Graphical abstract
Keywords: Bactrocera dorsalis, Chromosome-level genome assembly, Invasion routes and history, Resequencing, Species delimitation, Thermal adaptation
Highlights
-
•
The present study achieves a large-scale SNP dataset, a total of 512 genomes comprising 487 B. dorsalis and 25 B. carambolae.
-
•
B. dorsalis originates from the Southern India with three invasion routes worldwide, mainly facilitated by human activities.
-
•
CYP6a9 is identified that enhance the thermal adaptation of B. dorsalis and thus boost its invasion to temperate regions.
-
•
The gene function is further verified using the RNAi technology.
Abstract
Introduction
The oriental fruit fly Bactrocera dorsalis is one of the most destructive agricultural pests worldwide, with highly debated species delimitation, origin, and global spread routes.
Objectives
Our study intended to (i) resolve the taxonomic uncertainties between B. dorsalis and B. carambolae, (ii) reveal the population structure and global invasion routes of B. dorsalis across Asia, Africa, and Oceania, and (iii) identify genomic regions that are responsible for the thermal adaptation of B. dorsalis.
Methods
Based on a high-quality chromosome-level reference genome assembly, we explored the population relationship using a genome-scale single nucleotide polymorphism dataset generated from the resequencing data of 487 B. dorsalis genomes and 25 B. carambolae genomes. Genome-wide association studies and silencing using RNA interference were used to identify and verify the candidate genes associated with extreme thermal stress.
Results
We showed that B. dorsalis originates from the Southern India region with three independent invasion and spread routes worldwide: (i) from Northern India to Northern Southeast Asia, then to Southern Southeast Asia; (ii) from Northern India to Northern Southeast Asian, then to China and Hawaii; and (iii) from Southern India toward the African mainland, then to Madagascar, which is mainly facilitated by human activities including trade and immigration. Twenty-seven genes were identified by a genome-wide association study to be associated with 11 temperature bioclimatic variables. The Cyp6a9 gene may enhance the thermal adaptation of B. dorsalis and thus boost its invasion, which tended to be upregulated at a hardening temperature of 38 °C. Functional verification using RNA interference silencing against Cyp6a9, led to the specific decrease in Cyp6a9 expression, reducing the survival rate of dsRNA-feeding larvae exposed to extreme thermal stress of 45 °C after heat hardening treatments in B. dorsalis.
Conclusion
This study provides insights into the evolutionary history and genetic basis of temperature adaptation in B. dorsalis.
Introduction
Invasive pests present considerable threats to global agriculture [1], and the challenge has become more severe with increasing international tourism and trade. Attacking >250 species of fruits and vegetables [2], the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), poses a serious threat to agricultural products and represents one of the most detrimental invasive pests worldwide. It presently spreads to 75 countries across Asia, Africa, and Oceania [3]. Facilitated by the biological features of high prolificacy, short life history, broad host range [1], and adaptability [4], B. dorsalis is classified as the top member in the competitive hierarchy of fruit flies [5] and could replace and drive various fruit fly species to extinction, including other highly invasive fruit flies such as Ceratitis capitata, Ceratitis cosyra, Bactrocera tryoni, and Bactrocera zonata [4].
To prevent or alleviate the impact of future invasions, a global-scale population structure of B. dorsalis should be developed. This may serve as a prerequisite to identifying the effective formulation and implementation of preventive and mitigation agents by inspecting the sources of different invading populations and their corresponding invasion routes. Previous studies have attempted to trace the spread routes and invasion history of the fly at different temporal and spatial scales, but have yielded conflicting results due to inadequate sampling efforts [6], [7], [8] or by utilizing limited informative makers (microsatellites or a limited number of genes) [9]. In this study, we attempted to resolve this issue by addressing it at the genomic level for the first time. Most studies have speculated that China [6], the southeastern coastal areas of China [7], Southeast Asia [8], or South Asia [9] may be the evolutionary origin of B. dorsalis. In addition, the question of whether the largely indistinguishable morphologies of Bactrocera carambolae and B. dorsalis, whose hybridization have been observed under laboratory conditions and in the wild [10], are actually indicative of them being the same species, has long been a focus of controversy and hindered our understanding of the invasion history of the fly.
Originating from tropical Asia [11], invasions of B. dorsalis have occurred within a timespan of two decades across sub-Saharan Africa and the western Indian Ocean. These are tropical areas with high temperatures, which demonstrates the extremely high temperature tolerance of B. dorsalis. Recent surveillance records of established B. dorsalis populations from areas where have been previously considered climatically unsuitable due to overwintering cold stress, such as the central regions of China, specifically the Hubei and Henan provinces, have also been reported, indicating an intensified invasion capacity of B. dorsalis to thrive. This poses a great concern, not only for China, but also for climatically similar temperate regions in Europe and North America. Hence, there is an urgent need to understand the mechanisms and role of temperature tolerance in the success of the invasive process of B. dorsalis.
In this study, we obtained a chromosome-level genome of B. dorsalis and systematically sampled a total of 487 B. dorsalis with various geographical populations for genome sequencing with the aim of (i) revealing the population structure and global invasion routes of this species, and (ii) identifying genomic regions that are responsible for its thermal adaptation. Samples of the closely related B. carambolae were also included in this research so to (a) compare the extent of intra- and interspecific differentiation in B. dorsalis and (b) further verify the taxonomic status of B. dorsalis and B. carambolae by investigating possible introgression between these two species.
Materials and methods
Development of an inbred B. dorsalis strain for genome assembly
B. dorsalis individuals used for genome sequencing and assembly were collected from infested citrus fruits in an orchard in Guangzhou, Guangdong Province, China. A laboratory colony was established and reared on artificial diets in an artificial climate chamber at 25 ± 1 °C, relative humidity of 70 ± 5 %, and a light:dark photoperiod of 14:10 h. Two male adults were collected from an inbred strain (at least 100 generations from the colony) for subsequent genomic sequencing.
Genome survey
Genome size and heterozygosity were estimated using a k-mer analysis [12] and visualized using GenomeScope 1.0 [13]. DNA was extracted from an individual male adult thorax using the Promega Wizard SV Genomic DNA Purification System. A library with an average insert size of 350 bp was constructed using the Illumina TruSeq Nano DNA Library Prep Kit and sequenced on the Illumina NovaSeq platform with paired-end 150 (PE150) bp reads. After performing quality control using fastp [14], filtered sequences were used to generate a 19-mer distribution map using Jellyfish 2.11 [12].
Draft genome sequencing by PacBio, assembly, and polishing
For long-read sequencing, two PCR-free SMRTbell libraries, with an average insert size of 20 kb, were constructed using the PacBio SMRTbell Express Template Prep Kit 3.0. Two cells were sequenced on the PacBio Sequel II system under a continuous long read model. Canu v1.6 [15] was used to correct and trim the polymerase reads using default parameters to generate high-quality polymerase subreads. The trimmed polymerase subreads were assembled to a contig-level genome using WTDBG2 [16] with the parameter --tidy-reads 5000. To reduce the sequence error rate, we first used pbmm2 (https://github.com/PacificBiosciences/pbmm2) to align the PacBio subreads to the raw contigs and corrected the contigs by gcpp (https://github.com/PacificBiosciences/pbbioconda). The Illumina short reads for the genome survey were then mapped to the first round of corrected contigs by BWA-mem [17] and further polished using Pilon 1.16 [18]. Redundans [19] was used to remove the redundant contigs from the second round of corrected contigs with a parameter --overlap of 0.73 to obtain the final version of the contig genome.
Hi-C sequencing and scaffolding
We used the 3rd instar larvae after 3 days of starvation treatment for cellular protein cross-linking in 2 % formaldehyde. MobI was selected as a restriction enzyme for chromatin digestion. Treated Hi-C sample DNA was extracted and fragmented to 350 bp for Hi-C library preparation. The Hi-C library was sequenced using the Illumina NovaSeq platform with the PE150 read strategy. After filtering by fastp [14], the clean Hi-C data were mapped to the contigs-level genome using JUICER [20] to generate the contracted matrices. After polishing, splitting, sealing, and merging using a 3D de novo assembly [3D-DNA] pipeline, a chromosome-length assembly-level genome was generated.
Full-length transcript sequencing and analysis
Total RNA was extracted from the mixed samples (larvae, pupae, and adults) using the Promega SV Total RNA Isolation System kit. RNA samples with RIN ≥ 7.5 were evaluated with the Agilent 2100 RNA 6000 Nano kit to construct two libraries (1–4 kb and 1–10 kb) for IsoSeq. Full-length cDNA sequencing was performed on the PacBio Sequel II system using the circular consensus sequencing (CCS) model. Raw polymerase read data were filtered and analyzed using the IsoSeq software in SMRT® Analysis v3.0 (https://smrt-analysis.readthedocs.io/en/latest/SMRT-Analysis-Software-Installation-v2.3.0/) including building the CCS, classification, and clustering.
Completeness and quality assessment of genome assembly and annotation
Repeat sequences, non-coding RNA genes, and protein coding genes were annotated respectively (Supplementary Material and Methods). BUSCO version 4.1.4 [21] was used to evaluate the completeness and quality of the contig-level genome, Hi-C scaffold-level genome, and annotated coding sequence based on the insecta-odb10 database (1,367 genes).
Whole-genome synteny
The genome of Drosophila melanogaster (GCA_000001215.4 Release 6 plus ISO1 MT) was selected for whole-genome synteny with B. dorsalis. The protein sequences of B. dorsalis and D. melanogaster were aligned using blastp with 1e-10 value. Syntenic blocks against each chromosome were detected using MCScanX [22]. Synteny visualization was performed using SynVisio [23].
