Abstract
The emergence of antimicrobial resistance in commensal bacteria poses a serious public health burden worldwide. Commensals can disseminate the resistance genes to pathogenic bacteria causing life-threatening infections. This cross-sectional study was designed to investigate the antimicrobial resistance pattern and molecular mechanism(s) of ciprofloxacin resistance in commensal E. coli from three major one health components (humans, animals and the environment) in Bangladesh. Samples were randomly collected from broiler chickens, broiler farm environments and hospitalized human patients from the same geographical area. Isolation and identification of E. coli were performed following standard bacteriological techniques. Antimicrobial susceptibility testing (AST) was performed by disk diffusion and broth microdilution methods. Mutation at the quinolone-resistance determining region (QRDR) was analyzed by sequencing. Of 450 samples, a total of 287 (63.8%; 95% CI 59.2–68.1%) E. coli strains was isolated, where 240 (83.6%; 95% CI 78.9–87.5%) strains were phenotypically resistant to ciprofloxacin. The prevalence of ciprofloxacin-resistant E. coli in broiler chicken, broiler farm environments and hospitalized human patients are 77.6%, 88.8% and 89% respectively. In AST against nine antimicrobials, all the isolates were found to be multidrug-resistant (MDR). The minimum inhibitory concentration (MIC) of ciprofloxacin was ranged from 4 to >128mg/L. Point mutations were detected in several sites of QRDR, specifically at 83 and 87 amino acid positions in gyrA gene, and 56, 57, 78, 80 and 84 amino acid positions in parC gene. Mutations resulted in amino acid substitutions. Phylogenetic analysis of gyrA and parC gene sequences showed a close relationship between the strains isolated from different sources. This study demonstrates a high prevalence of ciprofloxacin resistance in commensal E. coli in humans, animals and environment interface and their genealogically similarity poses an alarming public health consequence.
Introduction
Antimicrobial resistance (AMR) is an urgent public health concern worldwide. The emergence and dissemination of AMR is an inevitable side effect of the irrational use of antimicrobials in animals and humans. The resistance phenotype disseminates not only into the pathogen but also into the commensal bacteria, constituting a reservoir of resistance genes for potentially pathogenic ones [1, 2]. E. coli is one of the versatile commensals in warm-blooded animals’ intestinal tract. Commensal E. coli has shown resistance against numerous antimicrobials due to their widespread use in animals [3, 4] and humans [5, 6]. This organism can transfer resistance traits to other closely related bacterial pathogens via plasmids, making them difficult to kill by common antimicrobials [7].
Quinolones are broad-spectrum antibiotics used in human and veterinary medicine to treat Gram-negative bacterial infections. Ciprofloxacin is one of the quinolone antibiotics commonly used to treat critical bacterial infections in humans. Therefore, WHO listed this antibiotic as one of the most effective and safe medicines needed in a health system [8]. However, inappropriate use of ciprofloxacin results in bacterial resistance development against this valuable antibiotic.
When ciprofloxacin was first introduced, resistance was practically non-existent. However, the widespread use of this antibiotic in both humans and animals as either disease prevention or feed additives results in the emergence of ciprofloxacin-resistant strains of E. coli. Ciprofloxacin resistance has been reported as high or extremely high in human and food-producing animal isolates from several countries [9]. Although the use of antimicrobials is strictly regulated in developed countries, there is no strict regulation of antimicrobial use in developing countries like Bangladesh. As a result, the emergence of antimicrobial resistance to critically important antibiotics like ciprofloxacin is common in these countries [10, 11].
Although little works have been carried out with E. coli to detect the ciprofloxacin resistance profile with molecular confirmation, more concise work has yet to be done to evaluate the status of resistance genes in both humans and animals. Therefore, we aimed to detect the resistance pattern and molecular mechanisms of ciprofloxacin resistance in commensal E. coli from three major one health components (humans, animals and the environment) in Bangladesh.
Materials and methods
Study area and population
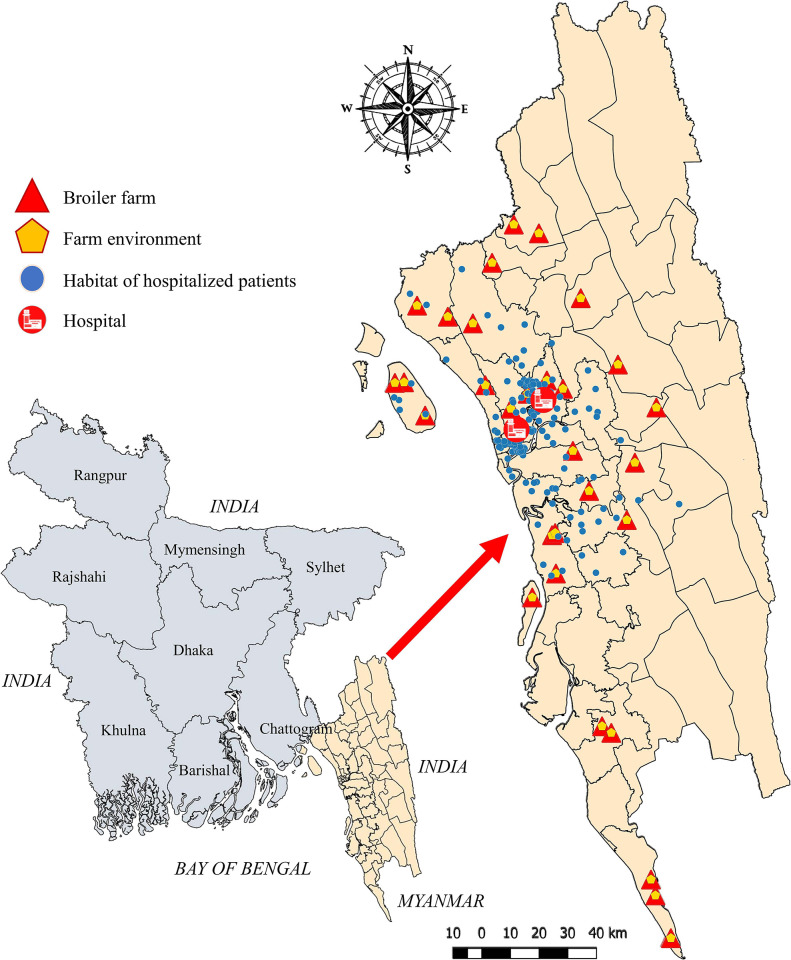
A cross-sectional study was carried out through January to July 2021 in the Chattogram division, located in the southeast region of Bangladesh. Samples from broiler chicken and broiler farm environments were collected from 30 randomly selected broiler farms of eighteen upazilas (administrative sub-district) in Chattogram, Bangladesh. The random selection of broiler farms was obtained from the government livestock office of each upazila. The human samples were collected from two randomly selected human hospitals in the same geographic area (one from a regional area and another from Chattogram metropolitan area). Samples were collected from the hospitalized human patients admitted into both hospitals with gastrointestinal disorders. The geographical distribution of the study area and samples from three different sources are demonstrated in Fig 1. The coordinates of broiler farms and habitat of the hospitalized patients were collected during sampling.
