Abstract
Microbial communities are a key part to tackling global challenges in human health, environmental conservation, and sustainable agriculture in the coming decade. Recent advances in synthetic biology to study and modify microbial communities have led to important insights into their physiology and ecology. Understanding how targeted changes to microbial communities result in reproducible alterations of the community’s intrinsic fluctuations and function is important for mechanistic reconstruction of microbiomes. Studies of synthetic microbial consortia and comparative analysis of communities in normal and disrupted states have revealed ecological principles that can be leveraged to engineer communities towards desired functions. Tools enabling temporal modulation and sensing of the community dynamics offer precise spatiotemporal control of functions, help to dissect microbial interaction networks, and improve predictions of population temporal dynamics. Here we discuss recent advances to manipulate microbiome dynamics through control of specific strain engraftment and abundance, modulation of cell-cell signaling for tuning population dynamics, infiltration of new functions in the existing community with in situ engineering, and in silico modeling of microbial consortia to predict community function and ecology.
Introduction
Microbial communities are complex and dynamic ecosystems that play a crucial role in a variety of important ecologies from soil to marine and host-associated environments. The physiology and ecology in microbial communities are dependent on the spatial organization and temporal dynamics of their members. Spatial structuring can promote microbial interactions, enabling metabolic co-dependencies that strengthen community robustness, resiliency and homeostasis [1]. Microbial communities undergo temporal dynamics where fluctuations in community composition, metabolism, and function can lead to community trajectories that manifest complex phenotypes [2–4]. Dissecting the governing spatiotemporal principles within a microbiome is fundamental to our understanding of its physiology and ecology.
Temporal dynamics in microbial communities reflect constant fluctuations and recurrent variations in the community structure, composition or function, and are governed by both intrinsic and extrinsic factors [4]. Intrinsic factors include the metabolism and colonization potential of individual species as well as intra- and interspecies interactions, while extrinsic factors are associated with periodic changes in environmental conditions such as pH and nutritional availability. Intrinsic factors can potentially be engineered through modulation of community composition or genetic alterations of specific member species. For example, a microbiota can be engineered with metabolic capacities to modulate the fitness of other community members. Extrinsic factors can be more easily tuned in a time dependent manner by introducing growth-promoting or inhibiting metabolites or changing the biochemistry of the environment. For instance, antibiotic exposure or nutritional changes can result in alterations to the composition and temporal dynamics of microbiomes in soil and the gut [5,6].
Controlling temporal dynamics through alteration of intrinsic and extrinsic factors can therefore serve as an important route to engineer microbial communities for a variety of applications (Fig. 1a) [4]. For example, changing temporal dynamics of communities that have detrimental effects on the host during dysbiosis can rescue healthy homeostasis. For instance, in patients with irritable bowel disease, shifts in temporal dynamics could prevent increased abundance (or presence) of pro-colitogenic strains and thus avoid inflammation flare-ups[7]. On the other hand, engineering temporal population dynamics in the soil community could directly affect plants’ growth, health state, and life cycle. For example, modulating the abundance of nitrogen-fixing bacteria or bacteria that regulate the phytohormones balance can lead to significant physiological changes in the plant growth and life cycle[8].
Figure 1:
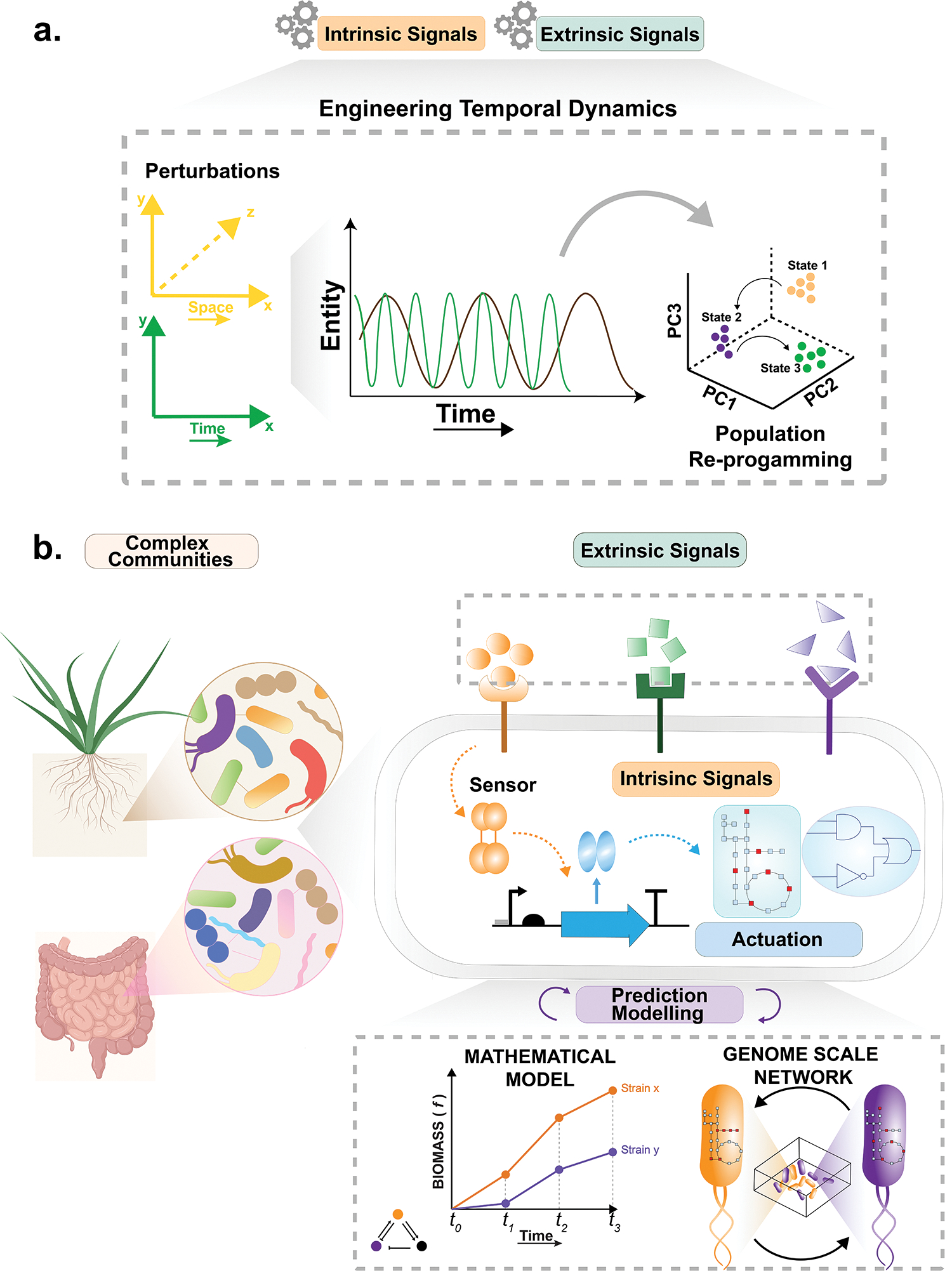
Schematic of the fundamental principle of spatiotemporal community dynamics
