Abstract
Alzheimer’s disease (AD) is a devastative disease, the 1st most frequent neurodegenerative disease worldwide. Its prevalence is increasing and early detection methods as well as potential genomic based therapeutics are urgently needed. Objectives: To better characterize recent seq studies of AD and site recent relevant literature. Using single-cell RNA sequencing, the characteristics of neuronal cell populations in Alzheimer’s disease (AD) have not been completely elucidated. Methods: We conducted a dynamic and longitudinal bibliometric analysis to investigate existing studies on Single-cell RNA sequencing analysis and Alzheimer’s Disease and identify data gaps and possible new research avenues. Results: All AD papers concentrating on Single-cell RNA sequencing analysis were found using the search terms “Alzheimer’s Disease”, and “Single-cell RNA sequencing analysis” in the PubMed/MEDLINE database. Only English publications published between 2015 and 2023 were chosen using filters. Conclusions: Original English-language research publications disclosing Single-cell RNA sequencing analysis and Alzheimer’s Disease were examined for inclusion. Two sets of independent reviewers discovered and extracted pertinent data. The bibliometric study was carried out using the R software packages Bibliometrix and Biblioshiny. The narrowed search yielded 158 publications, all published between 2015 and 2023. Yet, after applying filters and considering the inclusion requirements, the search results comprise just 51 original articles out of 158 articles. There were 107 articles eliminated. The importance of the discovery of Single-cell RNA sequencing analysis and Alzheimer’s Disease a decade ago only grows with time. Our results have important implications for future studies of AD and may help researchers across the world better understand the global context of the Single-cell RNA sequencing analysis and Alzheimer’s Disease link. This study puts emphasis on the critical need for more diverse participant demographics in Alzheimer’s disease investigations.
Keywords: Alzheimer’s disease, single-cell RNA sequencing analysis, bibliometric analysis
Introduction
Alois Alzheimer initially defined Alzheimer’s disease (AD) in the early twentieth century [1,2]. AD is a neurological illness characterised by gradual cognitive loss. Every year, a rising number of people are diagnosed with Alzheimer’s disease (AD), making it the most common form of neurodegeneration globally. Women are disproportionately affected by dementia, which is estimated to impact 44 million individuals globally today.
There is a favourable correlation between increased age and the number of persons diagnosed with Alzheimer’s disease (AD), which affects over 50 million people worldwide. It is important to keep in mind that most Alzheimer’s disease patients have a dismal outlook, with a median survival span of just 5-10 years owing to a lack of early diagnosis and therapy that is successful [3].
There is an urgent need for effective therapeutic therapy to halt or abolish illness development since the disease is now the sixth largest cause of mortality in the United States. Even though various FDA-approved pharmacological therapies, including as donepezil, rivastigmine, galanthamine, and other medicines, have been utilised to slow the development of AD in the last decades [4,5], the heterogeneity of AD patients restricts the therapeutic effectiveness of these drugs [6]. Different molecular properties have been identified as the primary driver of AD heterogeneity, which is also linked to disparities in clinical outcomes [7,8]. Nonetheless, the potential molecular mechanisms underlying Alzheimer’s disease heterogeneity are largely unknown. To guide the individualised therapy of AD patients, it is vital to understand the heterogeneity of the AD and precisely differentiate the molecular features of each patient.
Biochemical lesions of amyloid plaques and Tau tangles characterise the illness at the molecular level. Rare mutations in amyloid protein, amyloid precursor protein, presenilin 1 and 2, ADP-ribosyl adenosine monophosphate-activated metalloproteinase 10, and metalloproteinase 17 have been demonstrated to promote amyloid-beta build up and are sufficient to produce complete biochemical and morphological markers of AD. As stated by Lyketsos et al. [9], the primary pathogenic hallmarks of AD are neuronal death and synaptic degradation. Even though neurons contain defences against inflammatory assaults and neurological illnesses, some of these cells are flawed in AD. The pathological hallmarks of AD includes the formation of a neurofibrillary tangle inside the cell by hyperphosphorylated tau protein and the deposition of amyloid plaques outside the cell by these faulty neurons [10].
It is believed that the course of AD requires, in addition to the action of amyloid beta, a wide variety of neuronal cell types and intracellular mechanisms. It has been shown that non-neuronal genes, such as microglia (MG) cell markers, play a significant part in the pathophysiology of AD. This is best demonstrated by Genome Wide Association Studies (GWAS), in which linked variations have primarily glial cell limited expression. The results of a recent investigation of single-cell RNA-Sequencing (scRNA-Seq) datasets derived from the human AD post mortem and controls brain samples using Chromium machine, further reveal a link between microglia and astrocyte circuit disruptions. The analysis included mapping to the human genome, statistical and classification methods. Even though animal models of AD and MG provide more evidence that these cells are involved in AD-related pathways, one third of suspected Alzheimer’s risk genes do not have appropriate mouse orthologs [11]. In addition, the presence of oligomeric amyloid - in human brains, as opposed to the brains of mice without MG, causes a biological reaction [12].
Methodology
The single cell data [13] was produced from 2 AD and 2 post mortem control fronal cortex (FCTX) brain tissues using Chromium machine. iCell8 software was sued to map the data to the human genome. About 24,000 single cell nuclei were computationally analysed.
