ABSTRACT
Cilia and flagella are ancient structures that achieve controlled motor functions through the coordinated interaction based on microtubules and some attached projections. Radial spokes (RSs) facilitate the beating motion of these organelles by mediating signal transduction between dyneins and a central pair (CP) of singlet microtubules. RS complex isolation from Chlamydomonas axonemes enabled the detection of 23 radial spoke proteins (RSP1–RSP23), although the roles of some radial spoke proteins remain unknown. Recently, RSP15 has been reported to be bound to the stalk of RS2, but its homolog in mammals has not been identified. Herein, we show that Lrrc23 is an evolutionarily conserved testis-enriched gene encoding an RSP15 homolog in mice. We found that LRRC23 localizes to the RS complex within murine sperm flagella and interacts with RSPH3A and RSPH3B. The knockout of Lrrc23 resulted in male infertility due to RS disorganization and impaired motility in murine spermatozoa, whereas the ciliary beating was not significantly affected. These data indicate that LRRC23 is a key regulator that underpins the integrity of the RS complex within the flagella of mammalian spermatozoa, whereas it is dispensable in cilia.
This article has an associated First Person interview with the first author of the paper.
Keywords: LRRC23, Radial spoke, Sperm, Flagella, Male infertility
Summary: Lrrc23 knockout mouse studies reveal that LRRC23 is a RS protein that plays key roles in regulating sperm motility and sperm flagellar ultrastructural integrity.
INTRODUCTION
Flagella are essential mediators of male gamete motility in eukaryotic species. In mammals, the sperm flagella are characterized by the middle, principle and end segments through which the axonemal structure runs (Lindemann and Lesich, 2016). Most earlier studies of axonemal motility focused on sea urchin spermatozoa or on ciliated/flagellar unicellular organisms to gain insight into mammalian sperm functionality. Dynein motors serve as essential mediators of axonemal motility, and are composed of outer and inner arms lining along the longitudinal axis of the axonemal cylinder. These axonemal dyneins exhibit ATP-insensitive anchoring to the A-tubule in each doublet, whereas they undergo an ATP-dependent stepping motion along the neighboring doublet B-tubules, resulting in sliding between these doublets. Restriction of such sliding by structures that include nexin links converts such motion into axonemal bending (Satir, 1968; Summers and Gibbons, 1971). A central pair (CP) of singlet microtubules, radial spokes (RSs), the I1 inner arm dynein (IDA) and the dynein regulatory complex (DRC) are essential mediators of dynein motility within this context (Bower et al., 2009; Piperno et al., 1994; Smith, 2002; Smith and Sale, 1992; Witman et al., 1978). In a model, it is suspected that signal from the CP complex is passed through RSs to the IDAs, and the I1 dynein intermediate chain–light chain (IC–LC) complex and the DRC signal through IDAs and outer dynein–inner dynein (OID) linkers to outer dynein arms (ODAs). In addition, to mediate the beating motion of flagella by relaying signals between the CP and dynein proteins (Smith and Yang, 2004), the RS complex is also important for maintaining the stability of the ‘9+2’ axonemal structure (Abbasi et al., 2018; Shinohara et al., 2015). Through studies on Chlamydomonas reinhardtii, purification of the RS complex led to the identification of 23 Chlamydomonas flagellar RS proteins (RSP1–RSP23) (Huang et al., 1981; Piperno et al., 1981; Yang et al., 2001; Yang et al., 2006).
Although RS complexes exhibit a similar T-shaped morphological orientation across species, RS evolutionary divergence has been noted, with many evolutionary changes having favored the simplification of this multi-protein complex (Zhu et al., 2017). In line with the predicted partial redundancy of spoke head proteins, RSP1 and its putative binding partner NDK5, for example, are not present in Tetrahymena or other ciliates, whereas RSP4/6 are encoded by a single gene in Ciona intestinalis and sea urchins (Zhu et al., 2017). Indeed, the radial spoke proteins RSP4/6, RSP9, and one MORN (membrane occupation and recognition nexus) protein, MORN40, are detected in purified Ciona RS samples (Satouh and Inaba, 2009). MORN proteins are found in the RS head and RSP1 is one of them. Similar changes may have also influenced RS complex development in humans, although humans do possess orthologs of RSP1, RSP4, RSP6 and RSP10. The expression of some of these genes is tissue specific in humans, with RSP1 (RSPH1) and RSPH10 exhibiting inverse expression patterns in airway and testis tissues (Kott et al., 2013). We have recently demonstrated that RSPH6A is enriched in murine testis wherein it localizes to the sperm flagella. Male Rsph6a knockout (KO) mice exhibit infertility attributable to their short immotile sperm flagella (Abbasi et al., 2018). Overall, these data suggest that RS structures in mammals are distinct from those in other species, and differ between cilia and flagella. However, the latter differences emphasize the importance of conducting further experiments to verify the localizations and functions of different RS component proteins in mammalian species.
In Chlamydomonas, these RSPs are assembled to yield the RS complex in two primary stages. Cell body extract fractionation experiments have revealed that a 12S RS precursor complex is assembled in the cell body (Qin et al., 2004). Intraflagellar transport facilitates the delivery of these 12S precursors into flagella, wherein they undergo conversion to yield the mature 20S RS complex. RSP1-7 and RSP9-12 compose the 12S precursor complex, with the remaining RSPs being assembled following transportation to the axoneme (Diener et al., 2011). Nevertheless, the specific roles of RSP13, RSP15, RSP18, RSP19 and RSP21 in the maturely assembled RS complex are less understood. Recently in Chlamydomonas, RSP15 was found to be located to the stalk of RS2, and RSP18, also known as FAP91, is the adaptor of RS2 and part of the calmodulin and spoke-associated complex (CSC) (Gui et al., 2021). Chlamydomonas RSP15 is known to be a leucine-rich repeat (LRR) protein that is thought to be homologous to the LRR37 protein found within the RSs in the spermatozoa of Ciona intestinalis (Padma et al., 2003; Yang et al., 2006). Mutations in the LRR37 homolog LRRC23 result in defective ciliary motility in the cilia of the otic vesicle in zebrafish (Han et al., 2018). Mutations in LRRC23 also cause defective phagocytosis and reduced swimming velocity in Tetrahymena, consistent with the ciliary defects (Han et al., 2018). The functional role of LRRC23 in mammals, however, remains to be evaluated.
Herein, we show that Lrrc23 is an evolutionarily conserved gene preferentially enriched in testis and required for male fertility in mice. LRRC23 localized to the RS and interacted with RSPH3A/B, and the knockout of Lrrc23 resulted in RS complex disorganization and impaired murine sperm motility, whereas morphological features and ciliary motility in these mice were not apparently affected. Together, these data suggest that LRRC23 plays an essential role in stabilizing the RS complex in sperm flagella, but is dispensable in respiratory cilia.
RESULTS
Lrrc23 is an evolutionarily conserved gene enriched in the testis
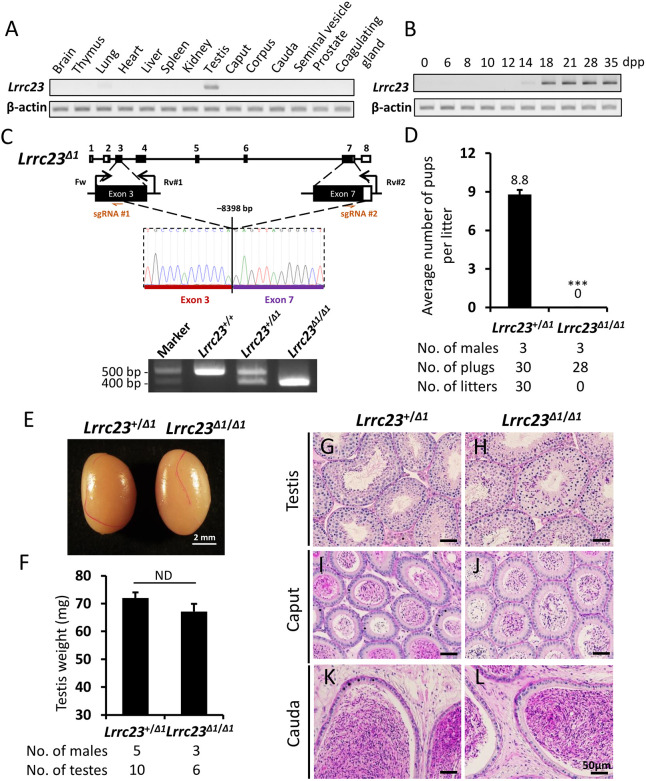
Lrrc23 is encoded by the genomes of known basal eukaryotic species that use flagella during at least one life cycle stage (Fig. S1A). In humans, the LRRC23 protein contains eight leucine-rich repeat (LRR) domains and a coiled-coil domain in its N-terminal region, with the LRRs of this protein being conserved among species (Fig. S1B). We began the present study by profiling Lrrc23 tissue-specific expression patterns via RT-PCR in adult mice, revealing a distinct band in the testis and a weaker band in lung tissues (Fig. 1A). We then evaluated the expression of Lrrc23 in postnatal testis to follow the leading edge of the first wave of spermatogenesis. This analysis revealed Lrrc23 expression initiated on postnatal day 14, which is roughly consistent with the first appearance of pachytene spermatocytes (Fig. 1B).
Fig. 1.
Generation and analysis of male Lrrc23Δ1/Δ1 mice. (A) Murine Lrrc23 expression in the indicated organs was assessed via RT-PCR, with Actb serving as a control. (B) Murine Lrrc23 expression in the testis tissue samples collected on the indicated day postpartum (dpp) was analyzed via RT-PCR with Actb as a normalization control. (C) The genomic structure of Lrrc23 and the CRISPR/Cas9 targeting approach. Dual sgRNAs (sgRNA#1 and sgRNA#2) were, respectively, used to target exons 3 and 7. The deletion of an 8398 bp fragment of Lrrc23 between exons 3 and 7 was confirmed via Sanger sequencing and PCR. The coding region is indicated using black rectangles. Genotyping primers (Fw, Rv#1, and Rv#2) were as shown. (D) Numbers of pups born per vaginal plug detected in the indicated groups. n=3 males each for Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice. Data are mean±s.d. ***P<0.001 (Student's t-test). (E) Testes of Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice. (F) A comparison of the weights of testes from Lrrc23+/Δ1 (n=5) and Lrrc23Δ1/Δ1 (n=3) mice. Data are mean±s.d. P>0.05 (Student's t-test). (G,H) Testis sections from Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice were subjected to periodic acid and Schiff’s reagent (PAS) staining. (I-L) Epididymides from Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice were subjected to hematoxylin-eosin (HE) staining. Scale bars: 50 μm.
Lrrc23 is essential for male fertility and sperm motility
To test the functional importance of Lrrc23 in mice, we employed the CRISPR/Cas9 genome editing system to generate two Lrrc23 mutant mouse lines. We first prepared a stable Lrrc23 mutant mouse line harboring a deletion of the exons 3-7 of this gene (Lrrc23Δ1/Δ1; Fig. 1C). Male Lrrc23Δ1/Δ1 mice did not exhibit any overt developmental or behavioral abnormalities. We assessed their fertility by housing individual Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 males with wild-type females and counting the numbers of offspring. Lrrc23Δ1/Δ1 males were unable to sire any offspring despite successful copulation with wild-type females confirmed by vaginal plugs (Fig. 1D), suggesting the loss of Lrrc23 expression results in male infertility in mice.
