Abstract
Genetic manipulation of fluorescent pseudomonads has provided major insight into their production of antifungal molecules and their role in biological control of plant disease. Burkholderia cepacia also produces antifungal activities, but its biological control activity is much less well characterized, in part due to difficulties in applying genetic tools. Here we report genetic and biochemical characterization of a soil isolate of B. cepacia relating to its production of an unusual antibiotic that is very active against a variety of soil fungi. Purification and preliminary structural analyses suggest that this antibiotic (called AFC-BC11) is a novel lipopeptide associated largely with the cell membrane. Analysis of conditions for optimal production of AFC-BC11 indicated stringent environmental regulation of its synthesis. Furthermore, we show that production of AFC-BC11 is largely responsible for the ability of B. cepacia BC11 to effectively control the damping-off of cotton caused by the fungal pathogen Rhizoctonia solani in a gnotobiotic system. Using Tn5 mutagenesis, we identified, cloned, and characterized a region of the genome of strain BC11 that is required for production of this antifungal metabolite. DNA sequence analysis suggested that this region encodes proteins directly involved in the production of a nonribosomally synthesized lipopeptide.
Each year, fungal diseases cause millions of dollars worth of crop damage all over the world despite the extensive use of pesticides (11). Also, environmental concerns and development of resistance in target populations have reduced the availability of effective fungicides. Nonetheless, the vast array of antimicrobial molecules produced by diverse soil microbes remains as a reservoir of new and potentially safer biopesticides. For these and other reasons, a heightened interest in so-called biological control or biocontrol (i.e., the use of natural microorganisms or their products to limit attack and damage by phytopathogens) has arisen (2, 4, 19, 63, 65).
There are many well-documented cases (reviewed in references 19 and 65) where the efficacy of a biological control microbe in the greenhouse and in the field depends on the production of single or multiple fungal growth antagonists (antifungal compounds [AFCs]). Many cases involve fluorescent pseudomonads that produce antibiotics such as phenazines (64), 2,4-diacetylphloroglucinol (12, 28), pyrrolnitrin (21, 22), pyoluteorin (23, 36, 43), or siderophores (10). Recently, gene clusters encoding biosynthetic pathways for many of these antibiotics have been cloned and characterized (18, 21, 36, 44; reviewed in reference 63). Mutants overexpressing the biosynthetic genes, and hence overproducing the AFCs, have shown increased efficacy and potential in biological control (40, 57).
In contrast, much less work has been done with Burkholderia (Pseudomonas) cepacia (3, 67), even though B. cepacia is a ubiquitous soil organism (5, 38) and various strains have been reported to produce a large variety of AFCs such as cepacin (49), altericidins (32), pyrrolnitrin (26, 27), xylocandins (also called cepacidines) (6, 37, 46), and siderophores (60). B. cepacia is one of the most nutrionally diverse bacteria known (i.e., it can use >200 different organic compounds as its carbon source [38]), a trait that probably contributes to its ability to compete for root exudates and very effectively colonize roots and the rhizosphere (5, 31, 47). Studies suggest that B. cepacia can be an effective biocontrol agent for Pythium-induced damping off and Aphanomyces-induced root rot of pea (30, 47, 48), Botrytis-induced gray mold of apple (25), Rhizoctonia solani-induced root rot of Poinsettia (7), and other fungal diseases (13). However, in all these cases very little is known about the genetics or biochemistry of the biocontrol ability of B. cepacia.
A major limitation for molecular studies of the biological control ability of B. cepacia is that unlike many other pseudomonads, tools for its genetic manipulation and analysis are much less well developed, largely due to its inherently high levels of resistance to many antibiotics and low frequencies of electroporation and conjugative plasmid transfer. Furthermore, the B. cepacia genome is very large (>7 Mb), is composed of multiple replicons, and contains a large number of insertion sequences, resulting in extensive genomic and physiological variability and heterogeneity (39). However, as we report here, it is possible to apply some standard molecular genetic techniques to B. cepacia, which, in conjunction with classical biochemical approaches, can provide new insights into the physiology and ecology of B. cepacia. We describe the discovery of a new antifungal metabolite that inhibits the growth of numerous fungi. We also demonstrate that this AFC (herein called AFC-BC11) is largely responsible for the ability of B. cepacia BC11 to effectively control a damping-off disease caused by R. solani. Purification and characterization of this AFC, as well as analysis of several genes required for its biosynthesis, provided good evidence that it is a novel lipopeptide antibiotic which is synthesized nonribosomally.
MATERIALS AND METHODS
Microorganisms, growth conditions, and media.
The bacterial strains used in this study are listed in Table 1. B. cepacia was routinely grown at 30°C on plates containing either nutrient agar (Difco) supplemented with 0.5% glucose (NAG) or potato dextrose agar (PDA [pH 5.6]; Difco). Escherichia coli was grown at 37°C in Luria-Bertani (LB) medium (42). BSM minimal medium was prepared as described previously (54). Fungi were obtained from G. Michaels (Microbiology Department, University of Georgia) and D. Sumner and R. Roncadori (Plant Pathology Department, University of Georgia) and cultured at 30°C on PDA plates. Pseudomonas aeruginosa PAO1, Pseudomonas putida ATCC 12633, and Acinetobacter calcoaceticus BND1 were from our laboratory collection. The antibiotic levels used to select for B. cepacia constructs were 350 μg/ml for kanamycin, 100 μg/ml for rifampin, and 200 μg/ml for tetracycline. The levels used to select for E. coli constructs were 50 μg/ml for kanamycin, 20 μg/ml for tetracycline, and 100 μg/ml for ampicillin.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| B. cepacia | ||
| BC11 | Wild-type B. cepacia soil isolate | 16 |
| BC11R | Spontaneous Rifr mutant of BC11 | This work |
| AFC143, AFC145, AFC202, AFC666 | afc::Tn5 derivatives of BC11R Rifr Kmr | This work |
| AFC666M2 | afcA::nptI derivative of BC11R made by allelic exchange | This work |
| E. coli | ||
| DH5α | endA1 hsdR trecA1 gyrA relA φ80lacZΔM15 | 42 |
| SM10 | RP4-2-Tc::Mu recA thi pro his | 58 |
| HB101 | recA rpsL ara galK leu hsdS Δ(gpt-proA) supE xyl | 42 |
| Plasmids | ||
| pTZ19U | ColE1 replicon, Apr | 45 |
| pRK415 | IncP1 replicon, Mob+ Tcr | 29 |
| pTOK2 | ColE1 replicon, suicide plasmid, Tcr | 33 |
| pSUP2021 | ColE1 replicon, suicide plasmid with Tn5, Kmr | 58 |
| pSB315 | pUC4K derivative with nonpolar nptI cartridge, Kmr Apr | 14 |
| pRK2013 | ColE1 replicon, Tra+-mobilizing plasmid, Kmr | 9 |
| pKW666 | 10-kb EcoRI genomic fragment with afc::Tn5 of AFC666 cloned into the EcoRI site of pTZ19U, Kmr Apr | This work |
| pT666 | 10-kb EcoRI fragment of pKW666 cloned into the EcoRI site of pTOK2, Tcr Kmr | This work |
| pKW666-1 | 5.6-kb BamHI-EcoRI fragment of pKW666 cloned in the BamHI and EcoRI sites of pTZ19U, Kmr Apr | This work |
| pKW666-2 | 2.8-kb PstI-EcoRI fragment of pKW666-1 cloned in the PstI and EcoRI sites of pTZ19U, Apr | This work |
| pKW666-4 | 3.0-kb HpaI-EcoRI fragment of pKW666-1 cloned in pTZ19U, Apr | This work |
| pKW666-5 | 0.9-kb SalI fragment with nonpolar nptI from pSB315 cloned into the XhoI site of pKW666-4, Kmr Apr | This work |
| pT666-1 | 3.9-kb XbaI-EcoRI fragment of pKW666-5 cloned into the XbaI and EcoRI sites of pTOK2, Tcr Kmr | This work |
| pR666 | 2.2-kb StyI-EcoRI fragment of pKW666-1 cloned into pRK415, Tcr | This work |
Tcr, Kmr, Rifr, and Apr denote resistance to tetracycline, kanamycin, rifampin, and ampicillin, respectively.
