Abstract
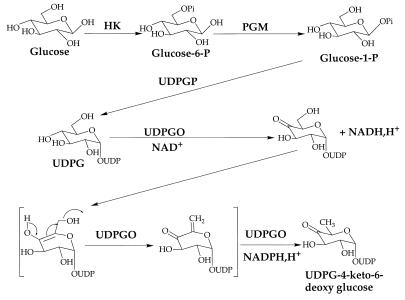
All of the 2,6-dideoxy sugars contained within the structure of chromomycin A3 are derived from d-glucose. Enzyme assays were used to confirm the presence of hexokinase, phosphoglucomutase, UDPG pyrophosphorylase (UDPGP), and UDPG oxidoreductase (UDPGO), all of which are involved in the pathway of glucose activation and conversion into 2,6-dideoxyhexoses during chromomycin biosynthesis. Levels of the four enzymes in Streptomyces spp. cell extracts were correlated with the production of chromomycins. The pathway of sugar activation in Streptomyces spp. involves glucose 6-phosphorylation by hexokinase, isomerization to G-1-P catalyzed by phosphoglucomutase, synthesis of UDPG catalyzed by UDPGP, and formation of UDP-4-keto-6-deoxyglucose by UDPGO.
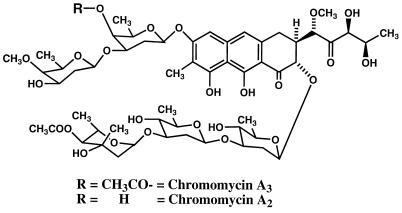
Dideoxy sugars occur commonly in the structures of cardiac glycosides from plants, in antibiotics like chromomycin A3 (Fig. 1), and in macrolides produced by microorganisms. On the basis of stable isotope-labeling experiments, biosynthetic studies conducted in Rosazza’s laboratory have indicated that all the deoxy sugars of chromomycin A3 are derived from d-glucose (21). While the assembly of the polyketide aglycone is reasonably well understood, relatively little is known of the details of 2,6-dideoxy sugar biogenesis in streptomycetes. Earlier studies with Streptomyces rimosus indicated that TDP-mycarose is synthesized from TDP-d-glucose (TDPG) and S-adenosyl-l-methionine (10, 23). The reaction requires NADPH as a cofactor, and TDP-4-keto-6-deoxy-d-glucose is an intermediate. Formation of TDP-4-keto-6-deoxy-d-glucose was catalyzed by the enzyme TDPG oxidoreductase (TDPG-4,6-dehydratase; EC 4.2.1.46). Similar 4-keto sugar nucleotides are intermediates for the biosynthesis of polyene macrolide antibiotic amino sugars (18). Similar pathways have been elaborated for the formation of 2,6-dideoxy-d-threo-4-hexulose of granaticin in Escherichia coli (6, 25) and 2,6-dideoxy-d-arabino-hexose of chlorothricin (12). The initial 6-deoxygenation of glucose during 3,6-dideoxy sugar formation involves a similar mechanism (32). In all of these processes, glucose is first activated by conversion into a sugar nucleotide such as UDPG followed by NAD+ oxidation of the 4 position to the corresponding 4-oxo derivative. Position 6 deoxygenation involves a general tautomerization, dehydration, and NADH,H+-catalyzed reduction process (6, 12, 25). A similar tautomerization and dehydration followed by reduction may produce C-3-deoxygenated products, such as CDP-3,6-dideoxyglucose (27). The pathway for formation of 3,6-dideoxyhexoses from CDPG in Yersinia pseudotuberculosis was clearly elucidated by Liu and Thorson (14). However, none of this elegant work was focused on the earlier steps of hexose nucleotide formation.
FIG. 1.
Structures of chromomycins A2 and A3.