In order to determine whether the male infertility was due to arrested spermatogenesis, we next investigated the sperm formation and production in Lrrc23Δ1/Δ1 male animals. However, no differences in testis appearance and weight were detected when comparing Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 littermates (Fig. 1E,F). Likewise, periodic acid and Schiff's reagent (PAS) staining, and counterstaining with Mayer Hematoxylin solution of testicular sections failed to reveal any differences in the spermatogenesis between Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 males (Fig. 1G,H). Epididymal ducts filled with spermatozoa were observed in both the cauda and caput regions (Fig. 1I-L).
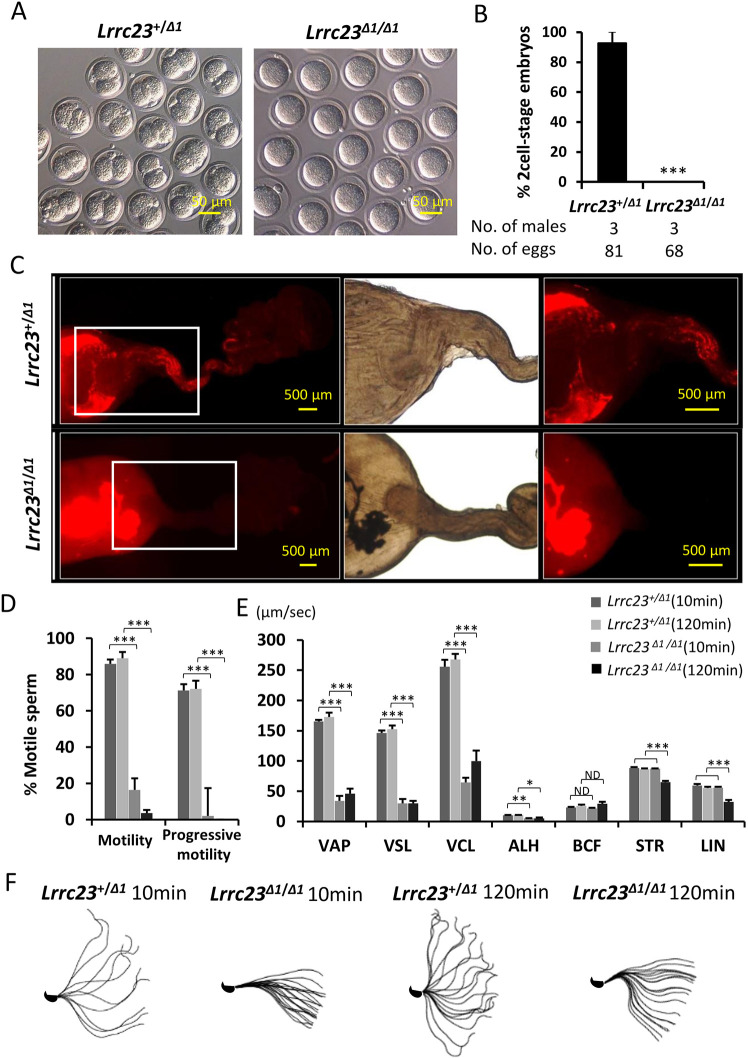
To further determine the cause of male infertility in Lrrc23 KO mice, we conducted an in vivo fertilization assay. No fertilized eggs were found in Lrrc23+/+ females after mating with Lrrc23Δ1/Δ1 males, suggesting the male sterility originated instead from impaired fertilization of defective embryogenesis (Fig. 2A,B). To investigate whether the impaired fertilization was derived from problems in sperm migration or downstream zona pellucida (ZP) penetration or gamete fusion, we then carried out a uterotubal junction (UTJ) penetration assay. Lrrc23 KO spermatozoa exhibited inability to pass through the UTJ (Fig. 2C). Computer-assisted sperm analysis (CASA) was further carried out to assess the sperm motility in Lrrc23Δ1/Δ1 males, revealing a significant reduction in motile spermatozoa and spermatozoa exhibiting progressive motility, a condition known as asthenozoospermia (Fig. 2D,E). Although progressive motility was impaired, the flagella of spermatozoa from Lrrc23Δ1/Δ1 mice did beat, albeit across a more limited range (Fig. 2F, Movies 1 and 2). We additionally conducted in vitro fertilization (IVF) and determined that Lrrc23 KO spermatozoa were unable to fertilize cumulus-intact or cumulus-free ZP-intact oocytes (Fig. S2A,B). However, following ZP removal, Lrrc23 KO spermatozoa were able to fuse with oocytes (Fig. S2C), indicating that Lrrc23 KO spermatozoa were defective in passing through the UTJ and penetrating the cumulus cell layer and/or the ZP due to impaired motility.
Fig. 2.
Assessment of the motility and fertility of spermatozoa from Lrrc23Δ1/Δ1mice. (A) Images of eggs retrieved from the oviducts of wild-type females mated with Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice during an in vivo fertilization test. (B) Percentages of two-cell embryos in cumulus-intact oocytes inseminated with spermatozoa from Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice, n=3. Data are mean±s.d. ***P<0.001 (Student's t-test). (C) Images of sperm migration through the female reproductive tract. Wild-type females were mated with Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice carrying the RBGS transgene such that spermatozoa were fluorescently tagged, revealing the failure of Lrrc23 KO spermatozoa to pass through the UTJ. White rectangles indicate magnified regions on the right. (D) Relative percentages of motile and progressively motile sperm from Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice after 10 and 120 min of incubation in the TYH medium. Data are mean±s.d. ***P<0.001 (Student's t-test). (E) Different motility parameters for sperm from Lrrc23+/Δ1 and Lrrc23Δ1/Δ1 mice, as determined via CASA following incubation for 10 or 120 min in TYH medium. VCL, curvilinear velocity; VAP, average path velocity; ALH, amplitude of lateral head; VSL, straight-line velocity; STR, straightness; LIN, linearity. Data are mean±s.d. ***P<0.001, **P<0.01, *P<0.05 (Student's t-test). (F) Flagellar waveforms were assessed after incubation for 10 and 120 min, with motility being imaged at 200 frames/s and with individual frames from a single beating cycle being superimposed.
To confirm these observations, we generated another stable Lrrc23 mutant mouse line harboring a 49 bp deletion in the exon 4 (Lrrc23Δ2/Δ2; Fig. S3A). Likewise, gross examinations revealed no significant differences in testis weight or sperm counts when comparing Lrrc23Δ2/Δ2 and Lrrc23+/+ males (Fig. S3B-E), and Lrrc23Δ2/Δ2 male mice also exhibited infertility (Fig. S3F). No abnormality in sperm morphology was detected in the Lrrc23Δ2/Δ2 males under a light microscope (Fig. S3G) and a scanning electron microscope (SEM; Fig. S3H). Consistent with the spermatozoa from Lrrc23Δ1/Δ1 males, CASA revealed a reduction in sperm motility and progressive movement in Lrrc23Δ2/Δ2 mice (Fig. S3I-K). In summary, these findings together confirm that Lrrc23 regulates sperm motility and is required for male fertility in mice.
LRRC23 is a radial spoke component that localizes to sperm flagella
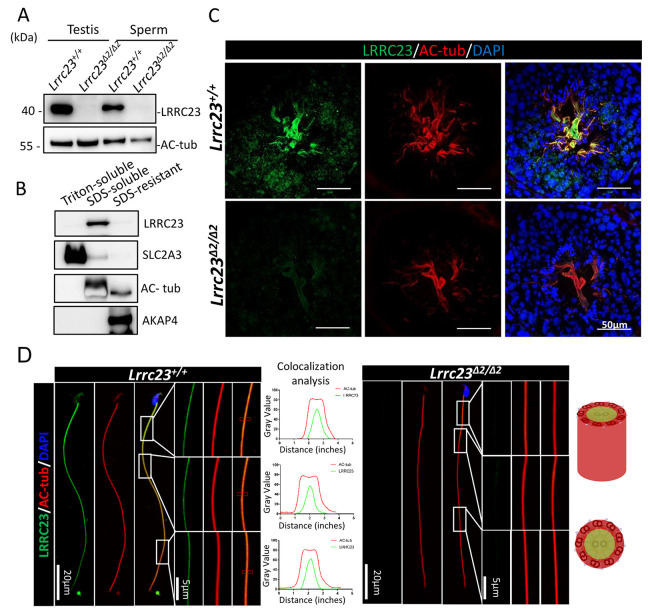
We assessed the subcellular localization of LRRC23 using internally generated antibodies. Western blotting revealed LRRC23 was ∼40 kDa in size and was absent in Lrrc23Δ2/Δ2 mice (Fig. 3A). Immunoblot analyses of sperm protein extracts indicated that LRRC23 was present within the Triton X-100-resistant, SDS-soluble fraction (Fig. 3B), which is the fraction associated with the axoneme (Cao et al., 2006). These results suggested that LRRC23 could be the component of axoneme in the sperm flagella.
Fig. 3.
LRRC23 localizes to the sperm tails. (A) LRRC23 expression in the testis and cauda epididymal spermatozoa from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice. No difference in the acetylated tubulin (Ac-tub) staining was detected between the two groups of mice. (B) Murine sperm fractionation revealed the presence of LRRC23 in the SDS-soluble fraction. SLC2A3, acetylated tubulin and AKAP4 were used, respectively, to mark the fractions that were Triton soluble, SDS soluble and SDS resistant. (C) Testis cross-sections from wild-type and Lrrc23Δ2/Δ2 mice were stained with immunofluorescent antibodies specific for AC-tub (red) and LRRC23 (green), with DAPI (blue) being used to detect nuclei. (D) Spermatozoa from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice were subjected to immunofluorescent staining using antibodies specific for AC-tub (red) and LRRC23 (green), with Hoechst 33342 (blue) being used to detect nuclei. AC-tub axonemal localization was evident in the sperm of both wild-type and Lrrc23Δ2/Δ2 mice. Magnified regions are marked with white rectangles; panels on the right demonstrate LRRC23 localization to the center of these axonemes. The line charts show quantitative colocalization analysis of LRRC23 and AC-tub in red rectangles.