Data sources
All AD publications focusing on Single-cell RNA sequencing analysis of AD genetics were retrieved from the PubMed/MEDLINE database using the search strings “Alzheimer’s Disease” and “Single-cell RNA sequencing analysis”. Using filters and the following inclusion criteria: 1) only English articles published between 2015 and 2023 were selected, 2) documents type include case report and journal article revealing Single-cell RNA sequencing analysis and AD only, 3) core source of Bradford’s law zone. The metadata were downloaded and saved as .csv format.
Data analysis
Bibliometrix (http://www.bibliometrix.org) is an R based package for quantitative bibliometric analysis. Applied together with this package is a web-interface of Bibliometrix, known as Biblioshiny (https://www.bibliometrix.org/home/index.php/layout/biblioshiny). The Bibliometrix package was installed and loaded through R Studio Version 2022.07.2+576 (RStudio, PBC, Boston, MA, USA). Consequently, the Biblioshiny package was loaded by digited “biblioshiny()” command in the R console. The detailed methods and algorithms for both packages were explained elsewhere [14,15]. The downloaded metadata were uploaded on Biblioshiny interface to be analyzed. Analysis was performed on three factors: general outputs (publication trends and collaborations), publication sources, and authors keywords. In the case of keywords having similar meanings, these words were merged prior to the analysis. Visual outputs from the analysis were downloaded and saved as .png format. There were two rounds of independent reviewers that found and retrieved relevant data. The Bibliometrix and Biblioshiny packages from R software were used to conduct the bibliometric analysis. Bradford’s Law of Scattering is a law pertaining to declining returns and dispersion. For each single topic or subject area, the top third (Zone 1 or core) reflects the journals that are most often referenced in the relevant literature and are hence most likely to be of interest to researchers in that field. The middle third (Zone 2) consists of journals with an average number of citations, and the bottom third (Zone 3 or tail) consists of publications that are seldom referenced and considered to be of minimal significance to the topic. Thus, we used core source of Bradford’s law zone in our search criteria to focus on the most cited articles that are of great interest to researchers.
Results
Analysis of publication outputs, growth trends, and scientific collaborations
As summarized in Tables 1 and 2, the initial search identified 158 articles, all of which were published between 2015 and 2023. However, after filter application and considering the inclusions criteria, the search results include only 51 original articles out of 158 articles. One hundred and seven articles were omitted. The primary grounds for elimination were as follows: the studies were not published in English and based on their titles and abstracts, none of these studies were original article with original data. In addition, no Single-cell RNA sequencing analysis was conducted, and the initial findings were not publicized.
Table 1.
Main information about data
| Description | Results |
|---|---|
| MAIN INFORMATION ABOUT DATA | |
| Timespan | 2015:2022 |
| Sources (Journals, Books, etc.) | 11 |
| Documents | 51 |
| Annual Growth Rate % | 44.26 |
| Document Average Age | 2.67 |
| Average citations per doc | 0 |
| References | 1 |
| DOCUMENT CONTENTS | |
| Keywords Plus (ID) | 208 |
| Author’s Keywords (DE) | 160 |
| AUTHORS | |
| Authors | 540 |
| Authors of single-authored docs | 0 |
| AUTHORS COLLABORATION | |
| Single-authored docs | 0 |
| Co-Authors per Doc | 11.9 |
| International co-authorships % | 0 |
| DOCUMENT TYPES | |
| Journal article | 51 |
Table 2.
Characteristics of the studies according to main inclusion criteria
| AUTHERS | TITLE | JOURNAL | YEAR | DI | |
|---|---|---|---|---|---|
| 1 | LAI et al. | Integration Of Bulk RNA Sequencing And Single-Cell Analysis Reveals A Global Landscape Of DNA Damage Response In The Immune Environment Of Alzheimer’s. | Frontiers In Immunology | 2023 | 10.3389/fimmu.2023.1115202 |
