Graphical abstract
Obacunone (OB) targets macrophage migration inhibitory factor (MIF) to impede osteoclastogenesis and alleviate osteoporosis.
Keywords: Obacunone, Macrophage migration inhibitory factor (MIF), Osteoclast, Reactive oxygen species (ROS), Bone loss
Highlights
-
•
Obacunone (OB) effectively inhibits osteoclast formation and function in vitro.
-
•
OB attenuates RANKL-induced osteoclast differentiation signaling pathways.
-
•
Macrophage migration inhibitory factor (MIF) is identified as the molecular target of OB.
-
•
OB alleviates estrogen deficiency-induced bone loss by impeding excessive osteoclast activity.
-
•
OB may serve as an alternative therapeutic candidate for osteoporosis.
Abstract
Introduction
Osteoporosis is the most common bone disorder where the hyperactive osteoclasts represent the leading role during the pathogenesis. Targeting hyperactive osteoclasts is currently the primary therapeutic strategy. However, concerns about the long-term efficacy and side effects of current frontline treatments persist. Alternative therapeutic agents are still needed.
Objectives
Obacunone (OB) is a small molecule with a broad spectrum of biological activities, particularly antioxidant and anti-inflammatory effects. This study aims to examine OB’s therapeutic potential on osteoporosis and explore the rudimentary mechanisms.
Methods
Osteoclast formation and osteoclastic resorption assays were carried out to examine OB’s inhibitory effects in vitro, followed by the in-vivo studies of OB’s therapeutic effects on ovariectomy-induced osteoporotic preclinical model. To further study the underlying mechanisms, mRNA sequencing and analysis were used to investigate the changes of downstream pathways. The molecular targets of OB were predicted, and in-silico docking analysis was performed. Ligand-target binding was verified by surface plasmon resonance (SPR) assay and Western Blotting assay.
Results
The results indicated that OB suppressed the formation of osteoclast and its resorptive function in vitro. Mechanistically, OB interacts with macrophage migration inhibitory factor (MIF) which attenuates receptor activator of nuclear factor kappa B (NF-κB) ligand (RANKL)-induced signaling pathways, including reactive oxygen species (ROS), NF-κB pathway, and mitogen-activated protein kinases (MAPKs). These effects eventually caused the diminished expression level of the master transcriptional factor of osteoclastogenesis, nuclear factor of activated T cells 1 (NFATc1), and its downstream osteoclast-specific proteins. Furthermore, our data revealed that OB alleviated estrogen deficiency-induced osteoporosis by targeting MIF and thus inhibiting hyperactive osteoclasts in vivo.
Conclusion
These results together implicated that OB may represent as a therapeutic candidate for bone disorders caused by osteoclasts, such as osteoporosis.
Introduction
As a self-renewal connective tissue, bone undergoes a continuous remodeling process, including osteoblastic bone deposition and osteoclastic bone removal. The precise coordination of bone resorption and formation is crucial for maintaining bone homeostasis in physiological conditions [1], [2]. Postmenopausal osteoporosis is known as the most common skeletal disorder that is featured by the imbalanced activity between osteoblast and osteoclast. The osteoclast is known as the major type of bone-resorbing cell, playing a leading role in estrogen deficiency-induced bone loss [3], [4], [5]. The list of agents targeting aberrant osteoclasts has been remarkably expanding over the past decades; however, the enormous and growing prevalence of osteoporosis and osteoporotic fractures worldwide remains challenging for healthcare workers [6]. The search for novel alternative therapies is still urgently needed due to the ‘off-target’ effects of current treatments [7], [8].
Multinucleated osteoclasts differentiate from the macrophage/monocyte lineage following the stimulations of two indispensable factors, receptor activator of nuclear factor kappa B (NF-κB) ligand (RANKL) and macrophage-colony stimulating factor (M-CSF) [9], [10]. Upon binding to RANK, RANKL initiates multiple downstream signaling pathways, including mitogen-activated protein kinases (MAPKs) and NF-κB pathways, eventually triggering the expression of the nuclear factor of activated T cells 1 (NFATc1) [9]. Embryonic stem cells lacking NFATc1 expression failed to differentiate into mature osteoclasts under RANKL’s stimulation; however, the ectopic expression of NFATc1 can induce osteoclast formation and function without RANKL [11]. Therefore, NFATc1 acts as a master transcriptional factor during RANKL-induced osteoclastogenesis. Accumulating studies also suggested that reactive oxygen species (ROS) are essential for osteoclastogenesis by facilitating RANKL-induced signaling [12], [13]. Besides, osteoclast differentiation is also regulated by macrophage migration inhibitory factor (MIF) via NF-κB and NFATc1 transcriptional activation in vitro and bone erosion in vivo [14].
Traditional herbal medicine has a long-standing role in the treatment and prevention of bone diseases [15], [16]. Obacunone (OB) is a small molecule originally from citrus fruits and noted with a wide range of pharmacological activities. For instance, it was identified that OB was able to trigger the activity of nuclear factor erythroid 2-related factor 2 (Nrf2) and protect against oxidative stress-related lung fibrosis [17]. By modulating gut microbiota and NF-κB signaling, OB ameliorated ulcerative colitis [18]. Besides, OB also exhibited anti-cancer [19], [20] and anti-inflammation effects [21] by affecting the MAPK pathway. More intriguingly, OB was recently also found to enhance osteoblastic function and might be promising for osteoporosis treatment [22]. Nevertheless, how OB affects osteoclast activity remained unknown. Given these osteoclast-related pathways were effectively suppressed by OB, we hypothesized that OB could impede osteoclastogenesis and thus alleviate osteoporosis. In this study, our data implicated that OB inhibited RANKL-induced osteoclast generation in vitro. Furthermore, we found that OB prevented estrogen deficiency-induced osteoporosis, as indicated in an ovariectomy (OVX) preclinical mice model. Mechanistically, MIF was predicted and validated as the critical target that OB interacts with during osteoclast formation. MIF was recently identified as closely related to osteoclast-related bone disorders and is thought to be a potential therapeutic target [23], [24]. Hence, we proposed that OB may target MIF to attenuate RANKL-induced downstream signaling in osteoclast differentiation, thereby protecting against osteoporosis.
Materials and methods
Reagents
OB, obtained from MUST BIO-TECHNOLOGY CO., Ltd (Sichuan, China), was prepared with DMSO at a stock concentration of 100 mM. Further working concentrations were diluted in the culture medium accordingly. For the animal experiment, OB was firstly dissolved in DMSO at a concentration of 120 mg/mL and further diluted in saline prior to injections. DMSO of the same dilution was used as the vehicle control in this study. Fetal bovine serum (FBS), DAPI, Rhodamine Phalloidin, a-MEM, H2DCFDA (2′,7′-dichlorodihydrofluorescein diacetate), and ProLong Gold Antifade Mountant were purchased from Thermo Fisher Scientific (Waltham, MA, US). Luciferase analysis kit and MTS assay kit were obtained from Promega (Madison, WI, US). Antibody for vinculin was purchased from Sigma-Aldrich (St. Louis, MO, US).Primary antibodies for NFATc1, V-ATPase-d2, Integrin αV, cathepsin K, actin, IκB-α, phosphor(p)-ERK, ERK, p-P38, and P38 were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Primary antibodies for HO-1, catalase, JNK, p-JNK, p-P65, P65, and c-Fos were obtained from Cell Signaling Technology (Danvers, MA, USA). Antibody for MIF and recombinant MIF protein were purchased from Abcam (Cambridge, United Kingdom). Recombinant M-CSF was from R&D Systems (Minneapolis, MIN, USA) and we prepared recombinant glutathione S-transferase (GST)-RANKL proteins as previously described [32]. Assay Kits for Blood Urea Nitrogen (BUN) (Cat. No. C013-2-1), Creatinine (CRE) (Cat. No. C011-2-1), Alanine Aminotransferase (ALT) (Cat. No. C009-2-1), Aspartate Aminotransferase (AST) (Cat. No. C010-2-1), Glutathione (GSH) (Cat. No. A006-2-1) were purchased from Jiancheng Bioengineering Institute (Nanjing, China). Malondialdehyde (MDA) Assay Kit (Cat. No. S0131) was purchased from Beyotime Biotechnology (Shanghai, China). Mouse MIF ELISA kit (Cat. No. KE10027) was purchased from Proteintech (Wuhan, China).
