Abstract
Drought is a major abiotic stress that severely limits sustainable wheat (Triticum aestivum L.) productivity via morphological and physio-biochemical alterations of cellular processes. The complex nature and polygenic control of drought tolerance traits make breeding tolerant genotypes quite challenging. However, naturally occurring variabilities among wheat germplasm resources could potentially help combating drought. The present study was conducted to assess the drought tolerance of 18 Bangladeshi hexaploid wheat genotypes, focusing on the identification of potent sources of diversity by combining microsatellite markers, also known as single sequence repeat markers, and morpho-physiological characteristics that might help accelerating wheat crop improvement programs. Initially, the genotypes were evaluated using 25 microsatellite markers followed by an on-field evaluation of 7 morphological traits (plant height, spike number, spike length, grains per spike, 1000-grain weight, grain yield, biological yield) and 6 physiological traits (SPAD value, membrane stability index, leaf relative water content, proline content, canopy temperature depression, and leaf K+ ion content). The field-trial was conducted in a factorial fashion of 18 wheat genotypes and two water regimes (control and drought) following a split-plot randomized complete block design. Regardless of genotype, drought was significantly damaging for all the tested traits; however, substantial variability in drought stress tolerance was evident among the genotypes. Spike length, 1000-grain weight, SPAD value, leaf relative water content, canopy temperature depression, proline content, and potassium (K+) ion content were the most representative of drought-induced growth and yield impairments and also correlated well with the contrasting ability of genotypic tolerance. Microsatellite markers amplified 244 alleles exhibiting 79% genetic diversity. Out of 25 markers, 23 was highly polymorphic showing 77% average polymorphism. Morpho-physiological trait-based hierarchical clustering and microsatellite marker-based neighbor-jointing clustering both revealed three genotypic clusters with 71% co-linearity between them. In both cases, the genotypes Kanchan, BAW-1147, BINA Gom 1, BARI Gom 22, BARI Gom 26, and BARI Gom 33 were found to be comparatively more tolerant than the other tested genotypes, showing potential for cultivation in water-deficit environments. The findings of this study would contribute to the present understanding of drought tolerance in wheat and would provide a basis for future genotype selection for drought-tolerant wheat breeding programs.
Keywords: Crop improvement, Genetic variability, Single sequence repeat, Stress tolerance, Triticum aestivum L.
1. Introduction
Wheat (Triticum aestivum L.), an annual, herbaceous plant belonging to the grass family Poaceae is one of the leading cereals satisfying the carbohydrate requirement for 35% of the global population [1]. However, abiotic stresses, drought in particular is significantly affecting wheat productivity worldwide by altering the plant's morphological, physiological, and biochemical attributes [2]. Recent reports highlighted drought as a permanent constraint to wheat cropping for at least 40 million hectors and 25 million hectors of croplands belonging to the developing and industrialized nations respectively [3]. Moreover, drought is yet to become more frequent in occurrence, longer in duration, and acuter in intensity in many parts of the world as a result of rapid exploitation of freshwater and groundwater resources, unproductive water loss, as well as climate change driving factors like global warming and greenhouse gas emission [4,5]. Under these circumstances, wheat production could drop as low as 50% or even more due to irrigation water deficit [6,7]. Developing a suitable wheat genotype tolerant to drought is thus essential to protect the crop loss from occurring [8,9]. Understanding germplasm diversity could play a significant part in this aspect as availability of desirable genes are prerequisite for any crop improvement program [10]. Particularly for wheat, greater variability within the cultivated varieties and breeding stocks could be observed because of extensive genetic recombination and wider variability during its domestication and development from its center of origin [11,12] which can be further manipulated to achieve adaptive/tolerance mechanisms against water deficit stress [10]. Bread wheat is a hexaploid crop with three sets of chromosomes (A, B, and D) for a total of 42 chromosomes making its genome size approximately 17 giga base pairs [13]. Wheat genome is comparatively larger than many other cereals and it contains about 107,000 genes in its genome [14]. This large genome size on one hand makes it challenging to work with [13,15]; however, on the other hand also indicates higher possibility of finding diverse and novel genes for desirable traits such as disease resistance, abiotic stress tolerance, and improved nutritional content [7,10,16,17].
Drought stress negatively impacts wheat growth and development by restricting water availability, leading to impaired cellular metabolic activities, membrane instability, increased free radical generation, altered hormonal balance, reduced photosynthetic activity, and ultimately low yield [18,19]. Conversely, these traits in association with other morphological and physiological traits (e.g., plant height, yield components, canopy temperature, water content, water use efficiency, osmolyte synthesis) are also excellent contrastable indicator for assessing the extent of drought tolerance within wheat genotypes [20,21]. Morphological, physiological, and yield contributing parameters-based phenotyping had been very effective for evaluating the genetic diversity of crops for drought tolerance, despite its polygenic hereditary nature and variable environmental restrictions [11]. However, relying only on phenotyping approach can prove to be highly resource-intensive, time-consuming and financially draining and might be hardly conclusive when a big number of genotypes are involved [6,7,16]. According to Lopes et al. [22], the predictability of the genotypic variability under stressful environments was about 34% when only agronomic and physiological traits were focused; although phenological, physiological, and yield attributes were reported extensively reliable in measuring genetic variation within a gene pool [23,24]. These shortcomings could easily be complemented by the integration of advanced tools and techniques such as marker-assisted selection (MAS) using deoxyribonucleic acid (DNA)-based molecular markers [25,26]. Molecular markers like random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and microsatellites are highly efficient for providing comparatively rapid, accurate and direct estimate of genetic diversity [27,28]. In the context of drought tolerance, molecular markers can help examining the presence or absence of novel genomic regions associated with drought tolerance and assist in MAS by allowing focus towards the most promising candidates [[29], [30], [31]]. Microsatellite markers also known as simple sequence repeat (SSR) markers are one of the best choices for wheat because of their chromosomal specificity, multi-allelic composition, co-dominant nature, high polymorphism rate and widespread prevalence within the genome [32,33]. Because of these unique characteristics, a few markers can draw contrastable relationship among small to large population of wheat genotypes [34]. Recent reports had highly recommended using microsatellite markers for genetic diversity evaluation of wheat germplasms for drought-tolerance breeding programs [24,32,35]. Both the morpho-physiological trait-based and microsatellite marker-based germplasm evaluation methodologies had been extensively used individually or combinedly on wheat population across continents; however, in the context of Bangladesh such approaches had rarely been adopted even though the country harbors quite big germplasm resource [36,37]. Thus, keeping the research gap in mind as well as considering the potential negative impacts on drought on wheat cultivation the objective of the current study was to investigate the genetic diversity of Bangladeshi wheat genotypes for drought tolerance combining two different approaches (i) microsatellite markers-based screening of genetic diversity for drought tolerance and (ii) morpho-physiological trait-based on-field performance evaluation of genotypes under drought exposure for aiding in drought tolerant wheat crop improvement programs.
2. Materials and methods
2.1. Experimental conditions and plant materials
A laboratory experiment followed by a field trial was conducted for the study at Bangabandhu Sheikh Mujibur Rahman Agricultural University (BSMRAU), Gazipur 1706, Bangladesh to assess the extent of drought tolerance among wheat (T. aestivum L.) genotypes. In total, 18 wheat genotypes consisting of 17 domestic cultivars and an advanced line were evaluated during this study (Table 1). The laboratory experiment aimed at evaluating the genetic diversity of the genotypes using microsatellite molecular markers and the field trial focused on assessing the drought tolerance of the genotypes via morphological and physiological characteristics. For efficient inter-genotype comparison, a well-known drought tolerant check Sonoita F-81 was included with the genotypes during the laboratory experiment which was generously provided by International Maize and Wheat Improvement Center (CIMMYT), Mexico.
Table 1.
Wheat (Triticum aestivum L.) genotypes used in this study.
| Serial | Name of the genotype | Pedigree | Source |
|---|---|---|---|
| 1 | BARI Gom 19 (Sourav) | NAC/VEE (NL 560) | BWMRI |
| 2 | BARI Gom 20 (Gourav) | TURACO/CHIL | BWMRI |
| 3 | BARI Gom 21 (Shatabdi) | MRNG/BVC//BLO/PVN/3/PJB-81 | BWMRI |
| 4 | BARI Gom 22 (Sufi) | KAN/6/COQ/F61.70//CNDR/3/OLN/4/PHO/5/MRNG/ALDAN//CNO | BWMRI |
| 5 | BARI Gom 23 (Bijoy) | NL297*2/LR25 | BWMRI |
| 6 | BARI Gom 24 (Prodip) | G. 162/BL 1316//NL 297 | BWMRI |
| 7 | BARI Gom 25 | ZSH 12/HLB 19//2*NL297 | BWMRI |
| 8 | BARI Gom 26 | ICTAL 123/3/RAWAL 87//VEE/HD 2285 | BWMRI |
| 9 | BARI Gom 27 | WAXWING*2/VIVISTI | BWMRI |
| 10 | BARI Gom 28 | CHIL/2*STAR/4/BOW/CROW//BUC/PVN/3/2*VEE#10 | BWMRI |
| 11 | BARI Gom 29 | SOURAV/7/KLAT/SOREN//PSN/3/BOW/4/VEE#5. 10/5/CNO 67/MFD//MON/3/SERI/6/NL297 | BWMRI |
| 12 | BARI Gom 30 | BAW 677/Bijoy | BWMRI |
| 13 | BARI Gom 31 | KAL/BB/YD/3/PASTOR | BWMRI |
| 14 | BARI Gom 32 | SHATABDI/GOURAV | BWMRI |
| 15 | BARI Gom 33 | KACHU/SOLALA | BWMRI |
| 16 | BARI advanced wheat 1147 (BAW-1147) | – | BWMRI |
| 17 | BINA Gom 1 | L-880-43 | BINA |
| 18 | Kanchan | UP301/C306 | BWMRI |
BARI: Bangladesh Agricultural Research Institute; BWMRI: Bangladesh Wheat and Maize Research Institute; BINA- Bangladesh Institute of Nuclear Agriculture
2.2. Laboratory experiment
A laboratory experiment was conducted at the Advanced Plant Breeding Laboratory of Department of Genetics and Plant Breeding, BSMRAU; to characterize the genetic diversity among the wheat genotypes (18 domestic genotypes and Sonoita F-81 as tolerant check) using microsatellite markers also known as SSR (single sequence repeat) markers. Highly polymorphic 25 SSR markers covering most of the chromosomes of the hexaploid wheat genome were chosen for the study (Table 1S). Marker selection were done based on their high polymorphism, genetic diversity and substantial genome coverage reported in previous investigations. Wheat seeds were surface sterilized with 3% sodium hypochlorite for 10 min, rinsed properly with distilled water and sown in pots containing 250 g of soil. Genomic DNA was extracted from leaves of 15 days old wheat seedlings.
2.2.1. Genomic DNA extraction
Genomic DNA was extracted following the modified cetyl-trimethyl ammonium bromide (CTAB) method as described by Yu et al. [38]. Briefly, 100 mg of fresh leaf samples were grounded to powder with liquid nitrogen using an ice-cold mortar and pestle. The powder was then mixed with 1000 μL CTAB extraction buffer and incubated in a water bath for 60 min at 65°C. Afterwards, 1000 μL of 24:1 (v/v) chloroform:isoamyl alcohol was added and mixed vigorously to form a homogenous emulsion. Then the samples were centrifuged at 2500G for 15 min and the supernatant was separated. Upon separation, 1500 μL absolute ethanol (99.9% v/v) was added to the supernatant, mixed carefully by rocking the tube back and forth sideways and centrifuged at 12000G for 15 min to obtain DNA pallet. The DNA pallet was then separated and suspended in 1x TE buffer [Tris-EDTA (ethylenediamine tetra-acetic acid)]. Ribonuclease A was added to the sample at 10 μg mL−1 rate and incubated at 37°C for 60 min. Afterwards 24:1 (v/v) chloroform:isoamyl alcohol was added again to the samples, mixed vigorously and centrifuged. The supernatant was separated, absolute ethanol was added, mixed carefully and recentrifuged to obtain DNA pallet. The DNA pallet was then carefully washed with 70% ethanol and dried overnight in an orbital shaker at room temperature. Finally, the pallet was suspended in 50 μL TE buffer and stored at −20°C.
2.2.2. Polymerase chain reaction amplification and gel electrophoresis
Polymerase chain reaction (PCR) was performed in 10 μL reaction mixture containing 3 μL DNA (50 ng μL−1), 1 μL 10x reaction buffer, 2 μL 25 mM MgCl2, 0.8 μL 25 mM dNTP (deoxynucleoside triphosphate), 0.5 μL each of 10 μM forward and reverse microsatellite primers, and 0.2 μL of Taq DNA polymerase following the procedures of Mohi-Ud-Din et al. [7]. The PCR amplification was performed on an Eppendorf thermocycler (Mastercycler® nexus, Scientific Laboratory Supplies Ltd., United Kingdom) with the following conditions: initial denaturation at 95°C for 5 min followed by 36 cycles where denaturation was performed at 95°C for 45 s, annealing for 45 s at 55°C, and extension for 5 min at 72°C. The DNA amplicons of the resultant PCR product were separated on 3.5% metaphor agarose gel in a 0.5x TBE (Tris Borate EDTA) system with a voltage of 100 V for 2 h [39]. Ethidium bromide was used for gel staining and the separated PCR products were visualized in G:Box F3 gel documentation system (Syngene, Synoptic Ltd., United Kingdom). Clear and explicit bands were used for scoring in the form of binary data (1 for presence of band and 0 for absence of band).
2.3. Field experiment
A field trial was conducted at the experimental field of Department of Crop Botany, BSMRAU; (24.03866° N, 90.39795° E) involving 18 wheat genotypes (17 domestic cultivars and an advanced line). The experimental site falls under AEZ (agro-ecological zone) number 28 of Bangladesh that features a sub-tropical climate with a temperature of 20.6 ± 3.8°C and scanty rainfall during winter season (October to March) which is the dominant wheat cultivation period (available from https://en.climate-data.org/asia/bangladesh/dhaka-division/gazipur accessed at 1st March 2023). Edaphically, the experimental site is characterized as upland and the soil was of silty loam textural class that attained full field capacity at 30.6% volumetric soil water content. The physical and chemical properties of the soil have been presented in Table 2S.