Fig 1. Geographical distribution of sampling sites and population samples.
Sample types are marked in the top left legends.
Sampling
Cloacal swabs from broiler chickens and rectal swabs from hospitalized human patients were collected using sterile cotton swabs. Environmental samples from each broiler farm were collected from feed, water, soil, farm floor litter, and dumping sites. Swab samples were immediately kept in Stuart’s transport medium (Oxoid, Basingstoke, UK) and transferred into the Poultry Research and Training Centre, CVASU, Bangladesh laboratory. Finally, all swab samples were stored at −80°C for further use.
E. coli isolation
Isolation and identification of E. coli were performed following standard bacteriological procedure. Swab samples were placed into 5mL MacConkey broth. Simultaneously, 5g of solid samples (e.g., feed, litter, soil) and 5mL of water samples were taken in separate sterile falcon tubes containing 45 mL MacConkey broth (Oxoid Ltd, Basingstoke, UK) and incubated at 37°C overnight for pre-enrichment. After incubation, one loopful broth was inoculated onto MacConkey agar (Oxoid Ltd, Basingstoke, UK) and incubated at 37°C overnight. Any suspected large pink colony from the plate was subcultured onto Eosin-methylene blue (EMB) agar (Scharlab, Spain) and incubated at 37°C overnight for biochemical confirmation. Colonies with a characteristic greenish metallic sheen from EMB agar were further subcultured onto blood agar (Oxoid Ltd, Basingstoke, UK) with 5% bovine blood and incubated at 37°C for 24 hours. All culture-positive isolates were subjected to E. coli species confirmation by polymerase chain reaction (PCR) assay targeting a house-keeping gene adenylate kinase (adk) using specific primers (AdkF: 5’-ATTCTGCTTGGCGCTCCGGG-3’ and AdkR: 5’- CCGTCAACTTTCGCGTATTT-3’). The thermal condition maintained as initial denaturation at 95°C for 2 minutes and final extension at 72°C for 5 minutes with the 35 cycles of denaturation at 95°C for 1 minute, annealing at 54°C for 1 minute and extension at 72°C for 2 minutes [2].
Antimicrobial susceptibility testing of E. coli isolates
Antimicrobial susceptibility testing for all E. coli isolates was carried out using disk diffusion technique recommended by the Clinical Laboratory and Standards Institute (CLSI) [12, 13]. The ATCC 25922 was used for quality control during the disk diffusion technique. A total of nine antimicrobials from eight different groups were used at the given concentration: colistin sulphate (10 μg), ciprofloxacin (5μg), tetracycline (30μg), ampicillin (10μg), gentamycin (10μg), enrofloxacin (5μg) ceftriaxone (30μg), chloramphenicol (30μg) and sulfamethoxazole/trimethoprim (23.75 + 1.25 μg) (Oxoid, Basingstoke, UK). The results of susceptibility testing were interpreted according to the CLSI guidelines [12, 13].
Detection of Minimum Inhibitory Concentration (MIC) of ciprofloxacin
All ciprofloxacin-resistant E. coli isolates were subjected to MIC determination by broth microdilution (BMD) method following ISO standard (20776–2) [14]. Cation-adjusted Mueller Hinton II Broth (Sigma-Aldrich, St Louis, MO, USA) and pure ciprofloxacin powder (Sigma-Aldrich, Saint Louis, MO, USA) were used. A ciprofloxacin resistant E. coli in-house strain and ATCC 25922 strain were used as the positive and negative control, respectively.
Gene sequencing and mutation analysis
All phenotypically ciprofloxacin resistant E. coli isolates were further investigated for the presence of gyrA and parC genes. The oligonucleotide primers used for the amplification of the genes by PCR are mentioned in Table 1. The thermal cyclic conditions for both genes consisted of an initial denaturation step of 95°C for 3 minutes, followed by 30 cycles of 95°C for 45 sec 56°C for 45 sec and 72°C for 90 sec with a final step of 72°C for 10 minutes. The PCR products were visualized on a gel documentation system (UVP UVsolo touch–Analytik Jena AG) after electrophoresis with 1.5% agarose gel (Thermo Scientific).
Table 1. Oligonucleotide primer sequences used to detect the gyrA and parC genes in the E. coli isolates.
| Gene | Primer name | Primer sequence (5΄- 3΄) | Annealing temperature (°C) | Amplicon size (bp) | References |
|---|---|---|---|---|---|
| gyrA | gyrA-F | TGGATTATGCGATGTCGGTCAT | 56°C | 696 | This study |
| gyrA-R | TTTTGGCGTCAACTTCCACTTC | ||||
| parC | parC-F | TGCCGTTTATTGGTGATGGTCT | 56°C | 477 | This study |
| parC-R | GAGCCACTTCACGCAGGTTAT |
Twenty E. coli isolates carrying both gyrA and parC genes were randomly selected for sequencing of both genes. The purified PCR products were Sanger-sequenced with the BigDye terminator v3.1 sequencing kit and a 3730xl automated sequencer (Applied Biosystems, Foster City, CA, USA) using the similar primer sets (Table 1). All sequences were checked for BLASTn and submitted to NCBI for accession number. Geneious prime (version 2022.1.1, Biomatters, New Zealand) software was used for multiple alignments of the sequences from this study with the reference sequence to detect the mutation in QRDR.
