Abstract
Stachydrine, a betaine released by germinating alfalfa seeds, functions as an inducer of nodulation genes, a catabolite, and an osmoprotectant in Sinorhizobium meliloti. Two stachydrine-inducible genes were found in S. meliloti 1021 by mutation with a Tn5-luxAB promoter probe. Both mutant strains (S10 and S11) formed effective alfalfa root nodules, but neither grew on stachydrine as the sole carbon and nitrogen source. When grown in the absence or presence of salt stress, S10 and S11 took up [14C]stachydrine as well as wild-type cells did, but neither used stachydrine effectively as an osmoprotectant. In the absence of salt stress, both S10 and S11 took up less [14C]proline than wild-type cells did. S10 and S11 appeared to colonize alfalfa roots normally in single-strain tests, but when mixed with the wild-type strain, their rhizosphere counts were reduced more than 50% (P ≤ 0.01) relative to the wild type. These results suggest that stachydrine catabolism contributes to root colonization. DNA sequence analysis identified the mutated locus in S11 as putA, and the luxAB fusion in that gene was induced by proline as well as stachydrine. DNA that restored the capacity of mutant S10 to catabolize stachydrine contained a new open reading frame, stcD. All data are consistent with the concept that stcD codes for an enzyme that produces proline by demethylation of N-methylproline, a degradation product of stachydrine.
Plants release a variety of compounds that affect soil microorganisms and contribute to structuring microbial communities in the rhizosphere. More than 400 different molecules identified in alfalfa (Medicago sativa L.) may eventually reach soil bacteria through exudation or decomposition (38). Some of these compounds serve as transcriptional regulators of genes in Rhizobium meliloti (29), which was recently reclassified as Sinorhizobium meliloti (8). Other compounds are cofactors for microbial enzymes (40), and a few function as osmoprotectants (33). Any plant-derived compound present in sufficient concentration probably can serve as an energy substrate for rhizosphere microbes (5). However, defining molecules that have selective or specific effects on bacterial growth in the alfalfa rhizosphere and identifying rhizobial genes that are involved in responding to these factors is essential to understanding how communities of microorganisms develop on plant roots.
Several genetic tools have been used to define molecular plant-microbe interactions in the rhizosphere. Mutagenesis experiments have located a number of bacterial genes affecting root colonization (23, 31). In addition, enrichment for bacterial transconjugants (3) or for DNA amplifications (30) has found genes that enhance rhizosphere growth. The importance of a specific molecule in the rhizosphere can also be assessed by transforming plants to overproduce the compound (17) or by measuring the competitiveness of bacterial mutants incapable of catabolizing that molecule (10, 24).
Stachydrine, also known as N,N-dimethylproline or proline betaine, is a quaternary ammonium derivative of proline that occurs widely in Medicago species (39) but not in other genera of the Leguminosae (48). Most stachydrine-accumulating plants are found in the Capparidaceae (caper) and Labiatae (mint) families (48). Stachydrine was identified as an inducer of S. meliloti nodulation (nod) genes in alfalfa seed rinses (37), and many Medicago species nodulated by S. meliloti deposit this compound on developing seeds, where it is available to soil bacteria during seed germination (39). Stachydrine probably is a general osmoprotectant in alfalfa, because it is found not only on dried seed coats and in cured hay (43) but also in water-stressed alfalfa root nodules (20). Thus, S. meliloti may be exposed to stachydrine both during colonization of the young alfalfa root and later as N2-fixing cells in the root nodules.
Because S. meliloti uses stachydrine as a carbon and nitrogen source (16) and as an osmoprotectant (14), this molecule may be ecologically important for root colonization by this microorganism. In the absence of osmotic stress, S. meliloti cells catabolize stachydrine by sequential demethylations to N-methylproline and proline, but osmotically stressed cells accumulate stachydrine while catabolism is strongly reduced (14). To examine the role of stachydrine in root colonization, we mutagenized S. meliloti with the transposon Tn5-luxAB (47), which generates both insertional mutations and transcriptional luciferase reporter gene fusions. Here we describe the identification of two stachydrine-inducible genes in S. meliloti and show that the corresponding Tn5-luxAB insertional mutants are impaired in the use of stachydrine as a catabolite and as an osmoprotectant. The data also suggest that both mutants are impaired in competition for colonization of the rhizosphere. One of the stachydrine-inducible genes is identified as putA, a gene that was recently shown to promote root colonization (19). The other stachydrine-inducible locus (stcD) has not been previously identified and is shown to encode a putative demethylase that converts N-methylproline to proline. Unlike other genes involved in stachydrine catabolism (15), stcD is not located on the symbiotic plasmid of S. meliloti.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. S. meliloti and Escherichia coli were maintained, respectively, in TY medium (4) and Luria-Bertani LB medium (41). S. meliloti was grown in GTS (22), LAS (14), MAS (42), or VM (46) medium as specified. NYG agar plates (peptone, 5 mg/ml; yeast extract, 3 mg/ml; glycerol, 20 mg/ml; agar, 15 mg/ml) were used for mating experiments. Antibiotics, unless otherwise stated, were used at the following concentrations: kanamycin, 10 μg/ml for E. coli and 200 μg/ml for S. meliloti; chloramphenicol, 50 μg/ml; tetracycline, 5 μg/ml for S. meliloti and 10 μg/ml for E. coli; ampicillin, 50 μg/ml. Growth temperatures were 28°C for S. meliloti and 37°C for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant trait(s) | Source or reference |