Osteoclast differentiating from macrophages in vitro
Primary mice bone marrow macrophages (BMMs) were isolated according to the ethics policies and procedures approved by the Animal Ethics Committee of The University of Western Australia (RA/3/100/1601). Mice long bones were used to flush out the bone marrow, which was then incubated in complete α-MEM containing both 10 % FBS and M-CSF at 50 ng/mL till cell confluency. 6 × 103 BMMs were seeded into each well of a 96-well plate for osteoclastogenesis assay. We treated the cells with RANKL (50 ng/mL) to induce differentiation, with the supplement of OB at the following concentrations: 0, 1, 5, 10, and 20 μM. The fresh medium was exchanged every two days to allow the formation of multinucleated cells. We fixed the cells for osteoclast identification using tartrate-resistant acid phosphatase (TRAP) staining. The multinucleated cell with 3 or more nuclei was counted as an osteoclast.
MTS assay
MTS assay was carried out to examine whether OB exhibits cytotoxicity on BMMs. We isolated primary BMMs and seeded the cells into 96-well plates to allow adherence overnight. OB at different working concentrations (1, 5, 10, and 20 μM) were then added to the cells for 48 h. We then added 10 μL MTS solution into each well and incubated it for 2 h. Plates were inserted into a BMG LABTECH microplate reader (Ortenberg, Germany) to measure the absorbance (490 nm) and obtain the readouts of the optical density (OD).
Podosome belt staining
To visualize the podosome belts, we fixed the mature osteoclasts with 4 % paraformaldehyde (PFA). Permeabilization was completed using 0.1 % (v/v) Triton X-100. This was followed by blocking which was performed in 3 % bovine serum albumin (BSA). Next, the cells were processed for the incubation with primary antibody against vinculin. Next, a secondary antibody (Alexa Fluor 488) and Rhodamine Phalloidin were used to treat the cells for 60 mins. DAPI was used to stain the nuclei for 10 mins. The osteoclast podosome belts were imaged using NIKON A1Si confocal microscope (Minato, Tokyo, Japan).
Hydroxyapatite resorption assay
To examine whether OB inhibits osteoclastic bone resorption, we performed the hydroxyapatite resorption assay as previously reported [25], [26]. In brief, BMMs were stimulated with RANKL till osteoclasts began to form. Cells were then separated from the plate using cell dissociation solution (Sigma-Aldrich, St. Louis, MO, US). Next, we re-seeded the cells into a 96-well plate coated with hydroxyapatite (Corning, NY, US) in the differentiation medium with or without OB for an additional 48 h. Half of the cells in each group were bleached and left air-dry to visualize the resorbed area. Cells in the rest of wells were fixed and performed with TRAP staining. The average resorbed area per well and per osteoclast were quantified.
Western blot assay
We incubated the cells with or without OB for the time as indicated, followed by cell lysis in radioimmunoprecipitation assay (RIPA). Proteins were first separated using SDS-polyacrylamide gel electrophoresis (SDS-PAGE). We then transferred the protein in the gel to a GE Healthcare nitrocellulose membrane (Chicago, IL, US). Next, the blocking of membrane was performed in 5 % skim milk (in TBST) for one hour. The primary antibodies were incubated with the membrane at 4 °C till the next day. Following 1xTBST washing for three times, the membrane was incubated using a secondary antibody conjugated by horseradish peroxidase (HRP) for 1 h at room temperature. To show the protein bands, the membrane was treated with PerkinElmer chemiluminescence substrate (Waltham, MA, US) and imaged using an GE Healthcare Image-quant LAS 4000. To quantitively evaluate the protein expression levels, the band intensities were analyzed using ImageJ.
mRNA sequencing and analysis
Primary BMMs were isolated and then treated with 50 ng/mL RANKL. OB (20 μM) was added or not in the culture medium for 24 h. Total RNA extraction was carried out using a QIAGEN Extraction Kit (QIAGEN, Hilden, Germany). The RNA purity was assessed using the kaiaoK5500 Spectrophotometer (Beijing, China). Meanwhile, RNA integrity was examined by the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) using Nano 6000 Assay Kit. Before fragmentation, mRNA was purified using poly-T oligo-attached magnetic. Next, QiaQuick PCR kit was used for library construction of cDNA and HiSeq PE Cluster Kit v4-cBot-HS (Illumina) was used for Library clustering, which was followed by an Illumina platform sequencing and the generation of 150 bp paired-end reads. Clean data was filtered from raw data and then mapped to reference genome using Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) [27]. Reads Count was counted and Fragments Per Kilobase Millon Mapped Reads (FPKM) [28] was then analyzed. DESeq2 [29] was used to analyse, and Wald test was used to calculate the p-value. Differentially expressed genes (DEGs) were defined by the adjusted p-value < 0.05 and |log2FoldChange| > 1. DEGs enrichment in different pathways were visualized using KEGG (Kyoto Encyclopedia of Genes and Genomes, https://www.kegg.jp/) analysis [30].
Intracellular ROS detection
We examined the intracellular ROS level as described previously [25]. Briefly, following RANKL’s treatment with or without OB, 5 mM H2DCFDA in Hank's balanced salt solution was added to cells for 60 mins. Upon oxidation, H2DCFDA is transformed to 2′,7′-dichlorofluorescein (DCF) which has fluorescence and can be detected under a NIKON A1Si confocal microscope. In the meantime, cells were imaged under the digital interference contrast (DIC). ROS-positive area was analyzed (Image J software).
Transcriptional activity of NF-κB
RAW264.7 cells, stably transfected with an NF-κB responsive luciferase construct, were used to assess NF-κB activity at the transcriptional level [31]. Cells (1.5 × 105 per well) were plated into a 48-well plate. Next, OB of different concentrations was added to pre-treat the cells for 1 h before the RANKL (50 ng/mL) stimulation. After 6 h-incubation, cell lysate was prepared using luciferase reporter assay kit and then measured using a BMG LABTECH microplate reader.
Target prediction and molecular docking
To further estimate the potential macromolecular targets of OB, a ligand-based target prediction process was carried out by using a web-based tool – SwissTargetPrediction (https://www.swisstargetprediction.ch/) [32]. The Simplified Molecular Input Line Entry System (SMILES) of OB was obtained and input to initiate the computation process. Following submission, a list of protein targets was presented and ranked according to the probability. The most likely target was selected for further investigations. To determine the binding mode of OB to the protein target, molecular docking was carried out in PyRx Virtual Screening Tool [33] using the Autodock Vina algorithm [34]. The MIF’s 3D crystal structure was acquired from Protein Data Bank (PDB) [35] (PDB code 3DJH) and the SDF file of OB was retrieved from PubChem [36]. Both structures were converted to PDBQT format prior to molecular docking. The protein structure was prepared for docking by adding hydrogens, removing the water molecules, and minimizing energy. A grid box centering on the active site residues was selected for docking. The results was visualized on UCSF Chimera [37]. A 2D interaction of protein and OB was generated in BIOVIA Discovery Studio Visualizer (2020, Dassault Systèmes). The pose with the lowest energy was presented and the potential interactive residues were also displayed.
Surface plasmon resonance (SPR) assay
To evaluate the binding affinity between OB and recombinant MIF protein, SPR was carried out in the study. OB was dissolved in DMSO and printed using a BioDot™ AD-1520 Array Printer (Irvine, CA, USA) on a label-free Photo-cross-linker SensorCHIP™ (BetterWays Inc., China). Next, sample was evaporated in a nitrogen atmosphere and then immediately proceeded for photo-cross-linking reaction using a UV spectroirradiator. UV-excited azo radicals attack the C—H bonds, which established the connections to the surface of chip via hydrogen substitution reactions. The real-time SPR experiment was conducted in a Label-free Microarray System (bScreen LB 991, BERTHOLD TECHNOLOGIES GmbH & Co. KG). HBS-EP running buffer was prepared as the following: 10 mM HEPES, 150 mM NaCl, 0.005 % (v/v) P20 surfactant, and 3 mM EDTA. The buffer was firstly primed through the chip surface for 3 times (2 μL·s−1, 40 s). Next, 1 × PBS containing 5 % DMSO was primed through. MIF protein was prepared using running buffer at various concentrations at 300 nM, 600 nM, 1200 nM, 2400 nM, and 4800 nM. The small molecule and protein interaction was determined during the association phase for 600 s (flow rate = 0.5 μL·s−1), and dissociation phase for 360 s.
Ethics statement
The ethical policies and experimental procedures with mice were approved by the Institutional Animal Care and Use Committee of Guangzhou University of Chinese Medicine (TCMF1-2020031).