Phylogenetic analysis
Maximum likelihood (ML) based phylogenetic reconstruction was used to determine the genealogical relatedness of both gyrA and parC amplicons within the study sequences rooting with the E. coli strain as a reference sequence (NC_007779). For both genes, selected individual sequences were annotated with accession number, strain name, host, country of circulation, collection year and sample ID. A multiple alignment of twenty-one sequences was generated in Geneious with Muscle v3.8.31 alignment algorithm implemented in the Geneious package. For both gyrA and parC sequences alignments, jModelTest 2.1.3 favoured a general-time-reversible model with gamma distribution rate variation and a proportion of invariable sites (GTR+G+I) for phylogeny. ML trees were reconstructed with 100 bootstrap replicates in PhyML 3.3 implemented in Geneious using the above parameters. The final trees were visualized and edited in FigTree 1.4.
Statistical analysis
Laboratory data were entered into Microsoft Excel 2016 spreadsheets. The prevalence of all categorical variables and 95% confidence intervals were calculated by the modified Wald method using GraphPad Quickcals (https://www.graphpad.com/quickcalcs/confInterval1/). The heatmap and dendrogram of the antimicrobial susceptibility testing phenotype of E. coli was generated using the R package ‘gplots’ version 3.0.1 [15].
Ethics statement
This study was approved [Memo# CVASU/Dir(R&E) EC/2019/39(2/6/6)] by the Ethics Committee of Chattogram Veterinary and Animal Sciences University (CVASU), Bangladesh.
Results
Demographic data
A total of 450 samples were collected from the study population. Among them, 150 cloacal swabs from broiler chickens, 150 samples of farm environments and 150 rectal swabs from hospitalized humans. Samples from different environmental matrices are described in Table 2. Although human samples were collected from two hospitals, the hospitalized patients were inhabited in 19 upazilas (Fig 1). Around 43% (65/150) of broiler chicken samples were from the smallholding farms (less than 2500 farm size) as well as the age group of 14 to 24 days (S1 Table). Surprisingly all the farms sampled had a record of antibiotic use, especially ciprofloxacin. The majority of the farms sampled use sawdust as litter materials (90%) and reuse them (S2 Table). About 53.3% of tested farms dump litter materials in nearby open places to the farms. Dumped litters were primarily connected with either canal (23.3%) or crop field (23.3%). Regarding hospitalized human samples, the highest proportion was from the age group 18 to 55 years. Around 59.3% of the sampled patients were male, and almost half of all patients were suffering from diarrhoea (S3 Table). Surprisingly, 71.3% of the human patients sampled had a history of using ciprofloxacin.
Table 2. Distribution of E. coli and ciprofloxacin resistant E. coli isolated from poultry, poultry farm environment and hospitalized human samples.
| Sample source | N | E. coli (%; 95% CI) | Ciprofloxacin resistant E. coli (%; 95% CI) |
|---|---|---|---|
| Poultry (broiler chicken) | 150 | 134 (89.3; 83.3–93.4) | 104 (77.6; 69.8–83.9) |
| Poultry farm environment | 150 | 80 (53.3; 45.4–61.1) | 71 (88.8; 79.8–94.1) |
| Litter | 30 | 12 (40; 24.6–57.7) | 12 (100; 71.8–100) |
| Soil | 30 | 14 (46.7; 30.2–63.9) | 9 (64.3; 38.6–83.8) |
| Feed | 30 | 15 (50; 33.2–66.9) | 12 (80; 54.1–93.7) |
| Water | 30 | 23 (76.7; 58.8–88.5) | 22 (95.6; 77.3–99.9) |
| Dumped litter | 30 | 16 (53.3; 36.1–69.7 | 16 (100; 77.3–100) |
| Hospitalized human | 150 | 73 (48.7; 40.8–56.6) | 65 (89; 79.6–94.6) |
| Total | 450 | 287 (63.8; 59.2–68.1) | 240 (83.6; 78.9–87.5) |
Here, N = number of samples, % = percentages and CI = confidence interval.
Prevalence of E. coli
A total of 287 (63.8%; 95% confidence interval (CI) 59.2–68.1%) samples were tested positive for the presence of E. coli (Table 2). The highest prevalence of E. coli was found in broiler chickens (89.3%, 95% CI 83.3–93.4%) followed by farm environments and hospitalized humans. as observed for broilers, farm environment (53.3%, 95% CI 45.4–61.1%) and hospitalized humans (48.7%, 95% CI 40.8–56.6%).
The age group of more than 28 days in broiler chicken has the highest prevalence of E. coli (98.3%) and ciprofloxacin-resistant E. coli (S1 Table). Drinking water collected from the water pan has the highest prevalence of E. coli (76.7%) and ciprofloxacin E. coli (73.3%) among other matrices (S2 Table). The prevalence of E. coli (54.1%) and ciprofloxacin-resistant E. coli (47.4%) were high in the farm environments of those farms reused litter materials. In hospitalized humans, the highest prevalence of E. coli (71.5%) and ciprofloxacin E. coli (67.3%) were isolated from the age group of more than 55 years. Male patients had the highest prevalence of E. coli (53.9%) and ciprofloxacin-resistant E. coli (46.1%). Simultaneously, the prevalence of E. coli (50.6%) and ciprofloxacin-resistant E. coli (44.3%) were high in non-diarrhoeic patients. A total of 48.6% samples from hospitalized humans those used ciprofloxacin for medication, were positive for ciprofloxacin resistant E. coli (S3 Table).
