Graphical abstract
Keywords: Brassica crops, Leaf morphology, Leaf polarity, Cell division and proliferation, Epigenetics, Environmental cues
Highlights
-
•
The heading leaves play an important role as nutrient storage organs, enhancing the high quality and commercial benefit of leafy heads.
-
•
Primary and secondary morphogenesis determines leaf shape.
-
•
Leaf incurvature is established by downregulation or knockdown of abaxial regulators and upregulation of adaxial regulators.
-
•
In heading vegetables, leaf incurvature is prerequisite for the development of leaf heads.
-
•
Understanding Arabidopsis leaf development enables us to predict the important regulators of leafy head formation in Brassica crops like Chinese cabbage and cabbage.
Abstract
Background
Heading is an important agronomic feature for Chinese cabbage, cabbage, and lettuce. The heading leaves function as nutrition storage organs, which contribute to the high quality and economic worth of leafy heads. Leaf development is crucial during the heading stage, most genes previously predicted to be involved in the heading process are based on Arabidopsis leaf development studies.
Aim of review
Till date, there is no published review article that demonstrated a complete layout of all the identified regulators of leaf curvature and heading. In this review, we have summarized all the identified physiological and genetic regulators that are directly or indirectly involved in leaf curvature and heading in Brassica crops. By integrating all identified regulators that provide a coherent logic of leaf incurvature and heading, we proposed a molecular mechanism in Brassica crops with graphical illustrations. This review adds value to future breeding of distinct heading kinds of cabbage and Chinese cabbage by providing unique insights into leaf development.
Key scientific concepts of review
Leaf curvature and heading are established by synergistic interactions among genes, transcription factors, microRNAs, phytohormones, and environmental stimuli that regulate primary and secondary morphogenesis. Various genes have been identified using transformation and genome editing that are responsible for the formation of leaf curvature and heading in Brassica crops. A range of leaf morphologies have been observed in Brassica, which are established because of the mutated determinants that are responsible for cell division and leaf polarity.
Introduction
Leaves are formed through primary and secondary morphogenesis [1]. Primary morphogenesis includes the development of the lamina, midrib, petiole, leaf base, leaflets, lobes, and serrations [2]. Secondary morphogenesis determines the ultimate shape of the leaf, which is regulated by precisely defined cell proliferation and expansion that is affected by cell number and cell size and is an irreversible, synchronized genetic process that can be altered by multiple factors. [3], [4]. Out of all altered leaf shapes, leaf curvature (LC) and leaf heading (LH) are of particular research interest, because they are crucial in determining the texture, increasing the shelf life, and improving the ease of harvest in Brassica crops. Leaf curvature (LC) is inward or outward curling of leaves that reflects the potential of plant photosynthesis, storage and edible organ in commonly cultivated leafy vegetables, especially Brassica species, which include mustard greens (Brassica juncea), Chinese cabbage (Brassica rapa ssp. pekinensis), and Cabbage (Brassica oleracea var. capitata) [5]. LC can be categorized into two types [6]: positive Gaussian curvature, or upward/inward curling, and negative Gaussian curvature, or downward/outward curling [7]. Leaf incurvature (Positive Gaussian curvature), characterized by curling, crinkling and folding of leaves is an essential prerequisite for LH formation [8], [9]. Incurvature (inward curling) begins at folding and is completed at the heading stage, at which point the leaves serve as storage organs in Brassica crops [10]. Changes in LC determine the angle of light absorption, efficiency of photosynthesis, and biomass yield in all leafy vegetables. The establishment of a spherical mass of edible leaves (LH), particularly in Chinese cabbage and cabbage, that is high in ascorbic acid, tocopherol, and vitamin A [11], [12]. The final LH phenotype is determined by events that occur at four developmental stages: the seedling, rosette, folding, and heading stages. Brassica leaves typically show a flat shape at the seedling and rosette stages and are actively involved in photosynthesis and respiration [13], [14]. LH shape is created by the overlapping of leaves [6] based on positive Gaussian curvature [7] and the folding of leaves around the shoot apexes to form compact and cylindrical heads or hearts in Chinese cabbage [15]. Many heading cultivars are high in nutrients and commercially valuable, such as Wakefield, Danish Ballhead, and Savoy [16].
Some Brassica species possess LC and LH e.g., Brassica rapa (B. rapa), and Brassica oleracea (B. oleracea), are polyploid, and their genome sequences have not only provided insight into evolution, domestication, and diversification but have also revealed the complexity of genetic LC and LH regulation [17]. B. rapa and B. oleracea both exhibit LH morphology, and their resequencing has provided evidence for their convergent domestication [18]. Studies on the regulation of leaf morphology have been reported more often in Arabidopsis thaliana than in Brassica. Identification of A. thaliana homologs in Brassica presents new challenges to researchers for finding leaf morphology regulators [19], [20].
Leaf morphogenesis events are regulated by a complex network of genetic, physiological, and environmental factors that are regulated by different phytohormone signaling pathways. Phytohormones are chemical messengers that are generated in one region of the plant and then translocated to other parts of the plant to regulate plant growth and development [21], [22]. A combined B. rapa and B. oleracea gene enrichment analysis revealed gibberellic acid (GA) biosynthesis as well as auxin-, cytokinin (CK), brassinosteriods (BR) and jasmonic acid (JA)-mediated signaling pathways. These pathways have been linked to the start and development of leaves [18]. These phytohormonal signaling pathways are also known to affect leaf polarity (primary morphogenesis) and cell division (secondary morphogenesis) that triggers LC and LH establishment [23], [24].
Nonetheless, an integrated understanding of the genetic, physiological, and environmental regulation of LC and LH is still needed. Therefore, this review provides a detailed summary of the developmental mechanisms by which the above-mentioned factors influence LC and LH. Arabidopsis regulators of leaf developmental stages, with a focus on orthologs genes in Brassica crops, provide the foundation for identifying the LC and LH regulators. We describe recent advances in our understanding of leaf shape development (LC and LH), and we outline how genetic mechanisms regulate this process, alone or in combination with other factors. We discuss physiological regulation through the effects of hormones and provide a brief account of environmental influences on leaf shape. We include graphical schematic model to enable rapid understanding of these regulatory mechanisms. This detailed overview can suggest new research avenues for investigating the coordination among regulators of leaf curvature and heading and their corresponding pathways.
Leaf formation: Primary and secondary morphogenesis
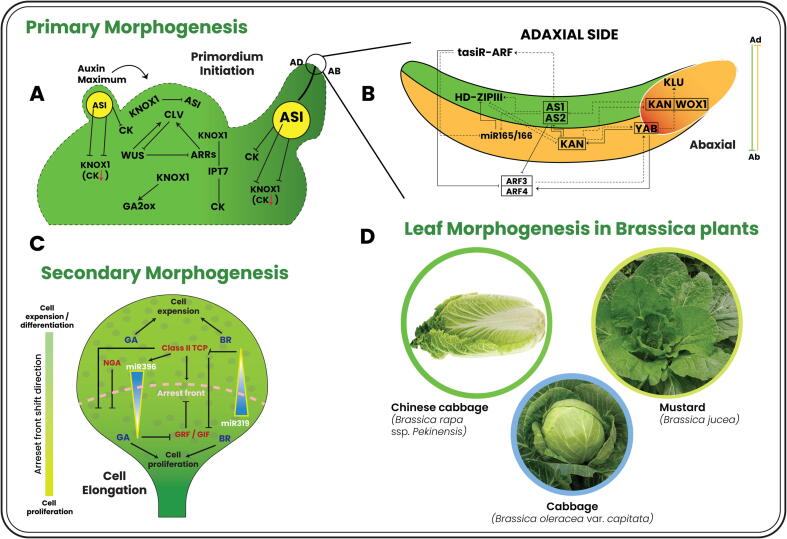
Leaves show dynamic differences in thickness, size, and shape, which distinguishes the plant species [25]. Nonetheless, leaf morphology is not fully predetermined. Thus, primary and secondary morphogenesis play a major role in leaf development, influencing the final shape of leaves [1], [2]. Primary morphogenesis consists of primordium initiation (Fig. 1-A) and the establishment of abaxial/adaxial (ad-ab) polarity (Fig. 1-B) [26]. The process involves several stages: auxin maxima activate the callus to initiate primordium formation; the second stage leads to an adaxial-abaxial polarity; and the third stage leads to a leaf blade that is detached from the petiole, which is the final step of the process [27], [28]. The formation of ad-ab patterns requires precise coordination between hundreds of cells during primordium development. Adaxial-abaxial polarity is responsible for defining medial–lateral polarity and leaf blade production. The medial–lateral axis of the leaf blade is flattened through perpendicular cell division [20]. Leaf primordium begin to expand once they have established polarity and reached their final shape that is controlled by coordination of several transcription factors. The majority of these transcription factors such as HD-ZIPIII, KANADI (KAN), Auxin response factor (ARFs) and YABBY are classified into various classes and thus are activated in ways that are simultaneously conserved and sometimes redundant [20].
Fig. 1.
. Primary and secondary leaf morphogenesis, together with LC and LH morphology, in Brassica plants The general mechanism of primary morphogenesis, from leaf initiation to lamina formation, is shown. A. The molecular mechanism of leaf primordium initiation. B. The adaxial/abaxial patterning mechanism. C. Secondary morphogenesis and leaf lamina formation by cell proliferation and expansion. D. Brassica plants showing LC and LH leaf morphologies.