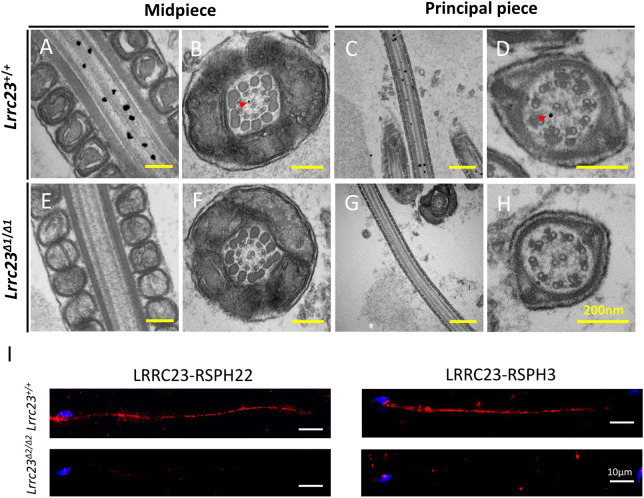
To analyze the localization of LRRC23 in the spermatozoa, we conducted immunofluorescence staining. Confocal microscopic observation revealed LRRC23 presented within the flagella and cytoplasm in murine testicular spermatids (Fig. 3C). Furthermore, high-resolution microscopy confirmed LRRC23 localized to the flagella of murine spermatozoa, and the signal of LRRC23 was at the center of the acetylated tubulin (AC-tub) via the quantitative colocalization analysis (Fig. 3D). The detailed localization of LRRC23 was further analyzed at the ultrastructural level by immunoelectron microscopy. The LRRC23 antibody-conjugated gold particles were mostly localized to the axonemal matrix between the central pair and microtubule doublets (Fig. 4A-H), which is consistent with the localization of a radial spoke head protein, RSPH6A (Fig. S5), implying that LRRC23 is also localized to the radial spokes.
Fig. 4.
LRRC23 localizes to axonemal radial spokes within sperm flagella. (A-H) Immunoelectron microscopy was conducted to evaluate sperm flagella from Lrrc23+/+ and Lrrc23Δ1/Δ1 mice. (A,C) Longitudinal and (B,D) cross-sections of the midpiece and principal piece labeled with anti-LRRC23. Gold particles (red arrowheads) were found to localize within the axoneme between the CP and the outer doublet microtubules, consistent with the location of the radial spokes. (E-H) Sperm flagella from Lrrc23Δ1/Δ1 mice served as controls. (I) Representative images of an in situ proximity ligation assay using antibodies directed against LRRC23, RSPH22 and RSPH3 in sperm flagella from Lrrc23+/+ and Lrrc23Δ2/Δ2.
In Chlamydomonas, RSP15, the homolog of LRRC23, has previously been shown to interact with RSP3 and RSP22 (Gui et al., 2021). In Chlamydomonas, RSP22 was confirmed to be localized to the base of RSs. RSP3 spans and plays a key role in uniting the spoke head and stalk. Dimeric RSP3 is a core scaffold of the entire RS complex, and provides localization signals for a variety of radial spoke proteins to facilitate the assembly of the RS (Gui et al., 2021; Sivadas et al., 2012). Thus, to analyze whether LRRC23 interacts with RSPH22 and RSPH3, we conducted co-immunoprecipitation experiments using HEK293T cells. Although we did not detect any interaction between LRRC23 and RSPH22 (Fig. S4A), we did find that it was able to interact with RSPH3A/B (Fig. S4B,C). LRRC23 may interact with RSPH22 indirectly and cannot be recapitulated in non-ciliated cells. To further analyze the interactions between LRRC23 and RS proteins in vivo, we performed in situ proximity ligation assays. Cells were post-stained with DAPI to visualize the nucleus. The red signals indicate the recognition of two proteins in close proximity (<40 nm) (Alam, 2018). The specificity of the assay was confirmed by staining Lrrc23 KO spermatids in parallel. Consistent with the co-immunoprecipitation data, the signals of LRRC23-RSPH3 and LRRC23-RSPH22 were detected along the sperm flagella (Fig. 4I). These results indicated that the physical distances between LRRC23 and the components of RSs are within 40 nm, implying that LRRC23 could be the component of RS.
Lrrc23 knockout causes abnormal RS formation in sperm flagella but does not adversely impact respiratory cilia
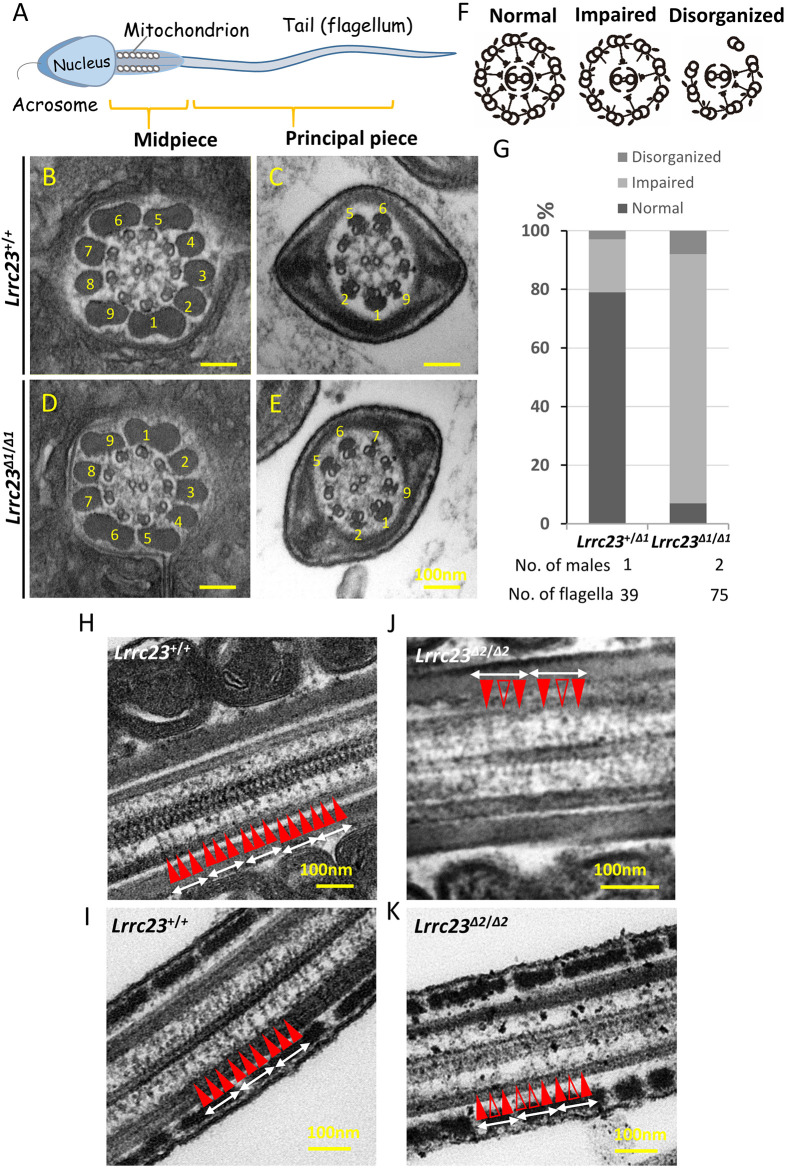
To test the impact of LRRC23 depletion on RS assembly, we next examined the ultrastructure of the sperm axonemes in wild-type and Lrrc23Δ1/Δ1 males via transmission electron microscopy (TEM; Fig. 5A-E), revealing about 85% showed impaired RS structures (‘impaired’ in Fig. 5F,G) and about 8% showed disorganized microtubule structures (‘disorganized’ in Fig. 5F,G) in the KO sperm cross-sections, whereas the percentages of RS impaired and microtubule disorganized axonemes were 18% and 3%, respectively, in wild-type cross-sections (see also Fig. S6). In the axonemal cross-sections lacking normal RS assembly, irregular electron density was observed between the central and peripheral microtubules, potentially indicating a disorder of the RS complex. Partial RS absence was similarly detected in the spermatozoa of Lrrc23Δ2/Δ2 mice (Fig. S7). To clarify the morphological heterogeneity of RSs in Lrrc23 KO mice, we observed the axonemal longitudinal sections under TEM and found that RS1, RS2 and RS3 were arranged in 96 nm repeats in wild-type sperm (Fig. 5H,I), while sperm flagella from Lrrc23 KO mice displayed one or two RSs per 96 nm repeat with one or two absent or truncated RSs (Fig. 5J,K), indicating that LRRC23 was needed for the assembly of specific radial spokes in mammalian sperm flagella. Owing to the insufficient resolution of our EM, we could not clearly identify the absent or incomplete radial spokes as RS2 or RS3. However, given that RSP15 (the homology of LRRC23 in Chlamydomona) is the specific component of RS2 in Chlamydomona, it seems reasonable to assume that the RS consistently observed at 96 nm distance in Lrrc23 KO mouse sperm is RS1.
Fig. 5.
Ultrastructural analysis of cauda epididymis spermatozoa from Lrrc23Δ1/Δ1 mice. (A) An overview of the structure of a mature spermatozoon. (B-E) Electron microscopy images demonstrating normal radial spoke structures in wild-type spermatozoa (B,C) and radial spokes that were partially formed or absent in Lrrc23Δ1/Δ1 sperm (D,E). Outer dense fibers are marked with numbers. (F) Schematic representation of three axonemal structures of spermatozoa. (G) Percentages of the different axonemal structures in spermatozoa from Lrrc23+/+ and Lrrc23Δ1/Δ1 mice. (H-K) Longitudinal sections of the axonemal structures in spermatozoa from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice. Radial spokes (red arrowheads, RS1-3) appeared in a repetitive pattern (average 96 nm repeat) in control (H,I). In Lrrc23 KO sperm flagella (J,K), the radial spokes were rather irregular and/or incomplete (unfilled red arrowheads) and missing.
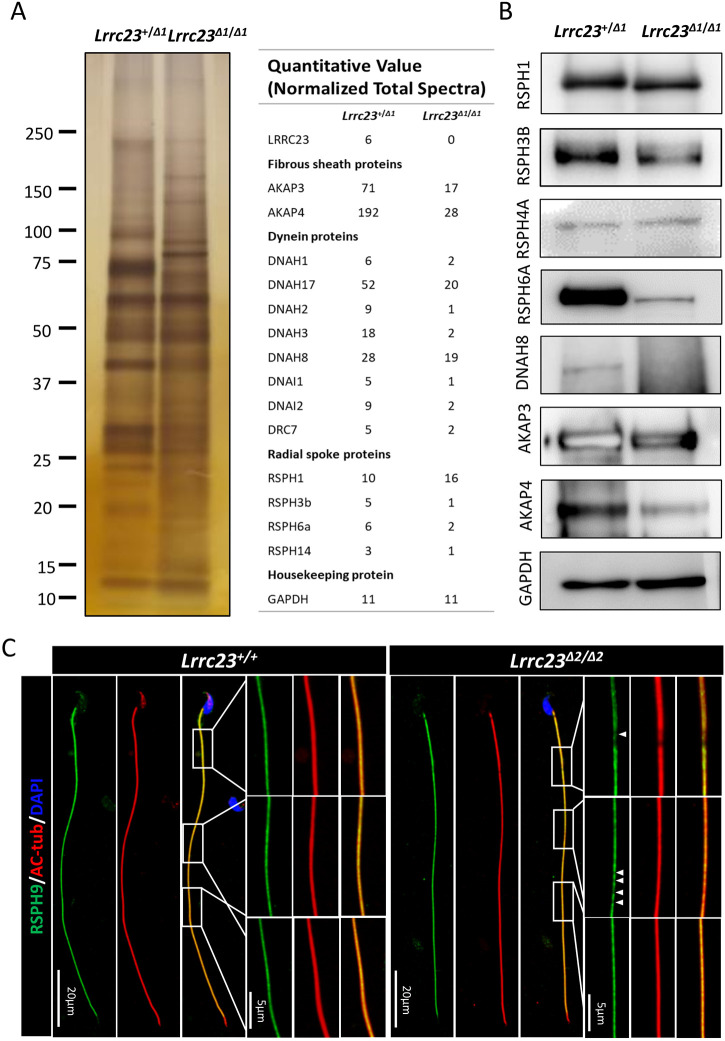
To evaluate the alterations of proteins in spermatozoa of control and Lrrc23 KO mice, a proteomic analysis was conducted and revealed downregulation of RSPH3A/B and RSPH6A in the spermatozoa of Lrrc23Δ2/Δ2 mice. Similarly, we observed a decrease of proteins associated with sperm motility, including AKAP3, AKAP4 (Luconi et al., 2011) and DNAH8 (Fig. 6A), as confirmed by western blotting (Fig. 6B). However, we could not find obvious abnormality of ODA in the cross-sections that show ‘Impaired’ RS. We speculated that the downregulation of outer dynein arm components could be caused by the disordered axonemal structures. In the Lrrc23 KO and wild-type sperm cross-sections, the percentages of microtubule disorganized axonemes were 18% and 3%, respectively. Additionally, high-resolution microscopy revealed a discontinuous or fragmented RSPH9 signal along the sperm flagella of Lrrc23Δ2/Δ2 mice (Fig. 6C), suggesting that the loss of flagellar LRRC23 resulted in RS structural defects.
