Abstract
The forward light scatter intensity of bacteria analyzed by flow cytometry varied with their dry mass, in accordance with theory. A standard curve was formulated with Rayleigh-Gans theory to accommodate cell shape and alignment. It was calibrated with an extinction-culture isolate of the small marine organism Cycloclasticus oligotrophus, for which dry weight was determined by CHN analysis and 14C-acetate incorporation. Increased light scatter intensity due to formaldehyde accumulation in preserved cells was included in the standard curve. When differences in the refractive indices of culture media and interspecies differences in the effects of preservation were taken into account, there was agreement between cell mass obtained by flow cytometry for various bacterial species and cell mass computed from Coulter Counter volume and buoyant density. This agreement validated the standard curve and supported the assumption that cells were aligned in the flow stream. Several subpopulations were resolved in a mixture of three species analyzed according to forward light scatter and DNA-bound DAPI (4′,6-diamidino-2-phenylindole) fluorescence intensity. The total biomass of the mixture was 340 μg/liter. The lowest value for mean dry mass, 0.027 ± 0.008 pg/cell, was for the subpopulation of C. oligotrophus containing cells with a single chromosome. Calculations from measurements of dry mass, Coulter Counter volume, and buoyant density revealed that the dry weight of the isolate was 14 to 18% of its wet weight, compared to 30% for Escherichia coli. The method is suitable for cells with 0.005 to about 1.2 pg of dry weight at concentrations of as low as 103 cells/ml and offers a unique capability for determining biomass distributions in mixed bacterial populations.
Measurements of bacterial biomass are essential for determining growth rates and cell yields and provide a base for formulations that relate rates of growth and nutrient accumulation to substrate concentration (12, 13). Accurate values are difficult to establish for dilute or mixed populations or for very small bacteria. Among the available methods (61), optical density measurement is commonly used and can be related to dry weight with light scatter theory (35). However, the method is insufficiently sensitive for analyzing very dilute concentrations of cells and cannot differentiate subpopulations or discriminate organisms from debris. Concentrations of some bacteria can be determined from plate counts or most-probable-number determinations (31), but many bacteria resist growth in laboratory cultures (64). The biomasses of such populations are often determined from cell number and size by microscopy (55, 56, 71), but the optical sizing of small cells appears to lack precision (45) and the dry matter content used is often inappropriately assumed to be similar to that measured for Escherichia coli (48). Coulter Counter impedance (40, 42) provides a fairly accurate estimate of cell volume, but the method lacks sensitivity for small bacteria, does not account for changes in the density of the cell material (28), and is subject to interference from debris.
Flow cytometry can determine biomass from the intensity of light scattered by single cells (52). Advantages include simultaneous multiparameter analysis, adequate sensitivity for organisms <0.1 μm3 in size at <1,000 cells/ml, favorable statistics due to analysis rates approaching 104 particles/min (14), resolution of individual species in a mixture (67), and ability to determine organism concentrations (14). Bacterium-sized particles scatter light mainly in the forward direction (35), within the detection range of most flow cytometers. Small size and an index of refraction only 3 to 6% higher than that of the surrounding medium minimize phase change and allow the use of Rayleigh-Gans theory to relate bacterial biomass to light scatter intensity in flow cytometric analyses (37).
The size of bacterial cells has been computed from flow cytometry data by Mie theory (8), with the results being reported in terms of diameters of polystyrene spheres (1). Davey et al. (20) related forward light scatter intensity from polystyrene spheres to particle size and calibrated the resulting polynomial with microscopic measurements of bacteria. They attributed an underestimate of cell volume by this method to a difference in the ways in which cells and polystyrene standards scatter light. Other attempts to relate forward light scatter to cell size have depended on measurements of cell volume by electrical impedance (3, 50). However, calibration of the light scatter intensity from the very small bacteria resolvable by flow cytometry with the signal from larger organisms measurable with a Coulter Counter required a long extrapolation, and variation in dry weight content was not addressed. Light scatter was reported to reflect the protein content (9, 62) of E. coli because of a constant protein-to-dry weight ratio, but the range of values examined was too small to establish the shape of a standard curve. Most flow cytometry data for cell size are reported in arbitrary units, such as channel number (2, 6, 22), which is nonlinear with cell volume or mass.
An earlier investigation (37) applied Rayleigh-Gans theory to flow cytometry by integrating the predicted intensity of light scattered at a specified angular range from the path of the laser beam by cells of a specified size, axial ratio, and orientation in the flow stream and illuminated at a particular frequency. Here, we explore limitations of the theory and compare predictions with data from standard spheres. A theoretical curve is produced and calibrated with measurements of dry weight obtained for a small marine isolate. Dry mass values of various bacterial species are interpreted in terms of their relative refractive index m and are verified by independent measurements of buoyant density and Coulter Counter volume. Forward light scatter and DNA-bound DAPI (4′,6-diamidino-2-phenylindole) fluorescence intensity are used to obtain the biomass of each of several subpopulations resolved from a mixture of species.
MATERIALS AND METHODS
Cultures.
An extinction-culture isolate (16) of the marine organism Cycloclasticus oligotrophus (68) (formerly oligobacterium RB1 [16]) was grown in synthetic seawater medium (54). Growth rate and cell size range were controlled by acetate concentrations ranging from 12 to 200 mg/liter. Enrichment culture isolates of Marinobacter (formerly Pseudomonas [49]) sp. strain T2 and RB95-4 were grown in synthetic seawater medium with amino acids and glucose, respectively, as the sole carbon sources. E. coli DH1 (ATCC 33849) and Pseudomonas diminuta (ATCC 19146) were grown in M9 medium (23) supplied with glucose.
Standards.
Fluorescent polystyrene microspheres (Fluoresbrite Microparticles; Polysciences, Inc.), 0.3 to 2.02 μm in diameter, with a refractive index of 1.6 and with a density of 1.05 g/cm3 (47), were used. A mixture of 0.90- and 0.60-μm spheres provided internal standards (14). The concentration of the larger spheres was adjusted at 105 cells/ml with a Coulter Counter, and the ratio of their frequency to that of bacteria was used to compute the populations of bacteria. Forward light scatter and blue fluorescence intensity of the smaller spheres were used to normalize intensities among samples.
Sample preparation for analysis by flow cytometry.
Samples were preserved with filtered formaldehyde (0.2-μm-pore size; as formalin; 0.5% [wt/vol]), stored at 5°C in the dark for at least 16 h unless otherwise noted, vortexed, diluted to about 106 cells/ml, treated with Triton X-100 (0.1%), stained with DAPI (0.5 μg/ml) at 10°C in the dark for 1 h, amended with the standard microspheres, and kept at 10°C during analysis (14).
Flow cytometry.
Instrumentation consisted of an Ortho Cytofluorograf IIS equipped with a flat-sided quartz flow cell, a 5-W argon laser tuned to UV emission (351.1 and 363.8 nm) at 100 mW of power, and a focusing lens to reduce the beam width to 44 μm for better resolution of small bacteria (50). Light scattered by bacteria in the forward direction past a vertical 1.5-mm beam blocker bar was reflected by a 424-nm long-pass dichroic filter through a 310- to 370-nm band-pass filter and focused onto the plane of a shielded fiber-optic cable leading to the photomultiplier detector. Orthogonal light scatter, used to evaluate the frequency of internal 0.9-μm-diameter standard spheres for population counts (14), was isolated with similar optical filters. Blue fluorescence from DNA-bound DAPI was collected orthogonally through the 424-nm dichroic filter and a 450- to 490-nm band-pass filter.
Logarithmic amplifiers with a dynamic range of 3.5 decades were calibrated (53) to establish the range of linearity between signal input and numeric response (14). Data acquisition, analysis, and storage were done with a PC-based Cicero system and Cyclops software (Cytomation, Inc.). Acquisition was triggered by DAPI-DNA fluorescence to eliminate the forward light scatter signals from nonfluorescent debris.
Conversion of 256 channels resolved by the logarithmic amplifiers to 103.5 linear channels was accommodated by the software, and mean values (linear) for the forward light scatter and DNA-bound DAPI fluorescence intensity of each population were recorded. The ratio of the mean forward light scatter or fluorescence intensity of a population to that of the 0.6-μm internal standard spheres was used to normalize among samples for calculations of cell mass and DNA content.