As a leaf blade develops, its cells must proliferate and expand in a balanced manner, coordinated by molecular networks. The process of cell division at the distal end of the leaf ceases as it grows, and instead of proliferating at the base, a process of expansion takes place. Eventually, older cells fall out of the proliferation zone due to displacement by the base. Additionally, the dynamic cell proliferation regions develop rapidly at the “arrest front” boundary between proliferation and expansion for several days before disappearing rapidly. In other words, the two main processes of cell proliferation and cell expansion determine the size of the leaf at maturity, and any changes in either process may affect leaf development [29]. At leaf lamina establishment, the timing of the transition from division to expansion is essential in determining final size, flatness, and shape [30]; this process of leaf blade formation is called secondary morphogenesis (Fig. 1-C). The class II TCPs are the key regulators of timing from division to expansion [31]. There are many genes that positively regulate and control the transition from proliferation to expansion, for example, AINTEGUMENTA (ANT) [32], KLUH/CYP78A5 [33] and GROWTH REGULATING FACTORS (GRFs) [34], whereas the negative regulators include CIN-TCPs and DAI [33]. In secondary morphogenesis, mature leaves of different sizes and shapes (such as., flat, curved, folded, or headed) develop by cell expansion are regulated by a well-integrated set of genes [2], [35]. Any perturbation in the genetic regulators of these morphogenesis events, which can be triggered by phytohormones or environmental regulators, has the potential to transform the flat leaf into a variety of leaf morphologies (Fig. 1-D).
Primary morphogenesis establishes LC and LH
Leaf developmental mechanisms in Arabidopsis provides a foundation to predict the candidate regulators of LC and LH in Brassica crops such as Chinese cabbage and cabbage [9]. It is shown that the B. rapa and B. oleracea leaf head formation is linked to a stable genetic network established through precise lineage analysis of the adaxial and abaxial progenitors in Arabidopsis [18]. Chinese cabbage and Arabidopsis are member of Cruciferae, most genes involved in leaf ad-ab polarity establishment are concluded to be conserved between these two genomes [18]. Furthermore, 45 genes (Version 1.5) involved in the founding of leaf adaxial-abaxial polarity were observed in Chinese Cabbage [36]. Adaxial-abaxial leaf polarity (primary morphogenesis) is regulated by a complex regulatory network of genetic regulators, which is profoundly influenced by phytohormones.
Interplay of phytohormones develops LC and LH via leaf polarity
Plant hormones are produced in small amounts but have the ability to regulate a wide range of plant cellular activities. They serve as chemical messengers, connecting cellular processes and playing critical roles in harmonizing numerous signal transduction pathways, which ultimately impact leaf shape [22]. Auxin has an extensive role on plant development and establishment of organ morphology by regulating the signal transduction pathways. Auxin maxima points are established at shoot apical meristem (SAM) for organ initiation, PIN-FORMED1(PIN1) which is a polar auxin transporter, defines the positions of leaf primordia initiation and shapes leaf margins [35], [37], [38]. Auxin regulates the expression of BrPIN recognized as leaf patterning regulator that is involved in leaf heading in B. rapa. Auxin also regulates the expression of BrLUX, a leaf patterning regulator that is also identified as LH regulator in Chinese cabbage [39].
Auxin response factor (ARFs) that regulate the expression of auxin response genes, such as AtARF2, AtARF3, AtARF4, and MONOPTEROS (MP)/ARF5, have been found to influence leaf development in Arabidopsis (AtARFs) [40]. Double mutant plants arf3arf4 showed narrow abaxial outgrowth with upward curved leaves in Arabidopsis [41] whereas in Chinese cabbage BrARFs are elucidated as a candidate regulator of LC formation [42], [43], [44], [45]. Several investigations have found that variations in leaf polarity genes are strongly linked with the establishment of LH in B. rapa [14], [18], [46]. After primordium initiation, primary morphogenesis proceeds to develop a flat lamina through a juxtaposition of abaxial and adaxial tissues which is controlled by multiple genetic regulators that might be controlled via phytohormones signaling pathways [47], [48].
In Arabidopsis, Auxin signaling pathway is involved into the determination of adaxial polarity that is regulated by the class III HD-ZIP family, including PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV). These adaxial regulated are also controlled with the activation of miR165 and miR166 [49], [50], [51], [52], [53]. All three mutated genes (phb, phv, and rev) culminated in radial abaxial leaves, a phenotype comparable to that of dominant flat leaf morphology [49]. Overexpression of REV in the hyl1 Arabidopsis mutant caused double-leaf curvature downward and upward (longitudinally and transversely) [54]. Moreover, downregulation of BrpREV-1, BrpREV-2, and BrpPHB-1 produced downward leaf curling in B. rapa [55].
A polar auxin transporter named PIN affects the activity of adaxial regulator KAN to negatively regulate the expression of WOX1/ PRESSED FLOWER (PRS). Narrow leaves with unaffected lengths (leaf length same as of wild type leaf) were showed in kan mutants of Arabidopsis [56], [57]. In Brassica heading crops, the ortholog of ARABIDOPSIS THALIANA HOMEOBOX 15 in B. oleracea; BoKAN2.2 and in B. rapa; BrKAN2.1 and BrKAN2.3 are suggested as possible genes of adaxial-abaxial patterning establishing the in LC and LH [18], [36].
In lettuce, LsKN1 has been discovered as an LH regulator that represses the adaxial gene LsAS1, which is partially involved in the production of leafy heads [46]. Besides that, LsKN1 inhibits the expression of SAWTOOTH 1 (LsSAW1), an Arabidopsis homolog. The functionally redundant SAWTOOTH (SAW1) and SAW2 proteins promote leaf serrations, and saw1saw2 mutants promote leaf serrations in Arabidopsis [58], [59]. Transcriptomic results demonstrate that LsSAW1 modulates leaf dorsiventrality and its silencing downregulates adaxial genes. By binding to the promoter region of the adaxial gene ASYMMETRIC LEAVES 1 (LsAS1), LsSAW1 enhances the expression of LsAS1. Overexpression of LsAS1, decrease the impact of Lssaw1 on heading. In contrast, LsSAW1 binds to the promoter of YABBY 1 (LsYAB1) to inhibit its expression. LsYAB1 overexpression caused curving of leaves in LsSAW1 genotypes. LsSAW1 interacts with LsKN1, which is requisite for the establishment of lettuce LH [60]. The role of SAW1 in downregulating the abaxial gene and upregulating the adaxial gene for the development of LC and LH in B. rapa and B. oleracea also needs to be investigated.
Auxin together with adaxial polarity regulator ASYMMETRIC LEAVES1/2 (AS1/2) play an essential role to repress expression of other genes such as KNOX gene BREVIPEDICELLUS (BP) in the establishment of leaf morphology [61]. A mutation in AS2 causes a petiole to curl upwards, resulting in a small leaflet on the petiole [62], [63]. The overexpression of pak choi BcAS2 in an Arabidopsis transgenic line causes the AS2 to be engaged in the production of curved leaves [64], [65]. Moreover, the AS1-AS2 complex negatively regulates YABBY, ARF, and KANADI gene families (abaxial regulators) [66]. The AS1-AS2 complex also directly suppresses the production of miR166A and ETTIN (ETT) in the adaxial region, as well as binding the promoter of the ta-siRNA precursor TAS3A activation [66]. By suppressing the ARF family members (ARF2, ARF3, and ARF4), TAS3 tasiRNAs and miR390 expand a pathway that determines leaf patterning and developmental time [67], [68], [69]. The findings of the study provide the basis for the assumption that downregulation of abaxial regulators could be a cause of LC and LH.
Auxin homeostasis is also affected by the WUS-related homeobox (WOX) genes (WOX1, WOX3) that is linked to adaxial/abaxial patterning and the formation of flat leaves [57], [70], [71]. Furthermore, studies suggest that auxin signaling acts upstream of WOX genes, demonstrating the feedback loops between auxin signaling and CK biosynthesis that assist to WOX function [40]. BrWOX1.1 (BraA05g029540.3C), BrWOX1.2 (BraA03g038230.3C), BrWOX3.1 (BraA04g020450.3C), and BrWOX3.2 (BraA03g024810.3C) were discovered as AtWOX orthologues in B. rapa LH study by specific SSR markers that are associated with LH establishment [36]. However, complete understanding of BrWOXs in LC and LH establishment has not been reported yet.
Exogenous auxin spray and transcriptomic analysis of the heading mutant flat growth-1 (fg-1) demonstrated that auxin is essential for the establishment of LH in Chinese cabbage [15], [72]. These findings also suggest that auxin continues to play a conserved role in establishing leaf polarity. Six adaxial identity genes, BrREV, BrHB8.1, BrHB8.2, BrHB9, BrHB14.1, and BrHB14.2, and six abaxial identity genes, BrKAN1, BrBOP2, BrYAB1.1, BrYAB1.2, BrYAB2, and BrYAB3 were significantly expressed in fg-1 mutant plants, indicating the involvement of primary morphogenesis regulators in the LH development of Chinese cabbage. Moreover, the exogenous application of auxin spray to the dorsal side of Brassica leaves causes upregulation of BcpLH/BrHYL, leading to head formation by controlling the abaxial miR165/166 [15]. The expression level of the adaxial identity gene REV was increased and the expression position was restricted on both sides of growing leaves near the margins in curved leaves of hyl1 mutants, whereas expression of the miR165 gene was significantly reduced. These findings suggest that HYL1 orthologue of BcpLH maintains growing leaf polarity by regulating the level of microRNA [54]. Auxin is also involved into the upregulation BREVIS RADIX (BRX) genes; BrBRX.1, BrBRX.2 and BrBRX.3 that are identified as LC and LH regulator in Chinese cabbage [73]. The functional analysis of BrBRX revealed their role in petiole length, leaf number, and leaf angle that are associated with leaf curvature and heading [73]. BRX orthologues, AtBRX showed epinastic leaf phenotype in Arabidopsis brx mutants [74]. BRX ortholog BoBRX.2 was identified under selection in leaf-heading accessions of B.oleracea in crop domestication of leaf-heading morphotype [18]. Furthermore, BRX is involved in crosstalk between cytokinin signaling, brassinosteroid biosynthesis and auxin signaling that regulates cell proliferation and cell expansion in the leaf [75], [76]. It will be interesting to determine if BrBRXs are involved in cross-talk between auxin and BR signaling pathways in Chinese cabbage LH development [73].