Dipteran orthology identification, phylogenetic analysis and divergence time estimation
Twenty dipteran species and the cotton leafworm, Spodoptera litura (Lepidoptera: Noctuidae), were selected as outgroups for a comparative genomic analysis (Table S12). OrthoFinder [24] was used to identify orthologs and orthogroups with parameters -S diamond, -M msa, and -T fasttree. The phylogenetic tree was automatically generated in the OrthoFinder pipeline using an approximately maximum-likelihood method based on a concatenated alignment of single-copy genes. The divergence time was estimated using MCMCTree in PAML [25]. We set the divergence time between Drosophila and Scaptodrosophila as 70–74 Ma and Diptera and Lepidoptera as 243–317 Ma, the values of which were acquired from the TIMETREE database (https://timetree.org/) as calibrations for estimation.
Dipteran gene family expansion and contraction analysis
A computational analysis of gene family evolution (CAFE) [26] was used for dipteran gene family expansion and contraction analyses with the parameters -p 0.05, -t 10, and -r 10,000. The input file was generated from the Orthogroups GeneCount.tsv in OrthoFinder. The results were visualized using the CAFE_fig (https://github.com/LKremer/CAFE_fig). The expanded and contracted genes were visualized using the R package clusterProfiler.
Samples collection, DNA extraction, and resequencing
We collected 487 male adults of B. dorsalis using methyleugenol traps in 50 populations from 29 countries, roughly covering the entire distribution range of B. dorsalis (Fig. 2a, Table S15). The 50 populations were divided into six geographical groups: China (CN) (N = 160), Northern Southeast Asia (NSA) (N = 40), Southern Southeast Asia (SSA) (N = 60), South Asia (SA) (N = 84), Africa (AF) (N = 133), and Hawaii (HW) (N = 10). The sampling number of each group used depend on the distribution range of B. dorsalis in the corresponding regions.
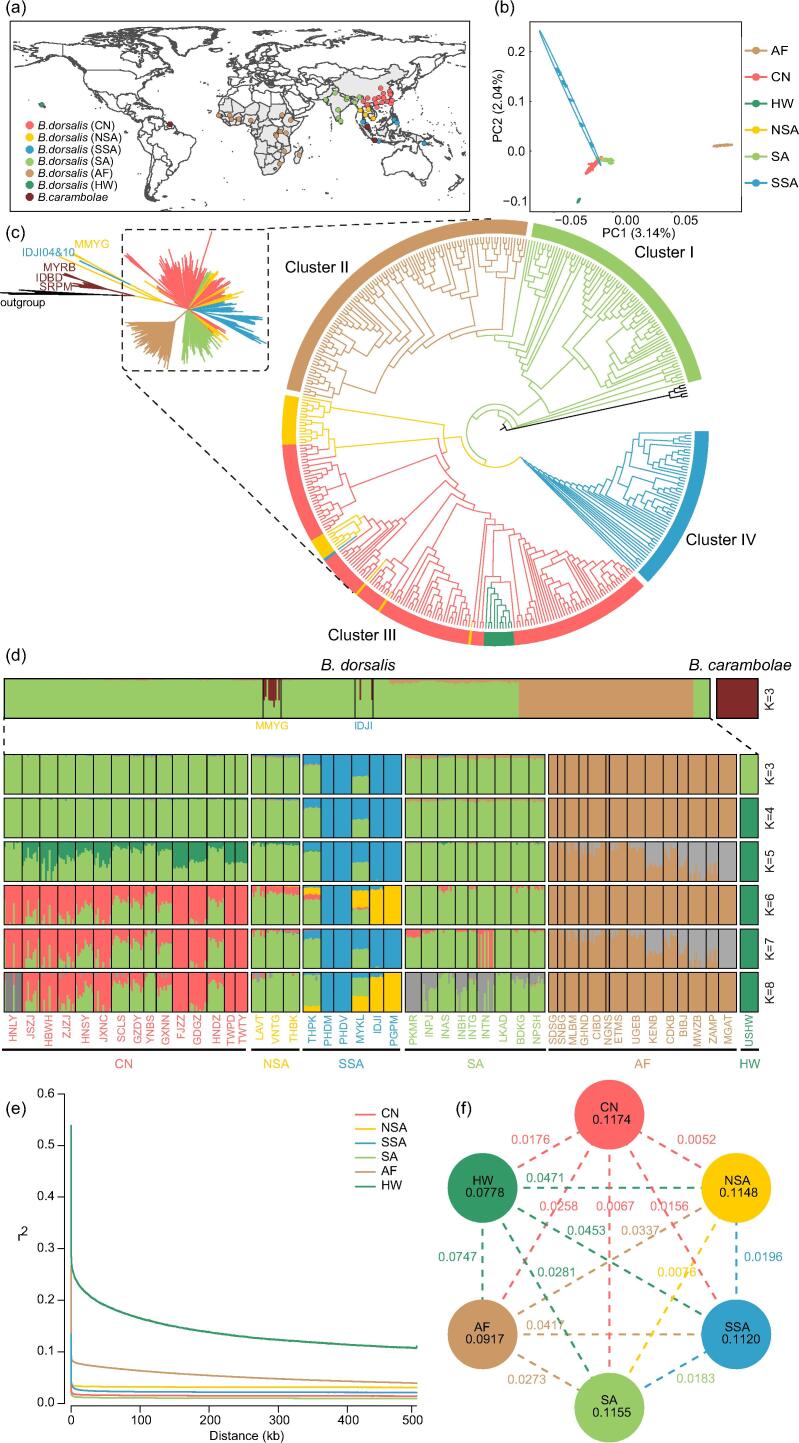
Fig. 2.
Geographic distribution, species delimitation, and population structure of resequenced Bactrocera dorsalis samples. (a) Sampling collection map of B. dorsalis and B. carambolae in which that specimens of B. dorsalis were divided into 6 groups: China (CN), Northern Southeast Asia (NSA), Southern Southeast Asia (SSA), South Asia (SA), Africa (AF), and Hawaii (HW). The countries in light grey indicate where B. dorsalis has distributed. (b) Principal component analysis plot of 458 B. dorsalis samples with PC1 plotted against PC2. (c) The neighbor-joining phylogenetic tree of B. dorsalis and B. carambolae based on whole-genome SNPs using other Bactrocera species as the outgroup. (d) Population genetic structure of B. carambolae and B. dorsalis based on ancestry fractions inferred by ADMIXTURE when K = 3 to 8. Each color represents one ancestral population. Each sample is represented by a vertical bar, and the length of each colored segment in each vertical bar represents the proportion contributed by ancestral populations. (e) Linkage disequilibrium (LD) between SNPs as measured using the squared coefficient of correlation (r2) of alleles at any two loci (y-axes). (f) The genetic diversity (π) and population differentiations (FST) across the six groups. Values in each circle represent nucleotide diversity for the group, and values between pairs indicate population divergence (FST). Different colors represent the six designated groups, CN: pink, NSA: yellow, SSA: blue, SA: light green, AF: light brown, and HW: dark green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Compared to B. dorsalis, B. carambolae has a more restricted distribution. In this study, 25 adult male samples were collected from Malaysia (N = 10), Indonesia (N = 5), and Suriname (N = 10), which nearly covered the global distribution of the species (Fig. 2a). B. dorsalis and B. carambolae have very similar morphologies, and sympatric distributions, with possible hybridization both in the laboratory and wild in Malaysia [10] and Indonesia. Samples from known sympatric distributions of these species were identified at the species level by Wee S.L. and Tati Suryati S., respectively. Malaysian strains were collected from infested wax apples (Syzygium spp.) near a forest fringe in the Raub district, Pahang state, Peninsular Malaysia. Five closely related Bactrocera species (B. correcta, B. zonata, B. tuberculata, B. nigrotibialis, and B. tryoni), not within the B. dorsalis complex, were selected as the outgroup.
All samples were preserved in an 95 % ethanol solution after collection and stored at −80 °C. DNA was extracted from the thoracic muscle of each fly using a Promega Wizard SV Genomic DNA Purification System. For each sample, a library with an average insert size of 350 bp was constructed using the Illumina TruSeq Nano DNA Library Prep Kit and sequenced on the Illumina NovaSeq platform with PE150 bp reads. Every sample was sequenced at a capacity of 6 Gb data to guarantee a sequencing depth of at least 10 × coverage.
Read mapping, variant calling, and filtration
The raw sequencing data were filtered using fastp with default parameters before mapping. The filtered data were mapped to the new B. dorsalis chromosome-level genome assembly using the Burrows-Wheeler Aligner (BWA) - mem algorithm [17] with default parameters. Samtools [27] was used to sort the bam output and calculate the sequencing coverage and depth for each sample. Duplicates were removed using Picard (https://sourceforge.net/projects/picard/). Variants were called using the GATK [28]. The following steps were executed in consecutive order: HaplotypeCaller (calling per sample single nucleotide polymorphisms (SNPs) and InDels), CombineGVCFs (combining per-sample gVCF files), GenotypeGVCFs (joint genotyping of all samples), SelectVariants (extracting SNPs and InDels), and VariantFiltration (hard-filtering variant calls based on the criteria: quality-by-depth ratio (QD) < 2.0 || read mapping quality (MQ) < 40.0 || probability of strand bias (FS) > 60.0 || symmetric odds ratio (SOR) > 3.0 || MQRankSum < –12.5 || ReadPosRankSum < –8.0).
Population genetic structure analysis
Before the analysis, PLINK [29] was used to remove SNPs with a missing genotype rate > 0.1 and minor allele frequency < 0.01. To avoid the effect of linkage disequilibrium (LD) on the results (especially for recently admixed populations), SNPs were pruned using PLINK with the following parameters: a window size of 50 kb, step size of 10, and a LD coefficient (r2) threshold of 0.2.
To construct a neighbor-joining tree, the program DNADIST of the Phylip package [30] was used to generate a distance matrix using multisample sequence alignments, after which the NEIGHBOR function of the Phylip package was used to build the neighbor-joining tree itself.