2.3.1. Experimental design and treatments
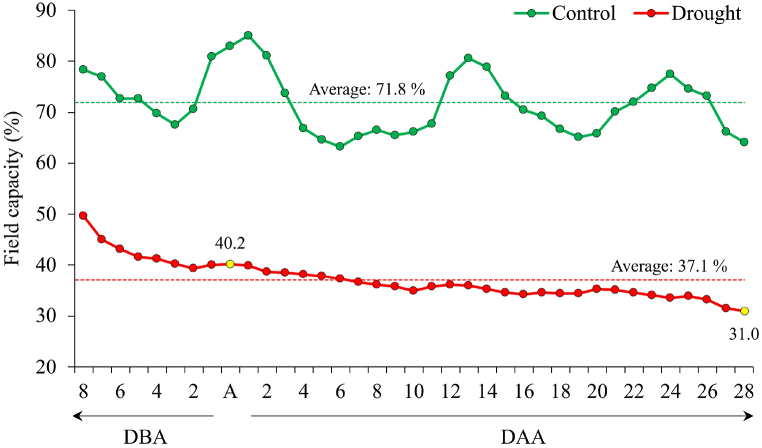
The experiment was laid out in a factorial combination of 18 wheat genotypes and two water regimes (control and drought) following a randomized complete block design (split plot) with three replications (Fig. 1S). The entire plot was divided into two main plots among which one was regularly irrigated (control) and in another irrigation was discontinued after 45 days of seed sowing (drought). The genotypes were assigned randomly in the sub-plots within the main plots. Soil moisture content was regularly monitored at 15 cm depth using a handheld digital soil moisture meter (PMS-714, Lutron Electronic Enterprise Co., Ltd., Taipei, Taiwan) till maturity. Withdrawing irrigation caused gradual dropping of soil moisture resulting in water scarcity (average field capacity 37.1%) in drought treated plots whereas, the controlled plots had ample available soil moisture (average field capacity 71.8%) during the experimental period (Fig. 1).
Fig. 1.
Pattern of changes in soil field capacity of the control and drought treated plots during pre-anthesis, anthesis and post-anthesis period. DBA: days before anthesis, A: anthesis, DAA: days after anthesis.
2.3.2. Determination of morphological traits, yield and yield components
Data on plant height (cm), spike number (m−2), spike length (cm), grains per spike, 1000-grain weight (g), grain yield (ton ha−1), and biological yield (ton ha−1) was collected as morphological, yield and yield contributing traits for the study. Plant height (cm) was measured from the collar region to the tip of spike using a centimeter scaled measuring tape. Spike number (m−2) was recorded by manual counting with help of a quadrat (2500 cm2) sampler. Spike length (cm) was measured from the base to the tip of a spike using a centimeter scale. Grains per spike was manually counted. 1000-grain weight (g) was measured by weighing thousand wheat grains in an electrical balance. Grain yield (ton ha−1) was recorded by weighing the harvested grains per sub-plot. Both the 1000-grain weight (g) and grain yield (ton ha−1) were recorded at 12% grain moisture content. Harvested wheat shoots were sun dried and then oven dried till constant weight for recording straw yield (ton ha−1) in an electrical balance. Biological yield (ton ha−1) was then calculated using the formula: [Biological yield (ton ha−1) = grain yield (ton ha−1) + straw yield (ton ha−1)].
2.3.3. Quantification of SPAD value and membrane stability
Leaf greenness measurement by SPAD (soil plant analysis development) value is a non-destructive quantification method of leaf chlorophyl content which was measured from the middle portion of the flag leaf using a hand held SPAD meter (SPAD-502 Plus, Minolta Co. Ltd., Osaka, Japan) [40]. Measurements were taken during heading (70 days after sowing), anthesis, 7 days after anthesis (DAA), 14 DAA and 21 DAA in between 10.00 AM and 12.00 PM from randomly selected ten flag leaves. For each sub-plot all the measured values were averaged and recorded. Heading and anthesis of the genotypes were ascertained by observing the visibility and blooming of 50% of the spikes in a plot, respectively.
Membrane stability index (%) was calculated from the middle portion of the flag leaf during heading according to Ahmed et al. [41]. Immediately after collection, leaf samples were wiped with tissue paper, stored in airtight zip-lock bags and transported to the laboratory. Leaf discs of 1 cm diameter were prepared and rinsed in deionized water. Thirty well-washed leaf discs were placed in a closed vial containing 10 mL deionized water and incubated for 30 min at 40°C in a hot water bath. Upon incubation, the electrical conductivity of the solutions was measured (EC1). Another set of samples containing the same number of leaf disks and the same volume of deionized water were boiled (100°C) in a hot water bath for 20 min and the electrical conductivity (EC2) was measured. Afterwards, membrane stability index (%) was calculated using the following formula: Membrane stability index (%) = [1 − (EC1/EC2)] × 100.
2.3.4. Measurement of leaf relative water content and proline content
Leaf relative water content (%) was determined from the middle portion of flag leaf during heading (70 days after sowing) according to Das et al. [42]. Harvested leaves were wiped with tissue paper, stored in airtight zip-lock bags and taken to the laboratory. Leaf disks of 1 cm diameter were prepared and thirty disks were weighed in an analytical balance for recording their fresh weight (Fw). Afterwards, those disks were immersed in distilled water and kept in room temperature for 12 h and turgid weight (Tw) was recorded. Surface moisture on the leaf disks were removed gently with blotting paper before measuring Tw. Then the leaf disks were dried in an oven (72°C) till constant weight and their dry weight (Dw) was measured. Finally leaf relative water content (%) was calculated using the formula: [Leaf relative water content (%) = {(Fw−Dw)/(Tw−Dw)} × 100].
Proline content (μmol g−1 fresh weight) of flag leaf samples was measured following the original methods of Bates et al. [43] as described in Ahmed et al. [44]. Freshly collected leaf samples were wiped with tissue paper, stored in airtight zip-lock bags and transported to the laboratory. Leaf sample (0.1 g) was homogenized in 2.0 mL of 10% aqueous sulfosalicylic acid. The homogenate was then centrifuged at 10,000 rpm for 10 min and 1 mL of the supernatant was transferred into a test tube containing 2 mL of 1:1 (v/v) acid ninhydrin solution:glacial acetic acid. The mixture was then incubated in a boiling water bath for 1 h, and the reaction was terminated rapidly in an ice bath. Afterwards, 2 mL toluene was added, vortexed vigorously at 6000 rpm for 5 min and kept at room temperature for precipitation. The upper layer of the mixture was separated for measuring spectrophotometer absorbance at 520 nm (Genesys 10S UV-VIS, Thermo Fisher Scientific, Waltham, MA, USA). Finally, the free proline levels in leaf samples were determined from a standard curve prepared with known concentrations of proline solutions and was expressed as μmol g−1 fresh weight.
2.3.5. Determination of canopy temperature depression and potassium ion content
Canopy temperature depression (°C) was determined following the procedures of Balota et al. [45]. Temperature of the crop canopy (CT) and instantaneous air temperature (AT) was measured simultaneously using a hand-held infrared thermometer (Model: IR-818, URCERI, USA; the distance-spot ratio of 13:1). Measurements were taken during heading (70 days after sowing), anthesis, 7 days after anthesis (DAA), 14 DAA and 21DAA in between 10.00 AM and 12.00 PM from five randomly selected locations within a sub-plot. All the measured values within a subplot were averaged and recorded. Finally canopy temperature depression (°C) was calculated using the equation: [Canopy temperature depression (°C) = AT (°C) − CT (°C)]. While taking a temperature measurement, the infrared thermometer was held at 2 m distance from the pointing surface and about 30° pointing angle with the horizontal soil surface were maintained.
Leaf K+ ion content (mg g−1 fresh weight) was estimated using the LAQUAtwin-B-731 potassium ion meter (HORIBA instruments, CA, USA) as described by Apon et al. [46]. Briefly, 0.1 g plant sample was homogenizing (taken from the middle portion of fully expanded flag leaf) with 10 mL of deionized water. The homogenate was then incubated in a hot water bath for 20 min at 120°C for complete ion extraction into the solution. Afterwards, the tubes were allowed to cooldown and centrifuged at 10,000 rpm for 10 min. The supernatant was then used for K+ ion content determination (mg L−1) and the values were expressed as mg g−1 fresh weight.
2.4. Data analysis
Data obtained from the laboratory experiment were subjected to analysis of major allele frequency, polymorphic information content (PIC) and gene diversity using PowerMarker 3.23 [47]. Genetic distances among the genotypes were measured through Euclidean distance matrix, and an unweighted pair group method with arithmetic means (UPGMA) based neighbor-jointing dendrogram was constructed to categorize the genotypes into different tolerance groups using Darwin 6.0.21 for windows (http://darwin.cirad.fr/) (accessed on 28 December 2021) [48].
Data obtained from field experiment were fitted to a linear model for the factorial randomized complete block design in R-4.3.0 for Windows. The analysis of variance (ANOVA) was performed using the following equation to determine if genotype, water regime, and their interactions had a significant influence on the parameters:
where, Yijk is observation for the i-th genotype in the j-th water regime and k-th replication (block); μ is the grand mean effect; Gi is the effect of the i-th genotype; Wj is the effect of the j-th water regime; Bk(j) is the effect of the k-th replication (block) within the j-th water regime; (GW)ij is the interaction effect of the i-th genotype with the j-th water regime; and εijk is the effect of the pooled error associated with the i-th genotype, j-th water regime and k-th replication (block).
Means of significant treatment effects were separated via post hoc analysis using Tukey's honest significant difference test. In all analyses, differences were considered significant at P ≤ 0.05. Descriptive statistics of the traits was presented as box and whisker plots prepared using the packages ggplot2 along with reshape2 [49] in R-4.3.0 for windows (https://cran.r-project.org/) in RStudio-2023.03.0–386 (https://posit.co/) (accessed on 30th April 2023). The degree of association of the studied traits was determined by correlation coefficients among them. The correlation coefficient matrix and correlation heatmap were visualized using the web-based MVApp application [50]. The magnitude of stress tolerance of the genotypes was quantified as [stress tolerance = performance under drought/performance under control]. The obtained data were normalized and used for categorizing the genotypes into genotype clusters using hierarchical cluster algorithm in the library pheatmap [16,51]. The extracted clusters were based on Eucledian distance matrix where genotypes grouped within same cluster were broadly similar to each other and genotypes belonging to different clusters are distinct to each other. The magnitude of tolerance for the studied traits was visualized for every genotype using a conditional tri-color scaled heatmap. Relative change in performance of genotype clusters under drought compared to control were extracted as percent change using the following equation: [% change over control = {(performance under drought−performance under control)/performance under control} × 100] and was presented as stacked dot-plot [7,49]. Principal component analysis (PCA) was carried out to reduce the dimensionality of the dataset without losing important information using the packages ggplot2, factoextra and FactoMineR [49,52]. Using the online interactive application Venny 2.1, the co-linearity between morpho-physiological trait-based hierarchical (HR) clusters and UPGMA-based neighbor-joining (NJ) clusters was determined [53].
3. Results
3.1. Microsatellite amplification and genetic diversity among wheat genotypes
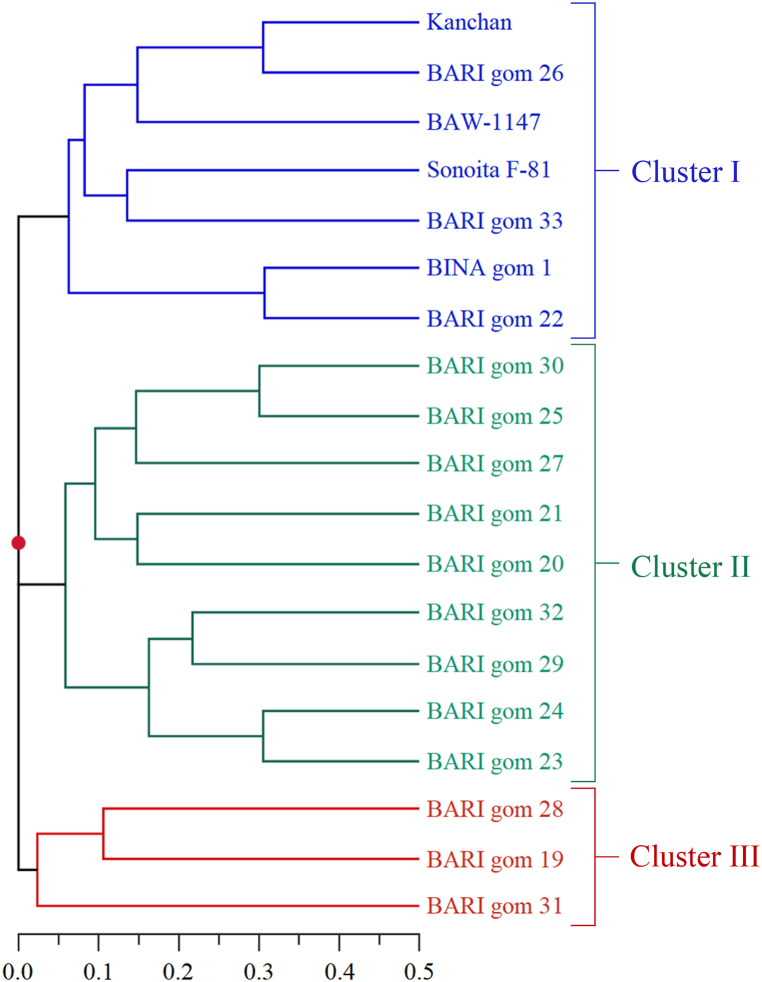
A total of 25 microsatellite markers were used to determine the genetic variability related to drought tolerance patterns within 19 wheat genotypes. Among the genotypes 18 were Bangladeshi wheat genotypes (Table 1) and a well-recognized drought tolerant variety Sonoita F-81 which was included to aid in drawing reliable and contrastable comparison. Major allele frequency, allele number, gene diversity and PIC were determined for all the microsatellite markers. The 25 microsatellite loci amplified 244 alleles in total although the number of amplified alleles by individual microsatellite was variable (Fig. 2S). Allele detection ranged from 3 (wms186) to 16 (wmc177) alleles with an average of 9.76 alleles per locus. Frequencies of major alleles amplified from the 25 microsatellite loci ranged between 0.16 (wmc179 and wms484) to 0.84 (wms186) with an average of 0.35. An average PIC of 0.77 was evident among the used microsatellites within a range between 0.27 (wms186) to 0.90 (wmc179 and wms484). Fifteen microsatellites were found showing PIC value ≥ 0.80 those amplified ≥10 alleles. The microsatellite wms136 also amplified 10 alleles although its PIC value was <0.80 (Table 2). Gene diversity of the genotypes lied between 0.28 (wms186) to 0.90 (wmc179, wmc177 and wms484) with an average of 0.79. Microsatellites of D genomes produced the highest mean allele per locus (10.3) and gene diversity (0.83) followed by B genome; whereas, A genome had the lowest mean allele per locus (9.2) and gene diversity (0.75) (Table 2). Based on the genetic diversity revealed by the microsatellites, an unweighted pair group method with arithmetic means (UPGMA)-based neighbor-jointing dendrogram was constructed to classify the genotypes in different clusters using the Euclidean distance matrix. The dendrogram revealed three genotype clusters where genotypes from same cluster group were genetically similar and genotypes belonging to different clusters were genetically distinct from each other (Fig. 2). The cluster I comprised of BARI Gom 22, BARI Gom 26, BARI Gom 33, BINA Gom 1, BAW-1147, Kanchan, and the drought tolerant check Sonoita F-81. This indicated genetic similarity of the genotypes among each other and also their closeness with Sonoita F-81. Moreover, the genotype BARI Gom 33 and BAW-1147 respectively shared the highest and second highest genetic similarity with Sonoita F-81. Furthermore, the cluster III comprising of 3 genotypes (BARI Gom 19, BARI Gom 28 and BARI Gom 31) had the least genetic similarity with the drought tolerant check Sonoita F-81 (Fig. 2).