| 2 | PARK et al. | Single-Cell RNA-Sequencing Identifies Disease-Associated Oligodendrocytes In Male App Nl-G-F And 5XFAD Mice. | Nature Communications | 2023 | 10.1038/s41467-023-36519-8 |
| 3 | TAGUCHI and TURKI | A Tensor Decomposition-Based Integrated Analysis Applicable To Multiple Gene Expression Profiles Without Sample Matching. | Scientific Reports | 2022 | 10.1038/s41598-022-25524-4 |
| 4 | QI et al. | Alzheimer’s Disease Alters The Transcriptomic Profile Of Natural Killer Cells At Single-Cell Resolution. | Frontiers In Immunology | 2022 | 10.3389/fimmu.2022.1004885 |
| 5 | LI et al. | Identification Of Diagnostic Genes For Both Alzheimer’s Disease And Metabolic Syndrome By The Machine Learning Algorithm. | Frontiers In Immunology | 2022 | 10.3389/fimmu.2022.1037318 |
| 6 | BLANCHARD et al. | APOE4 Impairs Myelination Via Cholesterol Dysregulation In Oligodendrocytes. | Nature | 2022 | 10.1038/s41586-022-05439-w |
| 7 | HA et al. | Neurotoxicity Of Diesel Exhaust Extracts In Zebrafish And Its Implications For Neurodegenerative Disease. | Scientific Reports | 2022 | 10.1038/s41598-022-23485-2 |
| 8 | WANG et al. | Integrative Cross-Species Analysis Of Gabaergic Neuron Cell Types And Their Functions In Alzheimer’s Disease. | Scientific Reports | 2022 | 10.1038/s41598-022-21496-7 |
| 9 | FU et al. | Single-Nucleus RNA Sequencing Reveals The Shared Mechanisms Inducing Cognitive Impairment Between COVID-19 And Alzheimer’s Disease. | Frontiers In Immunology | 2022 | 10.3389/fimmu.2022.967356 |
| 10 | HEMONNOT GIRARD et al. | Comparative Analysis Of Transcriptome Remodeling In Plaque-Associated And Plaque-Distant Microglia During Amyloid-β Pathology Progression In Mice. | Journal Of Neuroinflammation | 2022 | 10.1186/s12974-022-02581-0 |
| 11 | SHI et al. | Regulating microglial miR-155 transcriptional phenotype alleviates Alzheimer’s-induced retinal vasculopathy by limiting Clec7a/Galectin-3+ neurodegenerative microglia. | Acta Neuropathologica Communications | 2022 | 10.1186/s40478-022-01439-z |
| 12 | ZHAO et al. | Accelerated Aging-Related Transcriptome Alterations In Neurovascular Unit Cells In The Brain Of Alzheimer’s Disease. | Frontiers In Aging Neuroscience | 2022 | 10.3389/fnagi.2022.949074 |
| 13 | PANDEY et al. | Disease-Associated Oligodendrocyte Responses Across Neurodegenerative Diseases. | Cell Reports | 2022 | 10.1016/j.celrep.2022.111189 |
| 14 | ALI et al. | Single-Cell Transcriptional Profiling And Gene Regulatory Network Modeling In Tg2576 Mice Reveal Gender-Dependent Molecular Features Preceding Alzheimer-Like Pathologies. | Molecular Neurobiology | 2022 | 10.1007/s12035-022-02985-2 |
| 15 | MIEDEMA et al. | Distinct Cell Type-Specific Protein Signatures In GRN And MAPT Genetic Subtypes Of Frontotemporal Dementia. | Acta Neuropathologica Communications | 2022 | 10.1186/s40478-022-01387-8 |
| 16 | ZHOU et al. | Molecular Landscapes Of Human Hippocampal Immature Neurons Across Lifespan. | Nature | 2022 | 10.1038/s41586-022-04912-w |
| 17 | KARAAHMET et al. | Repopulated Microglia Induce Expression Of Cxcl13 With Differential Changes In Tau Phosphorylation But Do Not Impact Amyloid Pathology. | Journal Of Neuroinflammation | 2022 | 10.1186/s12974-022-02532-9 |
| 18 | NTTI et al. | Microglial Amyloid Beta Clearance Is Driven By PIEZO1 Channels. | Journal Of Neuroinflammation | 2022 | 10.1186/s12974-022-02486-y |
| 19 | CHEN et al. | Exercise Modifies The Transcriptional Regulatory Features Of Monocytes In Alzheimer’s Patients: A Multi-Omics Integration Analysis Based On Single Cell Technology. | Frontiers In Aging Neuroscience | 2022 | 10.3389/fnagi.2022.881488 |
| 20 | BELONWU et al. | Bioinformatics Analysis Of Publicly Available Single-Nuclei Transcriptomics Alzheimer’s Disease Datasets Reveals APOE Genotype-Specific Changes Across Cell Types In Two Brain Regions. | Frontiers In Aging Neuroscience | 2022 | 10.3389/fnagi.2022.749991 |
| 21 | ZHOU et al. | Discovery Of Novel Drug Candidates For Alzheimer’s Disease By Molecular Network Modeling. | Frontiers In Aging Neuroscience | 2022 | 10.3389/fnagi.2022.850217 |
| 22 | WANG et al. | Guidelines For Bioinformatics Of Single-Cell Sequencing Data Analysis In Alzheimer’s Disease: Review, Recommendation, Implementation And Application. | Molecular Neurodegeneration | 2022 | 10.1186/s13024-022-00517-z |