A principal component analysis (PCA) was performed on the variance-standardized relationship matrix. We plotted PC1 against PC2, PC1 against PC3, and PC2 against PC3.
ADMIXTURE [31] is an accurate, efficient, and versatile tool for ancestry estimation that utilizes maximum likelihood estimates, which could automatically rationalize the best value of ancestral populations K (number of ancestral groups) through cross-validation. We used ADMIXTURE to estimate ancestry proportions and population structures under K values ranging from 2 to 18. Cross-validation criteria were used to determine the optimal K.
TreeMix [32] was used to infer patterns of historical splits and admixture events among populations and six geographical groups by reconstructing the maximum-likelihood population tree with 100 bootstrap replicates. PLINK [29] was used to filter LD sites and generate a Freq file. The migration edges were set from 0 to 1, 2, …, until the variance of relatedness between populations explained by the model reached 99.8 %.
Linkage-disequilibrium analysis
PopLDdecay [33] was used to calculate the r2 between pairwise high-quality SNPs of six geographical groups (the maximum distance between two SNPs was set at 500 kb) and plot the LD decay graphs.
Genetic diversity and differentiation statistics
The Genetic diversity index (π), observed heterozygosity (Ho), expected heterozygosity (He), and pairwise genetic differentiation (FST) were calculated at the population level and at the level of six geographical groups using the populations function in the software Stacks 2.2 [34] with the parameters --hwe, --fstats, -k --smooth-fstats, and --smooth-popstats. The comparison of pairwise FST follows the criterion: FST ≤ 0.05 signifies negligible genetic differentiation; 0.05 < FST ≤ 0.15 signifies small genetic differentiation; 0.15 < FST ≤ 0.25 signifies moderate genetic differentiation; and FST > 0.25 signifies very large genetic differentiation.
Demographic history and effective population sizes [Ne] estimation
Seqbility (https://bit.ly/snpable) was used to mask the positions of the missing data and uncalled regions. The effective population sizes (Ne) were estimated using SMC++ [35]. For each group, we selected 10 representative individuals as composite likelihood samples to improve the precision of the estimates. The mutation rate was set to 2.9 × 10−8 per generation [36], and the generation time was set to 0.083 years (approximately 1 month).
Genome-wide association study (GWAS) and identification of the candidate genes associated with 11 temperature bioclimatic variables
To identify the loci that were potentially associated with environmental variables, a GWAS was performed using all 407 samples of B. dorsalis and 11 temperature bioclimatic variables (Table S23) using EMMAX [37] and TASSEL [38]. The 11 bioclimatic variables are available in WorldClim version 2 [39] and are the averages for the years 1970–2000 (Table S24). For EMMAX, we tested two models: an expedited mixed linear model and expedited mixed linear model with a Q-matrix as incorporating covariates. For TASSEL, we tested three models: the general linear model (glm), mixed linear model (mlm), and compressed linear mixed model (cmlm). The genome-wide significance threshold was determined using the Bonferroni correction as –log10 (0.05/SNP number) and –log10 (0.01/SNP number). The results for further analysis were compared to determine the best model using the corresponding quantile–quantile plots of observed versus expected log10 (P) of the GWAS results.
Quantitative Real-time PCR (qRT-PCR) for transcriptome verification of Cyp6a9 in B. dorsalis
Previous studies regarding thermal tolerance of B. dorsalis have shown that the most temperature-sensitive development stage of B. dorsalis was 7-day-old 3rd early-instar larvae [40]. For this reason, we subjected two groups of 7-day-old 3rd early-instar larvae to different temperatures during hardening. Following Gu et al. [41], we subjected one group to a temperature of 38 °C for 4 h while a control group was kept at a temperature of 25 °C. Both groups consisted of three biological replicates, containing 60 larvae, each contained in a 2 ml centrifuge tube.
RNA extraction and cDNA synthesis was completed per the instructions of Gu et al. [41]. The expression level of Cyp6a9 after heat hardening at 38 °C was quantified by qRT-PCR using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus), with 18S rRNA as reference gene (The primers are listed in Table S27). Three technical replicates were established for each biological replicate. Each reaction included 1 μl cDNA template, 12.5 μl TB Green Mix, 1 μl forward primers (10 pm), 1 μl reverse primers (10 pm), 0.5 μl ROX Reference Dye II, and 9 μl ddH2O. The thermocycler conditions were as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s, and 55.4 °C for 34 s. The following melting curve condition was 95 °C for 15 s, followed by 55.4 °C for 60 s with a decreasing rate of 1 °C/s from 95 °C. The relative expression level was calculated using the 2−ΔΔCT method.
Functional validation of Cyp6a9 in thermal adaptation through RNAi silencing
Double-stranded RNA (dsRNA) of Cyp6a9 (dsCyp6a9) was used to knock down the expression of Cyp6a9, with the double-stranded RNA of green fluorescent protein (dsGFP) as the negative control (The primers are listed in Table S27). The dsRNAs were synthesized with the T7 RiboMAX Express RNAi system (Promega, United States) using specific primers containing a T7 promotor sequence.
The 3-day-old 1st instar larvae of B. dorsalis were collected and placed into a 50 ml tube with 3 holes in the lid. Five biological replications were performed for each treatment, with 50 larvae for each replicate. The larvae were fed 3 g artificial diet material, comprised of 30 μl dsCyp6a9 solution, dsGFP solution, and ddH2O, respectively, with the primary concentration of 1 μg/μl. After 96 h, the larvae developed into 3rd instar. Five larvae of each biological replication were used for the expression level detection of Cyp6a9. Three technical replicates were established for each biological replication. The rest of the 3rd late-instar larvae (N = 45) were transferred to a 2 ml tube with 4 g diet material for the heat hardening treatment in a 38 °C circulating water bath for 4 h. Then, all the tubes were moved to 25 °C for 1 h, and exposed heat stress treatment at 45 °C for 1 h. After heat stress treatment, the tubes were kept at 25 °C for 4 h, after which the survival rate was calculated (Fig. 5a).
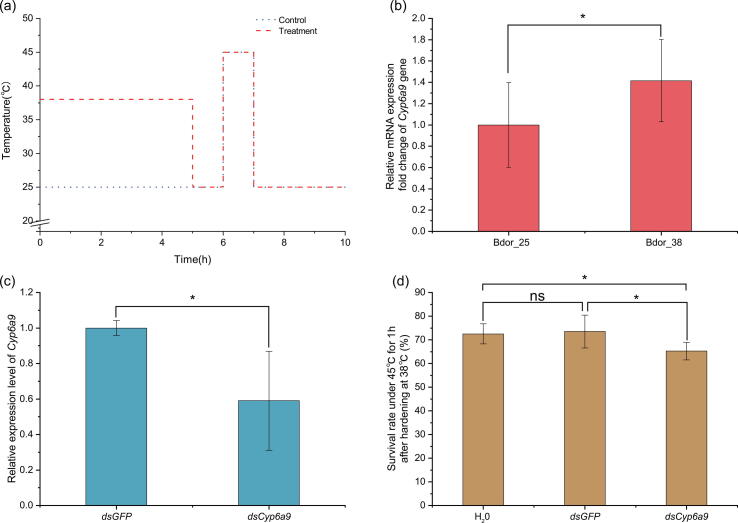
Fig. 5.
Functional analysis of Cyp6a9 on thermal adaptation using RNA interference (RNAi) in B.Bactrocera dorsalis. (a) Experimental design. The blue line represents temperature treatments in the control group. The red line represents the treatments under heat hardening and heat stress temperature. (b) The relative expression level of Cyp6a9 at hardening temperature of 38 °C and the 25 °C as control in B. dorsalis (Bd). Three biological replicates with three technical replicates each biological replicate were studied, and the data are presented as mean ± SE. (c) The relative expression level of Cyp6a9 after RNAi silencing in B. dorsalis. Three biological replicates with three technical replicates each biological replicate were studied, and the data are presented as mean ± SE. (d) Survival rate of dsRNA-feeding larvae exposed to extreme thermal stress 45 °C after heat hardening treatments of 38 °C in B. dorsalis. Five biological replicates were studied, and the data are presented as mean ± SE. The symbol “*” above the bars represents statistically significant differences at P < 0.05 determined by a t-test or LSD test; The letters “ns” above the bars represents no statistically significant difference (P > 0.05) determined by a t-test or LSD test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Statistical analysis
The qRT-PCR results and survival rate are presented as the mean ± standard error (SE) with three independent biological replicates. Comparisons between the means of two independent samples were determined by Student’s t-test, and multiple comparisons were performed with a one-way ANOVA followed by Least Significant Difference (LSD) test in SPSS 26 (IBM Corporation, USA. Statistically significance was set at P < 0.05. Graphs were generated using OriginPro 8 (OriginLab Corporation, USA).
Results
Chromosome-level genome sequencing, assembly, and annotation of the B. dorsalis genome
The 19-mer analysis based on Illumina sequencing data (31.08 Gb) estimated the genome size to be approximately 538.31 Mb with a high degree of duplication (0.91 %) and heterozygosity (2.85 %) (Fig. S1). For long-read genome sequencing, 200.05 Gb of data were obtained with an average length of 11,256 bp and N50 of 16,822 bp, corresponding to approximately 350-fold coverage of the B. dorsalis genome (Table S1). The genome assembly generated 3,334 contigs with an N50 of 1,514,667 bp and total length of 565,511,047 bp (Fig. S2, Table S2). Chromosome-level assembly (Fig. 1a) was achieved with the assistance of Hi-C libraries (Table S4), generating six pseudo-chromosomes that consisted of 97.58 % (2,241 contigs) of the contigs and reached a scaffold N50 of 86.23 Mb (Fig. S4). The Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis revealed a high proportion of complete orthologous genes (95.8 %) (Fig. S5). The genome annotation identified 255.60 Mb of repeat elements (45.11 %) in the genome (Table S6), 630 non-coding RNAs (ncRNAs) (Table S7), and 20,777 genes (Tables S8-S10) with an average length of 11,165 bp. The BUSCO analysis identified 93.70 % (single-copy genes: 92.1 %, duplicated genes: 1.6 %) of the 1,367 gene orthologues as complete (Fig. S5). Both the assembly and annotation processes demonstrated that we obtained an assembly with qualities that were significantly higher than those of other published genome versions of B. dorsalis (Table S11).