Table 2.
Allele frequency, number of alleles, genetic diversity and polymorphic information content (PIC) for 25 microsatellite loci.
| Marker | Location on chromosome | Major allele frequency | Number of alleles | Gene diversity | PIC |
|---|---|---|---|---|---|
| wms136 | 1A | 0.42 | 10 | 0.78 | 0.77 |
| wmc177 | 2A | 0.21 | 16 | 0.90 | 0.89 |
| wms304 | 2A | 0.21 | 10 | 0.87 | 0.86 |
| wms369 | 3A | 0.21 | 12 | 0.88 | 0.86 |
| wms165 | 4A | 0.47 | 5 | 0.73 | 0.70 |
| wms186 | 5A | 0.84 | 3 | 0.28 | 0.27 |
| wms169 | 6A | 0.16 | 9 | 0.88 | 0.86 |
| barc108 | 7A | 0.74 | 6 | 0.44 | 0.43 |
| wmc9 | 7A | 0.37 | 11 | 0.82 | 0.81 |
| wms260 |
7A |
0.21 |
10 |
0.87 |
0.86 |
| Mean (genome A) |
0.38 |
9.2 |
0.75 |
0.73 |
|
| wms11 | 1B | 0.37 | 10 | 0.81 | 0.80 |
| wms257 | 2B | 0.21 | 10 | 0.86 | 0.85 |
| wms389 | 3B | 0.21 | 11 | 0.87 | 0.86 |
| wms149 | 4B | 0.26 | 10 | 0.85 | 0.84 |
| wms375 | 4B | 0.21 | 11 | 0.88 | 0.86 |
| wms118 | 5B | 0.53 | 9 | 0.69 | 0.67 |
| wmc105 | 6B | 0.42 | 8 | 0.75 | 0.73 |
| wms46 |
7B |
0.47 |
9 |
0.74 |
0.72 |
| Mean (genome B) |
0.34 |
9.8 |
0.81 |
0.79 |
|
| wmc179 | 1D | 0.16 | 13 | 0.90 | 0.90 |
| wms484 | 2D | 0.16 | 13 | 0.90 | 0.90 |
| wms161 | 3D | 0.21 | 10 | 0.88 | 0.86 |
| wms192 | 4D | 0.42 | 9 | 0.76 | 0.73 |
| wms292 | 5D | 0.37 | 10 | 0.81 | 0.80 |
| psp3200 | 6D | 0.53 | 8 | 0.69 | 0.67 |
| wms295 |
7D |
0.32 |
11 |
0.85 |
0.83 |
| Mean (genome D) |
0.31 |
10.6 |
0.83 |
0.81 |
|
| Mean (total) | 0.35 | 9.76 | 0.79 | 0.77 | |
Fig. 2.
Unweighted pair group method with arithmetic means-based neighbor jointing dendrogram showing genetic relationship among wheat genotypes based on SSR allele data.
3.2. Morpho-physiological evaluation of drought tolerance
Genotypic evaluation for drought tolerance based on morphological traits, yield and yield components, and physiological traits (all together referred as ‘morpho-physiological traits’ hereafter) revealed a high level of significance among the 18 Bangladeshi wheat genotypes due to drought exposure. The two-way ANOVA between the genotypes (G) and water regimes (control and drought) (T) reviled significant individual and interactive effects for all the studied parameters except for the interactive effect (G × T) of spike number (m−2). The main effect of genotype or water regimes were significant at P ≤ 0.01 across all the tested traits; however, their interactive effect (G × T) for plant height (cm), grains per spike, grain yield (ton ha−1), and biological yield (ton ha−1) were significant at P ≤ 0.05 (Table 3, Table 4).
Table 3.
Effect of different water regimes (control and drought) on morphological, yield and yield contributing traits (plant height, spike number, spike length, grains per spike, 1000-grain weight, grain yield and biological yield) of tested wheat genotypes.
| Factor | Plant height (cm) | Spike number (m−2) | Spike length (cm) | Grains per spike | 1000-grain weight (g) | Grain yield (ton ha−1) | Biological yield (ton ha−1) |
|---|---|---|---|---|---|---|---|
| Genotype | |||||||
| BARI gom 19 | 77.7 ± 0.54 b-g | 225 ± 11.9 f | 10.2 ± 0.20 fgh | 51.7 ± 1.19 c | 34.9 ± 0.20 hi | 3.97 ± 0.03 g | 9.7 ± 0.49 efg |
| BARI gom 20 | 72.3 ± 0.49 gh | 239 ± 4.2 ef | 9.8 ± 0.20 gh | 40.5 ± 1.20 gh | 38.3 ± 0.67 fg | 4.41 ± 0.06 d-g | 9.8 ± 0.47 d-g |
| BARI gom 21 | 78.9 ± 1.12 a-e | 307 ± 8.7 abc | 10.6 ± 0.28 efg | 35.8 ± 0.63 h | 36.5 ± 0.42 gh | 5.05 ± 0.07 bcd | 10.9 ± 0.25 a-f |
| BARI gom 22 | 76.9 ± 1.11 c-g | 300 ± 8.1 a-d | 10.7 ± 0.14 ef | 49.8 ± 0.75 cde | 37.1 ± 0.38 gh | 4.64 ± 0.07 def | 11.7 ± 0.12 a-d |
| BARI gom 23 | 83.5 ± 1.25 a | 300 ± 11.3 a-d | 12.6 ± 0.09 abc | 41.3 ± 0.59 gh | 43.1 ± 0.81 bcd | 5.29 ± 0.10 abc | 11.8 ± 0.25 abc |
| BARI gom 24 | 80.3 ± 0.98 a-d | 212 ± 7.2 f | 12.2 ± 0.10 bc | 52.7 ± 0.70 c | 44.4 ± 0.43 abc | 3.90 ± 0.18 g | 9.9 ± 0.34 d-g |
| BARI gom 25 | 70.6 ± 0.69 h | 247 ± 6.5 def | 10.2 ± 0.08 e-h | 44.0 ± 0.93 efg | 38.4 ± 0.40 fg | 4.12 ± 0.07 efg | 11.2 ± 0.36 a-e |
| BARI gom 26 | 76.1 ± 0.49 d-h | 283 ± 8.9 bcde | 9.3 ± 0.13 h | 44.2 ± 1.08 efg | 42.0 ± 0.34 cde | 4.75 ± 0.05 cde | 10.0 ± 0.13 b-g |
| BARI gom 27 | 77.7 ± 0.90 b-g | 326 ± 10.9 ab | 10.2 ± 0.05 fgh | 42.8 ± 0.59 fg | 31.1 ± 0.39 j | 4.02 ± 0.08 fg | 10.5 ± 0.21 b-f |
| BARI gom 28 | 73.1 ± 1.13 fgh | 214 ± 5.5 f | 11.1 ± 0.12 def | 51.3 ± 1.13 c | 38.4 ± 0.58 fg | 3.92 ± 0.02 g | 8.2 ± 0.07 g |
| BARI gom 29 | 73.4 ± 0.86 e-h | 233 ± 9.3 ef | 10.5 ± 0.14 efg | 51.0 ± 0.88 cd | 40.7 ± 0.61 def | 4.97 ± 0.14 cd | 9.9 ± 0.38 d-g |
| BARI gom 30 | 75.5 ± 1.04 d-h | 258 ± 6.1 c-f | 10.9 ± 0.08 def | 47.0 ± 1.67 c-g | 40.7 ± 0.21 def | 4.03 ± 0.08 fg | 10.0 ± 0.18 c-g |
| BARI gom 31 | 78.2 ± 0.62 a-f | 255 ± 13.1 c-f | 12.3 ± 0.13 bc | 50.3 ± 0.52 cde | 31.3 ± 0.76 j | 4.24 ± 0.17 efg | 10.6 ± 0.27 a-f |
| BARI gom 32 | 77.2 ± 0.28 b-g | 302 ± 4.0 abc | 11.0 ± 0.01 def | 36.2 ± 0.68 h | 44.6 ± 0.13 abc | 4.73 ± 0.11 cde | 10.1 ± 0.28 b-f |
| BARI gom 33 | 82.1 ± 0.67 a-c | 266 ± 7.6 c-f | 13.3 ± 0.10 a | 70.2 ± 1.37 a | 46.4 ± 0.68 ab | 5.91 ± 0.04 a | 12.5 ± 0.12 a |
| BAW-1147 | 79.7 ± 1.03 a-d | 258 ± 7.3 c-f | 12.8 ± 0.10 ab | 48.8 ± 1.78 c-f | 47.1 ± 0.33 a | 5.64 ± 0.17 ab | 11.9 ± 0.25 ab |
| BINA gom 1 | 82.9 ± 0.65 ab | 238 ± 8.8 ef | 11.1 ± 0.01 de | 60.0 ± 1.01 b | 31.7 ± 0.12 ij | 3.80 ± 0.06 g | 9.2 ± 0.26 fg |
| Kanchan | 82.0 ± 0.83 abc | 342 ± 8.2 a | 11.7 ± 0.12 cd | 44.5 ± 0.52 d-g | 39.5 ± 0.29 efg | 4.70 ± 0.12 cde | 11.3 ± 0.19 a-e |
| Treatment | |||||||
| Control | 81.1 ± 0.85 a | 288 ± 8.32 a | 11.6 ± 0.11 a | 51.8 ± 1.02 a | 41.8 ± 0.42 a | 5.12 ± 0.09 a | 11.6 ± 0.24 a |
| Drought | 74.2 ± 0.78 b | 246 ± 8.08 b | 10.7 ± 0.12 b | 44.0 ± 0.90 b | 36.7 ± 0.44 b | 3.99 ± 0.09 b | 9.4 ± 0.28 b |
| Significance | |||||||
| Genotype (G) | ** | ** | ** | ** | ** | ** | ** |
| Treatment (T) | ** | ** | ** | ** | ** | ** | ** |
| G × T | * | ns | ** | * | ** | * | * |
Data are means of three replications ± standard errors. Means within a column followed by different letters are significantly different based on Tukey's honest significant difference test at P ≤ 0.05. **, *, and ns indicates significance at P ≤ 0.01, P ≤ 0.05 and non-significant respectively.
Table 4.
Effect of different water regimes (control and drought) on physiological parameters (SPAD value, leaf relative water content, membrane stability index, canopy temperature depression, proline content and potassium (K+) ion content) of tested wheat genotypes.