On the basis of previous work (7), it is reasonable to postulate that the biosynthesis of 2,6-dideoxyglucose in Streptomyces griseus involves phosphorylation to glucose-6-phosphate by hexokinase (HK; E.C.2.7.7.1), as in glycolysis; conversion to glucose-1-phosphate by phosphoglucomutase (PGM; EC 2.7.5.1); reaction with UTP to form UDPG in a reaction catalyzed by UDPG pyrophosphorylase (UDPGP) (glucose-1-phosphate uridylyltransferase; EC 2.7.7.9), and C-6 deoxygenation catalyzed by UDP-d-glucose-4,6-dehydratase with NAD+ as a cofactor (Fig. 2). UDPG and GDPG have been detected in cell extracts of S. griseus and Streptomyces sp. strain MRS202, suggesting that these compounds are active sugar nucleotides involved in the formation of dideoxyhexoses (15). UDPGP genes from several bacteria have been cloned and sequenced (1, 3, 4, 11, 29, 30). Although nucleotidyl diphosphohexose-4,6-dehydratases (NDP-hexose-4,6-dehydratases) have been purified and characterized from several sources (5, 8, 9, 13, 19, 25, 26, 31, 33), the occurrence of the glucose-activating enzymes HK, PGM, UDPGP, and UDPG oxidoreductase (UDPGO) involved in 2,6-dideoxyhexose formation has not been established in streptomycetes. This work provides evidence for the presence of these enzymes involved in the biosynthetic activation of glucose to the 2,6-dideoxyhexoses in chromomycin A3.
FIG. 2.
Proposed pathway for the formation of 2,6-dideoxy sugars in streptomycetes involving HK, PGM, UDPGP, and UDPGO.
MATERIALS AND METHODS
General material and instrumentation.
High-performance liquid chromatography (HPLC) was performed with a Rheodyne injector type 7125 with a 100-μl loop connected to a model LC-6A pump, an SPD-6AV module UV-VIS detector, a CR-501 Chromatopac recording integrator, and an SCL-6B system controller, all from Shimadzu Co. (Osaka, Japan). The analytical column (250- by 4.6-mm inside diameter) was packed with 5-μm partisil octyldecyl silane/C18 (Whatman Inc., Clifton, N.J.) and preceded by a guard column of the same composition (Alltech Inc., Deerfield, Ill.). UV and visible light spectroscopy was performed with a Shimadzu UV-160 spectrophotometer. Low-resolution fast atom bombardment-mass spectrometry spectra were measured on a ZAB-HF instrument by triethanolamine as a matrix solvent recorded in the negative-ion mode, and also with a Trio-III instrument in the University of Iowa Core Mass Spectrometry Facility.
Centrifugations were conducted with a Sorvall RC-5 refrigerated centrifuge (GSA or SS 35 rotors), or a Beckman model L8-55 refrigerated ultracentrifuge (type 35 or type 40 rotors), or a bench top Eppendorf microcentrifuge. Cell disruption was performed with a French press (SLM Instruments, Urbana, Ill.). A lyophilizer (The Virtis Co., Inc., Gardiner, N.Y.) was used for all sample lyophilizations.
HPLC-grade KH2PO4 and H3PO4 (85%) were both from Fisher Scientific (Fair Lawn, N.J.). Acetonitrile (HPLC grade) was from E. M. Science (Gibbstown, N.J.) and was filtered through type HV 0.45-μm-pore-size Millipore (Bedford, Mass.) membranes before use.
All nucleotide standards, tetrabutyl ammonium hydroxide (TBAH), dithiothreitol, HK (from baker’s yeast), PGM (from rabbit muscle), UDPGP (from baker’s yeast), glucose-6-phosphate dehydrogenase (G-6-PDH; EC 1.1.1.49), and UDPG dehydrogenase (EC 1.1.1.22) (from bovine liver) were purchased from Sigma Chemical Company (St. Louis, Mo.). d-Glucose (U-14C) (50 μCi; 313 mCi/mmol) was obtained from ICN Radiochemicals, Inc. (a division of ICN Biochemicals, Inc., Irvine, Calif.), and UDPG(1"-3H) (50 μCi; 15.3 Ci/mmol) was purchased from DuPont-New England Nuclear (Boston, Mass.). Budget-Solve liquid scintillation cocktail was purchased from Research Products International (Mount Prospect, Ill.).
Growth of microorganisms and preparation of cell extracts.
S. griseus ATCC 13273 was stored on Sabouraud maltose agar (Difco, Detroit, Mich.) slants. Streptomyces sp. strain MRS202 (obtained from Abbott Laboratories, Abbott Park, Ill.) was maintained on International Streptomyces Project medium 2 (ISP 2; yeast extract-malt extract agar [2% agar, 1% malt extract, 0.4% yeast extract, 0.4% glucose]).