Bioassay for AFC activity.
Samples to be assayed were dried in a vacuum centrifuge, and the residue was dissolved in acidic methanol (85% methanol adjusted to pH 3 with concentrated HCl). Serial dilutions (1:3) were made in acidic methanol, and 10-μl aliquots were added to wells of 96-well-format microtiter dishes containing 0.3 ml of solidified PDA. After being dried for 30 min at 37°C, the wells were inoculated with 10 μl of a hyphal suspension of R. solani AG4 prepared by scraping aerial hyphae from a 4-day-old PDA plate culture with a loop and resuspending them in sterile water. After incubation for 2 to 3 days at 30°C, the greatest dilution giving complete growth inhibition was recorded and defined as containing 1 U of activity; 10 μl of acidic methanol alone had no observable effect on the growth of R. solani. For a qualitative assay of AFC production by growing cells, the hyphal suspension was spread onto PDA plates and dried for 15 min before being subjected to spot inoculation with a loopful of test bacteria. After 3 days at 30°C, the size of the fungal growth inhibition zone around each bacterial patch was used as a measure of AFC production.
Purification of AFC-BC11.
A 5-ml volume of water containing 109 cells of B. cepacia BC11 was spread onto 500 ml of PDA in each of 10 disposable aluminum baking trays (40 by 25 cm). After 48 h at 30°C, the cells were scraped from the surface by using a glass plate spreader and 500 ml of water. Cells (ca 15 g [wet weight]) were collected by centrifugation and extracted twice by vigorous vortexing for 1 min with 50 ml of 80% acetone. Insoluble material was removed by centrifugation at 2,700 × g for 15 min and discarded. Acetone was evaporated with a stream of nitrogen at 50°C, and the remaining aqueous solution was centrifuged at 2,700 × g for 15 min. The pelleted insoluble material was washed with water three times and then dried in a vacuum centrifuge. The dried residue was extracted three times each with 50 ml of 100% acetone and then 1-butanol. After being dried, the residue was dissolved in 5 ml of dimethyl sulfoxide (DMSO) and applied to a C18 reverse-phase high-pressure liquid chromatography (HPLC) column (Z18TP54; Vydac Corp.). The column was eluted for 5 min with methanol-water-DMSO (50:45:5), after which the methanol concentration was increased to 75% over a 10-min period. Elution with methanol-water-DMSO (75:22:3) was continued until the AFC peak had completely eluted, usually after 10 to 20 min. Elution of BC11 AFC was monitored by measurement of the absorbance at 320 nm (A320) and/or bioassay.
Structural analyses of AFC-BC11.
Positive-mode fast atom bombardment mass spectrometry (FAB-MS) was performed with a JEOL SX/SX 102A tandem mass spectrometer at an accelerating potential of 10 kV (53). Equal volumes of sample and thioglycerol FAB matrix were mixed on the probe tip and then bombarded with ions generated with xenon and a JEOL FAB gun at 6 kV. The spectra consisted of averaged profile data of three scans recorded by a JEOL XMS data system and acquired from 200 to 2,000 m/z at a rate that would scan the mass range from 0 to 2,500 in 1 min; the filtering rate was 100 Hz. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) was performed on an LDI 1700XP spectrometer at ∼10−6 torr (accelerating voltage, 30 kV; extractor voltage, 9 kV) (1, 53). A sample–2,4-dihydroxybenzoic acid matrix mixture was serially vacuum dried on the probe and then ionized with a nitrogen laser (λ = 337 nm) by using a 3-ns, 6-mJ pulse. Spectra were recorded from m/z 400 to 10,000 and are the averages of ∼150 acquisitions.
For nuclear magnetic resonance spectroscopy (NMR), samples were deuterium exchanged by repeated suspension and lyophilization in D2O, dissolved in deuterated DMSO with 5% D2O, and analyzed at 298 K by using a Bruker AM500 spectrometer. Chemical shifts were measured relative to the DMSO resonance at δ2.49. Glycosyl composition analysis was performed by combined gas chromatography-mass spectrometry of trimethylsilyl methyl glycosides prepared by methanolysis and trimethylsilylation (Tri-Sil reagent; Pierce Chemical). For amino acid analysis, the samples were hydrolyzed in vacuo at 110°C for 20 h with HCl vapors. Released amino acids were then converted to phenylthiocarbamyl amino acids with phenylisothiocyanate in an ABI 420H derivatizer/130A phenylthiocarbamyl analyzer. PTC amino acids were separated by HPLC on a C18 silica column and quantified as specified by the manufacturer.
Assay for biological control of R. solani-induced damping-off of cotton.
Cotton seeds (Nucotin 35B) were immersed in 5% bleach for 10 min and then washed three times with sterile water. They were then immersed in bacterial suspensions of various concentrations (104 to 107 cells/ml) for 15 min and dried at 25°C for 30 min. The number of cells coating the seeds at each concentration was determined by vortexing portions of seed lots in sterile water and determining the number of viable cells in supernatants by dilution plating. Seed treatment with BC11 AFC was conducted by multiple applications of 10-μl aliquots of purified antibiotic dissolved at 0.3 mg/ml in methanol. After being dried, the seeds were planted in 9- by 8-cm pots containing R. solani-infested vermiculite (made by resuspending the aerial hyphae of a 5-day old PDA plate culture of R. solani in 50 ml of water and pouring the suspension onto the vermiculite). Disease development was monitored at 30°C for 3 weeks by scoring germination, seedling emergence, and survival.
Tn5 mutagenesis and screening for AFC-deficient B. cepacia mutants.
E. coli SM10, containing the Tn5 delivery plasmid pSUP2021 (58), and B. cepacia BC11R were grown to an A600 of 0.5 by shaking in 18 ml of LB medium or nutrient broth with 0.5% glucose (NBG), respectively. The cells were washed twice with sterile water by centrifugation, resuspended in 0.3 ml of sterile water, and combined, and 30 0.02-ml aliquots were spotted on NAG plates. After 24 h at 30°C, the cells were scraped up, suspended in water, and adjusted to an A600 of 2, and 0.1-ml aliquots were spread on NAG plates plus kanamycin and rifampin. After incubation at 37°C for 2 days, Kmr Rifr colonies arose at a frequency of ∼10−6 per donor. These were individually transferred into 96-well microtiter dishes filled with LB medium–7% glycerol–7% DMSO, incubated for 16 h at 37°C, and frozen at −80°C or transferred en masse with a 96-well format replicator array to a large (15- by 150-mm) PDA plate previously spread with a suspension of R. solani hyphae. After incubation for 2 to 3 days at 30°C, BC11 cells from patches showing reduced fungal growth inhibition were picked and purified to single colonies.
Cloning of genes involved in the production of BC11 AFC.
