originate from a single hybridization event. While the nuclear backgrounds of the parents involved in both crosses were probably very similar, the mitochondrial backgrounds were likely to be different.
It appears that among Saccharomyces yeasts used in fabrication of wine, cider, and beer, stable interspecies hybrids are quite common. Whether such hybrids originate from events having taken place in the production environments or in nature is not known. As the genetic constitution of these yeasts seems to be mirrored in at least one phenotype of importance to the wine industry, production of certain esters and higher alcohols, the specific properties of S. cerevisiae-S. bayanus hybrid strains can present an advantage in wine making, especially for white wines, which are fermented at a low temperature and for which middle amounts of β-phenylethyl alcohol and its acetate are a synonym of quality.
ACKNOWLEDGMENTS
We thank LALLEMAND Inc. (Canada) for the strain S6U and H. V. Nguyen (CLIB, Paris) for strains from Pr. Naumov.
A part of this work was supported by grants of the Danish Research Council, SNF, and Carlsberg Foundation to Jure Piskur and by SARCO (Bordeaux, France).
REFERENCES
- 1.Banno I, Kaneko Y. A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast. 1989;5:373–377. [PubMed] [Google Scholar]
- 2.Barnett J A. The taxonomy of the genus Saccharomyces Meyen ex Rees: a short review for non-taxonomist. Yeast. 1992;8:1–23. [Google Scholar]
- 3.Bellis M, Pages M, Roizes G. A simple and rapid method for preparing yeast chromosomes for pulsed field electrophoresis. Nucleic Acids Res. 1987;15:6749. doi: 10.1093/nar/15.16.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardinalli G, Martini A. Electrophoretic karyotypes of authentic strains of the sensu stricto group of the genus Saccharomyces. Int J Syst Bacteriol. 1994;44:791–797. doi: 10.1099/00207713-44-4-791. [DOI] [PubMed] [Google Scholar]
- 5.Dubourdieu D, Sokol A, Zucca J, Thalouarn A, Dattec C, Aigle M. Identification de souches de levures isolées de vins par l’analyse de leur ADN mitochondrial. Connaiss Vigne Vin. 1987;21:267–278. [Google Scholar]
- 6.Frézier V, Dubourdieu D. Ecology of yeast strain Saccharomyces cerevisiae during spontaneous fermentation in a Bordeaux winery. Am J Enol Vitic. 1992;43:375–380. [Google Scholar]
- 7.Groth C. Saccharomyces sensu stricto yeasts: characterization of mitochondrial DNA. Masters thesis. Copenhagen, Denmark: University of Copenhagen; 1998. [Google Scholar]
- 8.Hansen J, Kielland-Brandt M C. Saccharomyces carlsbergensis contains two functional MET2 alleles similar to homologues from S. cerevisiae and S. monacensis. Gene. 1994;140:33–40. doi: 10.1016/0378-1119(94)90727-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko Y, Banno I. Reexamination of Saccharomyces bayanus strains by DNA-DNA hybridization and electrophoretic karyotyping. IFO Res Commun. 1988;15:30–41. [Google Scholar]
- 11.Kielland-Brandt M, Nilsson-Tillgren T, Gjermansen C, Holmberg S, Pedersen M B. Genetics of brewing yeasts. In: Rose A H, Wheals A E, Harrison J S, editors. The yeasts. Vol. 6. London, England: Academic Press; 1995. pp. 223–254. [Google Scholar]
- 12.Kishimoto M. Fermentation characteristics of hybrids between the cryophilic wine yeast Saccharomyces bayanus and the mesophilic wine yeast Saccharomyces cerevisiae. J Ferm Bioeng. 1994;77:432–435. [Google Scholar]
- 13.Kishimoto M, Goto S. Growth temperatures and electrophoretic karyotyping as tools for practical discrimination of Saccharomyces bayanus and Saccharomyces cerevisiae. J Gen Appl Microbiol. 1995;41:239–247. [Google Scholar]
- 14.Kurtzman C P, Robnett C J. Phylogenetic relationship among species of Saccharomyces, Schizosaccharomyces, Debaryomyces and Schwanniomyces determined from partial ribosomal RNA sequences. Yeast. 1991;7:61–72. doi: 10.1002/yea.320070107. [DOI] [PubMed] [Google Scholar]
