Graphical abstract
Keywords: DNA methylome, Fruit flavor, Cold storage, Regulatory network, Transcriptome, Transcription factors (TFs)
Highlights
-
•
Multi-omics data unravels the regulatory control of flavor volatiles in fruit.
-
•
Total aroma volatiles decreased rapidly after cold storage of peach fruit.
-
•
PpMADS2 regulates fruit ester contents via affecting PpAAT1 expression.
-
•
Chilling caused fruit volatile loss is associated with induced DNA methylation.
Abstract
Introduction
Flavor is a major contributor to consumer preference. Despite being effective at extending the fruit’s commercial life, cold storage also results in a significant loss of flavor volatiles. To date, there has been few studies on the metabolic dynamics and the mechanism underlying the regulatory networks that modulate flavor loss during cold storage for fruit.
Methods
The volatile contents were detected by Gas Chromatography–Mass Spectrometer (GC–MS). Weighted gene co-expression network analysis (WGCNA) was used to identify structure genes and transcription factors (TFs). DNA methylation was analyzed by whole-genome methylation sequencing during cold storage.
Results
We generated a temporal map, over hourly to weekly timescales, for the effects of chilling on flavor volatiles by combining metabolome, transcriptome, and DNA methylome in peach fruit. Based on the big data analysis, we developed a regulatory network for volatile formation and found that a decrease in volatiles during cold storage was significantly correlated with a decrease in the expression of synthesis genes. Moreover, TFs associated with these structure genes were identified. Expression of genes involved in ethylene biosynthesis was reduced while cold tolerance pathway was activated in response to low temperature. Functions of those genes were confirmed through transgenic experiments and across peach cultivars, suggesting our dataset is a useful tool for elucidating regulatory factors that have not yet been clarified in relation to flavor and cold tolerance. Genome wide DNA methylation was induced by chilling and peaked at 7 d followed by a decline during 28 d cold storage. Reduction of gene expression was accompanied by major changes in the methylation status of their promoters, including PpACS1, PpAAT1, PpTPS3 and PpMADS2.
Conclusion
Our study revealed the mechanism for chilling-induced flavor loss of peach fruit through time-course transcriptome and DNA methylome analysis.
Introduction
Cold stress is a major environmental factor affecting distribution, growth, yield and quality of horticultural crops. Besides great impacts on whole plants, an additional effect of low temperature involves the cold stress occurring during storage. Cold storage is the most popular and reliable way to prolong the shelf life of perishable plant products including fresh fruits. Suitable storage temperatures can extend storage life by several months for fruit such as apple and kiwifruit, and up to 2–4 weeks for sweet cherries, apricots, and peaches [1]. However, fruit suffer the loss of flavor quality if storage at inappropriate temperature and/or storage time is extended at low temperature [2]. Chilling-induced flavor loss is a symptom of chilling injury and is complained by consumers for decades. Therefore, it is economically and scientifically interesting to understand the mechanism for such chilling-induced flavor loss.
The sensory quality of fruit is a complex property, mainly composed of soluble sugars, organic acids, and volatile organic compounds (VOCs). Changes in sugar contents during cold storage were observed for kiwifruit and banana, because these fruits have high contents of starch which could be converted to sugars after harvest [3]. For tomato fruit, chilling-induced flavor loss was associated with reduction of VOCs [2], [4], [5], [6], and no significant change was observed for sugars and acids [2], [7], [8]. Significant changes in VOC profiles caused by cold storage were also reported for fruit species such as mandarin [5], grapefruit [9], apple [10], kiwifruit [11], grape [12] and peach [13], [14]. Although there have been many reports on how the flavor quality of fruit changes when it is stored in the cold, most of them focus on the physiological and biochemical levels and the study of molecular mechanisms has lagged over the past three decades.
Multi-omics techniques, including metabolome, transcriptome, and DNA methylome have been applied in model fruit tomato to reveal mechanisms governing gene expression in response to cold stress. For instance, effects of storage temperatures on tomato fruit VOC production were compared and genes related to chilling-induced reduction of VOCs were identified through the combination of Gas Chromatography-Mass Spectrometer (GC–MS) and RNA sequencing (RNA-Seq) [6]. Furthermore, integration of metabolome and transcriptome with DNA methylome established a foundation for elucidating the molecular mechanisms of chilling-induced flavor loss in tomato fruit [2]. Low temperatures repressed the transcript of DNA demethylase SlDML2, which contributed to increase DNA methylation levels at the promoter of genes involved in biosynthesis of flavor volatiles and suppressed the expression of those genes [2]. However, whether such regulation of low temperature on flavor-related VOCs in tomato also fits to other fruits remains unknown.
Peach (Prunus persica L.) is an economically important fruit crop originated in China and is now widely distributed in the world. The global planting area of peaches was 1.527 million hectares, and the yield was 26.433 million tons in 2020 (Food and Agriculture Organization, https://www.fao.org/faostat/). Peach fruit are widely consumed due to their distinctive flavor and nutritional value. Characteristic aroma is one of the key aspects in determining the quality of peach flavor. Approximately 100 VOCs and flavor-related volatiles were identified [15], [16], [17]. Changes in VOC profiles as well as identification of key enzymes and related biosynthetic genes during peach fruit development and ripening were investigated [18], [19]. The sweet and floral aroma of peach fruit is largely made up of the monoterpene linalool, whose biosynthesis is catalyzed by terpene synthase 1 (PpTPS1) and PpTPS3 [20], [21]. Lipoxygenase (LOX) pathway enzyme alcohol acyltransferase 1 (PpAAT1) is involved in the biosynthesis of ester (Z)-3-hexenyl acetate and lactone γ-decalactone, which contribute to fruity note of ripe peach fruit [19], [22]. PpNAC1 was recently discovered as a transcription factor (TF) that activates PpAAT1 expression. [19]. TF PpbHLH1 and PpERF61 were associated with linalool synthesis via activating PpTPS3 expression during development and ripening of peach fruit at ambient temperature [21], [23]. Furthermore, effects of storage temperatures on peach fruit VOC profiles and expression of LOX pathway genes have been investigated [13], [18], [24], [25]. In addition, transcriptome has been applied to study the mechanism of cold tolerance during cold storage of peach fruit [26], [27], [28]. Previous study showed that anthocyanin accumulation in peach fruit induced by different cold storage temperature was related to DNA methylation [29]. However, it remains unclear how peach fruit flavor-related metabolites change dynamically during cold storage. Moreover, regulators associated with flavor loss caused by postharvest handling system that chills fruit need to be investigated.
Here, we provide a comprehensive analysis of the effect of low temperature on fruit flavor metabolome, transcriptome and DNA methylome. We selected samples based on hour, day, week, and month after cold storage, including fruits at harvest without cold storage (0 h), within cold storage of one day (1 h, 3 h, 6 h, 12 h, 1 d), one week (the 3rd day and the 7th day), and one month (the 28th day) at low temperature of white-fleshed melting peach fruit. A regulatory network associated with VOC synthesis and key transcription factors (TFs) that directly activate structural genes to produce VOCs were generated through the combination of flavor-related metabolites and transcriptome analysis. In addition, genes related to cold tolerance were identified and their functions were validated through overexpressing experiments. Integrating the transcriptome and DNA methylome allowed for further analysis of the relationship between DNA methylation and gene expression.
Materials and methods
Plant materials and treatments
The white-fleshed melting peach (Prunus persica L. Batsch cv. Hujingmilu) were harvested at commercial maturity. Fruit were harvested from an orchard (30°40′N, 120°49′E) in Jiaxing, Zhejiang Province, China, and brought to the laboratory within 2 h of being harvested. Fruit that was uniform and free of visual flaws were kept at 0 ℃ and relative humidity of 90 % for 0, 1, 3, 6, 12 h and 1, 3, 7, 28 days. At each time point, three biological replicates were set, and five fruits were chosen from each replicate. The flesh tissue slices with a thickness of about 5 mm were frozen in liquid nitrogen and kept at −80 ℃ for biochemical and molecular analysis.
Fruit temperature, ethylene production and firmness analysis
Dynamic changes in fruit temperature were recorded by inserting a sensor into peach flesh tissue (∼1 cm) using temperature and humidity recorder (ZDR-20 J, Hangzhou, China). Fruits were sealed for 1 h in a 1.8 L air-tight box for ethylene production analysis. 1 mL of gas sample was injected into a gas chromatography (GC) (Agilent Technologies 6890 N GC System, CA, USA) with a flame ionization detector. Oven, injector, and detector temperatures were 100, 140, and 230 °C, respectively. Using the previously described procedure, fruit firmness was assessed using a texture analyzer (TA-XT2i plus, Stable Micro System, UK) with a 7.9 mm diameter head. [18]. The final puncture depth was 10 mm, and the puncture speed was 1 mm s−1.
Soluble sugars and organic acids analysis
The previously described method was used to detect soluble sugars and organic acids. [30]. 0.1 g of fruit samples ground into powder were extracted by adding 1.4 mL methanol. After vortexing, centrifugation, add pyridine methoxylamine hydrochloride and Bis (trimethylsilyl) trifluoroacetamide (BATFA): Trimethylchlorosilane (TMCS) at 99:1 (V/V) to carry out derivatization reaction, Agilent 6890 N GC (Agilent, Palo Alto, CA, USA) fitted with a HP-5 column was used for detection. The oven, injector, and detector temperatures were set at 100, 250, and 280 °C, respectively. The concentrations of sucrose, sorbitol, glucose, fructose, quinic acid, and malic acid were determined based on standard curves.
Volatile analysis
Following the method that we previously described, VOCs were measured [20]. Briefly, 5 g of powdered frozen flesh tissue was integrated with 3 mL of 200 mM Ethylene Diamine Tetraacetic Acid (EDTA) and 3 mL of 20 % calcium chloride (CaCl2). Then, 30 μL of 2-octanol (0.8 mg mL−1) was added as an internal standard before sealing the vial. Volatiles were extracted using a 65 μm polydimethylsiloxane and divinylbenzene (PDMS/DVB) fiber (Supeclo Co., Bellefonte, PA). Volatiles were analyzed by GC–MS (Agilent, 7890 N-5975, CA, USA) equipped with a CTC-PAL2 (Combi PAL, CTC Analytics, Aligent Technologies, USA) autosampler system. DB-WAX column (30 m × 0.25 mm i.d. × 0.25 μm film thickness; J & W Scientific, Folsom, CA) was used to separate volatile chemicals. The oven temperature of GC–MS was incrased from 40 °C to 100 °C at a rate of 3 °C min−1 and then to 245 °C at 5 °C min−1. Helium was used as carrier gas at flow rate of 1.0 mL min−1. The column effluent was ionized by electron ionization (EI) at 70 eV with a transfer temperature of 250 °C and a source temperature of 230 °C. Mass spectrometer was scanned in the range of 35–350 m z−1. NIST Mass Spectral Library (NIST-08) and the retention time of authentic standards were used for volatile identification. The total ion chromatogram (TIC) area was used to calculate the substance content, and the internal standard was applied for relative quantification.