Antimicrobial resistance profile of E. coli
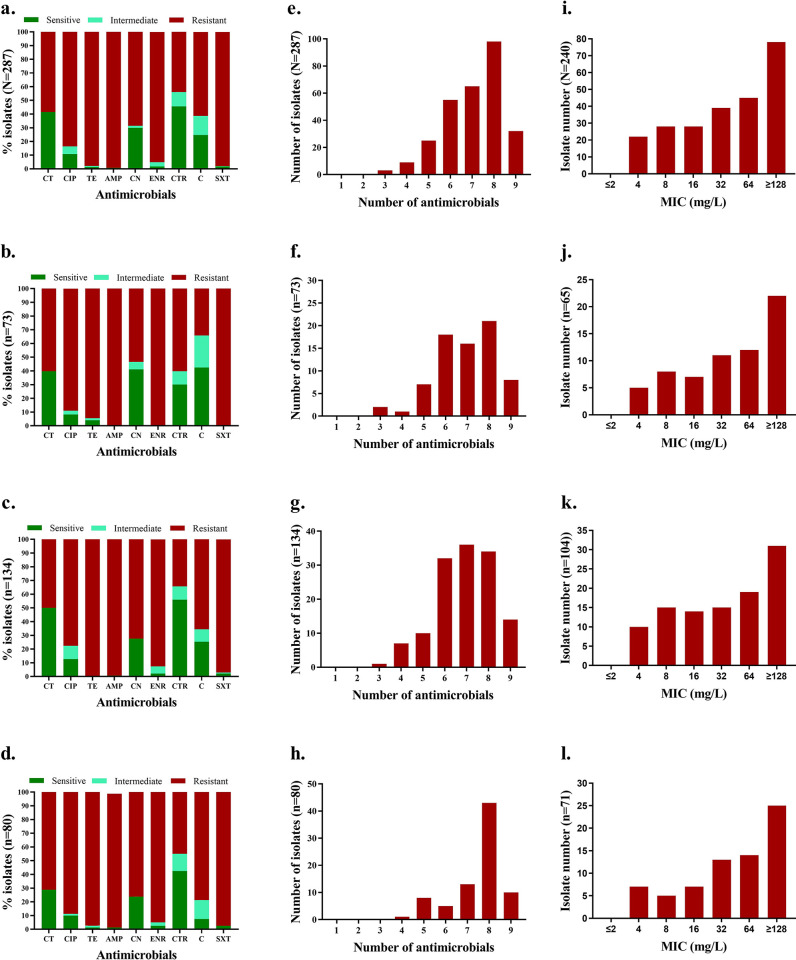
Overall antimicrobial susceptibility patterns of the E. coli strains to nine antimicrobials are presented in Fig 2. The highest resistance rate of E. coli was to ampicillin (99.3%), followed by sulfomethoxazole (97.9%), and tetracycline (97.9%) (Fig 2A). E. coli strains isolated from the human patients show the highest resistance against ampicillin (100%), enrofloxacin (100%) and sulfamethoxazole (100%) (Fig 2B). In contrast, strains isolated from the broiler chickens and the farm environment show the highest resistance against tetracycline (100% and 98.8%, respectively) (Fig 2C and 2D). On the other hand, the overall highest sensitivity was found to ceftriaxone (45.6%) followed by colistin sulphate (41.5%) and gentamycin (30%).
Fig 2. Antibiogram profile of E. coli isolates isolated from different sources.
(a) Overall susceptibility pattern of E. coli by disc diffusion method; (b) Susceptibility pattern in E. coli from human patients; (c) Susceptibility pattern in E. coli from broiler chickens; (d) Susceptibility pattern in E. coli from farm environment; (e) Overall multidrug-resistant pattern; (f) Multidrug-resistant pattern of E. coli from human patients; (g) Multidrug-resistant pattern of E. coli from broiler chickens; (h) Multidrug-resistant pattern of E. coli from farm environments; (i) Overall MIC of E. coli; (j) MIC of E. coli from human patients; (k) MIC of E. coli from broiler chickens; (l) MIC of E. coli from farm environments. Here, CT = Colistin Sulphate, CIP = Ciprofloxacin, TE = Tetracycline, AMP = Ampicilin, CN = Gentamycin, ENR = Enrofloxacin, CTR = Ceftriaxone, C = Chloramphenicol, SXT = Sulfamethoxazole/Trimethoprim.
Substantially, 83.6% (95% CI 78.9–87.5%) isolates were resistant to ciprofloxacin. The highest proportion of ciprofloxacin resistant E. coli isolates were detected in hospitalized humans (89%) (Fig 2B), where the proportion of ciprofloxacin resistant E. coli isolates in broiler and broiler farm environments were 77.6% and 88.8%, respectively (Fig 2C and 2D).
All the isolates were found to be multidrug-resistant (MDR) as they showed resistance to more than two antimicrobials tested (Fig 2E). Most isolates were resistant to more than six antimicrobials tested (Fig 2E–2H). A detailed outline of antimicrobial resistance phenotypes in E. coli strains from different sources is illustrated in heatmaps (S1 Fig). The dendrogram on the left side revealed the clustering of the isolates according to their antimicrobial resistance phenotypes.
MIC of ciprofloxacin
The MIC of all ciprofloxacin-resistant isolates (n = 240) was tested. Overall, 32.5% of the tested isolates show extremely high MIC values (≥128mg/L) against ciprofloxacin (Fig 2I). Alarmingly, 33.8% of ciprofloxacin-resistant E. coli strains from human patients showed very high MIC (≥128mg/L) (Fig 2J). The E. coli strains from broiler chickens show moderately lower MIC values compared with the environmental samples (Fig 2K and 2I).
Gene sequencing and mutations at QRDR
A total of 240 phenotypically ciprofloxacin-resistant isolates were selected for PCR assays to detect gyrA and parC genes. All the isolates were positive for gyrA and parC genes. As mutation(s) in the gyrA and parC genes, specifically in QRDR results in the development of resistance to the ciprofloxacin, we detected point mutation(s) in these two genes. Twenty isolates were selected randomly, covering all types of sample sources for sequencing the gyrA and parC genes. The gene sequences were submitted to the NCBI nucleotide sequence database, which are available under the accession number ON463777-96 for the gyrA gene and ON479036-65 for parC gene.
We detected point mutations at several sites of QRDR, specifically 83 and 87 amino acid positions in gyrA gene (Table 3). At 83 position gyrA genes of all sequences, serine (TCG) was substituted by leucine (TTG) due to a single point mutation in the amino acid codon. However, at 87 position aspartic acid (GAC) was replaced by either histidine (CAC) or asparagine (AAC) or glycine (GGC) or tyrosine (TAC).