| Factor | SPAD value | Leaf relative water content (%) | Membrane stability index (%) | Canopy temperature depression (°C) | Proline content (μmol g−1 fresh weight) | K+ ion content (mg g−1 fresh weight) |
|---|---|---|---|---|---|---|
| Genotype | ||||||
| BARI gom 19 | 43.1 ± 0.11 fg | 67.1 ± 0.24 fg | 60.6 ± 0.14 fg | 5.14 ± 0.09 fg | 4.35 ± 0.07 d-g | 49.5 ± 0.33 cde |
| BARI gom 20 | 42.6 ± 0.25 g | 68.4 ± 0.45 ef | 75.3 ± 0.49 cd | 4.07 ± 0.01 jk | 3.86 ± 0.09 g | 60.7 ± 0.93 a |
| BARI gom 21 | 43.5 ± 0.22 efg | 65.3 ± 0.52 g | 56.7 ± 0.69 gh | 5.71 ± 0.06 de | 4.14 ± 0.04 fg | 51.8 ± 0.67 cd |
| BARI gom 22 | 45.2 ± 0.23 bcd | 71.2 ± 0.45 cde | 62.5 ± 0.60 f | 6.09 ± 0.05 bcd | 4.91 ± 0.11 cde | 47.0 ± 0.48 d-g |
| BARI gom 23 | 42.2 ± 0.31 g | 67.4 ± 0.35 fg | 55.5 ± 0.87 h | 5.14 ± 0.10 fg | 4.40 ± 0.06 d-g | 45.5 ± 0.19 efg |
| BARI gom 24 | 44.4 ± 0.12 c-f | 67.2 ± 0.58 fg | 63.0 ± 0.59 f | 5.44 ± 0.05 ef | 4.09 ± 0.13 fg | 45.2 ± 0.68 efg |
| BARI gom 25 | 45.6 ± 0.10 bc | 58.8 ± 1.07 h | 44.2 ± 0.92 j | 5.84 ± 0.12 b-e | 4.60 ± 0.01 def | 50.2 ± 0.67 cde |
| BARI gom 26 | 46.5 ± 0.18 ab | 73.5 ± 0.34 bc | 81.1 ± 0.12 ab | 3.83 ± 0.03 kl | 4.99 ± 0.02 cd | 60.8 ± 0.53 a |
| BARI gom 27 | 44.7 ± 0.36 c-f | 68.7 ± 0.32 def | 50.8 ± 0.79 i | 6.16 ± 0.12 bc | 5.84 ± 0.20 ab | 54.2 ± 0.54 bc |
| BARI gom 28 | 43.8 ± 0.17 d-g | 64.7 ± 0.26 g | 57.7 ± 0.36 gh | 4.50 ± 0.06 hij | 5.54 ± 0.04 abc | 48.3 ± 0.84 c-f |
| BARI gom 29 | 43.2 ± 0.44 fgg | 71.7 ± 0.34 bcd | 48.6 ± 0.45 i | 8.00 ± 0.06 a | 4.48 ± 0.18 d-g | 58.3 ± 1.23 ab |
| BARI gom 30 | 44.4 ± 0.25 c-f | 74.4 ± 0.40 ab | 55.5 ± 0.99 h | 6.27 ± 0.05 b | 6.01 ± 0.23 ab | 49.5 ± 1.26 cde |
| BARI gom 31 | 44.7 ± 0.21 c-f | 68.4 ± 0.44 ef | 79.4 ± 0.47 bc | 4.14 ± 0.04 jk | 4.42 ± 0.02 d-g | 42.5 ± 0.19 fg |
| BARI gom 32 | 44.4 ± 0.33 c-f | 73.0 ± 0.37 bc | 73.1 ± 0.96 d | 4.83 ± 0.08 gh | 4.21 ± 0.06 efg | 51.2 ± 1.06 cde |
| BARI gom 33 | 47.3 ± 0.29 a | 70.8 ± 0.16 cde | 82.1 ± 0.28 ab | 3.57 ± 0.03 l | 6.23 ± 0.03 a | 60.8 ± 0.86 a |
| BAW-1147 | 44.9 ± 0.07 b-e | 76.7 ± 0.30 a | 71.9 ± 0.28 d | 5.73 ± 0.02 cde | 6.16 ± 0.06 a | 49.3 ± 0.96 cde |
| BINA gom 1 | 46.3 ± 0.25 ab | 70.7 ± 0.53 cde | 84.6 ± 0.71 a | 4.32 ± 0.05 ij | 4.09 ± 0.13 fg | 58.8 ± 1.04 ab |
| Kanchan | 44.6 ± 0.20 c-f | 72.1 ± 0.19 bc | 67.3 ± 0.55 e | 4.59 ± 0.01 hi | 5.35 ± 0.07 bc | 41.7 ± 1.94 g |
| Treatment | ||||||
| Control | 46.3 ± 0.20 a | 74.1 ± 0.30 a | 70.3 ± 0.49 a | 6.27 ± 0.05 a | 3.25 ± 0.06 b | 48.2 ± 0.49 b |
| Drought | 42.7 ± 0.26 b | 64.8 ± 0.51 b | 59.7 ± 0.65 b | 4.10 ± 0.06 b | 6.49 ± 0.11 a | 54.6 ± 1.12 a |
| Significance | ||||||
| Genotype (G) | ** | ** | ** | ** | ** | ** |
| Treatment (T) | ** | ** | ** | ** | ** | ** |
| G × T | ** | ** | ** | ** | ** | ** |
Data are means of three replications ± standard errors. Means within a column followed by different letters are significantly different based on Tukey's honest significant difference test at P ≤ 0.05. ** indicates significance at P ≤ 0.01. SPAD: soil plant analysis development.
3.2.1. Mean variability in studied traits
The descriptive statistics of the studied traits in response to control and drought treatments have been presented in the boxplots (Fig. 3S). The performance of the genotypes significantly decreased under drought compared to control for all the tested morpho-physiological traits except for proline content (μmol g−1 fresh weight) and K+ ion content (mg g−1 fresh weight) where the performance of the genotypes significantly increased under drought compared to control (Table 3, Table 4, Fig. 3). Regardless of genotype, the tested morphological traits, yield and yield components showed a steep performance downfall compared to control. For example, drought treatment led to 8.5, 15, 8, 15, 12, 22 and 19% decrease in plant height (cm), spike number (m−2), spike length (cm), grains per spike, 1000-grain weight (g), grain yield (ton ha−1), and biological yield (ton ha−1), respectively compared to control (Table 3). Likewise, in case of physiological parameters, we recorded 7.8, 12.5, 24.5, and 34.6% decrease in SPAD value, leaf relative water content (%), membrane stability index (%), and canopy temperature depression (°C) respectively compared to control. Contrariwise, we have observed that proline content (μmol g−1 fresh weight) was about 2-folds and K+ ion content (mg g−1 fresh weight) increased 13.4% under drought compared to control (Table 4). Apart from the differences due to different water regimes, contrastable variations were also present among the genotypes in terms of their individual performances on the studied traits. The genotype BARI Gom 33 followed by BAW-1147 showed overall outstanding performance compared to the other tested genotypes and stood out of the crowd. However, some other genotypes showed quite distinctive performance in some particular parameters. Notable mentions are BARI Gom 23 for plant height (cm), Kanchan for spike number (m−2), BINA Gom 1 for membrane stability index (%), and BARI Gom 20 and BARI Gom 26 for K+ ion content (mg g−1 fresh weight) (Table 3, Table 4). Similar trends were also evident for the interactive effects while genotype performances were compared across the two water regimes and vice versa (Tables 3S and 4S).
Fig. 3.
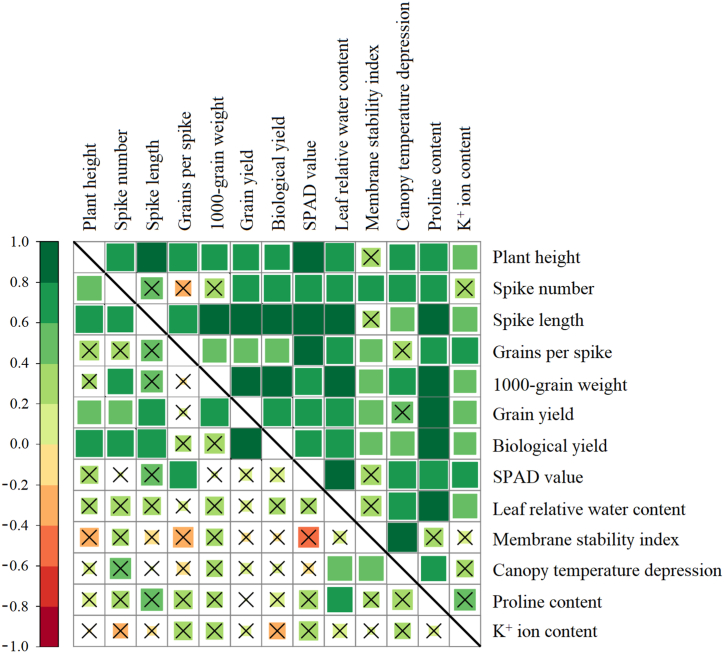
Correlation and pair-wise associations among the studied traits of wheat genotypes. Lower and upper diagonal shows correlation coefficients under control and drought respectively. The size and color intensity of squares indicates the magnitude of correlation based on the provided scale. Crossmark ( × ) indicates a non-significant correlation at P ≤ 0.05.
3.2.2. Association among the examined traits
The relationship between all the examined traits were assessed to highlight the nature and the magnitude of association among the traits. Under control condition, most of the traits showed a non-significant positive correlation; however, grain yield (ton ha−1) and biological yield (ton ha−1) showed significant positive correlation with plant height (cm), spike number (m−2) and spike length (cm) (Fig. 3). On the contrary, most of the tested morpho-physiological traits showed strong and significant positive correlation among them under drought stress. Under water deficit condition, the grain yield (ton ha−1) and biological yield (ton ha−1) was positively correlated to all the tested traits particularly with the physiological traits and yield components. Among the physiological traits, SPAD value, leaf relative water content (%), canopy temperature depression (°C) and proline content (μmol g−1 fresh weight) showed the strongest positive correlation with the other tested traits in most cases. Moreover, plant height (cm), spike number (m−2), spike weight, grains per spike, and 1000-grain weight (g) had stronger positive correlation with grain yield (ton ha−1) and biological yield (ton ha−1). Such responses indicated a high relatedness of the tested traits under drought compared to control.
3.2.3. Morpho-physiological trait-based hierarchical clustering of genotypes
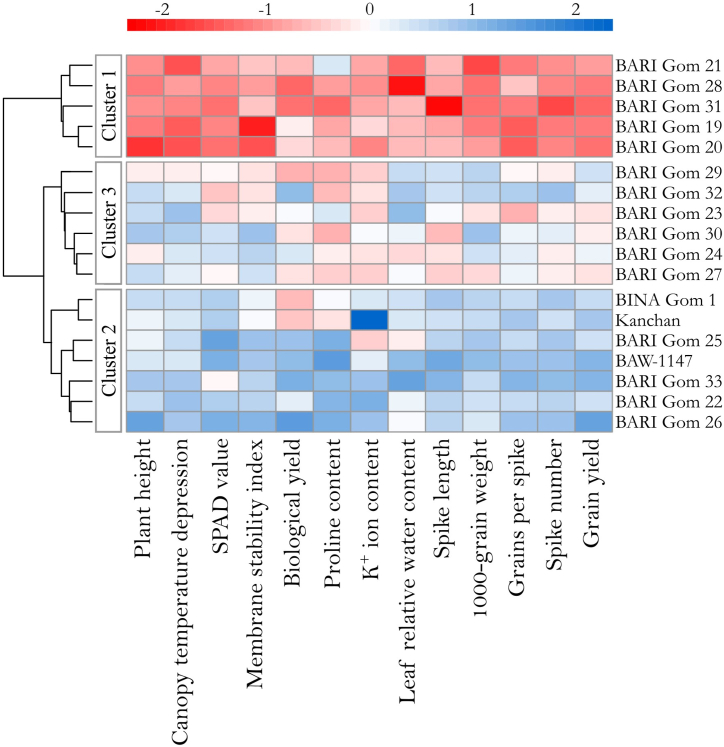
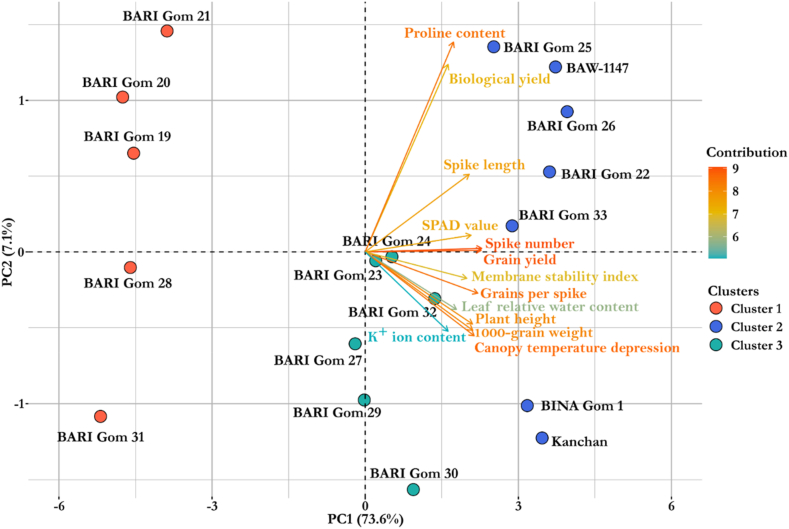
Based on the performance of genotypes a hierarchical cluster analysis was conducted using Euclidean distant matrix and the magnitude of tolerance for the tested traits were presented by a tri-colored heatmap along with the clusters (Fig. 4). Eighteen genotypes were grouped into three distinct clusters where genotypes belonging to same cluster shared close similarity among themselves. Thus, genotypes belonging to different clusters were distinct from each other. Cluster 1 assembled 5 genotypes (BARI Gom 19, BARI Gom 20, BARI Gom 21, BARI Gom 28, BARI Gom 31), cluster 2 grouped 7 genotypes (BARI Gom 22, BARI Gom 25, BARI Gom 26; BARI Gom 33, BAW-1147, BINA Gom 1, and Kanchan), and cluster 3 gathered 6 genotypes (BARI Gom 23, BARI Gom 24, BARI Gom 27, BARI Gom 29, BARI Gom 30, BARI Gom 32). The heatmap also revealed that, among the genotype clusters, cluster 2 genotypes showed the highest relative tolerance and the cluster 1 genotypes showed the lowest relative tolerance regarding the individual performance of the morpho-physiological traits (Fig. 4).
Fig. 4.
Hierarchical clustering and heatmap analysis of wheat genotypes.
3.2.4. Principal component analysis
Principal component analysis (PCA) revealed a total of 13 principal components (PCs) (Fig. 5, Table 5S); however, only one PC was found significant as they exhibited eigenvalues >1. The PC1 and PC2 could jointly explain >80% of the genotypic variability among which PC1 exhibited 73.6% of the variability, largely contributed by all the studied traits (Table 5S). The PC2 explained only 7.1% of the variability mostly explained by biological yield (ton ha−1) and proline content (μmol g−1 fresh weight) (Table 5S). The rest three PCs (PC3–PC13) jointly explained <20% of the total variability. Importantly, the PCA-biplot also classified wheat genotypes into 3 distinct groups similar to those from the hierarchical cluster analysis (Fig. 4, Fig. 5). The genotypes of cluster 2 were positioned towards the vectors of PC1 implying that the superiority of those genotypes under drought was positively contributed by the traits belonging to PC1 (Fig. 5).
Fig. 5.
PCA-Biplot of wheat genotypes and studied traits constructed from relative trait values. Based on their dissimilarity, genotypes were distributed in distinct ordinates.