| 23 | TSAI et al. | PLCG2 Is Associated With The Inflammatory Response And Is Induced By Amyloid Plaques In Alzheimer’s Disease. | Genome Medicine | 2022 | 10.1186/s13073-022-01022-0 |
| 24 | JOHNSON et al. | Diagnostic Evidence GAuge Of Single Cells (DEGAS): A Flexible Deep Transfer Learning Framework For Prioritizing Cells In Relation To Disease. | Genome Medicine | 2022 | 10.1186/s13073-022-01012-2 |
| 25 | CAKIR et al. | Expression Of The Transcription Factor PU.1 Induces The Generation Of Microglia-Like Cells In Human Cortical Organoids. | Nature Communications | 2022 | 10.1038/s41467-022-28043-y |
| 26 | SHAO et al. | Characterization of Alzheimer’s Disease-Associated Excitatory Neurons via Single-Cell RNA Sequencing Analysis. | Frontiers In Aging Neuroscience | 2021 | 10.3389/fnagi.2021.742176 |
| 27 | LU et al. | Expression Of Immune Related Genes And Possible Regulatory Mechanisms In Alzheimer’s Disease. | Frontiers In Immunology | 2021 | 10.3389/fimmu.2021.768966 |
| 28 | BELONWU et al. | Sex-Stratified Single-Cell RNA-Seq Analysis Identifies Sex-Specific And Cell Type-Specific Transcriptional Responses In Alzheimer’s Disease Across Two Brain Regions. | Molecular Neurobiology | 2022 | 10.1007/s12035-021-02591-8 |
| 29 | JIAN et al. | Microglia Mediate The Occurrence And Development Of Alzheimer’s Disease Through Ligand-Receptor Axis Communication. | Frontiers In Aging Neuroscience | 2021 | 10.3389/fnagi.2021.731180 |
| 30 | CHOI et al. | Hippocampal Glucose Uptake As A Surrogate Of Metabolic Change Of Microglia In Alzheimer’s Disease. | Journal Of Neuroinflammation | 2021 | 10.1186/s12974-021-02244-6 |
| 31 | XU and JIA | Single-Cell RNA Sequencing Of Peripheral Blood Reveals Immune Cell Signatures In Alzheimer’s Disease. | Frontiers In Immunology | 2021 | 10.3389/fimmu.2021.645666 |
| 32 | JONES et al. | Modest Changes In Spi1 Dosage Reveal The Potential For Altered Microglial Function As Seen In Alzheimer’s Disease. | Scientific Reports | 2021 | 10.1038/s41598-021-94324-z |
| 33 | LODDER et al. | CSF1R Inhibition Rescues Tau Pathology And Neurodegeneration In An A/T/N Model With Combined AD Pathologies, While Preserving Plaque Associated Microglia. | Acta Neuropathologica Communications | 2021 | 10.1186/s40478-021-01204-8 |
| 34 | JIN et al. | scGRNom: A Computational Pipeline Of Integrative Multi-Omics Analyses For Predicting Cell-Type Disease Genes And Regulatory Networks. | Genome Medicine | 2021 | 10.1186/s13073-021-00908-9 |
| 35 | WANG et al. | scGNN Is A Novel Graph Neural Network Framework For Single-Cell RNA-Seq Analyses. | Nature Communications | 2021 | 10.1038/s41467-021-22197-x |
| 36 | BHATTACHERJEE et al. | The CD33 short isoform is a gain-of-function variant that enhances Aβ1-42 phagocytosis in microglia. | Molecular Neurodegeneration | 2021 | 10.1186/s13024-021-00443-6 |
| 37 | ZHENG et al. | Exploring The Genetic Association Of The ABAT Gene With Alzheimer’s Disease. | Molecular Neurobiology | 2021 | 10.1007/s12035-020-02271-z |
| 38 | OLAH et al. | Single Cell RNA Sequencing Of Human Microglia Uncovers A Subset Associated With Alzheimer’s Disease. | Nature Communications | 2020 | 10.1038/s41467-020-19737-2 |
| 39 | PETITPREZ et al. | The Murine Microenvironment Cell Population Counter Method To Estimate Abundance Of Tissue-Infiltrating Immune And Stromal Cell Populations In Murine Samples Using Gene Expression. | Genome Medicine | 2020 | 10.1186/s13073-020-00783-w |
| 40 | THRUPP et al. | Single-Nucleus RNA-Seq Is Not Suitable For Detection Of Microglial Activation Genes In Humans. | Cell Reports | 2020 | 10.1016/j.celrep.2020.108189 |
| 41 | AGARWAL et al. | A Single-Cell Atlas Of The Human Substantia Nigra Reveals Cell-Specific Pathways Associated With Neurological Disorders. | Nature Communications | 2020 | 10.1038/s41467-020-17876-0 |
| 42 | SRINIVASAN et al. | Alzheimer’s Patient Microglia Exhibit Enhanced Aging And Unique Transcriptional Activation. | Cell Reports | 2020 | 10.1016/j.celrep.2020.107843 |
| 43 | GATE et al. | Clonally Expanded CD8 T Cells Patrol The Cerebrospinal Fluid In Alzheimer’s Disease. | Nature | 2020 | 10.1038/s41586-019-1895-7 |