Light scatter theory.
Expected intensities of light scattered by bacteria of a given mass, shape, and orientation were obtained from computer programs (37) based on Rayleigh-Gans theory. To verify the formulations, calculations for spherical particles were compared with those based on the more complete but complex Mie theory (34) as programmed by Tsay and Stephens (63), since the Mie theory is not easily applied to nonspherical particles. Input for all programs included an excitation wavelength of 360 nm and a forward light scatter collection angle of 0.5 to 20°. Error from signals excluded by the beam blocker bar was assumed to be minimal (37). Mie calculations were performed for m values of 1.04 and 1.19 to compare the curves obtained for bacteria with those for polystyrene standards.
Standard curve for dry mass.
A curve relating dry weight to forward light scatter intensity was calculated for particles modeled as ellipsoids of revolution with an axial ratio of three and aligned perpendicular to the laser beam (37). Curves were also computed for particles with different axial ratios and random or perpendicular alignment for comparison to validate the use of the input parameters. The curves were fitted with the Marquardt-Levenberg algorithm and a volume weighting factor of 1/V2.3 (where V is volume) (SigmaPlot; Jandel Scientific) to give the dry weight of a particle as a function of its forward light scatter intensity.
The proportionality constant required to normalize forward light scatter intensities obtained from the flow cytometer with values calculated from theory (37) was computed by determining the dry weight and the forward light scatter intensity of C. oligotrophus grown on 14C-acetate as the sole carbon source and by determining the ratio of carbon to dry weight. 1,2-14C-acetate (58.2 mCi/mmol; DuPont NEN) was diluted with unlabeled anhydrous sodium acetate (analytical reagent grade; Mallinckrodt) to a specific activity of 2.19 × 106 dpm/mg of sodium acetate or 6.83 × 106 dpm/mg of carbon. Populations were grown to attain at least a 14-fold increase in biomass so that, assuming an endogenous metabolism rate of 0.007 h−1, more than 98% of the cell carbon would be labeled. Dry weights were determined at a range of acetate concentrations and computed from the radioactivity (∼104 to 106 dpm) collected on a polycarbonate membrane filter (0.2-μm-pore size; Nuclepore Corp.) and counted with a scintillation spectrometer, the specific activity of the substrate, and the cell population determined by flow cytometry. To determine the ratio of cell carbon to dry weight, a culture was grown on acetate and pelleted by centrifugation. The 27-mg pellet was washed with 8 ml of saline to reduce the carryover of medium solutes to 28 μg (<0.8 μg of acetate), transferred into tared tin boats, dried along with cell-free controls overnight at 100°C in a vacuum oven, weighed, and analyzed for carbon content with a CHN600 analyzer (Leco Corp.) calibrated with a coal standard.
DNA content.
DNA was determined from DAPI-DNA fluorescence intensity and standardized with the signal from E. coli with a known genome size of 4.7 Mbp (38, 51), or 5.17 fg, and a GC content of 50 mol% (39). To account for the effects of medium salinity on DAPI fluorescence intensity (70), which amounted to a 10 to 30% reduction with increasing salt concentration, depending on the species measured (unpublished data), preserved E. coli was stained and analyzed in both M9 and seawater media. Linear regression analysis of modal values of DAPI-DNA fluorescence intensity of E. coli subpopulations containing cells with integral numbers of chromosome copies gave the intensity associated with a single chromosome copy relative to the intensity of the internal-standard 0.6-μm fluorescent microspheres. Cellular DNA content for the other strains or subpopulations within a culture was obtained from mean fluorescence intensity with a correction for the AT bias of DAPI (70) based on their G+C contents: 52.7 mol% for Marinobacter sp. strain T2 (25) and 41.6 mol% for C. oligotrophus (68).
Cell size.
The volumes of the organisms were measured with a Coulter Counter (model ZBI with a Channelyzer; Coulter Electronics, Inc.) with 16- and 30-μm apertures. Instrument calibration was done with 1.942-μm-mean-diameter spheres (Coulter lot no. 6179; National Institute of Standards and Technology traceable standard) and corroborated by data from a Coulter Multisizer II. Volumes were corrected for cell shape according to theory (27, 32). Cell dimensions were determined from electron micrographs. For scanning electron microscopy, cells were harvested by centrifugation, washed with basal medium, concentrated on polycarbonate filters, dehydrated in an ethanol series, and critical point dried. For transmission electron microscopy, harvested cells were applied to a Formvar grid stabilized with carbon on 100-Mbar copper (1 bar is 105 pascals) (Ted Pella, Inc.), stained with 2% uranyl acetate, rinsed, and air dried.
Buoyant density.
Cell densities were determined by equilibrium centrifugation in Percoll gradients made iso-osmotic with culture medium by the addition of sodium chloride. Mixtures of Percoll (Sigma Chemical Co.) and culture fluid at 70:30 for M9 medium and 40:60 for synthetic seawater medium were centrifuged at 30,000 × g in a fixed-angle rotor for 1 to 2 h at 4°C. Values were determined from the positions of cells and density marker beads (Sigma). The densities of the marker beads were corrected for medium salinity based on their position in a gradient of Percoll-synthetic seawater solution and the assumption that the density of the green beads of 1.099 g/cm3 remained constant (46). The refractive index of the medium at the location of the beads was determined from fluid sampled above and below the band by measurement with a refractometer (Abbe model 3L; Bausch & Lomb, Inc.). The densities of the remaining standard markers were computed from assumed linearity between the refractive index and the medium density (46) at the location of the green beads and the refractive index and measured density (by hydrometer) of Percoll-free medium.
Refractive index.
Organisms were suspended at approximately 107 to 108 cells/ml in basal medium containing 0 to 30% bovine serum albumin (66) and with adjustments of the sodium chloride concentration to maintain osmolarity. The refractive index of the organisms was taken as the refractive index of the medium adjusted with bovine serum albumin to give a minimal absorbance (UV-1201; Shimadzu) of the bacteria at 700 nm. The refractive index of cells as measured by a refractometer was normalized to that of basal medium to obtain m.
Calculations based on cell composition.
For comparison with dry mass determined from forward light scatter intensity, dry mass was computed from the product of cell volume, buoyant density, and the ratio of dry weight to wet weight (Xdry/Xwet). Buoyant density (ρcell) is given as the density of dry matter (ρdry), Xdry/Xwet, and the density of water (ρwater) (4):
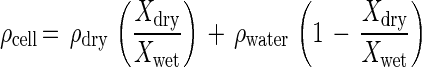 |
1 |
The value of ρdry was computed from the composition of the organisms and the density of each component taken from the literature. A value of 1.35 g/cm3 was obtained for E. coli from the composition of cells growing at a rate of 0.011 h−1 on glucose in M9 medium (17): protein at 54% of total dry weight, with a density of 1.3 g/cm3 (44); nucleic acid at 18.3%, with 1.7 g/cm3 (60); lipid at 9.1%, with 0.9 g/cm3 (60); and the remaining constituents at 13%, with 1.4 g/cm3. A ρdry value of 1.39 g/cm3 was used for C. oligotrophus, with protein at 44% of dry weight, nucleic acid at 36%, lipid at 14%, and others at 6%. Organisms of a smaller size and a higher surface-to-volume ratio, compared to E. coli, were expected to have a higher proportion of nucleic acids and lipids in their dry matter.
The refractive index of the organisms relative to their medium (m) was computed as follows (35):
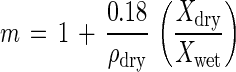 |
2 |
In this equation, 0.18 is the specific refractive index increment factor (66), based on the weighted average of values for protein (0.186), nucleic acid (0.16), and other dry-matter constituents (0.178).
For comparison with Coulter Counter volume measurements, cell volume (V) was computed as follows:
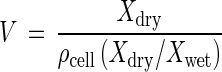 |
3 |
RESULTS
Theoretical predictions.