3.2.5. Cluster-based changes in the tested traits under drought
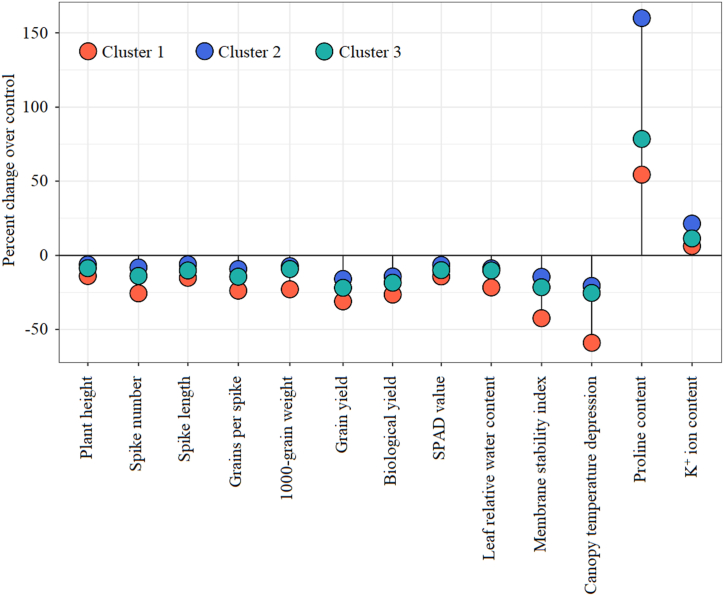
The magnitude of change in trait performances under drought in respect to control was quantified for each cluster groups as percent change over control and presented in stacked dot plot (Fig. 6). Cluster 2 and cluster 1 respectively showed the least and highest reduction in plant height (cm), spike number (m−2), spike length (cm), grains per spike, 1000-grain weight (g), grain yield (ton ha−1), biological yield (ton ha−1), SPAD-value, leaf relative water content (%), membrane stability index (%), and canopy temperature depression (°C). On the other hand, cluster 2 and cluster 1 respectively showed the highest and lowest increase of proline content (μmol g−1 fresh weight) and K+ ion content (mg g−1 fresh weight). Cluster 3 showed intermediate change in trait performances in between cluster 2 and cluster 1; however, performance of cluster 3 remained comparatively closer to cluster 2 than cluster 1 (Fig. 6).
Fig. 6.
Changes in the studied traits of different cluster groups under drought stress. Values are expressed as percent change due to drought over control.
3.3. Co-linearity between morpho-physiology and microsatellite-based clusters
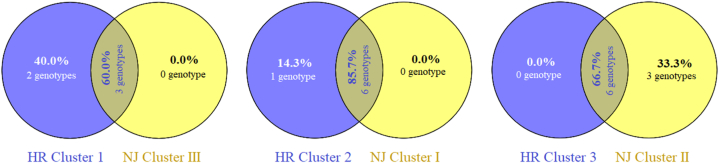
Similarity between morpho-physiological trait-based hierarchical (HR) clusters and UPGMA-based neighbor-jointing (NJ) clusters extracted from microsatellite data were compared using co-linearity of the model-based analysis and presented as Venn diagrams (Fig. 7). Both of the clusters had three distinct genotype groups among which HR cluster 1: NJ cluster III (60% co-linearity), HR cluster 2: NJ cluster I (85.7% co-linearity), and HR cluster 3: NJ cluster I (66.7% co-linearity) showed well alignment based on maximum possible similarity. Overall, both clusters shared 71% co-linearity between them with 15 genotype matches.
Fig. 7.
Venn diagram showing co-linearity between morpho-physiological trait-based hierarchical (HR) and microsatellite marker data-based neighbor-joining (NJ) clusters.
4. Discussion
Knowledge of morpho-physiological attributes contributing towards yield as well as physio-biochemical and genetic modes of crop adaptability under stressful environments are fundamental for proper execution of crop improvement programs against complex abiotic stresses like drought that could result from the interactive effects of multiple edaphic and environmental factors [10]. Drought itself is a complex trait that could result from the interactive effects of multiple edaphic and environmental factors [54]. Moreover, the heritability of drought tolerance is very low in wheat due to comparatively large genome size (approximately 17 giga base-pairs) and polygenic control [13,15]. Thus, evaluation of wheat genotypes based on either morpho-physiological traits or the genetic diversity might not translate towards effective conclusion. Previous studies extensively explored the hexaploid bread wheat gene-pool utilizing either agronomic/phenotypic or physiological traits and considerable studies have already been conducted to evaluate the genetic diversity among the wheat germplasms using various molecular markers [11,17,24,36,55,56]. However, the combined approach utilizing morpho-physiological trait-based evaluation and microsatellite-based evaluation of genetic diversity analysis of wheat is highly limited in Bangladesh, although a big number of native wheat germplasms are available here and she has yet to achieve much contribution from genome wide studies and MAS [7,37]. Furthermore, combined phenotyping and genotyping approach could potentially offer more precise genomic characterization, selection, marker and candidate gene discovery, and quanititative trait loci (QTL) mapping [[57], [58], [59]].
In our present study, we employed highly polymorphic 25 microsatellite markers covering the whole genome of hexaploid wheat to assess the genetic diversity (Table 1 and 1S). We obtained an average of 9.76 alleles per locus, 0.77 PIC-value, and 0.79 gene diversity (Table 2) which could be comparable to the reports of previous studies on wheat using microsatellite markers [24,32]. Allelic diversity in a population results from the existing diversity of genetic compositions; whereas, PIC-value is a reliable indicator for plant genetic diversity [24]. A PIC-value >0.5 indicates higher diversity and <0.25 denotes a lower diversity [60,61]. Our results are thus not only consistent with the existing literature but also indicates a high diversity among the tested genotypes [17,24,32,55,62]. We have also noticed, 15 of the 25 microsatellite markers amplified ≥10 alleles and scored PIC-value ≥0.8 (Table 2). This implied that, the microsatellites used in this study was robust, highly informative and effective to aid in parent selection for molecular plant breeding against drought [17,24,32]. While comparing among the genomes we observed that, D genome had the highest mean allele per locus, PIC value, and gene diversity followed by B and A genome (Table 2) implying the presence of higher diversity for drought tolerance in D genome compared to the other two. This result is in partial agreement with some of the recent studies where B genome was reported to harbor considerable genetic diversity for drought tolerance [17,63,64].
In our current study we employed UPGMA-based neighbor-jointing cluster analysis that is a highly useful technique in screening studies for grouping target genotypes against one or more factors [41,65]. In total, three cluster groups were revealed among which cluster I comprised of 7 genotypes including the tolerant check Sonoita F-81 implying that the genotypes BARI gom 22, BARI gom 26, BARI gom 33, BINA gom 1, BAW-1147 and Kanchan shares close genetic similarity to a naturally tolerant genotype and hence the cluster one can be referred as tolerant cluster (Fig. 2). The genotype Sonoita F-81 was an ideal choice for comparison as drought tolerant genotypes developed by CIMMYT possessed very high genetic diversity mostly due to the integration of genetic materials from diverse sources like synthetic varieties and landraces [62,66,67]. The genotype BARI gom 33 followed by BAW-1147 was the closest to the tolerant check Sonoita F-81 which made them genetically superior compared to the other genotypes of cluster I. Contrariwise, the genotypes of cluster III (BARI gom 19, BARI gom 28, BARI gom 31) shared the least genetic similarity with Sonoita-F81 and hence the cluster III could be referred as sensitive cluster (Fig. 2). Performance observed for these genotypes under field trial of the present study also supported the genotypic grouping based on genetic similarity (Fig. 5, Table 3, Table 4).
Productivity of wheat is comparatively low under arid and semi-arid regions due to both water deficit and degree of susceptibility of cultivated genotypes [68,69]. Water scarcity leads to physiological and metabolic impairments like membrane instability, high free radical generation, hormonal imbalance, chlorophyll damage, reduced photosynthesis thereby resulting in low productivity [3,18,19,70]. Tolerant genotypes on the other hand could mitigate the water stress to suffer as less injury as possible through variable means involving physiological and biochemical adjustments thereby ensuring better yield and biomass [71]. Therefore, crop improvement against drought for wheat can be achieved via genotype selection targeting enhanced traits related to morphology, yield, yield components, physiology and biochemical attributes [22,62,66,72,73]. The traits we have considered for this current study such as plant height, number of productive tillers (spikes), spike length, number of grains per spike, 1000-grain weight, above-ground biomass, leaf greenness, relative water content, bio-membrane stability, canopy temperature, free proline concentration, and K+ ion accumulation had been previously utilized in wheat crop improvement programs against numerous abiotic stress tolerance [10,22,72,[74], [75], [76], [77], [78], [79]]. Furthermore, our results agree with these previous findings via exhibiting a strong association among these traits as well as with grain yield and plant biomass (Fig. 3). We employed Pearson's correlation test to draw relationship among the tested morpho-physiological test and found significant positive correlation mostly under drought (Fig. 3). According to Würschum et al. [80] and Zhang et al. [78] genotypes with plant height around 60–80 cm was reported to well correlate with better biomass which was consistent with our results although we observed comparatively higher biomass with increased plant height (Table 3, Fig. 4). Similar to the report of Islam et al. [81], plant height was also found highly positively correlated with SPAD value (leaf greenness) that could explain the significant positive correlation of plant height with tested yield components and grain yield (Fig. 4). Similar to Beche et al. [79] and Gummadov et al. [68] we have also observed that both the grain yield and biological yield had strong positive correlation with yield components and physiological traits (Fig. 4). Parallel to our findings, previous investigations also reported spike number, spike length, grains per spike, 1000-grain weight as important agronomic traits positively influencing biomass and grain yield potential in wheat [22,76,77,[82], [83], [84], [85], [86], [87], [88], [89], [90]]. However, we did not record any significant correlation between grains per spike, and 1000-grain weight with spike number (Fig. 3). According to Bustos et al. [91], enhanced yield and biomass resulted from higher photosynthesis and radiation-use efficiency. Water and chlorophyll are the two essential prerequisite for photosynthesis that results in yield and biomass production [92]. In the present study, both the grain and biological yield was strongly and positively correlated with SPAD value, leaf relative water content, and membrane stability index (Fig. 4). SPAD value is an indicator of chlorophyll content and leaf relative water content acts as the medium aiding normal metabolic reactions within the cellular system [93,94]. Drought in turn reduced relative water content in leaves leading to metabolic disequilibrium and over-production of free radicals which was detrimental for bio-membranes, photosynthetic pigments, cell organelles, nucleic acids, proteins and other important cellular biomolecules [95,96]. Damage to chlorophyll ultrastructure, photosynthetic inhibition via impairing ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activity was reported as one of the most devastating consequences of drought mediated free radical overproduction [[97], [98], [99]]. Water deficit also reduced photosynthesis via stomatal closure as well as photosynthate translocation impairing yield and biomass production substantially [70,100,101]. These reports helped explaining the significant positive correlation among the traits themselves as well as with both grain and biological yield under drought. Canopy temperature depression measured from difference between air temperature and canopy temperature is an integrative trait that indicated water status of plant under drought [102,103]. Previously canopy temperature depression was reported to show strong correlation with traits related to plant water status and yield components [104,105]. Our results also support the previous reports except the fact that we did not record any significant correlation between canopy temperature depression and grain yield; however, we reported a very strong positive correlation between canopy temperature depression and leaf relative water content of wheat (Fig. 4). Proline content and K+ ion content is two of the most desired traits within a tolerant genotype. Proline is a cyclic amino acid that has osmoregulation, free radicle scavenging, protein and membrane structure stabilizing, and signaling roles in plants [[106], [107], [108], [109]]. Potassium ion is involved in ion homeostasis, protein synthesis, electrolytes balance, activation of various enzymes for a range of metabolic reactions, and free radical scavenging apart from its osmoregulatory roles [110,111]. Similar to our results, recent reports have shown that drought stress stimulates proline synthesis and K+ ion accumulation in wheat which is directly dependent on the degree of genotypic tolerance and positively correlates with plant growth, yield and biomass production [[112], [113], [114], [115]]. Apart from the traits association we noticed that, the overall performance among the genotypes on tested morpho-physiological traits varied greatly under drought compared to control (Tables 3S and 4S). However, genotypes showing better performance in physiological traits and yield components exhibited comparatively better grain yield and biological yield (Table 3, Table 4). Our results are also in parallel with reports from previous studies which could be due to the genetic superiority of the genotypes resulting in enhanced mitigation effects via morpho-physiological attributes [20,36,[116], [117], [118]].
We used a hierarchical cluster analysis to classify the genotypes in different tolerance groups based on their overall performance in the tested morpho-physiological traits under drought compared to control (relative tolerance). We found three distinct clusters among which the cluster 2 and cluster 1 genotypes showed the highest and the lowest relative tolerance, respectively. Hence, the genotype clusters could be ranked as cluster 2 > cluster 3 > cluster 1 and could be referred as tolerant, moderately tolerant, and susceptible clusters, respectively (Fig. 5). The magnitude of performance change in the three genotype clusters for all the tested morpho-physiological traits was also examined, where cluster 2 and cluster 1 genotypes, respectively, showed the lowest and highest impairments that strongly supported the hierarchy of the clusters (Fig. 6). Similar results were also reported by Mohi-Ud-Din et al. [16] and Grzesiak et al. [119] while examining the drought tolerance potential of diverse wheat genotypes. Principal component analysis is a strong statistical tool for representing highly correlated data that effectively draws constructive feedback by reducing the dimensionality of variables [120]. In a PCA biplot, cosine of the angles between vectors represents their correlation, where an angle <90°, >90°, and = 90°, respectively represents positive, negative and no correlation. Similar to our present study, PCA biplot had been previously employed by other researchers for effective screening of wheat genotypes [7,121]. Similar to the HR cluster analysis, PCA biplot also grouped the tested genotypes into three distinct clusters (Fig. 7). Furthermore, the direction and cosine angles between vectors also strongly represented the correlations among the tested morpho-physiological parameters (Fig. 4, Fig. 7); although a previous report stated that, the angles between two vectors of a PCA biplot might not match precisely with their correlation [122]. Among the two different genotypic clustering (morpho-physiological trait-based HR clusters and UPGMA-based NJ clusters) similarity index was assessed using co-linearity of the model-based analysis. Both the clusters had 71% co-linearity with 15 exact genotype matches. The tolerant genotype clusters from both analysis (HR cluster 2 and NJ cluster I) had 6 genotypes in common and the susceptible cluster from both analysis (HR cluster 1 and NJ cluster III) had 3 genotypes in common (Fig. 7). High similarity between both clusters greatly enhanced the experimental reliability and reproducibility [7,16,123]. Performance variations on the tested morpho-physiological traits were evident among the genotypes; however, genotypes belonging to UPGMA-based NJ cluster I showed comparatively better performance than the genotypes from the other two clusters (Table 3, Table 4). Similarly, the genotypes belonging to HR cluster 2 were found combatively superior in genetic diversity and had better genetic similarity with the drought tolerant check Sonoita F-81 (Fig. 2) explaining and supporting the thumb rule, genetically superior genotypes tend to perform significantly better than inferior genotypes under stressful environments [124,125].