Engineering permanent changes in the community and/or its members can shift the intrinsic community fluctuations, thus resulting in long-lasting alterations of temporal dynamics. Through advances in synthetic biology, microbial communities can now be engineered to carry out a variety of novel functions such as sensing dynamic signals and actuating tailored responses. The ability to modulate and sense the community’s intrinsic fluctuations enables transient modifications in the community function and dynamics that can help to elucidate the fundamental principles that govern the overall temporal dynamics. In this article, we discuss emerging approaches to rationally engineer temporal modulations and sensing in microbial communities. We focus on new emerging tools including rewiring signal transduction systems, modulating biophysical characteristics, engineering metabolism and cell-cell interactions, and quantitative modeling of community dynamics (Fig. 1b). We highlight mostly work involving temporal dynamic modulation in the human microbiome as an example community. Temporal modulation and sensing of intrinsic factors are a subset of perturbations that can affect the overall temporal dynamics in microbial communities. Using synthetic biology to modify these factors will enable a more accurate prediction and specific long-lasting intervention of the community temporal dynamics. Furthermore, temporal modulations and sensing offer a deeper understanding of the environmental context that other forms of engineering temporal dynamics can leverage to alter extrinsic factors. Due to space constraints, we refer the reader to other excellent reviews focused on systems and computational biology aspects of the topic [9,10].
Molecular signaling mediated temporal dynamics
Bacteria utilize a variety of mechanisms to sense the environment and modulate population dynamics in response to specific stimuli. Quorum sensing (QS) is one strategy to gain precise spatiotemporal control in an environment and regulate cell functions through coordinate gene expression at a population level (Fig. 2a). Quorum sensing signaling relies on small molecule inducers such as acyl-homoserine lactones (AHLs) or autoinducers (AIs) that regulate genetic outputs. QS systems have been repurposed in many ways, such as for controlled release of a therapeutic in a population density-dependent manner or for coordinating the geography of cells into specific spatial patterns [11–13]. In order to increase the tunability of QS, inducible QS (iQS) can be used to couple a gene of interest with QS and allow external control of gene expression outputs [14]. For instance, a lysis gene can be developed with iQS for temporal and spatial control of population death and release of a protein cargo [15]. These approaches can be extended with well-characterized and orthogonal QS systems with minimal cross-talk to enable control of multiple strains in a community [14]. To maintain the stability of synthetic QS genetic circuits over time, a strategy that leverages ecological interactions and cyclical population control has been devised using strains that could kill or be killed by one another [16]. This approach provides a way to control synthetic ecosystems and maintain gene circuits without the use of antibiotic selection [16]. In addition, more complex genetic circuits using CRISPRi or other inducers[17] can be used to expand the communication capacity towards engineering more sophisticated temporal community dynamics such as programmed cellular differentiation, multicellular pattern formation, and the coordination of multiple metabolic pathways between strains in a community.
Figure 2.
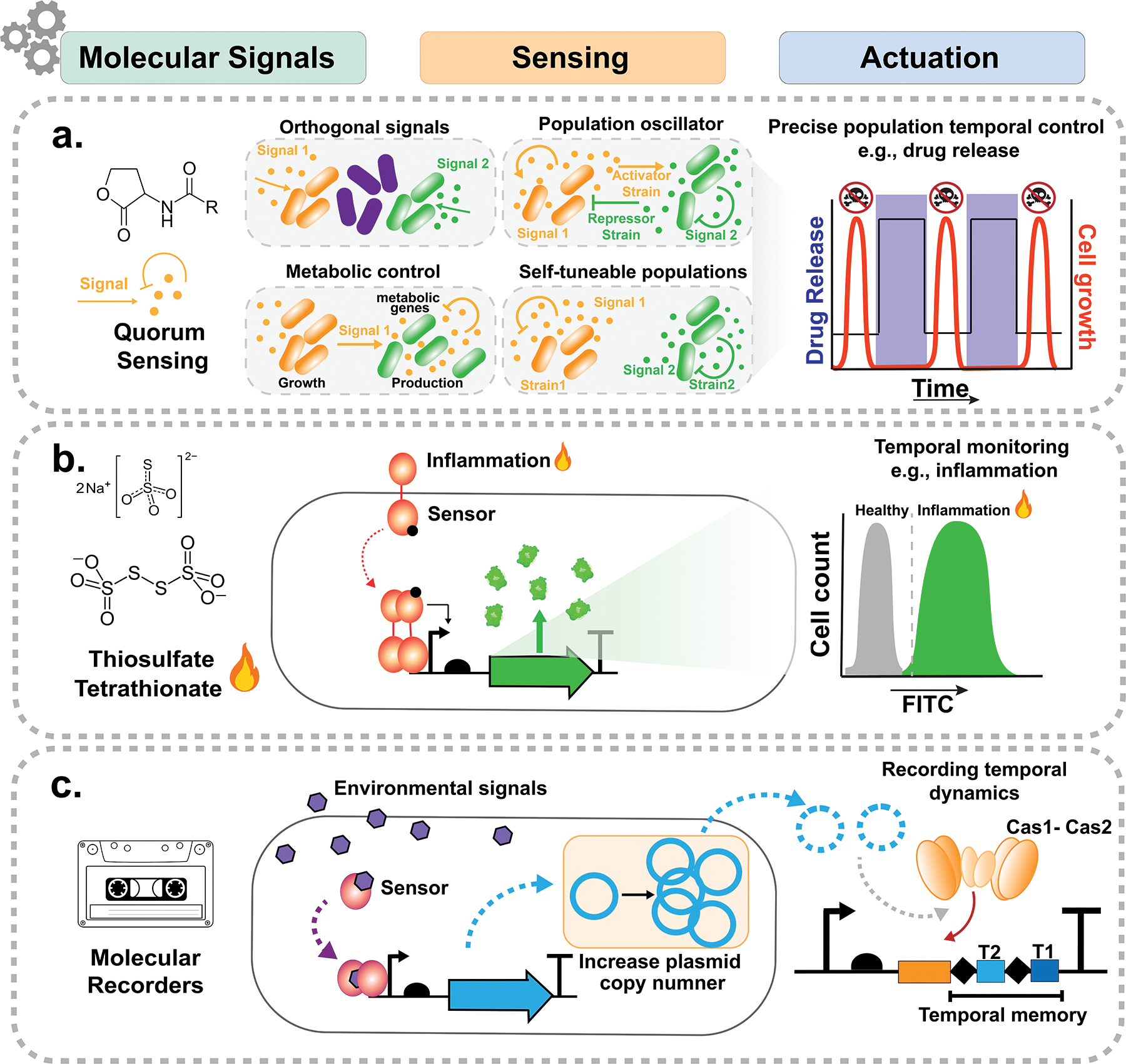
Molecular signal-mediated temporal dynamics. (a) Quorum sensing (QS) harnessed to gain precise population spatiotemporal control and regulate cell functions through coordinated gene expression at the population level. (b) Two-component signal transduction pathways leveraged to rewire and record population dynamics. The thiosulfate (ThsSR) and tetrathionate (TtrSR) TCS combined with a reporter gene have been used as biosensor to detect inflammation in the mammalian gut. (c) CRISPR systems engineered to record information about temporally fluctuating signals in the population. The natural CRISPR adaptation based on spacer acquisition (Cas1-Cas2) has been used to record environmental signals.