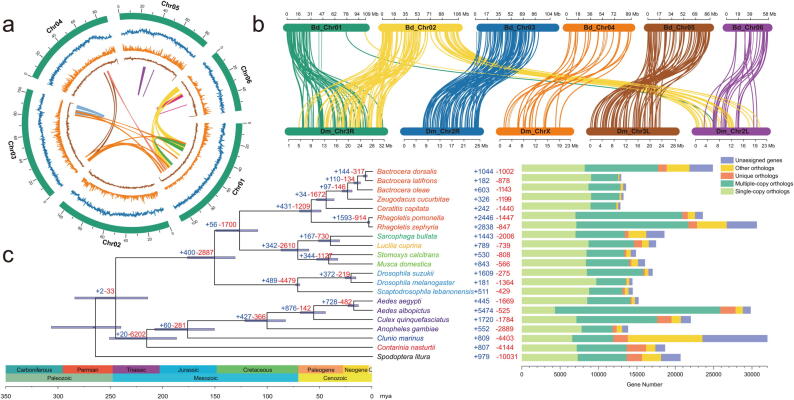
Fig. 1.
Chromosome-level genome assembly features of Bactrocera dorsalis and the Diptera evolutionary analysis. (a) Genomic landscape of the six B. dorsalis chromosomes including basic genomic features, shown in Circos plots from outer to inner circles: I, six chromosomes at the Mb scale; II, repeat density (blue) across the genome per 0.1-Mb non-overlapping windows; III, gene density (orange) across the genome per 0.1-Mb non-overlapping windows; IV, CG contents (brown) across the genome per 0.1-Mb non-overlapping windows; center: intra genomic synteny of B. dorsalis. Colored arcs indicate homologous genomic blocks within B. dorsalis. (b) Whole-genome synteny between B. dorsalis (Bd) and Drosophila melanogaster (Dm). (c) Divergence time, gene orthology, and expanded and contracted gene family analysis of the Diptera species with Spodoptera litura as the outgroup. Divergence times were estimated by the MCMCTree algorithm: mean age is given for each node with gray bars indicating 95 % posterior densities. The horizontal blue bars on the nodes show divergence times with 95 % highest posterior density (HPD) intervals. Gene orthology comparison among 17 species. The bar charts on the right show the five subdivided orthology relationships, including single-copy orthologs, multiple-copy orthologs, unique orthologs, other orthologs, and unassigned genes. The numbers of expanded gene families (colored in red) and contracted gene families (colored in blue) are shown on the branch and at each node. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Synteny and comparative genomics analysis of dipterans
The synteny analysis showed that B. dorsalis and D. melanogaster shared a highly conserved gene order with small-scale rearrangements and translocations (Fig. 1b). Chr04 could be the X chromosome for its conserved synteny with the ChrX of D. melanogaster. We only identified two Y-linked short scaffolds (scaffold1 and scaffold2) based on several Y-specific genes (MoY [42] and spermless [43]). This could be attributed to the heterochromatin of ChrY, which consists of rich repeats and limited protein-coding genes [44].
Based on the phylogenetic tree of the genome scale datasets, including 20 dipteran species (Table S12), the gene family evolution analysis showed that (Fig. 1c, Tables S13 and S14, Fig. S6), in contrast to the overall tendency of gene family contraction in the tephritids, B. dorsalis showed a trend of expansion for 1,044 genes in 238 gene families, including odorant-binding proteins (OBPs), chemosensory proteins (CSPs), olfactory receptors (ORs), gustatory receptors (GRs), cytochrome P450, carboxyl/cholinesterase (CCEs), and glutathione-S-transferase (GST). The expanded gene family could have played an essential role in polyphagy and invasive behaviors, such as host recognition and selection [45], feeding habitats [46], and insecticide resistance and detoxification [47]. In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was mainly enriched in the pathways of lipid metabolism, xenobiotic biodegradation, and metabolism of cofactors and vitamins, which may facilitate the absorption of nutrients from plant hosts and the detoxification of natural xenobiotics from unripened fruits [48].
Whole-genome resequencing and variant detection
To elucidate the origin, invasion history, and genetic basis of thermal adaptation in B. dorsalis, we created the most extensive collections to date. These comprised samples from 50 populations around the world, including group CN (N = 160), group NSA (N = 30), group SSA (N = 70), group SA (N = 84), group Africa (AF) (N = 133), and group HW (N = 10) (Table S15). This sampling covered all of the geographical regions where the species occurs originally and has invaded (Fig. 2a). We acquired a total of 14.15 TB of data for 518 samples, including 487 B. dorsalis, 25 B. carambolae, and five other Bactrocera-related species. This resulted in an average genome coverage of approximately 12 × per sample. In total, 59 samples of B. dorsalis were excluded from further analyses because of the low coverage rate (<70 %) (Table S16). The GATK pipeline identified 278,357,047 SNPs and 46,121,915 small InDels (shorter than 50 bp) (Table S17). The whole-genome heterozygosity estimation showed that the oriental fruit fly had a high level of polymorphisms (the genetic diversity index [π] of B. dorsalis was 0.099–0.131) (Table S18, Figs. S14a and S14b) compared to other invasive pests, such as, B. carambolae (0.039–0.070), Drosophila athabasca (0.002–0.009) [49], Crapholita molesta (0.066–0.116), and Carposina sasakii (0.063–0.088) [50]. This could have empowered the fly to invade and thrive in new habitats.
Species delimitation and evidence of hybridization between B. dorsalis and B. carambolae
The population structure analyses using PCA (Fig. S7), phylogenetic tree (Fig. 2c, Fig. S12), and admixture (Fig. 2d, Fig. S10) revealed that B. dorsalis and B. carambolae formed two distinct lineages. In addition, most of the B. dorsalis individuals from Myanmar and several from Indonesia (MMYG01, MMYG02, MMYG04, MMYG05, MMYG06, MMYG08, MMYG10, IDJI04, and IDJI06) were hybrids of B. dorsalis and B. carambolae. Evidence based on the genome-scale datasets revealed that they were phylogenetically distinct from each other, but could naturally hybridize in the wild. Further investigations of the fertility of hybrids are needed to clarify their isolation and speciation. In addition, the phylogenetic tree demonstrated that B. carambolae was at a basal position and displayed a topological relationship between ((Suriname population + Indonesia population) + Malaysia population), implying that the population that invaded Suriname was from Indonesia.
Genetic structure of the population and demographic history of B. dorsalis
We characterized the genetic relationships among all populations and groups of B. dorsalis using a PCA (Fig. 2b, Fig. S8), neighbor-joining tree (Fig. 2c, Fig. S13), and ancestry estimation (Fig. 2d [with K values ranging from 3 to 8]). Both the phylogeny and ancestry estimation analyses identified four major distinct clusters for the non-hybrid B. dorsalis samples, which exhibited strong geographical separation with shared properties in the CN and NSA groups. The phylogenetic tree provided strong support for a basal position (Cluster I) of populations in SA, suggesting that SA may be where B. dorsalis originated. The AF clade (Cluster II) was monophyletic and branched off from the SA populations, indicating that the initial propagule population that invaded the African continent was most likely from South Asia. This was validated by the detected gene flow in the ancestry estimation analysis at K = 3 and 4. The FST score also showed that SA had the lowest level of differentiation with AF compared to the other groups (Fig. 2f, Fig. S14, Tables S19 and S20). Cluster III encompassed all individuals from NSA, CN and HW, with most NSA individuals being basal. The remaining NSA individuals were mixed with Southwestern China individuals and nested into Cluster III, which showed the existence of gene flow between the southwestern border regions of China and neighboring Laos and Thailand. The exclusive SSA populations formed Cluster IV, which was consistent with the ancestry estimation analysis. The maximum-likelihood tree generated by the treemix analysis (Fig. 3a and 3b) largely recapitulated the NJ tree (Fig. 2c).
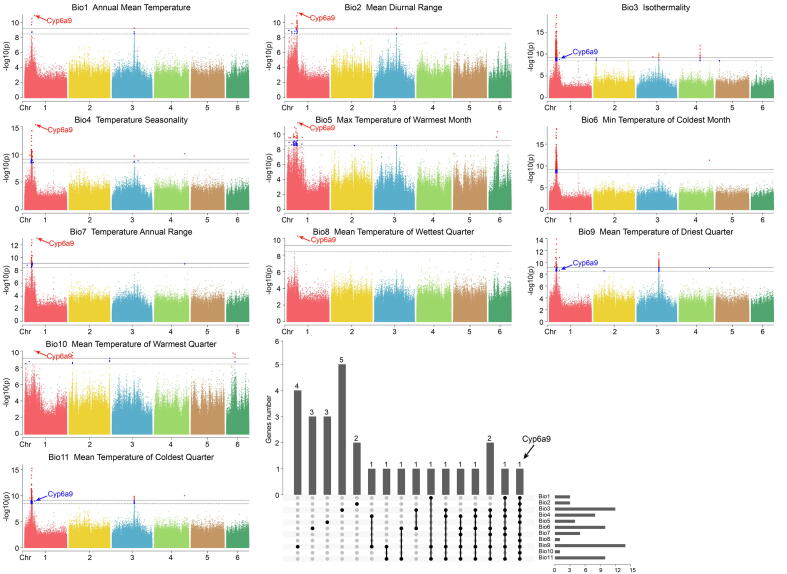
Fig. 4.