Fig. 6.
Lrrc23 knockout causes the downregulation of other flagella component proteins. (A) Left: SDS-PAGE and silver staining were conducted to compare the whole-sperm proteome in Lrrc23+/+ and Lrrc23Δ2/Δ2 mice. Right: mass spectrometry revealed the downregulation of certain flagella components in the spermatozoa of Lrrc23Δ2/Δ2 mice. (B) Western blotting was used to assess flagellum components in protein extracts from spermatozoa. (C) Immunofluorescence staining was conducted for spermatozoa obtained from wild-type and Lrrc23Δ2/Δ2 males using antibodies specific for AC-tub (red) and RSPH9 (green). Nuclei were identified through Hoechst staining (blue). While rectangles indicate regions that have been magnified, with areas of RSPH9 signal discontinuity being marked with white arrowheads.
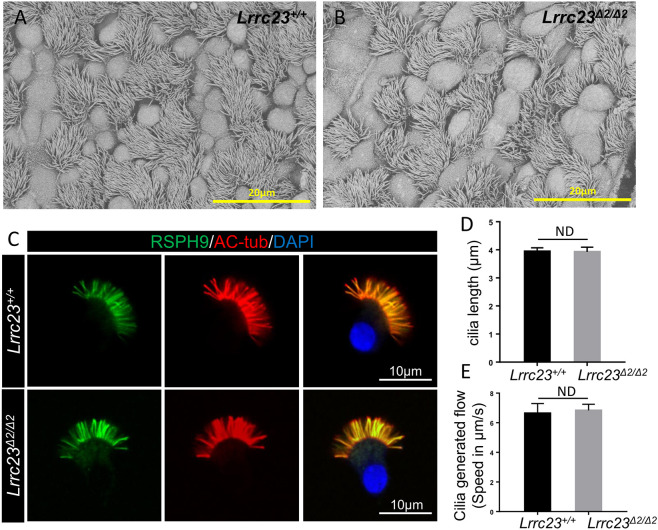
As Lrrc23 shows minor expression in lung, we investigated the functionality of the respiratory cilia in KO mice by analyzing the expression of RS and the central pair (CP) components, such as LRRC23, NME5, DYDC1, RSPH9 and HYDIN in tracheal cilia via high-resolution immunofluorescence microscopy. NME5 is the homolog of RSP23 in Chlamydomonas and individuals with mutant NME5 were considered for primary ciliary dyskinesia (PCD) with RS/CP defects (Cho et al., 2020). DYDC1 or DYDC2 is considered to be the homolog of RSP2 in Chlamydomonas (Zhu et al., 2017). HYDIN is a component of a CP projection and the knockout of HYDIN in mice leads to the specific loss of a projection from one of the central microtubules (Lechtreck et al., 2008). However, no apparent differences were observed in the expression or distributions of these proteins between the samples from Lrrc23Δ2/Δ2 and Lrrc23+/+ mice (Fig. S8). Likewise, SEM analysis of lung sections showed no obvious alterations in the morphological features of the respiratory cilia in Lrrc23Δ2/Δ2 and Lrrc23+/+ animals (Fig. 7A,B). Additionally, cilia length was unaltered in the isolated Lrrc23Δ2/Δ2 murine tracheal epithelial cells (mTECs) when compared with the ones in wild-type mice (Fig. 7C,D). Furthermore, comparable tracheal ciliary beating was also detected in Lrrc23Δ2/Δ2 and Lrrc23+/+ mice (Movies 3 and 4). Meanwhile, we conducted the cilia-generated flow (CGF) assay to analyze tracheal ciliary beat. The analysis of bead velocity revealed no significant alterations of CGF in the Lrrc23Δ2/Δ2 respiratory cilia compared with those in wild-type mice (Movies 5 and 6 and Fig. 7E). Together, these data indicate that LRRC23 is an essential mediator of RS stability in mammalian sperm flagella, whereas it is dispensable for normal ciliary function.
Fig. 7.
No significant differences in tracheal cilia components are evident when comparing Lrrc23+/+ and Lrrc23Δ2/Δ2 mice. (A,B) Tracheal cilia from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice were examined via SEM. (C) Immunofluorescence staining was conducted for isolated mouse tracheal epithelial cells (mTECs) from wild-type and Lrrc23Δ2/Δ2 males using antibodies specific for AC-tub (red) and RSPH9 (green). Nuclei were identified through Hoechst staining (blue). (D) Analysis of cilia length in mTECs isolated from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice via the signals of AC-tub (red). Data are mean±s.d. (n=3, P>0.05, Student's t-test). (E) Analysis of cilia generated flow in trachea cilia from Lrrc23+/+ and Lrrc23Δ2/Δ2 mice. Data are mean±s.d. (n=3, P>0.05, Student's t-test).
Sterility of KO males is rescued by transgenic expression of LRRC23
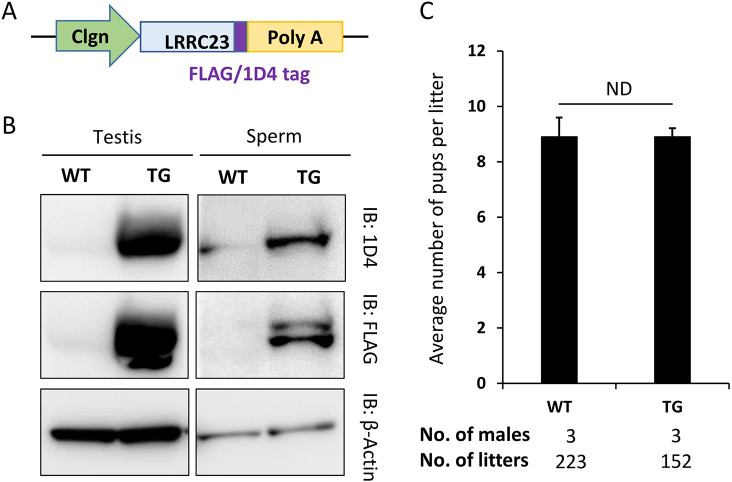
To determine whether the absence of Lrrc23 was responsible for the male infertility, we produced a transgenic (Tg) mouse line expressing FLAG/1D4-tagged LRRC23 driven by the testicular germ cell-specific Clgn promoter on a Lrrc23 KO background (Fig. 8A). Western blot analysis confirmed that both FLAG and 1D4-tagged LRRC23 was detected in testis and sperm lysates, whereas no signal was detected in the testis or spermatozoa of wild-type males that did not carry the transgene (Fig. 8B). Lrrc23 KO-Tg males were housed with wild-type females and resulted in normal litter sizes (8.9±2.9; Fig. 8C), indicating that the KO phenotype was rescued by the transgene. These results confirm that LRRC23 is required for normal sperm motility and male reproduction.
Fig. 8.
Sterility of KO males is rescued by transgenic expression of LRRC23. (A) Schematic representation of generating the Lrrc23 transgene (Tg) mouse line. Transgene designed in which FLAG or 1D4-tagged LRRC23 are expressed as a fused protein under the testicular germ cell-specific Clgn promoter. (B) FLAG or 1D4-tagged LRRC23 expression in wild-type and KO mice carrying the Lrrc23 transgene was detected by western blotting. β-Actin was analyzed as a loading control. (C) Average litter size of Lrrc23 KO male mice expressing FLAG or 1D4-tagged Lrrc23 Tg. The average litter size (mean±s.d.) was 8.9±1.6 in wild-type males and 8.9±2.9 in FLAG or 1D4-tagged Lrrc23 rescued KO male mice. Data are mean±s.d. ND, P>0.05.
DISCUSSION
PCD is a genetic condition that arises as a consequence of ciliary and flagellar motility defects in multiple organ systems (Afzelius and Eliasson, 1983; Munro et al., 1994), affecting about 1 in 10,000 of the human population globally (Lucas et al., 2014; Rubbo and Lucas, 2017). Infertility is one of clinical phenotypes of PCD. Almost all men with PCD are infertile. However, some men with PCD are azoospermic, and a few have normal sperm motility and have fathered children. There are cell type-specific differences in axonemal assembly and in the outer dynein arm components (Fliegauf et al., 2007), although gross axonemal structures in sperm flagella and motile cilia appear similar. Herein, we report a distinct form of PCD. The depletion of LRRC23 led to male infertility due to impaired sperm motility but did not compromise respiratory ciliary motility. Light microscopy revealed that asthenospermia occurred in Lrrc23 KO mice, but no morphological abnormalities were observed. We have previously reported a similar form of PCD in Tcte1 KO mice (Castaneda et al., 2017). Although there have been no reports to date linked LRRC23 or TCTE1 mutations to asthenospermia in humans, we nonetheless believe that these findings offer important insights into the tissue-specific differences in disease-related phenotypes that manifest in individuals with PCD.