|---|---|---|
| E. coli | ||
| HB101 | pro leu thi lacY Strr, endoI recA r−, m− | 9 |
| YMC11 | pro thi endA hsr Δ(glnA-glnD) | 2 |
| S. meliloti | ||
| Rm1021 | 2011, Smr | 32 |
| Rm1021-S10 (S10) | 1021::Tn5pRL1063a | This work |
| Rm1021-S11 (S11) | 1021::Tn5pRL1063a | This work |
| Plasmids | ||
| pBSK+ | Multicopy cloning vector | Stratagene |
| pRL1063a | Tn5-luxAB, Smr Kmr OriV | 47 |
| pLAFR1 | Tcr, Cos Tra− Mob+, IncP | 13 |
| pRK2013 | pRK212.2 conjugation helper plasmid | 11 |
| pSIF10 | Circularized 10-kb EcoRI fragment of Rm1021-S10::Tn5-1063 | This work |
| pSIF11 | Circularized 12-kb EcoRI fragment of Rm1021-S11::Tn5-1063 | This work |
| pLES1 | pLAFR1 cosmid of Rm1021 containing stcD | This work |
| pLEB1 | 6.3-kb EcoRI-BamHI fragment carrying stcD in pBSK+ | This work |
| pLBR55 | 6.8-kb EcoRI fragment carrying nodD2 in pBSK+ | 16 |
| pLBR56 | 6.5-kb EcoRI fragment near nodD2 in pBSK+ | 16 |
Generation of mutants and reporter gene insertions.
Plasmid pRL1063a, which contains the transposable promoter probe Tn5-1063 with a promoterless luxAB reporter and the oriV of E. coli (47), was used to generate 5,000 insertional mutants of S. meliloti 1021 (27). The mutants were screened for stachydrine-mediated induction of lux expression after being grown for 3 days on Rec-85 filters (Nuclepore Corp., Pleasanton, Calif.) on GTS agar medium. The filters were moved to fresh GTS agar with or without 100 μM stachydrine for 6 h and then exposed to the luciferase reporter substrate n-decanal (Sigma Co., St. Louis, Mo.). Luminescence was recorded for 4 h on sensitized (25) XAR X-ray film (Eastman Kodak, Rochester, N.Y.). Mutants showing stachydrine-dependent bioluminesence were retested twice and compared with a positive-control mutant in which luxAB was expressed constitutively. The stachydrine used in these studies was synthesized by standard procedures (35). Responses of mutant strains S10 and S11 to different concentrations of stachydrine and l-proline (Sigma), were monitored in GTS liquid medium for 1 min with a luminometer (Lumat LB9501; Wallace Inc., Gaithersberg, Md.) after various times of exposure to potential inducers. Data were recorded as relative light units (RLU) as specified by the manufacturer, using medium without cells as a zero standard and using the optical density at 600 nm (OD600) to monitor cell density.
DNA manipulations.
Genomic DNA was isolated by standard methods (41) and digested with restriction enzymes (Promega, Madison, Wis.) as specified by the manufacturer. In some experiments, large plasmids were separated by published methods (21). Restriction fragments were separated by electrophoresis in 0.8% agarose gels, transferred to nylon membranes (MSI, Westboro, Mass.), and cross-linked with UV light. Hybridizations were performed overnight with digoxigenin-labeled DNA probes under high-stringency conditions (68°C). Hybridization signals were detected with the chemoluminescent substrate CDP* as specified by the manufacturer (Boehringer, Mannheim, Germany). Recombinant plasmids pSIF10 and pSIF11 (stachydrine-inducible fragments) were recovered from S10 and S11 genomic DNA, which was cut with EcoRI, treated with T4 DNA ligase to form circular DNA, and electroporated into E. coli HB101, where oriV in pRL1063a maintained the DNA as a plasmid. Plasmid DNA from pSIF10 and pSIF11 was isolated by using the SNAP miniprep system (Invitrogen Corp., San Diego, Calif.) and used to determine the DNA sequence of the flanking regions of the Tn5 insertion in each mutant. DNA sequences were determined by the Sanger method (41) with an automatic ABI377 sequencer (Applied Biosystems Perkin-Elmer, Foster City, Calif.).
Probes for DNA hybridizations were generated by random-primed labeling with digoxigenin (Boehringer) during PCR amplification with primers Tn5out (5′GAA AGG TTC CGT TCA GGA CGC TAC3′) (GenBank, National Centre for Biotechnology Information [NCBI], and National Institutes of Health [NIH]) and either LJ1 (5′CCG ACA TGG GCG ACC AGT TCA TT3′) or LJ2 (5′ACG AGC CAG TTC AGG CAA TAG GCG3′) from the Tn5-flanking regions in pSIF10 and pSIF11, respectively. PCRs were carried out with a PTC-100 thermal cycler (MJ Research, Watertown, Mass.) with 25- or 35-cycle reactions consisting of denaturing (1 min at 94°C), annealing (1 min at 62 to 64°C), and extension (2 min at 72°C). A nodC-specific probe was generated by PCR with primers LJ3 (5′CTC TGC CAG CCG TGG ATG TTA TCG3′) and LJ4 (5′TCA CTC GAC CGG AGG TTT GAA TTG G3′) (GenBank, NCBI, and NIH). PCR products were analyzed on a 1.5% agarose gel relative to a 1-kb DNA marker ladder (Bethesda Research Laboratories, Gaithersburg, Md.).