Genome-wide association analysis of 11 bioclimatic variables in the 407 samples of Bactrocera dorsalis by EMMAX with Q-matrix as incorporating covariates. For each bioclimatic variable, black horizontal dashed lines represent the thresholds of –log10 (0.05/SNP number), and black horizontal solid line represents the thresholds of –log10 (0.01/SNP number). The histogram in the lower right-hand corner shows the distribution of genes associated with the 11 bioclimatic variables in the 407 samples of B. dorsalis. Horizontal bars to the right represent genes in each bioclimatic variable. Vertical bars indicate the number of genes exclusively associated with one or multiple bioclimatic variables.
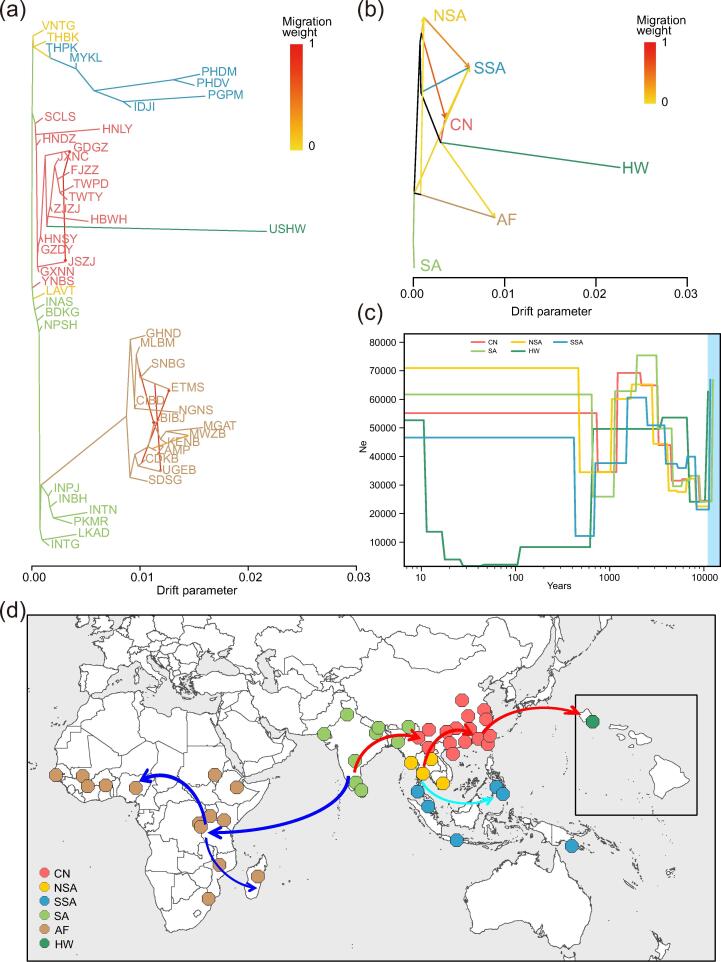
Fig. 3.
Demographic history and global spread routes of Bactrocera dorsalis. (a) Population splits and mixtures using the maximum-likelihood method with six possible migration events inferred from TreeMix analysis. The graph shows the topology and branch lengths according to the drift parameter. Migration arrows were colored according to their weight. (b) Group splits and mixtures using the maximum-likelihood method with six possible migration events inferred from TreeMix analysis. (c) Demographic history of B. dorsalis; five groups illustrated by the effective population sizes (Ne) inferred by SMC++, scaled with a generation time of 0.0833 years and a mutation rate of 8.4 × 10−9 per site per generation. The light blue shade denotes the period of the last glacial maximum (LGM, approximately 20,000 years ago). The horizontal and vertical coordinates are logarithmically scaled. (d) A proposed scenario of global spread routes of B. dorsalis. Arrows in different colors denote the proposed independent dispersal routes of B. dorsalis from India toward other areas. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The three features of the preliminarily inferred structures were special interest. First, the populations of group CN were subdivided into two subclusters: populations distributed in coastal areas and those populations distributed in southwestern and central areas. The Hawaii clade nested in the subcluster consisted of coastal individuals, implying that the origin population that invaded Hawaii could be sourced from the coastal areas of China. This was consistent with the ancestry estimation under K = 5, in which high-level genetic admixture was detected between groups CN and HW. Compared to SA, SSA, NSA and AF, HW had a lower FST score with CN, further supporting this hypothesis (Fig. 2f). Second, the populations distributed in SSA also showed a clear stepwise invasion and spread route from NSA to Thailand, then to Malaysia and the Philippines, Indonesia, and Papua New Guinea. This may have been due to the limited gene flow between island populations and between island and mainland populations, owing to the geographic water barrier. Third, the phylogenetic tree further recovered four main clusters: SA, AF, NSA + CN + HW, and SSA, whereas the ancestry estimation under K = 4 was divided into four clusters: AF, SA + NSA + CN, HW, and SSA. One possible cause of the conflict may have been that the absence of geographic isolation among groups SSA, NSA, SA, and CN facilitated the repetitive invasions of different lineages and subsequent gene flow. Most individuals in the four groups exhibited a high-level admixture detected under K = 3–8. In addition, we detected greater similarity in the genetic background and existence of gene flow among groups SA, NSA, SSA, and CN, as revealed by the PCA. An extremely low level of differentiation was also found among the CN, NSA and SA groups (FST < 0.01), which further testified to the limited differentiation within these three groups despite their frequent gene flow (Fig. 2f). This corresponded with the high π values in SA (0.1155), CN (0.1174), NSA (0.1148), and SSA (0.1120) compared to the lower π values in AF (0.0917) and HW (0.0778) populations (Fig. 2f). Alternatively, the results of the Hawaii and Africa groups could have been due to two independent invasion events with different genetic sources and separate founder events: that is, from South Asia to China to Hawaii over at least 100 years ago and from South Asia to Africa in the 21st century. This was further supported by the PCA in which the AF and HW groups were positioned on opposite sides of the SA group. This was in agreement with the hypothesis presented by Qin et al. regarding multiple global pathway scenarios under the ABC framework [9]. These fine separations between genetically closed groups demonstrated a high level of resolution in our study.
The LD decay patterns with physical distances between the SNPs of the HW and AF groups decreased sharply compared to that in the other groups (Fig. 2e), suggesting that the HW and AF groups had a significant reduction in genetic diversity and underwent a more severe bottleneck during their invasion history. We also observed distinct demographic trajectories for AF and HW compared to the other populations, of which the AF and HW showed recent bottlenecks at a time scale of <100 years.
We then reconstructed the evolutionary scenario and divergence times along the invasion histories for the different groups of B. dorsalis using SMC++ (Fig. 3c). We found that all of the groups shared similar and multiple substantial demographic fluctuations throughout history, which may have led to the variant pattern of B. dorsalis. The most ancient bottleneck event occurred during the last glacial maximum (LGM) period in all groups and suffered a sharp decrease in population size. The NSA, SSA, and CN groups diverged from a common ancestor at a similar time approximately 1,000 years ago, whereas the HW group manifested a later divergence time when splitting off from Asian ancestral populations. The second bottleneck may have occurred at the beginning of the invasion of new regions for each group, owing to the founder effect. B. dorsalis displayed a strong pre- or post-introduction adaptation as it could rapidly establish stable populations and increase the population size after the two main previously mentioned bottleneck events, which mainly contributed to its frequent gene flow, high reproductive and biotic potential, and broad host range.
Based on the whole-genome resequencing data analysis, we found that India was most probably the ancestral origin of B. dorsalis. With Southern India being the center, three distinct and subsequent independent invasion routes are also speculated: (i) from Northern India to Northern Southeast Asia, then to Southern Southeast Asia, (ii) from Northern India to Northern Southeast Asian, then to China and Hawaii, and (iii) from Southern India toward the African mainland, and then to Madagascar (Fig. 3d).
Identification of the candidate genes associated with local thermal adaptation
B. dorsalis has a strong adaptability to newly invaded environments and rapidly expands its population size. The GWAS was undertaken with 11 temperature bioclimatic variables (Tables S23 and S24) under five different models, of which the expedited mixed linear model with Q-matrix as incorporating covariates (EMMAX + Q) fitted the best (Fig. 4, and Figs. S17–S27). After Bonferroni correction, the associated SNP numbers ranged from 298 (in Bio6) to 854 (in Bio3), and 1,192 SNPs were significantly associated with the 11 bioclimatic variables. The Gene Ontology (GO) enrichment analysis of the 27 candidate genes showed significant functional representations in the categories of cellular processes (14/27), cellular anatomical entities (13/27), catalytic activities (11/27), and metabolic processes (11/27) (Fig. S28). The KEGG pathway analysis revealed enrichment in the xenobiotic biodegradation and metabolism, lipid metabolism, and digestive system pathways (Fig. S29). Cyp6a9 was the most pleiotropic candidate gene involved in environmental adaptation, which was associated with 10 of the 11 temperature bioclimatic variables (Fig.4, Tables S25 and S26).
Functional validation of Cyp6a9 in thermal adaptation in B. dorsalis
The fold change in the group subjected to the 38 °C hardening treatment (1.416 ± 0.385) was 1.42 times that in the control group (1.000 ± 0.399), suggesting the expression level of B. dorsalis Cyp6a9 could significantly increase after exposure to the heat hardening temperatures (P = 0.039) (Fig. 5b). After exposure to dsCyp6a9 solution at 1 μg/µl for 96 h, the mRNA expression level of Cyp6a9 (0.590 ± 0.279) was significantly reduced by 0.59 times in the 3rd early instar larvae stage of B. dorsalis (P = 0.015), compared to the control dsGFP group (1.000 ± 0.042) (Fig. 5c). After the heat hardening treatment and heat stress treatment, the survival rate of dsCyp6a9-feeding group, dsGFP-feeding group, and ddH2O-feeding group was 65.40 % ± 3.85 %, 73.60 % ± 6.66 %, and 72.40 % ± 4.16 %, respectively. Compared to the negative control (P = 0.025) and blank control (P = 0.049), the survival rate significantly decreased after dsCyp6a9 exposure (Fig. 5d), suggesting that constitutively expressed Cyp6a9 could be beneficial to the survival of B. dorsalis under heat stress. The expression of Cyp6a9 plays an important role in the heat hardening of B. dorsalis, which can enhance the tolerance and increase the survival rate under extremely high temperatures.