| 44 | MATHYS et al. | Single-Cell Transcriptomic Analysis Of Alzheimer’s Disease. | Nature | 2019 | 10.1038/s41586-019-1195-2 |
| 45 | SALA FRIGERIO et al. | The Major Risk Factors For Alzheimer’s Disease: Age, Sex, And Genes Modulate The Microglia Response To Aβ Plaques. | Cell Reports | 2019 | 10.1016/j.celrep.2019.03.099 |
| 46 | ZUCCHELLI et al. | Antisense Transcription In Loci Associated To Hereditary Neurodegenerative Diseases. | Molecular Neurobiology | 2019 | 10.1007/s12035-018-1465-2 |
| 47 | REN et al. | TMEM106B Haplotypes Have Distinct Gene Expression Patterns In Aged Brain. | Molecular Neurodegeneration | 2018 | 10.1186/s13024-018-0268-2 |
| 48 | MATHYS et al. | Temporal Tracking Of Microglia Activation In Neurodegeneration At Single-Cell Resolution. | Cell Reports | 2017 | 10.1016/j.celrep.2017.09.039 |
| 49 | BARBASH S and SAKMAR TP | Length-Dependent Gene Misexpression Is Associated With Alzheimer’s Disease Progression. | Scientific Reports | 2017 | 10.1038/s41598-017-00250-4 |
| 50 | WANG et al. | Integrative Network Analysis Of Nineteen Brain Regions Identifies Molecular Signatures And Networks Underlying Selective Regional Vulnerability To Alzheimer’s Disease. | Genome Medicine | 2016 | 10.1186/s13073-016-0355-3 |
| 51 | SRINIVASAN et al. | Untangling The Brain’s Neuroinflammatory And Neurodegenerative Transcriptional Responses. | Nature Communications | 2016 | 10.1038/ncomms11295 |
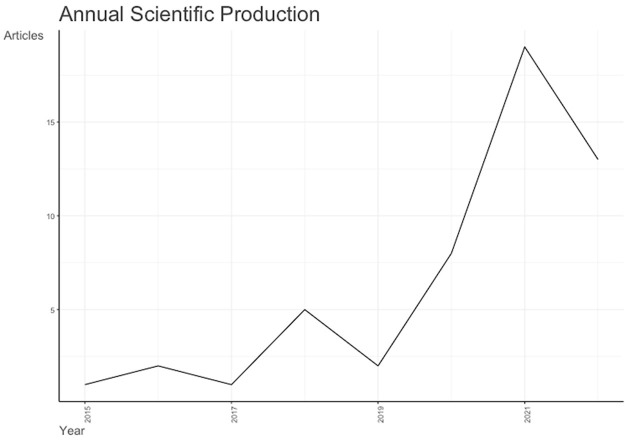
The first paper was published in 2016 by Srinivasan et al. [16] to untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. They acutely purified neurons, astrocytes and microglia from single adult mouse brains and analysed their transcriptomes by RNA sequencing. Using peripheral endotoxemia to establish the method, they revealed highly specific transcriptional responses and altered RNA processing in each cell type, with Tnfr1 required for the astrocytic response. There is an annual growth rate of publication of 44.26% with the average document age of 2.67 without any single author (Tables 1 and 2; Figure 1). Supplementary Table 1 denotes alternatively spliced genes in a previous junction microarray study. The most recent published article was written by Lai et al. [17] in Frontier Immunology to study the integration of bulk RNA sequencing and single-cell analysis that revealed a global landscape of DNA damage response in the immune environment of AD. They concluded that immunological microenvironment and disease progression in AD patients were significantly predicted by DNA Damage Response (DDR)-associated genes and lncRNAs. They also provided a theoretical underpinning for the individualized treatment of AD patients by the suggested genetic subtypes and risk model based on DDR (Table 2).
Figure 1.
Annual scientific production of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
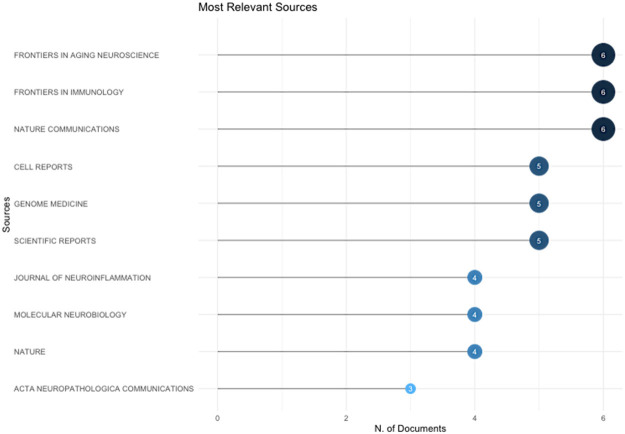
Analysis of publication sources
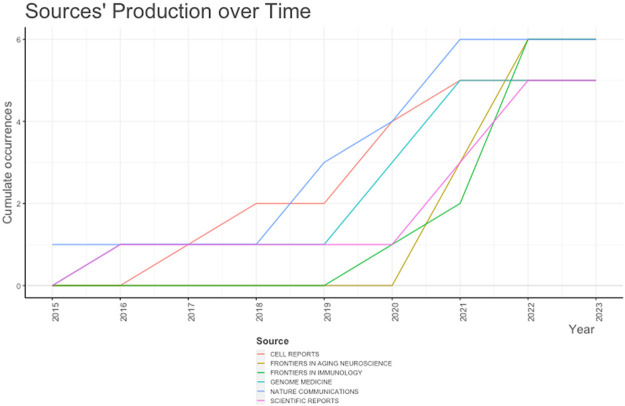
The most relevant source for publication is Frontiers in aging neuroscience, Frontiers in Immunology and Nature communication with 6 published articles per each (Figure 2). While the least published journal was ACA neuropathologica communications with only 3 published articles. Similarly, Figure 3 summarized the cumulative publication source production over time from 2015 till 2023.
Figure 2.
The most relevant journal for publications of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Figure 3.
Source production over time for publications of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Analysis of keywords
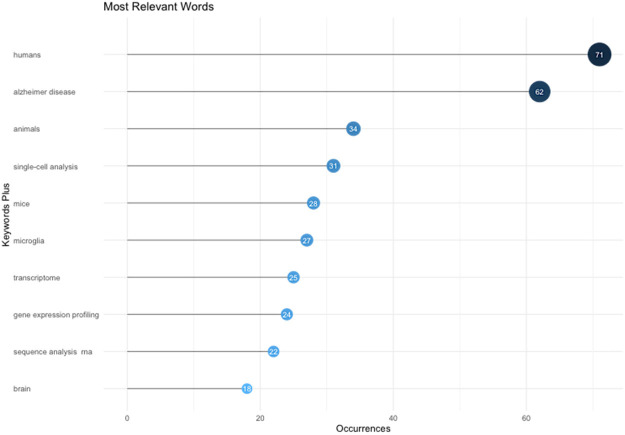
Keywords assigned by the authors in their articles are especially useful for bibliometric analysis when investigating the hotspots and trends of a specific research field. By performing keyword analysis, we revealed top 10 most frequent keywords used by the authors for the Single-cell RNA sequencing analysis and Alzheimer’s Disease research (Figure 4). The most frequent keyword was humans with 71 mentions across the results followed by AD with 62 mentions while the least mentioned keyword was brain with only 18 mentions. This is confirmed with the word’s frequency over time (Figure 5) and word cloud (Figure 6).