The plasmids used are listed in Table 1; the physical and genetic maps of some of these are shown in Fig. 1. Genomic DNA from AFC666 (afc666::Tn5) was isolated as described previously (54), digested with EcoRI, ligated with EcoRI-digested pTZ19U (45), and transformed into E. coli DH5α. A plasmid from one of the resultant Ampr Kmr transformants was designated pKW666 and further characterized. pKW666-1 was constructed by digesting pKW666 with BamHI, ligating under dilute conditions, and transforming E. coli to Ampr. Analogously, pKW666-1 was digested with PstI, ligated, and transformed into E. coli to obtain pKW666-2. pKW666-4 was constructed by digesting pKW666-1 with HpaI and BamHI, filling in cohesive ends with deoxynucleoside triphosphates (dNTPs) and Klenow polymerase, and ligating under dilute conditions. pKW666-5 was constructed by ligating the 0.9-kb SalI fragment of pSB315 (containing nptI [14]) into XhoI-digested pKW666-4. pT666-1 was constructed by ligating the 3.3-kb XbaI-EcoRI fragment of pKW666-5 into EcoRI- and XbaI-digested pTOK2 (33). pR666 was constructed by ligating the 2.2-kb StyI-EcoRI fragment of pKW666-1 into EcoRI- and XbaI-digested pRK415 (29). pT666 was made by ligating a 10-kb EcoRI fragment of pKW666 into EcoRI-digested pTOK2.
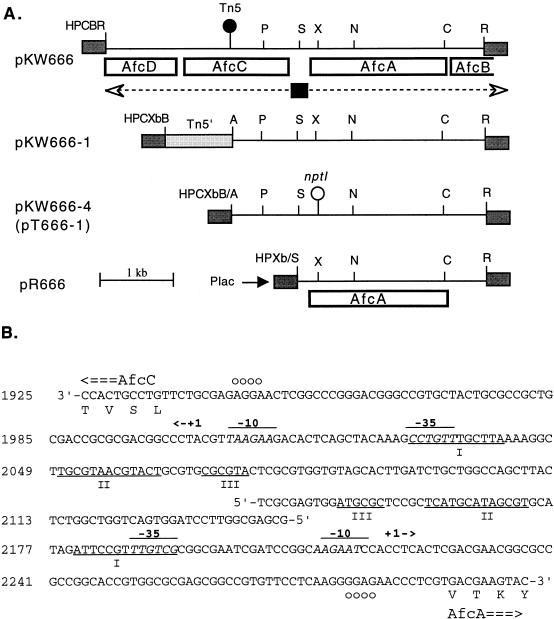
FIG. 1.
(A) Physical and genetic map of plasmids containing afc genes of B. cepacia BC11. The solid circle shows the position of the Tn5 insertion in the genome of strain AFC666. Dotted arrows show putative transcripts of afc genes, and the solid box represents the putative divergent promoter between afcA and afcC (see below). Open boxes represent the afc ORFs detected by DNA sequence analysis. The open circle shows location of the nonpolar nptI insertion in pT666-1 that was marker-exchanged into the BC11 genome to generate strain AFC666M2. Shaded boxes represent polylinkers of vectors; Plac, lac promoter of the pRK415 vector. Restriction sites: A, HpaI; B, BamHI; C, HincII; H, HindIII; N, NotI; P, PstI; R, EcoRI; S, StyI; X, XhoI; Xb, XbaI. The BamHI site in tn5 used to construct pKW666-1 is not shown. (B) DNA sequence of the divergent promoter region between afcA and afcC. The region between afcA and afcC is shown (nucleotides 1925 to 2300 from GenBank accession no. AF076477). Sequences resembling −35 and −10 E. coli consensus promoter sequences (overlined and italicized) are shown. +1->, putative transcription start sites; ○, RBS. Pairs of segments of the promoter region that are underlined and labelled I, II, and III are highly homologous in the DNA sequence.
B. cepacia strain constructions.
Derivatives of pTOK2 and pRK415 were mobilized from E. coli DH5α into B. cepacia by triparental mating with E. coli HB101 containing the helper plasmid pRK2013 (9). B. cepacia AFC666M1 was constructed by mobilizing the pTOK2 derivative pT666 into BC11R (a spontaneous rifampin-resistant mutant of BC11); since pTOK2 is unable to replicate in B. cepacia, selection for Kmr, Rifr, and Tcs yields allelic-exchange recombinants. Strain AFC666M2 was similarly constructed by transferring pT666-1 into BC11R and selecting Kmr Rifr Tcs allelic-exchange recombinants.
Recombinant DNA techniques and DNA sequence analysis.
Transformation of E. coli, restriction digests, ligations, electrophoresis, and DNA isolation were performed by standard methods as described previously (42, 54). Southern blots were prepared and hybridized with DNA labeled by random priming with [α-32P]dATP (42). Double-stranded plasmid DNAs were sequenced with commercial and custom primers on an ABI 380 sequencer. DNA sequence analysis and comparisons with sequences contained in the GenBank and EMBL databases were performed with Genetics Computer Group programs and the BLAST algorithm.
Nucleotide sequence accession number.
The DNA sequence of the 4.5-kb B. cepacia genomic DNA on pKW666 (Fig. 1) has been deposited in GenBank under accession no. AF076477.
RESULTS
Characterization of AFC-BC11.
Strain BC11 was isolated from agricultural soil in south Georgia and identified as B. cepacia by both fatty acid analysis and polyphasic taxonomic analysis (16). When grown on PDA plates next to R. solani, it produced a small, impenetrable growth inhibition zone. When fivefold-concentrated culture supernatants of BC11 were spotted on PDA plates, a similarly compact fungal growth inhibition zone was observed, suggesting limited aqueous solubility and diffusion. The levels of AFC activity found in preparations of filter-sterilized culture supernatants varied widely, but extraction of PDA-grown whole cells with 80% acetone consistently produced an organic-solvent-soluble cell-free preparation with high antifungal activity. When the cells were disrupted by sonication and centrifuged at 100,000 × g to yield a cytosolic fraction (supernatant) and membrane fraction (pellet), >80% of the AFC activity was found in the membrane fraction, suggesting that the AFC is lipophilic.
Preliminary experiments designed to find optimal growth conditions for AFC production revealed that its synthesis is stringently controlled. The AFC activity from cultures grown on PDA (initial pH, 5.5) plates for 3 days was 20 times higher than that from cultures grown for 24 h (580 and 30 U/109 cells, respectively), despite relatively minor differences in growth yields. If cells were grown for 3 days with shaking in 250-ml Erlenmeyer flasks containing 50 ml of potato dextrose broth (PDB), they usually contained <10% of the AFC activity of those grown on PDA plates (40 and 500 U/109 cells, respectively). However, if the cultures were grown in unshaken 250-ml flasks containing 50 ml of PDB, the AFC levels were at least ninefold higher than in identical shaken cultures (480 and 52 U/109 cells, respectively). AFC production by cells grown on BSM minimal-glucose plates at pH 7 or 8 was undetectable; in contrast, that of cells grown on analogous plates at pH 6 was 210 U/109 cells. These data suggest that acidic pH, low aeration, and/or growth on a surface to stationary phase are important conditions for maximal AFC production.
Purification and structural analyses of AFC-BC11.
A dried 80% acetone extract from 15 g of BC11 cells (58 mg) was sequentially extracted with various solvents (see Materials and Methods), removing 85% of the mass but only 10% of the AFC activity. Fractionation of the residue (8 mg) by HPLC gave a single peak of AFC activity that contained ca. half of the loaded mass. This purified AFC, like the AFC activity in concentrated culture supernatants, was reasonably soluble in DMSO, 80% acetone, or 70 to 95% methanol but poorly soluble in water, butanol, chloroform, or hexane. Similarly, it was also resistant to overnight treatment with protease or 30 min of treatment with 0.1 M acid or 0.1 M base or by boiling.