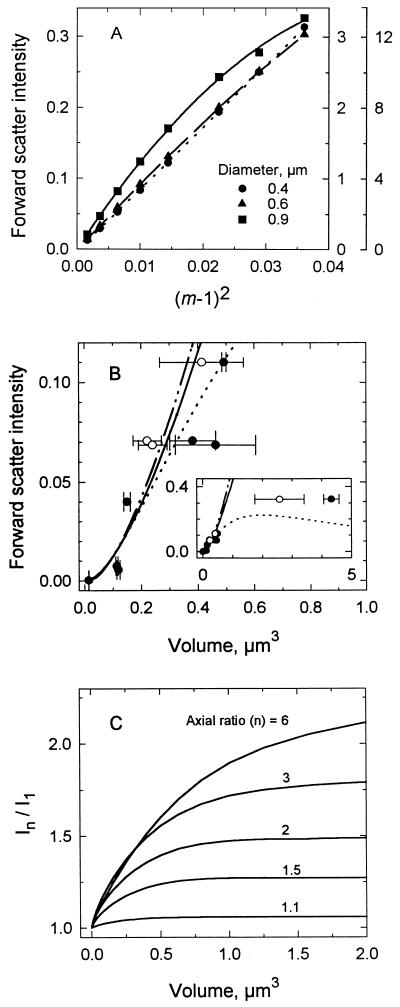
Mie theory specifies an increase in forward light scatter intensity with particle size and refractive index. The intensity of light scattered from small particles is a linear function of refractive index when expressed as (m − 1)2 (35) (Fig. 1A). This relationship is particularly useful for anticipating the effects of changes in m on forward light scatter intensity (see below). Curvature results at large particle sizes because of a greater proportion of light being scattered at angles broader than 20°. For very small particles and those such as bacteria for which m is 1.04 (35), forward light scatter intensity can be predicted by the Rayleigh-Gans approximation, as shown by the similarity between the results of Mie and Rayleigh-Gans calculations for spheres in Fig. 1B. Due to the low refractive index of bacteria, signal from cocci as large as 1.5 μm3 would be expected to lie in the region where the two curves appear identical, justifying the use of the simpler Rayleigh-Gans theory in calculations for cells of this size.
FIG. 1.
Dependence of forward light scatter intensity on particle characteristics. (A) Intensities calculated from Mie theory for three sizes of spheres as a function of m. Ordinate scales increase with particle size. (B) Intensities for latex microspheres of various sizes as predicted by Mie theory when m is 1.04 (—··—) or 1.19 (······) and by Rayleigh-Gans (37) theory (——). Data are measured intensities for microspheres with volumes and indicated standard deviations (error bars) computed according to diameters from electron microscopy reported by the supplier (•) and volumes determined with a Coulter Counter (○). The inset extends the particle size range. (C) Effect of axial ratio on forward light scatter intensity over a range of cell volumes as determined by Rayleigh-Gans theory. In/I1 compares the intensity of a particle with an axial ratio of n to that of a sphere of the same volume.
Figure 1B (inset) shows the expected difference between curves calculated for bacteria (m, 1.04) and polystyrene particles (m, 1.19) larger than 0.3 μm3 (0.83-μm diameter). Data for standard polystyrene spheres were difficult to interpret due to differences between the volumes computed from measurements by electron microscopy as specified by the supplier and those measured here with a Coulter Counter. However, they were consistent with Mie predictions at volumes greater than 1 μm3.
The forward light scatter intensity of spheres of a specified size analyzed over a wide range of photomultiplier gain settings varied less than 9% (n = 18) when normalized to that of an internal standard (data not shown). This fact allowed extension of the 3.5-decade dynamic range of the amplifier to include the full range of signals encountered from various particles.
Forward light scatter intensity increased with axial ratio (Fig. 1C). Since nonspherical particles tend toward alignment in the flow stream because of hydrodynamic forces (33), greater forward light scatter for the same volume was expected due to decreased interference and smaller phase shifts for light passing through smaller-diameter particles (37). The effect increased with size, but calculations showed that for particles of 0.05 μm3, there was only a 13% increase in forward light scatter intensity with elongation from an axial ratio of one to three, whereas for cells of 2 μm3, there was a 79% increase.
Effects of preservatives.
Formaldehyde treatment of the cells during sample preparation resulted in additional light scatter, as expected from reports of increased refractive index with preservation of Streptococcus faecalis (19) and E. coli (66). The increase in intensity for C. oligotrophus was 68%, which corresponded to a 35% increase in dry weight (Table 1). The effect was the same for cells containing a single chromosome (see below), so clumping was not a contributing factor. Other data (not shown) indicated that overnight fixation at 5°C was sufficient to stabilize light scatter properties for about 2 months. Effects on E. coli were less pronounced, amounting to a 9 to 15% increase in apparent biomass (Table 1).
TABLE 1.
Effect of formaldehyde on forward light scatter intensity and dry mass determined from flow cytometry and on Coulter Counter volume
| Organism | Preservation time (days) | Forward light scatter intensitya | Apparent mean dry mass (pg/cell)b | Coulter Counter volume (μm3) |
|---|---|---|---|---|
| C. oligotrophus | 0c | 115 | 0.032 | 0.26 |
| 0.1 | 159 | 0.038 | 0.26 | |
| 2 | 197 | 0.043 | 0.28 | |
| 49 | 200 | 0.044 | 0.28 | |
| E. coli | 0c | 7,363 | 0.383 | 1.13 |
| 0.1 | 7,502 | 0.388 | 1.03 | |
| 2 | 7,245 | 0.379 | 1.10 | |
| 14 | 7,442 | 0.386 | 1.07 | |
| 49 | 8,427 | 0.413 | 1.08 |
Mean intensity normalized with respect to the signal from the 0.6-μm internal standard. The value for E. coli has been multiplied by 0.8 to correct for the refractive index of the medium.
Dry mass computed from light scatter data with equation 6 of Koch et al. (37) and with K determined from the dry mass of C. oligotrophus.
No formaldehyde treatment.
Buoyant density measurements showed an apparent 50% increase in dry material for C. oligotrophus following preservation (Table 2) but only a 10% increase for E. coli, in agreement with the light scatter data. Similar increases were computed for m. Effects on Coulter Counter volumes were negligible over the 49 days observed. This finding indicates that preservation by formaldehyde can affect the cell mass and light scatter properties of organisms in a manner that is dependent on the species observed but that determination of the apparent sizes of cells from electrical impedance is less sensitive to the addition. The differences in the apparent addition of mass to the two organisms by formaldehyde may be related to differences in media and sample dilution. Binding is pH sensitive, maximal at pH 7.5 to 8.0, and reversible (29). The slightly lower pH of M9 medium (pH 7.5 versus 8) and the higher dilutions used for high-biomass cultures before staining to obtain appropriate populations for flow cytometry and for equilibration with DAPI may favor greater formaldehyde loss from E. coli.
TABLE 2.
Effect of formaldehyde on buoyant density
| Organism | Treatment | Buoyant density (g/cm3) | Dry wt/ wet wta | ma | Forward light scatter intensityb |
|---|---|---|---|---|---|
| C. oligotrophus | None | 1.044 | 0.12 | 1.016 | 1 |
| Formaldehyde (0.5%) | 1.067 | 0.18 | 1.024 | 2.2 | |
| E. coli | None | 1.100 | 0.27 | 1.035 | 1 |
| Formaldehyde (0.5%) | 1.112 | 0.30 | 1.039 | 1.1 |
Computed from buoyant density (equation 2).
Arbitrary units, computed from I = (m − 1)2 and normalized to the intensity of the untreated sample.
Preservation with 0.5% glutaraldehyde for 2 h gave a sixfold increase in the apparent biomass of C. oligotrophus (data not shown). Half of this increase can be accounted for by the larger molecular mass of the preservative. Differences in response to the fixation process among species may also affect observations, since formaldehyde penetrates rapidly as methylene glycol but reacts slowly (24), while the opposite has been reported for glutaraldehyde (29). Valkenburg and Woldringh (66) showed that glutaraldehyde treatment caused an increase in the buoyant density of E. coli from 1.093 to 1.138, which amounted to a 50% increase in apparent cell mass, according to equations 1 and 2 and as shown in Fig. 1A.
Standard curves for dry mass.