A pLAFR1 cosmid bank of Rm1021 DNA in E. coli YMC11 (7) was analyzed with the mutant-specific probes described above to identify cosmids carrying wild-type genes corresponding to the mutated genes in S10 and S11. Cosmid pLES1 was identified as containing the wild-type gene mutated in S10 and was subsequently transferred into S10 by conjugation, using the helper strain E. coli HB101(pRK2013). Transconjugants were selected on GTS agar containing kanamycin and tetracycline. Growth on VM agar without mannitol or potassium nitrate and with 0.3% stachydrine as the sole carbon and nitrogen source was tested. Plasmid pLEB1 was produced by digesting the Rm1021 DNA in cosmid pLES1 with EcoRI and BamHI and cloning the gene of interest into pBSK+ (Stratagene, La Jolla, Calif.). Plasmid pLEB1 was used for additional sequencing of the region mutated in S10. All sequences were examined for nucleic acid and deduced amino acid homologies in the GenBank (NCBI, NIH) database by using the BLAST program (1). Putative transcriptional start and stop sites were identified with the PC/GENE DNA analysis program (IntelliGenetics, Geneva, Switzerland) and used to define open reading frames (ORFs).
Growth experiments.
Mutant strains were tested for growth on 0.3% stachydrine, N-methylproline, or proline as the sole carbon and nitrogen source, using VM agar lacking mannitol and potassium nitrate. Complete VM agar was used as a positive control. Additional growth tests were conducted in VM liquid cultures containing 0.3% stachydrine, N-methylproline, or proline as the sole carbon and nitrogen source. Growth was monitored by measuring the OD600 in three replicates.
Mutant strains were examined for growth in LAS liquid medium with or without 650 mM NaCl and also in MAS medium with 550 mM NaCl and 1 mM stachydrine in the presence of kanamycin. Uptake studies were performed at 28°C as described previously (36), using cells that were grown in liquid TY and then transferred to LAS medium until the protein concentrations reached 100 μg/ml. [14C]proline (264 mCi/mmol) and [14C]stachydrine (126 mCi/mmol) were obtained from the Commissariat à l’Energie Atomique (Gif sur Yvette, France). After the long-term (18-h) stachydrine uptake experiment, bacterial cells were harvested by centrifugation, washed three times, and extracted with 70% ethanol to determine the amount of 14C in the ethanol-soluble and -insoluble fractions.
Rhizosphere colonization experiments.
Root colonization tests were conducted with Moapa 69 alfalfa plants in axenic vermiculite by recovering bacteria from roots of individual plants as described previously (45), except that sterilized seeds were planted directly to minimize stachydrine loss and inocula were grown in TY medium. Each seed was inoculated with 300 to 500 bacterial cells at the time of planting. CFU were determined by plating cells on TY agar with Congo red (10 μg/ml) and appropriate antibiotics. The roots were rinsed in sterile water, and then more than 95% of the viable bacteria were removed from the roots by vortexing and sonication (45). Treatments consisted of at least three replicate jars, each containing five plants. Data obtained with the wild-type and mutant strains (CFU/root) were transformed and compared by using an unpaired t test for single-strain trials and a paired t test for competitive colonization trials. All experiments were repeated at least once.
Nucleotide sequence accession number.
Sequence data for the 2,898-bp DNA fragment containing ORF I, here termed stcD, were submitted to the GenBank (NCBI) database as accession no. AF016307.
RESULTS
In vitro phenotypes.
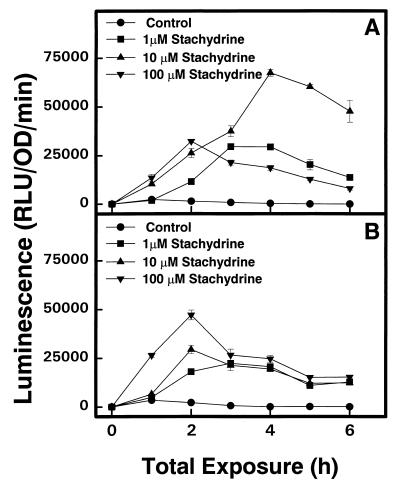
Twelve mutants containing stachydrine-inducible genes were isolated from S. meliloti 1021. Each mutant showed a single Tn5 insertion in hybridization tests after digesting total DNA with EcoRI or ClaI, which cut outside Tn5-1063 (reference 6 and data not shown). All 12 mutants formed effective nodules on Moapa 69 alfalfa plants; 9 mutants had an impaired capacity to use stachydrine as an osmoprotectant in LAS medium containing 500 mM NaCl. Only strains S10 and S11 failed to grow on stachydrine as the sole carbon and nitrogen source, and they were selected for further studies. Maximum induction of the luxAB reporter in strain S10 (Fig. 1A) was produced with a lower concentration of stachydrine (10 μM) than in S11 (100 μM) (Fig. 1B). Mutant S11, but not S10, also expressed luciferase in response to proline (Table 2).