Table 3. Mutations patterns in ciprofloxacin resistant E. coli isolates with AST zone and MIC value.
| Isolate name | Inhibition zone (mm) of ciprofloxacin | MIC mg/L of ciprofloxacin | Amino acid (codon) at indicated position of Quinolone-Resistance Determining Region | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrA accession | gyrA 83 | gyrA 87 | parC accession | parC 56 | parC 57 | parC 78 | parC 80 | parC 84 | |||
| E. coli ATCC strain | 34 | 0.02 | NC_007779 | Ser(TCG) | Asp(GAC) | NC_007779 | Ala(GCC) | Ser(AGC) | Gly(GGC) | Ser(AGC) | Glu(GAA) |
| EC/Chicken-CS009 | 6 | >128 | ON463777 | Leu(TTG) | His(CAC) | ON479036 | – | – | – | Ile(ATC) | Gly(GGA) |
| EC/Chicken-CS031 | 13 | 32 | ON463778 | Leu(TTG) | Asn(AAC) | ON479037 | – | Thr(ACC) | Cys(TGC) | Ile(ATC) | – |
| EC/Chicken-CS045 | 6 | >128 | ON463779 | Leu(TTG) | His(CAC) | ON479038 | Thr(ACC) | – | Cys(TGC) | Ile(ATT) | – |
| EC/Chicken-CS078 | 6 | >128 | ON463780 | Leu(TTG) | Gly(GGC) | ON479039 | – | – | – | Arg(AGA) | – |
| EC/Chicken-CS093 | 6 | >128 | ON463781 | Leu(TTG) | His(CAC) | ON479040 | – | – | – | Ile(ATT) | Gly(GGA) |
| EC/Chicken-CS108 | 6 | >128 | ON463782 | Leu(TTG) | Asn(AAC) | ON479041 | – | – | Cys(TGC) | Ile(ATC) | – |
| EC/Chicken-CS119 | 6 | 64 | ON463783 | Leu(TTG) | His(CAC) | ON479042 | – | Thr(ACC) | – | Ile(ATC) | – |
| EC/Chicken-CS133 | 6 | >128 | ON463784 | Leu(TTG) | His(CAC) | ON479043 | – | – | – | Thr(ACC) | Ala(GCA) |
| EC/Feed-CS140 | 13 | 32 | ON463785 | Leu(TTG) | His(CAC) | ON479044 | Gly(GGC) | – | Cys(TGC) | Arg(AGA) | – |
| EC/Soil-CS149 | 9 | 64 | ON463786 | Leu(TTG) | Tyr(TAC) | ON479045 | – | – | – | Arg(AGA) | – |
| EC/Water-CS158 | 11 | 64 | ON463787 | Leu(TTG) | His(CAC) | ON479046 | – | – | – | Ile(ATT) | Gly(GGA) |
| EC/Water-CS159 | 6 | >128 | ON463788 | Leu(TTG) | His(CAC) | ON479047 | – | – | – | Ile(ATC) | – |
| EC/Feed-CS177 | 6 | >128 | ON463789 | Leu(TTG) | Asn(AAC) | ON479048 | – | Ile(ATC) | – | Ile(ATT) | – |
| EC/Litter-CS184 | 14 | 16 | ON463790 | Leu(TTG) | Gly(GGC) | ON479049 | – | – | – | Arg(AGA) | – |
| EC/Litter-CS192 | 6 | 16 | ON463791 | Leu(TTG) | His(CAC) | ON479050 | – | – | – | Ile(ATC) | Lys(AAA) |
| EC/Human-CS199 | 6 | >128 | ON463792 | Leu(TTG) | Tyr(TAC) | ON479051 | – | – | Cys(TGC) | Ile(ATC) | – |
| EC/Human-CS207 | 6 | 64 | ON463793 | Leu(TTG) | Gly(GGC) | ON479052 | – | – | – | Ile(ATC) | Gly(GGA) |
| EC/Human-CS208 | 6 | >128 | ON463794 | Leu(TTG) | His(CAC) | ON479053 | – | Gly(GGC) | – | Ile(ATT) | – |
| EC/Human-CS217 | 14 | 32 | ON463795 | Leu(TTG) | Asn(AAC) | ON479054 | – | Ile(ATC) | – | Arg(AGA) | – |
| EC/Human-CS226 | 11 | 16 | ON463796 | Leu(TTG) | His(CAC) | ON479055 | – | – | – | Ile(ATC) | Ala(GCA) |
Here, AST = Antimicrobial susceptibility testing, MIC = Minimum inhibitory concentration, Ser = Serine, Asp = Aspartic acid, Leu = Leucine, His = Histidine, Gly = Glycine, Asn = Asparagine, Thr = Threonine, Cys = Cystine, Ile = Isoleuocine, Glu Glutamate, Arg = Argenine, Ala = Alanine
In QRDR of parC genes, mutations were detected in 56, 57, 78, 80 and 84 amino acid positions (Table 3). Single- or double-point mutation was detected at position 80 in all the sequences of the study. These mutations result in the substitution of serine (AGC) into either isoleucine (ATC/ATT) or arginine (AGA), or threonine (ACC). Simultaneously, two of the sequences processed mutations at 56 amino acid positions resulting in changing of alanine (GCC) into either threonine (ACC) or glycine (GGC). However, at 57 position, serine (AGC) was replaced by either threonine (ACC) or isoleucine (ATC) or glycine (GGC). At the 78 position, glycine (GGC) was swapped into cysteine (TGC) in five sequences. Glutamate (GAA) at the 84 position was replaced by either glycine (GGA) or alanine (GCA), or lysine (AAA) in some of the parC sequences.
A Common similarity between isolates from three types of sources was the substitution of serine (TCG) by leucine (TTG) with a single point mutation in the 83 amino acid codon of gyrA gene. On the other hand, those isolates had the mutation in the 78 amino acid codon of parC gene due to a single point mutation, namely glycine (GGC) was swapped into cysteine (TGC).