- 15.Langin T, Faugeron G, Goyon C, Nicolas A, Rossignol J-L. The MET2 gene of Saccharomyces cerevisiae: molecular cloning and nucleotide sequence. Gene. 1986;49:283–293. doi: 10.1016/0378-1119(86)90364-1. [DOI] [PubMed] [Google Scholar]
- 16.Masneuf I. Recherches sur l’identification génétique des levures de vinification; applications oenologiques. Thesis no. 423. Bordeaux, France: Bordeaux II University; 1996. [Google Scholar]
- 17.Masneuf I, Dubourdieu D. Comparaison de deux techniques d’identification des souches de levures de vinification basées sur le polymorphisme de l’ADN génomique: réaction de polymérisation en chaine (PCR) et analyse des caryotypes (electrophorèse en champ pulsé) J Int Sci Vignes Vin. 1994;28:153–160. [Google Scholar]
- 18.Masneuf I, Aigle M, Dubourdieu D. Development of a polymerase chain reaction/restriction fragment length polymorphism method for Saccharomyces cerevisiae and Saccharomyces bayanus identification in enology. FEMS Microbiol Lett. 1996;138:239–244. doi: 10.1111/j.1574-6968.1996.tb08164.x. [DOI] [PubMed] [Google Scholar]
- 19.Naumov G I. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol. 1996;17:295–302. [Google Scholar]
- 20.Naumov G I, Naumova E, Gaillardin C. Genetic and karyotypic identification of wine Saccharomyces bayanus yeasts isolated in France and Italy. Syst Appl Microbiol. 1993;16:274–279. [Google Scholar]
- 21.Nicoletti L, Laveder P, Pellizzari R, Cardazzo B, Carignani G. Comparative analysis of the region of the mitochondrial genome containing the ATPase subunit 9 gene in the two related yeast species Saccharomyces douglasii and Saccharomyces cerevisiae. Curr Genet. 1994;25:504–507. doi: 10.1007/BF00351669. [DOI] [PubMed] [Google Scholar]
- 22.Ollivier C, Stonestreet T, Larue F, Dubourdieu D. Incidence de la composition colloidale des moûts blancs sur leur fermentescibilité. Connaiss Vigne Vin. 1987;21:59–70. [Google Scholar]
- 23.Ooi B G, McMullen G L, Linnane A W, Nagley P, Novitski C E. Biogenesis of mitochondria: DNA sequence analysis of mit− mutations in the mitochondrial OLI1 gene coding for mitochondrial ATPases subunit 9 in Saccharomyces cerevisiae. Nucleic Acids Res. 1985;13:1327–1339. doi: 10.1093/nar/13.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen M B. DNA sequence polymorphism in the genus Saccharomyces. IV. Homologous chromosomes III in Saccharomyces bayanus, S. carlsbergensis and S. uvarum. Carlsberg Res Commun. 1986;51:185–202. doi: 10.1007/BF02904408. [DOI] [PubMed] [Google Scholar]
- 25.Piskur J. Respiratory-competent yeast mitochondrial DNAs generated by deleting intergenic regions. Gene. 1989;81:165–168. doi: 10.1016/0378-1119(89)90347-8. [DOI] [PubMed] [Google Scholar]
- 26.Piskur J. Inheritance of the yeast mitochondrial genome. Plasmid. 1994;31:229–241. doi: 10.1006/plas.1994.1025. [DOI] [PubMed] [Google Scholar]
- 27.Piskur, J., S. Smole, C. Groth, R. F. Petersen, and M. B. Pedersen. Structure and genetic stability of mitochondrial genomes vary among yeasts of the genus Saccharomyces. International J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 28.Shinohara T, Saito K, Yanagida F, Goto S. Selection and hybridization of wine yeasts for improved winemaking properties: fermentation rate and aroma productivity. J Ferm Bioeng. 1994;77:428–431. [Google Scholar]
- 29.Vaughan-Martini A, Martini A, Cardinali G. Electrophoretic karyotyping as a taxonomic tool in the genus Saccharomyces. Antonie Leeuwenhoek. 1993;63:145–156. doi: 10.1007/BF00872389. [DOI] [PubMed] [Google Scholar]
- 30.Vezinhet F, Blondin B, Hallet J N. Chromosomal DNA patterns and mitochondrial DNA polymorphism as tool for identification of enological strains of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1990;32:568–571. [Google Scholar]