FIG. 1.
Stachydrine induction of luxAB reporter gene expression in two S. meliloti mutant strains containing Tn5-luxAB. Luciferase activity was measured in Rm1021-S10 (A) and Rm1021-S11 (B) cells growing in liquid GTS medium containing 0, 1, 10, or 100 μM stachydrine.
TABLE 2.
Proline uptake and inducing effects in S. meliloti strains
| Strain | Uptake (mean ± SE)
|
luxAB induction (RLU/OD unit/min) (mean ± SE)b
|
||
|---|---|---|---|---|
| Rate (nmol of proline/min/mg)a | Decrease (%) | − Proline | + Proline | |
| Rm1021 | 27 ± 1 | 0 | NAc | NA |
| Rm1021-S10 | 16 ± 4 | 41 | 3,070 ± 584 | 1,390 ± 446 |
| Rm1021-S11 | 6 ± 1 | 78 | 2,490 ± 40 | 26,500 ± 3,180 |
Uptake measured with 10 μM [14C]proline.
Maximum response observed during a 6-h induction test with 10 or 100 μM proline for Rm1021-S10 and Rm1021-S11, respectively.
NA, not applicable.
Strains S10 and S11 differed in their capacity to grow on catabolic products of stachydrine (Table 3). S10 grew on proline but not on N-methylproline, while S11 grew on neither compound. The proline uptake capacity in strains S10 and S11, as measured with 10 μM [14C]proline, was impaired by 41 and 78%, respectively, relative to that in parent cells (Table 2).
TABLE 3.
Growth rates of S. meliloti strains on different carbon and nitrogen sources
| Strain | Doubling time (h) on:
|
||
|---|---|---|---|
| Stachydrine | N-Methylproline | Proline | |
| Rm1021 | 7.1 | 7.5 | 5.6 |
| Rm1021-S10 | NGa | NG | 10.0 |
| Rm1021-S11 | NG | NG | NG |
NG, no growth.
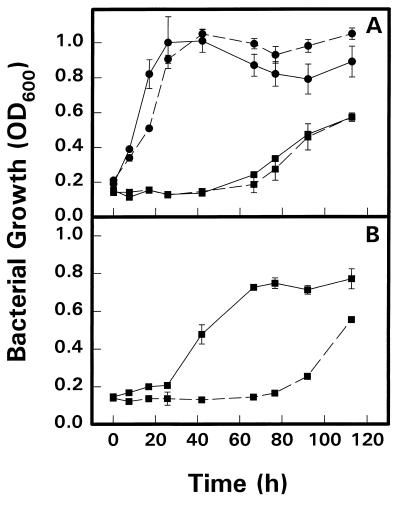
Tests for growth under salt stress showed that strains S10 (Fig. 2) and S11 (data not shown) behaved similarly. In the absence of salt stress, both strains grew as well as the parent strain, Rm1021. Stachydrine markedly protected parent cells from inhibition by 500 mM NaCl but had no such positive effect on S10 cells (Fig. 2B). Stachydrine (1 mM) protected S11 cells slightly from the inhibitory effects of 500 mM NaCl but not to the extent that it protected the parent cells (data not shown). Further tests showed that the mutations had no effect on [14C]stachydrine uptake and indicated that both mutant and parent cells had an increased total stachydrine uptake approximately 10-fold when grown in the presence of 300 mM NaCl (Table 4). Analysis of cells exposed to [14C]stachydrine for 18 h showed that both the parent strain Rm1021 and the mutant strain S11 transformed the 14C into ethanol-insoluble materials whereas the capacity of mutant S10 to do this was significantly impaired (Table 4).
FIG. 2.
Effects of NaCl and stachydrine on growth of Rm1021 and mutant strain Rm1021-S10 in MAS medium. (A) Growth of Rm1021 (——) or Rm1021-S10 (–––) in the absence (•) or presence (■) of 550 mM NaCl. (B) Growth of Rm1021 (——) or Rm1021-S10 (–––) in the presence of 550 mM NaCl and 1 mM stachydrine.
TABLE 4.
Stachydrine uptake and catabolism by S. meliloti
| Strain | NaCl addeda | Stachydrine uptakeb after:
|
% of 14C incorporatedc
|
||
|---|---|---|---|---|---|
| 10 min | 18 h | Soluble | Insoluble | ||
| Rm1021 | − | 2 | 2 | 54 | 46 |
| + | 18 | 30 | 63 | 37 | |
| Rm1021-S10 | − | 1 | 3 | 87 | 13 |
| + | 16 | 49 | 95 | 5 | |
| Rm1021-S11 | − | 1 | 1 | 61 | 39 |
| + | 20 | 25 | 70 | 30 | |
NaCl added to LAS medium at 300 mM 24 h prior to supplementation with [14C]stachydrine.
Uptake measured as nanomoles per milligram of protein per minute at 10 min and 18 h.
Ethanol-soluble and -insoluble cell fractions determined in the 18-h uptake sample.
Rhizosphere phenotypes.