Phylogeny
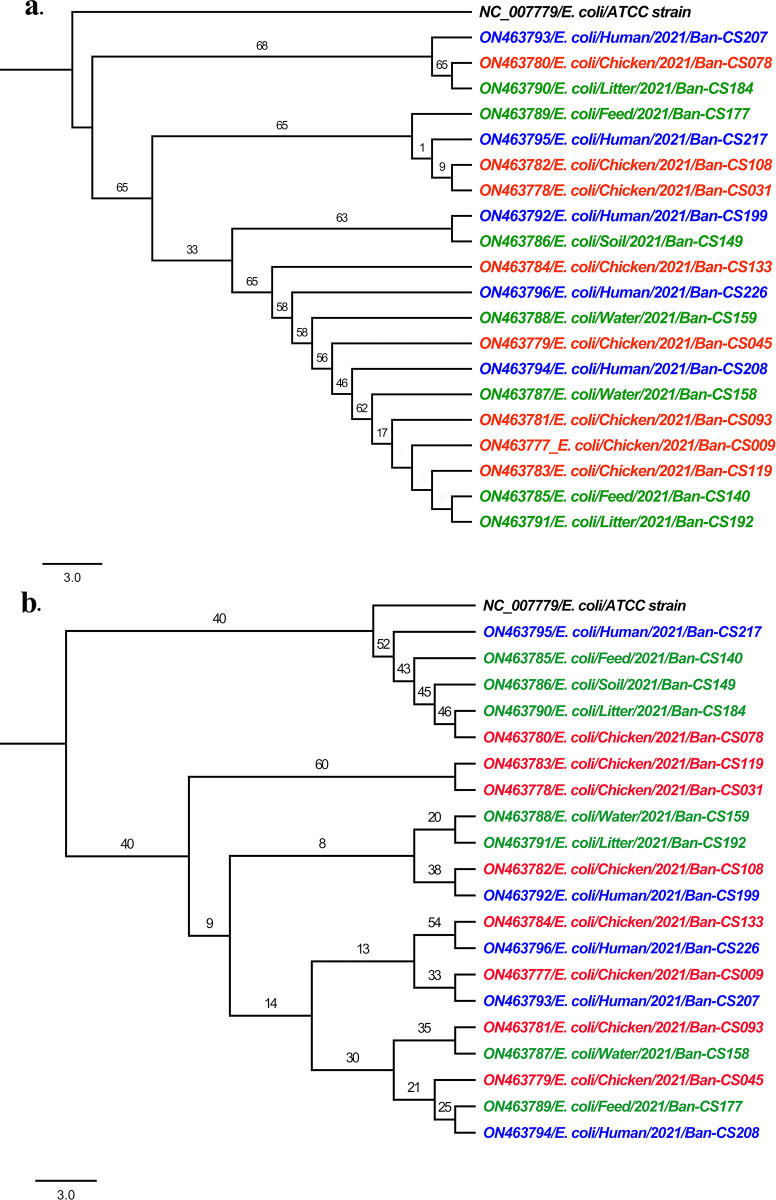
Maximum likelihood (ML) phylogenetic tree of twenty gyrA gene sequences revealed that all the sequences were grouped into three monophyletic clades (Fig 3A). Each of the clades has an admixture of samples of poultry, farm environment and hospitalized human patient origin indicating their phylogenetic similarities. Conversely, the ML phylogenetic tree of twenty parC gene sequences clustered into four monophyletic clades (Fig 3B), where three clades formed with the samples from all three sources and another clade formed with exclusively broiler chicken origin.
Fig 3. Phylogenetic tree of gyrA and parC sequences.
(a) Maximum likelihood (ML) phylogenetic tree of gyrA gene of study sequences rooted with ATCC reference sequence. (a) Maximum likelihood (ML) phylogenetic tree of gyrA gene of study sequences rooted with E. coli reference strain (NC_007779). Here, sequences from poultry (broiler chicken), poultry farm environments, and hospitalized human patients’ origin are highlighted in red, green, and blue, respectively.
Discussion
AMR is an alarming public health burden worldwide, and it is concerning when commensal bacteria develop resistance to vital antimicrobial agents. Our study generated evidence of a high prevalence of AMR in commensal E. coli isolated from hospitalized humans and animal sources. This study primarily focuses on the emergence of ciprofloxacin resistance in commensal E. coli followed by characterization of the mechanism of ciprofloxacin resistance phenomenon by mutation analysis in QRDR.
A high prevalence of E. coli from broiler chicken (89.3%) and farm environment (53.3%) was recorded, which can be influenced by factors such as age, strains and husbandry system of broiler chicken. For instance, the prevalence of E. coli is higher in free-range chickens than caged chickens [16]. However, similar to this study, a high prevalence of E. coli was recorded in chickens in Bangladesh [2–4, 17]. We observed a moderately high prevalence of E. coli (48.5%) in human diarrhoeal patients compared with previous studies in other countries, including India [18], Myanmar [19], Egypt [20], Iran [21], Mexico [21, 22]. This prevalence can be varied with geographical places due to multiple factors like age, disease condition, the onset of illness, etc.
We detected a high prevalence of ciprofloxacin-resistant E. coli in broiler chickens as well as in broiler farm environments and hospitalized human patients in Bangladesh. Previous studies in Bangladesh, India, Nepal, Pakistan and Myanmar have shown a comparatively lower prevalence of ciprofloxacin-resistant E. coli in chickens and hospitalized human patients [4, 19, 23–25]. However, several factors might regulate the prevalence among the geographical locations. The prevalence of ciprofloxacin resistance in commensal E. coli is alarming because these commensal bacteria can transfer their resistant traits among other enteric pathogens, which might have remarkable public health consequences. Ciprofloxacin is a crucial antimicrobial used to treat enteric infections in humans [8]. The high level of resistance developed in bacteria against this antibiotic indicates the consequence of over and indiscriminate use of this drug not only in the poultry industry of Bangladesh but also in human sectors [4, 26].
Microbes that are resistant to at least three groups or classes of antimicrobials are considered MDR [3, 27]. Alarmingly, all of the E. coli isolates in this study were found to be MDR. The widespread use of antimicrobials in the poultry and human sectors may be to blame for the high prevalence of MDR in the study area [28]. Previous studies have shown a wide range of MDR E. coli prevalence from different sources, including 85.7% in humans [29], 62.9% in drinking water [30] and 100% in poultry [26]. The prevalence of MDR E. coli varied in different age groups. For instance, young children aged between 3–24 years had a moderately low prevalence of E. coli than adult humans [23]. The common antimicrobials like ciprofloxacin, ampicillin, and tetracycline, used in both humans and broiler chickens found to be resistant against E. coli. The pattern of MDR E. coli isolated from humans and broiler chickens was remarkably similar, indicating possible transmission between the sources. Once these MDR isolates can produce any pathogenicity to the host, it will be a horrible situation to tackle the infection using the antimicrobials mentioned above.
Surprisingly, a significant proportion of all phenotypically ciprofloxacin-resistant E. coli had higher MIC values. According to the CLSI, the breakpoint of ciprofloxacin is 1mg/L. In contrast, most of the isolates in this study, had MIC values of more than 16mg/L, indicating a higher spectrum of resistance to ciprofloxacin by E. coli. A Similar pattern of high MIC values for ciprofloxacin-resistant E. coli from samples of diverse origin has been reported in several geographical areas [31–33], which indicates a widespread distribution of ciprofloxacin-resistant E. coli. The possible factors of such increasing frequency of MIC might be a horizontal and vertical transmission of the genotype within and among the hosts. Another significant predisposing factor, like irrational use of ciprofloxacin might trigger the phenomenon of high MIC value.