The research progress on wheat drought tolerance worldwide has witnessed significant strides in recent years, owing to the growing concerns about climate change-induced water scarcity and its impact on global food security. Researchers have conducted comprehensive studies to decipher the genetic basis of drought tolerance in wheat [[126], [127], [128], [129], [130], [131]]. Numerous genes and molecular pathways have been identified across diverse wheat germplasms (Table 6S) revealing a complex interplay of genetic elements in conferring resilience to drought. Among hundreds of identified drought tolerance contributor candidates only a notable few such as TaDREB2, TaSnRK2.4, and TaMYB44 are at the forefront [[132], [133], [134]]. Advancement of sequencing technologies and functional genomics could accelerate the process; however, understanding the genetic basis of wheat germplasms from diverse geographic regions would fuel the ongoing efforts of breeding drought-tolerant wheat cultivars [[135], [136], [137], [138]] which is pivotal for ensuring food security in a changing climate.
5. Conclusion
Drought stress induced water scarcity negatively influenced the tested morpho-physiologial traits as well as reduced wheat productivity for all tested wheat genotypes although the magnitude of drought tolerance among them were considerably diverse which was evident via microsatellite marker-based and morpho-physiological trait-based assessments. Although drought stress resulted in significant performance drop in all the traits, the genotypes BARI Gom 22, BARI Gom 26, BARI gom 33 and BAW-1147 sharing close genetic similarities with the tolerant check Sonoita F-81 showed distinctively low impairments compared to the other tested genotypes. Among the used 25 microsatellite markers, most were robust for wheat genetic evaluation and effective for screening genetically diverse wheat germplasm resources. Tested morpho-physiological traits were strongly and positively corelated under drought, hence are important target traits for drought tolerance studies. Both microsatellite marker-based and morpho-physiological trait-based approaches were individually effective; however, combining both approaches would aid in more efficient screening and coming up to more reliable conclusion. The reported findings should improve the present understanding on drought tolerance responses of wheat and should provide a basis for future genotype selection for enhancing drought tolerance in wheat. Furthermore, the selected tolerant genotypes particularly BARI Gom 22, BARI Gom 26, BARI gom 33 and BAW-1147 showed potentials for cultivation in drought affected lands for better productivity.
Author contribution statement
Sheikh Faruk AHMED: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Jalal Uddin AHMED: Contributed reagents, materials, analysis tools or data; Wrote the paper. Mehfuz HASAN: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data. Mohammed Mohi-Ud-Din: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21629.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Liu Q., Liu Z., Li W., Song X. Comparative transcriptome analysis indicates conversion of stamens into pistil-like structures in male sterile wheat (Triticum aestivum L.) with Aegilops crassa cytoplasm. BMC Genom. 2020;21:1–17. doi: 10.1186/s12864-020-6450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iqbal M.S., Singh A.K., Ansari M.I. In: New Frontiers in Stress Management for Durable Agriculture. Rakshit A., Singh H., Singh A., Singh U., Fraceto L., editors. Springer; Singapore: 2020. Effect of drought stress on crop production; pp. 35–47. [DOI] [Google Scholar]
- 3.Abdolshahi R., Nazari M., Safarian A., Sadathossini T.S., Salarpour M., Amiri H. Integrated selection criteria for drought tolerance in wheat (Triticum aestivum L.) breeding programs using discriminant analysis. Field Crops Res. 2015;174:20–29. doi: 10.1016/j.fcr.2015.01.009. [DOI] [Google Scholar]
- 4.Cook B.I., Ault T.R., Smerdon J.E. Unprecedented 21st century drought risk in the American southwest and central plains. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1400082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwalm C.R., Anderegg W.R., Michalak A.M., Fisher J.B., Biondi F., Koch G., Litvak M., Ogle K., Shaw J.D., Wolf A., Huntzinger D.N. Global patterns of drought recovery. Nature. 2017;548:202–205. doi: 10.1038/nature23021. [DOI] [PubMed] [Google Scholar]
- 6.El-Hendawy S., Al-Suhaibani N., Salem A.E.A., Rehman S.U., Schmidhalter U.R.S. Spectral reflectance indices as a rapid and nondestructive phenotyping tool for estimating different morphophysiological traits of contrasting spring wheat germplasms under arid conditions. Turk. J. Agric. For. 2015;39:572–587. doi: 10.3906/tar-1406-164. [DOI] [Google Scholar]
- 7.Mohi-Ud-Din M., Hossain M.A., Rohman M.M., Uddin M.N., Haque M.S., Dessoky E.S., Alqurashi M., Aloufi S. Assessment of genetic diversity of bread wheat genotypes for drought tolerance using canopy reflectance-based phenotyping and SSR marker-based genotyping. Sustainability. 2022;14:9818. doi: 10.3390/su14169818. [DOI] [Google Scholar]
- 8.Mir R.R., Zaman-Allah M., Sreenivasulu N., Trethowan R., Varshney R.K. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor. Appl. Genet. 2012;125:625–645. doi: 10.1007/s00122-012-1904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuberosa R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012;3:347. doi: 10.3389/fphys.2012.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mwadzingeni L., Shimelis H., Dube E., Laing M.D., Tsilo T.J. Breeding wheat for drought tolerance: progress and technologies. J. Integr. Agric. 2016;15:935–943. doi: 10.1016/S2095-3119(15)61102-9. [DOI] [Google Scholar]
- 11.Dodig D., Zorić M., Kobiljski B., Šurlan-Momirović G., Quarrie S.A. Assessing drought tolerance and regional patterns of genetic diversity among spring and winter bread wheat using simple sequence repeats and phenotypic data. Crop Pasture Sci. 2010;61:812–824. doi: 10.1071/CP10001. [DOI] [Google Scholar]
- 12.Dvorak J., Luo M.C., Akhunov E.D. NI Vavilov's theory of centres of diversity in the light of current understanding of wheat diversity, domestication and evolution. Czech J. Genet. Plant Breed. 2011;47:S20–S27. [Google Scholar]
- 13.Khakwani A.A., Dennett M.D., Munir M., Abid M. Growth and yield response of wheat varieties to water stress at booting and anthesis stages of development. Pakistan J. Bot. 2012;44:879–886. [Google Scholar]
- 14.Walkowiak S., Gao L., Monat C., Haberer G., Kassa M.T., Brinton J., Ramirez-Gonzalez R.H., Kolodziej M.C., Delorean E., Thambugala D., Klymiuk V. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020;588:277–283. doi: 10.1038/s41586-020-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkman P.J., Lai K., Lorenc M.T., Edwards D. Next‐generation sequencing applications for wheat crop improvement. Am. J. Bot. 2012;99:365–371. doi: 10.3732/ajb.1100309. [DOI] [PubMed] [Google Scholar]
- 16.Mohi-Ud-Din M., Hossain M.A., Rohman M.M., Uddin M.N., Haque M.S., Ahmed J.U., Hossain A., Hassan M.M., Mostofa M.G. Multivariate analysis of morpho-physiological traits reveals differential drought tolerance potential of bread wheat genotypes at the seedling stage. Plants. 2021;10:879. doi: 10.3390/plants10050879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Rawy M.A., Hassan M.I. Assessment of genetic diversity in durum and bread wheat genotypes based on drought tolerance and SSR markers. Plant Breed. Biotechnol. 2021;9:89–103. doi: 10.9787/PBB.2021.9.2.89. [DOI] [Google Scholar]
- 18.Chaudhry S., Sidhu G.P.S. Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep. 2022;41:1–31. doi: 10.1007/s00299-021-02759-5. [DOI] [PubMed] [Google Scholar]
- 19.Salehi-Lisar S.Y., Bakhshayeshan-Agdam H. In: Hossain M., Wani S., Bhattacharjee S., Burritt D., Tran L.S., editors. vol. 1. Springer; Cham: 2016. Drought stress in plants: causes, consequences, and tolerance; pp. 1–16. (Drought Stress Tolerance in Plants). [DOI] [Google Scholar]
- 20.Abdolshahi R., Safarian A., Nazari M., Pourseyedi S., Mohamadi-Nejad G. Screening drought-tolerant genotypes in bread wheat (Triticum aestivum L.) using different multivariate methods. Arch. Agron Soil Sci. 2013;59:685–704. doi: 10.1080/03650340.2012.667080. [DOI] [Google Scholar]
- 21.Ahmed H.G.M.D., Zeng Y., Shah A.N., Yar M.M., Ullah A., Ali M. Conferring of drought tolerance in wheat (Triticum aestivum L.) genotypes using seedling indices. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.961049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes M.S., Reynolds M.P., Jalal-Kamali M.R., Moussa M., Feltaous Y., Tahir I.S.A., Barma N., Vargas M., Mannes Y., Baum M. The yield correlations of selectable physiological traits in a population of advanced spring wheat lines grown in warm and drought environments. Field Crops Res. 2012;128:129–136. doi: 10.1016/j.fcr.2011.12.017. [DOI] [Google Scholar]
- 23.Gizaw S.A., Garland-Campbell K., Carter A.H. Evaluation of agronomic traits and spectral reflectance in Pacific Northwest winter wheat under rain-fed and irrigated conditions. Field Crops Res. 2016;196:168–179. doi: 10.1016/j.fcr.2016.06.018. [DOI] [Google Scholar]
- 24.Belete Y., Shimelis H., Laing M., Mathew I. Genetic diversity and population structure of bread wheat genotypes determined via phenotypic and SSR marker analyses under drought-stress conditions. J. Crop Improv. 2021;35:303–325. doi: 10.1080/15427528.2020.1818342. [DOI] [Google Scholar]
- 25.Mulualem T., Mekbib F., Shimelis H., Gebre E., Amelework B. Genetic diversity of yam (Dioscorea spp.) landrace collections from Ethiopia using simple sequence repeat markers. Aust. J. Crop. Sci. 2018;12:1222–1230. doi:10.3316/informit.991341360391074 [Google Scholar]
- 26.Salem K.F., Röder M.S., Börner A. Assessing genetic diversity of Egyptian hexaploid wheat (Triticum aestivum L.) using microsatellite markers. Genet. Resour. Crop Evol. 2015;62:377–385. doi: 10.1007/s10722-014-0159-5. [DOI] [Google Scholar]
- 27.Azizi M.M.F., Lau H.Y., Abu-Bakar N. Integration of advanced technologies for plant variety and cultivar identification. J. Biosci. 2021;46:1–20. doi: 10.1007/s12038-021-00214-x. [DOI] [PubMed] [Google Scholar]
- 28.Adhikari S., Saha S., Biswas A., Rana T.S., Bandyopadhyay T.K., Ghosh P. Application of molecular markers in plant genome analysis: a review. Nucleus. 2017;60:283–297. doi: 10.1007/s13237-017-0214-7. [DOI] [Google Scholar]
- 29.Ghazy M.I., Salem K.F., Sallam A. Utilization of genetic diversity and marker-trait to improve drought tolerance in rice (Oryza sativa L.) Mol. Biol. Rep. 2021;48:157–170. doi: 10.1007/s11033-020-06029-7. [DOI] [PubMed] [Google Scholar]
- 30.Jespersen D., Ma X., Bonos S.A., Belanger F.C., Raymer P., Huang B. Association of SSR and candidate gene markers with genetic variations in summer heat and drought performance for creeping bentgrass. Crop Sci. 2018;58:2644–2656. doi: 10.2135/cropsci2018.05.0299. [DOI] [Google Scholar]
- 31.Kage U., Kumar A., Dhokane D., Karre S., Kushalappa A.C. Functional molecular markers for crop improvement. Crit. Rev. Biotechnol. 2016;36:917–930. doi: 10.3109/07388551.2015.1062743. [DOI] [PubMed] [Google Scholar]
- 32.Ateş Sönmezoğlu Ö., Terzi B. Characterization of some bread wheat genotypes using molecular markers for drought tolerance. Physiol. Mol. Biol. Plants. 2018;24:159–166. doi: 10.1007/s12298-017-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma H., Borah J.L., Sarma R.N. Variability assessment for root and drought tolerance traits and genetic diversity analysis of rice germplasm using SSR markers. Sci. Rep. 2019;9:1–19. doi: 10.1038/s41598-019-52884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vieira M.L.C., Santini L., Diniz A.L., Munhoz C.D.F. Microsatellite markers: what they mean and why they are so useful. Genet. Mol. Biol. 2016;39:312–328. doi: 10.1590/1678-4685-GMB-2016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semahegn Y., Shimelis H., Laing M., Mathew I. Evaluation of bread wheat (Triticum aestivum L.) genotypes for yield and related traits under drought stress conditions. Acta Agric. Scand. - B Soil Plant Sci. 2020;70:474–484. doi: 10.1080/09064710.2020.1767802. [DOI] [Google Scholar]
- 36.Haque M.S., Saha N.R., Islam M.T., Islam M.M., Kwon S.J., Roy S.K., Woo S.H. Screening for drought tolerance in wheat genotypes by morphological and SSR markers. J. Crop Sci. Biotechnol. 2021;24:27–39. doi: 10.1007/s12892-020-00036-7. [DOI] [Google Scholar]