Analysis of the HPLC-purified AFC preparation by either FAB-MS or MALDI-TOF MS showed a single species with a molecular mass of 733. Proton NMR analysis gave three resonances between δ6.4 and δ7.2, indicative of a conjugated double-bond system and consistent with the results of UV-visible spectroscopy, which showed a single absorption maximum for the AFC at 322 nm. A triplet was found at δ0.85, indicating aliphatic methyl protons. A broad resonance from δ1.1 to δ1.35 and others at δ1.4 to δ1.7 denote the presence of aliphatic methylene protons, some on carbons adjacent to carbonyl and/or vinyl carbons. A triplet at δ5.3 is coupled to the methylene protons at δ1.4 to δ1.7, indicating protons of a cis double bond bordered by aliphatic methylene protons. In summary, the NMR results indicate that the AFC may contain an aromatic component and a fatty acyl component with one double bond in a cis configuration.
Amino acid analysis of the purified AFC showed three major components, possibly glycine, lysine, phenylalanine, and/or diaminobutyric acid, suggesting that the AFC contains a peptide moiety. The cepacidines (also called xylocandins), a family of AFCs previously found in B. cepacia (6, 37, 46), contain similar types of components. However, unlike the cepacidines, glycosyl composition analysis of our AFC showed only trace levels of any carbohydrate; moreover, the target specificity (see below) and molecular mass of the AFC from BC11 dramatically differ from those of any of the cepacidines. Therefore, we provisionally call this potentially novel compound AFC-BC11, pending completion of definitive structural determination.
Selectivity of AFC-BC11: determination of MICs for various fungi.
Using the microtiter dish dilution assay, we quantified the potency (MICs) of purified AFC-BC11 against various fungi and bacteria. Concentrations of <1 μg/ml strongly inhibited the growth of some soil-borne phytopathogenic fungi, especially Pythium ultimum and Colletotrichium sp. (Table 2). In contrast, it had little effect on Candida albicans, several other yeasts, and some pathogenic and nonpathogenic filamentous fungi (Table 2). The sensitivity pattern of various fungi to growth inhibition by purified AFC-BC11 was identical to that exhibited by growing B. cepacia BC11 cells on PDA plates, suggesting that AFC-BC11 is a primary factor responsible for fungal growth inhibition by this bacterium on agar plates.
TABLE 2.
Target specificity of purified AFC-BC11
| Target fungus | MICa (μg/ml) |
|---|---|
| Rhizoctonia solani | 0.40 |
| Pythium ultimum | 0.01 |
| Colletotrichum sp. | 0.04 |
| Helminthosporium maydis | 0.10 |
| Botrytis cinerea | 0.40 |
| Fusarium sp. | 0.90 |
| Rhizopus stolonifer | 1.20 |
| Rhodotorula glutinis | 2.50 |
| Sclerotium rolfsii | 3.60 |
| Scopulariopsis brevicaulis | 10.80 |
Measured by serial dilution on PDA agar as described in Materials and Methods. At 50 μg/ml, no growth inhibition was observed for Saccharomyces cerevisiae, Candida albicans, Aspergillus flavus, A. niger, A. fumigatus, Penicillium expansum, P. chrysogenum, Dipodascus uninucleatus, Eremascus fertilis, Schizosaccharomyces octosporus, Pichia membranaefaciens, or Lipomyces starkeyi.
We tested several prokaryotes (Pseudomonas aeruginosa, P. putida, E. coli, and A. calcoaceticus) for sensitivity to AFC-BC11 and found that none of them were inhibited by concentrations up to 50 μg/ml. When the same bacteria were tested for production of AFC against R. solani in microtiter dishes by using 80% acetone extracts, no growth inhibition (i.e., AFC activity) was observed. Surprisingly, 80% acetone extracts of the B. cepacia type strain ATCC 25416 and laboratory strain 249-2 (39) grown on PDA showed no AFC activity. However, extracts of four different environmental isolates of B. cepacia (TOd1, TOd2, TOd5, and TOd63 [66]) had AFC levels 20 to 100% of that produced by BC11. The reason for the lack of AFC production by B. cepacia laboratory strains is unclear, although phenotypic changes in bacteria after extensive laboratory culture are not uncommon.
Efficacy of B. cepacia BC11 and AFC-BC11 in biological control of damping-off of cotton caused by R. solani.
Cotton seeds were coated with various numbers of BC11 cells and then planted into vermiculite infested with R. solani. After 3 weeks, we found that as few as 100 BC11 cells per seed dramatically increased germination rates (Table 3). At 105 cells per seed, 100% survival and complete disease protection were observed. To evaluate the importance of AFC-BC11 in the observed biocontrol, different amounts of purified AFC-BC11 were applied to cotton seeds, which were subsequently planted into R. solani-infested vermiculite. As little as 5 μg of purified AFC-BC11 dramatically increased cotton seedling emergence and survival (Table 3). The disease control provided by 10 μg of AFC was the same as that provided by 104 BC11 cells. Taken together, these results show that B. cepacia BC11 can be very effective in controlling damping-off and imply that the AFC it produces is a primary determinant of its biological control ability.
TABLE 3.
Determination of the number of B. cepacia BC11 cells or the amount of purified AFC-BC11 needed to control damping-off of cotton caused by R. solani
| Antifungal treatmenta | Emergence (%)b,d | Survival (%)c,d |
|---|---|---|
| No. of BC11 cells/seed | ||
| 0 | 10 | 0 |
| 102 | 40 | 0 |
| 103 | 100 | 30 |
| 104 | 100 | 60 |
| 105 | 100 | 95 |
| Amt (μg) of AFC/seed | ||
| 0 | 3 | 0 |
| 1 | 15 | 8 |
| 5 | 70 | 52 |
| 10 | 100 | 85 |
| 20 | 100 | 95 |
Cotton seeds were coated with the indicated number of BC11 cells or indicated amount of purified AFC-BC11 and then planted into R. solani-infested vermiculite.
Percentage of cotton seeds that germinated in 3 weeks. Values are the means from two independent experiments with 20 seeds for each treatment. Without R. solani, 100% germination was observed.
Percentage of seedlings living after 3 weeks. Values are the means from two independent experiments with 20 seeds for each treatment. Without R. solani, there was 100% survival.
Analysis of variance (statistical analysis package version 6.3 [SAS Institute]) indicated a significant correlation between the number of B. cepacia cells or the amount of AFC and both emergence (cells, P = 0.041 and R2 = 0.8; AFC, P = 0.012 and R2 = 0.88) and survival (cells, P = 0.007 and R2 = 0.94; AFC, P = 0.005 and R2 = 0.93).
Generation and characterization of AFC-deficient mutants of B. cepacia BC11.
To confirm the primary importance of AFC-BC11 in biocontrol and gain insight into its biosynthesis, we isolated and characterized Tn5 insertion mutants defective in AFC-BC11 production. Matings between B. cepacia BC11R and E. coli SM10(pSUP2021) produced 2,000 putative Tn5 insertion mutants of B. cepacia, which were picked and replica plated onto PDA plates spread with R. solani hyphae. After 3 days, 20 mutants showed reduced fungal growth inhibition. However, after rigorous retesting, only four mutants (AFC143, AFC145, AFC202, and AFC666 [Table 1]) showed both a wild-type growth rate and a bona fide loss of ability to inhibit growth of R. solani. AFC activity in 80% acetone extracts of PDA-grown cells of all four mutants was less than 1% of that in extracts from wild-type cells. When extracts from mutants AFC202 and AFC666 were fractionated by HPLC, the major peak associated with AFC activity was missing (data not shown). Moreover, both extracts showed 10-fold less A322, the absorbance maximum of AFC-BC11. These results strongly suggested that these B. cepacia mutants have Tn5 inserted in genes required for production of AFC-BC11.