Two-component systems (TCSs), a large family of bacterial signal transduction pathways [18], can also be leveraged to rewire and record population dynamics [19]. By swapping TCS components from different bacterial species, it is possible to create new sensing modules that can coordinate novel signal transduction pathways to environmental stimuli such as pH, nitrate and different metabolites [19]. For example, a biosensor to detect inflammation in the mammalian gut was developed by linking thiosulfate sensor (ThsSR) and tetrathionate sensor (TtrSR) with a reporter gene (Fig. 2b) [20,21]. We and others have utilized these natural and engineered biosensor systems to record information about temporally fluctuating signals in the population, using DNA-based cellular recorders [26]. To record environmental signals, these systems either leveraged natural CRISPR adaptation based on Cas1-Cas2 spacer acquisition (Fig. 2c) [22] or used Cas9 endonuclease proprieties to deplete DNA molecules in a sequence-specific manner [24]. Biosensor outputs can trigger a DNA-recording module to chronicle oscillatory states in the population. Furthermore, TCSs can be interfaced with synthetic gene circuits for more complex tuning of signal transformation or to add more sophisticated functionality, such as signal integration and computation [25]
Biophysical mechanisms for controlling population dynamics
Cells exist in complex environments with diverse sets of biochemical and biophysical factors that can be exploited for population engineering. Control of localization and retention of microbiota in a complex environment, such as the gastrointestinal (GI) tract with spatiotemporally dynamic and heterogeneous niches, requires genetic circuits that can detect and respond to a myriad of chemical and environmental gradients [26]. Numerous approaches have been developed for engineering populations by leveraging these environmental gradients. Recent advances in the use of non-biochemical stimuli such as light, heat or electricity could drastically expand the cellular capacity to temporally regulate functions in an environment. TCS have been engineered to create light-responsive optogenetic systems [27,28] that link a light stimulus to the activation of metabolic functions or expression of synthetic genetic circuits to precisely deliver a target metabolite. For example, a green light-activated, red light de-activated two-component system CcaSR has been used to spatially and temporally induce a gut bacterium to produce colanic acid, which increased longevity in a C. elegans model of aging (Fig. 3a) [29]. Gene regulation using temperature offers several advantages over chemicals or light because temperature changes can be applied to biological samples globally by heat or electromagnetic radiation. Exquisite spatial and temporal patterns with penetrating depth can be generated with heat using techniques such as focused ultrasound. For instance, TlpA, a temperature-sensitive transcriptional repressor from Salmonella typhimurium, was engineered into a modular protein–protein dimerization system to transduce heat inputs into regulated gene expression (Fig. 3b) [30]. This platform could be safely translated clinically because high-intensity focused ultrasound is a non-invasive, FDA-approved therapeutic procedure that can be used to regulate blood and lymph flow and to treat cancers by ablating localized tumors. Beyond heat, electrical signals have also been used to modulate community dynamics. Redox responsive genetic circuits using the SoxRS regulon have been engineered to control gene expression using external electronic inputs [31]. In combination with QS systems, population-level bioelectronic circuits have been developed to relay electrical signals between cells to form engineered microbial communication networks (Fig. 3c) [32]. Redox imbalance is often associated with gut dysbiosis [33,34] thus, these systems could be customized to monitor the redox state within the gut microbiome and produce antioxidant metabolites able to rescue homeostasis in response.
Figure 3.