For cultivation of S. griseus (ATCC 13273), the composition of medium A was (wt/vol) 2.5% dextrose (stage I) or 5% dextrose (stage II), 0.3% sodium chloride, 0.3% calcium carbonate, and 1.5% soybean meal. For Streptomyces sp. strain MRS202, the composition of seed medium B was (wt/vol) 1.5% glucose monohydrate, 1.5% soy flour, 0.1% yeast extract (Difco, Detroit, Mich.), 0.1% NaCl, 0.1% CaCO3, and distilled water to 1 liter; the composition of cultivation medium C was (wt/vol) 2% starch, 1% glucose monohydrate, 0.5% dried distiller’s solubles (Sigma), 0.5% yeast extract, 0.2% CaCO3, water to 1 liter, and pH adjusted to 7.0 with NaOH or HCl. The media were autoclaved at 121°C and 0.7- kg/cm2 pressure for 15 min for 125-ml flasks containing 25 ml of medium, and for 20 min for 1,000-ml flasks holding 200 ml of medium. Cultures were incubated by our standard two-stage protocol (15) at 27°C with shaking at 250 rpm on New Brunswick Scientific Co. G-25 Gyrotory shakers. A 10%, 72-h-old stage I inoculum was used to initiate stage II cultures, which were incubated as before. Stage II cultures were harvested by filtration through cheesecloth and subsequent centrifugation of the filtrate in a Sorvall RC-5 Superspeed refrigerated centrifuge at 13,200 × g for 10 min. Cell pellets were washed twice with 30 ml of chilled 0.12 M KH2PO4 buffer (pH 6) and centrifuged again.
Cell extracts were prepared by suspending pellets in cold 0.12 M phosphate buffer (pH 7.0) to a final concentration of 0.5 g/ml. Lysozyme was added to a concentration of 1 mg/ml of cell suspension, and the mixture was incubated on ice for 2 h. Cell preparations were made by passing cell suspensions twice through a chilled French press at 17,000 lb/in2, and the resulting homogenates were centrifuged at 50,000 or 100,000 × g for 40 min.
Growth curves of streptomycetes were determined by cell dry-weight determinations and by measurements of turbidity (optical density at 600 nm).
Protein assay.
Protein concentrations were determined by using the Bio-Rad protein assay as originally developed by Bradford (2). Bovine serum albumin was used as the standard.
Enzyme assays for HK, PGM, UDPGP, and UDPGO.
HK, PGM, and UDPGP were all determined by measuring the changes in absorbance at 340 nm, and specific activities are expressed as micromoles of products formed per minute per milligram of protein under the conditions described. Each assay was performed in triplicate. HK was assayed based on the method of Magnani et al. (16) with the following modification. The assay is based on the coupled-enzyme reaction, in which G-6-P formed by HK is oxidized by G-6-PDH. G-6-PDH requires NADP+, which is stoichiometrically reduced to NADH as G-6-P is oxidized to gluconate-6-phosphate. Since HK activity is based on the amount of G-6-P generated, endogenous G-6-P in cell extracts was removed by dialysis before enzyme determinations were made. Cell extracts were dialyzed against 60 mM Tris-HCl buffer (pH 8.0), using dialysis membranes with a molecular weight cutoff of 10,000. Reaction mixtures contained 42 mM Tris-HCl buffer (pH 7.6), 222 mM glucose, 6.7 mM MgCl2, 2.7 mM ATP, 0.73 mM NADP+, and 0.5 U of G-6-PDH per ml in a total volume of 2 ml and were incubated at room temperature. The reaction was started by adding 50 μl of enzyme solution, and optical density changes were measured at 340 nm for 2 min. Blanks contained no glucose.
The assay for PGM is essentially the same as that for HK and is mainly based on the method of Marechal et al. (17). G-1-P is converted into G-6-P by PGM, and G-6-P is measured as described above. The PGM reaction was conducted at 25°C with a reaction mixture containing 84 mM Tris-HCl (pH 7.6), 3.4 mM G-1-P, 0.02 mM G-1,6-DP, 0.87 mM EDTA · 2Na+, 1.67 mM MgCl2, 0.18 mM NADP+, and 0.5 U of G-6-PDH per ml in a total volume of 2.5 ml. The reaction was initiated by the addition of 400 μl of enzyme solution, and the change in optical density at 340 nm was recorded for 2 min. Blanks contained all components but G-1-P.