Southern analysis (42) of genomic DNA from the four mutants with the 3.3-kb HindIII fragment of Tn5 as a probe showed that the Tn5 insertion in three of the mutants (AFC202, AFC145, and AFC143) was apparently in the same region of the genome, because they all showed the same sizes of hybridizing EcoRI and SmaI fragments (data not shown). Moreover, all the mutants appeared to have only a single Tn5 insertion, since only one EcoRI fragment hybridized, a 10-kb fragment for AFC666 and a 6-kb fragment for the others. Since AFC202 and AFC666 showed different sizes of hybridizing fragments when EcoRI-, BamHI-, KpnI-, or SmaI-digested genomic DNA was used, we selected them as unique for further analysis.
To confirm that biocontrol of R. solani depends strongly on production of AFC-BC11, 106 cells of the AFC-deficient BC11 mutants AFC202 and AFC666 were applied to cotton seeds and their biocontrol activity was compared to that of the wild type (Table 4). Both mutants had completely lost their ability to protect cotton seeds and seedlings from R. solani, even though we used >10-fold more cells than the minimum number of the wild-type cells needed for full control. Although these results strongly suggest that B. cepacia must produce AFC-BC11 for effective biological control, reduced soil survival or root colonization by the afc::Tn5 mutants could exaggerate the effect. To rule this out, a mixture of equal amounts of wild-type and AFC666 cells was inoculated onto cotton seeds, which were then planted in clean vermiculite. When the bacteria were isolated from the roots of seedlings 4, 8, and 12 days later, no significant change in the ratio of the two strains was observed, suggesting that AFC deficiency does not dramatically affect the ability of BC11 to colonize cotton roots in vermiculite. These data further support the claim that AFC-BC11 production is a primary determinant of biological control by B. cepacia BC11.
TABLE 4.
Biological control of damping-off of cotton by B. cepacia BC11 and Tn5::afc mutants
| Treatmenta
|
Emergence (%)b | Survival (%)b | |
|---|---|---|---|
| Seeds | Vermiculite | ||
| None | None | 100 A | 100 A |
| None | R. solani | 5 B | 0 B |
| BC11 | R. solani | 95 A | 90 A |
| AFC666 | R. solani | 15 B | 8 B |
| AFC202 | R. solani | 10 B | 5 B |
Cotton seeds were coated with 106 cells of the indicated B. cepacia strains and then planted into vermiculite and monitored for 3 weeks.
See Table 3, footnotes b and c. Means (n = 5) within a column followed by the same letter are not significantly different according to Duncan’s multiple-range test (d = 0.05).
Cloning of genes involved in biosynthesis of AFC-BC11.
In previous genetic studies of biocontrol bacteria, DNA sequence analysis of cloned genomic regions harboring Tn5 insertions that reduce the biocontrol activity has helped elucidate AFC biosynthetic pathways and their regulation (65). Thus, we cloned the EcoRI fragment of the AFC666 genome that had Tn5 inserted into an AFC-BC11 production gene. Plasmid DNA from a Kmr transformant (designated pKW666) harbored a 10-kb insert whose restriction endonuclease cleavage map indicated that it contained a 4.5-kb genomic DNA fragment with Tn5 inserted 1.5 kb from one end (Fig. 1).
To confirm that the cloned fragment harbored sequences involved in AFC-BC11 synthesis, we recloned it into the Tcr suicide plasmid pTOK2 (49) to produce pT666. Triparental mating of E. coli(pT666) with B. cepacia BC11R followed by selection for the Kmr Tcs phenotype produced allelic exchange recombinants of B. cepacia at a frequency of 10−9 per donor. No AFC activity was detected when 80% acetone extracts of cells from four colonies were bioassayed for AFC, confirming that they were probably marker exchanged double-crossover afc::Tn5 mutants and that pKW666 contains genes involved in AFC production. Analogous results were obtained with genomic DNA cloned from AFC202; characterization of a cloned afc::Tn5 fragment from AFC202 will be reported elsewhere.
DNA sequence analysis of a genomic region involved in AFC production.
The nucleotide sequence of the 4.5-kb genomic DNA on pKW666 was determined. Based on analysis of codon usage, putative ribosome-binding sites (RBS), and BLAST searches, four genes (afcA, afcB, afcC, and afcD) containing three complete and one partial open reading frame (ORF) were tentatively identified (Fig. 1). afcA and afcB appeared to be transcribed from one strand, while afcC and afcD were seemingly transcribed from the other. The ca. 200-bp intergenic region between afcA and afcC did not appear to contain an ORF and had a G+C content 20% lower than that of the flanking coding regions. Further inspection identified a putative divergent promoter region having two very similar sets of −35 and −10 consensus sequences separated by 150 bp (Fig. 1B). In fact, the DNA sequence upstream of the two putative −35 sequences showed extensive (60%) dyad symmetry centered around position 2130, suggesting that it may have arisen by duplication.
The afcA gene product (AfcA) appears to start at position 2282 with a GTG translation initiation codon that is preceded by a sequence with strong complementarity to the 3′ end of B. cepacia 16S rRNA (41), i.e., a strong RBS (Fig. 1B). This suggests that afcA encodes a 587-residue protein. Database searches revealed that over its entire length, AfcA has strong (ca. 35%) amino acid sequence identity to portions of saframycin MxI synthetase (SafB1) of Myxococcus xanthus (51, 52), pristinamycin synthetase (SnbC) of Streptomyces pristinaespiralis (8, 62), and several other very large proteins, all of which are involved in ATP-mediated activation and/or polymerization of amino acid building blocks of peptide antibiotics (34). In particular, residues 171 to 196 and 430 to 478 of AfcA show >50% identity to the most highly conserved regions of these proteins, i.e., the amino acid-activating domains (34, 61) (Fig. 2A). This region is characterized by two sequence motifs, TSGsTshPK and RTGD, the putative ATP-binding site and ATPase sites (I and II; Fig. 2A) (61).
FIG. 2.
Amino acid sequence similarities between afc gene products and proteins of known function. (A) Alignment of a portion of AfcA with the amino acid activating domains of several peptide synthetases. Residues similar or identical in at least five are marked #; residues identical or similar in at least three are marked ∗ and +, respectively. I, ATP-binding domain; II, ATPase domain. SafB1, saframycin Mx1 synthetase (U24657); SnbC, pristinamycin synthetase (X98690); Bsu1, polyketide synthetase of Bacillus subtilis (U00024); EntF, enterobactin synthetase component F (P11454); GrsB, gramicidin S synthetase (P14688). (B) Alignment of the predicted amino acid sequences of two portions of AfcC with two segments of the ω-3 fatty acid desaturase (ω-desat) of Brassica napus (L22963). Conserved histidine boxes believed to be associated with the active site are marked (∧∧∧∧∧).