BR is also responsible for the transfer of OCTOPUS (BrOPS) to the nucleus and interacts with BrBIN2 to suppress the phosphorylation of BRINSENSITIVE1 (BRI1)-EMS-SUPPRESSOR1 (BrBES1). This suppression leads to the downregulation of the leaf polarity transcription factor BrAS1, influencing LC and LH in Chinese cabbage [9]. Chinese cabbage inward curling mutant (ic1) boosted the phosphorylation of BrBES1, with the accumulation of BrBIN2 in the nucleus in the absence of BrOPS when the level of BR is low. Phosphorylated BrBES1 stimulates the repression of BrAS1, culminating the overexpression of the adaxial regulator BrAS1, which triggers leaf inward curling in Chinese cabbage ic1 mutants [9]. All these finding strongly consistent with the statement that primary morphogenesis regulators highly influenced the formation of LC and LH in Brassica crops (Fig. 2).
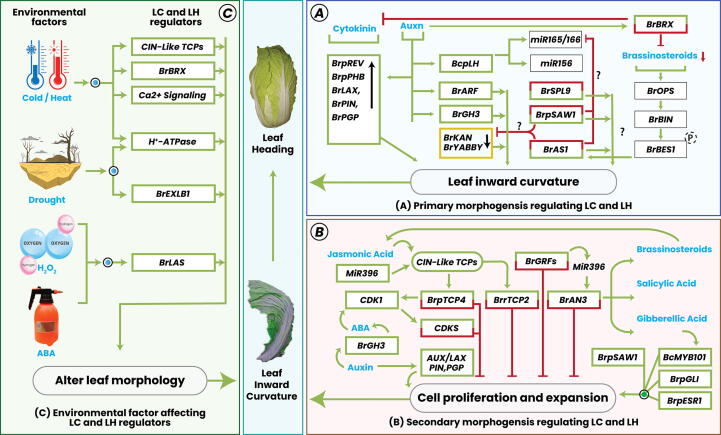
Fig. 2.
. Genetic and environmental regulators of LC and LH morphology in Brassica plants. A. Expression pattern of primary morphogenesis regulators to induce LC and LH. Leaf abaxial-adaxial polarity regulators involved in leaf curvature and heading, the regulation of leaf curvature and heading and the influence of polarity mutations are shown. Adaxial and abaxial genes that are responsible for the regulation of leaf curvature and heading through their overexpression are indicated in green box (BsAS2, BrpPHB1, BrpREV1, BrpREV2, and BrpSAW1). Genes whose downregulation induces leaf curvature and heading are indicated in yellow box (BrKan and BrYABBY). Green arrows indicate enhancement, and red bars indicate inhibition. B. Expression pattern of secondary morphogenesis regulators to induce LC and LH. Cell division and proliferation regulators involved in the formation of leaf curvature and heading, BrpTCP4, BrpTCP2 (regulated by miR319), BrGRFs, BrAN3 (regulated by miR396), and CDKs (downregulated by CDKI) have been identified as candidate genes for leaf curvature and heading because of their negative effects (represented by red T lines) on cell division. BrpGL1, BrpESR1, and BrpSAW1 have a positive effect on leaf curvature and heading (represented by green arrows) through the regulation of cell division and are considered to be candidate genes for these traits. C. Environmental cues culminate LC and LH. Temperature, drought, H2O2 and exogeneous phytohormonal spray affects the genetic regulators (CIN-LIKE TCPs, BrBRX, AHA2, BrEXLB1, and BrLAS) and signaling pathways (Ca+2 signaling) that cause the LC and LH.
Role of posttranscriptional regulators and DNA methylation
Gene expression is tightly controlled at both the transcriptional and post-transcriptional levels. Primary morphogenesis (adaxial-abaxial polarity) is also regulated by posttranscriptional mechanism [36]. Pre-messenger RNA (mRNA) processing (capping, splicing, and polyadenylation), mRNA stability, and mRNA translation are all instances of post-transcriptional gene expression control [77]. The production of microRNAs and tasiRNAs requires a number of proteins, which include ARGONAUTE1 (AGO1), ARGONAUTE7 (AGO7), ARGONAUTE10 (AGO10), SUPPRESSOR OF GENE SILENCING 3 (SGS3), RNADEPENDENT RNA POLYMERASE 6 (RDR6), DICERLIKE1 (DCL1), DICER-LIKE4 (DCL4) and SERRATE (SE) [78].
Orthologs of Arabidopsis AGO1, AGO7, AGO10, SGS3, RDR6, DCL1, DCL4, and SE that are associated with the leaf adaxial abaxial polarity, were determined in Chinese cabbage as BrAGO1.1 (BraA05g020200.3C), BrAGO7 (BraA07g030370.3C), BrAGO10.1 (BraA06g043560.3C); BrAGO10.2 (BraA09g019960.3C), BrSGS (BraA06g031000.3C), BrRDR6 (BraA01g023640.3C), BrDCL1 (BraA10g000840.3C), BrDCL4 (BraA10g020250.3C), and BrSE1 (BraA07g018000.3C); BrSE2 (BraA04g019460.3C) respectively. It is strongly hypothesized by previous study that these posttranscriptional polarity regulators are associated with the LC and LH formation in Chinese cabbage [56].
Hyponastic leaves 1 (HYL1) encodes a protein that regulates the leaf abaxial determinant miR166, and Arabidopsis hyl1 mutants showed inward-curling leaves [79]. BcpLH (BraA06g006590.3C), a homolog of Arabidopsis HYL1, was isolated from Chinese cabbage and shown to influence the inward curvature of folding leaves [54]. leaf Ad–Ab patterning candidate genes and BrHYL1.1 (also known as BcpLH) are also linked to head formation by regulating the expression of miR156, miR165/6, and miR319 [80], [81]. BcpLH and BcpLH2 regulate the BrpSPL9-2 (a homolog of Arabidopsis SQUAMOSA PROMOTER BINDING-LIKE (SPL), which control the timing (developmental stage of plant) of leaf incurvature and heading [82]. Tasi-RNAs have also been identified as the regulators of leaf polarity determinants such as ARF3/4 abaxial genes is regulated by tasiR-ARF [83]. Genetic linkage map screening of polarity regulators determined that BrARFs, leaf abaxial regulator is associated with the LC and LH in Chinese cabbage [36]. These findings prompted us to hypothesize that tasiR-ARF induce any mutation in the abaxial polarity regulator BrARFs to enhance or repress the LC and LC.
Furthermore, DNA methylation appears to be a key regulator of the mechanisms underlying the formation and maintenance of the LC and LH. DNA methylation is essential for epigenetic control of the genome and gene expression [84]. DNA methylation, in particular with regard to histone modifications (acetylation and methylation), causes genetically inherited and/or transient alterations in chromatin structure and gene expression that do not make a significant difference in nucleotide sequence, and leads to genomic imprinting [85] and gene silence [86]. DNA methylation-regulated auxin pathways contribute to the establishment of LC and LH by suppressing DNA methylation in promoters, which might also regulate the expression of genes involved in LC and LH [87]. Auxin signaling pathway was negatively affected with the methylated promoters of BrPIN1 and BrARFs and also these genes were downregulated in the detorted LH phenotype of Chinese cabbage [87]. Furthermore, a DNA methylation approach known as methylationsensitive amplification polymorphism (MSAP) indicated that B. oleracea's (heading crop) methylation polymorphism is the cause of morphological polymorphism as well as substantial structural genomic rearrangements in its related species [88]. Nonetheless, the mechanisms behind these impacts are unknown.
These alterations in gene expression most likely altered the adaxial-abaxial polarity of leaves, inducing inward leaf curling. Adaxial gene downregulation or loss of function, as well as abaxial gene overexpression, can indeed stimulate LH growth [60].
Secondary morphogenesis establishes LC and LH
In secondary morphogenesis, genes carry crucial instructions that determine leaf shape by specifying cellular growth patterns, cell cycling, cell differentiation, cell enlargement, and hormonal transport, via complex regulatory systems [34]. Any abnormal behavior of cell proliferation and expansion regulators may have a direct effect on LC and LH [80]. It has been speculated that genetic factors involved in cell division and expansion may influence the cabbage leafy head phenotype [89]. QTL mapping has been used for the efficient identification of genes responsible for different leaf traits in multiple crops (tomato, maize, grape, and Arabidopsis) [90], [91], [92]. In Chinese cabbage, three genes (BrpGL1, BrpESR1, and BrpSAW1) were identified as LH candidate genes (represented with green arrows in Fig. 2), and they also influence the development of trichomes, petioles, and leaf serration through their functions in the regulation of cell division [14].
Interplay of phytohormones develops LC and LH via cell proliferation and expansion
The auxin signal is essential for the formation of vascular system and the intake of nutrients for leaf cells. It might be difficult to discern the direct effect of auxin on cell proliferation/expansion from its indirect effect on cellular metabolism [20].
In the megaintegumenta (mnt) that are Arabidopsis mutant plants developed by creating mutations in auxin-inducible gene AUXIN RESPONSE FACTOR 2 (ARF2). Extra cell proliferation characteristics in the vegetative and floral organs of mnt indicated that ARF2 could function as a fundamental repressor of cell division in leaves [93]. These data suggested that the balance of auxin-inducible genes ARF2 is required for proper auxin function in cell proliferation and cell expansion [94], its orthologue BrARFs also speculated as LC and LH regulators in Brassica crop [36].