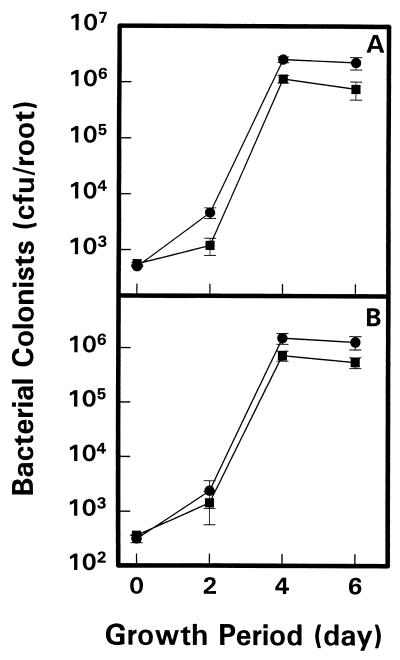
Mutant strains S10 and S11 colonized roots as well as parent Rm1021 cells when they were inoculated separately on seeds (data not shown). In all cases, an inoculum of several hundred cells increased to nearly 106 CFU/root by day 6. However, when the mutant strains were coinoculated with the parent strain in a competition experiment, both mutants showed impaired root colonization (Fig. 3). Strain S10 colonized roots with significantly fewer cells (P ≤ 0.01) than did Rm1021 on days 2, 4, and 6 (Fig. 3A). A 1.01 ratio of S10 to Rm1021 cells was measured on day 0, compared with significantly lower values (P ≤ 0.01) of 0.26, 0.45, and 0.34 on days 2, 4, and 6, respectively (Fig. 3A). The 1.16 ratio of S11 to Rm1021 on day 0 declined to significantly lower values (P ≤ 0.01) of 0.48 and 0.43 on days 4 and 6, respectively (Fig. 3B). Tests with other stachydrine-inducible Tn5-luxAB mutants from this study showed that some, but not all, were impaired in competitive colonization tests with the parent, Rm1021 (data not shown).
FIG. 3.
Alfalfa root colonization by S. meliloti mutant strains in competition with wild-type strain Rm1021. (A) Rm1021 (550 CFU/seed) (•) and Rm1021-S10 (557 CFU/seed) (■) were coinoculated on day 0. (B) Rm1021 (311 CFU/seed) (•) and Rm1021-S11 (361 CFU/seed) (■) were coinoculated on day 0. In both cases, mutant-to-Rm1021 ratios measured on the roots 6 days later were significantly lower (P ≤ 0.01) than were those predicted for equally competitive cells.
Molecular analyses.
DNA hybridization analyses located the Tn5 insertions of strains S10 and S11 in 10-kb and 12-kb EcoRI restriction fragments, respectively (Table 1). DNA sequence data from pSIF11 showed that the Tn5-luxAB insertion in strain S11 resided in a gene having nearly complete DNA nucleotide identity to the proline dehydrogenase gene (putA) of S. meliloti GR4 (19). Of 842 nucleotides, 813 (96.5%) totally matched those previously reported for strain GR4. The deduced amino acid homology was 96%.
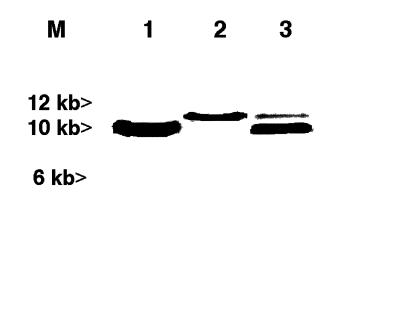
The PCR-generated hybridization probe corresponding to the flanking regions of the Tn5-tagged locus in strain S10 identified cosmid pLES1 as containing the corresponding wild-type Rm1021 DNA. Transconjugants of strain S10 containing cosmid pLES1 grew on stachydrine as the sole carbon and nitrogen source (data not shown), and DNA hybridization analysis of the complemented mutant showed that both the original mutated gene and the wild-type locus were present (Fig. 4). Electrophoretic analyses of plasmid versus total genomic DNA from the wild-type strain Rm1021 showed that the probe for the mutated locus in strain S10 hybridized to DNA which was physically distinct from the symbiotic plasmid that hybridized to the nodC-specific probe (data not shown). Other tests showed the probe produced from the mutated locus hybridized with neither pLBR55 nor pLBR56, both of which contain stachydrine catabolism genes from the nodC-containing symbotic plasmid of S. meliloti (16) (data not shown).
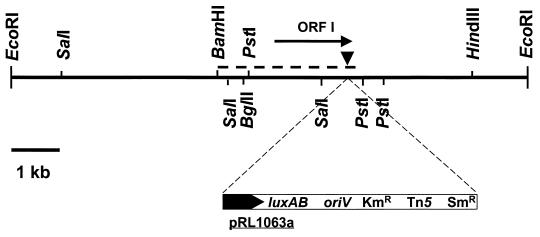
FIG. 4.
DNA gel blot analysis of the Rm1021-S10 mutant strain and a stachydrine-catabolizing transconjugant. Total genomic DNA from Rm1021 (lane 1), Rm1021-S10 (lane 2), and transconjugant Rm1021-S10(pLES1) (lane 3) was probed with DNA sequences derived from the Tn5-flanking region in Rm1021-S10. The transconjugant clearly contains both mutated and wild-type loci. Size markers are shown in lane M.