Upstream of the proposed afcA start codon and running in the opposite direction was the AfcC ORF (Fig. 1). While there are several potential translational start codons in the first 80 residues, only the TTG at nucleotide 1936 is preceded by a very strong RBS (Fig. 1B). Moreover, the region upstream of the TTG has a very atypical codon utilization frequency and high A+T content for an organism with a 65% G+C content. This region appears more likely to be part of the divergent promoter mentioned above and shown in Fig. 1B. Two segments of the putative 334-residue AfcC ORF show 40% amino acid sequence identity to portions of ω-3 fatty acid desaturases, a family of plant enzymes that introduce a double bond into 18:3 fatty acids (68) (Fig. 2B). The homologous regions contain several histidine box motifs proposed to be metal ion-binding sites involved in the activities of these enzymes (68). Following AfcC (starting with ATG at position 893) is the 283-residue AfcD ORF. Residues 64 to 88 and 139 to 182 of this ORF show ca 35% amino sequence identity to segments of several plant stearoyl acyl carrier protein desaturases, enzymes that introduce a double bond at the Δ9 position of a C18 fatty acyl chain (59). Interestingly, NMR analysis of AFC-BC11 suggests that it contains an unsaturated fatty acyl chain with at least one cis double bond. Moreover, the AFC-deficient strain AFC666 has Tn5 inserted in afcC, further implicating AfcC and/or AfcD as potential desaturases involved in biosynthesis of AFC-BC11.
Overlapping the afcA stop codon is the beginning of a fourth putative ORF, afcB; it begins with an ATG at position 4044, is preceded by a good RBS, and extends beyond the end of the cloned genomic fragment. Residues 40 to 70 of this 93-residue partial ORF show very strong amino acid sequence identity to a conserved region of PotG, a membrane-bound putrescine transport protein of E. coli (50) and a glutamine transporter of Methanococcus. The homologous region harbors the very highly conserved ATP-binding site motif (P-loop signature sequence) found in all members of the ATP-binding cassette transporter superfamily (20), especially the one found in amino acid transporters (reference 34a and results not shown).
Evidence for a direct role of the afc gene cluster in AFC-BC11 production.
In strain AFC666, Tn5 is in afcC, suggesting a role for the putative AfcC desaturase in AFC production, but possible polar effects by Tn5 complicate the interpretation. Based on its sequence homology to enzymes involved in synthesis of peptide antibiotics, it is plausible that AfcA is involved in synthesis of a peptide moiety of AFC-BC11. To directly assess this possibility, we inserted a nonpolar nptI cartridge (14) into the single XhoI site of afcA and in the same transcriptional orientation (Fig. 1). The resultant afcA::nptI fragment was recloned into the pTOK2 suicide vector, and the resultant plasmid (pT666-1) was transferred into BC11R by conjugative mobilization. Double-crossover marker-exchanged recombinants were selected by their Kmr Tcs phenotype and arose at a frequency of 10−9 per donor. Bioassay of 80% acetone extracts of cells from four such mutants (e.g., AFC666M2 [Table 1]) showed that they produced >300-fold-reduced levels of AFC-BC11 (data not shown). When afcA alone (i.e., the StyI-EcoRI fragment of pKW666-1) was cloned downstream of the lac promoter of pRK415 and the resultant plasmid (pR666 [Fig. 1]) was transferred into the afcA::nptI BC11 mutant, AFC production was increased >40-fold, arguing against strong polar effects by the nptI insertion. These results prove that AfcA is directly involved in the biosynthesis of AFC-BC11.
DISCUSSION
We previously screened 15 different soil isolates with biocontrol potential for consistent production of extracellular AFCs and amenability to genetic manipulation (56). One promising strain, B. cepacia BC11, strongly inhibited the growth of R. solani and Sclerotium rolfsii in artificial soils and, in later field tests, appeared to enhance the yield of peanuts (55). However, the inconsistency of the performance of BC11, and biocontrol systems in general (19, 65), prompted us to seek a more detailed understanding of B. cepacia BC11 and its production of AFCs. We found that the major antifungal antibiotic activity of BC11 resided in a relatively insoluble, membrane-associated compound, although sometimes it was detected in the culture supernatant. After purification of this AFC by differential solvent extraction and reverse-phase HPLC, MS analysis showed that it had a molecular mass of 733 Da. NMR suggested that it contains an aromatic component and a fatty acyl substituent with a cis double bond. Analysis of acid hydrolysates detected four amino acids, suggesting the presence of a peptide moiety. These characteristics imply that this AFC produced by B. cepacia BC11 (called AFC-BC11) is a lipopeptide different from most other previously purified and structurally characterized antibiotics from biocontrol pseudomonads (19, 65). While some of its characteristics are reminiscent of members of the cepacidine (xylocandin) family of lipopeptide antibiotics from some B. cepacia strains (6, 37, 46), AFC-BC11 is much smaller (733 versus 1100 Da) and lacks xylose. Moreover, cepacidines are very active against Candida spp., Saccharomyces cerevesiae, Aspergillus niger, and several other fungal dermatophytes whereas AFC-BC11 was not (Table 2). Nonetheless, AFC-BC11 was very active against many other fungi, most of which were soil isolates and plant pathogens. The in vitro growth of many fungi was completely inhibited by as little as 1 μg/ml, similar to the potency of amphotericin B (37), pyrrolnitrin (22), and pyoluteorin (23) but more potent than at least one phenazine derivative (64).
Production of a potent AFC does not always guarantee good biological control performance (35, 43). However, with B. cepacia BC11, we found that in the presence of R. solani as few as 104 cells (or 5 μg of purified AFC-BC11) per seed increased the germination and survival rates of cotton seedlings from 0 to >50%. In comparison to what has been reported for many biocontrol strains (21, 23, 30, 35, 40, 43, 57), the minimum number of BC11 cells per seed required for full disease control is 2 orders of magnitude lower than that for most strains. However, unlike some other tests, our biocontrol assays were not done in natural soils, and so the efficacy of BC11 relative to other strains remains to be clarified.
Using Tn5 tagging, we isolated a genomic fragment encoding proteins essential for biological control by B. cepacia BC11. DNA sequence analysis of this fragment provided insight into the structure and biosynthesis of AFC-BC11. Part of the putative amino acid sequence of the product of one of the resident ORFs, AfcA, showed high similarity to a domain conserved in proteins which use ATP to convert amino acids into aminoacyl adenylates (34, 61), in particular to the amino acid activation domains of two large multifunctional peptide synthetases, SafB1 (51, 52) and SnbC (8, 62), that are involved in nonribosomal peptide synthesis of the antibiotics saframycin (M. xanthus) and pristinamycin (S. pristinaespiralis). However, AfcA is much smaller than peptide synthetases, since it has only one activation domain instead of the more typical three or four and lacks domains for elongation and attachment of the 4-phosphopantetheinyl arm (62). This architecture is reminiscent of SnbA, the 3-OH picolinic acid:AMP ligase that catalyzes the adenylation and activation of 3-OH picolinic acid during the synthesis of pristinamycin but does not subsequently bind it as a thioester. Instead, SnbA is thought to “deliver” activated 3-OH picolinic acid to a peptide synthetase for incorporation into pristinamycin (8, 62). Since AfcA shows such high homology to these amino acid activation domains and since a nonpolar insertion of nptI in afcA completely knocked out AFC production, it is plausible that AfcA activates an amino acid before its incorporation into a nonribosomally synthesized peptide component of AFC-BC11, consistent with physiochemical data suggesting that AFC-BC11 is a lipopeptide.
Unlike most other reports about B. cepacia, we were able to use reverse genetics to confirm the function of a cloned DNA sequence in a physiological process. Insertion of a nonpolar nptI cartridge into the genomic copy of afcA of B. cepacia BC11 completely eliminated the production of AFC-BC11, the ability to inhibit growth of R. solani in vitro, and the biocontrol activity. These data strongly suggest that BC11 produces only one major AFC that is a primary determinant of its biocontrol ability for R. solani-induced damping-off of cotton. This is in contrast to many other biological control pseudomonads, which commonly produce multiple AFCs that additively contribute to their biocontrol activity (65).