An exogenous application of GA3 to leaves of Chinese cabbage nhm non-heading mutants restored the heading phenotype, demonstrating the crucial role of GA in B. rapa heading [95]. The high level of GA (gibberellin) treatment, elevated the BcMYB101 regulated by miR159 expression in pak choi. Overexpression of BcMYB101 accelerated leaf quantity and induced downward curling of leaves. Silencing of GAMYB transcription factor from pak choi BcMYB101, a of an Arabidopsis MYB gene (first identified in barley), in pak choi (Brassica rapa ssp. chinensis) produced upward leaf curling [96]. MYB genes involved into the leaf development by controlling the cell cycle [97], that represents the secondary morphogenesis contribution into the LH establishment.
In leaf development, cytokinin influences the balance of differentiation and expansion. cytokinin degradation promotes cell expansion and early termination of cell proliferation in Arabidopsis leaf primordia, suggesting that cytokinin delays the initiation of cell differentiation [98]. The Arabidopsis Class I KNOTTED1-like homeobox (KNOXI) gene is overexpressed in lettuce (Lactuca sativa) leaves, resulting in unpredictable growth and cytokinin deposition [99]. CIN-TCPs, on the contrary hand, enhance leaf maturation in Arabidopsis by decreasing leaf sensitivity to cytokinin. As a corollary, the antagonistic functions of KNOXI and TCPs on leaf blade formation are dependent on GA/cytokinin balance regulation [32], [100]. In cin mutants, the leaf margin is rumpled as a result of the slower arrest of cell divisions at margins relative to the middle of the leaf as highlighted by mutations in CINCINNATA (CIN) in Antirrhinum [101]. BrpTCP4 regulates the head shape of Chinese cabbage by miR319a BrTCP1c of CYC/TB1 was apparently upregulated. CIN-like TCPs, named BrrTCP2 in turnip, BrpTCP4 in Chinese cabbage, and LsTCP4 in lettuce, are responsible for the formation of wrinkles, bulges, serrations, and heading leaf morphologies by affecting on cell division [102], [103], [104], [105], [106]. The cin and triple tcp2/4/10 mutations alter leaf shape from flat to unevenly curved because of excessive cell proliferation at the leaf margins, and overexpression of BrrTCP2 restores the wild-type leaf morphology in these Arabidopsis mutants [107], [108]. Furthermore, Mao et al. (2014) discovered that rosette leaf shape and leafy head shape in Chinese cabbage are associated, and BrpTCP4 mutation at the miR319a recognition region altered cell division and proliferation, resulting in different rosette leaf and head morphologies [102].
CIN-Like TCPs is negatively regulated by CYCLIN-DEPENDENT KINASE INHIBITOR (ICKI)/ KIP RELATED PROTEIN1 (KRP1), which is involved in the regulation of organ shape through the control of cell division (secondary morphogenesis) [109]. A large number of cyclin-dependent kinases (CDKs) are involved in heading formation. Two CDKs, one mitogen–activated protein kinase (MAPK), four MAPKKs, three MAPKKKs, eight receptor-like kinases (RLKs), and one calcium-dependent protein kinase (CDPK) participate in protein phosphorylation events in the rosette and folding leaves of Chinese cabbage [10], CDKs as key regulators for an accurate cell cycle progression, to guarantee that plants develop with the correct form, these kinases also participate in the modulation of cell divisions to confer plasticity to leaf development [110], [111]. CDKs are also involved in regulation of the cell cycle and cell proliferation. Similarly, ICK1/KIP-Related Protein (KRP1) mutations repress cell proliferation and alter leaf morphology in Arabidopsis [112], [113]. Moreover, plants overexpressing ICK2/KRP2 also showed an altered root system and modifications in organs morphology [114] that are induced with the ABA treatments [115]. Based on previous findings CDKs and CDKI can be hypothesized to play a crucial role in any undetermined state (such as reduced cell number or cell expansion) of secondary morphogenesis that establish LC and LH in Brassica crops.
JA and BR signaling pathways are positively regulated by CIN-like TCP which are significant transcription factors for the cell division [116]. Through cell proliferation regulation, the miR319-TCP and miR396-GRF modules interact together to balance marginal and overall leaf growth [20]. TCP4 represses the expression of GROWTH-REGULATING FACTORs (GRF) genes and GIF1 that are not direct targets of miR396, via unknown mechanisms [117]. Thus, GRFs are negatively regulated by miR319 and miR396 [38], [55], which physically interact with GIF1/AN3. Silencing of BrAN3 produced leaf serration, curling, and heading in B. rapa through the repression of cell proliferation and striking of cell cycle [23]. Similarly, AtAN3/AtGIF1 has been shown to regulate cell division, and silencing of AtAN3 leads to the formation of narrow, serrated, and curling leaves in transgenic gif1 Arabidopsis and to smaller leaves than wild-type Arabidopsis because of reduced cell numbers [118]. GA, BR, and SA signaling pathways may be induced by the expression of BrAN3, an important heading candidate gene in Chinese cabbage, but these signaling pathways function negatively in heading formation [23] (Fig. 2). Gibberellins (GAs) and brassinosteroids (BRs) promote leaf growth by increasing cell proliferation and expansion [119]. BcMYB101 identified as LC gene speculated as modulators of BR signaling pathway to regulate leaf shape development in Brassica crops [120]. However, how secondary morphogenesis is coordinated to regulate LC and LH morphology remains to be elucidated in Brassica crops.
Environmental cues culminate LC and LH
Apart from the morphogenesis regulators within plant tissues, plant organs also experience physical restrictions from the outside, such as neighboring organs and more significantly by external environments which includes biotic and abiotic stresses [34]. Abiotic stress which includes; temperature [121], drought, and salinity all have a substantial impact on organ growth and development, as well as overall plant architecture [122]. Leaves are also sensitive to a wide range of internal and external stimuli, each of which initiates a specific signaling pathway that controls leaf size and leaf shape. Leaf morphological patterns vary markedly in response to temperature [123], light, and other external stresses [124], [125] and these effects have been documented for crops that exhibit LH and LC (Fig. 1-C). Primary and secondary morphogenesis can highly be affected with the influence of environmental stresses on phytohormonal signaling pathways. Modulation of phytohormones sets off a chain of events that trigger physiological processes in leaf architecture that aid in leaf development under inadequate growth conditions [126].
Temperature is a key factor that regulates leaf shape by controlling the ratio of midrib length to leaf width [24]. Low temperature activates the regulatory network that promotes head formation in Chinese cabbage [127]. CIN-Like TCPs are controlled by temperatures dynamics in Arabidopsis, leading to morphological changes in leaves [112]. The AtBRX homologs in B. rapa (BrBRX.1, BrBRX.2, and BrBRX.3) and in B. oleracea (BoBRX.2) influence epinastic leaf growth in which environmental factors are often involved [73]. BRX regulates leaf cell proliferation and epidermal cell size via modulating the crosstalk between cytokinin signaling, brassinosteroid production, and auxin signaling [76]. All these factors are linked with the leaf morphogenesis that further stimulates the establishment of LC and LH in Brassica crops.
A recent study of Brassica (Chinese heading cabbage and non-heading cabbage) revealed that a cluster of transcripts initiates the transition to the heading stage and suggested that temperature may be a conserved signal mediator or inducer of the leaf heading transition [128]. Lettuce is a low-temperature crop, stable head formation can be induced by complex factors such as light quality, soil fertility, and temperature [125]. Head weight is positively related to solar radiation and negatively related to mean temperature, indicating a temperature-weight association and solar radiation that affects not only head weight but also head shape and head density [129].
Levels of Ca2+ (a ubiquitous second messenger) is altered by external stimuli such as light or other stress conditions, leading to changes in plant growth and development [130]. Calmodulins (CaMs), CDPKs, and calcineurin B-like proteins are calcium sensor proteins, and calcium signaling has an important role in protein phosphorylation [131]. This phosphorylation affects head development by upregulating CDPK [132].
The H+-ATPase encoded by H+-ATPase 2 (AHA2, BraA01g007510.3C) that is hypothesized as a candidate gene for flat-leaf morphology and this gene is affected by environmental stress resulted into the leaf crinkled mutant (Bralcm) of B. rapa [133]. H + -ATPase is a transmembrane transporter enzyme and autoregulatory triggers the genes to alter their expression under any environmental stress which cause the different leaf morphologies [134]. Previous studies have shown that proton dynamics generated by H + -ATPase directly affect cell expansion [135]. During leaf morphogenesis, the GAs and BRs hormones promote both cell proliferation and expansion [30] that are associated with LC and LH.
Drought stress is a major environmental stress that influences the morphology of all Brassicaceae plants [136]. Under drought stress, the pleiotropic effects of B. rapa expansin-like B1 (BrEXLB1) on leaf were irregular and unstable [122]. BrEXLB1 under its specific promoters may participate in the regulation of leaf and plant growth and that it responds to hormone availability, light quality, dark periods, developmental stages, and drought conditions [137]. Similarly, exogenous application of ABA and abiotic stresses; polyethylene glycol (PEG), NaCl and H2O2 upregulated the B. rapa LATERAL SUPPRESSOR(BrLAS) expression and produced altered leaf morphology [138]. With the application of salt treatments on B. oleracea genotypes showed significant changes in its plant growth, leaf weight and leaf area [139]. That predict salt stress effect on the leaf development of Brassica crops [140], that might link with the establishment of LC and LH.