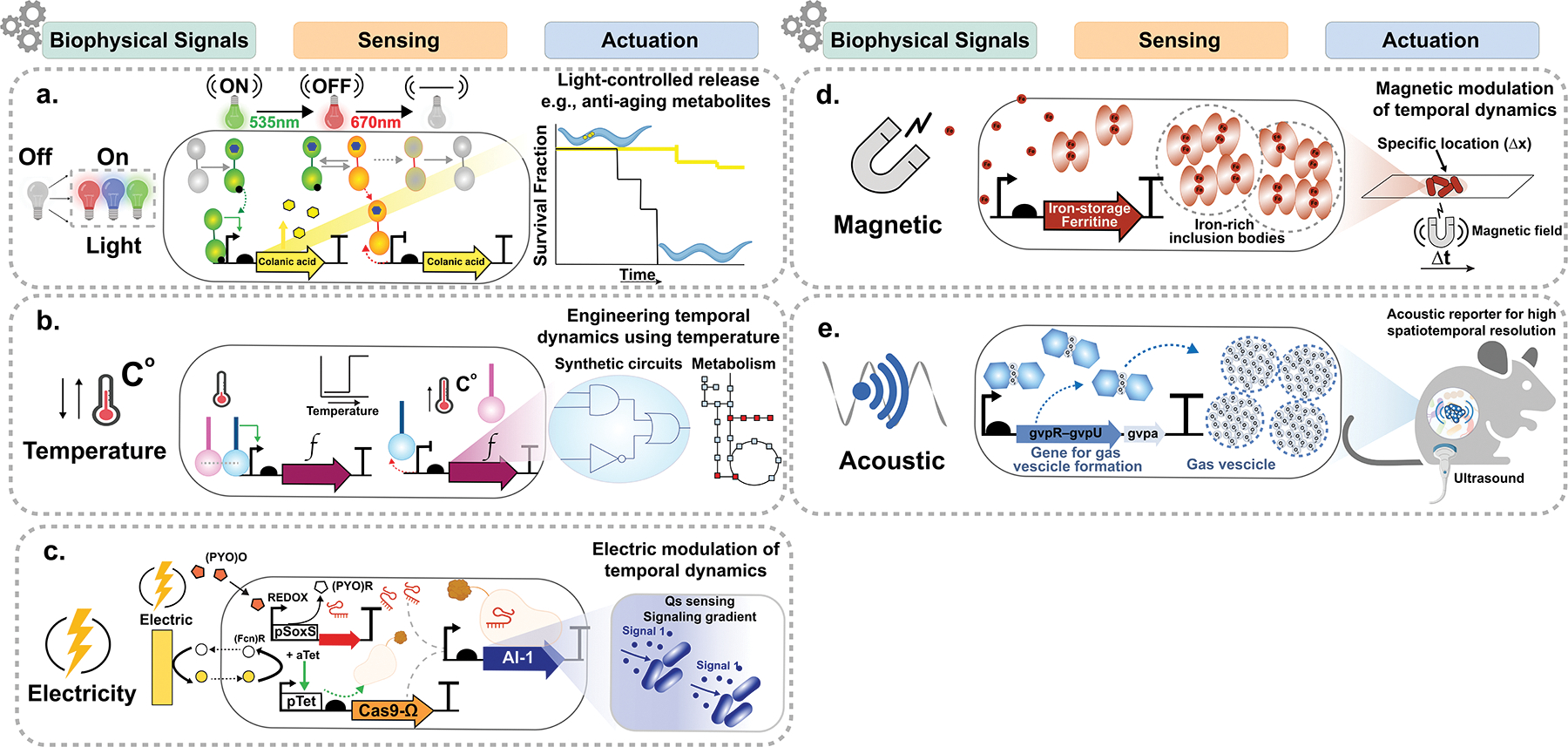
Biophysical mechanisms for controlling population dynamics (a) Light inducible system that activate metabolic pathways in vivo. Colanic acid was used to increase longevity in C. elegans. (b) Temperature dependent dimerization of the TlpA repressor from S. typhimurium used to modulate gene expression. (c) Redox responsive genetic circuits using the SoxRS regulon engineered to control gene expression using external electronic inputs in combination with QS for population-level bioelectronic control. (d) Magnetic responsive system employed for spatial localization of strains. (e) Acoustic signals used for both high temporal and spatial resolution of strains.
Other non-biochemical stimuli including magnetism and acoustics have also emerged as potential modulators of population dynamics. Magnetically-responsive genetic systems have been demonstrated where bacteria are engineered to produce iron-rich bodies by overexpressing iron-storage ferritins or iron-binding proteins inside their cytoplasm (Fig. 3d) [35]. A magnetic field or a ferromagnetic matrix (i.e. ferromagnetic beads) can then be used to capture these magnetically-tagged cells [35] for precise control of their localization in an environment. Another orthogonal system that leverages the generation of gas vesicles in bacteria enable both acoustic reporting and monitor of cellular function across a population with high temporal and spatial resolution using focused ultrasound [36]. The co-expression of structural gvpA genes from A. flos-aquae with the accessory genes gvpR–gvpU from B. megaterium enables the production of intracellular gas vesicles in bacteria and mammalian cells to allow the non-invasive imaging of acoustic reporter cells inside an animal (Fig. 3e) [36]. These and future non-biochemical modulation modalities are poised to have a significant impact on spatiotemporal control of community dynamics.
Cell-cell mediated strategies to engineer temporal dynamics
Numerous inter-microbial interactions mediated by direct cellular contact can result in population-level dynamics. Horizontal gene transfer (HGT) is an evolutionary strategy by which cells can alter their fitness through acquisition of new genetic material (i.e., antibiotic resistance or metabolic genes) in a changing environment. Transduction, conjugation and natural transformation are main routes to mediate microbial exchange of genetic material and have been engineered to provide community-wide control. Phage therapy relies on the life cycle of bacteriophages and their stringent host tropism to target specific members of a microbiome. This approach can be used to selectively eliminate target strains or transfer specific genes into defined species [37–39]. The narrow and specific tropism of phages makes this platform very appealing for its safety, but it reduces the power of this technology for broader applications. Different CRISPR systems can be loaded into a phage to allow programmable and sequence-specific modification of the host DNA and RNA to elicit cell death. For instance, Cas9/Cas3 has been used as a warhead in phages to target virulence genes in pathogens for selective killing (Fig. 3a) [40,41] and Cas13a has been used to degrade host mRNA and kill the host via “collateral” RNase activity (Fig. 3a) [42]. Endogenous Cas systems in target cells can also be leveraged to trigger cell death by delivering self-targeting crRNAs [43]. Community-wide modulation using phage therapy remains an open challenge in many applications since phages exhibit a very narrow host range and are difficult to reengineer [44]. CRISPR technology used in bacteria offers multiple levels of safety. Indeed, these systems (i) rely on sequence specificity, (ii) need to be delivered or endogenously re-purposed into the recipient cells, (iii) elicit cell death, thus eliminating any unwanted propagation of the systems within the community.