UDPGP activity was measured in both the synthetic (forward) and hydrolytic (reverse) directions. In the reverse assay, PGM and G-6-PDH were coupled in the reaction. The enzyme assay was modified based on the method of Nakano et al. (22). Incubations were conducted at room temperature, using reaction mixtures containing 50 mM Tris-HCl (pH 8.2), 3.2 mM sodium pyrophosphate, 2.1 mM UDPG, 14 mM MgCl2, 1.2 mM NADP+, 83 μM G-1,6-DP, 0.03 U of G-6-PDH [commercial enzyme in (NH4)2SO4 dialyzed against 50 mM Tris-HCl (pH 8.0) overnight before use]/3 μl, and 0.14 U of PGM/20 μl in a total volume of 0.9 ml. The reaction was initiated by the addition of 100 μl of enzyme solution, and the change in optical density at 340 nm was measured during the first minute of incubation. Blanks contained all components but UDPG. In the direction of UDPG synthesis, UDPGDH was used as an auxiliary enzyme, as described by Persat et al. (24). Their method was modified as follows: no mercaptoethanol or Triton X-100 was added, and 50 mM bicine buffer (pH 8.5) was replaced by 0.5 M Tris-HCl (pH 8.2). The assay medium contained 80 mM Tris HCl (pH 8.2), 2.4 mM UTP, 2 mM G-1-P, 4.5 mM MgCl2, 1.1 mM NAD+, and 0.03 U of UDPG dehydrogenase per 20 μl in a total volume of 0.9 ml. Again, the reaction was initiated by the addition of 100 μl of cell extract, and the absorbance at 340 nm was recorded at room temperature for 2 min.
The reaction catalyzed by UDPG-4,6-dehydratase (also known as UDPGO) forms UDP-4-keto-6-deoxy-D-glucose from UDPG. An enzymatic assay based on the determination of CDPG oxidoreductase (32) and TDPG oxidoreductase was adapted to these experiments with minor modifications. One unit of enzyme activity corresponds to the production of 1 μmol of UDP-4-keto-6-deoxy-d-glucose per h at 37°C. The standard 250-μl assay mixture contained 100 nmol of NAD+, 100 nmol of UDPG, 100 μl of 0.5 M Tris-HCl buffer (pH 8.0), and 50 μl of cell extract, which was added to initiate the reaction. Reaction mixtures were incubated at 37°C for 20 min. After quenching with 750 μl of 0.1 N NaOH, the absorbance at 320 nm was recorded following incubation for another 15 min at 37°C. Blanks were prepared in parallel by boiling the enzyme for 2 min prior to its addition to the assay mixture. Changes of absorbance between the samples and the blank at 320 nm (ɛ = 4,800 m−1 cm−1) were used to calculate enzyme activity. Similar extinction values were used to detect other 4-keto-6-deoxyglucose nucleotides (20, 28, 31, 33).
HPLC analyses of sugar nucleotide derivatives.
HPLC analyses were conducted over a reversed-phase, C18 column as described previously (15). Solvent system 1 consisted of mobile phase A (pH 5.3, 15 mM KH2PO4, containing 10 mM TBAH) and mobile phase B (pH 5.3, 35 mM KH2PO4, with 10 mM TBAH) in 30% (vol/vol) acetonitrile. Separations were obtained at flow rates of 1 ml/min with a concave gradient ranging from 5 to 100% of mobile phase B over a period of 58 min, while eluting peaks were monitored at 262 nm. Alternatively, for those nucleotides not well resolved by solvent system 1, separations were also obtained with solvent system 2 at a flow rate of 1.2 ml/min with a linear gradient ranging from 0 to 33.3% of solution B over a period of 30 min. HPLC of chromomycins A2 and A3 was done as previously reported (15, 21). Streptomyces sp. strain MRS202 produced mixtures of chromomycin A2 and A3 in a 1.6/1 ratio. Concentrations measured (see Fig. 3) represent the total amounts of chromomycins A2 and A3 formed at different times during the fermentation.
FIG. 3.
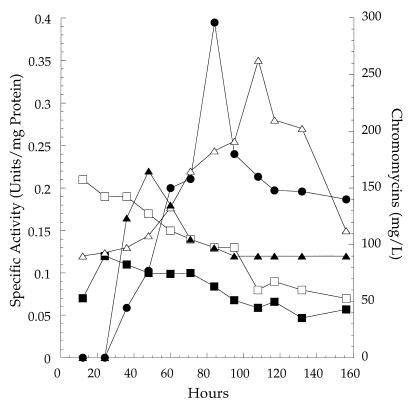
Relationships of HK (■), PGM (▵), UDPGP (×10) (□), and UDPGO (×10) (▴) to production of chromomycin A3 (•) by Streptomyces sp. strain MRS202. The results are the averages of duplicate assays within a deviation of no more than 4%.
Detection of nucleotide pyrophosphorylase activities by HPLC.