Conclusions and future perspectives
The leaf shape is regulated by leaf polarity (primary morphogenesis) as well as by cell proliferation and cell expansion (secondary morphogenesis). The genes and pathways involved in LC and LH establishment may be conserved among evolutionarily diverse crops, but specific mutations have led to variable heading phenotypes [18]. Although many regulators of LC and LH have been identified in Brassica crops, their identification alone is insufficient to explain their underlying mechanisms. Regulators of leaf curvature have been identified in Arabidopsis, but the number of corresponding Brassica is limited (Table 1).
Table 1.
Leaf curvature and heading regulators identified in Brassica crops.
| Species | Gene name | Gene ID | Function | Reference |
|---|---|---|---|---|
| B. rapa L. ssp. pekinensis | BrpSPL9-2 | BraA05g002720.3C | Interacts with BcpLH1/2, BrpREV, and TCP4; controls timing of leaf curvature | [82] |
| B. rapa L. ssp. pekinensis | BcpLH | BraA06g006590.3C | Interacts with other microRNAs, leading to the formation of leaf heading | [81] |
| B. rapa L. ssp. pekinensis | BrpTCP4 | BraA05g032060.3C | Excessive cell proliferation, uneven organ curvature, curved leaves | [105] |
| B. rapa L. ssp. rapa | BrrTCP2 | BraA02g012600.3C | Alters leaf morphology, recovers wavy leaf phenotype of Arabidopsis | [106] |
| B. rapa L. ssp. pekinensis | BrpREV-BrpREV-2 | BraA10g018460.3C BraA02g010200.3C |
Controls leaf curvature | [49] |
| B. rapa L. ssp. pekinensis |
BrWOX1.1 BrWOX1.2 |
BraA05g029540.3C BraA03g038230.3C |
Role in leaf curling; affects leaf heading through leaf polarity | [57] |
| B. rapa L. ssp. pekinensis |
BrWOX3.1 BrWOX3.2 |
BraA04g020450.3C BraA03g024810.3C |
Role in leaf curling; affects leaf heading through leaf polarity | [57] |
| B. rapa L. ssp. pekinensis | BrAN3 | BraA09g004620.3C | Effects head formation in Chinese cabbage | [23] |
| B. rapa ssp. chinensis | BcMYB101 | BraA05g012430.3C | Mutations cause upward leaf curling in pak choi | [120] |
| B. rapa ssp. chinensis | BcAS2 | BraA02g016770.3C | Mutations cause leaf curling in pak choi | [64] |
| B. rapa ssp. chinensis | BcYAB3 | BraA01g037320.3C | Involved in leaf morphology and responsible for leaf curling | [142] |
| B. rapa L. ssp. pekinensis | BrLAX3 | BraA02g000160.3C | Responsible for uneven auxin distribution leading to leafy head formation in Chinese cabbage | [15] |
| B. rapa |
Br-ARF3.1 Br-ARF3.2 |
BraA05g011080.3C BraA04g024390.3C |
Leaf abaxial determinants involved in leaf curling | [45] |
| B. rapa | Br-ARF4.1 | BraA10g018230.3C | Leaf abaxial determinants involved in leaf curling | [43] |
| B. rapa L. ssp. pekinensis | BrOPS | BraA01004411 | Inward curling and heading of leaves | [9] |
|
B. rapa L.ssp. pekinensis |
AHA2 | BraA01g007510.3C | flat-leaf morphology | [133] |
| B. rapa | BrEXLB1 | BraA03g047290.3C | Affect leaf growth and development under drought stress condition | [137] |
| B. rapa | BrLAS | BraA06g000510.3C | Involved in leaf morphology | [138] |
| B. rapa L. ssp. pekinensis | miR319a | GenBank ID KJ130320 | Wavy and bulging leaves | [143] |
| B. rapa | MiR156a | GenBank ID NR_128632.1 | Involved in leaf morphology | [144] |
Although transformation and functional verification of some candidate regulators have been performed in Arabidopsis transgenic lines and Brassica species, in most cases, the effects of gene functional loss and overexpression are still unknown. Based on Chang et al. (2016), leaf heading is a convergent morphotype in B. rapa and B. oleracea, confirming that these genes are involved in the regulation of cell division and leaf polarity during the formation of LC and LH [18]. An integrated understanding of the genetic, physiological, and environmental regulation of LC and LH is still needed to build an efficient and concise regulatory network of leaf incurvature and heading in Brassica crops. In particular, identification of the factors that affect BrTCPs, BrAN3, and BrGRFs and ad-ab regulators that alter leaf morphology is needed in order to construct the full molecular network that controls LC and LH.
In our study, we have found that leaf morphogenesis regulators might be adversely affect due to climatic changes such as heat, cold, light, humid, and nutrient level. Analysis of hormone levels, and inhibition treatments might confirm the leaf morphogenesis regulators that affect heading phenotype. Moreover, analysis of regulators utilizing functional genomics approaches and merging them with epigenetic technologies will thus be critical in identifying novel candidate genes for significantly improving Brassica crops [141]. Identifying leaf heading regulators and their interactions by using modern molecular techniques such as GWAS, MutMap, combined QTL and BSA analysis, Y2H and metabolomics can help to improve optimal leaf heading phenotypes as well as innovative vegetable varieties of Brassica crops. Moreover, different genome editing tools (ZFN, TALEN, and CRISPR/Cas system) to improve leaf heading trait in Brassica can lay foundation in the development of Brassica crops. All modern strategies would open up new avenues for manipulating leaf head shape in Chinese cabbage, cabbage and eventually other heading vegetables.
Compliance with ethics requirements
This review article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Javaria Tabusam: Conceptualization, Writing – original draft. Mengyang Liu: Conceptualization, Writing – original draft. Lei Luo: Writing – original draft. Sumer Zulfiqar: Writing – review & editing. Shuxing Shen: Supervision. Wei Ma: Conceptualization, Supervision. Jianjun Zhao: Conceptualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 32222076, 31902005, 32002054), the Natural Science Foundation of Hebei (Grant No. C2020204111, C2020204122, C2021204049), the International Cooperation Base Project in the Technology of Hebei (Grant No. 20592901D), the High-level Talents Funded Project in Hebei Province (Grant No. B2020003039), the Science and Technology Support Program of Hebei (Grant No. 21326344D), the Introduce of abroad talents funding project of Hebei (C20210363), and the Science and Technology Research Project of Universities in Hebei Province (BJ2021024, QN2021074). We are thankful to Asad Riaz for helping throughout the manuscript work and also in designing the graphical illustrations.
Biographies
Javaria Tabusam is pursuing her PhD in Vegetable Breeding and Genetics from Hebei Agriculture University, China. She received her M.Phil. Biotechnology degree from University of Agriculture Faisalabad, Pakistan. In masters she conducted her research on pea plant to check the efficiency of pea rhizobium for the nitrogen fixation. In doctoral degree, her research area is vegetable germplasm resources evaluation and utilization innovation, vegetable biotechnology and genetic breeding.
Liu Mengyang is an associate professor with doctoral degree. Her main research expertise is in vegetable molecular chromosome engineering, vegetable germplasm resource evaluation and utilization innovation. She participated in provisional research projects and won different awards. She published more than 10 SCI research articles.
Lei Luo is pursuing her PhD in Vegetable Breeding and Genetics from Hebei Agriculture University, China. She obtained her Bachelor’s degree in Horticulture from Hebei Agricultural University (HAU) in 2015. Then she applied successfully to a successive master-doctor program of HAU in 2017, researching on identification of genes regulating fruit color under calyx of eggplant and functional analysis of candidate genes.
Sumer Zulfiqar is pursuing her PhD in Vegetable Breeding and Genetics from Hebei Agriculture University, China. She received her MSc (Hons) Entomology degree from University of Agriculture Faisalabad, Pakistan. In masters she conducted her research, to check the efficacy of wood extractives of ziziphos muaritiana against sub-terranean termites under field conditions. In doctoral degree, her research area is vegetable germplasm resources evaluation and utilization innovations, vegetable biotechnology and genetic breeding.
Shen Shuxing is a professor with expertise on vegetable molecular chromosome engineering, vegetable germplasm resource evaluation and utilization innovation, vegetable biotechnology and genetic breeding. He is the director of the Chinese Horticultural Society, Vice President of the Molecular Breeding Branch of the Chinese Horticultural Society, Deputy Director of the Plant Production Professional Teaching Guidance Committee of the Ministry of Education, Chairman of the Horticulture (including Tea Science) Teaching Guidance Subcommittee, Chief of the Hebei Vegetable Industry System expert. He has also been the editor of three books. He has been the part of 20 research projects. He has been awarded with seven different awards. He patented the 3 Chinese cabbage, 1 pepper and 1 eggplant lines.
Ma Wei is a professor of Hebei Agriculture University. Main focus of his research is to study the enormous diversity displayed in the diploid Brassica rapa, Chinese cabbage. He has been engaged in molecular genetic analysis of important traits of Chinee cabbage for a long time, such as leaf morphology, leaf bulb development, etc., and focused on the research of genetic variation, molecular markers, gene mapping and gene function studies. To date, he has published more than 10 SCI papers, including Nature Genetics, PNAS, Molecular Plant, Cell Reports, Plant Biotechnology Journal.
Zhao Jianjun is a professor of Hebei Agriculture University, “Changjiang Scholars” distinguished professor. To unravel the genetic regulation of domestication traits in Chinese cabbage, segregating populations and core collections are developed, genetic maps are constructed, and large number of Chinese cabbage genotypes representing the different morphotypes have been re-sequenced. These data, combined with resequencing data generated of B. rapa core collections by Chinese partners, made it possible to detect signals of domestication for Chinese cabbage leafy head, flowering time and other agronomic traits formation. The focus of my research is to study these domestication events and understand the role of candidate genes. In addition, using the whole genome sequence, we developed a large number of gene-specific SSR and SNP markers and created a Chinese cabbage mutant library to obtain different types of mutants for Chinese cabbage breeding. Now, our research moves towards breeding crops that are better adjusted to sustainable agricultural systems, ranging from strip cropping to organic agriculture. To date, he has published more than 50 SCI papers, including PNAS, Molecular Plant, Cell Reports.