Mutations in specific domains of gyrA, parC, and parE cause single amino acid changes in either gyrase or topoisomerase IV that contribute to quinolone resistance. Multiple mutations in the QRDR of topoisomerase enzymes are usually associated with a high-level of fluoroquinolone resistance in E. coli strains [34]. We detected multiple mutations at QRDR in our study’s gyrA and parC genes sequences, which resulted in several amino acid substitutions in both genes. Most amino acid substitutions occurred due to single-point mutation at QRDR. This molecular confirmation supports the high level of phenotypic resistance found in this study. Regarding mutations in gyrA gene and parC gene, broiler chicken and human isolates had highly similar mutation patterns, indicating a commonality of the isolates. Although gyrA and parC genes of randomly selected isolates were sequenced, to understand the bigger picture of ciprofloxacin resistance, sequencing followed by mutation determination of all isolates might give a detailed mechanistic overview. Phylogenetic analysis also exhibited the genetic similarity of the isolates where they were clustered into similar monophyletic clades. These might indicate a link of transmission of resistant bacteria between humans and animal sources. Horizontal transmission of these resistance genes might be one of the considered factors for such resistance dissemination in one health interphases. Therefore, large scale-systematic study considering all one-health components is warranted to identify the detailed transmission route of antimicrobial resistance.
Conclusions
We isolated commensal E. coli from broiler chickens, broiler farm environments, and hospitalized human samples at a high frequency but with variable levels of prevalence. Most isolates obtained from the sources of investigations were found to be MDR. Surprisingly, more than 83% of ciprofloxacin-resistant commensal E. coli are circulating in one health interphase in Bangladesh carrying acquired mutations in gyrA, and parC genes. Therefore, it is important to develop awareness against the extensive use of antimicrobials in humans and animals to reduce the spread of MDR bacteria.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(a) cloacal swab poultry, (b) samples farm environment, (c) human patients [CT = Colistin Sulphate, CIP = Ciprofloxacin, TE = Tetracycline, AMP = Ampicilin, ENR = Enrofloxacin, CRT = Ceftriaxone, CN = Gentamycin, SXT = Sulfamethoxazole & Trimethoprim and C = Chloramphenicol]. (Here letters in the right side of the map represents the sample ID, red color represents resistance profile of the mentioned antimicrobials (lower axis of the map) and green color represents the sensitivity profile. All the intermediately susceptible isolates were considered as susceptible. The dendrogram in the left side clustered the isolates based on their phenotypic antibiogram pattern).
(TIF)
(DOCX)
(PDF)
Acknowledgments
The authors gratefully acknowledge all the poultry producers and officials of Veterinary Stations of participating Upazilas for providing cordial support to collect poultry and poultry farm environmental samples. The authors also acknowledge Dr. Md. Jashim Uddin, Dr. Md. Nuruddin, Dr. Md. Azizul Hoque, Dr. Nanda Barua and Jewel Dev for their administrative supports to collect the broiler chicken and farm environment samples. The authors also thank the authorities of Chattogram General Hospital, Bangladesh and J.K. Memorial Hospital, Bangladesh for providing the opportunity to collect samples and data from human patients for the study.
Data Availability
All data are available within the text and supplementary files. The gene sequencing data is available at NCBI nucleotide sequence database under the accession number of ON463777-ON463796 and ON479036-ON479055.
Funding Statement
This project work was funded by the University Grant Commission of the Peoples Republic of Bangladesh. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The salary of the author MZI was supported by the Lundbeck Foundation, Copenhagen, Denmark (Grant ID R288-2018-1123).
References
- 1.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18: 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 2.Das T, Islam MZ, Rana EA, Dutta A, Ahmed S, Barua H, et al. Abundance of Mobilized Colistin Resistance Gene (mcr-1) in Commensal Escherichia coli from Diverse Sources. Microb Drug Resist. 2021;27: 1585–1593. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed S, Das T, Islam MZ, Herrero-Fresno A, Biswas PK, Olsen JE. High prevalence of mcr-1-encoded colistin resistance in commensal Escherichia coli from broiler chicken in Bangladesh. Sci Rep. 2020;10: 18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das A, Dhar PK, Dutta A, Jalal MS, Ghosh P, Das T, et al. Circulation of oxytetracycline- and ciprofloxacin-resistant commensal Escherichia coli strains in broiler chickens and farm environments, Bangladesh. November-2020. 2020. pp. 2395–2400. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lushniak BD. Antibiotic resistance: a public health crisis. Public Health Rep. 2014;129: 314–316. doi: 10.1177/003335491412900402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Zowalaty ME, Al Thani AA, Webster TJ, El Zowalaty AE, Schweizer HP, Nasrallah GK, et al. Pseudomonas aeruginosa: arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 2015;10: 1683–1706. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, Chang W, Zhang H, Hu D, Wang X. The Role of Plasmids in the Multiple Antibiotic Resistance Transfer in ESBLs-Producing Escherichia coli Isolated from Wastewater Treatment Plants. Frontiers in Microbiology. 2019. doi: 10.3389/fmicb.2019.00633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO. World Health Organization model list of essential medicines for children-22nd list, 2021. World Health Organization; 2021. Available: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2021.02 [Google Scholar]
- 9.Authority EFS, European Food Safety Authority, European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA Journal. 2022. doi: 10.2903/j.efsa.2022.7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: A systematic review. Int J Infect Dis. 2019;80: 54–61. doi: 10.1016/j.ijid.2018.12.017 [DOI] [PubMed] [Google Scholar]
- 11.Hoque R, Ahmed SM, Naher N, Islam MA, Rousham EK, Islam BZ, et al. Tackling antimicrobial resistance in Bangladesh: A scoping review of policy and practice in human, animal and environment sectors. PLoS One. 2020;15: e0227947. doi: 10.1371/journal.pone.0227947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals Approved standard (5th ed.). 5th ed. Clinical Laboratory Standards Institute; Wayne, PA.; 2020. Available: https://clsi.org/standards/products/veterinary-medicine/documents/vet01/ [Google Scholar]