Incubation mixtures of 210 μl contained 30 μl of cell extract and 0.19 mM dithiothreitol, 0.48 mM MgAc2, 28.5 mM Tris-HCl buffer (pH 8.0), 0.57 mM G-6-P, 0.57 mM G-1-P, and 0.76 mM UTP. Concentrations of GTP, dTTP, ATP, and CTP were all the same as that of UTP. In addition to the above-mentioned substances, control incubations also contained 2 U each of inorganic pyrophosphatase (EC 3.6.1.1), UDPGP, and PGM. All enzyme mixtures were incubated for 1 h at 30°C and analyzed by HPLC as described above.
RESULTS AND DISCUSSION
Both Streptomyces species used in this study produce chromomycins A2 and A3, each of which contains four different 2,6-dideoxyhexoses linked to a polyketide aglycone. 13C-labeling studies revealed that all of the chromomycin A3 sugars are derived from [2-13C]glucose (21). HPLC analysis showed that UDPG was present in cell extracts of S. griseus, thus indicating that this sugar nucleotide is involved in the activation and metabolic conversion of glucose to the dideoxy sugars in the chromomycins (15). We initially confirmed the involvement of UDPG in the glucose activation pathway by incubating uniformly labeled [14C]glucose with Streptomyces sp. strain MRS202. When cells were harvested from labeled glucose containing incubations, disrupted by French press homogenization, and analyzed by HPLC, a radioactive peak was eluted at a retention volume of 26 ml, consistent with the formation of [14C]UDPG by Streptomyces sp. (15).
The formation of UDPG would typically involve the glucose activation pathway summarized in Fig. 2. The enzymes required include HK, PGM, UDPGP, and UDPGO. Assays were established in order to detect these enzyme activities in cell extracts of the Streptomyces species in this work. Results have been expressed in specific enzyme activities in order to correct for subtle differences in cell breakage and recovery. Growth curves for these two microorganisms were similar, each approaching stationary growth at 95 h (10.7 g [dry weight] of cells per liter). Typically, the amounts of protein obtained by cell breakage ranged between 15 and 35 mg/ml.
Table 1 shows the specific activities of HK, PGM, UDPGP, and UDPGO in cell extracts of both Streptomyces cultures taken at 98 h during the chromomycin-producing, stage II culture. In this experiment, all enzymes were detected in both culture extracts. However, strain MRS202 contained twice the levels of HK and PGM specific activities for S. griseus ATCC 13273. Comparisons of these same enzyme activities in chromomycin A3-negative mutants showed that cultures incapable of producing the antibiotics lacked measurable UDPGO activity. Interestingly, the specific activities of UDPGP in two antibiotic-negative mutants (45 and 55 mU/mg of protein) were more than twice as high as those observed in antibiotic-producing cultures. These results indicate a positive correlation between UDPGO activity and antibiotic formation and a negative correlation between UDPGP activity and antibiotic formation.
TABLE 1.
Specific activities of HK, PGM, UDPGP, and UDPGO in 98-h S. griseus ATCC 13273, Streptomyces sp. strain MRS202, and non-antibiotic-producing mutant culture extractsa
| Streptomyces strain | Sp act (U/mg of protein)
|
||||
|---|---|---|---|---|---|
| HK | PGM | UDPGP | UDPGO (UDPG) | UDPGO (GDPG) | |
| 13273 | 0.024 | 0.055 | 0.024 | 0.010 | 0.007 |
| MRS202 | 0.039 | 0.114 | 0.021 | 0.010 | 0.008 |
| A3− mutants | |||||
| ASFz | 0.098 | 0.247 | 0.055 | ||
| AMY | 0.137 | 0.542 | 0.045 | ||
Assays were conducted in duplicate, with a variation of no more than 3% for any sample.
One aim of this work was to determine possible relationships between these initial steps of glucose activation and antibiotic biosynthesis. Therefore, we compared the temporal relationships among HK, PGM, UDPGP, and UDPGO expressed activities and chromomycin biosynthesis for Streptomyces sp. strain MRS202. The results are shown in Fig. 3. Traces of chromomycins were evident in 24-h cultures, increased to 150 mg/liter at 60 h, and reached a peak concentration of 300 mg/liter at 84 h before decreasing to about 150 mg/liter thereafter. PGM specific activities were measured at 0.125 U/mg of protein at 12 h and gradually increased to a peak level of 0.35 U/mg of protein at 108 h before declining again. HK activity was highest at 24 h (0.13 U/mg of protein), declined to about 0.1 U/mg of protein by 48 h, and gradually decreased thereafter. Measured specific activities for UDPGP and UDPGO were much less than those for HK and PGM. UDPGP started at 0.02 U/mg of protein at 12 h, remained the same until 48 h, and then gradually declined to about half that level at 108 h and thereafter. UDPGO, however, was undetectable at 24 h, reached a peak of 0.025 U/mg of protein by 48 h, and then gradually declined to 0.017 U/mg of protein by 108 h, where it remained. Interestingly, UDPGO peak activity preceded antibiotic peak production by about 24 h, and the gradual decline in UDPGO activity likewise preceded the gradual decline in antibiotic levels by about 24 h. These results link expressed UDPGO activity to antibiotic production in this streptomycete. Furthermore, these results confirm the involvement of the enzymes indicated in Fig. 2 in glucose activation by Streptomyces sp. strain MRS202.