RNA-Seq and real time-quantitative PCR (RT-qPCR) for gene expression analysis
According to our previous study, total RNA from peach samples was extracted for RNA-Seq [2]. Agilent 2100 bioanalyser (Agilent, Palo Alto, CA, USA) was used to test the integrity of RNA. Fragments per kilobase per million reads (FPKM) were used to evaluate transcripts. The illustrated sequencing quality and statistics of related data, as well as the overall relative relationship between fruit samples and biological replicates were shown in Table S3. An instrument called a CFX96 (made by Bio-Rad, Hercules, California, USA) was used to perform RT-qPCR analysis of gene expression. Internal controls were implemented using PpTEF2. Table S4 contained a list of primers.
Weighted correlation network analysis and visualization
Differentially expressed genes (DEGs) between 0 h and other time points were identified using the DESeq R package (1.10.1), with cut-off value fold change > 2 and corrected P value < 0.05. Weighted gene co-expression network analysis (WGCNA) (package in R) was used to identify the highly co-expressed gene modules associated with volatile contents. WGCNA network construction and module detection used the topological overlap metric (TOMtype = unsigned, minModuleSize = 30, mergeCutHeight = 0.25, powerβ = 0). The initial clusters were merged on eigengenes. Each module’s eigengene value was calculated, and the eigenvalue was then used to look for associations with the volatiles produced during cold storage. Based on correlations (r > 0.8) between structure gene and TF modules, transcriptional regulatory networks were generated. TFs were obtained from plantTFDB [31]. The networks were visualized using Cytoscape_v.3.8.2 [32].
Whole genome bisulfite sequencing (WGBS) analysis
Genomic DNA of peach fruit was extracted using a DNeasy Plant Maxi Kit (Qiagen, Hilden, Germany) following the manufacturer’s procedure. Agar gel electrophoresis was used to verify the integrity of the DNA, and NanoDropTM One (Thermo, Waltham, MA, USA) was used to verify the concentration and purity of the DNA. After the genomic DNA had been qualified, it was mechanically interrupted and fragmented, which was then subjected to fragment purification, end repair, addition of A to the 3́-end, addition of a methylation linker, and agarose coagulation. Gel electrophoresis was used for fragment size selection, and then the DNA fragments were treated with bisulfite using ZYMO EZ DNA Methylation-Gold Kit (ZYMO, Irvine, CA, USA), and further PCR amplification was performed to prepare a sequencing library. Qualified libraries were sequenced using the Illumina HiSeq 2500 platform. Clean reads were mapped to the reference peach genome (https://phytozome-next.jgi.doe.gov/info/Ppersica_v2_1). Cytosine methylation (5-methylcytosine, 5mC) was detected by Bismark software. Different Methylation Regions (DMRs) between samples were identified according to Model based analysis of bisulfite sequencing data (MOABS) [33].
Transient dual luciferase reporter assays
Full-length sequences encoding the TFs were constructed into the pGreen II 0029 62-SK vector (EU048865), while the promoters of structure genes were constructed into the pGreen II 0800-LUC reporter vector with primers (Table S4). All recombinant plasmids were transformed into Agrobacterium tumefaciens (GV3101::pSoup) by electroshock. These strains were resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, 150 μM acetosyringone, pH 5.6) and injected into tobacco (Nicotiana benthamiana) leaves. The injected tobacco was cultured in a lighted incubator for 3 d. Then samples were analyzed with the Dual-luciferase Reporter Assay System kit (Promega, Madison, WI, USA). The firefly luciferase (LUC) and renilla luciferase (REN) values were detected on the Modulus Luminometer (Promega, Madison, WI, USA) and the ratio was calculated to analyze the regulatory relationship between TFs and promoters of target genes. The LUC/REN of the empty vector SK to the promoters were negative controls, at least three biological replicates were set.
Electrophoretic mobility shift assay (EMSA)
By using the primers in Table S4, the full-length coding sequence (CDS) of TF PpMADS2 was created and inserted into the pGEX-4 T-1 expression vector with a GST-tag, and the recombinant plasmid was transformed into E. coli BL21(DE3) pLysS (Promega, Madison, WI, USA). Recombinant protein was produced and purified in accordance with the manufacturer’s instructions (Beyotime, Shanghai, China). According to the previously described procedure, EMSA was carried out using LightShift® Chemiluminescent EMSA kit (ThermoFisher Scientific, Waltham, MA, USA) [19].
Transient overexpression analysis in peach
According to our previous study [20], the CDS of TF PpMADS2 was amplified and cloned into SK vector for transient overexpression in peach. The construct was electroporated into A. tumefaciens GV3101. Permeation buffer (10 mM MES, 10 mM MgCl2 and 150 μM acetosyringone, pH 5.6) was added. After soaking in sodium hypochlorite solution for 20 min, peach leaves were washed 3 times in sterile water. Half of each leaf’s section was used to overexpress PpMADS2, while the other half served as a negative control (pGreen-SK empty vector). The leaves were placed on Murashige and Skoog (MS) medium (20 °C, relative humidity 85 %) for culture. We set up 3 biological replicates, each with ten leaves. For gene expression and volatile analysis, leaves were frozen in liquid nitrogen and kept at −80 °C after 4 days of incubation.
Heterologous overexpression in Arabidopsis and tobacco plants
Transformation was conducted in four-week-old seedlings of Arabidopsis plants by dipping floral [34]. PpCAMTA1-pBI121 constructs were used to obtain overexpression (OE) mutant in Camta2/3 mutant genetic background [35]. Notably, CaMV 35S promoter was replaced by AtCAMTA3 promoter to drive PpCAMTA1 expression with primes listed in Table S4. On 1/2 MS medium with 0.8 % agar, Arabidopsis seedlings were sown and grown for two weeks at 22 °C. Beginning at 1 °C, the freezing program was reduced by 1 °C every hour until it reached −9 °C for 6 or 20 h. Following the freezing procedure, the seedlings were moved to 4 °C for 12 h in the dark before being returned to normal conditions for 3 days. After recovery, seedlings were judged to have survived if they continued to produce new leaves. Transgenic tobacco seeds of PpFAD3-1, PpFAD3-2 and PpFAD8 were obtained from Wang et al., 2016 [36]. Transgenic tobacco seeds were sterilized and sown in MS medium containing kanamycin, wild-type (WT) were sown in MS medium without resistance. Transgenic tobaccos with two cotyledons were placed at 0 °C for 10 d followed by additional 10 d at room temperature.
Statistical analysis
SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) was used to conduct the statistical analysis. Statistical analysis between two samples was performed by the unpaired Student t-test (* P ≤ 0.05, ** P < 0.01). One-way ANOVA (Analysis of Variance) with post-hoc Tukey HSD (Honestly Significant Difference) Test was performed for multiple group comparison, P ≤ 0.05 was considered to indicate statistical significance. Origin Pro 9.0 (Origin Lab Corporation., Northampton, MA, USA) was used to generate figures.
Accession numbers
Sequence data from this study were deposited at the National Center for Biotechnology Information’s Sequence Read Archive (SRA) under accession number PRJNA852345.
Results
Cold storage influenced flavor-related volatiles in peach fruit
To extend commercial life after harvest, peach fruit were kept at 0 ℃. We initiated our study to record time series of temperature during cold storage, where 11 time points (0 h, 1 h, 3 h, 6 h, 12 h, 1 d, 3 d, 7 d, 14 d, 21 d and 28 d) were selected. A rapid decline in temperature after chilling was observed, in which fresh temperature dropped from 20 ℃ to 0 ℃ within 1 d (Fig. 1A). Such decline in temperature is accompanied by significant inhibition of ethylene production of fruit during cold storage (Fig. S1A). Meanwhile, firmness decreased slowly throughout 28 d cold storage (Fig. S1B).
Fig. 1.
Changes in flavor compounds during peach fruit cold storage. (A) Changes of fruit internal temperature. (B) Changes in total volatiles after cold storage. (C) Heatmap display for individual volatiles. (D) PCA analysis of volatiles in response to cold storage. Error bars indicate means ± SE; three biological replicates.
To understand how flavor quality changes during cold storage, we analyzed soluble sugars, organic acids, and VOCs. A total of 28 flavor related chemicals were identified, including four soluble sugars (sucrose, glucose, fructose, and sorbitol), two organic acids (quinic acid and malic acid), and 22 flavor-related volatiles (composed of fatty acids, terpenes, carotenoids and branched amino acids). It’s interesting to note that during cold storage, the contents of soluble sugars and organic acids did not differ significantly (Fig. S1C and D). In contrast, cold storage induced a dramatic decline in volatile contents (Fig. 1B). After cold storage for 3 d and 28 d, content of total volatiles was decreased by approximately 50 % and 90 % when compared to that at harvest (0 h), respectively. Therefore, we investigated changes in content of individual volatiles of fruit during cold storage (Fig. 1C; Table S1).
As shown in Fig. 1C, volatile components of peach fruit were divided into two groups during cold storage. For group I, volatiles tended to decrease with extended cold storage period, including lactones (γ-hexalactone, γ-octalactone, γ-decalactone, δ-decalactone and γ-undecanolactone), esters (hexyl acetate, (Z)-3-hexenyl acetate and (E)-2-hexenyl acetate), apocarotenoids (damasenone, dihydro-β-Ionone, geranyl acetone and β-ionone) and terpenes (linalool). For instance, content of peach characteristic flavor γ-decalactone declined from 107.57 ng/g at harvest (0 h) to under detected limit level after 28 d cold storage (Table S1). For group Ⅱ, volatiles tended to increase after chilling, particular at 7 d of cold storage. Group Ⅱ includes defensed related β-myrcene and d-limonene (Fig. 1C). Principal Component Analysis (PCA) confirmed that peach fruit at 7 d and 28 d cold storage could be separated from those samples within 3 d chilling after harvest (Fig. 1D), indicating a shift in content of volatiles after 7 d cold storage.