Nucleotide sequence analysis of a 2,898-bp DNA fragment in pLEB1 identified ORF I, which in mutant S10 contained the Tn5-luxAB insertion (Fig. 5). Data from ORF I are consistent with the production of a protein containing 580 amino acids. Alignment analysis (1) of ORF I showed highly significant (P ≤ 4 × 10−60) amino acid homology to NADH-dependent oxidases reported from Thermoanaerobium (39%) (28) and Eubacterium (35%) (12), which are assigned to a family of flavoproteins characterized as α/β-barrel oxidoreductases.
FIG. 5.
Physical map of the stachydrine-inducible stcD locus in S. meliloti 1021. ORF I (→) was identified as stcD by nucleotide sequencing. Mutant strain Rm1021-S10 contained Tn5 at the indicated site (▾). The region indicated above the partial restriction map of pLES1 (––––––) corresponds to the pLES1 sequence, which was deposited in GenBank as accession no. AF016307. The promoterless luxAB transposable reporter contained in pRL1063a was oriented in the direction indicated in pSIF10.
DISCUSSION
Of 12 stachydrine-inducible Tn5-luxAB insertions found among 5,000 S. meliloti mutants, two could not use stachydrine as the sole carbon and nitrogen source. These mutant strains, S10 and S11, colonized alfalfa roots normally in single-strain tests but showed reduced competitiveness in colonization tests with the parent, Rm1021 (Fig. 3). Growth, uptake, and DNA sequence data established that S11 was mutated in the known S. meliloti putA gene (19), and evidence here is consistent with the conclusion that the mutated ORF in S10, stcD, is involved in the second demethylation of stachydrine (i.e., demethylation of N-methylproline to form proline).
Data obtained with mutant strain S11 (Fig. 3B) confirm the known contribution of putA to competitive alfalfa root colonization (19) and extend our understanding by showing that exogenous stachydrine induces that gene. The impairment of proline uptake in this mutant (Table 2) suggests that PutA in S. meliloti plays a regulatory role, as has been shown for this multifunctional protein in enteric bacteria (34) but not in Bradyrhizobium (44). Whether the putA gene in S. meliloti is induced directly by stachydrine or indirectly by degradation of stachydrine to proline, as suggested by proline effects on luxAB expression (Table 2), was not established by these experiments. However, the ecological significance of putA presumably is proportional to the sum total of stachydrine and proline released by the plant. Because high levels of stachydrine are released from germinating Medicago seeds (39), significant amounts of proline may be produced by catabolism. Preliminary results have shown that [14C]stachydrine is effectively catabolized to proline by young alfalfa plants (26) and that S. meliloti may therefore be exposed to N-methylproline as well as to stachydrine.
Data presented here indicate that the mutated locus in S10 is genetically and phenotypically distinct from stachydrine catabolism genes identified previously in S. meliloti (16). First, the DNA-DNA hybridization analysis of plasmid and total DNA indicates that DNA containing the mutated locus in S10 is not on the symbiotic plasmid and may be on the chromosome or another plasmid. Second, symbiotic plasmid fragments containing known stachydrine catabolism genes (16) did not hybridize with the mutated locus in S10. Finally, while previously reported mutants grew on both N-methylproline and proline (16), S10 failed to grow on N-methylproline (Table 2). These data and the identification here of a previously unreported ORF containing the Tn5 insertion in S10 indicate that a novel gene, referred to here as stcD, is mutated in S10.
The function of stcD is not completely established by this study, but three lines of evidence are consistent with the concept that stcD codes for a protein required for the demethylation of N-methylproline. First, the physiological data indicate that strain S10 is impaired in the second demethylation of stachydrine because it grows on proline but not on N-methylproline as the sole carbon and nitrogen source (Table 3). Second, while S10 is capable of taking up stachydrine at levels similar to the parent, it does not catabolize the stachydrine (Table 4). In S10 cells, most 14C remains as ethanol-soluble material and is not incorporated into the ethanol-insoluble cellular fraction. This result contrasts with results for both the parent strain and the S11 putA mutant, in which a large fraction of the 14C was incorporated into cellular material. The mutation in putA would prevent the catabolism of proline but not the direct use of proline for protein synthesis. Third, the Tn5 insertion in S10 (Fig. 5) interrupted an ORF (Genbank accession no. AF016307) with significant deduced amino acid homology to genes encoding α/β-barrel oxidoreductase flavoproteins. In Pseudomonas, at least two oxidases, 4-methoxybenzoate demethylase and vanillate demethylase, function as demethylases (18). Although previous work identified genes involved in the conversion of stachydrine to N-methylproline (16), a locus responsible for the demethylation of N-methylproline has not been reported. Complementation of S10 by a cosmid containing the sequenced ORF (Fig. 4) was also consistent with this interpretation.
Root colonization results with the putA mutant, S11, in this study differed markedly from those reported by others (19). The much smaller effect of the mutation on colonization observed here may have resulted from the test we used. Our assay measured the capacity of a small initial inoculum to colonize roots growing in vermiculite, while the previous work (19) used roots exposed to a large initial inoculum in a hydroponic medium. Under our conditions, both the Rm1021 parent and the S11 mutant grew significantly but the mutant cells achieved a final population on the root approximately half the size of that produced by the parent cells. Jimenez-Zurdo et al. (19) saw no growth of the putA mutant after inoculation while the number of wild-type cells increased 10-fold in 48 h. These results suggest that different factors were affecting bacterial growth and survival in the two studies.