Discussion
The presence of species complexes that cannot be adequately resolved by morphological or molecular characteristics leads to increased trade barriers and a lower efficiency of sterile insect technique (SIT) application [5]. In this study, a population structure analysis, based on genomics data, successfully supported the assertion that B. dorsalis and B. carambolae are two separate phylogenetic species that remain unresolved by fragmentary mitochondrial and nuclear genes (EF-1α, COI and period) [51], which could be in an incipient process of speciation that may or may not end up in two different species in the future. During recent or ongoing speciation, restricted markers have led to conflicting or misleading phylogenetic resolutions due to incomplete lineage sorting, introgression, and limited speciation time to accumulate fixed and adequate interspecific differences [52]. Multilocus sequencing data, such as genome-wide SNPs, can provide unprecedented and accurate insights into species delimitation and speciation. In addition, hybrids were found in the Myanmar and Indonesian populations of B. dorsalis by an ancestry estimation under any K value. Substantial gene flow was also found in the sympatric regions (Indonesia and Thailand) inferred from microsatellite DNA data [53], which further demonstrated the incomplete reproductive isolation between the two species in the wild. Given the sympatric distribution of B. dorsalis and B. carambolae in Indonesia, it is not surprising that hybrid individuals were found in the Indonesian population of B. dorsalis. However, B. carambolae has not been reported in Myanmar [54]. Considering that B. carambolae was already present in neighboring countries, such as Bangladesh and Thailand (the distribution points were located near the borders with Myanmar) [54], the hybridized individuals possibly disperse from these neighboring countries or the lack of geographical isolation allowed B. carambolae to disperse to Myanmar and mate with B. dorsalis. Aketarawong et al. [6] and Qin et al. [9] also previously found that the Myanmar population of B. dorsalis was unique and demonstrated low genetic diversity and high differentiation with all of the other Asian populations, as revealed by microsatellites. They speculated that geographic barriers resulted in the lack of long-distance fruit trade and gene flow between the Myanmar population and neighboring countries. Considering the hybridization evidence found in the present study, we speculate that their previous studies may have used hybrid individuals for the analysis to explain the low genetic diversity and high differentiation in Myanmar.
The ancestral origin and genetic structure of B. dorsalis have been the focus of research in recent years. Previous studies have explored the genetic structure of B. dorsalis at different geographic scales using microsatellites and fragmentary genes to determine the ancestral origin of the species. By providing integrative evidence based on whole-genome resequencing data of 428 B. dorsalis samples from 50 geographic populations, we identified the origin of B. dorsalis as India, with three independent spread routes around the world. Although an initial taxonomic record does not necessarily reflect a presumed origin [11], our results match the first record of B. dorsalis from “East India” (India orientali) under the synonymous name of Musca ferruginea by Fabricius in 1794 [11]. This hypothesis is further in accordance with that of Qin et al. [9] who speculated that South Asia (India and Bangladesh) is the most likely origin of B. dorsalis rather than Southeast Asia [8] or mainland China [6], [7]. Before clarifying the first record to be East India [11], the generally accepted origin of B. dorsalis was Taiwan province, China, where it was first detected in 1912 [11]. This misled previous demographic analysis. For example, Aketarawong et al. [6] used an approximate Bayesian computation analysis under the incorrect definition of scenarios, resulting in opposite invasion routes from Taiwan province to mainland China, and then to Southeast Asia between 1918 and 2000. This study did not consider that B. dorsalis was widely distributed in South and Southeast Asia before 1912 [11]. In addition, the limited sampling representation, which has mainly been focused on populations from Southeast Asia and China in previous studies, could not reveal the actual invasion history of B. dorsalis under the incomplete framework. However, the present study and that conducted by Qin et al. [9], which used a set of populations that better represent the global distribution of B. dorsalis, may contribute to a comprehensive understanding of the invasion history of B. dorsalis.
The recent expansion of B. dorsalis has been associated with human activities, such as trade and colonization. According to the divergence times estimated by SMC++, the SA, NSA, SSA, and CN groups diverged from a common ancestor approximately 1,000 years ago. This time period is associated with the ancient Maritime Silk Road during the rule of the Song Dynasty. The development of shipbuilding and navigation technology promoted communication and trade with South Asia and Southeast Asia, which further facilitated the spread and colonization of B. dorsalis in Asia. The HW group manifested a later divergence time when splitting from the Asian ancestral population approximately 100 years ago. Based on the findings that the Hawaiian population originated from China, we speculate that the invasion of Hawaii was associated with large-scale labor migration from the coastal areas of China for Hawaiian development in the 20th century. This speculated history is consistent with the first detection of B. dorsalis in Hawaii in 1945.
Geographical distances and dispersal limitation is an important driver of population genetic differentiation in nature [55]. Populations exhibit increasing genetic divergence as the geographic distance increasing. In contrast, high genetic diversity and frequent gene flow were observed in all four Asian groups, which is similar to previous studies [6], [7], [8], [9]. The high dispersal ability of B. dorsalis may have contributed to gene flow in regions without geographic isolation. For example, the frequent gene flow detected between populations caught on the Chinese border and populations caught in nearby countries (Laos, Thailand, and Vietnam), can be attributed to B. dorsalis being able to move and mate freely, if its host plants are continuously distributed throughout these areas. The polyphagous habits of B. dorsalis may facilitate gene flow and repeated introduction driven by frequent trades of fruits and vegetables. Previous studies have shown that gene flow, mediated by repeated introduction and admixture, provides an adaptive advantage for invasive species to overcome environmental constraints [56]. In contrast, restricted gene flow and reduced genetic diversity result in higher vulnerability to rapid environmental change [57]. This study suggests that long-distance dispersal and repeated introductions by humans promote the rapid spread and adaptation of invasive species to the local environment rapidly.
Emerging research indicates that host plant adaptation is a prominent factor that drives the shaping of genome-wide patterns in genetic differentiation, especially in polyphagous and oligophagous insects (e.g. Frankliniella occidentalis and Pseudatomoscelis seriatus) and is referred to as host-associated genetic differentiation [58]. Changes in host preference is presumed to raise assortative mating and consequently creates a barrier to gene flow. Among tephritids, host‐related restrictions to gene flow and associated elevation in genetic differentiation was detected among local Prunus and Lonicera populations of Rhagoletis cerasi [59]. Furthermore, Wan et al. [60] experimentally demonstrated that genetic differentiation between lab populations of B. dorsalis can occur after 20 generations when the host plant was shifted from banana to navel orange compared to when the host species remained the same. Due to the limitation of our sampling method which obtained from pheromone scented traps in open land and thus cannot tell their host plants by observation. While the host plant adaptation may play a prominent role in forging genomic diversities and thus contribute to genetic differentiation, we suppose further studies are needed to take characteristics of host plant into consideration to provide a better insight into the invasive route and evolutionary history of B. dorsalis.
During the invasion process, species have to adapt to their new environment through genetically evolutionary changes, phenotypic plasticity, or a combination of these two mechanisms [61]. A wide host range, high reproductive potential, adaptability to environmental stress, insecticide resistance, and immune priming most likely greatly contribute to the rapid adaptive capacity of B. dorsalis. Jiang et al. [62] identified several gene families that were associated with environmental adaptation by comparative genomic analysis, among which, genes encoding heat shock proteins (Hsps), mitogen-activated protein kinases (MAPKs), chemosensory receptors, and cytochrome P450 monooxygenase (CYP450s). For host preferences, Rh6 was found to play a uniquely essential role in the vision-mediated host preference, which was highly selected by the classic specialist species Bactrocera minax, rather than the derived generalist species B. dorsalis [63]. For immune regulation, Yao et al. [64] found that PGRP-LB and PGRP-SB establish a protective zone consisting of symbiotic bacteria colonies by diminishing Imd-pathway activation, and PGRP-LC and AMPs in the foregut allow increased antibacterial peptide production to efficiently filter the entry of pathogens, protecting the symbiotic bacteria. Additionally, epigenetic modifications can also contribute to adaptation responses underlying adaptive phenotypic variation and adaptive evolution. For example, numerous genes associated with the wing development and muscle energy supply were modified by H3K4me3 and H3K27me3 in B. dorsalis compared to D. melanogaster, implying the role of histone modification in facilitating phenotypic change during insect development as well as invasive range expansion [65]. In the present study, the GWAS analysis showed that Cyp6a9 was the most pleiotropic candidate gene involved in thermal adaptation. The P450 CYP6 family is specific to insects [66] and comprises a variety of enzymes that play vital roles in detoxifying numerous xenobiotic chemicals, including plant secondary metabolites (for example, larval adaptation to the unripened citrus fruits in Bactrocera minax [67]) and insecticide metabolism and resistance (for example, dichlorodiphenyltrichloroethane, phenobarbital and caffeine in D. melanogaster [68], and malathion, and beta-cypermethrin in B. dorsalis [69]). The P450 CYP6 family is also related to the thermal stress response, including acute stress (for example, cold stress in Solenopsis japonica [70] and heat stress in Monochamus alternatus [71]) and long-term stress (for example, the upregulated expression of CYP6s has been shown to aid the overwintering of Laodelphax striatellus at high latitudes [72]). Suffering from temperature stress can activate stress-responsive signal transduction pathways to regulate intracellular oxidative stress and cellular toxic substance metabolism [73]. CYP6a17 affects the temperature preference behavior (TPB) of D. melanogaster which is an important target of cAMP-dependent protein kinase signaling that mediates TPB in mushroom bodies [74]. In this study, Cyp6a9 is also proved that be associated with the adaptation to thermal temperatures in B. dorsalis. Interestingly, it is also positively associated with pupal diapause in B. minax [75]. Members of the P450 CYP6 family have further been found to be involved in diapause in Drosophila montana [76] and B. dorsalis [77], although the mechanisms remain unknown. Further study could improve our understanding of the mechanisms and regulatory roles of the P450 CYP6 family in the environmental adaptation and invasive process.