Dynamic changes in the transcriptome of peach fruit during cold storage
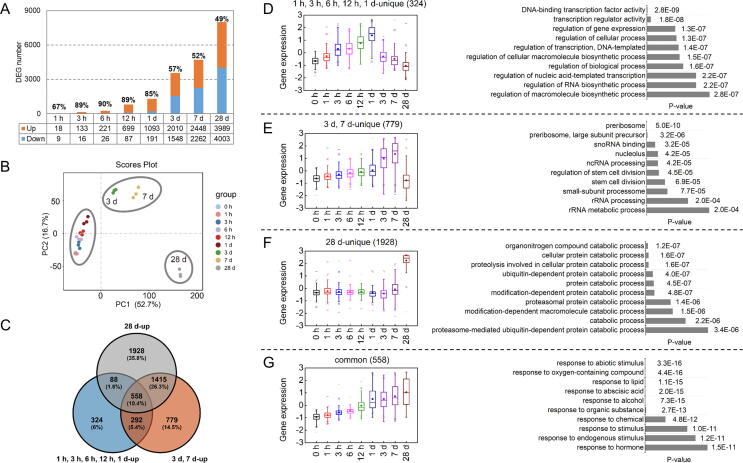
To explore the molecular mechanism for cold storage caused changes in flavor volatile contents, transcriptome analysis was performed. There were 27 RNA-Seq libraries for sequencing, producing 1.17 Gb unique reads (Table S2). Compared to fruit stored at 0 h (room temperature), a total of 9983 DEGs were identified, including 599 TFs derived from 52 families (Fig. S2). Both up-regulated and down-regulated DEG numbers were gradually increased along with extended cold storage period (Fig. 2A). It noteworthy that during cold storage from 1 h to 1 d, 67 %-90 % DEGs were up-regulated, similar numbers of up- and down-regulated DEGs were observed for 3 d, 7 d and 28 d storage. According to their expression patterns during cold storage, these DEGs could also be divided into three groups (Fig. 2B). DEGs from 1 h, 3 h, 6 h, 12 h and 1 d were grouped, DEGs from 3 d and 7 d were clustered as the second group, DEGs from 28 d were separated from the first two groups in a PCA model.
Fig. 2.
Changes in transcriptome of peach fruit during cold storage. (A) DEG numbers after cold storage. (B) PCA analysis of DEGs using FPKM as variables. (C) Venn diagram showing the overlapping and uniquely differentially up-regulated genes during cold storage. (D) Normalized box plots of up-regulated genes expression and enriched GO terms at 1 h, 3 h, 6 h, 12 h, 1 d uniquely. (E) Normalized box plots of up-regulated genes expression and enriched GO terms at 3 d, 7 d uniquely. (F) Normalized box plots of up-regulated genes expression and enriched GO terms at 28 d uniquely. (G) Normalized box plots of up-regulated common genes expression and enriched GO terms during cold storage.
As shown in Fig. 2C, Venn diagram revealed that 324 up-DEGs were unique for peach fruit within 1 d storage (Fig. 2C). The top 10 gene ontology (GO) terms were mainly associated with transcription activity (Fig. 2D; Table S5), suggesting that transcriptional regulation of gene expression was the early response to low temperature. For 779 up-DEGs identified for 3 d and 7 d (Fig. 2C and E; Table S5), genes were enriched in translation-related terms, such as preribosome and RNA processing, suggesting that protein synthesis boosted with extended cold storage of peach fruit, probably to make up for the reduced enzyme activity under low temperature. A total of 1928 up-DEGs identified at 28 d were enriched in protein degradation terms involving proteolysis and ubiquitin-dependent protein catabolic process, indicating that post-translation is involved in peach fruit cold storage (Fig. 2C and F; Table S5). Interestingly, we observed 558 DEGs whose expression was induced by low temperature throughout 28 d cold storage of peach fruit, in which response to abiotic stimulus, lipid and hormone were enriched (Fig. 2C and G; Table S5).
For down-regulated DEGs, only 69 unique down-DEGs were identified within 1 d cold storage (Fig. S3A). These genes were particularly enriched in proteolysis and acid chemical response according to GO analysis (Fig. S3B; Table S6), suggesting that fruit may maintain normal physiological activity by inhibiting proteolysis during short-term cold storage. GO analysis showed that down-regulated genes (7 5 8) in 3 d and 7 d were enriched in phosphorus metabolic process and protein modification (Fig. S3C; Table S6). After 28 d of cold storage, there were 1995 down-DEGs which were enriched in cytoplasm and organelle GO term, including chloroplast and ribosome (Fig. S3D; Table S6), suggesting that cell activities were inhibited during prolonged cold storage. A total of 98 down-DEGs were identified throughout 28 d cold storage, which were mainly enriched in GO term for cell wall (Fig. S3E; Table S6). These results indicated a time-course reprogramming of gene expression in peach fruit during postharvest cold storage.
Fig. 3.
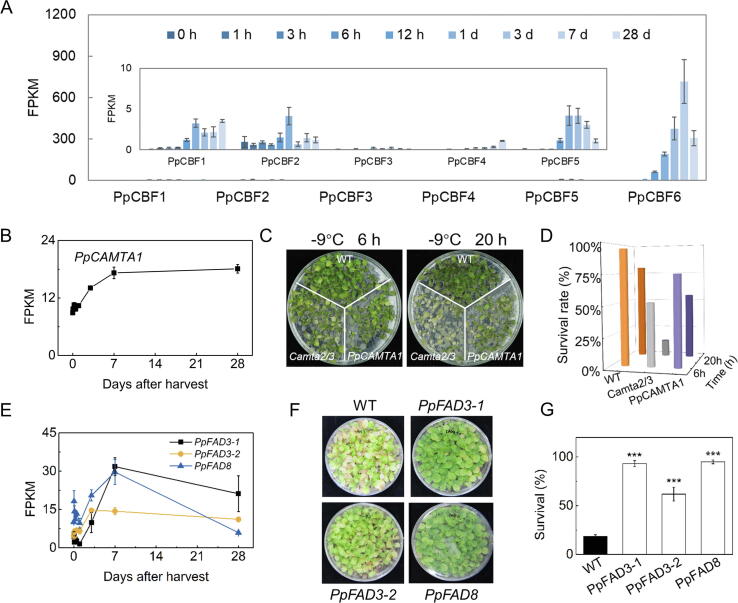
Expression of genes related to cold stress during peach fruit cold storage. (A) Expression profiles of PpCBF1-6 during cold storage. (B) Expression of PpCAMTA1 during cold storage. (C) Phenotype of Arabidopsis in response to cold stress. (D) Survival rates of Arabidopsis during cold storage after overexpressing PpCAMTA1. (E) Expression of PpFADs in response to cold storage. (F) Phenotype of tobacco plants in response to cold stress. (G) Survival rates of tobacco during cold storage after overexpressing PpFAD3-1, PpFAD3-2 and PpFAD8. Error bars indicate means ± SE; three biological replicates. ***P < 0.001.
It is conserved in plants that the C-repeat binding factor (CBF)/dehydration responsive-element binding factor mediates cold signaling regulatory cascade [37]. In peach fruit, six CBF genes were identified [38], whose transcript levels were immediately induced by chilling and showed different expression patterns. PpCBF6 had the highest transcript levels of these CBF genes, peaking at 7 days of cold storage. (Fig. 3A). Based on sequence homologs of Arabidopsis CBF regulons [39], a total of 172 homologs were identified in peach fruit. We found expressions of 108 genes (approximately 63 % CBF regulons) were induced to different extent by cold storage (Fig. S4A; Table S7). For example, transcript levels of PpRD29 and PpCOR47 were increased during cold storage, while PpCOR414 showed a reverse pattern (Fig. S4B). In Arabidopsis, TF Calmodulin Binding Transcription Activator 3 (CAMTA3) activates AtCBF2 which in turn contributes to cold tolerance [35]. Expression of peach PpCAMTA1, homology of AtCAMTA3, was rapidly induced by chilling and peaked at 7 d during cold storage of peach fruit (Fig. 3B). To investigate the role of PpCAMTA1 in cold stress, we overexpressed PpCAMTA1 driven by AtCAMTA3 promoter in Camta2/3 mutant, and then exposed Arabidopsis plants to freezing temperature at −9 ℃ (Fig. 3C). Enhanced freezing tolerance was observed in transgenic plants, and their survival rates were higher than that of mutants, closed to that of the WT (Fig. 3D). To test the relationship between CAMTA and CBFs in peach, we performed a dual luciferase assay. As shown in Fig. S5, PpCAMTA1 did not activate PpCBF6 at room temperature. Activation was observed for PpCAMTA1 toward PpCBF6 when temperature was declined to 4 ℃. These results indicated that TF PpCAMTA1 activates PpCBF6 during cold storage. Together, these findings revealed that PpCAMTA1 functionally complement AtCAMTA3 loss of function and may play an active role in modulating peach fruit chilling tolerance during cold storage.
Fig. 4.
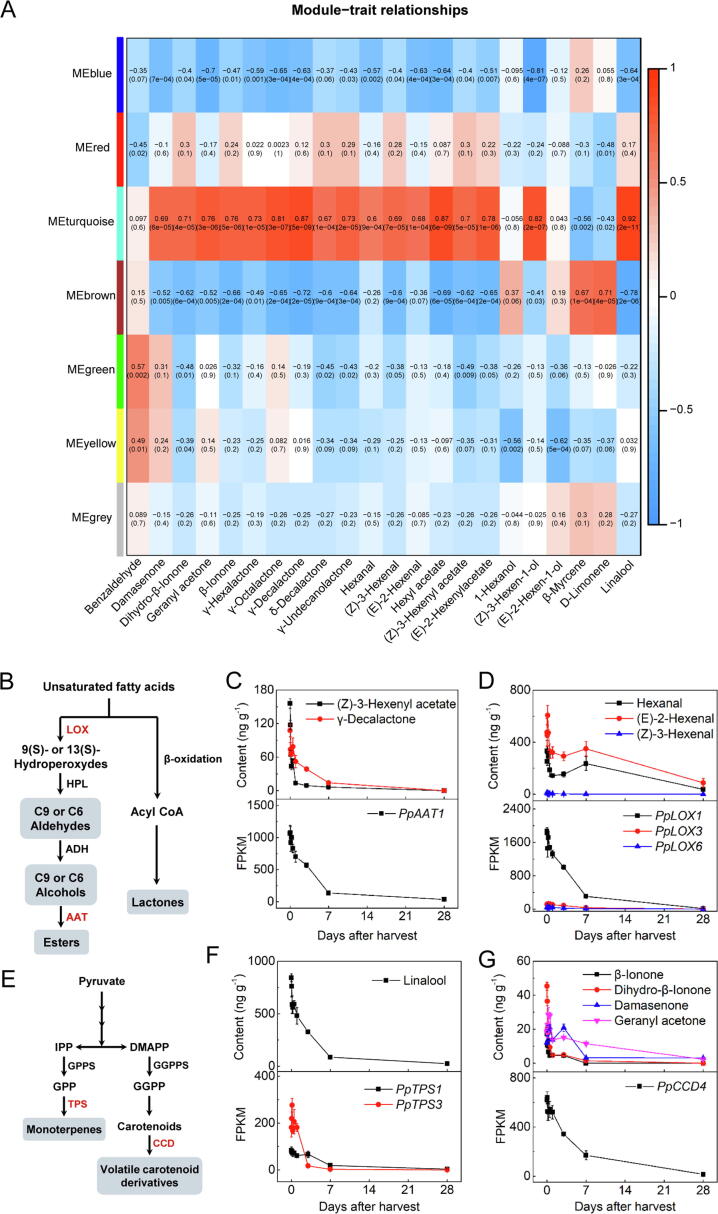
Identification of genes related to reduced volatiles during cold storage of peach fruit. (A) Correlation analysis between the module eigengene and physiological traits. Module-trait correlations and the corresponding P-values are in parentheses. The left panel presents the 7 modules. Colour scale on the right presents the module-trait correlations from −1 (blue) to 1 (red). (B) Synthesis pathway of fatty acid-derived volatiles. (C) Contents of ester and lactone and expression of PpAAT1. (D) Contents of aldehydes and expression of PpLOX1, PpLOX3 and PpLOX6. (E) Synthesis pathway of terpenoids. (F) Contents of linalool and expression of PpTPS1 and PpTPS3. (G) Contents of apocarotenoids and expression of PpCCD4. Error bars indicate means ± SE; three biological replicates.