Mice ovariectomy (OVX) model
An osteoporotic mice model was established as we previously reported [25]. The C57BL/6J female mice (11-week-old) were randomly assigned to three groups as following (n = 10 per group): sham group with only vehicle administration, OVX group with vehicle administration, and OVX group with OB administration. They were housed in animal facility (temperature 22 °C ± 2 °C, 12-hour dark/light cycle). After a successful intraperitoneal anesthetization with pentobarbital sodium, the mice in the OVX group were subjected to bilateral ovariectomy; in the control group, mice were only exposed the ovaries but without resecting. Following 1-week recovery, mice were injected intraperitoneally with saline (containing ∼ 41 μL /kg DMSO) as control or 5 mg/kg OB every-two days for six weeks. All mice were euthanized at the endpoint of the study, and the femur and lumber samples were dissected for bone histomorphometry analysis. To determine whether OB has toxicity in vivo, blood was collected for liver and kidney function test. Meanwhile, liver and kidney were also collected, fixed, paraffin embedded, and sectioned for histological analysis. To assess the MIF expression in vivo, blood MIF level was measured by ELISA kit and bone proteins were also extracted for WB assays as described above.
Micro-CT scanning and analysis
To evaluate the bone microstructure in different groups, long bones (femurs) and lumbar vertebrates (L5) were fixed (10 % neutral buffered formalin, 24 h) prior to scanning using a Bruker micro-CT (Skyscan 1176, Kontich, Belgium). The scanning settings were as the following: a pixel size of 9 μm, a source current of 500 μA, a source voltage of 50 kV, an AI filter of 0.5 mm, and a rotation step of 0.4°. The scanning images were reconstructed using NRecon software (Bruker micro-CT, Kontich, Belgium). The trabecular bone within the range of 0.5–1.5 mm over the femur distal growth plate and trabecular bone between vertebral endplate were selected as the volume of interest. To quantitively compare the difference of bone parameters between groups, we performed the data analysis using Bruker CTAn software (Kontich, Belgium). The parameters including, bone volume per tissue volume (BV/TV), connective denstity (Conn.Dn), trabecular separation (Tb.Sp) and trabecular number (Tb.N) were presented in this study.
Bone histomorphometry analysis
For decalcification, the bone samples were incubated in ethylenediaminetetraacetic acid (EDTA) (14 %, PH = 7.4) at 37 °C, with EDTA solution being replaced every day till the bone becomes soft. After decalcification, we used an automatic tissue processor (TP-1020, Leica Microsystems, Germany) to make the samples go through ethanol series for dehydration. Paraffin was used to embed the bone samples which were then sectioned using a Leica Microsystems Microtome (Leica RM 2035 Biocut, Wetzlar, Germany). Bone sections (5-μm-thickness) were collected for picro sirius red (PSR) staining and TRAP staining, followed by the imaging using a microscopy. Bone histomorphometry analysis was achieved using Image J software (NIH, US).
Statistical analysis
Mean ± standard deviation (SD) was displayed in all bar graphs. Bar graphs as well as data analysis were conducted using GraphPad Prism 6.0 software. Two-tailed Student’s t test was used to assess the differences between two groups. The differences among three or more groups were examined by the One-way ANOVA analysis. A P value<0.05 was considered as a significant difference.
Results
OB’s suppressive effects on RANKL-induced osteoclast formation in vitro
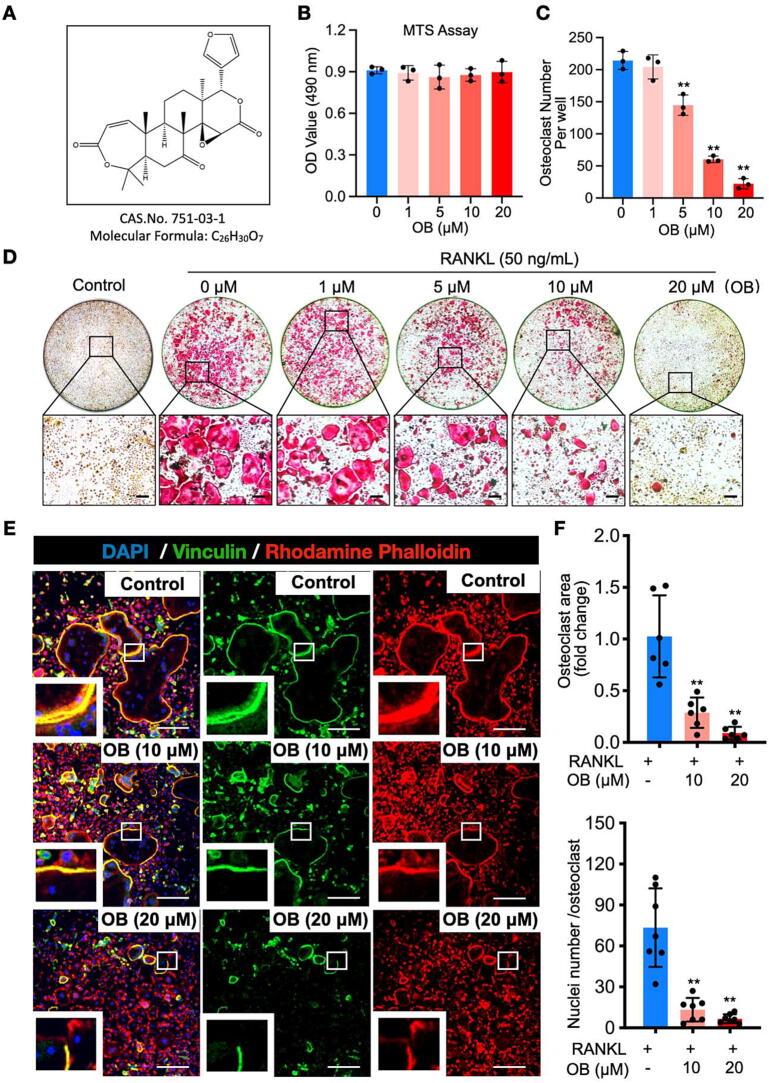
To determine whether OB (Fig. 1A) has cytotoxicity, the MTS assay was performed in BMMs following a 48-h treatment of OB at various concentrations. The optical density (OD) readouts revealed that OB, at the concentrations applied in this study, has little effect on BMMs proliferation (Fig. 1B). Next, we assessed OB’s inhibitory effects on osteoclast differentiation. Primary BMMs were seeded and induced differentiation with the stimulations of RANKL and M‐CSF. OB was added into the culture medium at various concentrations (0, 1, 5, 10, 20 µM). TRAP staining indicated that a large number of osteoclasts formed without OB treatment, while the addition of OB could dose-dependently inhibit osteoclastogenesis, with the least and most diminutive formation of osteoclasts at 20 µM of OB (Fig. 1C and D).
Fig. 1.
OB represses osteoclast formation induced by RANKL in vitro. (A) OB’s molecular formula and chemical structure. (B) The viability of BMMs following the 48-h treatment of OB as determined by the MTS assay (n = 3). (C) The number of TRAP-positive osteoclasts per well with or without the addition of OB at the indicated concentrations (n = 3). (D) Representative whole-well view of osteoclasts stained by TRAP. (E) Osteoclasts podosome belts are stained by rhodamine phalloidin (red), anti-vinculin (green), and DAPI (blue). (F) Quantitative analyses of osteoclast area (n = 6) and the number of nuclei per osteoclast (n = 7). Scale bar = 200 µm. Bar graphs are displayed as mean ± SD. *p < 0.05, **p < 0.01 compared with the control group without OB treatment. BMMs: bone marrow macrophages; OB: obacunone; RANKL: receptor activator of nuclear factor‐κB ligand; TRAP: tartrate-resistant acid phosphatase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The mature osteoclasts were incubated with both anti-vinculin and Rhodamine Phalloidin to investigate the effects of OB on the morphological changes of podosome belt formation (Fig. 1E). The podosome belt is the signature structure responsible for osteoclast function. We observed that well‐defined podosome belts formed in the “RANKL-only” group, characterized by a larger area and more nucleus on average for each cell than OB-treated groups (Fig. 1F). Next, we aimed to examine the most affected stage during osteoclast differentiation by OB treatment. A time-course based osteoclastogenesis assay was performed by exposing the cells to 20 µM OB at early (Day 1–3), middle (Day 3–5), or late (Day 5–6) stage of osteoclast formation. TRAP staining revealed that the osteoclast formation was disturbed following OB treatment at all stages, with the most significant impact at the early stage (Fig. S1). Taken together, we revealed that OB could significantly inhibit RANKL‐induced osteoclastogenesis in vitro without showing apparent cytotoxicity.