Dupuis et al. [78] filtrated 28 highly informative SNPs that could trace the geographic source of Anastrepha ludens, and succeeded in tracing back the intercepted specimens from Texas and California to Mexico. Popa-Báez et al. [79] also developed reference datasets of genome-wide markers for B. tryoni and found that the recent incursions into Tasmania and South Australia originated from the east coast of Australia. As climate change facilitates the invasion of B. dorsalis into other continents, B. dorsalis was intercepted in Italy in 2018, which was the first record of B. dorsalis occurring in a European orchard [80], although the origin of the population remains unknown. In this study, we built a global variant datasets of B. dorsalis which will be a valuable resource for extracting geography-specific SNPs. Accurate and rapid origin tracing of monitored samples at ports of entry or in the field can provide scientific evidence for the maintenance or further elaboration of phytosanitary protocols. Furthermore, it can aid in efficient decision-making (including quarantine restrictions, surveillance, and pest management response) in terms of allocation of funds and resources and target trade relationships at risk of importing pest species via fruit trade.
Conclusion
In summary, we assembled a high-quality chromosome-level genome of B. dorsalis as reference genome. Based on 512 resequencing accessions, we supported that B. dorsalis and B. carambolae as separated phylogenetic species and clarified that B. dorsalis originates from the Southern India region with three independent invasion and spread routes worldwide, which is mainly facilitated by human activities including trade and immigration. This study provides insights into the evolutionary history and invasion routes in B. dorsalis in detail as first time. Further, the Cyp6a9 gene was the most pleiotropic candidate gene involved in environmental adaptation, which was associated with 10 of the 11 temperature bioclimatic variables. Thus, we found the novel environmental adaptation mechanism, involving the P450 family, of insects. Further research on the mechanisms and regulatory roles of the Cyp6a9 gene could further reveal the environmental adaptation during invasion and establishment process. In addition, we built a global variant datasets of B. dorsalis, which will be a valuable resource for extracting geography-specific SNPs. Accurate and rapid origin-tracing of intercepted samples at ports or in the field can facilitate the decision-making in the prevention and mitigation of the invasive B. dorsalis.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Yue Zhang: Methodology, Data curation, Formal analysis, Software, Visualization, Writing-original draft, Resources. Shanlin Liu: Methodology, Supervision, Writing-original draft, Software, Validation. Marc De Meyer: Supervision, Writing-review & editing, Validation, Resources. Zuxing Liao: Methodology, Software, Visualization. Yan Zhao: Methodology, Software, Visualization. Massimiliano Virgilio: Supervision, Writing review & editing, Validation, Resources. Shiqian Feng: Methodology, Software, Supervision, Formal analysis. Yujia Qin: Writing-review & editing, Software, Supervision, Formal analysis. Sandeep Singh: Writing-review & editing, Validation, Resources. Suk Ling Wee: Writing-review & editing, Validation, Resources. Fan Jiang: Writing-review & editing, Validation. Shaokun Guo: Writing-review & editing, Validation. Hu Li: Writing review & editing, Validation. Pablo Deschepper: Writing-review & editing, Validation, Methodology. Sam Vanbergen: Writing-review & editing, Validation, Methodology. Hélène Delatte: Writing-review & editing, Validation, Methodology. Alies van Sauers-Muller: Validation, Resources. Tati Suryati Syamsudin: Validation, Resources. Anastasia Priscilla Kawi: Validation, Resources. Muo Kasina: Validation, Resources. Kemo Badji: Validation, Resources. Fazal Said: Validation, Resources. Lijun Liu: Validation, Writing-review & editing. Zihua Zhao: Validation, Writing-review & editing. Zhihong Li: Methodology, Investigation, Supervision, Writing review & editing, Conceptualization, Validation, Funding acquisition, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We are thankful to Dr. Yubing Huang from Taiwan Agricultural Research Institute, Dr. Ratovonomenjanahary Zelin Tefiarisoa from Direction de la Protection des Végétaux, Mr. Shine Shane Naing from Plant Quarantine Section Myanmar, Dr. Md. Shibly Noman, Dr. Xiaoliang Wang, Dr. Hao Li, Mr. Delin Kang, Mr. Jixiang Cui, Miss Yangming Lan, Mrs Jing Wei, Miss Guocai Lu, Miss Yan Zhao, Miss Yun Su and Miss Zhiying Zhou from China Agricultural University, Dr. Qianqian Yang from China Jiliang University, Dr. Yi Yang from Chinese Academy of Tropical Agricultural Sciences, Mr. Wengang Lv from Guangzhou Customs, Mrs Yan Zhang from Beijing Ecoman Biotechnology Co., LTD to provide related samples for us. We also thank Dr. Ying Wang and Dr. Xudong Zhang from Genek for the guidance on bioinformatics analysis.
This work was financially supported by National Natural Science Foundation of China (31972341) and this manuscript has been written within the collaborative framework of the EU H2020 FF-IPM project (818184).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.12.012.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Paini D.R., Sheppard A.W., Cook D.C., et al. Global threat to agriculture from invasive species. Proc Natl Acad Sci USA. 2016;113:7575–7579. doi: 10.1073/pnas.1602205113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White I.M., Elson-Harris M.M. CABI; Wallingford, UK: 1992. Fruit flies of economic significance: their identification and bionomics. [Google Scholar]
- 3.Zeng Y.Y., Reddy G.V.P., Li Z.H., et al. Global distribution and invasion pattern of oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae) J Appl Entomol. 2019;143:165–176. [Google Scholar]
- 4.Duyck P.F., David P., Quilici S. A review of relationships between interspecific competition and invasions in fruit flies (Diptera: Tephritidae) Ecol Entomol. 2004;29:511–520. [Google Scholar]
- 5.Clarke A.R. Biology and management of Bactrocera and related fruit flies. CABI; 2019. pp. 129–149. [Google Scholar]
- 6.Aketarawong N., Bonizzoni M., Thanaphum S., et al. Inferences on the population structure and colonization process of the invasive oriental fruit fly, Bactrocera dorsalis (Hendel) Mol Ecol. 2007;16:3522–3532. doi: 10.1111/j.1365-294X.2007.03409.x. [DOI] [PubMed] [Google Scholar]
- 7.Wan X.W., Nardi F., Zhang B., et al. The Oriental fruit fly, Bactrocera dorsalis, in China: origin and gradual inland range expansion associated with population growth. PLoS ONE. 2011;6:e25238. doi: 10.1371/journal.pone.0025238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y.L., Wu Y., Chen H., et al. Population structure and colonization of Bactrocera dorsalis (Diptera: Tephritidae) in China, inferred from mtDNA COI sequences. J Appl Entomol. 2012;136:241–251. [Google Scholar]
- 9.Qin Y.J., Krosch M.N., Schutze M.K., et al. Population structure of a global agricultural invasive pest, Bactrocera dorsalis (Diptera: Tephritidae) Evol Appl. 2018;11:1990–2003. doi: 10.1111/eva.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wee S.L., Tan K.H. Evidence of natural hybridization between two sympatric sibling species of Bactrocera dorsalis complex based on pheromone analysis. J Chem Ecol. 2005;31:845–858. doi: 10.1007/s10886-005-3548-6. [DOI] [PubMed] [Google Scholar]
- 11.Clarke A.R., Li Z.H., Qin Y.J., et al. Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) is not invasive through Asia: it's been there all along. J Appl Entomol. 2019;143:797–801. [Google Scholar]
- 12.Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vurture G.W., Sedlazeck F.J., Nattestad M., et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., Zhou Y., Chen Y., et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koren S., Walenz B.P., Berlin K., et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;7:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan J., Li H. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 2020;17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv e-prints; 2013.