Since UDPGP apparently is a centrally important enzyme in glucose activation in Streptomyces sp. strain MRS202, several experiments were developed to confirm the presence of the reaction product, UDPG, and to rule out the involvement of other nucleotide pyrophosphorylases in the glucose activation process. UDPG levels in cell extracts were determined with UDPG dehydrogenase (24), which oxidizes UDPG to UDP glucuronate and concomitantly reduces NAD+. The preparations were also analyzed by HPLC (solvent system 1). Incubation mixtures all contained cell extract plus G-6-P, G-1-P, and one of the following triphosphonucleotides: UTP, GTP, dTTP, ATP, or CTP. Incubations containing inorganic pyrophosphatase, UDPGP, and PGM were evaluated as controls. UDPG was formed in cell extract preparations amended with UTP and either G-1-P or G-6-P, showing that endogenous PGM and G-1,6-diP were present in cell extracts. However, when ATP, dTTP, CTP, or GTP was added to cell extracts and incubated with G-1-P, no corresponding ADPG, dTDPG, CDPG, or GDPG was detected. These results indicated that either there were no corresponding ADPG, GDPG, CDPG, and dTDPG pyrophosphorylases or that Streptomyces UDPGP could not use ATP, dTTP, CTP, and GTP as substrates.
Using 100,000 × g supernatants of MRS202 and S. griseus cell extracts, the optimum pH of UDPGO was determined to be pH 7.5. Crude UDPGO is stable below 45°C for 1 h without significant loss of activity, thus permitting analysis of its substrate range. The apparent Km and Vmax values of UDPG for UDPGO were determined to be 50 μM and 23 nM min−1 mg−1, respectively, whereas the Km (NAD+) was 100 μM. Vmax was lower than that for the E. coli enzyme (7 μM min−1 mg−1) (8). Comparison of the Streptomyces and E. coli Vmax/Km values for UDPGO (4.6 × 10−4 and 1.66 × 10−3, respectively) reveals that the Streptomyces sp. enzyme is 28 times less efficient than the E. coli enzyme and 33 times less efficient than TDPG oxidoreductase from Saccharopolyspora erythraea (28). Unlike UDPGO in other bacteria, the enzyme from Streptomyces sp. was active when CDPG, GDPG, or TDPG was substituted for UDPG. K′m values for GDPG, CDPG, and TDPG were 58, 74, and 118 μM, respectively, with the Streptomyces UDPGO. No activity could be detected with ADPG. The V′max values for CDPG, TDPG, and GDPG were 1.9, 2.7, and 3.2 nM min−1 mg−1, respectively. Comparison of V′max and K′m of UDPG (determined as 50 μM min−1 mg−1 and 23 nM, respectively) with those of GDPG (8.4 times slower), CDPG (18 times slower), and TDPG (20 times slower) indicates that UDPG is the preferred substrate.
In summary, this work has demonstrated that activities for HK, PGM, UDPGP, and UDPG-4,6-dehydratase (also known as UDPGO) occur in cell extracts of Streptomyces sp. strain MRS202 and S. griseus (ATCC 13273). G-6-P plays a central role in glycolysis, and it is a key intermediate in the oxidation and fermentation of glucose as an energy source. The nucleoside diphosphate derivative of glucose, UDPG, is an activated form of the sugar important in anabolic events, such as the polysaccharide and cell wall biosynthesis, and as a precursor for other nucleoside diphosphate sugars. Such sugar nucleotides are also implicated in the biosynthesis of secondary metabolites, such as the chromomycins. Both UDPGP and UDPGO were partially purified from MRS202 (results not shown here). Among the enzymes examined, it was observed that UDPGO activities appear to be correlated with the production of chromomycins. Thus, the pathway of sugar oxidation likely involves the formation of UDP-4-keto-6-deoxyglucose via G-6-P, G-1-P, and UDPG, catalyzed by HK, PGM, UDPGP, and UDPGO, respectively.