Although we did not unambiguously demonstrate a role in biosynthesis of AFC-BC11 for the two other complete ORFs on the cloned fragment, the fact that the initial Tn5-generated AFC-deficient mutant AFC666 had Tn5 inserted in afcC implies that one or both of these genes are required for AFC-BC11 biosynthesis. However, potential polar effects on downstream genes could also cause AFC deficiency. On the other hand, the similarity of the predicted amino acid sequences of segments of AfcC and AfcD to domains of desaturase enzymes which introduce cis double bonds into aliphatic chains (68), coupled with our NMR data indicating the presence of an acyl substituent with a cis double bond in AFC-BC11, further argues for a direct role for AfcC and/or AfcD in the biosynthesis of AFC-BC11. Genetic and biochemical experiments to confirm this are in progress.
For more effective field use of biocontrol microbes, we need a clearer knowledge of the field conditions that affect the biosynthesis of AFCs and hence biocontrol. The availability of cloned AFC-BC11 biosynthetic genes should allow the construction of reporter strains to study in situ rhizosphere conditions that increase expression of the biosynthetic genes and possibly the consistency of biocontrol. This type of approach has already been proven to be valuable (15, 36). Alternatively, addition of cloned AFC-BC11 biosynthetic genes to other biocontrol strains could lead to improved biocontrol agents (12, 17, 40, 57).
ACKNOWLEDGMENTS
This research was supported in part by a grant from the Biotechnology Program of the University of Georgia Research Foundation and the Georgia Commodity Commission for Peanuts. Support for instrumentation used in structural analysis was provided to the Complex Carbohydrate Research Center by Department of Energy Center grant DE-FG09-93ER20097.
We thank Tim Denny and Mark Wise for critical comments on the manuscript.
REFERENCES
- 1.An J, Carlson R W, Glushka J, Streeter J G. Structure of a novel polysaccharide produced by Bradyrhizobium within soybean nodules. Carbohydr Res. 1994;269:303–317. doi: 10.1016/0008-6215(94)00361-i. [DOI] [PubMed] [Google Scholar]
- 2.Andrews J H. Biological control in the phyllosphere. Annu Rev Phytopathol. 1992;30:603–635. doi: 10.1146/annurev.py.30.090192.003131. [DOI] [PubMed] [Google Scholar]
- 3.Ballard R W, Palleroni N J, Doudoroff M, Stanier R Y. Taxonomy of the aerobic Pseudomonads: P. cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970;60:199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- 4.Becker J O, Schwinn F J. Control of soil borne pathogens with living bacteria and fungi—status and outlook. Pestic Sci. 1993;37:355–363. [Google Scholar]
- 5.Bevivino A, Tabacchioni S, Chiarini L, Caruri M Y, Del Gallo M, Visca P. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology. 1994;140:1069–1077. doi: 10.1099/13500872-140-5-1069. [DOI] [PubMed] [Google Scholar]
- 6.Bisacchi G S, Hockstein D R, Koster W H, Parker W, Rathnum M L, Unger S E. Xylocandin: a new complex of antifungal peptides. II. Structural studies and chemical modifications. J Antibiot. 1987;40:1520–1529. doi: 10.7164/antibiotics.40.1520. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright, D. K., and M. D. Benson. February 1994. Pseudomonas cepacia strain 5.5B and method of controlling Rhizoctonia solani therewith. U.S. patent 5,288,633.
- 8.Crecy-Lagard V, Blanc V, Gil P, Naudin L, Lorenzon S, Famechon A, Bamas-Jacques N, Crouzet J, Thibaut D. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J Bacteriol. 1997;179:705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowling D N, O’Gara F. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 1994;12:133–141. [Google Scholar]
- 11.Farr D F, Bills G F, Chamuris G P, Rossman A Y. Fungi on plants and plant products in the United States. St. Paul, Minn: American Phytopathological Society; 1989. [Google Scholar]
- 12.Fenton A M, Stephens P M, Crowley J, O’Callaghan M, O’Gara F. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridlender M, Inbar J, Chet I. Biological control of soilborne plant pathogens by a β-1,3 glucanase-producing Pseudomonas cepacia. Soil Biol Biochem. 1993;25:1121–1221. [Google Scholar]
- 14.Galan J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgakopoulos D G, Henderson M, Panopoulos N J, Schroth M N. Analysis of expression of a phenazine biosynthesis locus of Pseudomonas aureofaciens PG12 on seeds with a mutant carrying a phenazine biosynthesis locus-ice nucleation reporter gene fusion. Appl Environ Microbiol. 1994;60:4573–4579. doi: 10.1128/aem.60.12.4573-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitaitis, R., and D. Sumner. Unpublished results.
- 17.Gutterson N C. Microbial fungicides: recent approaches to elucidating mechanisms. Crit Rev Biotechnol. 1990;10:69–91. [Google Scholar]
- 18.Hammer P E, Hill D S, Lam S, VanPee K, Ligon J. Four genes from the genome of Pseudomonas fluorescens that encodes the biosynthesis of pyrrolnitrin. Appl Environ Microbiol. 1997;63:2147–2154. doi: 10.1128/aem.63.6.2147-2154.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handelsman J, Stabb E V. Biocontrol of soilborne plant pathogens. Plant Cell. 1996;8:1855–1869. doi: 10.1105/tpc.8.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 21.Hill D S, Stein J I, Torkewitz N R, Morse A M, Howell C R, Pachlatko J P, Becker J O, Ligon J M. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl Environ Microbiol. 1994;60:78–85. doi: 10.1128/aem.60.1.78-85.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howell C R, Stipanovic R D. Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 23.Howell C R, Stipanovic R D. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens Pf-5 and its antibiotic, pyoluteorin. Phytopathology. 1980;70:712–715. [Google Scholar]
- 24.Howie W J, Suslow T. Role of antibiotic synthesis in the inhibition of Pythium ultimum in the cotton spermosphere and rhizosphere by Pseudomonas fluorescens. Mol Plant-Microbe Interact. 1991;4:393–399. [Google Scholar]
- 25.Janisiewicz W J, Roitman J. Biological control of blue mold and gray mold on apple and pear with Pseudomonas cepacia. Phytopathology. 1988;78:1679–1700. [Google Scholar]
- 26.Jayaswal R K, Fernandez M F, Visintin L, Upadhyay R S. Transposon Tn5-259 mutagenesis of Pseudomonas cepacia to isolate mutants deficient in antifungal activity. Can J Microbiol. 1992;38:309–312. doi: 10.1139/m92-051. [DOI] [PubMed] [Google Scholar]
- 27.Jayaswal R K, Fernandez M, Upadhyay R S, Visintin L, Kurz M, Webb J, Rinehart K. Antagonism of Pseudomonas cepacia against phytopathogenic fungi. Curr Microbiol. 1993;26:17–22. doi: 10.1007/BF01577237. [DOI] [PubMed] [Google Scholar]
- 28.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner P, Haas D, Defago G. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 29.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.King E B, Parke J L. Biocontrol of Aphanomyces root rot and Pythium damping-off by Pseudomonas cepacia AMMD on four pea cultivars. Plant Dis. 1993;77:1185–1188. [Google Scholar]
- 31.King E B, Parke J L. Population density of the biocontrol agent Burkholderia cepacia ammdr1 on four pea cultivars. Soil Biol Biochem. 1996;28:307–312. [Google Scholar]
- 32.Kirinuki T, Iwanuma K, Suzuki N, Fukami H, Ueno T. Altericidins, a complex of polypeptide antibiotics produced by Pseudomonas sp. and their effect for the control of black spot of pear caused by Alternaria Kikuchiana Tanaka. Sci Rep Fac Agric Kobe Univ. 1977;12:223–230. [Google Scholar]
- 33.Kitten T, Willis D K. Suppression of a sensor kinase-dependent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J Bacteriol. 1996;178:1548–1555. doi: 10.1128/jb.178.6.1548-1555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinkauf H, Von Dohren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochemistry. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 34a.Koonin, E. Personal communication.