Equation 4 (37) was used to express the relationship between the cell mass and forward light scatter intensity of bacteria as calculated from Rayleigh-Gans theory:
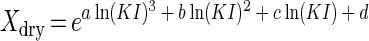 |
4 |
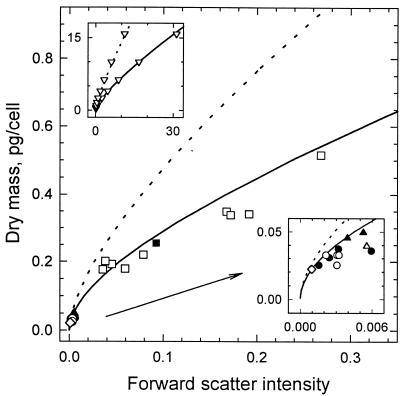
K is the proportionality constant converting instrument intensity (I) to intensity expected from theory (KI), and the constants a, b, c, and d are 1.62 × 10−4, 0.0144, 0.480, and 0.274, respectively, for ellipsoidal cells with an axial ratio of three and oriented in the direction of flow. An axial ratio of three was used for the calculation because it agreed with the dimensions of E. coli, Marinobacter sp. strain T2, and C. oligotrophus measured by electron microscopy (axial ratio, 2.7 ± 0.3; n = 7 samples; 168 cells). Equation 4 fits the computed values to within 5% for cells with up to 40 pg of dry mass (Fig. 2, upper inset; data shown to 15 pg/cell). The divergence of the theoretical curves for cells aligned in the flow stream and cells with a random orientation showed the anticipated increase (37) in light scatter intensity with orientation. Experimental values were 83% ± 12% (n = 22) predicted values for aligned cells, whereas values for bacteria with a large mass, presumed to be more strongly influenced by hydrodynamic forces, were only about 50% those expected for randomly oriented cells. The better fit of experimental data to the curve for oriented cells is consistent with the expectation (33) of alignment in the flow stream. The effects of orientation increase with cell mass, as shown by the difference between the shapes of the two curves.
FIG. 2.
Standard curves for dry mass. Curves were computed for cells with an axial ratio of three and with linear (——) and random (·····) orientations in the flow stream (37) and were calibrated with the dry mass measurement for C. oligotrophus (◊). Data are from light scatter intensities of C. oligotrophus (• and ○), Marinobacter sp. strain T2 (▴ and ▵), and E. coli (■ and □), with dry weights computed from Coulter Counter volumes corrected for cell density on the basis of equations 1 and 3. Closed symbols are for data in Table 4; open symbols are for additional data. The upper inset shows data generated from Rayleigh-Gans theory (▿) and fitted curves and extends computations to larger particle sizes. The lower inset expands data from small organisms.
Dry weights determined for cultures of C. oligotrophus grown on 14C-acetate were used to obtain K for calibrating the curve (Table 3). CHN analysis determined cell carbon content to be 47% ± 1% (n = 3) dry weight, similar to values reported for other bacterial strains (11). Agreement was within 6% between populations measured by flow cytometry (14) and with the Coulter Counter for various cultures of E. coli, Marinobacter sp. strain T2, and C. oligotrophus (r2 = 0.995; n = 15; data not shown), so that mean dry weight could be obtained on a per-cell basis. The carbon content calculated from 14C-acetate incorporated by C. oligotrophus was 10.3 ± 1.8 fg/cell (n = 4) over a range of populations, for a dry weight of 22 ± 4 fg/cell. Experimental data were fit to the theoretical curve at a K value of 1.61 × 10−5 (Fig. 2).
TABLE 3.
Dry weight of C. oligotrophus for determining the proportionality constant K
| 14C-acetate supplied (μg of C/ml) | Bacterial carbon produced (μg of C/ml)a | Population (106 cells/ml) | Carbon content (fg/cell) | Dry wt (fg/cell)b | Forward light scatter intensityc |
|---|---|---|---|---|---|
| 3.4 | 0.74 | 56.6 | 13.1 | 28.0 | 61.5 |
| 10.5 | 2.28 | 244 | 9.33 | 19.9 | 62.6 |
| 31.4 | 7.93 | 841 | 9.43 | 20.1 | 55.5 |
| 56.6 | 15.81 | 1,671 | 9.46 | 20.1 | 55.3 |
In the final sample for dry weight determination 185 h after inoculation.
Carbon content divided by 0.47; mean, 22 ± 4.
The channel number was normalized to the signal for 0.6-μm internal standard spheres; mean, 58.7 ± 3.9.
Since the standard curve was based on C. oligotrophus in seawater medium, for bacteria analyzed in freshwater medium, K was multiplied by 0.80 to correct for the difference between the refractive index of M9 medium (1.3331) and that of seawater medium (1.3380). For E. coli, for which m is 1.037 in M9 medium (see below) (Table 4), the expected m in seawater would be 1.037 × 1.3331/1.3380, or 1.033. Based on the linearity between forward light scatter intensity and (m − 1)2 from Fig. 1A, E. coli would produce (0.033)2/(0.037)2, or 0.80 as much forward light scatter intensity if it were in seawater. To account for the smaller effect of formaldehyde on the light scatter intensity of E. coli, K was further multiplied by 1.46 to give a KM9 of 1.88 × 10−5 for organisms grown in M9 medium.
TABLE 4.
Comparison of dry weights determined by flow cytometry with independent measurements
| Organism | Measured
|
Computed
|
Computed/ measured dry wt | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Dry wt (fg/cell)a | Vol (μm3/cell)b | Buoyant density (pg/μm3) | Dry wt/ wet wtc | Wet wt (fg/cell)d | Dry wt (fg/cell)e | Vol (μm3/cell)f | mg | ||
| C. oligotrophus | 35.7 | 0.16 | 1.07 | 0.179 | 199 | 30.7 | 0.186 | 1.023 | 0.86 |
| 28.3 | 0.13 | 1.07 | 0.179 | 158 | 25.0 | 0.148 | 1.023 | 0.88 | |
| 41.3 | 0.25 | 1.055 | 0.141 | 293 | 37.2 | 0.278 | 1.020 | 0.90 | |
| 57.7 | 0.24 | 1.055 | 0.141 | 409 | 35.7 | 0.388 | 1.020 | 0.62 | |
| Marinobacter sp. strain T2 | 54.0 | 0.24 | 1.075 | 0.192 | 281 | 49.6 | 0.261 | 1.025 | 0.92 |
| 46.2 | 0.22 | 1.075 | 0.192 | 240 | 45.5 | 0.214 | 1.025 | 0.98 | |
| E. coli | 322 | 0.81 | 1.10 | 0.286 | 1,128 | 255 | 1.03 | 1.037 | 0.79 |
From equation 4 and Fig. 2. Determined by flow cytometry.
Determined with a Coulter Counter.
Xdry/Xwet (equation 1), determined with measured buoyant densities and a ρdry of 1.39 g/cm3 for C. oligotrophus and Marinobacter sp. strain T2 and of 1.35 g/cm3 for E. coli.
Dry weight from flow cytometry divided by (Xdry/Xwet) from buoyant density.
Coulter volume × buoyant density × (Xdry/Xwet).
Wet weight divided by buoyant density (equation 3).
From equation 2.
Dry mass measurements.
Samples of bacteria for dry mass analysis represented a variety of species and culture conditions at the time of preservation to generate a range of mean cell sizes for method evaluation. Values for the dry masses of C. oligotrophus, Marinobacter sp. strain T2, and E. coli measured by flow cytometry were generally within 15% those computed from Coulter Counter volume and buoyant density and from Xdry/Xwet (equations 1 and 3). Agreement supported the use of an axial ratio of three in the calculations, which set the shape of the standard curve (37). Buoyant densities measured here were within the range reported for numerous bacterial strains, including E. coli (5, 28).