Bacterial conjugation is a widespread mechanism by which cells share DNA with one another through a contact-dependent manner over large phylogenetic distances [45]. Thus, conjugation is a highly flexible delivery platform for community-scale modulations. For example, a conjugation-based microbiome engineering approach, MAGIC, that uses modular mobile vectors was used to deliver genetic payloads to diverse members of the mammalian gut microbiome [46]. This system achieved high efficiency gene transfer in diverse bacterial species spanning multiple phyla, while minimally impacting the native microbiome. To improve host targeting, strategies leveraging genome targeting enzymes such as integrative and conjugative elements (ICE) [47] and programmable CRISPR-Cas based transposases have been developed to allow payload introduction at a nucleotide-level resolution in a specific recipient within a complex community (Fig. 4a) [48]. These powerful technologies allow alterations of metabolism and functional selection of species within the population that can offer spatial and temporal control at an unprecedented capacity. Systems for biocontainment and cargo stability such as sequence entanglement of the cargo gene with a toxin or an essential gene [49] and environmental dependency of the synthetic cargo stability can be employed to control the dissemination and the persistence of the engineered function.
Figure 4.
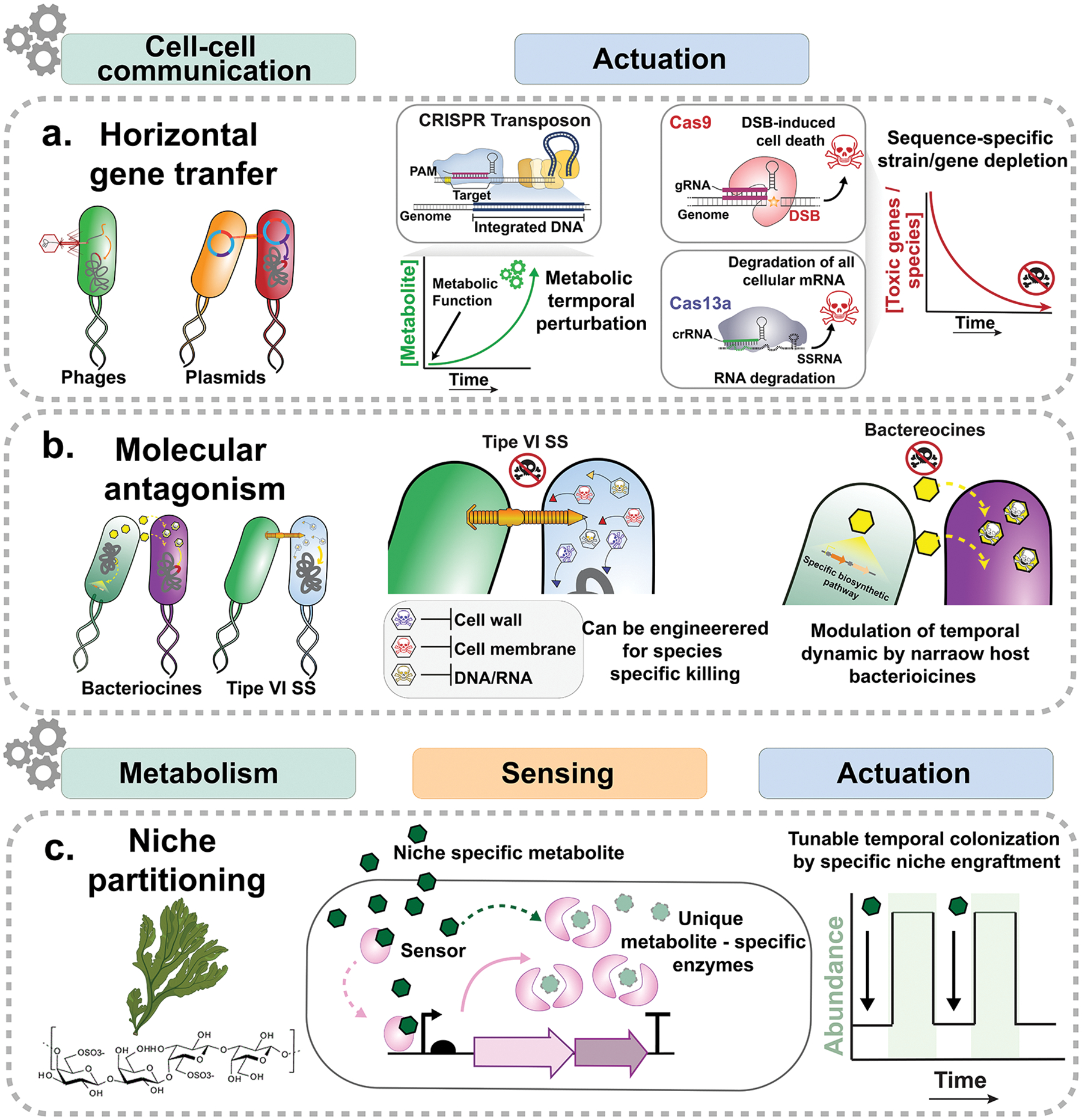
Engineering cell-cell communication. (a) Horizontal gene transfer paired with CRISPR technologies to genetically engineer microbial communities at sequence level resolution. CRISPR systems in engineering microbiome have been mostly used for sequence specific strain depletion. (b) Molecular antagonism provides a platform to modulate strain depletion in complex communities. (c) Niche partitioning leverages principles of microbial ecology by altering the metabolic interactions and introduces substrate exclusivity to enable temporal control of strain specific growth.
Various types of diffusible microbial inhibitors such as soluble small molecules, peptides, and proteins have evolved during the evolution of microbial warfare [50]. As such, these antagonistic systems can be repurposed to modulate community dynamics. Broad-spectrum inhibitors such as bacteriocins and microcins are effective against numerous gram-negative Enterobacteria pathogens by disrupting essential cellular machineries [50]. More narrow inhibitors include the type VI secretion system (T6SS), which is a contact-dependent, membrane-associated apparatus used by gram-negative bacteria to inject target-specific “effector” toxins into adjacent foreign cells [51]. Effector proteins determine the specificity of T6SS antagonism and can be reprogrammed for defined bacterial targeting (Fig. 4b) [52]. These systems can be harnessed to manipulate and modulate taxa presence and extinction within the microbial community to enable temporal and spatial control of interspecies dependencies.