- 35.Kraus J, Loper J E. Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of Pythium damping-off of cucumber. Phytopathology. 1992;82:264–271. [Google Scholar]
- 36.Kraus J, Loper J E. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 1995;61:849–854. doi: 10.1128/aem.61.3.849-854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C, Kim S, Hyun B, Suh J, Yon C, Kim C, Lim Y, Kim C. Cepacidine A, a novel antifungal antibiotic produced by Pseudomonas cepacia. J Antibiot. 1994;47:1402–1418. doi: 10.7164/antibiotics.47.1402. [DOI] [PubMed] [Google Scholar]
- 38.Lessie T G, Gaffney T. Catabolic potential of Pseudomonas cepacia. In: Sokatch J R, Ornston L N, editors. The bacteria: a treatise on structure and function. X. The biology of Pseudomonas. New York, N.Y: Academic Press, Inc.; 1986. pp. 439–481. [Google Scholar]
- 39.Lessie T G, Hendrickson W, Manning B, Devereux R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett. 1996;144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 40.Ligon J, Lam S, Gaffney T, Hill S, Hammer R, Torkewitz N. Biocontrol: modifications for enhanced antifungal activity. In: Stacey G, Mullin B, Gresshoff P, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interactions; 1996. pp. 457–462. [Google Scholar]
- 41.Ludwig W, Rossello-Mora R, Aznar R, Klugbauer S, Spring S, Reetz K, Beimfohr C, Brockmann E, Kirchhof G, Dorn S, Bachleitner M, Klugbauer N, Springer N, Lane D, Nietupsky R, Weizenegger M, Schleifer K H. Comparative sequence analysis of 23S rRNA from proteobacteria. Syst Appl Microbiol. 1995;18:164–188. [Google Scholar]
- 42.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 43.Maurhofer M, Keel C, Schinder U, Voisard C, Haas D, Defago G. Influence of enhanced antibiotic production in Pseudomonas fluorescens strain CHA0 on its disease suppressive capacity. Phytopathology. 1992;82:190–195. [Google Scholar]
- 44.Mavrodi D V, Ksenzenko V, Chatuev B, Thomashow L S, Boronin A M. Structural and functional organization of Pseudomonas fluorescens genes encoding enzymes of phenazine-1-carboxylic acid biosynthesis. Mol Biol. 1997;31:62–68. [PubMed] [Google Scholar]
- 45.Mead D A, Skorupa E S, Kemper B. Single stranded DNA promoter plasmids for engineering mutant RNA’s and protein: synthesis of a ’stretched’ parathyroid hormone. Nucleic Acids Res. 1985;13:1103–1108. doi: 10.1093/nar/13.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyers E, Bisacchi G S, Dean L, Liu W C, Minassian B, Slusarchyk D S, Sykes R B, Tanaka S K, Trejo W. Xylocandin: a new complex of antifungal peptides. I. Taxonomy, isolation, and biological activity. J Antibiot. 1987;40:1515–1529. doi: 10.7164/antibiotics.40.1515. [DOI] [PubMed] [Google Scholar]
- 47.Parke J L. Population dynamics of Pseudomonas cepacia in the pea spermosphere in relation to biocontrol of Pythium. Phytopathology. 1990;80:1307–1311. [Google Scholar]
- 48.Parke J L, Rand R, Joy A, King E B. Biological control of Pythium-damping off and Aphanomyces root rot of peas by application of Pseudomonas cepacia or Pseudomonas fluorescens to seed. Plant Dis. 1991;75:987–992. [Google Scholar]
- 49.Parker W L, Rathnum M L, Seiner V, Trejo W H, Principe P A, Sykes R B. Cepacin A and cepacin B, two new antibiotics produced by Pseudomonas cepacia. J Antibiot. 1984;37:431–440. doi: 10.7164/antibiotics.37.431. [DOI] [PubMed] [Google Scholar]
- 50.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobyashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes of the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 51.Posiech A, Cruzel B, Bietenhader J, Schupp T. A new Myxococcus xanthus gene cluster for the biosynthesis of the antibiotic saframycin Mx1 encoding a peptide synthetase. Microbiology. 1995;141:1793–1803. doi: 10.1099/13500872-141-8-1793. [DOI] [PubMed] [Google Scholar]
- 52.Posiech A, Bietenhader J, Schupp T. Two multifunctional peptide synthetases and an O-methyltransferase are involved in the biosynthesis of the DNA-binding antibiotic and antitumour agent saframycin Mx1 from Myxococcus xanthus. Microbiology. 1996;142:741–746. doi: 10.1099/00221287-142-4-741. [DOI] [PubMed] [Google Scholar]
- 53.Rhaman M, Guard-Petter J, Asokan K, Carlson R W. The structure of the capsular polysaccharide from a swarming strain of pathogenic Proteus vulgaris. Carbohydr Res. 1997;301:213–220. doi: 10.1016/s0008-6215(97)00093-1. [DOI] [PubMed] [Google Scholar]
- 54.Roberts D P, Denny T P, Schell M A. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J Bacteriol. 1987;170:1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schell, M., and T. Brenneman. 1992. Unpublished observations.
- 56.Schell, M., and Y. Kang. 1996. Unpublished observations.
- 57.Schinder U, Keel C, Blumer C, Troxler J, Defago G, Haas D. Amplication of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J Bacteriol. 1995;177:5387–5392. doi: 10.1128/jb.177.18.5387-5392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 59.Slocombe S, Cummins I, Jarvis R, Murphy D J. Nucleotide sequence and temporal regulation of a seed-specific Brassica napus cDNA encoding a stearoyl-acyl carrier protein (ACP) desaturase. Plant Mol Biol. 1992;20:151–155. doi: 10.1007/BF00029157. [DOI] [PubMed] [Google Scholar]
- 60.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 61.Stachelhaus T, Marahiel M A. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 62.Thibaut D, Bisch D, Ratet N, Maton L, Couder M, Debussche L, Blanche F. Purification of peptide synthetases involved in pristinamycin I biosynthesis. J Bacteriol. 1997;179:697–704. doi: 10.1128/jb.179.3.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomashow L S. Biological-control of plant-root pathogens. Curr Opin Biotechnol. 1996;7:343–347. doi: 10.1016/s0958-1669(96)80042-5. [DOI] [PubMed] [Google Scholar]
- 64.Thomashow L S, Weller D M. Role of a phenazine antibiotic from Pseudomonas fluorescens 2-97 in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomashow L S, Weller D. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 1. London, United Kingdom: Chapman & Hall, Ltd.; 1996. pp. 187–236. [Google Scholar]
- 66.Wise M G, Shimkets L, McArthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hahimoto Y, Ezaki T, Arakawa T. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas group II to the new genus with the type species Burkholderia cepacia Palleroni and Holmes 1981 comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 68.Yadav N S, Wierzbicki A, Aegerter M, Caster C, Perez-Grau L, Kinney A, Hitz W, et al. Cloning of higher plant omega-fatty acid desaturases. Plant Physiol. 1993;103:466–476. doi: 10.1104/pp.103.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]