The mean cell dry mass for additional cultures was computed from Coulter volumes and buoyant densities given in Table 4 (1.07 for C. oligotrophus) and plotted against forward light scatter intensity determined from flow cytometry (Fig. 2). For all of the samples, linear regression analysis between the computed values and cell dry mass determined from forward light scatter intensity gave an r2 of 0.979, with cell dry mass determined by flow cytometry being 20% larger. Error is expected due to changes in cell density with growth conditions and to Coulter Counter volume measurements of C. oligotrophus near the limits of the instrumentation. In addition, the assumption of cell alignment in the aperture of a Coulter Counter in which a sample is not hydrodynamically focused adds to the uncertainty of Coulter Counter volume measurements that are corrected for the axial ratio of these rod-shaped bacteria (27, 32). Measurements from electron micrographs of the organisms gave axial ratios of about three, subject to error from shrinkage during sample preparation (20). The error expected in mass measured by flow cytometry due to variation in axial ratios of two to four was less than 10% (Fig. 1C).
Variation in refractive index.
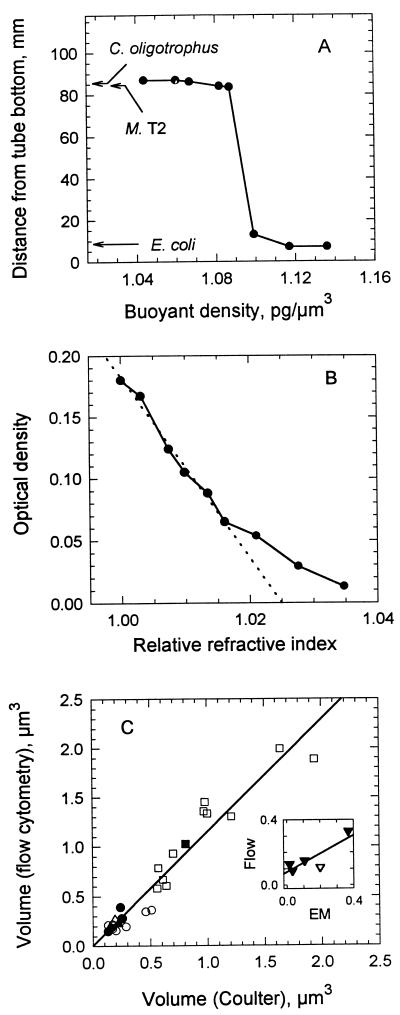
Buoyant density measurements demonstrated large differences in Xdry/Xwet and, therefore, m for the organisms (equations 1 and 2). The results in Fig. 3A show the separation between the marine isolates and E. coli. This distribution was in an 0.85 osM gradient to provide a direct comparison between the organisms. However, data were consistent with band locations of single species in separate iso-osmotic gradients (not shown) that gave Xdry/Xwet values of 14 to 18% for C. oligotrophus and 29% for E. coli. The diluteness of C. oligotrophus was corroborated by optical density (OD) measurements which gave Xdry/Xwet values of 19 to 21% from the proportionality between Xdry and OD0.75 (36), the biovolume (Coulter Counter volume × population) of the two cultures, and Xdry/Xwet values of 27 to 30% reported for E. coli (11, 72). Also, the values of m determined for C. oligotrophus from buoyant density measurements (Table 4) agreed with the measurement of 1.025 in Fig. 3B and were significantly lower than the m value of 1.037 determined here and reported elsewhere (66) for E. coli.
FIG. 3.
Physical properties of test organisms. (A) Buoyant densities of C. oligotrophus, Marinobacter sp. strain T2 (M. T2), and E. coli from their positions (arrows) in a 70:30 gradient of Percoll-seawater medium. Data are for standard density marker beads. (B) m of C. oligotrophus determined by the minimal-absorbance method (solid line). The broken line is the least squares fit to the data for an optical density of >0.06 and yields an m of 1.025. (C) Cell volume computed from dry mass by flow cytometry, buoyant density, and Xdry/Xwet (equation 3) with respect to Coulter Counter volume measured for the three species. The inset shows cell volumes of C. oligotrophus determined by flow cytometry compared with those determined by transmission (▾) and scanning (▿) electron microscopy (EM).
Cell volume.
Measurements of dry mass and buoyant density gave cell volumes (equation 3) which agreed with measurements made by Coulter impedance, as shown in Fig. 3C. These values were 1.14 times Coulter Counter volumes (r2 = 0.936; n = 28). Measurements made by electron microscopy gave volumes only half those obtained from forward light scatter intensity measurements (Fig. 3C, inset), consistent with significant shrinkage due to dehydration during sample preparation.
Biomass in a mixture of species.
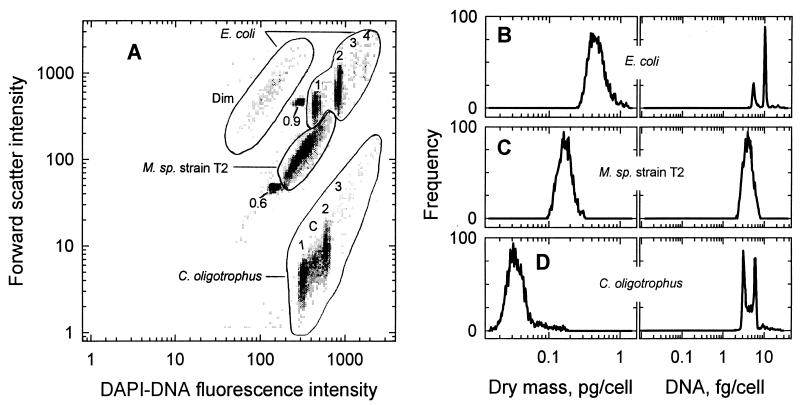
Nine subpopulations were resolved within three major clusters when preserved samples of E. coli, Marinobacter sp. strain T2, and C. oligotrophus were combined and analyzed by flow cytometry (Fig. 4A). Species were identified by comparison with data from analyses of pure cultures. Concentrations in the subpopulations were as low as 104 cells/ml, with a dry weight of 1 ng/ml, compared to a total of 106 cells/ml and 0.3 μg of dry mass/ml for the mixture (Table 5). Coefficients of variation (CV) for forward light scatter intensity were 40 to 55% for the species clusters and 27 to 36% for the subpopulations. Since CV for forward light scatter intensity were only 5% for the internal standard spheres and less than 2% for the spheres used for instrument alignment, distributions for cell mass (Fig. 4B to D) were thought to reflect real variation in the light scatter properties of the cells rather than analytical error. While the concentrations of cells in the mixture were about the same for E. coli and C. oligotrophus, the total biomass of the E. coli culture was 10-fold higher (Table 5), reflecting the higher dry weight of that organism.
FIG. 4.
Cell mass and DNA content of subpopulations in a mixture of three species. (A) Bivariate histogram of forward light scatter versus DAPI-DNA fluorescence intensity for the mixture along with the 0.6- and 0.9-μm-diameter microspheres. Whole numbers give the number of chromosome copies contained by cells in various subpopulations. Dim indicates cells of E. coli showing very low fluorescence, and C indicates cells of C. oligotrophus undergoing chromosome replication. M., Marinobacter. (B, C, and D) Distributions of dry mass and DNA content for E. coli, Marinobacter sp. strain T2, and C. oligotrophus, respectively.
TABLE 5.
Subpopulations in a mixture of three bacterial cultures according to biomass and DNA contenta
| Culture | Subpopulationb | Population (106 cells/ ml) | Cell mass (pg/cell)
|
Biomass (μg/liter)
|
DNA content (fg/cell) | ||
|---|---|---|---|---|---|---|---|
| Dryc | Wetd | Drye | Wetf | ||||
| C. oligotrophus | Total | 0.484 | 0.042 | 0.234 | 20.2 | 113.2 | 5.0 |
| 1 n | 0.191 | 0.027 | 0.151 | 5.2 | 28.8 | 3.2 | |
| 2 n | 0.173 | 0.040 | 0.223 | 7.0 | 38.6 | 5.8 | |
| 3 n | 0.015 | 0.073 | 0.408 | 1.1 | 6.1 | 8.9 | |
| C phase | 0.074 | 0.032 | 0.179 | 2.4 | 13.2 | 4.2 | |
| Marinobacter sp. strain T2 | Total | 0.434 | 0.172 | 0.896 | 74.6 | 388.8 | 4.1 |
| E. coli | Total | 0.492 | 0.497 | 1.74 | 244.5 | 856.1 | 8.7 |
| 1 n | 0.117 | 0.368 | 1.29 | 43.1 | 150.9 | 5.4 | |
| 2 n | 0.263 | 0.488 | 1.71 | 128.3 | 449.7 | 10.2 | |
| 3 n | 0.010 | 0.688 | 2.41 | 6.9 | 24.1 | 14.9 | |
| 4 n | 0.014 | 0.811 | 2.84 | 11.4 | 39.8 | 20.6 | |
| Dim | 0.048 | 0.500 | 1.75 | 24.0 | 84.0 | 1.7 | |
Mixture of C. oligotrophus, Marinobacter sp. strain T2, and E. coli, as shown in Fig. 4.