Modeling and engineering metabolism for analysis of population dynamics
Quantitative metabolic modeling of the microbiome can help to identify the core and accessory biochemical pathways that could be tuned, added, or removed to control community dynamics [53,54]. However, genome-scale modeling is limited by the quality of the functional gene annotations. As such, bottom-up approaches to build and characterize synthetic microcosms offer the opportunity to deconvolute complex community interactions. Synthetic microbial consortia from the human gut [55]and soil [56] have shown that dynamic models based on pairwise interactions could predict community assembly. These efforts can yield deeper insights into the impact of various environmental factors such as pH [57], nutrient availability [58], toxins [59], and temperature [60] on community dynamics. For example, in silico multi-level trophic models of the human gut microbiome led to mechanistic links between microbial abundances and specific metabolites [61]. This model aimed to approximate the metabolic flow through the intricate cross-feeding network of microbes in the human lower intestine and allowed the authors to simultaneously capture the metabolic activities of hundreds of species consuming and producing hundreds of metabolites contributing to the ever-changing ecosystem. This advancement enabled the prediction of the metabolic environment and the associated microbial abundances based on their metabolic capacities,
Combining experimental characterizations with mathematical modeling can help to dissect metabolite changes by individual species in a community [62]. However, models that can predict both community dynamics and functional outputs require integration of quantitative datasets from experimental measurements of microbiomes and interaction networks. Such a data-driven approach has been taken to model butyrate production by human gut communities in vitro [63]. Heuristic metabolic modeling approaches have also been used to predict cross-feeding interactions and dynamic population changes [64]. Other experimental platforms using microfluidic systems can further improve the throughput of data generation and investigation of spatially structured environments [65]. Such systems offer exquisite spatiotemporal control of various experimental parameters and enable systematic quantification of community properties, such as diffusion-mediated processes in governing interspecies interactions.
From an engineering perspective, altering metabolic interactions or resistances to environmental metabolites are useful strategies to modulate population growth. For instance, polysaccharide utilization enzymes can enhance microbial colonization in the GI tract [66] and bile salt hydrolases can mediate resistance to otherwise toxic primary bile acids in the chemical milieu of the gut [67]. Modifying a strain to have access to an exclusive metabolic niche enables precise temporal control over its engraftment capacity and abundance in the gut. Administration of the unique substrate that can be exclusively accessed by the engineered strain can shape the microbiota membership (Fig. 4c) [68,69]. For example, a Bacteroides species was engineered with a rare gene cluster for porphyran utilization that enabled nutrient-driven temporal control of its abundance in the mouse gut through varying the amount of porphyran available to the animal [68,69]. Thus, these approaches can be used to control and modulate site-specific engraftment and spatiotemporal abundance of natural probiotics and live bacterial therapeutics.
Conclusions and future prospects
A multidisciplinary approach combining synthetic and systems biology to study microbial community dynamics will offer new possibilities to engineer natural and defined microbiota. These advances are poised to propel engineered microbiomes into innovative applications for many different sectors. Outstanding challenges remain in these areas including 1) better methods to collect temporal datasets at higher resolution, 2) obtaining meaningful spatial biogeography information across a population, and 3) assessing transcriptional and metabolic changes at a species resolution across the entire community. Improved annotations of microbial genomes and higher accuracy and more efficient genomic tools and gene delivery technologies could transform our capacity to tune microbiomes at an unprecedented resolution in space and time.
Highlights.
The ecology and physiology of microbial communities are governed by their spatiotemporal dynamics.
Precision modulation of the microbiome can help to elucidate the role of specific microbes in community function and dynamics.
Various engineering approaches can be taken to modulate temporal community dynamics via characterization and control of (i) molecular and biophysical properties, (ii) interspecies interactions and gene transfer processes, and (iii) cellular and population-scale metabolism and metabolic modeling.
Acknowledgements
We thank A. Kaufman and members of the Wang laboratory for advice and comments on the manuscript. H.H.W. acknowledges funding support from the DOE (47879/SCW1710), NSF (MCB-2025515, MCB-2032259), NIH (1R01AI132403, 1R01DK118044, 1R21AI146817), ONR (N00014-17-1-2353) the Burroughs Wellcome Fund (1016691) and the Irma T. Hirschl Trust. C.R. is supported by a Junior Fellowship of the Simons Society of Fellows (#527896).
Footnotes
Conflict of interest
H.H.W. is a scientific advisor and equity holder of SNIPR Biome and Kingdom Supercultures. The authors declare no additional competing interests.
References and Recommended Reading
- 1.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG: Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A 2016, 113:E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buscardo E, Geml J, Schmidt SK, Freitas H, Da Cunha HB, Nagy L: Spatio-temporal dynamics of soil bacterial communities as a function of Amazon forest phenology. Sci Rep 2018, 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shenhav L, Furman O, Briscoe L, Thompson M, Silverman JD, Mizrahi I, Halperin E: Modeling the temporal dynamics of the gut microbial community in adults and infants. PLoS Comput Biol 2019, 15:e1006960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.**.Ryo M, Aguilar-Trigueros CA, Pinek L, Muller LAH, Rillig MC: Basic Principles of Temporal Dynamics. Trends Ecol Evol 2019, 34:723–733. [DOI] [PubMed] [Google Scholar]; This work discusses the basic principle of temporal dynamics necessary to understand the ecology and evolution of complex temporal dynamics in a given system, which can help to better predict the behaviour of complex communities and to design new engineering approaches.