It has been well characterized that increased expression ω-3 FADs significantly improve the cold resistance in plants through maintaining unsaturated fatty acids level [40]. Here, we found increased expression of peach fruit ω-3 PpFAD3-1, PpFAD3-2 and PpFAD8, peaking at approximately 7 d after cold storage (Fig. 3E). Transgenic tobacco plants generated in our previous study [36] were placed at 0 ℃ for 10 d followed by 10 d recovery at room temperature (Fig. 3F). Transgenic plants had significantly higher survival rates than WT plants. (Fig. 3G), suggesting a positive contribution of PpFAD3-1, PpFAD3-2 and PpFAD8 to increase chilling tolerance of peach during cold storage.
In agreement with reduced production of ethylene during cold storage (Fig. S1), expression of genes associated with the biosynthesis of ethylene was significantly inhibited in peach fruit, including PpACS1 and PpACO1 (Fig. S6). PpNAC1 is a crucial regulator involved in peach fruit ripening and ethylene production [19], [41]. Consistent with inhibited ethylene production during cold storage, great decrease in transcripts of PpNAC1 was observed after 3 d cold storage and maintained constant at low level (Fig. S6).
Generation of regulatory modules to identify genes related to flavor volatile synthesis
As mentioned above, both the metabolome and transcriptome analysis showed a temporal regulation manner during cold storage of peach fruit. To explore the molecular basis for flavor loss, WGCNA was used to identify genes related to volatiles (Fig. S7). Module-trait relationships revealed a strong correlation between expression of genes in module Turquoise (MEturquoise) and volatiles whose contents were significantly decreased during cold storage (Fig. 4A; Table S8). In MEturquoise, eight genes encoding enzymes involved in the biosynthesis pathways for flavor volatiles were identified. In agreement with previous studies where PpAAT1 is involved in biosynthesis of esters [19] and lactones [22] in peach fruit, expression of PpAAT1 was positively correlated with the decrease of ester and lactone contents during cold storage (Fig. 4B and C). Three PpLOXs (PpLOX1, PpLOX3, PpLOX6) expression correlated with contents of aldehydes (hexanal, (E)-2-hexenal, (Z)-3-hexenal) (Fig. 4B and D). Expression of PpADH3 was associated with (Z)-3-hexen-1-ol production (Fig. S8A). For volatile terpenes, cold storage induced decline of linalool was associated with reduced transcripts of PpTPS1 and PpTPS3 (Fig. 4E and F). Moreover, involvement of PpCCD4 in biosynthesis of carotenoid-derived volatiles [42] was further supported in the present study (Fig. 4E and G). Regarding to volatiles whose contents were induced by cold storage, candidate genes were identified in module Brown (MEbrown) (Fig. 4A; Table S9). For instance, high content of d-limonene at 7 d storage was associated with induced expression of PpTPS11 (Fig. S8 B).
Fig. 8.
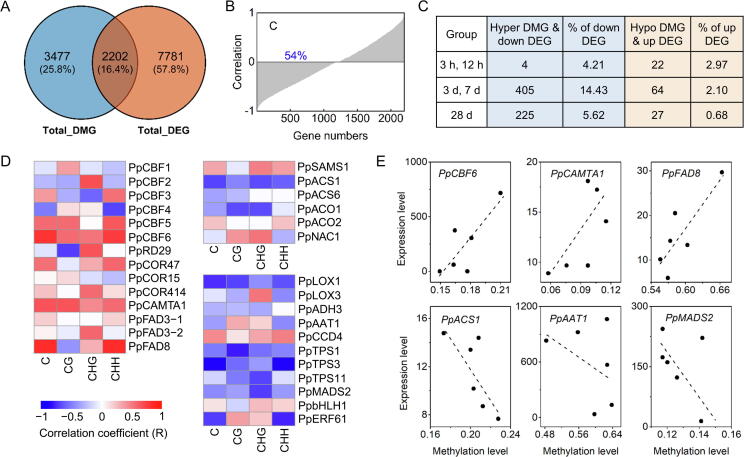
Correlation between DNA methylation in promoter region and gene expression levels during cold storage. (A) Venn diagram showing overlaps among the total_DMGs in promoter region and total_DEGs. (B) Correlation of methylation and gene expression. Percentage is the number of genes with negative correlation between methylation and transcripts. (C) Numbers and proportions of hyper DMG & down DEG and hypo DMG & up DEG. (D) Heatmap display for correlation between DNA methylation in promoter region and expression levels of gene related to cold tolerance, ethylene synthesis and volatile formation. (E) Correlation between DNA methylation and gene expression levels of PpCBF6, PpCAMTA1, PpFAD8, PpACS1, PpAAT1 and PpMADS2.
Based on the structure genes we identified for volatile biosynthesis, we next screened TFs by generating regulatory network based on gene co-expression analysis (Fig. 5A). A total of 21 TFs were identified, including members form family of ARF, ARR, bHLH, ERF, GATA, HB, MADS, MIF, NF-YC, REM, SBP, TCP, THX WRKY and ZFP. We previously identified that TF PpNAC1 contributed to ester production by regulating transcript levels of PpAAT1 during ripening at ambient temperature of peach fruit [19]. However, we did not find PpNAC1 in the regulatory network, suggesting there may exist extra TFs in peach fruit that could regulate PpAAT1 expression which in turn affect the synthesis of flavor esters and lactones during postharvest cold storage.
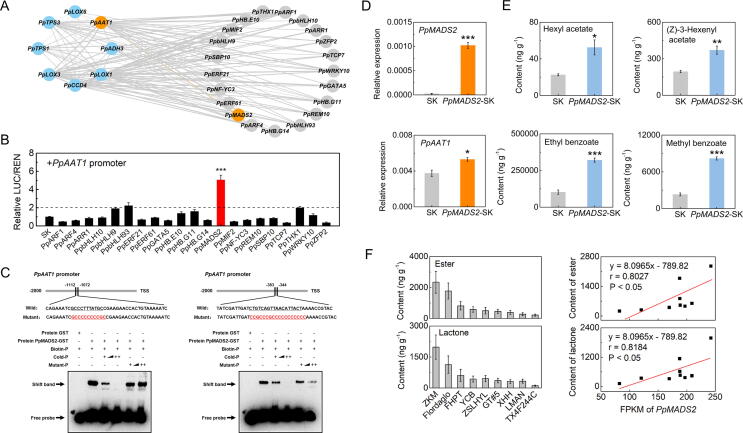
Fig. 5.
Identification of PpMADS2 as a transcription factor regulating peach fruit PpAAT1 expression. (A) The correlation network of module MEturquoise. (B) Regulatory effects of transcription factors on the promoter of PpAAT1. Error bars indicate means ± SE; three biological replicates, ***P < 0.001. (C) EMSA of PpMADS2 binding to the PpAAT1 promoter; (D) Expression of PpMADS2 and PpAAT1 caused by transient overexpression in peach leaves. (E) Content of esters in peach leaves. (F) Content of ester and lactone and expression levels of PpAAT1 and PpMADS2 across peach cultivars. ZKM, zaokuimi; FHPT, fenghuapantao; YCB, yuanchunbai; ZSLHYL, zaoshuliheyulu; GT#5, guantao#5; XHH, xiaohonghua; LMAN, luomianna. Correlation analysis between content of ester and lactone and expression levels of PpMADS2 across peach cultivars. Error bars indicate means ± SE; three biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001.
PpMADS2 regulates the synthesis of volatile esters
Given that PpAAT1 is involved in the biosynthesis of characteristic esters and lactones in peach fruit [19], [22], TFs that could activate the transcription of PpAAT1 was investigated. Candidate TFs in MEturquoise showing positive correlation with PpAAT1 (r > 0.8, P < 0.05) were cloned (Fig. 5A; Table S10). The transcriptional regulation of PpAAT1 by candidate TFs was investigated by dual-luciferase reporter assays. PpMADS2 had the strongest induction of PpAAT1 according to dual-luciferase assays, exhibiting a roughly 5-fold increase in LUC activity (Fig. 5B).
EMSA was performed to validate that TF PpMADS2 activates the PpAAT1 via binding to its promoter. Based on our analysis of the MADS binding sites, we designed two probes with 3′ biotin labeling. Recombinant protein of PpMADS2-GST was generated and purified for analysis. The EMSA assay reveals that PpMADS2 could bind these two biotin probes containing the MADS binding sites. With the increase of the cold probe as competitor, binding decreased. When the predicted binding sites were mutated, binding was eliminated, and a mutant probe had been mutated did not compete for binding (Fig. 5C). Collectively, these results demonstrated that TF PpMADS2 activates PpAAT1 expression through direct binding to its promoter.
To further test whether PpMADS2 could regulate the formation of volatiles by activating PpAAT1 expression in planta, PpMADS2 was overexpressed in peach. As expected, expression of PpAAT1 was significantly induced after overexpression of PpMADS2 (Fig. 5D). Consequently, contents of hexyl acetate, (Z)-3-hexenyl acetate, ethyl benzoate and methyl benzoate were significantly increased 2.3-, 1.9-, 3.1- and 3.5-fold, respectively (Fig. 5E). These results indicated that PpMADS2 can increase peach esters by increase transcript levels of PpAAT1. To further validate the role of PpMADS2 in affecting volatile formation, correlation was analyzed between gene expression and volatile contents across nine peach cultivars with different genetic background (Table S11). Linear regression analysis exhibited that expression of PpMADS2 significantly correlated with ester (r = 0.80, P < 0.05) and lactones (r = 0.82, P < 0.05) contents among different cultivars (Fig. 5F). Our observation demonstrated that PpMADS2 is essential for modulating the synthesis of volatile esters and lactones in peach fruit by activating the expression of structure gene PpAAT1.