OB inhibits osteoclast resorptive function and suppresses osteoclast-specific makers
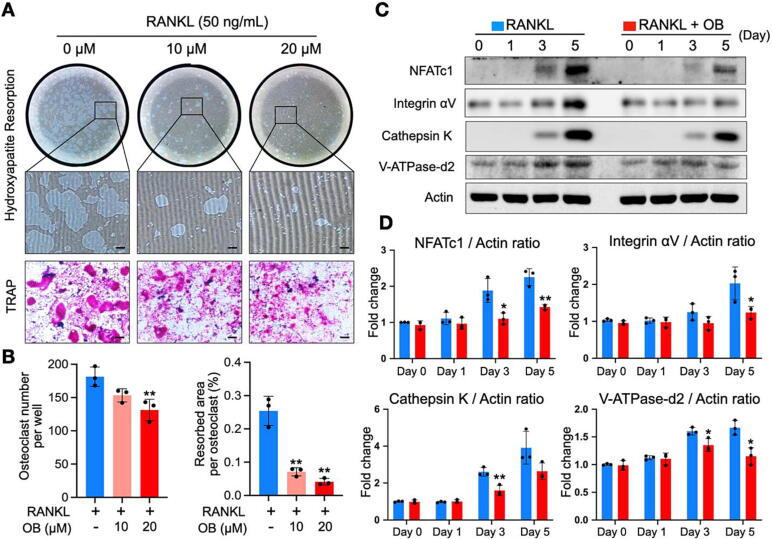
Given OB’s inhibitory effects on osteoclast formation, we next sought to determine whether OB affects the function of osteoclasts by using a hydroxyapatite resorption assay. The OB treatment could consistently inhibit osteoclastogenesis at the late stage with a smaller number of osteoclasts, as indicated by TRAP staining (Fig. 2A and B). Intriguingly, the resorption area per osteoclast was reduced substantially following the addition of OB at 10 and 20 µM (Fig. 2A and B), indicating the inhibition of OB on osteoclastic resorptive function. Meanwhile, we used western blot assays to determine the protein expression levels which were related to osteoclast formation and function. Our data indicated that OB significantly inhibited NFATc1 protein expression, particularly on day 3 and day 5 (Fig. 2C and D). The impaired NFATc1 expression caused the comprehensive downregulation of the downstream markers, including cathepsin K, integrin αV, and V‐ATPase‐d2 (Fig. 2C and D). Thus, OB has a suppressive effect on osteoclastogenesis and osteoclastic hydroxyapatite resorptive function through the NFATc1 signaling pathway.
Fig. 2.
OB inhibits osteoclastic resorption by suppressing osteoclast-specific proteins. (A) Representative whole-well images showing the osteoclastic hydroxyapatite resorption following OB treatment and osteoclast formation in the same group. (B) Quantitative analyses of osteoclast number in each well and averaged resorbed area of each osteoclast (n = 3). (C) Representative western blot images of OB’s effects on the expressions of NFATc1, integrin αV, cathepsin K, V-ATPase-d2, and actin during osteoclastogenesis. (D) Quantitative analyses of the relative band intensities of NFATc1, integrin αV, cathepsin K, V-ATPase-d2 to actin (n = 3). Bar graphs are displayed as mean ± SD. *p < 0.05, **p < 0.01 compared with the control group without OB treatment. Scale bar = 200 µm. NFATc1: nuclear factor of activated T cells 1; OB: obacunone; RANKL: receptor activator of nuclear factor‐κB ligand; TRAP: tartrate-resistant acid phosphatase.
OB alters RANKL-induced gene expression profiling in BMMs
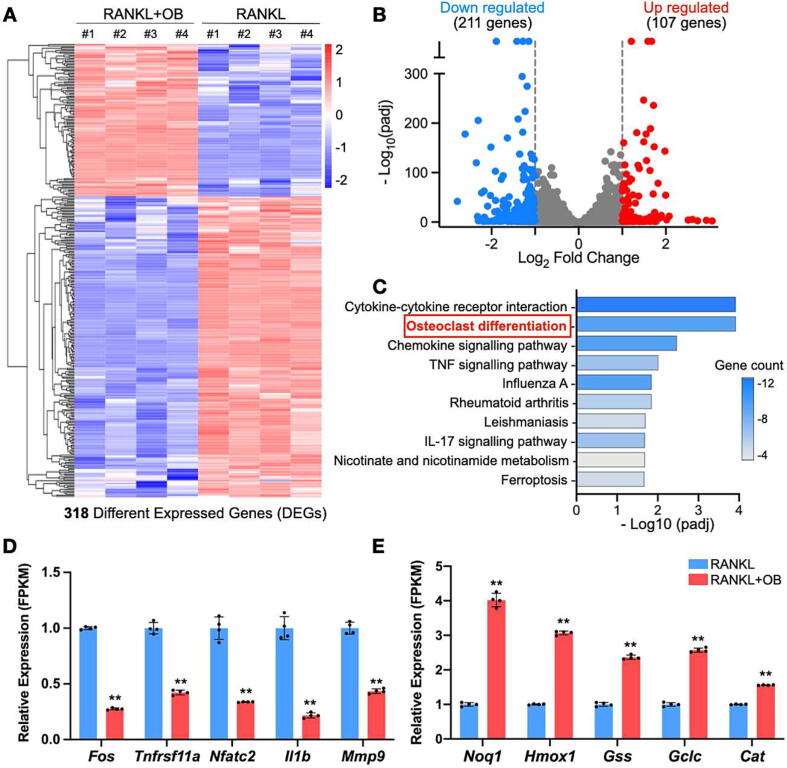
To explore the mechanisms underlying the impaired osteoclastogenesis, RNAseq was performed on RANKL-treated BMMs to profile the gene expression with or without the addition of OB. Given that OB predominantly affects osteoclast differentiation at early stage (Fig. S1), we treated the RANKL-induced BMMs with or without OB for 48 h prior to RNAseq. We found 318 differentially expressed genes (DEGs) in the cells following OB’s treatment (Fig. 3A, adjusted p-value < 0.05 and |log2FoldChange| > 1). Specifically, 211 genes were downregulated, and 107 genes were upregulated among these DEGs (Fig. 3B). Intriguingly, the KEGG signaling pathway enrichment analysis indicated that 10 DEGs were enriched in the osteoclast differentiation pathway, ranking one of the top pathways (Fig. 3C, Table S1). Other highly significant pathways, including cytokine-cytokine receptor interaction, chemokine signaling, and TNF signaling pathways (Fig. 3C, Table S1), were also well-known to promote osteoclast differentiation. In particular, our results suggested that pro-osteoclastogenesis genes, including Fos, Tnfrsf11a, Nfatc2, II1b, and Mmp9, were significantly downregulated by the treatment of OB (Fig. 3D). In contrast, the antioxidant genes Noq1, Hmox1, Gss, Gclc, and Cat were upregulated (Fig. 3E). These results align with our observation of OB’s inhibitory effects on osteoclasts and previous studies demonstrating OB’s antioxidant capability. Herein, we hypothesized that OB might affect osteoclasts by reducing ROS production induced by RANKL.
Fig. 3.
OB alters gene expression profiles in BMMs induced by RANKL. (A) Heatmap showing the DEGs between BMMs following a 24-h treatment with RANKL with or without the addition of OB. (B) Volcano-plot showing DEGs. (C) KEGG pathway enrichment analysis on DEGs. Relative gene expression levels of osteoclast-related genes (D) and antioxidant genes (E) indicated as FPKM between the two groups. DEGs were defined by adjusted p-value < 0.05 and |log2FoldChange| > 1. **p < 0.01 compared with the group without OB treatment by Student t-test. DEGs: differentially expressed genes; FPKM: fragments per kilobase per million mapped fragments; OB: obacunone; padj: adjusted p-value.