Figure 4.
The most relevant word for publications of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Figure 5.
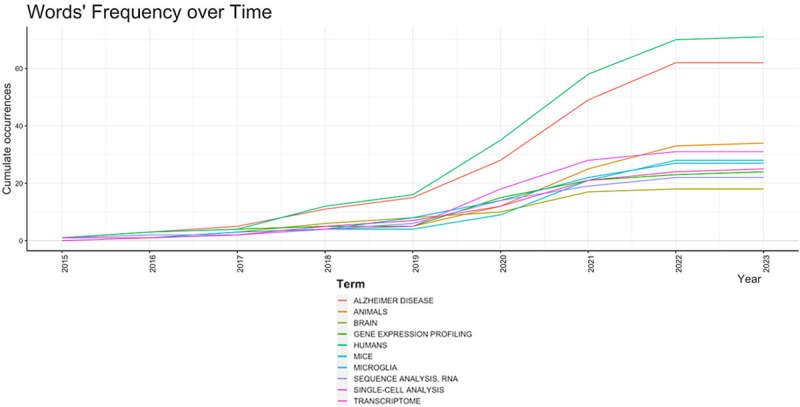
Word frequency over time for publications of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Figure 6.
Word cloud for publications of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
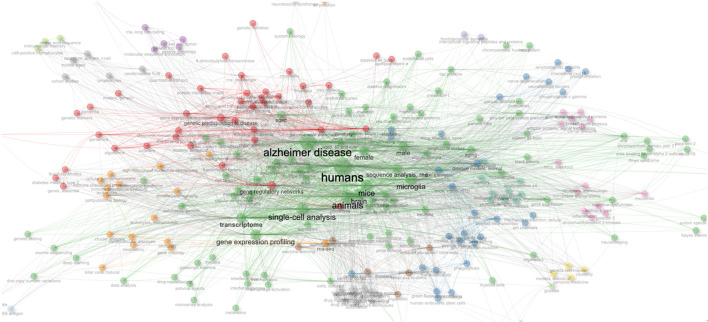
As shown in Figure 7 of the thematic map, each node inside a network represents a keyword object. wherein: (1) the size of the node indicates the occurrence of the keyword (i.e., the number of times that the keyword occurs), (2) the link between the nodes represents the co-occurrence between keywords (i.e., keywords that co-occur or occur together), (3) the thickness of the link indicates the occurrence of co-occurrences between keywords (i.e., the number of times that the keywords co-occur or occur together), and (4) the larger the node, the greater the Each color indicates a thematic cluster whose nodes and connections may be used to demonstrate the theme’s (cluster’s) coverage of subjects (nodes) and the linkages (links) between topics (nodes) falling under that theme (cluster).
Figure 7.
Thematic map of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
According to topic dendogram (Figure 8) created by the bibliometrix software based on the titles of the literature in the field. Multiple correspondence analysis methods have been adopted for the analysis. It indicates the research topics that correlate with the research on Single-cell RNA sequencing analysis and Alzheimer’s Disease from 2015 to 2023. As evident from the figure, we can conclude how research has been related to our research keywords.
Figure 8.
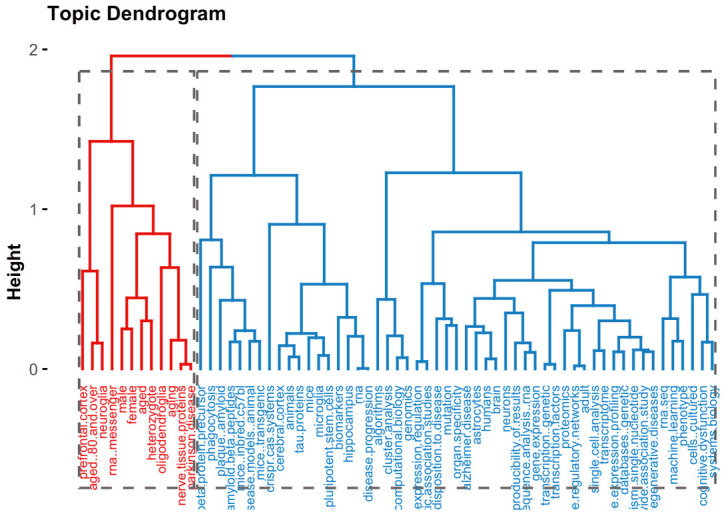
Topic dendrogram of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Analysis of research collaboration
All papers were done via international collaboration without any single author paper. The most frequent collaboration was between USA and China with 11 papers followed by USA and Canada with 6 papers then 3 papers between Australia and Italy (Figure 9). As co-authorship is a formal method of intellectual cooperation amongst academics, it is essential to comprehend how scholars communicate with one another (including author qualities such as connected institutions and nations). In truth, scholarly partnerships may result in advancements in research; for instance, inputs from many academics might contribute to better clarity and deeper insights. The analysis allows collaborations to be mapped across different time periods, allowing scholars to review the trajectory of intellectual development in relation to collaboration networks and equipping aspiring scholars with valuable information to reach out to established and trending scholars in the research field.
Figure 9.