- 13.CLSI. Performance standards for antimicrobial susceptibility testing. Clinical Laboratory Standards Institute; Wayne, PA.; 2018. Available: https://clsi.org/media/1930/m100ed28_sample.pdf [Google Scholar]
- 14.ISO. Clinical laboratory testing and in vitro diagnostic test systems-Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. International Standards Organisation; 2019. Report No.: 20776–1. Available: https://www.iso.org/obp/ui/#iso:std:iso:20776:-1:ed-2:v2:en
- 15.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. Package gplots: various R programming tools for plotting data. R Package Ver. 2015. [Google Scholar]
- 16.Adeboye OA, Kwofie MK, Bukari N. Campylobacter, Salmonella and Escherichia coli Food Contamination Risk in Free-Range Poultry Production System. Advances in Microbiology. 2020. pp. 525–542. doi: 10.4236/aim.2020.1010039 [DOI] [Google Scholar]
- 17.Dutta A, Islam MZ, Barua H, Rana EA, Jalal MS, Dhar PK, et al. Acquisition of Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli of Livestock Origin in Bangladesh. Microb Drug Resist. 2020;26: 1058–1062. [DOI] [PubMed] [Google Scholar]
- 18.Aggarwal P, Uppal B, Ghosh R, Prakash SK, Rajeshwari K. Highly-resistant E. coli as a common cause of paediatric diarrhoea in India. J Health Popul Nutr. 2013;31: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San T, Moe I, Ashley EA, San N. High burden of infections caused by ESBL-producing MDR E. coli in paediatric patients, Yangon, Myanmar. JAC Antimicrob Resist. 2021;3: dlab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zakaria AM, Aziz MHA, Selim SA. Multi-drug resistant (MDR) Escherichia coli originated from clinical and environmental sources in Ismailia-Egypt. European Journal of Advanced Research in Biological and Life Sciences. 2015;3: 8–20. [Google Scholar]
- 21.Heidary M, Momtaz H, Madani M. Characterization of Diarrheagenic Antimicrobial Resistant Escherichia coli Isolated from Pediatric Patients in Tehran, Iran. Iran Red Crescent Med J. 2014;16: e12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canizalez-Roman A, Flores-Villaseñor HM, Gonzalez-Nuñez E, Velazquez-Roman J, Vidal JE, Muro-Amador S, et al. Surveillance of Diarrheagenic Escherichia coli Strains Isolated from Diarrhea Cases from Children, Adults and Elderly at Northwest of Mexico. Frontiers in Microbiology. 2016. doi: 10.3389/fmicb.2016.01924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shakya P, Barrett P, Diwan V, Marothi Y, Shah H, Chhari N, et al. Antibiotic resistance among Escherichia coli isolates from stool samples of children aged 3 to 14 years from Ujjain, India. BMC Infect Dis. 2013;13: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shrestha A, Shrestha R, Koju P, Tamrakar S, Rai A, Shrestha P, et al. The Resistance Patterns in E. coli Isolates among Apparently Healthy Adults and Local Drivers of Antimicrobial Resistance: A Mixed-Methods Study in a Suburban Area of Nepal. Tropical Medicine and Infectious Disease. 2022. p. 133. doi: 10.3390/tropicalmed7070133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masood F. Occurrence and antibiogram of enteric bacterial isolates from stool samples of gastroenteritis children under 5 years of age in district Faisalabad, Pakistan. Pure and Applied Biology. 2019. doi: 10.19045/bspab.2019.80153 [DOI] [Google Scholar]
- 26.Hasan B, Faruque R, Drobni M, Waldenström J, Sadique A, Ahmed KU, et al. High prevalence of antibiotic resistance in pathogenic Escherichia coli from large- and small-scale poultry farms in Bangladesh. Avian Dis. 2011;55: 689–692. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Watanabe N, Xiao C, Harter T, McCowan B, Liu Y, et al. Antibiotic-resistant E. coli in surface water and groundwater in dairy operations in Northern California. Environ Monit Assess. 2014;186: 1253–1260. [DOI] [PubMed] [Google Scholar]
- 28.Shuford JA, Patel R. Antimicrobial growth promoter use in livestock- implications for human health. Reviews in Medical Microbiology. 2005. pp. 17–24. doi: 10.1097/00013542-200501000-00003 [DOI] [Google Scholar]
- 29.Leski TA, Taitt CR, Bangura U, Stockelman MG, Ansumana R, Cooper WH, et al. High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infectious Diseases. 2016. doi: 10.1186/s12879-016-1495-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patoli AA, Patoli BB, Mehraj V. High prevalence of multi-drug resistant Escherichia coli in drinking water samples from Hyderabad. Gomal Journal of Medical Sciences. 2010;8. [Google Scholar]
- 31.Begum YA, Talukder KA, Azmi IJ, Shahnaij M, Sheikh A, Sharmin S, et al. Resistance Pattern and Molecular Characterization of Enterotoxigenic Escherichia coli (ETEC) Strains Isolated in Bangladesh. PLoS One. 2016;11: e0157415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahoo KC, Tamhankar AJ, Sahoo S, Sahu PS, Klintz SR, Lundborg CS. Geographical Variation in Antibiotic-Resistant Escherichia coli Isolates from Stool, Cow-Dung and Drinking Water. International Journal of Environmental Research and Public Health. 2012. pp. 746–759. doi: 10.3390/ijerph9030746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman M, Shoma S, Rashid H, El Arifeen S, Baqui AH, Siddique AK, et al. Increasing spectrum in antimicrobial resistance of Shigella isolates in Bangladesh: resistance to azithromycin and ceftriaxone and decreased susceptibility to ciprofloxacin. J Health Popul Nutr. 2007;25: 158–167. [PMC free article] [PubMed] [Google Scholar]
- 34.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25: 358–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(a) cloacal swab poultry, (b) samples farm environment, (c) human patients [CT = Colistin Sulphate, CIP = Ciprofloxacin, TE = Tetracycline, AMP = Ampicilin, ENR = Enrofloxacin, CRT = Ceftriaxone, CN = Gentamycin, SXT = Sulfamethoxazole & Trimethoprim and C = Chloramphenicol]. (Here letters in the right side of the map represents the sample ID, red color represents resistance profile of the mentioned antimicrobials (lower axis of the map) and green color represents the sensitivity profile. All the intermediately susceptible isolates were considered as susceptible. The dendrogram in the left side clustered the isolates based on their phenotypic antibiogram pattern).
(TIF)
(DOCX)
(PDF)
Data Availability Statement
All data are available within the text and supplementary files. The gene sequencing data is available at NCBI nucleotide sequence database under the accession number of ON463777-ON463796 and ON479036-ON479055.