OB reduces RANKL-induced ROS production and NF-κB/MAPK pathway
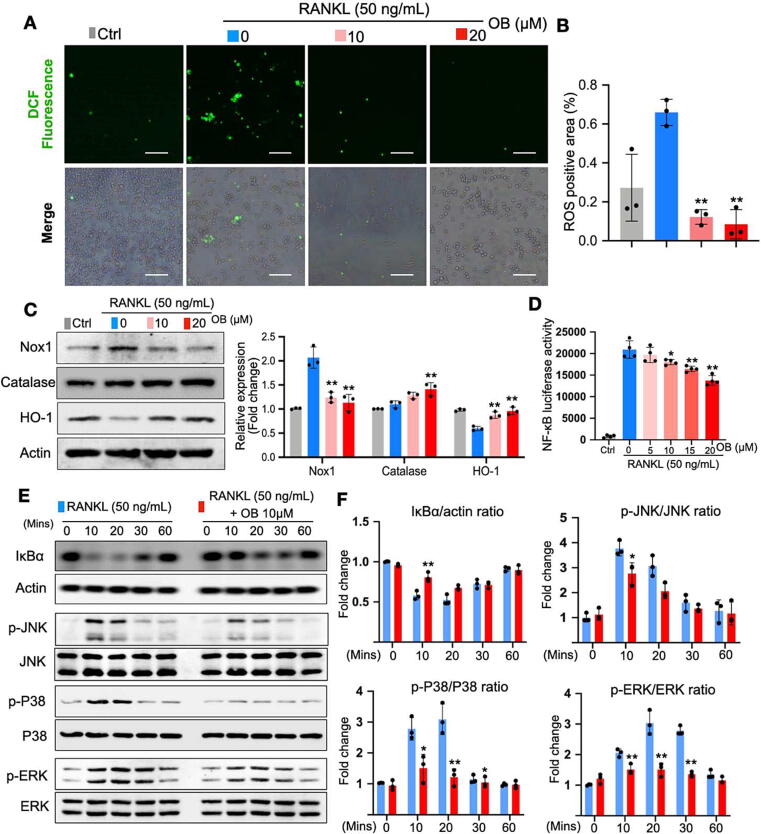
Following the clue of the profiling of antioxidant genes, we further verified OB’s effects on intracellular ROS level during osteoclast differentiation induced by RANKL. Upon the stimulation of RANKL, ROS generation was vastly enhanced as detected by H2DCFDA, a probe that can convert to fluorescent DCF after oxidation (Fig. 4A and B). Remarkably, OB efficiently impeded the DCF fluorescence (Fig. 4A and B). Western blot assay was then implemented to observe the ROS-related protein expressions. The results indicated that Nox-1, a protein that contributes to RANKL-induced ROS generation, was significantly enhanced by RANKL but effectively suppressed by OB treatment (Fig. 4C). Meanwhile, the antioxidant enzymes that scavenge ROS, including catalase and HO-1, were enhanced by OB treatment (Fig. 4C), consistent with RNAseq results above (Fig. 4C).
Fig. 4.
OB reduces RANKL-induced ROS production and NF-κB/MAPK pathways. (A) Representative images of RANKL-induced intracellular ROS generation in BMMs, in which ROS were detected in the form of fluorescent DCF. (B) Quantitative analyses of DCF fluorescence area in each group (n = 3). (C) Representative blotting images and quantifications of protein expressions including Nox1, Catalase, HO-1, and Actin (n = 3). (D) Luciferase assays showing the effect of OB on the activity of NF-κB transcription (n = 3). (E) Representative blotting images of NF-κB and MAPKs related markers, including IκBα, p-ERK, p-P38, and p-JNK. (F) Quantitative analyses of relative band intensities of proteins in (E) to actin, JNK, P38, and ERK (n = 3). *p < 0.05, **p < 0.01 compared with the group without OB treatment. Scale bar = 200 µm. BMMs: bone marrow macrophages; DCF: dichlorofluorescein; OB: obacunone; NOX: nicotinamide adenine dinucleotide phosphate (NADPH) oxidase; RANKL: receptor activator of nuclear factor‐κB ligand; ROS: reactive oxygen species.
NF-κB and MAPK pathways are essential for osteoclastogenesis and regulated by ROS. To this end, we examined OB’s effects on these two pathways. First, a luciferase reporter assay showed that RANKL enhanced the transcriptional activity of NF‐κB, which could be dose-dependently inhibited by OB (Fig. 4D). IκB‐α degrades to release the NF‐κB complex, which causes the activation of downstream signaling. The result of luciferase reporter assay was also supported by western blot, which exhibited that OB hindered the degradation of IκB‐α (Fig. 4E and F). Similarly, for the MAPK pathway, RANKL stimulated the phosphorylation of JNK, ERK, and P38, which peaked after 10 or 20 min, while the OB treatment could significantly hamper the phosphorylation of these proteins (Fig. 4E and F). Therefore, these results suggested that OB effectively inhibits osteoclast formation and bone resorption function via ROS, NF‐κB, and MAPK signaling pathways.
OB interacts with MIF to inhibit RANKL-induced osteoclastogenesis
To identify the probable targets that OB may interact with, we performed a prediction process via a webtool-SwissTargetPrediction. In total, 102 macromolecular targets were output as a list with varying probability values calculated from a combination of 2D and 3D similarity measures with known molecules [38]. Among all the predictions, MIF ranked the first in the target class of enzyme and remained top 3 of the whole list (Table S2), suggesting it may be highly involved in OB’s interaction in the context of osteoclasts. Indeed, our result was consistent with a study that previously predicated the protein targets of OB using Similarity Ensemble Approach (SEA) Search Server [21]. Growing evidence also suggested that MIF mediated osteoclast-related bone disorders [24], [39] and MIF inhibition efficiently alleviated the bone loss by targeting osteoclasts [23]. Thus, we hypothesized that MIF might be a key molecular target of OB when it interacts with osteoclasts. Next, to further explore the possible binding mode between OB and MIF, we performed the molecular docking using Autodock Vina. We showed that OB fits in the N-terminal pocket of the Pro-1 active site (Fig. 5A). The other vital residues include PHE-113, ILE-64, SER-63, TYR-36, and LYS-32 (Fig. 5A).
Fig. 5.
OB targets RANKL-induced signaling by binding to MIF. (A) In Silico docking analysis of OB in the active pocket of MIF (PDB code 3DJH). The potential interactive residues were displayed in 3D and 2D. (B) Surface plasmon resonance (SPR) assays showing the binding of OB to MIF protein. (C) Representative blotting images showing OB’s effect on MIF and its related osteoclast markers following the treatment of RANKL in BMMs. BMMs: bone marrow macrophages; MIF, macrophage migration inhibitory factor; OB: obacunone; RANKL: receptor activator of nuclear factor‐κB ligand.
To validate the binding affinity of OB to MIF, we further performed a SPR assay. OB was bound on a sensor chip surface where the recombinant MIF of different concentrations was primed through. The result indicated OB has a strong binding affinity to MIF protein and the binding signal increased with a higher MIF concentration (Fig. 5B). Then we found that OB treatment led to a lower expression of MIF as shown by western blotting assay, accompanied by downregulated signaling including c-Fos and NF-κB-p65 (Fig. 5C). Our data indicated that OB might interact with MIF to reduce its expression and downstream pathways, leading to the impaired osteoclastogenesis.
OB prevents ovariectomized (OVX)-induced bone loss and shows no drug toxicity
To examine OB’s therapeutic effects in vivo, we established an estrogen deficiency-induced osteoporosis mice model by performing bilateral ovariectomy (Fig. 6A). The mice were administrated intraperitoneally with vehicle or OB for six weeks separately. No fatalities or significant adverse effects were recorded throughout the study. Micro-CT scanning revealed a bone loss in the femurs that caused by OVX procedure (Fig. 6B). However, with OB treatment, the bone loss trend was significantly prevented (Fig. 6B). These changes of bone phenotypes were indicated by bone microstructure parameters including BV/TV, Tb.N, Conn.Dn, and Tb.Sp (Fig. 6C). Furthermore, bone histomorphometry analysis indicated that the hyperactive osteoclasts appeared in the femur following OVX, while OB treatment could inhibit osteoclast activity on the bone surface (Fig. 6D and E). The similar changes of osteoclast activity were also observed in calvaria (Fig. S2). Meanwhile, the bone microstructure of the fifth lumbar vertebrate (L5) was also examined by micro-CT. Consistently, we found a similar bone phenotype change between these groups (Fig. 6F and G). To assess the drug toxicity in vivo, mice body weights in each group were recorded and no significant weight loss was found following OB treatment (Fig. S3). Moreover, blood tests (Fig. 6H) and histological analysis (Fig. S4) showed OB treatment did not alter liver and kidney function and structures as compared with the control group, demonstrating the drug safety when administrated in vivo. In consistent with our in-vitro data, OB treatment can alleviate the oxidative stress caused by ovariectomy as evidenced by the elevated level of GSH (antioxidant) and the declined level of MDA (oxidative stress marker), which were accompanied by the diminished serum MIF level following OB treatment in the OVX mice (Fig. 6H). Moreover, proteins extracted from bone tissues showed MIF, together with osteoclast-related markers (c-Fos, CTSK, and TRAP), increased significantly following ovariectomy which can be reversed by the OB treatment (Fig. 6I). Therefore, our data showed that OB administration could effectively alleviate osteoporosis with little drug toxicity and this could be achieved by targeting MIF.