ACKNOWLEDGMENTS
We are indebted to Kerry Kulowski for determination of concentrations of chromomycins. We thank Randal Chen and Jim Karwowski for the gift of Streptomyces sp. strain 657-1985M-134-MRS202 from Abbott Laboratories. We thank Eric Zirbes for the supply of mutants of Streptomyces spp. and Lynn Teesch and Diane Lamb of the University of Iowa Core Mass Spectrometry facility for fast atom bombardment-mass spectrometry analysis.
REFERENCES
- 1.Becker A, Kleickmann A, Keller M, Arnold W, Puhler A. Identification and analysis of the Rhizobium meliloti exo AMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exo HKLAMONP fragment. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Brede G, Fjævik E, Valla S. Nucleotide sequence and expression analysis of the Acetobacter xylinum uridine diphosphoglucose pyrophosphorylase gene. J Bacteriol. 1991;173:7042–7045. doi: 10.1128/jb.173.21.7042-7045.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang H, Lee C, Peng H. Identification of the Pseudomonas UDPG pyrophosphorylase gene. 1993. From NCBI (accession no. U03751). [Google Scholar]
- 5.Elbein A D, Heath E C. The biosynthesis of cell wall lipopolysaccharide in Escherichia coli. II. Guanosine diphosphate 4-keto-6-deoxy-d-mannose, an intermediate in the biosynthesis of guanosine diphosphate colitose. J Biol Chem. 1965;240:1926–1931. [PubMed] [Google Scholar]
- 6.Floss H G. Biosynthesis of isochromanequinone antibiotics. I. Structures and some properties of isochromanequinones. In: Corcoran J W, editor. The antibiotics. Vol. 4. Berlin, Germany: Springer-Verlag; 1981. pp. 215–235. [Google Scholar]
- 7.Gabriel O. Biological mechanisms involved in the formation of deoxy sugars: enzymatic hydrogen mediation. In: Gould R F, editor. Carbohydrates in solution. Washington, D.C: American Chemical Society; 1973. pp. 387–410. [Google Scholar]
- 8.Gilbert J M, Matsuhashi M, Strominger J L. Thymidine diphosphate-4-acetamido-4,6-dideoxyhexoses. II. Purification and properties of thymidine diphosphate-d-glucose oxidoreductase. J Biol Chem. 1965;240:1305–1308. [PubMed] [Google Scholar]
- 9.Gonzalez-Porque P, Strominger J L. Enzymatic synthesis of cytidine diphosphate 3,6-dideoxyhexoses. V. Purification to homogeneity and some properties of cytidine diphosphate-d-glucose oxidoreductase, enzyme E1 and enzyme E3. J Biol Chem. 1972;247:6748–6756. [PubMed] [Google Scholar]
- 10.Gray P P, Bhuwapathanapun S. Biotechnology of industrial antibiotics. In: Vandamme E J, editor. Drugs and the pharmaceutical sciences. Vol. 22. New York, N.Y: Marcel Dekker Inc.; 1984. pp. 743–768. [Google Scholar]
- 11.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 12.Lee J J, Lee J P, Keller P J, Cottrel C E, Chang C J, Zahner H, Floss H G. Further studies on the biosynthesis of chlorothricin. J Antibiot. 1986;39:1123–1134. doi: 10.7164/antibiotics.39.1123. [DOI] [PubMed] [Google Scholar]
- 13.Liao T H, Barber G A. Purification of guanosine 5′-diphosphate-d-mannose oxidoreductase from Phaseolus vulgaris. Biochim Biophys Acta. 1972;276:85–93. doi: 10.1016/0005-2744(72)90010-1. [DOI] [PubMed] [Google Scholar]
- 14.Liu H W, Thorson J S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 15.Liu S Y, Rosazza J P N. Ion-pairing reversed-phase HPLC identification of sugar nucleotides in cell free extracts of Streptomyces griseus. J Liq Chromatogr. 1995;18:4081–4095. [Google Scholar]
- 16.Magnani M, Dacha M, Stocchi V, Ninfali P, Fornaini G. Rabbit red blood cell hexokinase. Purification and properties. J Biol Chem. 1980;255:1752–1756. [PubMed] [Google Scholar]