DNA methylation profiling of peach fruit during cold storage
To explore the relationship between DNA methylation and flavor loss during cold storage, a single-base resolution map of DNA methylation was generated by WGBS. We explored temporal changes in DNA methylation levels at 0 h, 3 h, 12 h, 3 d, 7 d and 28 d after cold storage of peach fruit. The WGBS conversion rates were > 99.6 %, and each library’s sequencing depth was > 26-fold coverage per DNA strand (Table S12). An analysis of the distribution of DNA methylation in three contexts (CG, CHG, and CHH) and gene density were presented in Fig. S9. Gene-rich areas characterized by methylation enrichment in the CG context across entire chromosomes had low 5mC densities (Fig. S9A). By comparing the methylation levels of three contexts of the gene, we found that CG had the highest methylation levels in the gene body and upstream and downstream regions (2 kb), and CHH context had the lowest methylation levels (Fig. S9B). Through the statistics of the distribution of methylation sites in the whole genome, we found that CG (34.6 %) and CHH (40.39 %) contexts are major contributors for methylation sites in whole genome of peach fruit (Fig. S9C). Similar percentage for CG, CHG and CHH contexts of peach fruit were observed in previous study [29], where the predominant context is CHH, followed by CG and CHG. Fig. S9D displayed the percentages of methylated mCs in three contexts (CG, CHG, and CHH) on each chromosome.
Fig. 9.
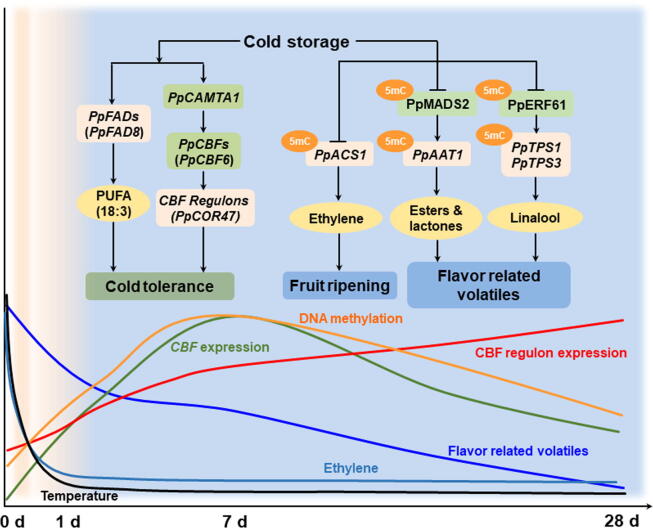
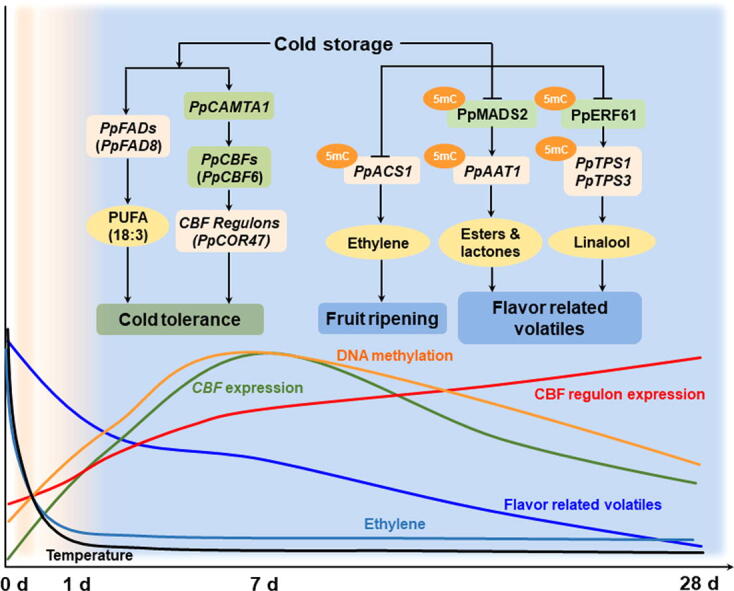
A proposed model for chilling-induced flavor loss in peach fruit.
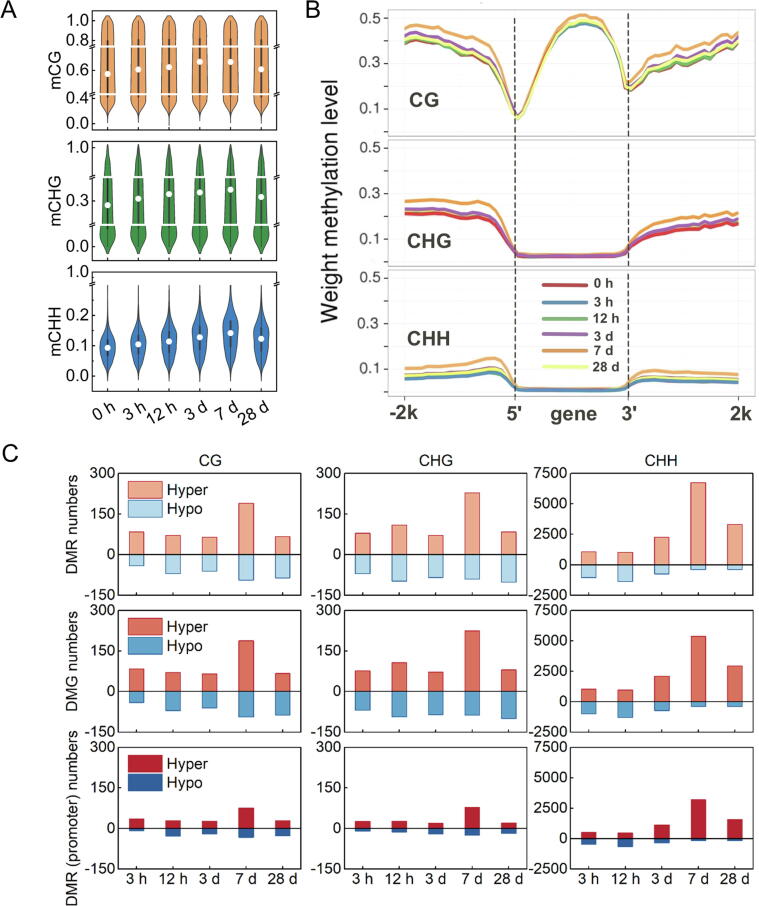
Violin plot showed that whole genome methylation levels of mCG, mCHG and mCHH increased by chilling treatment and peaked at 7 d after cold storage (Fig. 6A). We next analyzed DNA methylation levels of 5′ and 3′ flanking regions of genes and found cold storage-induced levels (Fig. 6B). Notably, fruit after 7 d cold storage exhibited the highest DNA methylation level and then declined with extended storage time. We next identified 20,246 DMRs associated with 18,014 differentially methylated genes (DMGs) during postharvest cold storage. There were 15,477 hyper DMRs and 4769 hypo DMRs. The overwhelming higher number of hyper DMRs was suggested to be associated with high DNA methylation level in genic caused by cold storage (Fig. 6C). Among the hyper DMRs identified in the present study, approximately 44 % (6738 hyper DMRs) was observed at 7 d after cold storage (Fig. 6C), consistent with the highest DNA methylation level induced by low temperature. It is noteworthy that DMRs are mainly observed in promoter regions and in intergenic of peach fruit, accounting for>90 % DMRs (Fig. S10). For instance, peach fruit exhibited 50 % DMRs in promoter and 43 % in intergenic after 7 d cold storage. Moreover, CHH DMRs are major types observed for peach fruit, accounting for 91 % of the total DMRs.
Fig. 6.
Changes in DNA methylation of peach fruit during cold storage. (A) Whole genome methylation levels of mCG, mCHG and mCHH in response to cold storage. (B) DNA methylation levels of genes in different storage time. Methylation levels of mCG, mCHG, mCHH are shown. (C) Numbers of cold-induced DMRs, DMGs and DMRs methylated in promoter region in 3 h, 12 h, 3 d, 7 d and 28 d relative to 0 h are shown for the mCG, mCHG and mCHH sequence contexts.
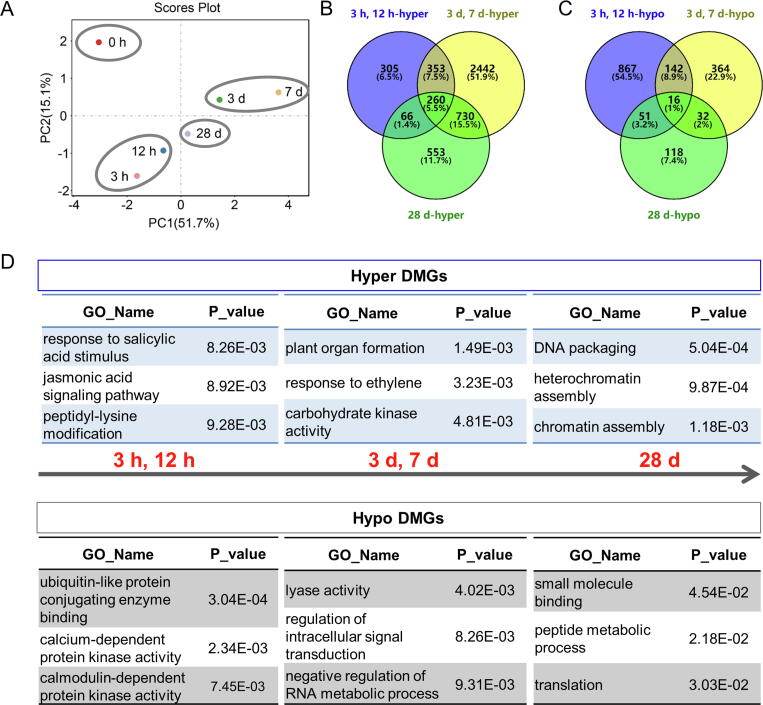
PCA analysis showed that genes associated with DMRs in promoter region were classified into four groups (Fig. 7A). For peach fruit after 3 h and 12 h cold storage, 305 hyper DMGs was uniquely detected (Fig. 7B). GO analysis for these hyper DMGs showed that the enriched terms were associated with stimulus response (Fig. 7D; Table S13), including response to salicylic acid and jasmonic acid. With extended cold storage time, numbers of hyper DMGs increased and genes related to development terms were enriched, including organ formation and response to ethylene that required for fruit ripening (Fig. 7B and D; Table S13). These may be associated with delayed fruit ripening process caused by postharvest cold storage. For 28 d, terms associated with chromatin assembly were significantly enriched for 553 hyper DMGs, including DNA packaging, heterochromatin assembly and chromatin assembly (Fig. 7B and D; Table S13). Thus, epigenetic regulation was involved in fruit response to postharvest cold storage.
Fig. 7.
Analysis of DMGs of peach fruit during cold storage. (A) PCA analysis of DMRs in promoter region. (B) Venn diagram showing the overlapping and uniquely hyper-DMGs. (C) Venn diagram showing the overlapping and uniquely hypo-DMGs. (D) GO analysis for peach fruit unique DMGs.