Through high-resolution fluorescence and immunoelectron microscopic analyses, we determined that LRRC23 localized near the center of the axonemal structure. Furthermore, the immunogold-EM of LRRC23 and co-immunoprecipitation analyses indicated that LRRC23 was able to interact with RSPH3A and RSPH3B: the core scaffold of RS. PLA revealed that the spatial distances between LRRC23 and two RS proteins in different parts were within 40 nm, suggesting that LRRC23 could be a component of RS complexes. Therefore, when Lrrc23 was knocked out, one or two radial spokes were disturbed in 96 nm repeats, exhibiting incompleteness or absence, while the remaining were complete; the consistently present radial spoke could be RS1. Although RS abnormalities were detected in spermatozoa, no significant changes in the length of LRRC23-null sperm flagella were detected, in marked contrast to the short-tail phenotype observed in Rsph6a KO spermatozoa. The homologue of RSPH6A in Chlamydomona, RSP6, functions as a key 12S precursor complex component (Pigino et al., 2011). The 12S precursor complex that composed of RSP1-7 and RSP9-12, is an intermediate form of the radial spoke, found in the cytoplasm and matrix of the flagella in transit to the tip, where it combines with other spoke proteins to form the completed 20S complex (Diener et al., 2011). These data indicate that the absence of 12S precursor components may have a profound adverse impact on the flagellar formation. In contrast, the loss of LRRC23 compromised the stability of sperm flagellar structures, such as dyneins, RSs and fibrous sheaths. As dyneins serve as molecular motors and the RS complex functions as a mechanochemical sensor underpinning the flagellar motility (Smith and Yang, 2004), the disruption of these axonemal components can therefore compromise the motility-related signal transmission and cause asthenospermia.
Although Lrrc23 is essential for male fertility due to its role in sperm motility in mice, it is dispensable for the formation and function of cilia in respiratory tract, confirmed by the SEM and immunofluorescent analyses of the trachea cilia. The trachea cilia length was unaltered in the absence of Lrrc23. The cilia-generated flow (CGF) assay also revealed no significant differences between Lrrc23 KO and wild-type mice. These results imply that the assembly processes of respiratory cilia and sperm flagella are not completely identical. In humans with TTC12 mutations, TTC12 defects lead to the complete absence of ODAs and IDAs in the sperm flagella, which was different from the phenotype in the cilia of respiratory cells, where only IDAs were missing (Thomas et al., 2020). In addition, mutations in the SPEF2 gene lead to misshapen sperm head and multiple morphological abnormalities of the sperm flagella (MMAF). Nonetheless, the morphological features of the respiratory cilia did not exhibit any apparent abnormalities (Tu et al., 2020). Similarly, our previous study revealed that mutations in the DRC1 gene, the component of the nexin-dynein regulatory complex (N-DRC), caused MMAF but maintained the stability of the ‘9+2’ axonemal structure in the respiratory cilia (Zhang et al., 2021). These studies suggest that the structures and compositions of sperm flagella and respiratory cilia are similar but not completely identical.
Moreover, the loss of LRRC23 caused the disorder of one RS in the sperm flagella, which resembles the Cfap206 mutant sperm. In contrast to the largely immotile sperm flagella, the respiratory cilia from Cfap206 mutant mice were motile and show normal ultrastructure but an enhanced beating frequency (Beckers et al., 2020). The phenotype of respiratory cilia in Lrrc23 KO mice is similar to that in Cfap206 KOs. However, the loss of some other radial spoke components lead to more severe phenotype in the cilia ultrastructure. For example, all the triplet spoke heads in the cilia of Rsph4a KO mice were deficient (Yoke et al., 2020). In the PCD patient with mutations in RSPH1, the primary RS1 and RS2 were both deficient in cilia, whereas RS3 was intact (Lin et al., 2014). Furthermore, the proportion of respiratory cilia with normal axonemal structure is 50% in human patients carrying mutations in RSPH4A (Daniels et al., 2013), whereas it is 80% in patients with RSPH1 mutations (Knowles et al., 2014). When two RSs were missing in axonemal structures, patients with RSPH1 mutations had PCD with a mild disease phenotype. However, Rsph4a KO mice with three RSs defective show hydrocephalus, which is a typical phenotype of PCD, suggesting that the extent of the RS structural defects is largely related to phenotype of cilia. This could explain the normal phenotype of respiratory cilia in Lrrc23 KO mice.
Sequence analysis showed that LRRC23 possesses six LRR motifs and belongs to the LRR protein family. The LRR motif is found in proteins with diverse intracellular and extracellular functions, and probably facilitates protein-protein interactions (Kajava, 1998). Many members of the LRR family are structural components of the motile cilial axoneme, such as LRRC48, LRRC50 and LRRC56 (Freshour et al., 2007; Kamiya, 1995; Lin et al., 2011). LRRC48, also known as DRC3, the Chlamydomonas ortholog of which is FAP134 or DRC3, is a component of the N-DRC and is required for normal flagellar waveform (Awata et al., 2015). To find the proteins similar to LRRC23, we analyzed the data on wild-type sperm that was collected from tandem mass tag (TMT) labeling and mass spectrometry analysis published in our previous study (Castaneda et al., 2017). The proteins that contain LRR motifs were picked out and their protein sequences were aligned against the sequence of LRRC23. We found these proteins were not similar to LRRC23, except for their LRR motifs. Thus, five similar proteins, LRRC28, LRRC59, LRRC69, LRRC46 and LRRC34 were analyzed regarding their expression patterns in various organs and tissues. Based on the NCBI database, we found that LRRC28 could be a candidate axonemal component expressed in the ciliated tissues or sperm because it has the same expression pattern as LRRC23. However, the amino acid sequences of LRRC28, LRRC23 and LRR37 (the homology of LRRC23 in Ciona) show low similarity. Therefore, LRRC23 may play an unique role in mammals.
Overall, we demonstrated that LRRC23 is a RS protein that plays key roles in regulating sperm motility and sperm flagellar ultrastructural integrity. These data offer a theoretical basis for the incidence of asthenospermia and highlight novel targets that can be studied for a better understanding of the mechanistic basis underlying sperm motility.
MATERIALS AND METHODS
Animals
All mice used in the present study were housed under standard conditions (20-22°C, 50-70% humidity, 12 h light/dark cycle) with free food and water access. The Institutional Animal Care and Use Committees of Nanjing Medical University and Osaka University approved all studies carried out by both laboratories (Approval No. IACUC-1810020; #Biken-AP-H30-01), and the Animal Ethical and Welfare Committee of both universities reviewed all animal protocols. Transgenic mice with Red Body Green Sperm [RBGS; B6D2-Tg(CAG/Su9-DsRed2, Acr3-EGFP)RBGS002Osb] prepared and housed in the laboratory of M.I. were used to assess spermatozoa migration through the uterotubal junction (UTJ).
In silico expression analysis
Murine testis transcriptome analyses were carried out using previously reported single-cell transcriptomic data (Hermann et al., 2018). Lrrc23 mRNA levels in spermatogenic cells and related somatic cells were evaluated using the Loupe Cell Browser 3.3.1 (10X Genomics).
RT-PCR
Total RNA was extracted from the tissues of adult ICR mice and from the testes of 1- to 5-week-old male mice, and a SuperScript III First Strand Synthesis Kit (Thermo Fisher) was used to prepare cDNA based on the manufacturer's instructions. The expression of Lrrc23 was assessed via PCR using the primers shown in Table S1.
Generation of Lrrc23Δ1 mutant mice by CRISPR/Cas9
The CRISPR/Cas9 system was used to generate Lrrc23Δ1 mice. Two single-guide RNAs (sgRNAs; 5′-CATCATGGCCTCCGTGATGG-3′ and 5′-GGCTGGGCACACGGGACGAG-3′) were designed to target the exon 3 and exon 7 of Lrrc23, and their potential for off-target genomic editing was assessed using the CRISPRdirect program (crispr.dbcls.jp) (Naito et al., 2015). Female B6D2F1 mice were superovulated via intraperitoneal injection with pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG; ASKA Animal Health, Tokyo, Japan), after which they were paired with wild-type B6D2F1 males. The resultant two-pronuclear zygotes were isolated from the oviducts of the superovulated female mice, and a NEPA21 electroporation instrument (NEPA GENE) was used to introduce crRNA/tracrRNA/Cas9 ribonucleoprotein complexes into the zygotes that were subsequently incubated in potassium simplex optimized medium (KSOM) medium (Ho et al., 1995) to the two-cell stage and transplanted into the ampullary segment of the oviducts in 0.5 days pseudopregnant ICR female mice. Founder animals were then obtained via natural delivery or Cesarean section after 19 days of pregnancy, with genotyping being conducted by Sanger sequencing and PCR. The genotyping primers are available in Table S1.
Generation of Lrrc23Δ2 mutant mice by CRISPR/Cas9
The CRISPR/Cas9 system was used to generate Lrrc23Δ2 KO mice using two sgRNAs targeted to knock out the exon 4 of Lrrc23 (5′-GCAATCAGCTTCGGAGTGCT-3′ and 5′-GATCTGGTTGTAGGAAAAGC-3′). Two complementary DNA oligos for each of these sgRNA targets were annealed and ligated to the BsaI-digested pUC57-T7-sgRNA vector, while sgRNA templates were amplified from sgRNA plasmids via PCR using primers (Fw trans and Rv trans, shown in Table S1). A MinElute PCR Purification Kit (QIAGEN) was then used to isolate the amplified template sequences, and sgRNAs were generated with the MEGAshortscript Kit (Ambion) and purified with a MEGAclear Kit (Ambion) as per the manufacturer's instructions. Following linearization with AgeI, a Cas9 plasmid (Addgene #44758) was purified using a MinElute PCR Purification Kit (QIAGEN), after which an mMESSAGE mMACHINE T7 Ultra Kit (Ambion) was used to transcribe Cas9 mRNA that was subsequently purified with an RNeasy Mini Kit (QIAGEN) as per the manufacturer's instructions. The Cas9 mRNA (50 ng/μl) and sgRNA (20 ng/μl) were then co-injected into murine zygotes that were transferred into pseudopregnant females. On postnatal day 7, toe clipping was conducted to tag the newborn mice, and DNA was extracted from these tissue samples with a Mouse Direct PCR Kit (Biotool, Shanghai, China). Sanger sequencing was performed after PCR amplification with appropriate primers (Fw#3 and Rv#4, Table S1) and PrimeSTAR HS DNA Polymerase (Takara).
Fertility testing
Three Lrrc23Δ1/Δ1 and three Lrrc23Δ2/Δ2 sexually mature male mice were individually housed with three 8-week-old wild-type B6D2F1 female mice for a minimum of 2 months, with wild-type male mice undergoing the same housing conditions as a control. Litter sizes were recorded at the date of birth, with average litter size being calculated by dividing total numbers of pups by total numbers of litters.
Antibody preparation
Antibodies were produced as detailed previously (Liu et al., 2014). The full-length murine Lrrc23 cDNA (amino acids 1-340) was expressed as His fusion protein in E. coli with the pET-28a(+) vector, and the Ni-NTA His Bind Resin (TransGen Biotech) was used to achieve the affinity purification of the resultant protein. The purified protein was then used to immunize two rabbits and four mice to yield anti-LRRC23 antisera.
LRRC23 monoclonal antibody used for Immunogold-EM and sperm protein fractionation analyses was generated as previously described. The sequence encoding mouse LRRC23 (residue 171-340 amino acids, CCDS20528.1) was cloned and inserted into pGEX6p-1 expression vector (GE Healthcare), followed by transformation into E. coli strain BL21 (de3) pLysS (C606003, Thermo Fisher Scientific). GST-fused LRRC23 recombinant protein was expressed and subjected to the treatment of PreScission Protease to remove the GST tag (Noda et al., 2020). The recombinant LRRC23 protein was then purified and injected into female rats in combination with a complete adjuvant. After 17 days of injection, lymphocytes were collected from iliac lymph nodes, and hybridomas were generated and cultured (Sado et al., 2006). The supernatants obtained from the hybridomas were used as antibodies. The candidates were screened by ELISA against LRRC23.