Total includes all cells from a region; n indicates the number of chromosomes per cell; C phase indicates cells undergoing chromosome replication; Dim indicates cells with very low fluorescence. The sum of values for the subpopulations is >91% the total for all organisms in regions electronically bounded as shown.
From flow cytometry as shown in Fig. 2, with corrections for E. coli in M9 medium.
Dry mass divided by (Xdry/Xwet) (Table 4; 17.9% for C. oligotrophus).
Population density × dry mass per cell.
Population density × wet mass per cell.
The mean cell volume of Marinobacter sp. strain T2 calculated from dry mass (Table 5) and from Xdry/Xwet and buoyant density (Table 4) was 0.172/(0.192 × 1.075), or 0.83 μm3; the Coulter Counter volume determined 11 years earlier, when the culture was preserved, was 0.73 μm3. However, according to equation 1, an increase in buoyant density to 1.17 g/cm3, consistent with the observed 56% decrease in Coulter Counter volume over the duration, gives a volume of 0.77 μm3, within 5% of the initial value. Contrary to reports that apparent populations of DAPI-stained cells decrease exponentially with preservation time (65), populations determined with the Coulter Counter remained unchanged at 2.3 × 109 cells/ml. The data suggest that during extended refrigerated storage of preserved cells, the changes in properties affecting electrical impedance are most likely related to cell shrinkage rather than significant changes in dry matter composition and ρdry.
Over 90% of the cells of each species were within a threefold range in biomass (Fig. 4B to D); the remainder appeared as large particles and extended the range to over 10-fold. Clumps of cells, which have been observed in some substrate-depleted cultures of C. oligotrophus and other bacteria (21, 26), would cause an underestimate of total biomass. However, an examination of data from off-scale signals (data not shown) indicated their insignificance here.
Most subpopulations had narrow distributions of DNA content. CV of only 6 to 9% for DAPI-DNA fluorescence intensity facilitated separation of the nine groups resolved. The predominance of cells containing two chromosomes and the very low incidence of cells with three or four chromosomes in the E. coli culture suggest a doubling time of 40 to 60 min based on cell cycle analysis (18, 59). However, the absence of cells in the C phase of the cell cycle, in which chromosome replication occurs, is consistent with chromosome runout with inhibition of protein synthesis (58). About 15% of C. oligotrophus cells were in the C phase and 70% were about equally apportioned between the B and D phases, respectively, in which chromosome replication has not been initiated and in which replication has been completed but cell division has not occurred. These data are consistent with cell cycle analysis of the organism at its maximal doubling time of 4 h on acetate.
About 10% of the population of E. coli was comprised of “dim” cells with DAPI-DNA fluorescence intensity only one-third that observed for cells containing a single chromosome. However, their light scatter properties were the same as those of the main population (Table 4). Dim cells are commonly observed in samples from natural aquatic systems (15) and may represent organisms undergoing nucleic acid degradation due to stress (69). The low fluorescence intensity and the lack of structure in the DNA profile of Marinobacter sp. strain T2 (Fig. 4C) were very likely a result of nucleic acid degradation during the extended duration of sample storage, since DNA profiles of the organisms observed within 6 months of preservation were similar to those seen here for C. oligotrophus (unpublished data).
Estimated buoyant densities of other bacteria analyzed by flow cytometry.
When the buoyant density of an organism was not measured, it was calculated from the dry mass determined by flow cytometry and Coulter Counter volume (equations 1 and 3) and taken to be the value which gave the smallest error in computed mass and volume. For P. diminuta and marine isolate RB95-4, values of 1.05 and 1.055 g/cm3, respectively, gave that agreement for each parameter to within 5% and appeared reasonable, since they were not very different from the buoyant density determined for C. oligotrophus.
DISCUSSION
Our data demonstrate that the biomass of small bacteria at low concentrations in a mixture can be determined from measurements of forward light scatter intensity by flow cytometry. Constraints imposed by Rayleigh-Gans theory on the theoretical curve (37) give an upper limit for the method of 1.2 pg (dry weight) per cell (about 6 μm3 in size) for bacteria with an axial ratio of three, assuming that m is 1.03 and that elongated cells are aligned in the flow stream, as the data suggest. The lower limit is set by instrument capability. For the Cytofluorograf IIS, it is about 0.005 pg (0.025 μm3) per cell at the highest gain setting that shows no evidence of photomultiplier saturation. Although the dynamic range of the amplifiers gives only about 2 decades of cell mass (Fig. 4B, C, and D), the data show that the full theoretical range can be accommodated with an error of only 5% when internal standards for signal normalization are used along with gain changes. Concordance between population counts determined by flow cytometry and those determined with a Coulter Counter allowed total biomass for each subpopulation to be computed on the basis of population, cell mass, and sample volume. Since 100 cells in a 0.1-ml sample can be resolved (14), biomass as low as 0.5 pg can be measured by this method.
Agreement between dry mass measurements obtained from forward light scatter intensity and values computed from Coulter Counter volume and buoyant density (ρcell) measurements (Fig. 2) depended on corrections of m for medium composition and species-specific effects of formaldehyde absorption and on estimates of the density of dry matter. The value of ρdry, 1.39 g/cm3, for C. oligotrophus reflects the greater contribution of high-density nucleic acids to the dry matter of small cells and is within the range reported by others for cultures of marine bacteria (57). The influence of growth conditions on the composition of dry matter (17) and ρdry was likely to have contributed to the differences between dry mass determined from flow cytometry and values computed from Coulter Counter volume and buoyant density. Buoyant density can vary with growth conditions (72) and is affected by cellular inclusions (4), capsules and gas vacuoles (28), and medium osmolarity (5) but not by growth rate, at least for E. coli in continuous cultures (41). Although cells were harvested at different stages of the growth cycle, intraspecies variations in density were expected to be modest in this study, since there was constancy in medium preparation and culture incubation conditions. Changes in the specific refraction increment factor based on variations in the compositions of bacteria were less than 1% and so should not have significantly influenced ρdry. Error due to variations in axial ratio from 2:1 to 6:1 among the rod-shaped bacteria was less than 10%, based on Rayleigh-Gans calculations (37).
A dry weight content of 15 to 20% determined for C. oligotrophus contrasts with values of 50 to 60% reported for marine bacteria (10, 43, 57). However, due to small cell size, cell shrinkage during preparation for microscopy, the presence of debris in natural aquatic samples, and the large error seen in measurements of standard microspheres observed here, there is general agreement that reliable data for the cell volumes needed for such assessments are not easily obtained (10, 30, 45). Agreement between the dry weight content determined here for E. coli and values in the literature and the differences between the buoyant densities observed here for E. coli and C. oligotrophus support the dilute nature of the marine isolate.
Greater precision in dry mass measurements from forward light scatter can be obtained by developing standard curves for specific bacterial strains. In this study, we used C. oligotrophus for standardization. Since there was little variation in the axial ratios of the organisms, the shape of their standard curves remained the same. K for E. coli was adjusted to accommodate differences in the absorption of formaldehyde, ρdry, and the growth medium. Reduced precision is to be expected in an analysis of undescribed organisms. Based on differences in cell shape, an overestimate of 30% for dry mass could occur if the standard curve used here were applied to spherical cells with an equivalent mean cell volume. For cocci larger than 1.5 μm3, it would be useful to compute a new curve with an axial ratio of one. The small size of bacteria in aquatic systems minimizes error due to differences in cell shape (Fig. 1C).