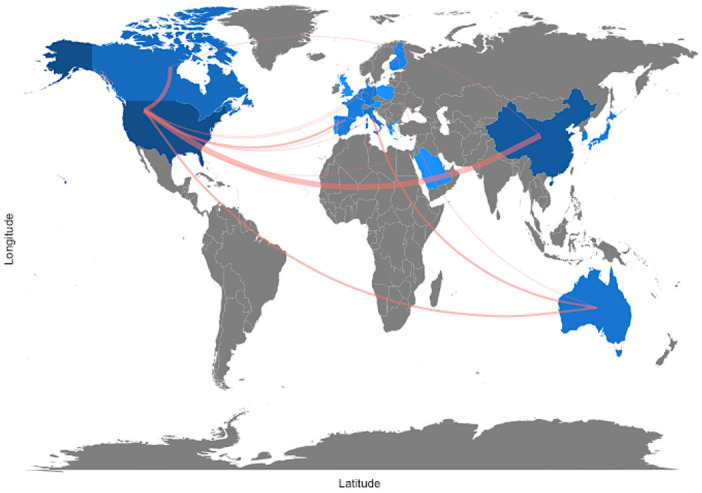
Collaborator world map of single-cell RNA sequencing analysis and Alzheimer’s disease from 2015 to 2023.
Discussion
The main purpose of this study is to investigate the application of single-cell RNA sequencing analysis in the diagnosis and treatment of Alzheimer’s disease (AD). In this work, a bibliometric analysis of studies on Single-cell RNA sequencing analysis and Alzheimer’s Disease was employed. To diagnose Alzheimer’s, physicians may use medical history, mental status tests, physical and neurological exams, diagnostic tests and brain imaging. Among treatments is neuromodulary treatment or medical one (e.g., using Brexpiprazole, Donepezil, Glantamine, Memantine, Rivastigmine). The goals were to discover emerging article trends, patterns of collaboration, and research components, as well as to explore the intellectual structure of a certain field in existing literatures. Bibliometric analysis is a useful technique to find out what scientists have learnt and how their disciplines have evolved over time by making sense of a large amount of unstructured data in a systematic fashion. As a result, the present bibliometric research may set the framework for furthering a discipline in novel and significant ways. This is because it permits and empowers academics to (1) acquire a comprehensive picture in one location, (2) identify knowledge gaps, (3) generate new research ideas, and (4) position their intended contributions to the subject.
Single-Cell RNA-Seq data analysis of AD and control samples may improve our knowledge on the disease underlying molecular mechanisms [13]. So far, several genes and molecular pathways have been linked as related to the disease [18], however still many others are to yet be discovered. Our bioinformatic analysis identified discrete glial, immune, neuronal, and vascular cell populations spanning AD and controls. Astrocytes and microglia displayed the greatest transcriptomic impacts, with the induction of both shared and distinct gene programs.
Soreq et al. [13] tried to identify distinct transcriptional networks impacted into distinct neuronal populations in AD. They surveyed gene expression differences in over 25,000 single-nuclei collected from the brains of two AD patients in Braak stage I and II and age- and gender-matched controls hippocampal brain samples. APOE status was not measured for this study samples (as well as CERAD and THAL scores). Their bioinformatic analysis identified discrete glial, immune, neuronal and vascular cell populations spanning AD and controls. Astrocytes and microglia displayed the greatest transcriptomic impacts, with the induction of both shared and distinct gene programs. This is a critical research with a tiny but possibly significant dataset. The main conclusion is that cell-specific glial cell markers are higher in Alzheimer’s samples than in controls, which is consistent with earlier research. Because of the single time point and small sample size, it is difficult to determine whether detected alterations are upstream in disease aetiology and which are the result of neurodegeneration per se. They also identified an increase in the frequency of microglia, astrocytes, and oligodendrocytes, which has previously been documented in Alzheimer’s disease, including genome-wide transcriptome investigations. However, Soreq et al. [13] were unable to determine whether the observed changes are downstream or [13] upstream of disease progression. This is a possible limitation of their study. In terms of glial cell alterations, their key finding is that they may be reactive rather than causative in the illness.
Study limitations
Bibliometric analysis is a common and thorough technique for examining and interpreting vast quantities of scientific data. It helps us to dissect the evolutionary subtleties of a certain topic and offer light on its developing areas. Notwithstanding its virtues, bibliometric analysis is still relatively new in business research, and in many situations, it is not used to its full potential. This happens when bibliometric research depends on a narrow collection of bibliometric data and procedures, resulting in a fragmented knowledge of the studied topic. It is important to note that there is no authoritative guide to bibliometric analysis in business research, which poses a significant challenge for business scholars who wish to learn more about the bibliometric methodology and its application to business research in a comprehensive, easily-digestible format.
Although while bibliometric analysis is a valuable technique for summarizing and synthesizing literature, its limitations must be acknowledged. Furthermore, since bibliometric data from scientific databases such as Scopus and Web of Science are not designed specifically for bibliometric analysis, they may include mistakes that may affect any research done using such data. To limit the chance of errors, we rigorously remove duplicates and inaccurate entries from the bibliometric data we’ve acquired. The second limitation is intrinsic to the bibliometric method. Due to the quantitative nature of bibliometric study, in which the relationship between quantitative and qualitative conclusions may be vague, the qualitative claims of bibliometrics can be particularly subjective. In this setting, we exercise extra care when making qualitative assertions based on bibliometric data, and where appropriate, we back these assertions with content analysis.