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Javaria Tabusam, Email: shensx@hebau.edu.cn.
Mengyang Liu, Email: mawei0720@163.com.
Shuxing Shen, Email: shensx@hebau.edu.cn.
Wei Ma, Email: mawei0720@163.com.
Jianjun Zhao, Email: jjz1971@aliyun.com.
References
- 1.Efroni I., Eshed Y., Lifschitz E. Morphogenesis of simple and compound leaves: a critical review. Plant Cell. 2010;22(4):1019–1032. doi: 10.1105/tpc.109.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar M., Ori N. Leaf development and morphogenesis. Development. 2014;141(22):4219–4230. doi: 10.1242/dev.106195. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez N., Vanhaeren H., Inzé D. Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 2012;17(6):332–340. doi: 10.1016/j.tplants.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Xu P., Ali A., Han B., Wu X. Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front Plant Sci. 2018;9:1528. doi: 10.3389/fpls.2018.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z., Xie B., Wu T., Xin X., Man L., Tan G., et al. Karyotyping and identifying all of the chromosomes of allopolyploid Brassica juncea using multicolor FISH. Crop J. 2016;4(4):266–274. [Google Scholar]
- 6.J.D. Krieger. Controlling for curvature in the quantification of leaf form. In: Morphometrics for nonmorphometricians. Springer, Berlin: Heidelberg; 2010. p. 27-71.
- 7.Nath U., Crawford B.C., Carpenter R., Coen E. Genetic control of surface curvature. Science. 2003;299(5611):1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y., Wu F., Yu X., Bai J., He Y. miR319a-targeted BrpTCP genes modulate head shape in Brassica rapa by differential cell division arrest in leaf regions. Plant Physiol. 2013;710–720 doi: 10.1104/pp.113.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.X. Zhang, W. Ma, M. Liu, X. Li, J. Li, Y. Lu, G. Li, S. Zhang, D. Feng, Y. Wang. OCTOPUS regulates BIN2 to control leaf curvature in Chinese cabbage, Proceedings of the National Academy of Sciences 119(34) (2022). e2208978119. [DOI] [PMC free article] [PubMed]
- 10.Wang F., Li L., Li H., Liu L., Zhang Y., Gao J., et al. Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics. 2012;99(5):299–307. doi: 10.1016/j.ygeno.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Mou B. Nutritional quality of lettuce. Curr Nutr Food Sci. 2012;8(3):177–187. [Google Scholar]
- 12.Šamec D., Pavlović I., Salopek-Sondi B. White cabbage (Brassica oleracea var. capitata f. alba): botanical, phytochemical and pharmacological overview. Phytochem Lett. 2017;16(1):117–135. [Google Scholar]
- 13.Caunii A., Cuciureanu R., Zakar A.M., Tonea E., Giuchici C. Chemical composition of common leafy vegetables, Studia Universitatis“ Vasile Goldis”. 2010;20(2):45. Arad. Seria Stiintele Vietii (Life Sciences Series. [Google Scholar]
- 14.Yu X., Wang H., Zhong W., Bai J., Liu P., He Y. QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa. PLoS One. 2013;8(10):e76059. doi: 10.1371/journal.pone.0076059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y.K., Xue W.X., Sun Y.D., Yu X.H., Liu P.L. Leafy head formation of the progenies of transgenic plants of Chinese cabbage with exogenous auxin genes. Cell Res. 2000;10(2):151–160. doi: 10.1038/sj.cr.7290044. [DOI] [PubMed] [Google Scholar]
- 16.Fahey J.W. Encyclopedia of food and health. Elsevier Inc; 2015. Brassica: characteristics and properties; pp. 469–477. [Google Scholar]
- 17.Cheng F., Wu J., Cai X., Liang J., Freeling M., Wang X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 2018;4(5):258–268. doi: 10.1038/s41477-018-0136-7. [DOI] [PubMed] [Google Scholar]
- 18.Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat Genet. 2016;48(10):1218–1224. doi: 10.1038/ng.3634. [DOI] [PubMed] [Google Scholar]
- 19.Barkoulas M., Galinha C., Grigg S.P., Tsiantis M. From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol. 2007;10(6):660–666. doi: 10.1016/j.pbi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Du F., Guan C., Jiao Y. Molecular mechanisms of leaf morphogenesis. Mol Plant. 2018;11(9):1117–1134. doi: 10.1016/j.molp.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Javid M.G., Sorooshzadeh A., Moradi F., Modarres Sanavy S.A.M., Allahdadi I. The role of phytohormones in alleviating salt stress in crop plants. Aust J Crop Sci. 2011;5(6):726–734. [Google Scholar]
- 22.Raza A., Salehi H., Rahman M.A., Zahid Z., Madadkar Haghjou M., Najafi-Kakavand S., et al. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front Plant Sci. 2022;13 doi: 10.3389/fpls.2022.961872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J., Gao L., Liu W., Song L., Xiao D., Liu T., et al. Transcription coactivator ANGUSTIFOLIA3 (AN3) regulates leafy head formation in Chinese cabbage. Front Plant Sci. 2019;10:520. doi: 10.3389/fpls.2019.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishijima T., Fukino N. Geometrical analysis of development of erect leaves as a factor in head formation of Brassica rapa L.:(II) Comparative analysis of headed and non-headed cultivars. Sci Hortic. 2005;104(4):421–431. [Google Scholar]
- 25.Royer D.L., McElwain J.C., Adams J.M., Wilf P. Sensitivity of leaf size and shape to climate within Acer rubrum and Quercus kelloggii. New Phytol. 2008;179(3):808–817. doi: 10.1111/j.1469-8137.2008.02496.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu P., Ali A., Han B., Wu X. Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front Plant Sci. 2018:1528. doi: 10.3389/fpls.2018.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichihashi Y., Kawade K., Usami T., Horiguchi G., Takahashi T., Tsukaya H. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 2011;157(3):1151–1162. doi: 10.1104/pp.111.185066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakata M., Okada K. The leaf adaxial-abaxial boundary and lamina growth. Plants. 2013;2(2):174–202. doi: 10.3390/plants2020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Kong F., Zhou C. From genes to networks: the genetic control of leaf development. J Integr Plant Biol. 2021;63(7):1181–1196. doi: 10.1111/jipb.13084. [DOI] [PubMed] [Google Scholar]
- 30.Ali S., Khan N., Xie L. Molecular and hormonal regulation of leaf morphogenesis in Arabidopsis. Int J Mol Sci. 2020;21(14):5132. doi: 10.3390/ijms21145132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazama T., Ichihashi Y., Murata S., Tsukaya H. The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 2010;51(6):1046–1054. doi: 10.1093/pcp/pcq051. [DOI] [PubMed] [Google Scholar]
- 32.Nicolas M., Cubas P. TCP factors: new kids on the signaling block. Curr Opin Plant Biol. 2016;33:33–41. doi: 10.1016/j.pbi.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Anastasiou E., Kenz S., Gerstung M., MacLean D., Timmer J., Fleck C., et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell. 2007;13(6):843–856. doi: 10.1016/j.devcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Czesnick H., Lenhard M. Size control in plants—lessons from leaves and flowers. Cold Spring Harb Perspect Biol. 2015;7(8) doi: 10.1101/cshperspect.a019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y., Dai X., Zhao Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19(8):2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J., Liu B., Wu J., Cheng F., Wang X. Genetic variation and divergence of genes involved in leaf adaxial-abaxial polarity establishment in Brassica rapa. Front Plant Sci. 2016;7:94. doi: 10.3389/fpls.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinon V., Prasad K., Grigg S.P., Sanchez-Perez G.F., Scheres B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(3):1107–1112. doi: 10.1073/pnas.1213497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez R.E., Debernardi J.M., Palatnik J.F. Morphogenesis of simple leaves: regulation of leaf size and shape. Wiley Interdisciplinary Reviews: Dev Biol. 2014;3(1):41–57. doi: 10.1002/wdev.115. [DOI] [PubMed] [Google Scholar]
- 39.Gao L.-W., Lyu S.-W., Tang J., Zhou D.-Y., Bonnema G., Xiao D., et al. Genome-wide analysis of auxin transport genes identifies the hormone responsive patterns associated with leafy head formation in Chinese cabbage. Sci Rep. 2017;7(1):1–13. doi: 10.1038/srep42229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan C., Wu B., Yu T., Wang Q., Krogan N.T., Liu X., et al. Spatial auxin signaling controls leaf flattening in Arabidopsis. Curr Biol. 2017;27(19) doi: 10.1016/j.cub.2017.08.042. pp. 2940–2950. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi N., Wu M.-F., Winter C.M., Berns M.C., Nole-Wilson S., Yamaguchi A., et al. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell. 2013;24(3):271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Guilfoyle T.J., Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Huang Z., Duan W., Song X., Tang J., Wu P., Zhang B., et al. Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families in Brassica rapa shed light on their evolution patterns in plants. Genome Biol Evol. 2016;8(2):302–316. doi: 10.1093/gbe/evv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mun J.-H., Yu H.-J., Shin J.Y., Oh M., Hwang H.-J., Chung H. Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution Mol. Genet Genom. 2012;287(10):765–784. doi: 10.1007/s00438-012-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pekker I., Alvarez J.P., Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17(11):2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu C., Yan C., Liu Y., Liu Y., Jia Y., Lavelle D., et al. Upregulation of a KN1 homolog by transposon insertion promotes leafy head development in lettuce. Proc Natl Acad Sci USA. 2020;117(52):33668–33678. doi: 10.1073/pnas.2019698117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manuela D., Xu M. Patterning a leaf by establishing polarities. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.568730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waites R., Hudson A. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. J Dev. 1995;121(7):2143–2154. [Google Scholar]