Further, GO analysis was also performed for hypo DMGs of peach fruit during cold storage. Among the 1590 hypo DMGs identified, 867 hypo DMGs were uniquely detected for 3 h and 12 h after cold storage (Fig. 7C). GO analysis for these hypo DMGs showed that terms related to calcium-dependent protein kinase activity and calmodulin-dependent protein kinase activity were enriched (Fig. 7D; Table S14). The observation is consistent with the fact that calcium signaling is the early response to cold stress. For peach fruit stored at low temperature for 3 d and 7 d, enrichment of hypo DMGs was observed for negative regulation of RNA processing, which is consistent with enrichment of RNA processing observed for DEGs (Fig. 7C and D; Table S14). With the prolonged storage time at 28 d, GO terms related to protein translation were enriched, including peptide metabolic process and macromolecule biosynthetic process (Fig. 7C and D; Table S14).
Influence of DNA methylation on gene expression
To investigate the impact of DNA methylation on gene expression changes in expression profiles of DMGs were examined during peach fruit cold storage. A total of 2202 DMGs in the promoter region displayed different expression (Fig. 8A), making up 22 % of the 9983 DEGs discovered in peach fruit during cold storage. For these DMG-associated DEGs, we then looked at the relationship between DNA methylation level and transcript abundance. In general, approximately 54 % DMGs showed a negative correlation between gene expression and promoter region methylation levels (Fig. 8B). For CG, CHG, and CHH contexts, approximately 48 %, 49 %, and 52 % of DMGs showed a negative correlation between transcript abundance and methylation level, respectively (Fig. S11A). These genes were associated with GO terms “light signaling pathway”, “cellular protein complex assembly”, “cellular respiration”, “terpene synthase activity”, and “fruit development” (Fig. S11B).
Furthermore, we compared differential (hyper/hypo) methylation and their direction of differential (up/down) expression for peach fruit during cold storage. For example, a total of 405 hyper DMGs were down regulated for peach fruit after 3 d and 7 d cold storage, accounting for approximately 14.43 % DEGs based on our RNA-Seq data (Fig. 8C; Fig. S12). Much lower percentage was observed for fraction of genes between hypo DMGs and up regulated DEGs. For instance, only 64 hypo DMGs were up regulated for peach fruit at 3 d and 7 d cold storage (Fig. 8C; Fig. S13), accounting for approximately 2.10 % of DEGs that were up regulated. Therefore, no obvious trend of correlation between the differential methylation and direction of different gene expression was observed during postharvest cold storage. Although overlap between DMGs and DEGs is relatively low, GO analysis for these genes were performed during peach fruit cold storage (Table S15; Table S16). Go terms such as negative regulation of transcription, regulation of gene expression, protein ubiquitination, response to stimulus and fatty acid metabolic process were observed.
We next looked correlation between transcript abundance and DNA methylation for genes related to cold tolerance and fruit ripening. Peach PpCBF6 had the highest transcript abundance among CBF gene family and increased expression during cold storage (Fig. 3A). No inverse pattern was observed for expression and DNA methylation of PpCBF6 and target genes PpCOR15, PpCOR47, PpCOR414 (Fig. 8D). Moreover, DNA methylation and gene expression did not exhibit a negative correlation for TF PpCAMTA1, which is located upstream of PpCBF6. Similarly, PpFAD3-1, PpFAD3-2 and PpFAD8 that contribute to cold tolerance also did not exhibit reverse pattern between methylation and transcript abundance during cold storage (Fig. 8D and E). Fruit ethylene synthesis is associated with PpACS1 expression in peach, whose reduced expression is correlated with hypermethylation in its promoter region throughout cold storage period (Fig. 8D and E).
DNA methylation affects fruit volatile synthesis during cold storage
To further investigate correlation between transcript abundance and methylation levels of genes associated with the synthesis of volatiles during cold storage. Correlation between differential methylation levels in different sequence contexts (C, CG, CHG, CHH) located in promoter regions and their expression levels were examined (Fig. 8D). Regarding to fatty acids derived C6 aldehydes, esters and lactones (Fig. 4), negative correlation (blue column) between gene expression and DNA methylation levels was observed for synthesis related structure genes, including PpLOX1, PpLOX3 and PpAAT1 (Fig. 8D). For PpLOX1, inverse patterns were observed for CG, CHG and CHH contexts. For PpAAT1, hyper methylation in CHH context was associated with reduced expression during cold storage, but not for CG and CHG contexts. Besides PpAAT1, inverse patterns between methylation levels and transcripts were observed for TF PpMADS2 which controls peach fruit ester production (Fig. 8E).
For volatile monoterpene linalool, reduced expression of synthesis related structure genes PpTPS1 and PpTPS3 was negatively related to increased DNA methylation levels during cold storage of peach fruit (Fig. 8D). TF PpbHLH1 and PpERF61 are associated with linalool formation through activating PpTPSs expression in peach fruit [21], [23]. No inverse pattern between expression and DNA methylation was observed for PpbHLH1. For PpERF61, hyper methylation in CHH was negatively correlated with transcript levels during cold storage. The biosynthesis of volatile carotenoid derivatives is catalyzed by PpCCD4 in peach fruit [42]. No inverse pattern between PpCCD4 expression and DNA methylation was observed (Fig. 8D). Collectively, our results indicated a major alternation in DNA methylation caused by cold storage often correlated with expression change of genes associated with fruit volatile synthesis.
To understand the function of DNA methyltransferases and DNA demethylases in determining DNA methylation during cold storage, their expression patterns in peach fruit were analyzed. There are at least six DNA methyltransferases in peach (Fig. S14A). PpDRM2 had the highest expression abundance and tended to increase with extended cold storage time (Fig. S15A). The maintenance of DNA methylation requires DNA methyltransferase 1 (MET1) at the CG context, chromomethylase 3 (CMT3) at the CHG context, and domains rearranged methylase 2 (DRM2) or chromomethylase 2 (CMT2) at the CHH context [43]. The relationship between gene expression and methylation was then examined in various contexts, and we discovered that PpDRM2 exhibited the highest correlation coefficient with CHH (Fig. S14B). Removal of DNA methylation is mediated by Repressor of Silencing 1 (ROS1), demeter (DME), demeter-like (DML) and increased DNA methylation 1 (IDM1) [44]. Peach has at least six DNA demethylases (Fig. S14B), and PpDML1 is the member showing the highest transcript abundance (Fig. S15C). Additionally, there was a negative correlation between PpDML1 expression and methylation contexts (CG, CHG, and CHH) during cold storage (Fig. S15D). Although we are unable to establish a causal relationship between gene expression and methylation alterations using the data at hand, the results showed that changes in PpDRM2 and PpDML1 were related to cold storage induced DNA methylation for postharvest peach fruit.
Discussion
Cold storage is the most common method to extend fruit shelf life. However, fruit quality deterioration occurs with prolonged cold storage, particular for chilling-sensitive fruit species such as peach. Here, we investigated the dynamic patterns of melting peach fruit on ‘hour-day-week’ time scales by combining flavor metabolome, transcriptome and DNA methylome during postharvest cold storage.
Significant changes in peach fruit quality caused by postharvest cold storage
For peach fruit, chilling injury develops faster and more intensely during storage at temperatures between 2.2 and 7.6 ℃ than those stored at 0 ℃ [45]. To prolong storage life and maintain fruit quality, peach fruit are suggested to be stored at 0 ℃. Our present data reveals that the internal temperature of peach fruit flesh decreased from 20 °C to 0 °C within 1 d of postharvest cold storage, accompanied by a rapid decrease in ethylene production. The inhibition of fruit ethylene synthesis during cold storage may be related to the delayed fruit ripening process. Consequently, firmness of peach fruit was maintained constant after 14 d postharvest chilling, followed by a slight decrease after 28 d cold storage. Similar delayed decline in fruit firmness during cold storage were in agreement with previous studies for peach fruit [14], [46].
Fruit flavor is determined by a combination of soluble sugars, organic acids and volatiles. We found no significant level of change in total sugar and acid contents of peach fruit during cold storage, but the total volatile contents showed a significant decrease. Similar findings have been reported in other fruits. For example, tomato fruit stored at 5 °C for 8 d after harvest maintained stable soluble sugar and organic acid contents, but the volatile contents decreased by 65 % [2]. Volatile chemicals belong to secondary metabolites which is a possible reason contributing to more sensitivity of volatiles to environmental factors than major metabolites such as soluble sugars and organic acids [47]. In addition, unlike apple, kiwifruit and potato with high content of starch which is converted to sugars during postharvest cold storage, sugar contents of peach fruit are mainly affected by harvesting maturity stage.
Cold storage significantly reduced the volatile contents, such as esters, lactones, apocarotenoids and terpenes. γ-hexalactone, γ-octalactone, γ-decalactone, δ-decalactone, γ-undecanolactone, hexyl acetate, (Z)-3-hexenyl acetate, damasenone and linalool are important substances in the formation of aromatic qualities of peach fruit [17]. It is assumed that the decrease in the content of these aroma volatiles is the main reason for the flavor deterioration of peach fruit during cold storage. It is worth noting that not all volatiles whose contents were reduced by low temperature. For instance, β-myrcene and d-limonene increased at 7 d cold storage, in agreement with reports in citrus [5], apple [48] and grapefruit [9]. Low temperature-induced terpenoids is suggested to be associated with fruit adaptation to cold stress by participating in oxidation–reduction reaction [2].
A time course of cold-induced transcriptional reprogramming
RNA-Seq data were collected for peach fruit samples at harvest without cold storage (0 h), with 1 h, 3 h, 6 h, 12 h, 1 d, 3 d, 7 d and 28 d storage at low temperature. A total of 9983 DEGs was found of peach fruit during cold storage, accounting for approximately 30 % of the total number of peach genes. The decrease in flesh temperature from 20 °C to 0 °C within 1 d was accompanied by overwhelming greater number of up-regulated DEGs. GO analysis revealed that transcriptional regulation of gene expression was significantly enriched for these DEGs occurred within 1 d cold storage. Expression of genes related to cold response and abiotic stresses were altered immediately for fruit transfer to cold storage. For instance, transcript levels of PpCBF6 and its upstream modulator PpCAMTA1 and downstream target PpCOR47 were significantly induced during cold storage. With extended cold storage time to 7 d, a greater number of DEGs was observed, in which approximately 50 % DEGs were up-regulated by cold storage. For fruit under cold storage for 28 d, expression of 7992 genes were significantly altered, which are associated with protein hydrolysis and ubiquitin GO terms, suggesting potential tissue damage occurred after prolonged cold storage. Overexpressing CBF in Arabidopsis resulted in upregulation of 133 cold-induced genes and the downregulation of 39 cold-induced genes [39]. In peach fruit, approximately 32 % of CBF regulons showed similar expression patterns to that in Arabidopsis. In tomato fruit, there are approximately 13 % of CBF regulons whose expression was affected by cold storage [2]. The above results suggest that CBF regulons and their functions may be influenced by plant species or be dependent on plant organs such as fruits and leaves in response to chilling stress.