- 17.Marechal L R, Oliver G, Veiga L A, Ruiz Holgado A A P. Partial purification and some properties of β-phosphoglucomutase from Lactobacillus brevis. Arch Biochem Biophys. 1984;228:592–599. doi: 10.1016/0003-9861(84)90027-4. [DOI] [PubMed] [Google Scholar]
- 18.Martin J F. Biosynthesis of polyene macrolide antibiotics. Annu Rev Microbiol. 1977;31:13–38. doi: 10.1146/annurev.mi.31.100177.000305. [DOI] [PubMed] [Google Scholar]
- 19.Matsuhashi S, Matsuhashi M, Brown J G, Strominger J L. Enzymatic synthesis of cytidine diphosphate 3,6-dideoxyhexoses. III. Cytidine diphosphate-d-glucose oxidoreductase. J Biol Chem. 1966;241:4283–4287. [PubMed] [Google Scholar]
- 20.Matsuhashi S, Matsuhashi M, Strominger J L. Enzymatic synthesis of cytidine diphosphate 3,6-dideoxyhexoses. I. Over-all reactions. J Biol Chem. 1966;241:4267–4274. [PubMed] [Google Scholar]
- 21.Montanari A, Rosazza J P N. Biogenesis of chromomycin A-3 by Streptomyces griseus. J Antibiot. 1990;43:883–889. doi: 10.7164/antibiotics.43.883. [DOI] [PubMed] [Google Scholar]
- 22.Nakano K, Omura Y, Tagaya M, Fukui T. UDP-glucose pyrophosphorylase from potato tuber: purification and characterization. J Biochem. 1989;106:528–532. doi: 10.1093/oxfordjournals.jbchem.a122886. [DOI] [PubMed] [Google Scholar]
- 23.Pape H, Brillinger U. Biosynthesis of thymidine diphospho-mycarose in cell free system of Streptomyces rimosus. Arch Microbiol. 1973;88:25–35. [PubMed] [Google Scholar]
- 24.Persat F, Azzar G, Martel M B, Got R. Properties of uridine diphosphate glucose pyrophosphorylase from golgi apparatus of liver. Biochim Biophys Acta. 1983;749:329–332. doi: 10.1016/0167-4838(83)90243-1. [DOI] [PubMed] [Google Scholar]
- 25.Snipes C E, Chang C J, Floss H G. Biosynthesis of the antibiotic granaticin. J Am Chem Soc. 1979;101:701–706. [Google Scholar]
- 26.Thompson M W, Strohl W R, Floss H G. Purification and characterization of TDP-d-glucose-4,6-dehydratase from anthracycline-producing streptomycetes. J Gen Microbiol. 1992;138:779–786. doi: 10.1099/00221287-138-4-779. [DOI] [PubMed] [Google Scholar]
- 27.Thorson J S, Lo S F, Liu H W, Hutchinson C R. Biosynthesis of 3,6-dideoxyhexoses: new mechanistic reflection upon 2,6-dideoxy, 4,6-dideoxy, and amino sugar construction. J Am Chem Soc. 1993;115:6993–6994. [Google Scholar]
- 28.Vara J A, Hutchinson C R. Purification of thymidine-diphospho-d-glucose-4,6-dehydratase from an erythromycin-producing strain of Saccharopolyspora erythraea by high resolution liquid chromatography. J Biol Chem. 1988;263:14992–14995. [PubMed] [Google Scholar]
- 29.Varón D, Boylan S A, Okamoto K, Price C W. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor ςB. J Bacteriol. 1993;175:3964–3971. doi: 10.1128/jb.175.13.3964-3971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker J E, Gay N J, Saraste M, Eberle A N. DNA sequence around the Escherichia coli unc operon completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glans and phos. Biochem J. 1984;224:799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S F, Gabriel O. Biological mechanisms involved in the formation of deoxy sugars. V. Isolation and crystallization of thymidine diphosphate-d-glucose oxidoreductase from Escherichia coli B. J Biol Chem. 1969;244:3430–3437. [PubMed] [Google Scholar]
- 32.Yu Y, Russel R N, Thorson J S, Liu L D, Liu H W. Mechanistic studies of the biosynthesis of 3,6-dideoxyhexoses in Yersinia pseudotuberculosis. J Biol Chem. 1992;267:5868–5875. [PubMed] [Google Scholar]
- 33.Zankowsky H, Glaser L. The mechanism of 6-deoxyhexose synthesis. III. Purification of deoxythymidine diphosphate-glucose oxidoreductase. J Biol Chem. 1969;244:4750–4756. [PubMed] [Google Scholar]