- 37.Amiruzzaman M., Sarker M.A.Z., Ahmed S., Hossain M.M., Mia M.A., Hakim M.A., Bazzaz M.M., Hossain A., Shah M.M.R., Mondal M.S.N., Kabir M.R., Alam M.A., Roy K.K. 2022. Annual Research Report 2020-21. Bangladesh Wheat and Maize Research Institute (BWMRI). Dinajpur, Bangladesh; pp. 39–60. [Google Scholar]
- 38.Yu G., Hatta A., Periyannan S., Lagudah E., Wulff B.B. In: Wheat Rust Diseases: Methods and Protocols. Periyannan S., editor. Springer, Humana Press; New York: 2017. Isolation of wheat genomic DNA for gene mapping and cloning; pp. 207–213. [DOI] [PubMed] [Google Scholar]
- 39.Huda M.N., Hasan M., Abdullah H.M., Sarker U. Spatial distribution and genetic diversity of wild date palm (Phoenix sylvestris) growing in coastal Bangladesh. Tree Genet. Genomes. 2019;15:3. doi: 10.1007/s11295-018-1310-9. [DOI] [Google Scholar]
- 40.Hussain F., Bronson K.F., Yadvinder S., Singh B., Peng S. Use of chlorophyll meter sufficiency indices for nitrogen management of irrigated rice in Asia. Agron. J. 2000;92:875–879. doi: 10.2134/agronj2000.925875x. [DOI] [Google Scholar]
- 41.Ahmed S.F., Ullah H., Aung M.Z., Tisarum R., Cha-Um S., Datta A. Iron toxicity tolerance of rice genotypes in relation to growth, yield and physiochemical characters. Rice Sci. 2023;30:321–334. doi: 10.1016/j.rsci.2023.02.002. [DOI] [Google Scholar]
- 42.Das D., Ullah H., Tisarum R., Cha-um S., Datta A. Morpho-physiological responses of tropical rice to potassium and silicon fertilization under water-deficit stress. J. Soil Sci. Plant Nutr. 2023;23:220–237. doi: 10.1007/s42729-021-00712-9. [DOI] [Google Scholar]
- 43.Bates L.S., Waldren R.P., Teare I.D. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- 44.Ahmed S.F., Biswas A., Ullah H., Himanshu S.K., Tisarum R., Cha-um S., Datta A. Interactive effects of silicon and potassium on photosynthesis and physio-biochemical traits of rice (Oryza sativa L.) leaf mesophyll under ferrous iron toxicity. Plant Stress. 2023;10 doi: 10.1016/j.stress.2023.100203. [DOI] [Google Scholar]
- 45.Balota M., Payne W.A., Evett S.R., Lazar M.D. Canopy temperature depression sampling to assess grain yield and genotypic differentiation in winter wheat. Crop Sci. 2007;47:1518–1529. doi: 10.2135/cropsci2006.06.0383. [DOI] [Google Scholar]
- 46.Apon T.A., Ahmed S.F., Bony Z.F., Chowdhury M.R., Asha J.F., Biswas A. Sett priming with salicylic acid improves salinity tolerance of sugarcane (Saccharum officinarum L.) during early stages of crop development. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu K., Muse S.V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 48.Perrier X., Jacquemoud-Collet J.P. Darwin software. 2010. http://darwin.cirad.fr/
- 49.Wickham H. In: ggplot2. Wickham H., editor. Springer; Cham: 2016. Data analysis; pp. 189–201. [DOI] [Google Scholar]
- 50.Julkowska M.M., Saade S., Agarwal G., Gao G., Pailles Y., Morton M., Awlia M., Tester M. MVApp—multivariate analysis application for streamlined data analysis and curation. Plant Physiol. 2019;180:1261–1276. doi: 10.1104/pp.19.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolde R., Kolde M.R. vol. 1. 2022. p. 790.https://cran.ms.unimelb.edu.au/web/packages/pheatmap/pheatmap.pdf (Package ‘pheatmap’. R Package). Avaiable online: [Google Scholar]
- 52.Lê S., Josse J., Husson F. FactoMineR: an R package for multivariate analysis. J. Stat. Software. 2008;25:1–18. doi: 10.18637/jss.v025.i01. [DOI] [Google Scholar]
- 53.Oliveros J.C. VENNY- an interactive tool for comparing lists with Venn diagrams. 2007. http://bioinfogp.cnb.csic.es/tools/venny/index.html
- 54.Prasad P.V.V., Pisipati S.R., Momčilović I., Ristic Z. Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF‐Tu expression in spring wheat. J. Agron. Crop Sci. 2011;197:430–441. doi: 10.1111/j.1439-037X.2011.00477.x. [DOI] [Google Scholar]
- 55.Slim A., Piarulli L., Chennaoui Kourda H., Rouaissi M., Robbana C., Chaabane R., Pignone D., Montemurro C., Mangini G. Genetic structure analysis of a collection of Tunisian durum wheat germplasm. Int. J. Mol. Sci. 2019;20:3362. doi: 10.3390/ijms20133362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X., Tan B., Liu H., Zhu W., Xu L., Wang Y., Fan X., Sha L., Zhang H., Zeng J., Wu D. Genetic diversity and population structure of Asian and European common wheat accessions based on genotyping-by-sequencing. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.580782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacLeod I.M., Bowman P.J., Vander Jagt C.J., Haile-Mariam M., Kemper K.E., Chamberlain A.J., Schrooten C., Hayes B.J., Goddard M.E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016;17:1–21. doi: 10.1186/s12864-016-2443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Y., Wang J., Crouch J.H., Xu Y. Efficiency of selective genotyping for genetic analysis of complex traits and potential applications in crop improvement. Mol. Breed. 2010;26:493–511. doi: 10.1007/s11032-010-9390-8. [DOI] [Google Scholar]
- 59.Alqudah A.M., Sallam A., Baenziger P.S., Börner A. GWAS: fast-forwarding gene identification and characterization in temperate cereals: lessons from barley–a review. J. Adv. Res. 2020;22:119–135. doi: 10.1016/j.jare.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagy S., Poczai P., Cernák I., Gorji A.M., Hegedűs G., Taller J. PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 2012;50:670–672. doi: 10.1007/s10528-012-9509-1. [DOI] [PubMed] [Google Scholar]
- 61.Ramadugu C., Keremane M.L., Hu X., Karp D., Federici C.T., Kahn T., Roose M.L., Lee R.F. Genetic analysis of citron (Citrus medica L.) using simple sequence repeats and single nucleotide polymorphisms. Sci. Hortic. 2015;195:124–137. doi: 10.1016/j.scienta.2015.09.004. [DOI] [Google Scholar]
- 62.Mkhabela S.S., Shimelis H., Mashilo J. Genetic differentiation of selected drought and heat tolerant wheat genotypes using simple sequence repeat markers and agronomic traits. S. Afr. J. Plant Soil. 2020;37:211–219. doi: 10.1080/02571862.2020.1718787. [DOI] [Google Scholar]
- 63.El-Rawy M.A. Assessment of genetic diversity for some Egyptian wheat varieties based on morphological characters and SSR markers. Sci. J. Agric. Sci. 2020;2:144–160. doi: 10.21608/SJAS.2020.37530.1033. [DOI] [Google Scholar]
- 64.Jaiswal S., Sheoran S., Arora V., Angadi U.B., Iquebal M.A., Raghav N., Aneja B., Kumar D., Singh R., Sharma P., Singh G.P. Putative microsatellite DNA marker-based wheat genomic resource for varietal improvement and management. Front. Plant Sci. 2017;8:2009. doi: 10.3389/fpls.2017.02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menkir A., Olowolafe M.O., Ingelbrecht I., Fawole I., Badu-Apraku B., Vroh B.I. Assessment of testcross performance and genetic diversity of yellow endosperm maize lines derived from adapted× exotic backcrosses. Theor. Appl. Genet. 2006;113:90–99. doi: 10.1007/s00122-006-0275-5. [DOI] [PubMed] [Google Scholar]
- 66.Crespo-Herrera L.A., Crossa J., Huerta-Espino J., Vargas M., Mondal S., Velu G., Payne T.S., Braun H., Singh R.P. Genetic gains for grain yield in CIMMYT's semi‐arid wheat yield trials grown in suboptimal environments. Crop Sci. 2018;58:1890–1898. doi: 10.2135/cropsci2018.01.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gordon E., Kaviani M., Kagale S., Payne T., Navabi A. Genetic diversity and population structure of synthetic hexaploid-derived wheat (Triticum aestivum L.) accessions. Genet. Resour. Crop Evol. 2019;66:335–348. doi: 10.1007/s10722-018-0711-9. [DOI] [Google Scholar]
- 68.Gummadov N., Keser M., Akin B., Cakmak M., Mert Z., Taner S., Ozturk I., Topal A., Yazar S., Morgounov A. Genetic gains in wheat in Turkey: winter wheat for irrigated conditions. Crop J. 2015;3:507–516. doi: 10.1016/j.cj.2017.04.004. [DOI] [Google Scholar]
- 69.Keser M., Gummadov N., Akin B., Belen S., Mert Z., Taner S., Topal A., Yazar S., Morgounov A., Sharma R.C., Ozdemir F. Genetic gains in wheat in Turkey: winter wheat for dryland conditions. The Crop J. 2017;5:533–540. doi: 10.1016/j.cj.2017.04.004. [DOI] [Google Scholar]
- 70.Farooq M., Wahid A., Kobayashi N., Fujita D., Basra S.M.A. In: Sustainable Agriculture. Lichtfouse E., Navarrete M., Debaeke P., Véronique S., Alberola C., editors. Springer; Dordrecht: 2009. Plant drought stress: effects, mechanisms and management; pp. 153–188. [DOI] [Google Scholar]
- 71.Tiwari A., Punetha S., Kesarvani K. Drought stress and its impact on plant mechanism. IJPS Int. J. Plant Sci. 2021;16:95–112. doi: 10.15740/HAS/IJPS/16.AAEBSSD/95-112. [DOI] [Google Scholar]
- 72.Sharma R.C., Crossa J., Velu G., Huerta‐Espino J., Vargas M., Payne T.S., Singh R.P. Genetic gains for grain yield in CIMMYT spring bread wheat across international environments. Crop Sci. 2012;52:1522–1533. doi: 10.2135/cropsci2011.12.0634. [DOI] [Google Scholar]
- 73.Joudi M., Ahmadi A., Mohammadi V., Abbasi A., Mohammadi H. Genetic changes in agronomic and phenologic traits of Iranian wheat cultivars grown in different environmental conditions. Euphytica. 2014;196:237–249. doi: 10.1007/s10681-013-1027-7. [DOI] [Google Scholar]
- 74.Chen X., Hao M.D. Low contribution of photosynthesis and water-use efficiency to improvement of grain yield in Chinese wheat. Photosynthetica. 2015;53:519–526. doi: 10.1007/s11099-015-0147-9. [DOI] [Google Scholar]
- 75.Chen X., Min D., Yasir T.A., Hu Y.G. Evaluation of 14 morphological, yield-related and physiological traits as indicators of drought tolerance in Chinese winter bread wheat revealed by analysis of the membership function value of drought tolerance (MFVD) Field Crops Res. 2012;137:195–201. doi: 10.1016/j.fcr.2012.09.008. [DOI] [Google Scholar]
- 76.Aisawi K.A.B., Reynolds M.P., Singh R.P., Foulkes M.J. The physiological basis of the genetic progress in yield potential of CIMMYT spring wheat cultivars from 1966 to 2009. Crop Sci. 2015;55:1749–1764. doi: 10.2135/cropsci2014.09.0601. [DOI] [Google Scholar]
- 77.Wu X., Cheng R., Xue S., Kong Z., Wan H., Li G., Huang Y., Jia H., Jia J., Zhang L., Ma Z. Precise mapping of a quantitative trait locus interval for spike length and grain weight in bread wheat (Triticum aestivum L.) Mol. Breed. 2014;33:129–138. doi: 10.1007/s11032-013-9939-4. [DOI] [Google Scholar]
- 78.Zhang Y., Xu W., Wang H., Dong H., Qi X., Zhao M., Fang Y., Gao C., Hu L. Progress in genetic improvement of grain yield and related physiological traits of Chinese wheat in Henan Province. Field Crops Res. 2016;199:117–128. doi: 10.1016/j.fcr.2016.09.022. [DOI] [Google Scholar]
- 79.Beche E., Benin G., da Silva C.L., Munaro L.B., Marchese J.A. Genetic gain in yield and changes associated with physiological traits in Brazilian wheat during the 20th century. Eur. J. Agron. 2014;61:49–59. doi: 10.1016/j.eja.2014.08.005. [DOI] [Google Scholar]
- 80.Würschum T., Langer S.M., Longin C.F.H. Genetic control of plant height in European winter wheat cultivars. Theor. Appl. Genet. 2015;128:865–874. doi: 10.1007/s00122-015-2476-2. [DOI] [PubMed] [Google Scholar]
- 81.Islam M.R., Haque K.S., Akter N., Karim M.A. Leaf chlorophyll dynamics in wheat based on SPAD meter reading and its relationship with grain yield. Sci. Agric. 2014;8:13–18. doi: 10.15192/PSCP.SA.2014.4.1.1318. [DOI] [Google Scholar]
- 82.Mwadzingeni L., Shimelis H., Tsilo T.J. Variance components and heritability of yield and yield components of wheat under drought-stressed and non-stressed conditions. Aust. J. Crop. Sci. 2017;11:1425–1430. doi: 10.3316/informit.434178254294039. [DOI] [Google Scholar]
- 83.Würschum T., Leiser W.L., Langer S.M., Tucker M.R., Longin C.F.H. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor. Appl. Genet. 2018;131:2071–2084. doi: 10.1007/s00122-018-3133-3. [DOI] [PubMed] [Google Scholar]
- 84.Yu M., Mao S.L., Chen G.Y., Pu Z.E., Wei Y.M., Zheng Y.L. QTLs for uppermost internode and spike length in two wheat RIL populations and their affect upon plant height at an individual QTL level. Euphytica. 2014;200:95–108. doi: 10.1007/s10681-014-1156-7. [DOI] [Google Scholar]
- 85.Liu D., Zhang L., Hao M., Ning S., Yuan Z., Dai S., Huang L., Wu B., Yan Z., Lan X., Zheng Y. Wheat breeding in the hometown of Chinese Spring. The Crop J. 2018;6:82–90. doi: 10.1016/j.cj.2017.08.009. [DOI] [Google Scholar]
- 86.Alonso M.P., Mirabella N.E., Panelo J.S., Cendoya M.G., Pontaroli A.C. Selection for high spike fertility index increases genetic progress in grain yield and stability in bread wheat. Euphytica. 2018;214:1–12. doi: 10.1007/s10681-018-2193-4. [DOI] [Google Scholar]
- 87.Tausz-Posch S., Dempsey R.W., Seneweera S., Norton R.M., Fitzgerald G., Tausz M. Does a freely tillering wheat cultivar benefit more from elevated CO2 than a restricted tillering cultivar in a water-limited environment? Eur. J. Agron. 2015;64:21–28. doi: 10.1016/j.eja.2014.12.009. [DOI] [Google Scholar]