These results offer additional evidence suggesting that stachydrine catabolism contributes to rhizobial colonization of alfalfa seedling roots. S. meliloti mutants incapable of growing on stachydrine as the sole carbon and nitrogen source are less efficient at forming root nodules on two Medicago species (16) and less competitive against isogenic wild-type strains (Fig. 3) (19). It is not surprising that utilization of a unique carbon resource such as stachydrine facilitates root colonization (10, 24). The moderate benefits measured in this study (Fig. 3), however, suggest either that stachydrine is not a major controlling factor in root colonization under the conditions tested or that higher levels of stachydrine are required for S. meliloti to benefit fully from the capacity to metabolize this compound. Experiments in nonsterile soil might help to assess the ecological significance of this trait in more complex microbial communities, and the use of plants modified to produce more stachydrine might overcome problems associated with limiting amounts of this compound.
ACKNOWLEDGMENTS
This work was supported by grants from the U.S. Department of Agriculture (NRICGP91-37305) and the U.S. National Science Foundation (IBN-92-18567). The Ministère de L’Enseignement Supérieur et de la Recherche (DREIF, France) is thanked for financial support for cooperative research.
We thank M. Thomashow for use of the luminometer and D. Tepfer for supplying pLBR55 and pLBR56.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Backman K, Chen Y M, Magasanik B. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie G A, Handelsman J. Evaluation of a strategy for identifying nodulation competitiveness genes in Rhizobium leguminosarum biovar phaseoli. J Gen Microbiol. 1993;139:529–538. doi: 10.1099/00221287-139-3-529. [DOI] [PubMed] [Google Scholar]
- 4.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 5.Bolton H, Fredrickson J K, Elliott L F. Microbial ecology of the rhizosphere. In: Metting F B, editor. Soil microbial ecology. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 27–63. [Google Scholar]
- 6.de Bruijn F J, Rossbach S. Transposon mutagenesis. In: Gerhardt P, et al., editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 387–405. [Google Scholar]
- 7.de Bruijn F J, Rossbach S, Schneider M, Ratet P, Messmer S, Szeto W W, Ausubel F M, Schell J. Rhizobium meliloti 1021 has three differentially regulated loci involved in glutamine biosynthesis, none of which is essential for symbiotic nitrogen fixation. J Bacteriol. 1989;171:1673–1682. doi: 10.1128/jb.171.3.1673-1682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, Collins M D, Dreyfus B, Kersters K, Gillis M. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol. 1994;44:715–733. [Google Scholar]
- 9.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrand S K, Wilson M, Lindow S E, Savaka M A. Modulating competition in the rhizosphere by resource utilization. Mol Ecol. 1994;3:619. [Google Scholar]
- 11.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklund C V, Baron S F, Hylemon P B. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J Bacteriol. 1993;175:3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 14.Gloux K, Le Rudulier D. Transport and catabolism of proline betaine in salt-stressed Rhizobium meliloti. Arch Microbiol. 1989;151:143–148. [Google Scholar]
- 15.Goldmann A, Boivin C, Fleury V, Message B, Lecoeur L, Maille M, Tepfer D. Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbotic region. Mol Plant-Microbe Interact. 1991;4:571–578. doi: 10.1094/mpmi-4-571. [DOI] [PubMed] [Google Scholar]
- 16.Goldmann A, Lecoeur L, Message B, Delarue M, Schoonejans E, Tepfer D. Symbiotic plasmid genes essential to the catabolism of proline betaine, or stachydrine, are also required for efficient nodulation by Rhizobium meliloti. FEMS Microbiol Lett. 1994;115:305–312. [Google Scholar]
- 17.Guyon P, Petit A, Tempé J, Dessaux Y. Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol Plant-Microbe Interact. 1993;6:92–98. [Google Scholar]
- 18.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenase. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 19.Jimenez-Zurdo J I, Garcia-Rodriguez F M, Toro N. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol Microbiol. 1997;23:85–93. doi: 10.1046/j.1365-2958.1997.1861555.x. [DOI] [PubMed] [Google Scholar]
- 20.Jones G P, Naidu B P, Starr R K, Paleg L G. Estimates of solutes accumulating in plants by 1H nuclear magnetic resonance spectroscopy. Aust J Plant Physiol. 1986;13:649–658. [Google Scholar]
- 21.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss G B, Vincze E, Kalman Z, Forrai T, Kondorosi A. Genetic and biochemical analysis of mutants affected in nitrate reduction in Rhizobium meliloti. J Gen Microbiol. 1979;113:105–118. [Google Scholar]
- 23.Lam S T, Ellis D M, Ligon J M. Genetic approaches for studying rhizosphere colonization. In: Keister D L, Creagan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 43–50. [Google Scholar]