If buoyant density is known and constant, biomass in terms of wet weight can be obtained from dry mass by flow cytometry and Xdry/Xwet or, as previously suggested (37), a standard curve calibrated according to Coulter Counter volume can give organism size for computing wet weight from cell volume and density.
The large difference between m values for bacteria and the latex microspheres often used as size standards as well as internal standards for comparison among samples is noteworthy. Due to much greater forward light scatter intensity from microspheres (Fig. 1A) and inconstancy in the dependence of light scatter intensity on the size of large particles with a high refractive index (Fig. 1B, inset), the use of latex particles as size standards for biological cells may be problematic, as suggested by others (7, 20).
The method presented here is an improvement over other flow cytometric methods for determining bacterial biomass (3, 20, 50) because it is more general, due to its foundation in light scatter theory, it accounts for differences in cell shape and relative refractive index (37), and it extends analysis to very small bacteria and to mixed populations. It is particularly valuable when biomass is low and cell numbers are limited, as in experiments for quantifying viability (16) and evaluating the nutritional requirements of new species isolated by extinction culture techniques (54). In combination with DNA analyses, biomass measurements for resolved subpopulations offer improved evaluation of bacterial growth according to cell cycle theory (18, 59). This ability to quantify light scatter intensity in terms of biomass enables inquiry toward an improved understanding of microbial processes with unprecedented detail.
ACKNOWLEDGMENTS
We thank Knut Stamnes and Yong-Xiang Hu of the Geophysical Institute, University of Alaska Fairbanks, for helpful discussions and the Mie calculations; Pham X. Quang of the Mathematics Department, University of Alaska Fairbanks, for help with formulating the standard curve; and Michael Dowling of the Mineral Industry Research Laboratory for CHN analyses.
This work was supported by grants from the Biological Oceanography and Metabolic Biochemistry sections of the National Science Foundation.
REFERENCES
- 1.Ackleson S G, Spinrad R W. Size and refractive index of individual marine particulates: a flow cytometric approach. Appl Optics. 1988;27:1270–1277. doi: 10.1364/AO.27.001270. [DOI] [PubMed] [Google Scholar]
- 2.Åkerlund T, Nordström K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allman R, Hann A C, Phillips A P, Martin K L, Lloyd D. Growth of Azotobacter vinelandii with correlation of Coulter cell size, flow cytometric parameters and ultrastructure. Cytometry. 1990;11:822–831. doi: 10.1002/cyto.990110708. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin W W, Kirkish M A, Koch A L. A change in a single gene of Salmonella typhimurium can dramatically change its buoyant density. J Bacteriol. 1994;176:5001–5004. doi: 10.1128/jb.176.16.5001-5004.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin W W, Myer R, Powell N, Anderson E, Koch A L. Buoyant density of Escherichia coli is determined solely by the osmolarity of the culture medium. Arch Microbiol. 1995;164:155–157. doi: 10.1007/s002030050248. [DOI] [PubMed] [Google Scholar]
- 6.Bernander R, Åkerlund T, Nordström K. Inhibition and restart of initiation of chromosome replication: effects on exponentially growing Escherichia coli cells. J Bacteriol. 1995;177:1670–1682. doi: 10.1128/jb.177.7.1670-1682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder B J, Chisholm S W, Olson R J, Frankel S L, Worden A Z. Dynamics of picophytoplankton, ultraphytoplankton and bacteria in the central equatorial Pacific. Deep-Sea Res. 1996;43:907–931. [Google Scholar]
- 8.Bohren C F, Huffman D R. Absorption and scattering of light by small particles. New York, N.Y: John Wiley & Sons, Inc.; 1983. [Google Scholar]
- 9.Boye E, Steen H B, Skarstad K. Flow cytometry of bacteria: a promising tool in experimental and clinical microbiology. J Gen Microbiol. 1983;129:973–980. doi: 10.1099/00221287-129-4-973. [DOI] [PubMed] [Google Scholar]
- 10.Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985;49:1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratbak G, Dundas I. Bacterial dry matter content and biomass estimations. Appl Environ Microbiol. 1984;48:755–757. doi: 10.1128/aem.48.4.755-757.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Button D K. Kinetics of nutrient-limited transport and microbial growth. Microbiol Rev. 1985;49:270–297. doi: 10.1128/mr.49.3.270-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Button D K, Robertson B R. Kinetics of bacterial processes in natural aquatic systems based on biomass as determined by high-resolution flow cytometry. Cytometry. 1989;10:558–563. doi: 10.1002/cyto.990100511. [DOI] [PubMed] [Google Scholar]
- 14.Button D K, Robertson B R. Use of high-resolution flow cytometry to determine the activity and distribution of aquatic bacteria. In: Kemp P F, Sherr B F, Scherr E B, Cole J J, editors. Handbook of methods in aquatic microbial ecology. Ann Arbor, Mich: Lewis Publishers; 1993. pp. 163–173. [Google Scholar]
- 15.Button D K, Robertson B R, Jüttner F. Microflora of a subalpine lake: bacterial populations, size, and DNA distributions, and their dependence on phosphate. FEMS Microbiol Ecol. 1996;21:87–101. [Google Scholar]
- 16.Button D K, Schut F, Quang P, Martin R, Robertson B R. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchward G, Bremer H. Macromolecular composition of bacteria. J Theor Biol. 1982;94:651–670. doi: 10.1016/0022-5193(82)90305-8. [DOI] [PubMed] [Google Scholar]
- 18.Cooper S. Bacterial growth and division: biochemistry and regulation of prokaryotic and eukaryotic division cycles. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 19.Daneo-Moore L, Dicker D, Higgins M L. Structure of the nucleoid in cells of Streptococcus faecalis. J Bacteriol. 1980;141:928–937. doi: 10.1128/jb.141.2.928-937.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey H M, Davey C L, Kell D B. On the determination of the size of microbial cells using flow cytometry. In: Lloyd D, editor. Flow cytometry in microbiology. New York, N.Y: Springer-Verlag; 1993. pp. 49–65. [Google Scholar]
- 21.Dawes E A. Starvation, survival and energy reserves. In: Fletcher M, Floodgate G D, editors. Bacteria in their natural environments. New York, N.Y: Academic Press, Inc.; 1985. pp. 43–79. [Google Scholar]
- 22.DeLeo P C, Baveye P. Enumeration and biomass estimation of bacteria in aquifer microcosm studies by flow cytometry. Appl Environ Microbiol. 1996;62:4580–4586. doi: 10.1128/aem.62.12.4580-4586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenstadt E, Carlton B C, Brown B J. Gene mutation. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 297–316. [Google Scholar]
- 24.Fox C H, Johnson F B, Whiting J, Roller P P. Formaldehyde fixation. J Histochem Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 25.Gauthier M J, LaFay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Bertrand J-C. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 26.Givskov M, Eberl L, Møller S, Poulsen L K, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grover N B, Naaman J, Ben-Sasson S, Doljanski F. Electrical sizing of particles in suspensions. I. Theory. Biophys J. 1969;9:1398–1414. doi: 10.1016/S0006-3495(69)86461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerrero R, Pedros-Alio C, Schmidt T M, Mas J. A survey of buoyant density of microorganisms in pure cultures and natural samples. Microbiologia (Madrid) 1985;1:53–65. [PubMed] [Google Scholar]
- 29.Hayat M A. Principles and techniques of electron microscopy. Biological applications. Vol. 1. Baltimore, Md: University Park Press; 1981. Fixation; pp. 1–106. [Google Scholar]
- 30.Heldal M, Norland S, Tumyr O. X-ray microanalytic method for measurement of dry matter and elemental content of individual bacteria. Appl Environ Microbiol. 1985;50:1251–1257. doi: 10.1128/aem.50.5.1251-1257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbert R A. Methods for enumerating microorganisms and determining biomass in natural environments. Methods Microbiol. 1990;22:1–39. [Google Scholar]