Testis weights and sperm motility analyses
Lrrc23 KO males were anesthetized and euthanized via cervical dislocation, after which testis and body weight were measured for both heterozygous and homozygous KO animals. Spermatozoa were extracted from cauda epididymides, after which the motility of these cells was assessed with a CEROS II computer-assisted sperm analysis (CASA) system (Hamilton Thorne Biosciences) at 10 min and 2 h after incubation in Toyoda, Yokoyama, Hoshi (TYH) medium. Sperm movement was additionally recorded at 200 frames/s using an Olympus BX-53 microscope equipped with a high-speed camera (HAS-L1, Ditect). The BohBoh sperm motion analysis software was then used to reconstruct sperm flagellar waveforms based on these videos (BohBohsoft, Tokyo, Japan).
After extraction, sperm derived from the epididymis of Lrrc23Δ2/Δ2 mice or littermate controls (Lrrc23Δ2/+ or wild-type males) were incubated in human tubal fluid (HTF) media (FUJIFILM Irvine Scientific) containing 10% FBS at 37°C, with Hamilton Thorne's Ceros II system (Hamilton-Thorne Research) being used to dilute and analyze these samples. Normal sperm motility was determined by the straight-line velocity (VSL) and average path velocity (VAP). The VSL indicates the straight-line distance between the first and last points of the trajectory, whereas the VAP refers to the distance the spermatozoon has traveled in the average direction of movement in the observation period (Si and Olds-Clarke, 2000). The criteria of normal sperm motility in our CASA system are that both VSL and VAP are >4 μm/s. The criteria of progressive motility are that progressive straightness (STR) is >80% and progressive VAP is >50 μm/s. STR is the ratio of VSL to VAP.
Sperm UTJ migration assay
Wild-type B6D2F1 females were superovulated by peritoneal injection of pregnant mare serum gonadotropin (PMSG) and hCG. After 12 h of hCG injection, the hormone-primed females were individually housed with a Lrrc23 KO male mouse expressing DsRed2/Acr3-EGFP. Success of copulation was confirmed by the formation of a vaginal plug. The female mice were sacrificed 2 h after copulation and the female reproductive tract was collected for imaging. Spermatozoa with red fluorescence in the midpiece and green fluorescence in the acrosome were observed directly under a Keyence BZ-X710 microscope.
Analyses of testis and epididymis histology and sperm morphology
Testes and epididymides were fixed in Bouin's solution (Polysciences) and embedded in paraffin wax. Paraffin wax sections were stained with periodic acid (Nacalai Tesque) and Schiff's reagent (Wako), and counterstained with Mayer Hematoxylin solution (Wako). The cauda epididymal spermatozoa were dispersed in the TYH medium and observed under an Olympus BX53 phase-contrast microscope.
Transmission electron microscopy (TEM)
Ultrastructural analyses of testes and spermatozoa using TEM were conducted as previously described (Shimada et al., 2019). Male wild-type and Lrrc23Δ1/Δ1 mice were anesthetized and perfused with 4% paraformaldehyde (PFA), after which the fixed cauda epididymis tissues were collected and fixed for an additional 6 h in 4% PFA at 4°C. These epididymides were then minced with razor blades to yield 2 mm cubes that were fixed overnight with 1% glutaraldehyde in 30 mM HEPES (pH 7.8) at 4°C. Post-fixation for 1 h with 1% osmium tetroxide (OsO4) and 0.5% potassium ferrocyanide in 30 mM HEPES was then performed at room temperature, after which an ethanol gradient was used to dehydrate samples, which were then embedded for 2 days using epoxy resin at 60°C for 2 days. An Ultracut Microtome was then used to prepare ultrathin sections that were stained with both uranyl acetate and lead citrate prior to mounting onto copper grids and evaluation with a JEM-1400 Plus electron microscope (JEOL) at 80 kV equipped with a Veleta 2K×2K CCD camera (Olympus).
Similarly, spermatozoa from Lrrc23Δ2/Δ2 male mice were fixed overnight with 2.5% glutaraldehyde, post-fixed with 2% OsO4, and embedded in Araldite for ultrastructural analyses. Ultrathin (80 nm) sections were then stained using uranyl acetate and lead citrate and were imaged via EM (JEM.1010, JEOL).
Scanning electron microscopy (SEM)
Spermatozoa samples were fixed for 2 h with 2.5% phosphate-buffered glutaraldehyde at 4°C. Spermatazoa were then allowed to attach to coverslips coated with poly-L-lysine. Both sample types were then washed with PBS, dehydrated with a chilled ethanol gradient (30%, 50%, 70%, 80%, 90% and 100%), and subjected to critical point drying with a Lecia EM CPD300 Critical Point Dryer. Samples were then attached to appropriate specimen holders and coated with gold particles via the use of an ion sputter coater (EM ACE200, Leica). A Helios G4 CX scanning electron microscope (Thermo Fisher Scientific) was then used to image samples.
In vitro fertilization (IVF)
Spermatozoa isolated from the cauda epididymides of wild-type and Lrrc23 KO male mice were suspended in TYH medium (Toyoda and Yokoyama, 2016). Cumulus-oocyte complexes (COCs) were collected from superovulated B6D2F1 female mice, and were treated for 5 min with 1.0 mg/ml hyaluronidase (Wako) at 37°C to remove the cumulus cells or with 1.0 mg/ml collagenase (Sigma) to remove the zona pellucida (ZP). Cumulus-intact and cumulus-free oocytes were then inseminated by combing them with 2.0×105 sperm/ml, while ZP-free oocytes were combined with 2.0×104 sperm/ml. Following a 6 h insemination period, the formation of two pronuclei was assessed to gauge fertilization success.
Sperm protein fractionation
The fractionation of sperm proteins was performed as in previous reports (Castaneda et al., 2017). Briefly, 1% Triton X–100 lysis buffer [50 mM NaCl and 20 mM Tris-HCl (pH 7.5)] was used to lyse isolated spermatozoa for 2 h at 4°C. Supernatants containing the Triton-soluble fraction were then collected after spinning for 10 min at 15,000 g, while the insoluble pellets were subjected to lysis for 1 h with 1% SDS lysis buffer [75 mM NaCl and 24 mM EDTA (pH 6.0)] at room temperature. After being spun down for an additional 10 min at 15,000 g, the SDS-soluble supernatant fraction was collected, while sample buffer (66 mM Tris–HCl, 2% SDS, 10% glycerol and 0.005% Bromophenol Blue) was used to lyse the SDS-resistant pellet, and samples were boiled for 5 min, with the SDS-resistant fraction being isolated following centrifugation.
Western blotting
Urea/thiourea lysis buffer [8 M urea, 50 mM Tris-HCl (pH 8.2) and 75 Mm NaCl] containing a 2% (v/w) protease inhibitor cocktail (Roche) was used to extract proteins from murine tissue samples. These proteins were then separated via SDS-PAGE and transferred to PVDF membranes that were blocked for 2 h with 5% non-fat milk in TBST at room temperature, following by overnight incubation with primary antibodies. These primary antibodies are shown in Table S2. The membranes were then washed thrice in TBST and probed for 2 h with appropriate secondary antibodies (Table S2). The High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China) Western Blotting Detection system was then used to detect protein bands.
Co-immunoprecipitation
Plasmids were transfected into HEK293T cells and then cell samples were lysed by RIPA buffer (Beyotime) containing a 2% proteinase inhibitor cocktail at 4°C for 20 min, after which samples were spun down for 25 min at 12,000 g. Supernatants were then mixed for 1 h with Protein A magnetic beads (Thermo Fisher Scientific), and lysates were then incubated with primary anti-FLAG at 4°C overnight. Samples were then mixed for 3 h with 50 μl of Protein A magnetic beads at 4°C, after which they were washed with PBST. Immunoprecipitated pellets and extract samples were used for western blotting after boiling in SDS loading buffer.
Immunofluorescence staining
The immunofluorescent staining of tissue sections was conducted as detailed previously (Castañeda et al., 2014). For analyses of sperm cells, these samples were smeared onto slides, air-dried, fixed for 40 min with 4% paraformaldehyde, washed thrice with PBS (5 min/wash) and antigen retrieval was then conducted by boiling slides in a microwave for 10 min in 10 mM citrate buffer (pH 6.0). Following three additional washes in PBST (PBS containing 0.05% Tween-20; 10 min/wash), 5% BSA in PBST was used to block slides for 2 h, after which they were probed overnight with appropriate primary antibodies (Table S2) at 4°C. After three additional PBST washes, slides were probed for 2 h with secondary antibodies (Table S2), counterstained for 5 min with Hoechst 33342 (Invitrogen), rinsed with PBST, mounted and imaged with a LSM800 confocal microscope (Carl Zeiss) and TCS SP8X confocal microscope (Leica).
Duolink in situ proximity ligation assays were carried as per the manufacturer's instructions (Sigma-Aldrich, DUO92007). Briefly, isolated sperm cells were smeared onto slides, air-dried, fixed for 40 min with 4% paraformaldehyde, washed thrice with PBS (5 min/wash), followed by blocking with Duolink blocking solution for 60 min at 37°C. Primary antibodies were diluted in Duolink antibody diluent and incubated overnight at 4°C. Appropriate secondary antibodies conjugated to synthetic oligonucleotides (rabbit PLA probe PLUS and mouse PLA probe MINUS) were applied for 1 h at 37°C. A ligation reaction was performed using the Duolink ligation solution and ligase (30 min at 37°C), which results in binding of the two PLA probes if they are less than 40 nm from one another. Rolling circle amplification and hybridization with fluorescently labelled nucleotides was achieved using the Duolink amplification solution and polymerase (100 min at 37°C). Slides were mounted with Duolink mounting medium with DAPI and imaged with a TCS SP8X confocal microscope (Leica).
For high-resolution immunofluorescence, a confocal laser microscope (Leica TCS SP8X) with a built-in LIGHTNING module, which offers deconvolution-based high-resolution, was used. For the capture of the images, the LIGHTNING module was used. The objective lens used was the HC PL APO CS2 63×/1.40 oil; scan speed was 700 Hz at a resolution at 3696×3696 pixels and normal acquisition of the hybrid detector was used. Excitation of the DAPI channel was achieved using a 405 nm diode laser. Emission signal was captured with standard PMT Channel 1 with an emission gap of 415 nm-511 nm. Excitation of Alexa fluor 488 and Alexa fluor 555 used the Leica Supercontinuum white light laser. Emission signal was captured with Leica HyD detector. Leica Application Suite X (LAS X) was used as a software platform. The quantitative colocalization analysis was performed using ImageJ software.