A combined transcriptome and metabolome analysis revealed that the transcript levels of genes related to volatile synthesis were significantly reduced during cold storage. Meanwhile, a large number of TFs were identified to be candidate regulators for production of volatile esters and lactones. PpAAT1 has been shown to be a key enzyme in peach fruit volatile ester synthesis [19] and is involved in lactone synthesis [22]. Combining the results of tobacco dual luciferase, EMSA and homologous transient expression, we identified that TF PpMADS2 activated PpAAT1 expression and is involved in ester production. MADS-box TFs play important roles during fruit ripening, including the biosynthesis of volatiles [49], [50], [51], [52]. Our previous study found that TF PpNAC1 regulates the synthesis of esters and lactones during peach fruit ripening at ambient temperature [19]. In the present study, PpNAC1 was not involved in modules associated with reduced content of volatile esters and lactones caused by postharvest cold storage. These results indicated that formation of esters and lactones were affected by multiple TFs in peach fruit. It is well known that secondary metabolites could be regulated by a range of TFs. For instance, multiple TFs have been found to influence how plants synthesize anthocyanin [53], [54], [55], [56]. While during peach fruit storage at low temperature, reduced esters and lactones are associated with decreased expression of TF PpMADS2 and its target gene PpAAT1.
Reduced volatile synthesis caused by low temperature is associated with increased DNA methylation
Besides TF, we further investigated association between gene expression and DNA methylation for chilling-induced reduction of volatiles in peach fruit. DNA methylation is an important epigenic modification that is involved in fruit development, ripening, and response to stress [41], [43], [44]. In this study, we analyzed the pattern of DNA methylation of peach fruit during postharvest cold storage by WGBS analysis. Our results revealed that low temperature induced DNA methylation levels and peaked at 7 d, followed by a decrease as prolonged storage period. Low temperature induced DNA methylation was consistent with previous findings in tomato fruit [2] and tea tree [57]. It is interesting to further investigate whether chilling-induced DNA methylation is conserved in other plant species or fruit species in future.
DNA methylation in promoter region is considered as a marker of transcriptional repression. Although low temperature induced expression of PpCBFs and their target genes, no negative correlations were observed between gene expression and DNA methylation levels. Studies indicated that epigenetic modifications such as histone methylation and histone acetylation were involved in plant responses to cold stress [58], [59]. Histone deacetylase (HDA) activates maize DREB1 expression and histone hyperacetylation after cold treatment [60]. Moreover, induced expression of CBF target genes COR15A and GOLS3 were associated with reduced H3K27me3 levels [61]. It has been reported that DNA demethylation can occur in the binding region of ZmICE1 [60], no similar results were observed in the present study. It is an open question of whether DNA methylation in the gene body region would be involved in gene regulation during cold storage needs to be further investigated.
In this study, we found a significant negative correlation between PpMADS2 and PpAAT1 transcript abundance and DNA methylation levels in the promoter region. These results suggest that the reduction of esters and lactones, important volatiles for peach fruit flavor, is associated with increased DNA methylation levels during cold storage. In addition, reduced linalool was associated with cold storage induced promoter hypermethylation and decrease in expression of PpTPS3. Our results indicated that chilling-induced reduction of flavor volatiles was associated with increased DNA methylation of peach fruit, in agreement with our previous study for tomato fruit [2]. Although an interaction between TFs and DNA methylation has been proposed [62], it is still unclear how methylation in TF binding sites affects the binding and dissociation of protein and DNA complexes. New approaches are needed to quantitatively measure the contribution of DNA methylation to TF binding [63], [64].
Conclusions
Cold storage caused flavor loss of peach fruit is associated with decrease in volatile contents, accompanying with delayed ripening process and induced cold tolerance (Fig. 9). Expression of TF PpCBF6 was induced and peaked at approximately 7 d storage at 0 °C of melting peach fruit Hujingmilu. Postharvest cold storage reduced expression of ethylene synthesis gene PpACS1, inhibited transcription of PpAAT1 for esters and lactones formation, and PpTPS1/3 for linalool synthesis of peach fruit. Combination of metabolome and transcriptome allowed us to identify that TF PpMADS2 is associated with synthesis of flavor volatiles. Genome wide DNA methylation was induced by low temperature and peaked at 7 d followed by a decline with extended cold storage period. Reduction of gene expression was accompanied by major changes in DNA methylation status of their promoters, including PpACS1, PpAAT1, PpTPS3, PpMADS2 and PpERF61. In brief, our observation demonstrated the power of using bioinformatics alongside multi-omics approach in unraveling the complex regulatory control of flavor related volatiles in fruit. The present study offers an alternative efficient way of identifying regulatory factors which may ultimately allow breeders to generate flavorsome fruit in future.
Compliance with Ethics Requirement
There was no use of any animals or human patients.
CRediT authorship contribution statement
Wenyi Duan: Methodology, Investigation, Data curation, Writing – original draft, Visualization. Can Yang: Investigation, Resources. Xiangmei Cao: Methodology, Resources. Chunyan Wei: Investigation, Resources. Kunsong Chen: Funding acquisition. Xian Li: Writing – review & editing, Supervision. Bo Zhang: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31972379, 32102445), Zhejiang Provincial Natural Science Foundation (LD22C150001), the Fundamental Research Funds for the Zhejiang Provincial Universities (2021XZZX026), and the Service Local Economic Development Program of Shandong (Linyi) Institute of Modern Agriculture, Zhejiang University (ZDNY-2020-FWLY01008A).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.12.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Brizzolara S., Manganaris G.A., Fotopoulos V., Watkins C.B., Tonutti P. Primary metabolism in fresh fruits during storage. Front Plant Sci. 2020;11:80. doi: 10.3389/fpls.2020.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B., Tieman D.M., Jiao C., Xu Y., Chen K., Fei Z., et al. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc Natl Acad Sci U S A. 2016;113(44):12580–21255. doi: 10.1073/pnas.1613910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J., Quan P., Liu H., Li L., Qi S., Zhang M., et al. Transcriptomic and metabolic analyses provide new insights into the apple fruit quality decline during long-term cold storage. J Agric Food Chem. 2020;68(16):4699–4716. doi: 10.1021/acs.jafc.9b07107. [DOI] [PubMed] [Google Scholar]
- 4.Maul F.S.S., Sims C.A., Baldwin E.A., Balaban M.O., Huber D.J. Tomato flavor and aroma quality as affected by storage temperature. J Food Sci. 2000;65(7):1228–1237. doi: 10.1111/j.1365-2621.2000.tb10270.x. [DOI] [Google Scholar]
- 5.Obenland D., Collin S., Sievert J., Arpaia M.L. Mandarin flavor and aroma volatile composition are strongly influenced by holding temperature. Postharvest Biol Technol. 2013;82:6–14. doi: 10.1016/j.postharvbio.2013.02.013. [DOI] [Google Scholar]
- 6.Zou J., Chen J., Tang N., Gao Y., Hong M., Wei W., et al. Transcriptome analysis of aroma volatile metabolism change in tomato (Solanum lycopersicum) fruit under different storage temperatures and 1-MCP treatment. Postharvest Biol Technol. 2018;135:57–67. doi: 10.1016/j.postharvbio.2017.08.017. [DOI] [Google Scholar]
- 7.Raffo A., Nicoli S., Nardo N., Baiamonte I., D’Aloise A., Paoletti F. Impact of different distribution scenarios and recommended storage conditions on flavor related quality attributes in ripening fresh tomatoes. J Agric Food Chem. 2012;60(42):10445–10455. doi: 10.1021/jf3028528. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Bel P., Egea I., Sanchez-Ballesta M.T., Sevillano L., Del Carmen B.M., Flores F.B. Proteome changes in tomato fruits prior to visible symptoms of chilling injury are linked to defensive mechanisms, uncoupling of photosynthetic processes and protein degradation machinery. Plant Cell Physiol. 2012;53(2):470–484. doi: 10.1093/pcp/pcr191. [DOI] [PubMed] [Google Scholar]
- 9.Lado J., Gurrea A., Zacarias L., Rodrigo M.J. Influence of the storage temperature on volatile emission, carotenoid content and chilling injury development in Star Ruby red grapefruit. Food Chem. 2019;295:72–81. doi: 10.1016/j.foodchem.2019.05.108. [DOI] [PubMed] [Google Scholar]
- 10.Lee J., Jang H.W., Jeong M.C., Yoo S., Ha J. Analysis of volatile compounds as quality indicators for Fuji apples after cold storage. J Food Biochem. 2017;41(6):1–11. doi: 10.1111/jfbc.12410. [DOI] [Google Scholar]
- 11.Günther C.S., Marsh K.B., Winz R.A., Harker R.F., Wohlers M.W., White A., et al. The impact of cold storage and ethylene on volatile ester production and aroma perception in ‘Hort16A’ kiwifruit. Food Chem. 2015;169:5–12. doi: 10.1016/j.foodchem.2014.07.070. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto H., Ikoma Y. Effect of postharvest temperature on the muscat flavor and aroma volatile content in the berries of ‘Shine Muscat’ (Vitis labruscana Baily × V. Vinifera L.) Postharvest Biol Technol. 2016;112:256–265. doi: 10.1016/j.postharvbio.2015.09.004. [DOI] [Google Scholar]
- 13.Yang C., Duan W., Xie K., Ren C., Zhu C., Chen K., et al. Effect of salicylic acid treatment on sensory quality, flavor-related chemicals and gene expression in peach fruit after cold storage. Postharvest Biol Technol. 2020;161 doi: 10.1016/j.postharvbio.2019.111089. [DOI] [Google Scholar]
- 14.Zhang B., Xi W.P., Wei W.W., Shen J.Y., Ferguson I., Chen K.S. Changes in aroma-related volatiles and gene expression during low temperature storage and subsequent shelf-life of peach fruit. Postharvest Biol Technol. 2011;60(1):7–16. doi: 10.1016/j.postharvbio.2010.09.012. [DOI] [Google Scholar]
- 15.Aubert C., Milhet C. Distribution of the volatile compounds in the different parts of a white-fleshed peach (Prunus persica L. Batsch) Food Chem. 2007;102(1):375–384. doi: 10.1016/j.foodchem.2006.05.030. [DOI] [Google Scholar]
- 16.Wang Y., Yang C., Li S., Yang L., Wang Y., Zhao J., et al. Volatile characteristics of 50 peaches and nectarines evaluated by HP-SPME with GC-MS. Food Chem. 2009;116(1):356–364. doi: 10.1016/j.foodchem.2009.02.004. [DOI] [Google Scholar]