Fig. 6.
OB prevents OVX-induced bone loss by suppressing osteoclast activity. (A) Schematic illustration of in vivo study of OB’s therapeutic effects. (B) Representative three-dimensional micro-CT images of femur. (C) Quantifications of bone parameters, including BV/TV, Tb.N, Conn.Dn, and Tb.Sp (n = 10). (D) Representative histological images of decalcified femur sections (PSR and TRAP staining). (E) Quantification of osteoclasts number and surface area in bone sections (n = 4). Representative three-dimensional micro-CT images of lumbar vertebrate (L5) (F) and analyses of bone parameters (n = 7) (G). (H) Blood tests showing liver and kidney function, oxidative stress as well as serum MIF level. (I) Representative western blotting images showing the protein expression of osteoclast-specific proteins and MIF protein in the bone tissue. Bar graphs are presented as mean ± SD.*p < 0.05, **p < 0.01 relative to the OVX group. ALT, Alanine Aminotransferase; AST, Aspartate Aminotransferase; BUN, Blood urea nitrogen; BV/TV, bone volume per tissue volume; CRE, Creatinine; Conn.Dn, connectivity density; GSH, Glutathione; CTSK, Cathepsin K; HE, hematoxylin and eosin; MDA, Malondialdehyde; MIF, macrophage migration inhibitory factor; N.Oc/B.Pm, number of osteoclasts per bone perimeter; ns, no significance; OB, obacunone; Oc.S/BS, osteoclast surface per bone surface; OVX, ovariectomized; PSR, picro sirius red; Tb.N, trabecular number; Tb.Sp, trabecular separation; TRAP, tartrate-resistant acid phosphatase. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
Osteoporosis is the most common bone disorder where the hyperactive osteoclasts represent the leading role during the pathogenesis. Thus, targeting excessive osteoclast formation and function is the primary strategy. Despite tremendous advances in therapeutics over the past decades, concerns about current frontline treatments' long-term efficacy and side effects persist. To this end, it is needed to search for alternatives to battle this disease. Given that OB exhibits a broad spectrum of therapeutic activities, we sought to elucidate whether OB could serve as a candidate for osteoporosis treatment.
We first showed that OB could inhibit osteoclast differentiation and resorption in vitro dose-dependently. Intriguingly, this suppressive effect was accompanied by the diminished expression of NFATc1, the master transcriptional factor for osteoclastogenesis [11]. NFATc1 mediates the downstream expression levels of osteoclast-specific proteins [40], [41], including integrin αV, V-ATPase-d2, and cathepsin K. Integrin αV is required for the formation of sealing zone which is essential for osteoclast’s attachment to bone surface. Both V-ATPase-d2 and cathepsin K are required to form an acidified resorptive lacuna. Cathepsin K is secreted through the ruffle border to degrade the bone matrix [42] and V-ATPase-d2 is linked to osteoclast fusion as well as proton transport [43]. Thus, our results indicated that OB impedes osteoclast differentiation by suppressing RANKL-induced NFATc1 activation and its downstream osteoclast-related markers.
RNAseq results further revealed the alterations of transcriptome profile of BMMs affected by OB treatment. The DEGs are significantly enriched in pathways, including osteoclast differentiation, cytokine-cytokine receptor interaction, chemokine signaling, and TNF signaling pathways. Of note, an array of osteoclast-related genes is downregulated, accompanied by the upregulation of a couple of antioxidant genes. These lines of evidence aligned with previous studies in which OB was reported to protect against inflammation [18] and oxidative stress [17]. Herein, we hypothesized that OB might revoke RANKL-induced signaling partly by its antioxidant property. Indeed, our data suggested that OB could effectively decrease RANKL-induced intracellular ROS generation by promoting antioxidant proteins, including catalase and HO-1; however, ROS generator (Nox1) was found suppressed by OB treatment.
Accumulating evidence has demonstrated that ROS are essential for osteoclastogenesis by enhancing NF-κB and MAPK pathways [13], [44]. Upon RANKL stimulation, IκB degradation leads to the release of the NF-κB complex, which then translocates into the nucleus and contributes to NFATc1 activation [45]. Similarly, MAPKs, including ERKs, JNKs, and p38, are also triggered by RANKL to drive osteoclastogenesis [9]. The present study revealed that OB comprehensively mitigated RANKL-induced MAPK and NF-κB signalings which may be partly due to the decreased ROS level. However, it remains elusive whether OB also directly affects these two pathways.
To determine the targets that OB may interact with, we used SwissTargetPrediction system, a web-based tool, to screen out the top-ranking enzyme, MIF. MIF is a pleiotropic pro-inflammatory factor engaged in a broad range of biological and pathological conditions, including sepsis [46], systemic lupus erythematosus, and rheumatoid arthritis (RA) [47]. Interestingly, growing evidence suggested that MIF is crucial in the pathogenesis of osteoclast-associated bone disorders. Gu et al. reported that MIF and its receptor CD74 are required for osteoclastogenesis in vitro via mediating NF-κB-p65 and ERK1/2 phosphorylation [14]. Moreover, MIF deficiency exhibits protection against bone loss caused by OVX [48] or periodontal disease [49], whereas MIF overexpression leads to high-turnover osteoporosis accompanied by elevated levels of matrix metalloproteinases (MMPs) [24]. Intriguingly, MIF produced locally at the bone lytic site supports homing of circulating osteoclast precursors, which can be inhibited by the administration of an anti-MIF neutralizing monoclonal antibody [39]. These findings together implicated that MIF may serve as an attractive therapeutic target. MIF inhibitors also showed promising effects on excessive osteoclast activation in pathological conditions such as wear-particles or OVX induced bone loss [23], [50]. In contrast, Jacquin et al. found that MIF knockout (KO) mice have increased osteoclastogenesis, contributing to a low bone volume [51]. The same group then identified that the deletion of MIF receptor CD74 also displayed a similar bone phenotype [52]. These contradictory results are still challenging to fully elucidate and will require further investigations.
Our data here are more aligned with the previous evidence indicating that MIF is a positive regulator of osteoclasts whereby targeting MIF may suppress osteoclast formation and function. We revealed that OB has a strong binding affinity to recombinant MIF, as indicated by the SPR assay. Moreover, the treatment with OB in BMMs led to a significant and dose-dependent reduction of MIF expression. As an upstream stimulus, MIF was previously indicated to induce NF-κB-p65 signaling [53] and c-Fos [54]. Consistently, we found that the diminished expression of MIF was accompanied by declined levels of c-Fos and phosphorylated NF-κB-p65 in the context of osteoclast differentiation. C-Fos acts as an essential component that activates NFATc1 and thus is required for osteoclastogenesis [45]. P65 is sequestered in the cytoplasm and can translocate into the nucleus to induce the expression of osteoclast-related genes upon RANKL-induced phosphorylation. Meanwhile, P65 also mediates the arrest of cell apoptosis and enhances osteoclast differentiation [55]. Therefore, OB may bind to MIF and abrogate its downstream signaling. As discussed above, OB also significantly suppressed the MAPK pathway. Given that MIF triggers the sustained activation of the MAPK pathway [56], we speculate that OB’s repressive effect on MAPK may be partly due to its binding to MIF. A previous study reported that lipopolysaccharide (LPS) activated p-P38, but not p-JNK, which can be hindered by OB in RAW 264.7 cells [21]. In comparison, it was indicated here that OB ablated both p-P38 and p-JNK in primary BMMs stimulated by RANKL. This difference could be related to the different conditions, including the type of cells and stimulus, that OB was investigated.
Nevertheless, the direct binding of OB to MIF seems not the only cause of the diminished expression of MIF. A couple of studies suggested that intracellular ROS production can trigger the MIF expression [57], [58], [59], [60]. However, it remains unknown whether ROS generation contributes to MIF expression in the context of RANKL-mediated osteoclastogenesis. This study found that OB efficiently blocked ROS, which raised the possibility that OB’s antioxidant effect might also indirectly mediate the downregulation of MIF. Interestingly, MIF was also demonstrated to induce ROS generation in multiple studies. Chuang et al. reported that MIF induces ROS production and autophagy in the hepatoma cell line [61]. Lv et al. showed that MIF activates ROS signaling which is partly responsible for breast cancer metastasis [62]. In contrast, MIF was also found to exhibit a cardioprotective effect by reducing oxidative stress [63], [64]. These contradictory findings may be due to the different experimental settings that were employed. Here, our data suggested that OB has a net inhibitory effect on intracellular ROS signaling, which might attribute to OB’s direct antioxidant property and indirect MIF blockade.