- 49.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13(20):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 50.Hay J.O., Moulia B., Lane B., Freeling M., Silk W.K. Biomechanical analysis of the Rolled (RLD) leaf phenotype of maize. Am J Bot. 2000;87(5):625–633. [PubMed] [Google Scholar]
- 51.Moulia B. Leaves as shell structures: double curvature, auto-stresses, and minimal mechanical energy constraints on leaf rolling in grasses. J Plant Growth Regul. 2000;19(1):19–30. doi: 10.1007/s003440000004. [DOI] [PubMed] [Google Scholar]
- 52.Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25(2):223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- 53.Prigge M.J., Clark S.E. Evolution of the class III HD-Zip gene family in land plants. Evol Dev. 2006;8(4):350–361. doi: 10.1111/j.1525-142X.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 54.Yu L., Yu X., Shen R., He Y. HYL1 gene maintains venation and polarity of leaves. Planta. 2005;221(2):231–242. doi: 10.1007/s00425-004-1439-7. [DOI] [PubMed] [Google Scholar]
- 55.Ren W., Wang H., Bai J., Wu F., He Y. Association of microRNAs with types of leaf curvature in Brassica rapa. Front Plant Sci. 2018;9:73. doi: 10.3389/fpls.2018.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Gao, Y. Lu, X. Li, N. Li, X. Zhang, X. Su, D. Feng, M. Liu, S. Xuan, A. Gu. Development and Application of SSR Markers Related to Genes Involved in Leaf Adaxial-Abaxial Polarity Establishment in Chinese Cabbage (Brassica rapa L. ssp. pekinensis), Front. Genet. 11 (2020). 773. [DOI] [PMC free article] [PubMed]
- 57.Nakata M., Matsumoto N., Tsugeki R., Rikirsch E., Laux T., Okada K. Roles of the middle domain–specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell. 2012;24(2):519–535. doi: 10.1105/tpc.111.092858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeon H.-W., Byrne M.E. SAW homeodomain transcription factors regulate initiation of leaf margin serrations. J Exp Bot. 2021;72(5):1738–1747. doi: 10.1093/jxb/eraa554. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S., Giri D. Does changing the pulling direction give better insight into biomolecules? Phys Rev Lett. 2007;98(4) doi: 10.1103/PhysRevLett.98.048101. [DOI] [PubMed] [Google Scholar]
- 60.An G., Yu C., Yan C., Wang M., Zhang W., Jia Y., et al. Loss-of-function of SAWTOOTH 1 affects leaf dorsiventrality genes to promote leafy heads in lettuce. Plant Cell. 2022 doi: 10.1093/plcell/koac234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. Hay, M. Barkoulas, M. Tsiantis. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis, (2006). [DOI] [PubMed]
- 62.Iwakawa H., Iwasaki M., Kojima S., Ueno Y., Soma T., Tanaka H., et al. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007;51(2):173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- 63.Lin W.-C., Shuai B., Springer P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES–domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 2003;15(10):2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin Y., Hou H., Zhang Y., Hou X. Overexpression of a Pak Choi Gene, BcAS2, Causes Leaf Curvature in Arabidopsis thaliana. Genes. 2021;12(1):102. doi: 10.3390/genes12010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsumura Y., Iwakawa H., Machida Y., Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant Sci J. 2009;58(3):525–537. doi: 10.1111/j.1365-313X.2009.03797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Husbands A.Y., Benkovics A.H., Nogueira F.T., Lodha M., Timmermans M.C. The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell. 2015;27(12):3321–3335. doi: 10.1105/tpc.15.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16(9):927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 68.Garcia D., Collier S.A., Byrne M.E., Martienssen R.A. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16(9):933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 69.C. Hunter, M.R. Willmann, G. Wu, M. Yoshikawa, M. de la Luz Gutiérrez-Nava, S.R. Poethig. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis, (2006). [DOI] [PMC free article] [PubMed]
- 70.Kidner C.A., Timmermans M.C. Mixing and matching pathways in leaf polarity. Curr Opin Plant Biol. 2007;10(1):13–20. doi: 10.1016/j.pbi.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 71.Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A.S., Gerats T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell. 2009;21(8):2269–2283. doi: 10.1105/tpc.109.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J., Zhang X., Lu Y., Feng D., Gu A., Wang S., et al. Characterization of non-heading mutation in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis), Front. Plant Sci. 2019;10:112. doi: 10.3389/fpls.2019.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y., Liang J., Cai X., Chen H., Wu J., Lin R., et al. Divergence of three BRX homoeologs in Brassica rapa and its effect on leaf morphology. Hortic Res. 2021;8:68. doi: 10.1038/s41438-021-00504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beuchat J., Scacchi E., Tarkowska D., Ragni L., Strnad M., Hardtke C.S. BRX promotes Arabidopsis shoot growth. New Phytol. 2010;188(1):23–29. doi: 10.1111/j.1469-8137.2010.03387.x. [DOI] [PubMed] [Google Scholar]
- 75.Scacchi E., Osmont K.S., Beuchat J., Salinas P., Navarrete-Gómez M., Trigueros M., et al. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development. 2009;136(12):2059–2067. doi: 10.1242/dev.035444. [DOI] [PubMed] [Google Scholar]
- 76.Mouchel C.F., Osmont K.S., Hardtke C.S. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443(7110):458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- 77.Floris M., Mahgoub H., Lanet E., Robaglia C., Menand B. Post-transcriptional regulation of gene expression in plants during abiotic stress. Int J Mol Sci. 2009;10(7):3168–3185. doi: 10.3390/ijms10073168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411(6838):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 79.Yang X., Ren W., Zhao Q., Zhang P., Wu F., He Y. Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res. 2014;42(19):12224–12236. doi: 10.1093/nar/gku907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Z., Jia L., Mao Y., He Y. Classification and quantification of leaf curvature. J Exp Bot. 2010;61(10):2757–2767. doi: 10.1093/jxb/erq111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren W., Wu F., Bai J., Li X., Yang X., Xue W., et al. BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis) Hortic Res. 2020;7(1):1–13. doi: 10.1038/s41438-019-0222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y., Wu F., Bai J., He Y. Brp SPL 9 (B rassica rapa ssp. pekinensis SPL 9) controls the earliness of heading time in C hinese cabbage. Plant Biotechnol J. 2014;12(3):312–321. doi: 10.1111/pbi.12138. [DOI] [PubMed] [Google Scholar]
- 83.Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16(9):939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 84.Saeed F., Chaudhry U.K., Bakhsh A., Raza A., Saeed Y., Bohra A., et al. Moving beyond DNA sequence to improve plant stress responses. Front Genet. 2022;929 doi: 10.3389/fgene.2022.874648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3(9):662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 86.Martienssen R.A., Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293(5532):1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y., Xu C., Tang X., Pei S., Jin D., Guo M., et al. Genomic methylation and transcriptomic profiling provides insights into heading depression in inbred Brassica rapa L. ssp. pekinensis. Gene. 2018;665:119–126. doi: 10.1016/j.gene.2018.04.047. [DOI] [PubMed] [Google Scholar]
- 88.Salmon A., Clotault J., Jenczewski E., Chable V., Manzanares-Dauleux M.J. Brassica oleracea displays a high level of DNA methylation polymorphism. Plant Sci. 2008;174(1):61–70. [Google Scholar]
- 89.J. Alemán-Báez, J. Qin, C. Cai, C. Zou, J. Bucher, M.-J. Paulo, R.E. Voorrips, G. Bonnema. Genetic dissection of morphological variation in rosette leaves and leafy heads in cabbage (Brassica oleracea var. capitata), (2022). [DOI] [PMC free article] [PubMed]
- 90.Chen Q., Song J., Du W.-P., Xu L.-Y., Jiang Y., Zhang J., et al. Identification, mapping, and molecular marker development for Rgsr8. 1: a new quantitative trait locus conferring resistance to Gibberella stalk rot in maize (Zea mays L.) Front Plant Sci. 2017;8:1355. doi: 10.3389/fpls.2017.01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skopelitis D.S., Benkovics A.H., Husbands A.Y., Timmermans M.C. Boundary formation through a direct threshold-based readout of mobile small RNA gradients. Dev Cell. 2017;43(3):265–273. doi: 10.1016/j.devcel.2017.10.003. e6. [DOI] [PubMed] [Google Scholar]
- 92.Foolad M.R. Genome mapping and molecular breeding of tomato. Int J Plant Genomics. 2007 doi: 10.1155/2007/64358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.M.C. Schruff, M. Spielman, S. Tiwari, S. Adams, N. Fenby, R.J. Scott. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs, (2006). [DOI] [PubMed]
- 94.Gray W.M. Hormonal regulation of plant growth and development. PLoS Biol. 2004;2(9):e311. doi: 10.1371/journal.pbio.0020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao Y., Huang S., Qu G., Fu W., Zhang M., Liu Z., et al. The mutation of ent-kaurene synthase, a key enzyme involved in gibberellin biosynthesis, confers a non-heading phenotype to Chinese cabbage (Brassica rapa L. ssp pekinensis), Hortic Res. 2020;7 doi: 10.1038/s41438-020-00399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millar A.A., Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17(3):705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.N. Haga, K. Kato, M. Murase, S. Araki, M. Kubo, T. Demura, K. Suzuki, I. Müller, U. Voß, G. Jürgens. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana, (2007). [DOI] [PubMed]