Immunoelectron microscopy
Cauda epididymides from wild-type and Lrrc23Δ1/Δ1 mice were dissected after perfusion fixation of the whole bodies with 4% PFA in 0.1 M phosphate buffer under anesthesia. The epididymides were then sliced into 3-4 mm sections and fixed in 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4) for 1 h at room temperature. After fixation, the samples were washed with 4% sucrose in 0.1 M phosphate buffer (pH 7.4) three times. Tissue samples were then incubated in 10%, 15% and 20% sucrose in 0.1 M phosphate buffer (pH 7.4) sequentially for 6 h each, embedded in OCT compound (Sakur) and frozen in liquid nitrogen. The samples were subsequently sectioned under −20°C using a cryostat (Thermo Fisher Scientific), and the cryosections were attached to MAS-coated glass coverslips (Matsunami Glass) and air-dried for 30 min. The coverslips were placed in 24-well culture plate and blocked with blocking solution (0.1 M phosphate buffer containing 0.1% saponin, 10% BSA, 10% normal goat serum and 0.1% cold water fish skin gelatin) for 30 min. Gold particle labeling was performed using rat anti-LRRC23 primary antibody (1:150; antibody was prepared in-house) in blocking solution and goat anti-rat IgG secondary antibody (1:300; 112-035-167, Jackson ImmunoResearch) coupled to 1.4 nm gold (Nanogold, Nanoprobes) in blocking solution followed the procedures as previously described (Shimada et al., 2021). After post-fixed in 1% OsO4 and 1.5% potassium ferrocyanide in 0.1 M phosphate buffer (pH 7.4) for 1 h, samples were dehydrated in a graded series of ethanol, substituted with propylene oxide and embedded in epoxy resin. Ultrathin sections were stained with 8% uranyl acetate and lead staining solution. The samples were examined using a JEM-1400 plus electron microscope (JEOL) at 80 kV with a CCD Veleta 2K×2K camera (Olympus).
Analysis of cilia length in multiciliated cells and cilia-generated flow in mouse trachea
Tracheas of each three adult wild-type and Lrrc23 KO male mice were dissected and mTECs isolated and stained using anti-acetylated α-tubulin and RSPH9 antibodies as described above. Cells were imaged using a confocal microscope (Leica TCS SP8 AOBS) and the cilia length was determined using LAS X (Leica) software by measuring the length of the ciliary tuft of each cell at three locations and averaging the measurements. For each experimental group, 20 cells were analyzed.
Tracheas of 10-day-old mice were dissected, added to high-glucose DMEM containing 10% FBS (Gibco), opened on the dorsal side, and minced under stereoscopic magnification to yield ∼5 mm tissue fragments. These tissues were then transferred to a confocal dish (BDD012035, BIOFIL) and a Scotch tape spacer was used to facilitate their imaging under a 40× objective (CFI S Plan Flour ELWD NAMC) with an inverted microscope (Eclipse Ti2-U, Nikon). Movies were recorded for 10 s at frame rates of 25 fps.
To analyze cilia-generated flow in mouse trachea. Beads (1:200, Thermo, FluoSpheres Fluorescent Microspheres 1 μm) were added to the trachea explants and the bead flow was recorded at 23 fps for 6 s at 24-27°C using fluorescent microscopy. Trajectories of fluorescent microspheres near the ciliary tuft were analyzed using the ImageJ.
Mass spectrometry
Whole-sperm proteomic analyses were performed as previously described (Shimada et al., 2021). Briefly, protein samples were extracted from spermatozoa using lysis buffer (6 M urea, 2 M thiourea and 2% sodium deoxycholate) and centrifuged at 15,000× g for 15 min at 4°C. The samples were processed and the resultant protein peptides were subjected to nanocapillary reversed-phase LC-MS/MS analysis using a C18 column (10 cm×75 μm, 1.9 µm, Bruker Daltoniks) on a nanoLC system (Bruker Daltoniks) connected to a timsTOF Pro mass spectrometer (Bruker Daltoniks) and a nano-electrospray ion source (CaptiveSpray, Bruker Daltoniks). The resulting data were processed using DataAnalysis (Bruker Daltoniks), and proteins were identified using MASCOT Sever (Matrix Science) compared with the SwissProt database. Quantitative value and fold exchange were calculated by Scaffold4 (Proteome Software). The raw data are accessible from the ProteomeXchange Consortium via the dataset identifier PXD025166.
Production of transgenic mice
Fertilized eggs were obtained from in vitro fertilization between the spermatozoa from Lrrc23 heterozygous males and the oocytes from homozygous or heterozygous KO females. The LRRC23 coding sequence with a C-terminal FLAG or 1D4 tag was individually cloned and inserted into the multiple cloning sites of a plasmid bearing a Calmergin (Clgn) promoter and a rabbit β-globin polyadenylation signal. The two plasmids encoding LRRC23-FLAG and LRRC23-1D4 were linearized, mixed and microinjected into the pronuclei of the zygotes. The treated zygotes were then cultured in KSOM medium to two-cell stage and transplanted into the ampullary segment of the oviducts in 0.5 days pseudopregnant ICR females. Founder animals were obtained by natural delivery or Cesarean section after 19 days of pregnancy. Primer sequences used for genotyping of transgenic mice are enumerated in Table S1 (Fw#4, FLAG and 1D4).
Statistical analysis
Data are given as mean±s.d. and were compared using a two-tailed unpaired Student's t-tests. *P<0.05, **P<0.01, ***P<0.001.
Supplementary Material
Acknowledgements
We thank the Center of Cryo-Electron Microscopy at Zhejiang University for technical assistance. We also thank the members of both the State Key Laboratory of Reproductive Medicine and Animal Core Facility of Nanjing Medical University for experimental assistance.
Footnotes
Author contributions
Conceptualization: M.I., M.L.; Methodology: J.S., J.Z., K.S., T.N., S. Zhao, T.K., M.M., J.W.; Formal analysis: Y.L., J.W.; Investigation: X.Z., J.S., Y.L., J.Z., K.S., T.N., S. Zhao, T.K., M.M., S. Zhou, J.W.; Data curation: X.Z., J.S., Y.L., J.Z., K.S., T.N., S. Zhao, T.K., M.M., J.W.; Writing - original draft: Y.L., M.L., J.S., X.Z., M.I.; Writing - review & editing: X.Z., Y.L., J.S., M.I., M.L.; Visualization: S. Zhou; Supervision: Y.L., M.I., M.L.; Project administration: M.I., M.L.; Funding acquisition: M.I., M.L.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFA0500902 to M.L.), Natural Science Foundation of China (31771654 and 32070842 to M.L.) and the Natural Science Foundation of Jiangsu Province (Grants No. BK20190081 to M.L.). Meanwhile, the work was also supported by Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI (Grants-in-Aid for Scientific Research) Grants (JP18K16735 to Y.L., JP20K16107 to K.S., JP18K14612 and JP20H03172 to T.N., JP17H01394 and JP19H05750 to M.I.), Takeda Science Foundation grants (to T.N. and M.I.), Japan Agency for Medical Research and Development Grant (JP20gm5010001 to M.I.), Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants (R01HD088412 and P01HD087157 to M.I.), and the Bill & Melinda Gates Foundation Grant (INV-001902 to M.I.).
Data availability
The raw MS data generated in this study are accessible from the ProteomeXchange Consortium via the dataset identifier PXD025166.
Contributor Information
Masahito Ikawa, Email: ikawa@biken.osaka-u.ac.jp.
Mingxi Liu, Email: mingxi.liu@njmu.edu.cn.
References
- Abbasi, F., Miyata, H., Shimada, K., Morohoshi, A., Nozawa, K., Matsumura, T., Xu, Z., Pratiwi, P. and Ikawa, M. (2018). RSPH6A is required for sperm flagellum formation and male fertility in mice. J. Cell Sci. 131, jcs221648. 10.1242/jcs.221648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzelius, B. A. and Eliasson, R. (1983). Male and female infertility problems in the immotile-cilia syndrome. Eur. J. Respir. Dis. Suppl. 127, 144-147. [PubMed] [Google Scholar]
- Alam, M. S. (2018). Proximity Ligation Assay (PLA). Curr. Protoc. Immunol. 123, e58. 10.1002/cpim.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata, J., Song, K., Lin, J., King, S. M., Sanderson, M. J., Nicastro, D. and Witman, G. B. (2015). DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol. Biol. Cell 26, 2788-2800. 10.1091/mbc.E15-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers, A., Adis, C., Schuster-Gossler, K., Tveriakhina, L., Ott, T., Fuhl, F., Hegermann, J., Boldt, K., Serth, K., Rachev, E.et al. (2020). The FOXJ1 target Cfap206 is required for sperm motility, mucociliary clearance of the airways and brain development. Development 147, dev188052. 10.1242/dev.188052 [DOI] [PubMed] [Google Scholar]
- Bower, R., VanderWaal, K., O'Toole, E., Fox, L., Perrone, C., Mueller, J., Wirschell, M., Kamiya, R., Sale, W. S. and Porter, M. E. (2009). IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol. Biol. Cell 20, 3055-3063. 10.1091/mbc.e09-04-0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, W., Gerton, G. L. and Moss, S. B. (2006). Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics 5, 801-810. 10.1074/mcp.M500322-MCP200 [DOI] [PubMed] [Google Scholar]
- Castañeda, J., Genzor, P., van der Heijden, G. W., Sarkeshik, A., Yates, J. R., III, Ingolia, N. T. and Bortvin, A. (2014). Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 33, 1999-2019. 10.15252/embj.201386855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda, J. M., Hua, R., Miyata, H., Oji, A., Guo, Y., Cheng, Y., Zhou, T., Guo, X., Cui, Y., Shen, B.et al. (2017). TCTE1 is a conserved component of the dynein regulatory complex and is required for motility and metabolism in mouse spermatozoa. Proc. Natl. Acad. Sci. USA 114, E5370-E5378. 10.1073/pnas.1621279114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, E. H., Huh, H. J., Jeong, I., Lee, N. Y., Koh, W. J., Park, H. C. and Ki, C. S. (2020). A nonsense variant in NME5 causes human primary ciliary dyskinesia with radial spoke defects. Clin. Genet. 98, 64-68. 10.1111/cge.13742 [DOI] [PubMed] [Google Scholar]
- Daniels, M. L. A., Leigh, M. W., Davis, S. D., Armstrong, M. C., Carson, J. L., Hazucha, M., Dell, S. D., Eriksson, M., Collins, F. S., Knowles, M. R.et al. (2013). Founder mutation in RSPH4A identified in patients of Hispanic descent with primary ciliary dyskinesia. Hum. Mutat. 34, 1352-1356. 10.1002/humu.22371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, D. R., Yang, P., Geimer, S., Cole, D. G., Sale, W. S. and Rosenbaum, J. L. (2011). Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 68, 389-400. 10.1002/cm.20520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf, M., Benzing, T. and Omran, H. (2007). When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880-893. 10.1038/nrm2278 [DOI] [PubMed] [Google Scholar]
- Freshour, J., Yokoyama, R. and Mitchell, D. R. (2007). Chlamydomonas flagellar outer row dynein assembly protein ODA7 interacts with both outer row and I1 inner row dyneins. J. Biol. Chem. 282, 5404-5412. 10.1074/jbc.M607509200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui, M., Ma, M., Sze-Tu, E., Wang, X., Koh, F., Zhong, E. D., Berger, B., Davis, J. H., Dutcher, S. K., Zhang, R.et al. (2021). Structures of radial spokes and associated complexes important for ciliary motility. Nat. Struct. Mol. Biol. 28, 29-37. 10.1038/s41594-020-00530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Xie, H., Wang, Y. and Zhao, C. (2018). Radial spoke proteins regulate otolith formation during early zebrafish development. FASEB J. 32, 3984-3992. 10.1096/fj.201701359R [DOI] [PubMed] [Google Scholar]