- 5.Robinson CJ, Young VB: Antibiotic administration alters the community structure of the gastrointestinal microbiota. Gut Microbes 2010, 1:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrinpar A, Chaix A, Yooseph S, Panda S: Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 2014, 20:1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengfelder I, Sava IG, Hansen JJ, Kleigrewe K, Herzog J, Neuhaus K, Hofmann T, Sartor RB, Haller D: Complex Bacterial Consortia Reprogram the Colitogenic Activity of Enterococcus faecalis in a Gnotobiotic Mouse Model of Chronic, Immune-Mediated Colitis. Front Immunol 2019, 0:1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL: Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front Plant Sci 2018, 0:1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, Löffler FE, O’Malley MA, García Martín H, Pfleger BF, Raskin L, Venturelli OS, et al. : Common principles and best practices for engineering microbiomes. Nat Rev Microbiol 2019, 17:725–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leggieri PA, Liu Y, Hayes M, Connors B, Seppälä S, O’Malley MA, Venturelli OS: Integrating Systems and Synthetic Biology to Understand and Engineer Microbiomes. Annu Rev Biomed Eng 2021, 23:annurev-bioeng-082120–022836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R: A synthetic multicellular system for programmed pattern formation. Nature 2005, 434:1130–1134. [DOI] [PubMed] [Google Scholar]
- 12.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J: A synchronized quorum of genetic clocks. Nature 2010, 463:326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott SR, Din MO, Bittihn P, Xiong L, Tsimring LS, Hasty J: A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol 2017, 2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miano A, Liao MJ, Hasty J: Inducible cell-to-cell signaling for tunable dynamics in microbial communities. Nat Commun 2020, 11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, Julio E, Atolia E, Tsimring LS, Bhatia SN, et al. : Synchronized cycles of bacterial lysis for in vivo delivery. Nature 2016, 536:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.**.Liao MJ, Din MO, Tsimring L, Hasty J: Rock-paper-scissors: Engineered population dynamics increase genetic stability. Science (80- ) 2019, 365:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work showcases an elegant example of engineering community dynamics by leveraging ecological interactions and cyclical population control using strains that could kill or be killed by one another. This system represents the first example of creating synthetic homeostasis within a community without any external intervention.
- 17.Sexton JT, Tabor JJ: Multiplexing cell‐cell communication. Mol Syst Biol 2020, 16:e9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao R, Stock AM: Biological insights from structures of two-component proteins. Annu Rev Microbiol 2009, 63:133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.**.Schmidl SR, Ekness F, Sofjan K, Daeffler KNM, Brink KR, Landry BP, Gerhardt KP, Dyulgyarov N, Sheth RU, Tabor JJ: Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat Chem Biol 2019, 15:690–698. [DOI] [PubMed] [Google Scholar]; This work describes a pipeline to discover new two component system (TCS) and demonstrated that the two largest families of response regulator DNA-binding domains can be interchanged with remarkable flexibility, which can accelerate new discoveries in the field of TCS and open up opportunities to harness new systems for more complex and tunable tools for microbiome engineering.
- 20.Daeffler KN, Galley JD, Sheth RU, Ortiz‐Velez LC, Bibb CO, Shroyer NF, Britton RA, Tabor JJ: Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol 2017, 13:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA: Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol 2017, doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheth RU, Yim SS, Wu FL, Wang HH: Multiplex recording of cellular events over time on CRISPR biological tape. Science (80- ) 2017, 358:1457–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheth RU, Wang HH: DNA-based memory devices for recording cellular events. Nat Rev Genet 2018, 19:718–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang W, Liu DR: Rewritable multi-event analog recording in bacterial and mammalian cells. Science (80- ) 2018, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brophy JAN, Voigt CA: Principles of genetic circuit design. Nat Methods 2014, 11:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. : Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147:1055–1063.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong NT, Olson EJ, Tabor JJ: Engineering an E. coli Near-Infrared Light Sensor. ACS Synth Biol 2018, 7:240–248. [DOI] [PubMed] [Google Scholar]
- 28.Ong NT, Tabor JJ: A Miniaturized Escherichia coli Green Light Sensor with High Dynamic Range. ChemBioChem 2018, 19:1255–1258. [DOI] [PubMed] [Google Scholar]
- 29.Hartsough LA, Park M, Kotlajich MV, Lazar JT, Han B, Lin CCJ, Musteata E, Gambill L, Wang MC, Tabor JJ: Optogenetic control of gut bacterial metabolism to promote longevity. Elife 2020, 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piraner DI, Wu Y, Shapiro MG: Modular Thermal Control of Protein Dimerization. ACS Synth Biol 2019, 8:2256–2262. [DOI] [PubMed] [Google Scholar]
- 31.Bhokisham N, VanArsdale E, Stephens KT, Hauk P, Payne GF, Bentley WE: A redox-based electrogenetic CRISPR system to connect with and control biological information networks. Nat Commun 2020, 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.*.Terrell JL, Tschirhart T, Jahnke JP, Stephens K, Liu Y, Dong H, Hurley MM, Pozo M, McKay R, Tsao CY, et al. : Bioelectronic control of a microbial community using surface-assembled electrogenetic cells to route signals. Nat Nanotechnol 2021, doi: 10.1038/s41565-021-00878-4. [DOI] [PubMed] [Google Scholar]; This work provided a method for bioelectronics interfacing with a living biological network (BioLAN) by using redox molecules to interconvert between electronic and biological input/output to achieve bidirectional information exchange.