Conclusions
In conclusion, the current bibliometric study can only give a short-term forecast of Single-cell RNA sequencing analysis in AD: A bibliometric analysis; hence, academics should refrain from making too ambitious assertions about the research field and its long-term significance. Notwithstanding these limitations, the bibliometric method may help neurologists to overcome their fear of dealing with large bibliometric datasets and perform ambitious big genetic data research retrospectives. Bibliometric analysis may facilitate the creation of data in genetic field. In this strategy, a quick but vital step is taken towards understanding the Single-cell RNA sequencing analysis and Alzheimer’s Disease: A bibliometric analysis. The advantage of single cell RNA-Seq is more accurate computational/bioinformatic analyses of patient’s samples data. Similar future studies may improve our understanding of some of the disease underlying molecular mechanisms.
Acknowledgements
L.S. was funded by Alzheimer’s society UK and RoseTrees UK.
Disclosure of conflict of interest
None.
Author contributions
L.S and W.M concieved and wrote the paper W.M prepared the figures and tables.
Abbreviations
- AD
Alzheimer’s disease
- lncRNAs
long non-coding RNAs
- RNA-Seq
RNA sequencing
Supporting Information
References
- 1.Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011;377:1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 2.Bondi MW, Edmonds EC, Salmon DP. Alzheimer’s disease: past, present, and future. J Int Neuropsychol Soc. 2017;23:818–831. doi: 10.1017/S135561771700100X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song X, Mitnitski A, Rockwood K. Nontraditional risk factors combine to predict Alzheimer disease and dementia. Neurology. 2011;77:227–34. doi: 10.1212/WNL.0b013e318225c6bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Z, Wang C, Yang W. Role of berberine in Alzheimer’s disease. Neuropsychiatr Dis Treat. 2016;12:2509–20. doi: 10.2147/NDT.S114846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs R, Kennelly SP, O’Neill D. Drug treatments in Alzheimer’s disease. Clin Med (Lond) 2016;16:247–53. doi: 10.7861/clinmedicine.16-3-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mancuso C, Siciliano R, Barone E, Butterfield DA, Preziosi P. Pharmacologists and Alzheimer disease therapy: to boldly go where no scientist has gone before. Expert Opin Investig Drugs. 2011;20:1243–61. doi: 10.1517/13543784.2011.601740. [DOI] [PubMed] [Google Scholar]
- 7.Canevelli M, Adali N, Voisin T, Soto ME, Bruno G, Cesari M, Vellas B. Behavioral and psychological subsyndromes in Alzheimer’s disease using the neuropsychiatric inventory. Int J Geriatr Psychiatry. 2013;28:795–803. doi: 10.1002/gps.3904. [DOI] [PubMed] [Google Scholar]
- 8.Lambon Ralph MA, Patterson K, Graham N, Dawson K, Hodges JR. Homogeneity and heterogeneity in mild cognitive impairment and Alzheimer’s disease: a cross-sectional and longitudinal study of 55 cases. Brain. 2003;126:2350–62. doi: 10.1093/brain/awg236. [DOI] [PubMed] [Google Scholar]
- 9.Lyketsos CG. Commentary on “Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. ” New criteria for a new era. Alzheimers Dement. 2011;7:328–9. doi: 10.1016/j.jalz.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Tober J, Maijenburg MMW, Li Y, Gao L, Hadland BK, Gao P, Minoura K, Bernstein ID, Tan K, Speck NA. Maturation of hematopoietic stem cells from prehematopoietic stem cells is accompanied by up-regulation of PD-L1. J Exp Med. 2018;215:645–659. doi: 10.1084/jem.20161594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunkle BW, et al. Author Correction: Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:1423–1424. doi: 10.1038/s41588-019-0495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elmore MRP, Hohsfield LA, Kramar EA, Soreq L, Lee RJ, Pham ST, Najafi AR, Spangenberg EE, Wood MA, West BL, Green KN. Replacement of microglia in the aged brain reverses cognitive, synaptic, and neuronal deficits in mice. Aging Cell. 2018;17:e12832. doi: 10.1111/acel.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soreq L, Bird H, Mohamed W, Hardy J. Single-cell RNA sequencing analysis of human Alzheimer’s disease brain samples reveals neuronal and glial specific cells differential expression. PLoS One. 2023;18:e0277630. doi: 10.1371/journal.pone.0277630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aria M, Cuccurullo C. Bibliometrix: an R-tool for comprehensive science mapping analysis. J Informetr. 2017;11:959–975. [Google Scholar]
- 15.Derviş H. Bibliometric analysis using bibliometrix an R package. J Scientometric Res. 2019;8:156–160. [Google Scholar]
- 16.Jeppsson S, Srinivasan S, Chandrasekharan B. Neuropeptide Y (NPY) promotes inflammation-induced tumorigenesis by enhancing epithelial cell proliferation. Am J Physiol Gastrointest Liver Physiol. 2017;312:G103–G111. doi: 10.1152/ajpgi.00410.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai YJ, Liu SH, Manachevakul S, Lee TA, Kuo CT, Bello D. Biomarkers in long COVID-19: a systematic review. Front Med (Lausanne) 2023;10:1085988. doi: 10.3389/fmed.2023.1085988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy J, Escott-Price V. Genes, pathways and risk prediction in Alzheimer’s disease. Hum Mol Genet. 2019;28:R235–R240. doi: 10.1093/hmg/ddz163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.