Considering OB’s remarkable suppressive effects on osteoclasts in vitro, we next sought to examine whether OB has a protective impact on the estrogen deficiency-induced osteoporosis preclinical mice model. The removal of the bilateral ovaries is a standardized procedure to study the bone phenotypes changes caused by the estrogen-deficiency. In this study, we found significant bone loss in both femur and lumbar vertebrate following the OVX procedure, which can be well prevented with OB treatment. Bone histomorphometry analysis further mechanistically demonstrated that hyperactive osteoclasts in OVX mice were suppressed when treated with OB. Furthermore, OB treatment is found to alleviate oxidative stress and inhibit MIF expression in vivo, which could lead to the downregulation of osteoclast activities. In the meantime, liver and kidney function tests, along with the histological analysis, demonstrated the drug safety in vivo.
Conclusions
To summarize, our study has elucidated for the first time that OB ameliorated estrogen deficiency-induced bone loss by impeding hyperactive osteoclasts. Mechanistically, the binding of OB to MIF attenuates RANKL-induced signaling, including ROS, MAPK and NF-κB pathways. These effects eventually cause the diminished expression of NFATc1 and osteoclast-related proteins. Therefore, we proposed that OB could serve as a therapeutic candidate for osteoporosis or osteoclast-related bone disorders.
Compliance with ethics requirements
All experimental procedures with mice were approved by the Institutional Animal Care and Use Committee of Guangzhou University of Chinese Medicine (TCMF1-2020031) and used in line with the NIH Guide for the Care and Use of Laboratory Animals.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by National Natural Science Foundation of China (82002279, 81960405, 82104898), Natural Science Foundation of Guangxi Province (2018GXNSFAA050092), Natural Science Foundation of Zhejiang Province (LY20H270015), and Wenzhou Major Science and Technology Project (2018ZY015). We acknowledge the facilities and technical assistance of the Centre for Microscopy, Characterization & Analysis, The University of Western Australia. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.01.003.
Contributor Information
Qian Liu, Email: liuqian@gxmu.edu.cn.
Jiake Xu, Email: jiake.xu@uwa.edu.au.
Kai Chen, Email: kai.chen@uwa.edu.au.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Clarke B. Normal bone anatomy and physiology. Clin J Am Soc Nephrol. 2008;3(Suppl. 3):S131–S139. doi: 10.2215/CJN.04151206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datta H.K., Ng W.F., Walker J.A., Tuck S.P., Varanasi S.S. The cell biology of bone metabolism. J Clin Pathol. 2008;61(5):577–587. doi: 10.1136/jcp.2007.048868. [DOI] [PubMed] [Google Scholar]
- 3.Edwards J.R., Mundy G.R. Advances in osteoclast biology: old findings and new insights from mouse models. Nat Rev Rheumatol. 2011;7(4):235–243. doi: 10.1038/nrrheum.2011.23. [DOI] [PubMed] [Google Scholar]
- 4.Charles J.F., Aliprantis A.O. Osteoclasts: more than ‘bone eaters’. Trends Mol Med. 2014;20(8):449–459. doi: 10.1016/j.molmed.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng X., Teitelbaum S.L. Osteoclasts: new insights. Bone Res. 2013;1(1):11–26. doi: 10.4248/BR201301003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khosla S., Hofbauer L.C. Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol. 2017;5(11):898–907. doi: 10.1016/S2213-8587(17)30188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi H., Chen X., Gao S., Yu X., Xiao J., Zhang B., et al. Key triggers of osteoclast-related diseases and available strategies for targeted therapies: a review. Front Med (Lausanne) 2017;4:234. doi: 10.3389/fmed.2017.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maraka S., Kennel K.A. Bisphosphonates for the prevention and treatment of osteoporosis. BMJ. 2015;351 doi: 10.1136/bmj.h3783. [DOI] [PubMed] [Google Scholar]
- 9.Boyle W.J., Simonet W.S., Lacey D.L. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 10.Levaot N., Ottolenghi A., Mann M., Guterman-Ram G., Kam Z., Geiger B. Osteoclast fusion is initiated by a small subset of RANKL-stimulated monocyte progenitors, which can fuse to RANKL-unstimulated progenitors. Bone. 2015;79:21–28. doi: 10.1016/j.bone.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 12.Kanzaki H., Shinohara F., Kanako I., Yamaguchi Y., Fukaya S., Miyamoto Y., et al. Molecular regulatory mechanisms of osteoclastogenesis through cytoprotective enzymes. Redox Biol. 2016;8:186–191. doi: 10.1016/j.redox.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyeon S., Lee H., Yang Y., Jeong W. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic Biol Med. 2013;65:789–799. doi: 10.1016/j.freeradbiomed.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Gu R., Santos L.L., Ngo D., Fan H., Singh P.P., Fingerle-Rowson G., et al. Macrophage migration inhibitory factor is essential for osteoclastogenic mechanisms in vitro and in vivo mouse model of arthritis. Cytokine. 2015;72(2):135–145. doi: 10.1016/j.cyto.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo H., Yin W., Zou Z., Zhang C., Sun M., Min L., et al. Quercitrin alleviates cartilage extracellular matrix degradation and delays ACLT rat osteoarthritis development: an in vivo and in vitro study. J Adv Res. 2021;28:255–267. doi: 10.1016/j.jare.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J., Li X., Wang Z., Bennett S., Chen K., Xiao Z., et al. Therapeutic anabolic and anticatabolic benefits of natural Chinese medicines for the treatment of osteoporosis. Front Pharmacol. 2019;10:1344. doi: 10.3389/fphar.2019.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu S., Chen W., Xie Q., Xu Y. Obacunone activates the Nrf2-dependent antioxidant responses. Protein Cell. 2016;7(9):684–688. doi: 10.1007/s13238-016-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo X., Yue B., Yu Z., Ren Y., Zhang J., Ren J., et al. Obacunone protects against ulcerative colitis in mice by modulating gut microbiota, attenuating TLR4/NF-kappaB signaling cascades, and improving disrupted epithelial barriers. Front Microbiol. 2020;11:497. doi: 10.3389/fmicb.2020.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J., Zhang A.H., Qiu S., Zhang T.L., Li X.N., Yan G.L., et al. Identification of the perturbed metabolic pathways associating with prostate cancer cells and anticancer affects of obacunone. J Proteomics. 2019;206 doi: 10.1016/j.jprot.2019.103447. [DOI] [PubMed] [Google Scholar]
- 20.Kim J., Jayaprakasha G.K., Patil B.S. Obacunone exhibits anti-proliferative and anti-aromatase activity in vitro by inhibiting the p38 MAPK signaling pathway in MCF-7 human breast adenocarcinoma cells. Biochimie. 2014;105:36–44. doi: 10.1016/j.biochi.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Gao Y., Hou R., Liu F., Liu H., Fei Q., Han Y., et al. Obacunone causes sustained expression of MKP-1 thus inactivating p38 MAPK to suppress pro-inflammatory mediators through intracellular MIF. J Cell Biochem. 2018;119(1):837–849. doi: 10.1002/jcb.26248. [DOI] [PubMed] [Google Scholar]
- 22.Park K.R., Kim S., Cho M., Yun H.M. Limonoid triterpene, obacunone increases runt-related transcription factor 2 to promote osteoblast differentiation and function. Int J Mol Sci. 2021;22(5) doi: 10.3390/ijms22052483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng L., Gao J., Jin K., Chen Z., Yu W., Zhu K., et al. Macrophage migration inhibitory factor (MIF) inhibitor 4-IPP suppresses osteoclast formation and promotes osteoblast differentiation through the inhibition of the NF-kappaB signaling pathway. FASEB J. 2019;33(6):7667–7683. doi: 10.1096/fj.201802364RR. [DOI] [PubMed] [Google Scholar]
- 24.Onodera S., Sasaki S., Ohshima S., Amizuka N., Li M., Udagawa N., et al. Transgenic mice overexpressing macrophage migration inhibitory factor (MIF) exhibit high-turnover osteoporosis. J Bone Miner Res. 2006;21(6):876–885. doi: 10.1359/jbmr.060310. [DOI] [PubMed] [Google Scholar]