- 33.Reese AT, Cho EH, Klitzman B, Nichols SP, Wisniewski NA, Villa MM, Durand HK, Jiang S, Midani FS, Nimmagadda SN, et al. : Antibiotic-induced changes in the microbiota disrupt redox dynamics in the gut. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.JW B, BJ S: Translational Approaches with Antioxidant Phytochemicals against Alcohol-Mediated Oxidative Stress, Gut Dysbiosis, Intestinal Barrier Dysfunction, and Fatty Liver Disease. Antioxidants (Basel, Switzerland) 2021, 10:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aubry M, Wang WA, Guyodo Y, Delacou E, Guigner JM, Espeli O, Lebreton A, Guyot F, Gueroui Z: Engineering E. coli for magnetic control and the spatial localization of functions. ACS Synth Biol 2020, 9:3030–3041. [DOI] [PubMed] [Google Scholar]
- 36.Bourdeau RW, Lee-Gosselin A, Lakshmanan A, Farhadi A, Kumar SR, Nety SP, Shapiro MG: Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 2018, 553:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva YJ, Costa L, Pereira C, Cunha Â, Calado R, Gomes NCM, Almeida A: Influence of environmental variables in the efficiency of phage therapy in aquaculture. Microb Biotechnol 2014, 7:401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yehl K, Lemire S, Yang AC, Ando H, Mimee M, Torres MDT, de la Fuente-Nunez C, Lu TK: Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179:459–469.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu BB, Plant IN, Lyon L, Anastassacos FM, Way JC, Silver PA: In situ reprogramming of gut bacteria by oral delivery. Nat Commun 2020 111 2020, 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA: Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 2014, 32:1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Citorik RJ, Mimee M, Lu TK: Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 2014, 32:1141–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiga K, Tan XE, Ibarra-Chávez R, Watanabe S, Aiba Y, Sato’o Y, Li FY, Sasahara T, Cui B, Kawauchi M, et al. : Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun 2020, 11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, Vridhambal GS, Rivera AJ, Montgomery SA, Fortier LC, Barrangou R, et al. : In vivo targeting of clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. MBio 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abedon ST, García P, Mullany P, Aminov R: Editorial: Phage therapy: Past, present and future. Front Microbiol 2017, 8:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redondo-Salvo S, Fernández-López R, Ruiz R, Vielva L, de Toro M, Rocha EPC, Garcillán-Barcia MP, de la Cruz F: Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun 2020, 11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronda C, Chen SP, Cabral V, Yaung SJ, Wang HH: Metagenomic engineering of the mammalian gut microbiome in situ. Nat Methods 2019, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brophy JAN, Triassi AJ, Adams BL, Renberg RL, Stratis-Cullum DN, Grossman AD, Voigt CA: Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol 2018, 3:1043–1053. [DOI] [PubMed] [Google Scholar]
- 48.Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH, Sternberg SH: CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat Biotechnol 2021, 39:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blazejewski T, Ho H-I, Wang HH: Synthetic sequence entanglement augments stability and containment of genetic information in cells. Science (80- ) 2019, 365:595–598. [DOI] [PubMed] [Google Scholar]
- 50.Simons A, Alhanout K, Duval RE: Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, Yang X, Shen X: Confirmed and potential roles of bacterial T6SSs in the intestinal ecosystem. Front Microbiol 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ting SY, Martínez-García E, Huang S, Bertolli SK, Kelly KA, Cutler KJ, Su ED, Zhi H, Tang Q, Radey MC, et al. : Targeted Depletion of Bacteria from Mixed Populations by Programmable Adhesion with Antagonistic Competitor Cells. Cell Host Microbe 2020, 28:313–321.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauer E, Zimmermann J, Baldini F, Thiele I, Kaleta C: BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol 2017, 13:e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faust K: Microbial Consortium Design Benefits from Metabolic Modeling. Trends Biotechnol 2019, 37:123–125. [DOI] [PubMed] [Google Scholar]
- 55.Venturelli OS, Carr AV, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP: Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 2018, 14:e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedman J, Higgins LM, Gore J: Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol 2017, 1:109. [DOI] [PubMed] [Google Scholar]
- 57.Ratzke C, Gore J: Modifying and reacting to the environmental pH can drive bacterial interactions. PLOS Biol 2018, 16:e2004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratzke C, Barrere J, Gore J: Strength of species interactions determines biodiversity and stability in microbial communities. Nat Ecol Evol 2020, 4:376–383. [DOI] [PubMed] [Google Scholar]
- 59.Piccardi P, Vessman B, Mitri S: Toxicity drives facilitation between 4 bacterial species. Proc Natl Acad Sci U S A 2019, 116:15979–15984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lax S, Abreu CI, Gore J: Higher temperatures generically favour slower-growing bacterial species in multispecies communities. Nat Ecol Evol 2020, 4:560–567. [DOI] [PubMed] [Google Scholar]
- 61.Wang T, Goyal A, Dubinkina V, Maslov S: Evidence for a multi-level trophic organization of the human gut microbiome. PLOS Comput Biol 2019, 15:e1007524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medlock GL, Carey MA, McDuffie DG, Mundy MB, Giallourou N, Swann JR, Kolling GL, Papin JA: Inferring Metabolic Mechanisms of Interaction within a Defined Gut Microbiota. Cell Syst 2018, 7:245–257.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clark RL, Connors BM, Stevenson DM, Hromada SE, Hamilton JJ, Amador-Noguez D, Venturelli OS: Design of synthetic human gut microbiome assembly and function. bioRxiv 2020, doi: 10.1101/2020.08.19.241315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.*.Diener C, Gibbons SM, Resendis-Antonio O: MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. mSystems 2020, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes a mathematical modeling framework that can recapitulate the growth rates of diverse bacterial species in the gut and can simulate metabolic interactions within microbial communities and is helpful for dissecting the ecological rules that shape the microbial community landscape and rational engineering of community dynamics.
- 65.*.Gupta S, Ross TD, Gomez MM, Grant JL, Romero PA, Venturelli OS: Investigating the dynamics of microbial consortia in spatially structured environments. Nat Commun 2020, 11:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a new microfluidic platform to quantify spatiotemporal parameters influencing diffusion-mediated interactions in microbial consortia, thus allowing high-throughput characterization studies to deconvolute microbial dynamics and complexity.
- 66.Crook N, Ferreiro A, Gasparrini AJ, Pesesky MW, Gibson MK, Wang B, Sun X, Condiotte Z, Dobrowolski S, Peterson D, et al. : Adaptive Strategies of the Candidate Probiotic E. coli Nissle in the Mammalian Gut. Cell Host Microbe 2019, 25:499–512.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P: Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 2002, 45:1095–1106. [DOI] [PubMed] [Google Scholar]
- 68.*.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL: An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 2018, 557:434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that modifying a specific strain for metabolic niche exclusivity enables precise temporal control over the engraftment and abundance of that strain. By providing a unique substrate that the strain can exclusively access, but is unusable by the rest of the community, this approach highlights the possibility of shaping microbiota membership by modulating the fitness of a specific strain.
- 69.Kearney SM, Gibbons SM, Erdman SE, Alm EJ: Orthogonal Dietary Niche Enables Reversible Engraftment of a Gut Bacterial Commensal. Cell Rep 2018, 24:1842–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]