- 24.Lam S T, Torkewitz N R, Nautiyal C S, Dion P. Impact of the ability to utilize a single substrate on colonization competitiveness. Phytopathology. 1991;81:1163–1164. [Google Scholar]
- 25.Laskey R A, Mills A D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- 26.Le Rudulier D, Gloux K, Poggi M C, Noel J P. Accumulation, biosynthesis and fate of proline betaine in nodulated alfalfa plants (Medicago sativa L.) subjected to salinity stress. In: Leigh R A, Blake-Kalff M M A M, editors. Stressnet. Brussels, Belgium: European Commission Directorate General VI; 1995. pp. 257–262. [Google Scholar]
- 27.Lim P O, Ragatz D, Renner M, de Bruijn F J. Environmental control of gene expression: isolation of Rhizobium meliloti gene fusions induced by N- and C-limitation. In: Guerrero R, Pedros-Alio C, editors. Trends in microbial ecology. Barcelona: Spanish Society for Microbiology; 1993. pp. 97–100. [Google Scholar]
- 28.Liu X L, Scopes R K. Cloning, sequencing and expression of the gene encoding NADH oxidase from the extreme anaerobic thermophile Thermoanaerobium brockii. Biochim Biophys Acta. 1993;1174:187–190. doi: 10.1016/0167-4781(93)90113-r. [DOI] [PubMed] [Google Scholar]
- 29.Long S R, Staskawicz B J. Prokaryotic plant parasites. Cell. 1993;73:921–935. doi: 10.1016/0092-8674(93)90271-q. [DOI] [PubMed] [Google Scholar]
- 30.Mavingui P, Flores M, Romero D, Martinez-Romero E, Palacios R. Generation of Rhizobium strains with improved symbiotic properties by random DNA amplification (RDA) Nat Biotechnol. 1997;15:564–569. doi: 10.1038/nbt0697-564. [DOI] [PubMed] [Google Scholar]
- 31.McLoughlin T J, Merlo A O, Satola S W, Johansen E. Isolation of competition-defective mutants of Rhizobium fredii. J Bacteriol. 1987;169:410–413. doi: 10.1128/jb.169.1.410-413.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller K J, Wood J M. Osmoadaptation by rhizosphere bacteria. Annu Rev Microbiol. 1996;50:101–136. doi: 10.1146/annurev.micro.50.1.101. [DOI] [PubMed] [Google Scholar]
- 34.Muro-Pastor A M, Maloy S. Proline dehydrogenase activity of the transcriptional repressor PutA is required for induction of the put operon by proline. J Biol Chem. 1995;270:9819–9827. doi: 10.1074/jbc.270.17.9819. [DOI] [PubMed] [Google Scholar]
- 35.Musich J A, Rapoport H. Reaction of O-methyl-N,N′-diisopropylisourea with amino acids and amines. J Org Chem. 1977;42:139–141. doi: 10.1021/jo00421a028. [DOI] [PubMed] [Google Scholar]
- 36.Perroud B, Le Rudulier D. Glycine betaine transport in Escherichia coli: osmotic modulation. J Bacteriol. 1985;161:393–401. doi: 10.1128/jb.161.1.393-401.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips D A, Joseph C M, Maxwell C A. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol. 1992;99:1526–1531. doi: 10.1104/pp.99.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips D A, Streit W R. Applying plant-microbe signalling concepts to alfalfa: roles for secondary metabolites. In: McKersie B D, Brown D C W, editors. Biotechnology and the improvement of forage legumes. Wallingford, England: CAB International; 1997. pp. 319–342. [Google Scholar]
- 39.Phillips D A, Wery J, Joseph C M, Jones A D, Teuber L R. Release of flavonoids and betaines from seeds of seven Medicago species. Crop Sci. 1995;35:805–808. [Google Scholar]
- 40.Rovira A D, Harris J R. Plant root excretions in relation to the rhizosphere effect V. The exudation of B-group vitamins. Plant Soil. 1961;14:199–214. [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Smith L T, Smith G M, Dsouza M R, Pocard J A, Le Rudulier D, Madkour M A. Osmoregulation in Rhizobium meliloti: mechanism and control by other environmental signals. J Exp Zool. 1994;268:162–165. [Google Scholar]
- 43.Steenbock H. Isolation and identification of stachydrin from alfalfa hay. J Biol Chem. 1918;35:1–13. [Google Scholar]
- 44.Straub P F, Reynolds P H S, Althomsons S, Mett V, Zhu Y X, Shearer G, Kohl D H. Isolation, DNA sequence analysis, and mutagenesis of a proline dehydrogenase gene (putA) from Bradyrhizobium japonicum. Appl Environ Microbiol. 1996;62:221–229. doi: 10.1128/aem.62.1.221-229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Streit W R, Joseph C M, Phillips D A. Biotin and other water-soluble vitamins are key growth factors for alfalfa rhizosphere colonization by Rhizobium meliloti 1021. Mol Plant-Microbe Interact. 1996;5:330–338. doi: 10.1094/mpmi-9-0330. [DOI] [PubMed] [Google Scholar]
- 46.Vincent J M. A manual for the practical study of root-nodule bacteria. Vol. 15. Oxford, England: Blackwell Scientific Publications; 1970. [Google Scholar]
- 47.Wolk C P, Cai Y, Panoff J M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyn Jones R G, Storey R. Betaines. In: Paleg L G, Aspinall D, editors. The physiology and biochemistry of drought resistance in plants. Sydney, Australia: Academic Press, Ltd.; 1981. pp. 171–204. [Google Scholar]