- 25.Chen K., Qiu P., Yuan Y., Zheng L., He J., Wang C., et al. Pseurotin A inhibits osteoclastogenesis and prevents ovariectomized-induced bone loss by suppressing reactive oxygen species. Theranostics. 2019;9(6):1634–1650. doi: 10.7150/thno.30206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K., Yuan Y., Wang Z., Song D., Zhao J., Cao Z., et al. Helvolic acid attenuates osteoclast formation and function via suppressing RANKL-induced NFATc1 activation. J Cell Physiol. 2019;234(5):6477–6488. doi: 10.1002/jcp.27385. [DOI] [PubMed] [Google Scholar]
- 27.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Steer J.H., Joyce D.A., Yip K.H., Zheng M.H., Xu J. 12-O-tetradecanoylphorbol-13-acetate (TPA) inhibits osteoclastogenesis by suppressing RANKL-induced NF-kappaB activation. J Bone Miner Res. 2003;18(12):2159–2168. doi: 10.1359/jbmr.2003.18.12.2159. [DOI] [PubMed] [Google Scholar]
- 32.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 34.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burley S.K., Berman H.M., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(Web Server issue):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Movila A., Ishii T., Albassam A., Wisitrasameewong W., Howait M., Yamaguchi T., et al. Macrophage migration inhibitory factor (MIF) supports homing of osteoclast precursors to peripheral osteolytic lesions. J Bone Miner Res. 2016;31(9):1688–1700. doi: 10.1002/jbmr.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song I., Kim J.H., Kim K., Jin H.M., Youn B.U., Kim N. Regulatory mechanism of NFATc1 in RANKL-induced osteoclast activation. FEBS Lett. 2009;583(14):2435–2440. doi: 10.1016/j.febslet.2009.06.047. [DOI] [PubMed] [Google Scholar]
- 41.Feng H., Cheng T., Steer J.H., Joyce D.A., Pavlos N.J., Leong C., et al. Myocyte enhancer factor 2 and microphthalmia-associated transcription factor cooperate with NFATc1 to transactivate the V-ATPase d2 promoter during RANKL-induced osteoclastogenesis. J Biol Chem. 2009;284(21):14667–14676. doi: 10.1074/jbc.M901670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everts V., Korper W., Hoeben K.A., Jansen I.D., Bromme D., Cleutjens K.B., et al. Osteoclastic bone degradation and the role of different cysteine proteinases and matrix metalloproteinases: differences between calvaria and long bone. J Bone Miner Res. 2006;21(9):1399–1408. doi: 10.1359/jbmr.060614. [DOI] [PubMed] [Google Scholar]
- 43.Duan X., Yang S., Zhang L., Yang T. V-ATPases and osteoclasts: ambiguous future of V-ATPases inhibitors in osteoporosis. Theranostics. 2018;8(19):5379–5399. doi: 10.7150/thno.28391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H.S., Nam S.T., Mun S.H., Lee S.K., Kim H.W., Park Y.H., et al. DJ-1 controls bone homeostasis through the regulation of osteoclast differentiation. Nat Commun. 2017;8(1):1519. doi: 10.1038/s41467-017-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202(9):1261–1269. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bozza F.A., Gomes R.N., Japiassu A.M., Soares M., Castro-Faria-Neto H.C., Bozza P.T., et al. Macrophage migration inhibitory factor levels correlate with fatal outcome in sepsis. Shock. 2004;22(4):309–313. doi: 10.1097/01.shk.0000140305.01641.c8. [DOI] [PubMed] [Google Scholar]
- 47.Bilsborrow J.B., Doherty E., Tilstam P.V., Bucala R. Macrophage migration inhibitory factor (MIF) as a therapeutic target for rheumatoid arthritis and systemic lupus erythematosus. Expert Opin Ther Targets. 2019;23(9):733–744. doi: 10.1080/14728222.2019.1656718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oshima S., Onodera S., Amizuka N., Li M., Irie K., Watanabe S., et al. Macrophage migration inhibitory factor-deficient mice are resistant to ovariectomy-induced bone loss. FEBS Lett. 2006;580(5):1251–1256. doi: 10.1016/j.febslet.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 49.Madeira M.F., Queiroz-Junior C.M., Costa G.M., Santos P.C., Silveira E.M., Garlet G.P., et al. MIF induces osteoclast differentiation and contributes to progression of periodontal disease in mice. Microbes Infect. 2012;14(2):198–206. doi: 10.1016/j.micinf.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Yamada C., Ho A., Akkaoui J., Garcia C., Duarte C., Movila A. Glycyrrhizin mitigates inflammatory bone loss and promotes expression of senescence-protective sirtuins in an aging mouse model of periprosthetic osteolysis. Biomed Pharmacother. 2021;138 doi: 10.1016/j.biopha.2021.111503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacquin C., Koczon-Jaremko B., Aguila H.L., Leng L., Bucala R., Kuchel G.A., et al. Macrophage migration inhibitory factor inhibits osteoclastogenesis. Bone. 2009;45(4):640–649. doi: 10.1016/j.bone.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 52.Mun S.H., Won H.Y., Hernandez P., Aguila H.L., Lee S.K. Deletion of CD74, a putative MIF receptor, in mice enhances osteoclastogenesis and decreases bone mass. J Bone Miner Res. 2013;28(4):948–959. doi: 10.1002/jbmr.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim M.J., Kim W.S., Kim D.O., Byun J.E., Huy H., Lee S.Y., et al. Macrophage migration inhibitory factor interacts with thioredoxin-interacting protein and induces NF-kappaB activity. Cell Signal. 2017;34:110–120. doi: 10.1016/j.cellsig.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 54.Onodera S., Kaneda K., Mizue Y., Koyama Y., Fujinaga M., Nishihira J. Macrophage migration inhibitory factor up-regulates expression of matrix metalloproteinases in synovial fibroblasts of rheumatoid arthritis. J Biol Chem. 2000;275(1):444–450. doi: 10.1074/jbc.275.1.444. [DOI] [PubMed] [Google Scholar]
- 55.Vaira S., Alhawagri M., Anwisye I., Kitaura H., Faccio R., Novack D.V. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest. 2008;118(6):2088–2097. doi: 10.1172/JCI33392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell R.A., Metz C.N., Peng T., Bucala R. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J Biol Chem. 1999;274(25):18100–18106. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 57.Zglinska K., Niemiec T., Lozicki A., Matusiewicz M., Szczepaniak J., Puppel K., et al. Effect of Elaeagnus umbellata (Thunb.) fruit extract on H2O2-induced oxidative and inflammatory responses in normal fibroblast cells. PeerJ. 2021;9 doi: 10.7717/peerj.10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J.H., Lee J., Bae S.J., Kim Y., Park B.J., Choi J.W., et al. NADPH oxidase 4 is required for the generation of macrophage migration inhibitory factor and host defense against Toxoplasma gondii infection. Sci Rep. 2017;7(1):6361. doi: 10.1038/s41598-017-06610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cotzomi-Ortega I., Rosas-Cruz A., Ramirez-Ramirez D., Reyes-Leyva J., Rodriguez-Sosa M., Aguilar-Alonso P., et al. Autophagy Inhibition induces the secretion of macrophage migration inhibitory factor (MIF) with autocrine and paracrine effects on the promotion of malignancy in breast cancer. Biology (Basel) 2020;9(1) doi: 10.3390/biology9010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parekh A., Das S., Parida S., Das C.K., Dutta D., Mallick S.K., et al. Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo. Oncogene. 2018;37(33):4546–4561. doi: 10.1038/s41388-018-0272-6. [DOI] [PubMed] [Google Scholar]
- 61.Chuang Y.C., Su W.H., Lei H.Y., Lin Y.S., Liu H.S., Chang C.P., et al. Macrophage migration inhibitory factor induces autophagy via reactive oxygen species generation. PLoS One. 2012;7(5):e37613. doi: 10.1371/journal.pone.0037613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lv W., Chen N., Lin Y., Ma H., Ruan Y., Li Z., et al. Macrophage migration inhibitory factor promotes breast cancer metastasis via activation of HMGB1/TLR4/NF kappa B axis. Cancer Lett. 2016;375(2):245–255. doi: 10.1016/j.canlet.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Koga K., Kenessey A., Powell S.R., Sison C.P., Miller E.J., Ojamaa K. Macrophage migration inhibitory factor provides cardioprotection during ischemia/reperfusion by reducing oxidative stress. Antioxid Redox Signal. 2011;14(7):1191–1202. doi: 10.1089/ars.2010.3163. [DOI] [PubMed] [Google Scholar]
- 64.Kimura H., Sato Y., Tajima Y., Suzuki H., Yukitake H., Imaeda T., et al. BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression. Chem Biol. 2010;17(12):1282–1294. doi: 10.1016/j.chembiol.2010.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.