- 18.Walker B.J., Abeel T., Shea T., et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pryszcz L.P., Gabaldón T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 2016;44:e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand N.C., Shamim M.S., Machol I., et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seppey M., Manni M., Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness. In: Kollmar M, editor. Gene prediction. New York, USA: Springer; 2019. p. 227–45. [DOI] [PubMed]
- 22.Wang Y.P., Tang H.B., Debarry J.D., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandi V.K. University of Saskatchewan; 2020. SynVisio: a multiscale tool to explore genomic conservation. Doctoral dissertation. [Google Scholar]
- 24.Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:1–14. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 26.Han M.V., Thomas G.W., Lugo-Martinez J., et al. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A., Hanna M., Banks E., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Retief J.D. Bioinformatics methods and protocols. Humana Press; Totowa, NJ: 2000. Phylogenetic analysis using PHYLIP; pp. 243–258. [Google Scholar]
- 31.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell J., Pritchard J. Inference of population splits and mixtures from genome-wide allele frequency data. Nat Preced. 2012;1:1. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., Dong S.S., Xu J.Y., et al. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- 34.Catchen J., Hohenlohe P.A., Bassham S., et al. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terhorst J., Kamm J.A., Song Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49:303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haag-Liautard C., Dorris M., Maside X., et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 37.Kang H.M., Sul J.H., Service S.K., et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradbury P.J., Zhang Z.W., Kroon D.E., et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 39.Fick S.E., Hijmans R.J. WorldClim 2: new 1km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–4315. [Google Scholar]
- 40.Hu J.T., Chen B., Li Z.H. Thermal plasticity is related to the hardening response of heat shock protein expression in two Bactrocera fruit flies. J Insect Physiol. 2014;67:105–113. doi: 10.1016/j.jinsphys.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Gu X.Y., Zhao Y., Su Y. A transcriptional and functional analysis of heat hardening in two invasive fruit fly species, Bactrocera dorsalis and Bactrocera correcta. Evol Appl. 2019;12:1147–1163. doi: 10.1111/eva.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meccariello A., Salvemini M., Primo P., et al. Maleness-on-the-Y (MoY) orchestrates male sex determination in major agricultural fruit fly pests. Science. 2019;365:1457–1460. doi: 10.1126/science.aax1318. [DOI] [PubMed] [Google Scholar]
- 43.Zheng W.P. Huazhong Agricultural University; 2019. Establishment of Crisper/Cas9 system and function research of Y-link gene spermless in Bactrocera dorsalis. PhD dissertation. [Google Scholar]
- 44.Chang C.H., Larracuente A.M. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics. 2019;211 doi: 10.1534/genetics.118.301765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrasco D., Larsson M.C., Anderson P. Insect host plant selection in complex environments. Curr Opin Insect Sci. 2015;8:1–7. doi: 10.1016/j.cois.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 46.Visser J.H. Host odor perception in phytophagous insects. Annu Rev Entomol. 1986;31:121–144. [Google Scholar]
- 47.Li X., Schuler M.A., Berenbaum M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 48.Xiao H., Ye X., Xu H., et al. The genetic adaptations of fall armyworm Spodoptera frugiperda facilitated its rapid global dispersal and invasion. Mol Ecol Resour. 2020;20:1050–1068. doi: 10.1111/1755-0998.13182. [DOI] [PubMed] [Google Scholar]
- 49.Miller W.K.M, Bracewell R.R., Eisen M.B., et al. Patterns of genome-wide diversity and population structure in the Drosophila athabasca species complex. Mol Biol Evol. 2017;34:1912–1923. doi: 10.1093/molbev/msx134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L.J., Li B.Y., Chen J.C., et al. Local climate adaptation and gene flow in the native range of two co-occurring fruit moths with contrasting invasiveness. Mol Ecol. 2021;30:4204–4219. doi: 10.1111/mec.16055. [DOI] [PubMed] [Google Scholar]
- 51.San Jose M., Leblanc L., Geib S.M., et al. An evaluation of the species status of Bactrocera invadens and the systematics of the Bactrocera dorsalis (Diptera: Tephritidae) complex. Ann Entomol Soc Am. 2013;106:684–694. [Google Scholar]
- 52.Weiss M., Weigand H., Weigand A.M., et al. Genome-wide single-nucleotide polymorphism data reveal cryptic species within cryptic freshwater snail species-the case of the Ancylus fluviatilis species complex. Ecol Evol. 2017;8:1063–1072. doi: 10.1002/ece3.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aketarawong N., Guglielmino C.R., Karam N., et al. The oriental fruitfly Bactrocera dorsalis s.s. in East Asia: disentangling the different forces promoting the invasion and shaping the genetic make-up of populations. Genetica. 2014;142:201–213. doi: 10.1007/s10709-014-9767-4. [DOI] [PubMed] [Google Scholar]
- 54.Leblanc L., Hossain M.A., Doorenweerd C., et al. Six years of fruit fly surveys in Bangladesh: a new species, 33 new country records and discovery of the highly invasive Bactrocera carambolae (Diptera, Tephritidae) ZooKeys. 2019;876:87. doi: 10.3897/zookeys.876.38096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orsini L., Vanoverbeke J., Swillen I., et al. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol. 2013;22(24):5983–5999. doi: 10.1111/mec.12561. [DOI] [PubMed] [Google Scholar]
- 56.Smith A.L., Hodkinson T.R., Villellas J., et al. Global gene flow releases invasive plants from environmental constraints on genetic diversity. Proc Natl Acad Sci USA. 2020;117:4218–4227. doi: 10.1073/pnas.1915848117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas L., Kennington W.J., Evans R.D., et al. Restricted gene flow and local adaptation highlight the vulnerability of high-latitude reefs to rapid environmental change. Global Change Biol. 2017;23:2197–2205. doi: 10.1111/gcb.13639. [DOI] [PubMed] [Google Scholar]
- 58.Evans L.M., Allan G.J., Meneses N., et al. Herbivore host-associated genetic differentiation depends on the scale of plant genetic variation examined. Evol Ecol. 2013;27(1):65–81. [Google Scholar]
- 59.Bakovic V., Schuler H., Schebeck M., et al. Host plant-related genomic differentiation in the European cherry fruit fly, Rhagoletis cerasi. Mol Ecol. 2019;28(20):4648–4666. doi: 10.1111/mec.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan X.W., Liu Y.H., Luo L.M., et al. Influence of host shift on genetic differentiation of the oriental fruit fly, Bactrocera dorsalis. J Integr Agr. 2014;13(12):2701–2708. [Google Scholar]
- 61.Gibert P., Hill M., Pascual M., et al. Drosophila as models to understand the adaptive process during invasion. Biol Invasions. 2016;18:1089–1103. [Google Scholar]
- 62.Jiang F., Liang L., Wang J., et al. Chromosome-level genome assembly of Bactrocera dorsalis reveals its adaptation and invasion mechanisms. Commun Biol. 2022;5(1):1–11. doi: 10.1038/s42003-021-02966-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Fang G., Xu P., et al. Behavioral and genomic divergence between a generalist and a specialist fly. Cell Rep. 2022;41(7) doi: 10.1016/j.celrep.2022.111654. [DOI] [PubMed] [Google Scholar]
- 64.Yao Z., Cai Z., Ma Q., et al. Compartmentalized PGRP expression along the dipteran Bactrocera dorsalis gut forms a zone of protection for symbiotic bacteria. Cell Rep. 2022;41(3) doi: 10.1016/j.celrep.2022.111523. [DOI] [PubMed] [Google Scholar]
- 65.Zhao Y., Hu J., Wu J., et al. Epigenetic signature of invasiveness: ChIP-seq profiling of H3K4me3 and H3K27me3 in an invasive insect. Authorea Preprints. 2022 [Google Scholar]
- 66.Scott J.G. Cytochromes P450 and insecticide resistance. Insect Biochem Molec. 1999;29:757–777. doi: 10.1016/s0965-1748(99)00038-7. [DOI] [PubMed] [Google Scholar]
- 67.Zhang G.J., Xu P.H., Wang Y.H., et al. New insights into the biological interaction between unripe citrus fruits and the tephritid fly Bactrocera minax based on omics. (2021). [Preprint (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-556841/v1].
- 68.Morra R., Kuruganti S., Lam V., et al. Functional analysis of the cis-acting elements responsible for the induction of the Cyp6a8 and Cyp6g1 genes of Drosophila melanogaster by DDT, phenobarbital and caffeine. Insect Mol Biol. 2010;19:121–130. doi: 10.1111/j.1365-2583.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- 69.Huang Y., Shen G.M., Jiang H.B., et al. Multiple P450 genes: identification, tissue-specific expression and their responses to insecticide treatments in the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidea) Pestic Biochem Phys. 2013;106:1–7. [Google Scholar]
- 70.Vatanparast M., Park Y. Comparative RNA-seq analyses of Solenopsis japonica (Hymenoptera: Formicidae) reveal gene in response to cold stress. Genes. 2021;12:1610. doi: 10.3390/genes12101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., Zhao X.Y., Qiao H., et al. Comparative transcriptome analysis of the heat stress response in Monochamus alternatus Hope (Coleoptera: Cerambycidae) Front Physiol. 2020;10:1568. doi: 10.3389/fphys.2019.01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang H.J., Xue J., Zhuo J.C., et al. Comparative analysis of the transcriptional responses to low and high temperatures in three rice planthopper species. Mol Ecol. 2017;26:2726–2737. doi: 10.1111/mec.14067. [DOI] [PubMed] [Google Scholar]
- 73.Xiong Y., Liu X.Q., Xiao P.A., et al. Comparative transcriptome analysis reveals differentially expressed genes in the Asian citrus psyllid (Diaphorina citri) upon heat shock. Comp Biochem Phys D. 2019;30:256–261. doi: 10.1016/j.cbd.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 74.Kang J., Kim J., Choi K.W. Novel cytochrome P450, cyp6a17, is required for temperature preference behavior in Drosophila. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0029800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong Y.C., Desneux N., Lei C., et al. Transcriptome characterization analysis of Bactrocera minax and new insights into its pupal diapause development with gene expression analysis. Int J Biol Sci. 2014;10:1051. doi: 10.7150/ijbs.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kankare M., Parker D.J., Merisalo M., et al. Transcriptional differences between diapausing and non-diapausing D. montana females reared under the same photoperiod and temperature. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0161852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen E.H., Hou Q.L., Dou W., et al. RNA-seq analysis of gene expression changes during pupariation in Bactrocera dorsalis (Hendel) (Diptera: Tephritidae) BMC Genomics. 2018;19:1–16. doi: 10.1186/s12864-018-5077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupuis J.R., Ruiz-Arce R., Barr N.B., et al. Range-wide population genomics of the Mexican fruit fly: toward development of pathway analysis tools. Evol Appl. 2019;12:1641–1660. doi: 10.1111/eva.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Popa-Báez Á., Lee S.F., Yeap H.L., et al. Tracing the origins of recent Queensland fruit fly incursions into South Australia, Tasmania and New Zealand. Biol Invasions. 2021;23:1117–1130. [Google Scholar]
- 80.Nugnes F., Russo E., Viggiani G., et al. First record of an invasive fruit fly belonging to Bactrocera dorsalis complex (Diptera: Tephritidae) in Europe. Insects. 2018;9:182. doi: 10.3390/insects9040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.