- 98.Holst K., Schmülling T., Werner T. Enhanced cytokinin degradation in leaf primordia of transgenic Arabidopsis plants reduces leaf size and shoot organ primordia formation. J Plant Physiol. 2011;168(12):1328–1334. doi: 10.1016/j.jplph.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 99.Frugis G., Giannino D., Mele G., Nicolodi C., Chiappetta A., Bitonti M.B., et al. Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 2001;126(4):1370–1380. doi: 10.1104/pp.126.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15(17):1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 101.Crawford B.C., Nath U., Carpenter R., Coen E.S. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 2004;135(1):244–253. doi: 10.1104/pp.103.036368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014;164(2):710–720. doi: 10.1104/pp.113.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Das Gupta M., Aggarwal P., Nath U. CINCINNATA in A ntirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 2014;204(4):901–912. doi: 10.1111/nph.12963. [DOI] [PubMed] [Google Scholar]
- 104.Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010;22(11):3574–3588. doi: 10.1105/tpc.110.075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lan J., Qin G. The regulation of CIN-like TCP transcription factors. Int J Mol Sci. 2020;21(12):4498. doi: 10.3390/ijms21124498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seki K., Komatsu K., Tanaka K., Hiraga M., Kajiya-Kanegae H., Matsumura H., et al. A CIN-like TCP transcription factor (LsTCP4) having retrotransposon insertion associates with a shift from Salinas type to Empire type in crisphead lettuce (Lactuca sativa L.) Hortic Res. 2020;7(1):1–14. doi: 10.1038/s41438-020-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Du J., Hu S., Yu Q., Wang C., Yang Y., Sun H., et al. Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa, Front. Plant Sci. 2017;8:1588. doi: 10.3389/fpls.2017.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6(9):e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H., Zhou Y., Gilmer S., Whitwill S., Fowke L.C. Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 2000;24(5):613–623. doi: 10.1046/j.1365-313x.2000.00899.x. [DOI] [PubMed] [Google Scholar]
- 110.Zulfiqar S., Zhao T., Liu Y., Wei L., Farooq M.A., Tabusam J., et al. Genome-Wide Identification, Characterization, and Transcriptomic Analysis of the Cyclin Gene Family in Brassica rapa. Int J Mol Sci. 2022;23(22):14017. doi: 10.3390/ijms232214017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carneiro A.K., Montessoro P.d.F., Fusaro A.F., Araújo B.G., Hemerly A.S. Plant CDKs—driving the cell cycle through climate change. Plants. 2021;10(9):1804. doi: 10.3390/plants10091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malinowski R., Kasprzewska A., Fleming A.J. Targeted manipulation of leaf form via local growth repression. Plant J. 2011;66(6):941–952. doi: 10.1111/j.1365-313X.2011.04559.x. [DOI] [PubMed] [Google Scholar]
- 113.Schnittger A., Weinl C., Bouyer D., Schöbinger U., Hülskamp M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell. 2003;15(2):303–315. doi: 10.1105/tpc.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanz L., Dewitte W., Forzani C., Patell F., Nieuwland J., Wen B., et al. The Arabidopsis D-type cyclin CYCD2; 1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell. 2011;23(2):641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang H., Qi Q., Schorr P., Cutler A.J., Crosby W.L., Fowke L.C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998;15(4):501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- 116.Reinhardt B., Hänggi E., Müller S., Bauch M., Wyrzykowska J., Kerstetter R., et al. Restoration of DWF4 expression to the leaf margin of a dwf4 mutant is sufficient to restore leaf shape but not size: the role of the margin in leaf development. Plant J. 2007;52(6):1094–1104. doi: 10.1111/j.1365-313X.2007.03304.x. [DOI] [PubMed] [Google Scholar]
- 117.Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137(1):103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.G. Horiguchi, G.T. Kim, H. Tsukaya. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana, Plant J. 43(1) (2005). 68-78. J. Dev. [DOI] [PubMed]
- 119.Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19(14):1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 120.Hou H., Zhang C., Hou X. Cloning and Functional Analysis of BcMYB101 Gene Involved in Leaf Development in Pak Choi (Brassica rapa ssp. Chinensis) Int J Mol Sci. 2020;21(8):2750. doi: 10.3390/ijms21082750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Raza A., Tabassum J., Kudapa H., Varshney R.K. Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol. 2021;41(8):1209–1232. doi: 10.1080/07388551.2021.1898332. [DOI] [PubMed] [Google Scholar]
- 122.Raza A., Charagh S., Razzaq A., Javed R., Khan R.S.A., Hasanuzzaman M. Springer; The plant family brassicaceae, In: 2020. Brassicaceae plants response and tolerance to drought stress: physiological and molecular interventions; pp. 229–261. [Google Scholar]
- 123.Tabusam J., Shi Q., Feng D., Zulfiqar S., Shen S., Ma W., et al. HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress. Cells. 2022;11(15):2316. doi: 10.3390/cells11152316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ge L., Peng J., Berbel A., Madueño F., Chen R. Regulation of compound leaf development by PHANTASTICA in Medicago truncatula. Plant Physiol. 2014;164(1):216–228. doi: 10.1104/pp.113.229914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee R.J., Bhandari S.R., Lee G., Lee J.G. Optimization of temperature and light, and cultivar selection for the production of high-quality head lettuce in a closed-type plant factory. Hortic Environ Biotechnol. 2019;60(2):207–216. [Google Scholar]
- 126.A. EL Sabagh, M.S. Islam, A. Hossain, M. Mubeen, M.A. Iqbal, M. Waleed, M. Reginato, M. Battaglia, S. Ahmed, A. Rehman. Phytohormones as growth regulators during abiotic stress tolerance in plants, (2022).
- 127.Zhang C.-W., Wei Y.-P., Xiao D., Gao L.-W., Lyu S.-W., Hou X.-L., et al. Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. J Proteomics. 2016;144:1–10. doi: 10.1016/j.jprot.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 128.Zhang K., Yang Y., Wu J., Liang J., Chen S., Zhang L., et al. A cluster of transcripts identifies a transition stage initiating leafy head growth in heading morphotypes of Brassica. Plant J. 2022 doi: 10.1111/tpj.15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wurr D., Fellows J.R. The influence of solar radiation and temperature on the head weight of crisp lettuce. J Hortic Sci. 1991;66(2):183–190. [Google Scholar]
- 130.Doo H.-S., Bae H.-J., Kwon B.-R., Shin Y.-J., Chung C.-H., Kwon T.-H. Heading according to LED modules and short length waves for cultivation of head lettuce (Lactuca sativa var. capitata) in plant factory. Symposium of Korean society of plant resources. 2012 [Google Scholar]
- 131.Batistič O., Kudla J. Analysis of calcium signaling pathways in plants. Biochim Biophys Acta Gen Subj. 2012;1820(8):1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 132.Hepler P.K. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17(8):2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang M., Huang S., Gao Y., Fu W., Qu G., Zhao Y., et al. Fine mapping of a leaf flattening gene Bralcm through BSR-Seq in Chinese cabbage (Brassica rapa L. ssp pekinensis), Sci Rep. 2020;10(1):1–10. doi: 10.1038/s41598-020-70975-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Portillo F. Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim Biophys Acta - Biomembr. 2000;1469(1):31–42. doi: 10.1016/s0304-4157(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 135.Haruta M., Tan L.X., Bushey D.B., Swanson S.J., Sussman M.R. Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol. 2018;176(1):364–377. doi: 10.1104/pp.17.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Raza A., Mubarik M.S., Sharif R., Habib M., Jabeen W., Zhang C., et al. Developing drought-smart, ready-to-grow future crops. J Plant Genome Sci. 2022:e20279. doi: 10.1002/tpg2.20279. [DOI] [PubMed] [Google Scholar]
- 137.Krishnamurthy P., Muthusamy M., Kim J.A., Jeong M.-J., Lee S.I. Brassica rapa expansin-like B1 gene (BrEXLB1) regulate growth and development in transgenic Arabidopsis and elicits response to abiotic stresses. J Plant Biochem Biotechnol. 2019;28(4):437–446. [Google Scholar]
- 138.Li P., Zhang B., Su T., Li P., Xin X., Wang W., et al. a GRAS transcription factor from Brassica rapa, is involved in drought stress tolerance in transgenic Arabidopsis. Front Plant Sci. 2018;9:1792. doi: 10.3389/fpls.2018.01792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lawal A. University of Warwick; 2019. Investigating salt stress resilience in Brassica oleracea. [Google Scholar]
- 140.Raza A., Tabassum J., Fakhar A.Z., Sharif R., Chen H., Zhang C., et al. Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol. 2022;1–28 doi: 10.1080/07388551.2022.2093695. [DOI] [PubMed] [Google Scholar]
- 141.Ali A., Wu T., Zhang H., Xu P., Zafar S.A., Liao Y., et al. A putative SUBTILISIN-LIKE SERINE PROTEASE 1 (SUBSrP1) regulates anther cuticle biosynthesis and panicle development in rice. J Adv Res. 2022 doi: 10.1016/j.jare.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hou H., Lin Y., Hou X. Ectopic Expression of a Pak-choi YABBY Gene, BcYAB3, Causes Leaf Curvature and Flowering Stage Delay in Arabidopsis thaliana. Genes. 2020;11(4):370. doi: 10.3390/genes11040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. The role of JAGGED in shaping lateral organs. Development. 2004;131(5):1101–1110. doi: 10.1242/dev.00949. [DOI] [PubMed] [Google Scholar]
- 144.Liu H., Yu H., Tang G., Huang T. Small but powerful: function of microRNAs in plant development. Plant Cell Rep. 2018;37(3):515–528. doi: 10.1007/s00299-017-2246-5. [DOI] [PubMed] [Google Scholar]