- 88.Tian Z., Jing Q., Dai T., Jiang D., Cao W. Effects of genetic improvements on grain yield and agronomic traits of winter wheat in the Yangtze River Basin of China. Field Crops Res. 2011;124:417–425. doi: 10.1016/j.fcr.2011.07.012. [DOI] [Google Scholar]
- 89.Morgounov A., Zykin V., Belan I., Roseeva L., Zelenskiy Y., Gomez-Becerra H.F., Budak H., Bekes F. Genetic gains for grain yield in high latitude spring wheat grown in Western Siberia in 1900–2008. Field Crops Res. 2010;117:101–112. doi: 10.1016/j.fcr.2010.02.001. [DOI] [Google Scholar]
- 90.Zheng T.C., Zhang X.K., Yin G.H., Wang L.N., Han Y.L., Chen L., Huang F., Tang J.W., Xia X.C., He Z.H. Genetic gains in grain yield, net photosynthesis and stomatal conductance achieved in Henan Province of China between 1981 and 2008. Field Crops Res. 2011;122:225–233. doi: 10.1016/j.fcr.2011.03.015. [DOI] [Google Scholar]
- 91.Bustos D.V., Hasan A.K., Reynolds M.P., Calderini D.F. Combining high grain number and weight through a DH-population to improve grain yield potential of wheat in high-yielding environments. Field Crops Res. 2013;145:106–115. doi: 10.1016/j.fcr.2013.01.01. [DOI] [Google Scholar]
- 92.Carvalho A.P., Silva S.O., Baptista J.M., Malcata F.X. Light requirements in microalgal photobioreactors: an overview of biophotonic aspects. Appl. Microbiol. Biotechnol. 2011;89:1275–1288. doi: 10.1007/s00253-010-3047-8. [DOI] [PubMed] [Google Scholar]
- 93.Huang H., Ran J., Ji M., Wang Z., Dong L., Hu W., Deng Y., Hou C., Niklas K.J., Deng J. Water content quantitatively affects metabolic rates over the course of plant ontogeny. New Phytol. 2020;228:1524–1534. doi: 10.1111/nph.16808. [DOI] [PubMed] [Google Scholar]
- 94.Ling Q., Huang W., Jarvis P. Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth. Res. 2011;107:209–214. doi: 10.1007/s11120-010-9606-0. [DOI] [PubMed] [Google Scholar]
- 95.Lubitz W., Chrysina M., Cox N. Water oxidation in photosystem II. Photosynth. Res. 2019;142:105–125. doi: 10.1007/s11120-019-00648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sallam A., Alqudah A.M., Dawood M.F., Baenziger P.S., Börner A. Drought stress tolerance in wheat and barley: advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019;20:3137. doi: 10.3390/ijms20133137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farooq M., Hussain M., Wahid A., Siddique K.H.M. In: Plant Responses to Drought Stress: from Morphological to Molecular Features. Aroca R., editor. Springer; Berlin, Heidelberg: 2012. Drought stress in plants: an overview; pp. 1–33. [DOI] [Google Scholar]
- 98.Zhang J., Lin Y., Zhu L., Yu S., Kundu S.K., Jin Q. Effects of 1-methylcyclopropene on function of flag leaf and development of superior and inferior spikelets in rice cultivars differing in panicle types. Field Crops Res. 2015;177:64–74. doi: 10.1016/j.fcr.2015.03.003. [DOI] [Google Scholar]
- 99.Vaghar M., Ehsanzadeh P. Comparative photosynthetic attributes of emmer and modern wheats in response to water and nitrogen supply. Photosynthetica. 2018;56:1224–1234. doi: 10.1007/s11099-018-0825-5. [DOI] [Google Scholar]
- 100.Zivcak M., Kalaji H.M., Shao H.B., Olsovska K., Brestic M. Photosynthetic proton and electron transport in wheat leaves under prolonged moderate drought stress. J. Photochem. Photobiol. B Biol. 2014;137:107–115. doi: 10.1016/j.jphotobiol.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 101.Bhattacharya A., Bhattacharya A. In: Soil Water Deficit and Physiological Issues in Plants. Bhattacharya A., editor. Springer; Singapore: 2021. Dry matter production, partitioning, and seed yield under soil water deficit: a review; pp. 585–702. [DOI] [Google Scholar]
- 102.Berger B., Parent B., Tester M. High-throughput shoot imaging to study drought responses. J. Exp. Bot. 2010;61:3519–3528. doi: 10.1093/jxb/erq201. [DOI] [PubMed] [Google Scholar]
- 103.Lepekhov S.B. Canopy temperature depression for drought-and heat stress tolerance in wheat breeding. Vavilov J. Genet. Breed. 2022;26:196. doi: 10.18699/VJGB-22-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gulnaz S., Zulkiffal M., Sajjad M., Ahmed J., Musa M., Abdullah M., Ahsan A., Rehman A. Identifying Pakistani wheat landraces as genetic resources for yield potential, heat tolerance and rust resistance. Int. J. Agric. Biol. 2019;21:520–526. doi: 10.17957/IJAB/15.0924. [DOI] [Google Scholar]
- 105.Bonari A., Edalat M., Ghadiri H., Kazemeini S.A., Modarresi M. The study of temperature depression and its association with grain yield in six wheat cultivars under heat stress conditions and salicylic acid application. Iran Agric. Res. 2020;39:99–108. doi: 10.22099/iar.2020.31975.1318. [DOI] [Google Scholar]
- 106.Kaur G., Asthir B.J.B.P. Proline: a key player in plant abiotic stress tolerance. Biol. Plant. (Prague) 2015;59:609–619. doi: 10.1007/s10535-015-0549-3. [DOI] [Google Scholar]
- 107.Surekha C.H., Kumari K.N., Aruna L.V., Suneetha G., Arundhati A., Kavi Kishor P.B. Expression of the Vigna aconitifolia P5CSF129A gene in transgenic pigeonpea enhances proline accumulation and salt tolerance. Plant Cell Tissue Organ Cult. 2014;116:27–36. doi: 10.1007/s11240-013-0378-z. [DOI] [Google Scholar]
- 108.Tang X., Mu X., Shao H., Wang H., Brestic M. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2015;35:425–437. doi: 10.3109/07388551.2014.889080. [DOI] [PubMed] [Google Scholar]
- 109.Per T.S., Khan N.A., Reddy P.S., Masood A., Hasanuzzaman M., Khan M.I.R., Anjum N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017;115:126–140. doi: 10.1016/j.plaphy.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 110.Johnson R., Vishwakarma K., Hossen M.S., Kumar V., Shackira A.M., Puthur J.T., Abdi G., Sarraf M., Hasanuzzaman M. Potassium in plants: growth regulation, signaling, and environmental stress tolerance. Plant Physiol. Biochem. 2022;172:56–69. doi: 10.1016/j.plaphy.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 111.Kumar P., Kumar T., Singh S., Tuteja N., Prasad R., Singh J. Potassium: a key modulator for cell homeostasis. J. Biotechnol. 2020;324:198–210. doi: 10.1016/j.jbiotec.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 112.Akter N., Brishty T.A., Karim M.A., Ahmed M.J.U., Islam M.R. Leaf water status and biochemical adjustments as a mechanism of drought tolerance in two contrasting wheat (Triticum aestivum L.) varieties. Acta Physiol. Plant. 2023;45:50. doi: 10.1007/s11738-023-03530-x. [DOI] [Google Scholar]
- 113.Ami K., Planchais S., Cabassa C., Guivarc’h A., Very A.A., Khelifi M., Djebbar R., Abrous-Belbachir O., Carol P. Different proline responses of two Algerian durum wheat cultivars to in vitro salt stress. Acta Physiol. Plant. 2020;42:1–16. doi: 10.1007/s11738-019-3004-9. [DOI] [Google Scholar]
- 114.Faisal S., Mujtaba S.M., Asma, Mahboob W. Polyethylene Glycol mediated osmotic stress impacts on growth and biochemical aspects of wheat (Triticum aestivum L.) J. Crop Sci. Biotechnol. 2019;22:213–223. doi: 10.1007/s12892-018-0166-0. [DOI] [Google Scholar]
- 115.Loutfy N., El-Tayeb M.A., Hassanen A.M., Moustafa M.F., Sakuma Y., Inouhe M. Changes in the water status and osmotic solute contents in response to drought and salicylic acid treatments in four different cultivars of wheat (Triticum aestivum) J. Plant Res. 2012;125:173–184. doi: 10.1007/s10265-011-0419-9. [DOI] [PubMed] [Google Scholar]
- 116.Bukhari M.A., Shah A.N., Fahad S., Iqbal J., Nawaz F., Manan A., Baloch M.S. Screening of wheat (Triticum aestivum L.) genotypes for drought tolerance using polyethylene glycol. Arabian J. Geosci. 2021;14:2808. doi: 10.1007/s12517-021-09073-0. [DOI] [Google Scholar]
- 117.Chowdhury M.K., Hasan M.A., Bahadur M.M., Islam M.R., Hakim M.A., Iqbal M.A., Javed T., Raza A., Shabbir R., Sorour S., Elsanafawy N.E. Evaluation of drought tolerance of some wheat (Triticum aestivum L.) genotypes through phenology, growth, and physiological indices. Agron. 2021;11:1792. doi: 10.3390/agronomy11091792. [DOI] [Google Scholar]
- 118.Pradhan G.P., Xue Q., Jessup K.E., Rudd J.C., Liu S., Devkota R.N., Mahan J.R. Cooler canopy contributes to higher yield and drought tolerance in new wheat cultivars. Crop Sci. 2014;54:2275–2284. doi: 10.2135/cropsci2013.11.0788. [DOI] [Google Scholar]
- 119.Grzesiak S., Hordyńska N., Szczyrek P., Grzesiak M.T., Noga A., Szechyńska-Hebda M. Variation among wheat (Triticum easativum L.) genotypes in response to the drought stress: I–selection approaches. J. Plant Interact. 2019;14:30–44. doi: 10.1080/17429145.2018.1550817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bahrami F., Arzani A., Karimi V. Evaluation of yield‐based drought tolerance indices for screening safflower genotypes. Agron. J. 2014;106:1219–1224. doi: 10.2134/agronj13.0387. [DOI] [Google Scholar]
- 121.Arifuzzaman M., Barman S., Hayder S., Azad M.A.K., Turin M.T.S., Amzad M.A., Masuda M.S. Screening of bread wheat (Triticum aestivum L.) genotypes under drought stress conditions using multivariate analysis. Cereal Res. Commun. 2020;48:301–308. doi: 10.1007/s42976-020-00039-8. [DOI] [Google Scholar]
- 122.Abdi H., Williams L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010;2:433–459. doi: 10.1002/wics.101. [DOI] [Google Scholar]
- 123.Al-Ashkar I., Al-Suhaibani N., Abdella K., Sallam M., Alotaibi M., Seleiman M.F. Combining genetic and multidimensional analyses to identify interpretive traits related to water shortage tolerance as an indirect selection tool for detecting genotypes of drought tolerance in wheat breeding. Plants. 2021;10:931. doi: 10.3390/plants10050931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fleitas M.C., Mondal S., Gerard G.S., Hernández-Espinosa N., Singh R.P., Crossa J., Guzmán C. Identification of CIMMYT spring bread wheat germplasm maintaining superior grain yield and quality under heat-stress. J. Cereal. Sci. 2020;93 doi: 10.1016/j.jcs.2020.102981. [DOI] [Google Scholar]
- 125.Li P., Chen J., Wu P. Agronomic characteristics and grain yield of 30 spring wheat genotypes under drought stress and nonstress conditions. Agron. J. 2011;103:1619–1628. doi: 10.2134/agronj2011.0013. [DOI] [Google Scholar]
- 126.Noman M.U., Azhar S. Metabolomics, a potential way to improve abiotic stresses tolerance in cereal crops. Int. J. Agric. Biosci. 2023;12:47–55. doi: 10.47278/journal.ijab/2023.043. [DOI] [Google Scholar]
- 127.Tardieu F. Any trait or trait-related allele can confer drought tolerance: just design the right drought scenario. J. Exp. Bot. 2012;63:25–31. doi: 10.1093/jxb/err269. [DOI] [PubMed] [Google Scholar]
- 128.Tattaris M., Reynolds M.P., Chapman S.C. A direct comparison of remote sensing approaches for high-throughput phenotyping in plant breeding. Front. Plant Sci. 2016;7:1131. doi: 10.3389/fpls.2016.01131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tester M., Langridge P. Breeding technologies to increase crop production in a changing world. Sci. 2010;327:818–822. doi: 10.1126/science.1183700. [DOI] [PubMed] [Google Scholar]
- 130.Gahlaut V., Jaiswal V., Tyagi B.S., Singh G., Sareen S., Balyan H.S., Gupta P.K. QTL mapping for nine drought-responsive agronomic traits in bread wheat under irrigated and rain-fed environments. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Voss-Fels K.P., Cooper M., Hayes B.J. Accelerating crop genetic gains with genomic selection. Theor. Appl. Genet. 2018;132:669–686. doi: 10.1007/s00122-018-3270-8. [DOI] [PubMed] [Google Scholar]
- 132.Shen Y.G., Zhang W.K., He S.J., Zhang J.S., Liu Q., Chen S.Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 2003;106:923–930. doi: 10.1007/s00122-002-1131-x. [DOI] [PubMed] [Google Scholar]
- 133.Mao X., Zhang H., Tian S., Chang X., Jing R. TaSnRK2. 4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xiong H., Li J., Liu P., Duan J., Zhao Y., Guo X., Li Y., Zhang H., Ali J., Li Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kulkarni M., Soolanayakanahally R., Ogawa S., Uga Y., Selvaraj M.G., Kagale S. Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 2017;5:106. doi: 10.3389/fchem.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nezhadahmadi A., Prodhan Z.H., Faruq G. Drought tolerance in wheat. Sci. World J. 2013;2013:1–12. doi: 10.1155/2013/610721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Placido D.F., Campbell M.T., Folsom J.J., Cui X., Kruger G.R., Baenziger P.S., Walia H. Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiol. 2013;161:1806–1819. doi: 10.1104/pp.113.214262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sehgal D., Autrique E., Singh R., Ellis M., Singh S., Dreisigacker S. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep41578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.