- 32.Hurley J. Sizing particles with a Coulter Counter. Biophys J. 1970;10:74–79. doi: 10.1016/S0006-3495(70)86286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kachel V, Fellner-Feldegg H, Menke E. Hydrodynamic properties of flow cytometry instruments. In: Melamed M R, Lindmo T, Mendelsohn M L, editors. Flow cytometry and sorting. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 27–44. [Google Scholar]
- 34.Kerker M. The scattering of light and other electromagnetic radiation. New York, N.Y: Academic Press, Inc.; 1969. [Google Scholar]
- 35.Koch A L. Some calculations on the turbidity of mitochondria and bacteria. Biochim Biophys Acta. 1961;51:429–441. doi: 10.1016/0006-3002(61)90599-6. [DOI] [PubMed] [Google Scholar]
- 36.Koch A L, Ehrenfeld E. The size and shape of bacteria by light scattering measurements. Biochim Biophys Acta. 1968;165:262–273. doi: 10.1016/0304-4165(68)90054-8. [DOI] [PubMed] [Google Scholar]
- 37.Koch A L, Robertson B R, Button D K. Deduction of the cell volume and mass from forward scatter intensity of bacteria analyzed by flow cytometry. J Microbiol Methods. 1996;27:49–61. [Google Scholar]
- 38.Kohara Y, Akiyame K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 39.Krawiec S, Riley M. Organization of the bacterial chromosome. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubitschek H E. Counting and sizing micro-organisms with the Coulter Counter. Methods Microbiol. 1969;1:593–610. [Google Scholar]
- 41.Kubitschek H E, Baldwin W W, Graetzer R. Buoyant density constancy during the cell cycle of Escherichia coli. J Bacteriol. 1983;155:1027–1032. doi: 10.1128/jb.155.3.1027-1032.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubitschek H E, Friske J A. Determination of bacterial cell volume with the Coulter Counter. J Bacteriol. 1986;168:1466–1467. doi: 10.1128/jb.168.3.1466-1467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Fuhrman J A. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morel A, Yu-Hwan A. Optical efficiency factors of free-living marine bacteria: influence of bacterioplankton upon the optical properties and particulate organic carbon in oceanic waters. J Mar Res. 1990;48:145–175. [Google Scholar]
- 45.Norland S M, Heldal M, Tumyr O. On the relation between dry matter and volume of bacteria. Microb Ecol. 1987;31:95–101. doi: 10.1007/BF02011246. [DOI] [PubMed] [Google Scholar]
- 46.Pharmacia Fine Chemicals. Percoll. Methodology and applications. Density marker beads for calibration of gradients of Percoll. Uppsala. Sweden: Pharmacia Fine Chemicals; 1980. [Google Scholar]
- 47.Polysciences, Inc. Polybead microparticles. Data sheet 238. Warrington, Pa: Polysciences, Inc.; 1987. [Google Scholar]
- 48.Rivkin R B, Legendre L, Deibel D, Tremblay J-E, Klein B, Crocker K, Roy S, Silverberg N, Lovejoy C, Mesplé F, Romero N, Anderson M R, Matthews P, Savenkoff C, Vézina A, Therriault J-C, Wesson J, Bérubé C, Ingram R G. Vertical flux of biogenic carbon in the ocean: is there food web control? Science. 1996;272:1163–1166. doi: 10.1126/science.272.5265.1163. [DOI] [PubMed] [Google Scholar]
- 49.Robertson B R, Button D K. Toluene induction and uptake kinetics and their inclusion in the specific-affinity relationship for describing rates of hydrocarbon metabolism. Appl Environ Microbiol. 1987;53:2193–2205. doi: 10.1128/aem.53.9.2193-2205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson B R, Button D K. Characterizing aquatic bacteria according to population, cell size and apparent DNA content by flow cytometry. Cytometry. 1989;10:70–76. doi: 10.1002/cyto.990100112. [DOI] [PubMed] [Google Scholar]
- 51.Rudd K E, Miller W, Ostell J, Benson D A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990;18:313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salzman G C, Singham S B, Johnston R G, Bohren C F. Light scattering and cytometry. In: Melamed M R, Lindmo T, Mendelsohn M L, editors. Flow cytometry and sorting. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 81–107. [Google Scholar]
- 53.Schmid I, Schmid P, Giorgi J V. Conversion of logarithmic channel numbers into relative linear fluorescence intensity. Cytometry. 1988;9:533–538. doi: 10.1002/cyto.990090605. [DOI] [PubMed] [Google Scholar]
- 54.Schut F, De Vries E, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sieracki M E, Johnson P W, Seiburth J M. Detection, enumeration, and sizing of planktonic bacteria by image-analyzed epifluorescence microscopy. Appl Environ Microbiol. 1985;49:799–810. doi: 10.1128/aem.49.4.799-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sieracki M E, Viles C L, Webb K L. Algorithm to estimate cell biovolume using image analyzed microscopy. Cytometry. 1989;10:551–557. doi: 10.1002/cyto.990100510. [DOI] [PubMed] [Google Scholar]
- 57.Simon M, Azam F. Protein content and protein synthesis rates of planktonic bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 58.Skarstad K, Bernander R, Wold S, Steen H B, Boye E. Cell cycle analysis of microorganisms. In: Al-Rubeai M, Emery A N, editors. Flow cytometry applications in cell culture. New York, N.Y: Marcel Dekker, Inc.; 1996. pp. 241–255. [Google Scholar]
- 59.Skarstad K, Steen H B, Boye E. Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J Bacteriol. 1983;154:656–662. doi: 10.1128/jb.154.2.656-662.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sober H A, editor. Handbook of biochemistry: selected data for molecular biology. Cleveland, Ohio: The Chemical Rubber Co.; 1968. [Google Scholar]
- 61.Sonnleitner B, Locher G, Fiechter A. Biomass determination. J Biotechnol. 1992;25:5–22. doi: 10.1016/0168-1656(92)90107-k. [DOI] [PubMed] [Google Scholar]
- 62.Steen H B, Boye E. Escherichia coli growth studied by dual-parameter flow cytophotometry. J Bacteriol. 1980;145:1091–1094. doi: 10.1128/jb.145.2.1091-1094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsay S-C, Stephens G L. A physical/optical model for atmospheric aerosols with application to visibility problems. Fort Collins, Colo: Colorado State University; 1990. [Google Scholar]
- 64.Tuomi P, Torsvik T, Heldal M, Bratbak G. Bacterial population dynamics in a meromictic lake. Appl Environ Microbiol. 1997;63:2181–2188. doi: 10.1128/aem.63.6.2181-2188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turley C M, Hughes D J. Effects of storage on direct estimates of bacterial numbers of preserved seawater samples. Deep-Sea Res. 1992;39:375–394. [Google Scholar]
- 66.Valkenburg J A C, Woldringh C L. Phase separation between nucleoid and cytoplasm in Escherichia coli as defined by immersive refractometry. J Bacteriol. 1984;160:1151–1157. doi: 10.1128/jb.160.3.1151-1157.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.VanDilla M A, Langlois R G, Pinkel D, Yajko D, Hadley W K. Bacterial characterization by flow cytometry. Science. 1983;220:620–621. doi: 10.1126/science.6188215. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Lau P C, Button D K. A marine oligobacterium harboring genes known to be part of aromatic hydrocarbon degradation pathways of soil pseudomonads. Appl Environ Microbiol. 1996;62:2169–2173. doi: 10.1128/aem.62.6.2169-2173.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weichart D, McDougald D, Jacobs D, Kjelleberg S. In situ analysis of nucleic acids in cold-induced nonculturable Vibrio vulnificus. Appl Environ Microbiol. 1997;63:2754–2758. doi: 10.1128/aem.63.7.2754-2758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson W D, Tanious F A, Barton H J, Jones R L, Fox K, Wydra R L, Strekowski L. DNA sequence dependent binding modes of 4′,6-diamidino-2-phenylindole (DAPI) Biochemistry. 1990;29:8452–8461. doi: 10.1021/bi00488a036. [DOI] [PubMed] [Google Scholar]
- 71.Winkler J, Timmis K N, Snyder R A. Tracking the response of Burkholderia cepacia G4 5223-PR1 in aquifer microcosms. Appl Environ Microbiol. 1995;61:448–455. doi: 10.1128/aem.61.2.448-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woldringh C L, Binnerts J S, Mans A. Variation in Escherichia coli buoyant density measured in Percoll gradients. J Bacteriol. 1981;148:58–63. doi: 10.1128/jb.148.1.58-63.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]