- 17.Eduardo I., Chietera G., Bassi D., Rossini L., Vecchietti A. Identification of key odor volatile compounds in the essential oil of nine peach accessions. J Sci Food Agric. 2010;90(7):1146–1154. doi: 10.1002/jsfa.3932. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B., Shen J.Y., Wei W.W., Xi W.P., Xu C.J., Ferguson I., et al. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J Agric Food Chem. 2010;58(10):6157–6165. doi: 10.1021/jf100172e. [DOI] [PubMed] [Google Scholar]
- 19.Cao X., Wei C., Duan W., Gao Y., Kuang J., Liu M., et al. Transcriptional and epigenetic analysis reveals that NAC transcription factors regulate fruit flavor ester biosynthesis. Plant J. 2021;106(3):785–800. doi: 10.1111/tpj.15200. [DOI] [PubMed] [Google Scholar]
- 20.Liu H., Cao X., Liu X., Xin R., Wang J., Gao J., et al. UV-B irradiation differentially regulates terpene synthases and terpene content of peach. Plant Cell Environ. 2017;40(10):2261–2275. doi: 10.1111/pce.13029. [DOI] [PubMed] [Google Scholar]
- 21.Wei C., Liu H., Cao X., Zhang M., Li X., Chen K., et al. Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol J. 2021;19(10):2082–2096. doi: 10.1111/pbi.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng B., Yu M., Zhang B., Xu J., Ma R. Differences in PpAAT1 activity in high- and low-aroma peach varieties affect γ-decalactone production. Plant Physiol. 2020;182(4):2065–2080. doi: 10.1104/pp.19.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei C., Li M., Cao X., Jin Z., Zhang C., Xu M., et al. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022;317 doi: 10.1016/j.plantsci.2022.111200. [DOI] [PubMed] [Google Scholar]
- 24.Cano-Salazar J., Echeverria G., Crisosto C.H., Lopez L. Cold-storage potential of four yellow-fleshed peach cultivars defined by their volatile compounds emissions, standard quality parameters, and consumer acceptance. J Agric Food Chem. 2012;60(5):1266–1282. doi: 10.1021/jf204126m. [DOI] [PubMed] [Google Scholar]
- 25.Muto A., Muller C.T., Bruno L., McGregor L., Ferrante A., Chiappetta A.A.C., et al. Fruit volatilome profiling through GC × GC-ToF-MS and gene expression analyses reveal differences amongst peach cultivars in their response to cold storage. Sci Rep. 2020;10(1):18333. doi: 10.1038/s41598-020-75322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanou G., Minas I.S., Scossa F., Belghazi M., Xanthopoulou A., Ganopoulos I., et al. Exploring priming responses involved in peach fruit acclimation to cold stress. Sci Rep. 2017;7(1):11358. doi: 10.1038/s41598-017-11933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K., Yin X.R., Zhang B., Grierson D., Xu C.J., Chen K.S. Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit. Plant Cell Environ. 2017;40(8):1531–1551. doi: 10.1111/pce.12951. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y., Wang K., Wu C., Hao Y., Zhang B., Grierson D., et al. DNA hypermethylation associated with the development of temperature-dependent postharvest chilling injury in peach fruit. Postharvest Biol Technol. 2021;181 doi: 10.1016/j.postharvbio.2021.111645. [DOI] [Google Scholar]
- 29.Zhu Y.C., Zhang B., Allan A.C., Wang K.L., Zhao Y., Wang K., et al. DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 2020;102(5):965–976. doi: 10.1111/tpj.14680. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C., Duan W., Chen K., Zhang B. Transcriptome and methylome analysis reveals effects of ripening on and off the vine on flavor quality of tomato fruit. Postharvest Biol Technol. 2020;162 doi: 10.1016/j.postharvbio.2019.111096. [DOI] [Google Scholar]
- 31.Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45(D1):D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otasek D., Morris J.H., Boucas J., Pico A.R., Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D.Q., Xi Y.X., Rodriguez B., Park H.J., Tong P., Meong M., et al. MOABS: Model based analysis of bisulfite sequencing data. Genome Biol. 2014;15(2):R38. doi: 10.1186/gb-2014-15-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y., Park S., Gilmour S.J., Thomashow M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013;75(3):364–376. doi: 10.1111/tpj.12205. [DOI] [PubMed] [Google Scholar]
- 36.Wang J.J., Liu H.R., Gao J., Huang Y.J., Zhang B., Chen K.S. Two ω-3 FADs are associated with peach fruit volatile formation. Int J Mol Sci. 2016;17(4):464. doi: 10.3390/ijms17040464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomashow M.F. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154(2):571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang L., Zhang B., Yin X.R., Xu C.J., Sun C.D., Chen K.S. Differential expression of the CBF gene family during postharvest cold storage and subsequent shelf-life of peach fruit. Plant Mol Biol Rep. 2013;31(6):1358–1367. doi: 10.1007/s11105-013-0600-5. [DOI] [Google Scholar]
- 39.Park S., Lee C.M., Doherty C.J., Gilmour S.J., Kim Y., Thomashow M.F. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015;82(2):193–207. doi: 10.1111/tpj.12796. [DOI] [PubMed] [Google Scholar]
- 40.Dominguez T., Hernandez M.L., Pennycooke J.C., Jimenez P., Martinez-Rivas J.M., Sanz C., et al. Increasing ω-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010;153(2):655–665. doi: 10.1104/pp.110.154815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lü P., Yu S., Zhu N., Chen Y.R., Zhou B., Pan Y., et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat Plants. 2018;4(10):784–791. doi: 10.1038/s41477-018-0249-z. [DOI] [PubMed] [Google Scholar]
- 42.Brandi F., Bar E., Mourgues F., Horvath G., Turcsi E., Giuliano G., et al. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 2011;11:24. doi: 10.1186/1471-2229-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu R., Lang Z.B. The mechanism and function of active DNA demethylation in plants. J Integr Plant Biol. 2020;62(1):148–159. doi: 10.1111/jipb.12879. [DOI] [PubMed] [Google Scholar]
- 44.Chang Y.N., Zhu C., Jiang J., Zhang H., Zhu J.K., Duan C.G. Epigenetic regulation in plant abiotic stress responses. J Integr Plant Biol. 2020;62(5):563–580. doi: 10.1111/jipb.12901. [DOI] [PubMed] [Google Scholar]
- 45.Lurie S., Crisosto C.H. Chilling injury in peach and nectarine. Postharvest Biol Technol. 2005;37(3):195–208. doi: 10.1016/j.postharvbio.2005.04.012. [DOI] [Google Scholar]
- 46.Wang Y., Deng L., Meng J., Niu L., Pan L., Lu Z., et al. Transcriptomic and metabolic analyses reveal the mechanism of ethylene production in stony hard peach fruit during cold storage. Int J Mol Sci. 2021;22(21):11308. doi: 10.3390/ijms222111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pichersky E., Noel J.P., Dudareva N. Biosynthesis of plant volatiles: Nature’s diversity and ingenuity. Science. 2006;311(5762):808–811. doi: 10.1126/science.1118510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leisso R.S., Gapper N.E., Mattheis J.P., Sullivan N.L., Watkins C.B., Giovannoni J.J., et al. Gene expression and metabolism preceding soft scald, a chilling injury of ‘Honeycrisp’ apple fruit. BMC Genomics. 2016;17(1):798. doi: 10.1186/s12864-016-3019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McAtee P.A., Richardson A.C., Nieuwenhuizen N.J., Gunaseelan K., Hoong L., Chen X., et al. The hybrid non-ethylene and ethylene ripening response in kiwifruit (Actinidia chinensis) is associated with differential regulation of MADS-box transcription factors. BMC Plant Biol. 2015;15:304. doi: 10.1186/s12870-015-0697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elitzur T., Yakir E., Quansah L., Zhangjun F., Vrebalov J., Khayat E., et al. Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 2016;171(1):380–391. doi: 10.1104/pp.15.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S., Chen K., Grierson D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019;221(4):1724–1741. doi: 10.1111/nph.15545. [DOI] [PubMed] [Google Scholar]
- 52.Qin G., Wang Y., Cao B., Wang W., Tian S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 2012;70(2):243–255. doi: 10.1111/j.1365-313X.2011.04861.x. [DOI] [PubMed] [Google Scholar]
- 53.An J.P., Wang X.F., Li Y.Y., Song L.Q., Zhao L.L., You C.X., et al. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018;178(2):808–823. doi: 10.1104/pp.18.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An J.P., Wang X.F., Zhang X.W., Xu H.F., Bi S.Q., You C.X., et al. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J. 2020;18(2):337–353. doi: 10.1111/pbi.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Y.G., Ma Y.Y., Bi P.P., Wei W., Liu J., Hu Y., et al. Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol Biochem. 2020;146:374–383. doi: 10.1016/j.plaphy.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Ma H., Yang T., Li Y., Zhang J., Wu T., Song T., et al. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell. 2021;33(10):3309–3330. doi: 10.1093/plcell/koab188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong W., Li R., Huang J., Zhao H., Ge R., Wu Q., et al. Divergent DNA methylation contributes to duplicated gene evolution and chilling response in tea plants. Plant J. 2021;106(5):1312–1327. doi: 10.1111/tpj.15237. [DOI] [PubMed] [Google Scholar]
- 58.Ding Y., Shi Y., Yang S. Molecular regulation of plant responses to environmental temperatures. Mol Plant. 2020;13(4):544–564. doi: 10.1016/j.molp.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H., Zhu J., Gong Z., Zhu J.K. Abiotic stress responses in plants. Nat Rev Genet. 2022;23(2):104–119. doi: 10.1038/s41576-021-00413-0. [DOI] [PubMed] [Google Scholar]
- 60.Hu Y., Zhang L., Zhao L., Li J., He S., Zhou K., et al. Trichostatin a selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS One. 2011;6(7):e22132. doi: 10.1371/journal.pone.0022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwon C.S., Lee D., Choi G., Chung W.I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009;60(1):112–121. doi: 10.1111/j.1365-313X.2009.03938.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31(2):154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 63.Sabatucci A., Berchet V., Bellia F., Maccarrone M., Dainese E., D’Addario C., et al. A new methodological approach for in vitro determination of the role of DNA methylation on transcription factor binding using AlphaScreen® analysis: Focus on CREB1 binding at hBDNF promoter IV. J Neurosci Methods. 2020;341:108720. doi: 10.1016/j.jneumeth.2020.108720. [DOI] [PubMed] [Google Scholar]
- 64.Khamis M.M., Adamko D.J., El-Aneed A. Strategies and challenges in method development and validation for the absolute quantification of endogenous biomarker metabolites using liquid chromatography-tandem mass spectrometry. Mass Spectrom Rev. 2021;40(1):31–52. doi: 10.1002/mas.21607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.