- Hermann, B. P., Cheng, K., Singh, A., Roa-De La Cruz, L., Mutoji, K. N., Chen, I.-C., Gildersleeve, H., Lehle, J. D., Mayo, M., Westernströer, B.et al. (2018). The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650-1667.e8. 10.1016/j.celrep.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., Wigglesworth, K., Eppig, J. J. and Schultz, R. M. (1995). Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol. Reprod. Dev. 41, 232-238. 10.1002/mrd.1080410214 [DOI] [PubMed] [Google Scholar]
- Huang, B., Piperno, G., Ramanis, Z. and Luck, D. J. (1981). Radial spokes of Chlamydomonas flagella: genetic analysis of assembly and function. J. Cell Biol. 88, 80-88. 10.1083/jcb.88.1.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava, A. V. (1998). Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277, 519-527. 10.1006/jmbi.1998.1643 [DOI] [PubMed] [Google Scholar]
- Kamiya, R. (1995). Exploring the function of inner and outer dynein arms with Chlamydomonas mutants. Cell Motil. Cytoskeleton 32, 98-102. 10.1002/cm.970320205 [DOI] [PubMed] [Google Scholar]
- Knowles, M. R., Ostrowski, L. E., Leigh, M. W., Sears, P. R., Davis, S. D., Wolf, W. E., Hazucha, M. J., Carson, J. L., Olivier, K. N., Sagel, S. D.et al. (2014). Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am. J. Respir. Crit. Care. Med. 189, 707-717. 10.1164/rccm.201311-2047OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott, E., Legendre, M., Copin, B., Papon, J.-F., Dastot-Le Moal, F., Montantin, G., Duquesnoy, P., Piterboth, W., Amram, D., Bassinet, L.et al. (2013). Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet. 93, 561-570. 10.1016/j.ajhg.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F., Delmotte, P., Robinson, M. L., Sanderson, M. J. and Witman, G. B. (2008). Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180, 633-643. 10.1083/jcb.200710162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Tritschler, D., Song, K., Barber, C. F., Cobb, J. S., Porter, M. E. and Nicastro, D. (2011). Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem. 286, 29175-29191. 10.1074/jbc.M111.241760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Yin, W., Smith, M. C., Song, K., Leigh, M. W., Zariwala, M. A., Knowles, M. R., Ostrowski, L. E. and Nicastro, D. (2014). Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat. Commun. 5, 5727. 10.1038/ncomms6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann, C. B. and Lesich, K. A. (2016). Functional anatomy of the mammalian sperm flagellum. Cytoskeleton (Hoboken) 73, 652-669. 10.1002/cm.21338 [DOI] [PubMed] [Google Scholar]
- Liu, M., Shi, X., Bi, Y., Qi, L., Guo, X., Wang, L., Zhou, Z. and Sha, J. (2014). SHCBP1L, a conserved protein in mammals, is predominantly expressed in male germ cells and maintains spindle stability during meiosis in testis. Mol. Hum. Reprod. 20, 463-475. 10.1093/molehr/gau014 [DOI] [PubMed] [Google Scholar]
- Lucas, J. S., Burgess, A., Mitchison, H. M., Moya, E., Williamson, M., Hogg, C. and National PCD Service, UK. (2014). Diagnosis and management of primary ciliary dyskinesia. Arch. Dis. Child. 99, 850-856. 10.1136/archdischild-2013-304831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luconi, M., Cantini, G., Baldi, E. and Forti, G. (2011). Role of a-kinase anchoring proteins (AKAPs) in reproduction. Front. Biosci. (Landmark Ed) 16, 1315-1330. 10.2741/3791 [DOI] [PubMed] [Google Scholar]
- Munro, N. C., Currie, D. C., Lindsay, K. S., Ryder, T. A., Rutman, A., Dewar, A., Greenstone, M. A., Hendry, W. F. and Cole, P. J. (1994). Fertility in men with primary ciliary dyskinesia presenting with respiratory infection. Thorax 49, 684-687. 10.1136/thx.49.7.684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito, Y., Hino, K., Bono, H. and Ui-Tei, K. J. B. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120-1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Lu, Y., Fujihara, Y., Oura, S., Koyano, T., Kobayashi, S., Matzuk, M. M. and Ikawa, M. (2020). Sperm proteins SOF1, TMEM95, and SPACA6 are required for sperm-oocyte fusion in mice. Proc. Natl. Acad. Sci. USA 117, 11493-11502. 10.1073/pnas.1922650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma, P., Satouh, Y., Wakabayashi, K., Hozumi, A., Ushimaru, Y., Kamiya, R. and Inaba, K. (2003). Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the Ascidian Ciona intestinalis. Mol. Biol. Cell 14, 774-785. 10.1091/mbc.02-06-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino, G., Bui, K. H., Maheshwari, A., Lupetti, P., Diener, D. and Ishikawa, T. (2011). Cryoelectron tomography of radial spokes in cilia and flagella. J. Cell Biol. 195, 673-687. 10.1083/jcb.201106125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Huang, B., Ramanis, Z. and Luck, D. J. (1981). Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 88, 73-79. 10.1083/jcb.88.1.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Mead, K., LeDizet, M. and Moscatelli, A. (1994). Mutations in the “dynein regulatory complex” alter the ATP-insensitive binding sites for inner arm dyneins in Chlamydomonas axonemes. J. Cell Biol. 125, 1109-1117. 10.1083/jcb.125.5.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Diener, D. R., Geimer, S., Cole, D. G. and Rosenbaum, J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbo, B. and Lucas, J. S. (2017). Clinical care for primary ciliary dyskinesia: current challenges and future directions. Eur. Respir. Rev. 26, 170023. 10.1183/16000617.0023-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado, Y., Inoue, S., Tomono, Y. and Omori, H. (2006). Lymphocytes from enlarged iliac lymph nodes as fusion partners for the production of monoclonal antibodies after a single tail base immunization attempt. Acta Histochem. Cytochem. 39, 89-94. 10.1267/ahc.06001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir, P. (1968). Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 39, 77-94. 10.1083/jcb.39.1.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satouh, Y. and Inaba, K. (2009). Proteomic characterization of sperm radial spokes identifies a novel spoke protein with an ubiquitin domain. FEBS Lett. 583, 2201-2207. 10.1016/j.febslet.2009.06.016 [DOI] [PubMed] [Google Scholar]
- Shimada, K., Kato, H., Miyata, H. and Ikawa, M. (2019). Glycerol kinase 2 is essential for proper arrangement of crescent-like mitochondria to form the mitochondrial sheath during mouse spermatogenesis. J. Reprod. Dev. 65, 155-162. 10.1262/jrd.2018-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, K., Park, S., Miyata, H., Yu, Z., Morohoshi, A., Oura, S., Matzuk, M. M. and Ikawa, M. (2021). ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc. Natl. Acad. Sci. USA 118, e2018355118. 10.1073/pnas.2018355118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, K., Chen, D., Nishida, T., Misaki, K., Yonemura, S. and Hamada, H. (2015). Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell 35, 236-246. 10.1016/j.devcel.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Si, Y. and Olds-Clarke, P. (2000). Evidence for the involvement of calmodulin in mouse sperm capacitation. Biol. Reprod. 62, 1231-1239. 10.1095/biolreprod62.5.1231 [DOI] [PubMed] [Google Scholar]
- Sivadas, P., Dienes, J. M., St. Maurice, M., Meek, W. D. and Yang, P. (2012). A flagellar A-kinase anchoring protein with two amphipathic helices forms a structural scaffold in the radial spoke complex. J. Cell Biol. 199, 639-651. 10.1083/jcb.201111042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. F. (2002). Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskeleton 52, 33-42. 10.1002/cm.10031 [DOI] [PubMed] [Google Scholar]
- Smith, E. F. and Sale, W. S. (1992). Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science 257, 1557-1559. 10.1126/science.1387971 [DOI] [PubMed] [Google Scholar]
- Smith, E. F. and Yang, P. (2004). The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton 57, 8-17. 10.1002/cm.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers, K. E. and Gibbons, I. R. (1971). Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA 68, 3092-3096. 10.1073/pnas.68.12.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, L., Bouhouche, K., Whitfield, M., Thouvenin, G., Coste, A., Louis, B., Szymanski, C., Bequignon, E., Papon, J.-F., Castelli, M.et al. (2020). TTC12 loss-of-function mutations cause primary ciliary dyskinesia and unveil distinct dynein assembly mechanisms in motile cilia versus flagella. Am. J. Hum. Genet. 106, 153-169. 10.1016/j.ajhg.2019.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda, Y. and Yokoyama, M. (2016). The early history of the TYH medium for in vitro fertilization of mouse ova. J. Mamm. Ova Res. 33, 3-10. 10.1274/032.033.0103 [DOI] [Google Scholar]
- Tu, C., Nie, H., Meng, L., Wang, W., Li, H., Yuan, S., Cheng, D., He, W., Liu, G., Du, J.et al. (2020). Novel mutations in SPEF2 causing different defects between flagella and cilia bridge: the phenotypic link between MMAF and PCD. Hum. Genet. 139, 257-271. 10.1007/s00439-020-02110-0 [DOI] [PubMed] [Google Scholar]
- Witman, G. B., Plummer, J. and Sander, G. (1978). Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 76, 729-747. 10.1083/jcb.76.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Diener, D. R., Rosenbaum, J. L. and Sale, W. S. (2001). Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J. Cell Biol. 153, 1315-1326. 10.1083/jcb.153.6.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Diener, D. R., Yang, C., Kohno, T., Pazour, G. J., Dienes, J. M., Agrin, N. S., King, S. M., Sale, W. S., Kamiya, R.et al. (2006). Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 119, 1165-1174. 10.1242/jcs.02811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoke, H., Ueno, H., Narita, A., Sakai, T., Horiuchi, K., Shingyoji, C., Hamada, H. and Shinohara, K. (2020). Rsph4a is essential for the triplet radial spoke head assembly of the mouse motile cilia. PLoS Genet. 16, e1008664. 10.1371/journal.pgen.1008664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., He, X., Wu, H., Zhang, X., Yang, S., Liu, C., Liu, S., Hua, R., Zhou, S., Zhao, S.et al. (2021). Loss of DRC1 function leads to multiple morphological abnormalities of the sperm flagella and male infertility in human and mouse. Hum. Mol. Genet. ddab171. 10.1093/hmg/ddab171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Liu, Y. and Yang, P. (2017). Radial spokes-a snapshot of the motility regulation, assembly, and evolution of cilia and flagella. Cold Spring Harb. Perspect. Biol. 9, a028126. 10.1101/cshperspect.a028126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.