Abstract
Four pairs of primers were designed for PCR amplification of known polymorphic regions of the mitochondrial genome of Phytophthora infestans. Digestion of the amplified products with restriction enzymes allows identification of previously identified haplotypes. Product P2 cut with MspI uniquely identifies haplotypes Ib and IIa, while types Ia and IIb are differentiated by digestion of product P4 with EcoRI. Digestion of products P1 and P3 gave results similar to that with digestion of P4, but amplification of these products was less robust. Thus, all four common haplotypes are identified by amplifying and digesting products P2 and P4. Identification of haplotypes was also possible from DNA extracted directly from small, late-blight lesions on both tomato and potato leaves, making isolation of the fungus unnecessary. A rapid and efficient method of monitoring changes in the pathogen population is facilitated. These PCR primers were also useful for differentiating other Phytophthora species.
Populations of Phytophthora infestans from Central Mexico are highly diverse, indicating that this area is the center of origin of the pathogen (14, 16, 20). In contrast, populations from other parts of the world were almost totally monomorphic for allozyme and restriction fragment length polymorphism (RFLP) loci. Over the last 20 years, new genotypes migrating from Mexico have become established and have replaced the old monomorphic genotype around the world (10, 14, 16, 20, 21, 30). As some new genotypes are more pathogenic or more resistant to curative systemic fungicides (6, 15, 23), rapid identification of the strains is crucial so that optimal methods of control can be implemented. The development of such identification and detection methods is considered a high priority by the agriculture industry (34).
Polymorphisms in mitochondrial DNA (mtDNA) of P. infestans are particularly useful for monitoring pathogen populations; they are easily detected and, because they are uniparentally (and probably maternally) inherited (35), ideal for tracing lines of descent. These polymorphisms have previously been studied by hybridization of digested total DNA with labelled, cloned mtDNA (8, 17, 28) and digestion of isolated mtDNA (12, 13, 24). Carter et al. (3, 4) defined two mitochondrial types, type I and type II, by digestion of total DNA with the frequently cutting restriction enzymes MspI or CfoI (which produce bands of mtDNA on a background smear of nuclear DNA upon separation). Type II differed from type I by an insert of 1.6 kb and rearrangement of flanking sequences. Type I was further differentiated into haplotypes Ia and Ib, the latter possessing an additional MspI site; similarly, type II was subdivided into haplotypes IIa and IIb, the latter possessing an additional CfoI site. The recent sequencing of the mitochondrial genome of P. infestans (5, 29) allows the design of primers to amplify by PCR the known polymorphic sequences of the genome.
Our objective in this study was to develop a PCR-based method for the rapid detection of mtDNA polymorphisms in P. infestans, using small amounts of fungus from agar cultures or infected host tissue. A secondary goal was to test whether the same primers can be used to amplify mtDNA from other species of Phytophthora to aid identification and diagnoses.
MATERIALS AND METHODS
Isolates of P. infestans and other Phytophthora spp.
The cultures used in this investigation were from the UW Bangor collection or imported to the United Kingdom via the International Mycological Institute. P. infestans was isolated on ryeA medium containing antibiotics (rifamycin, ampicillin, and nystatin), as described by Griffith et al. (22). Long-term stocks were maintained as agar plugs in 5% dimethyl sulfoxide under liquid nitrogen (31). Details of the origins of cultures are given in Table 1. Isolates were grown routinely on ryeA agar at 18°C and stored under liquid nitrogen. Cultures of other Phytophthora spp. were from the UW Bangor collection and were cultured as described above.
TABLE 1.
Isolates of P. infestans used in this study
| Country of origin | Isolate name | Hosta | Mating type | MtDNA | Field location | Collection yr | Supplierd |
|---|---|---|---|---|---|---|---|
| Canada | Can2 | T | A2 | Ia | Vancouver | 1994 | A |
| Can4 | P | A1 | IIb | Vancouver | 1994 | A | |
| Colombia | GF29 | T | A1 | Ib | Rionegro | 1990 | B |
| GF256 | T | A1 | IIa | NK | 1993 | B | |
| GF1361 | NKb | NK | IIa | NK | 1993 | B | |
| GF1368 | NK | NK | IIa | NK | 1993 | B | |
| Russia | 19 | P | A2 | IIa | Voskresensk, Moscow | 1991 | C |
| 31 | P | A1 | IIa | Voskresensk, Moscow | 1991 | C | |
| 45 | P | A2 | IIa | Voskresensk, Moscow | 1991 | C | |
| 52 | P | A2 | IIa | Voskresensk, Moscow | 1991 | C | |
| 112 | P | A2 | IIa | Voskresensk, Moscow | 1991 | C | |
| 113 | P | A1 | IIa | Voskresensk, Moscow | 1991 | C | |
| 154 | P | A2 | IIa | Voskresensk, Moscow | 1991 | C | |
| 14.91 | P | SFc | IIa | Belorussia | 1991 | C | |
| 31.91 | P | SF | Ib | Belorussia | 1991 | C | |
| 1.T1 | T | A1 | Ib | Moscow region | 1993 | C | |
| 2.T0.K | P | A2 | IIa | Moscow region | 1993 | C | |
| 3.T1 | T | A2 | IIa | Moscow region | 1993 | C | |
| 4.T1 | T | A1 | Ib | Moscow region | 1993 | C | |
| 6.T0.K | P | A1 | IIa | Moscow region | 1993 | C | |
| 7.T0.K | P | A1 | IIa | Moscow region | 1993 | C | |
| 8.T0.K | T | A1 | IIa | Moscow region | 1993 | C | |
| 9.T1 | T | A1 | Ia | Moscow region | 1993 | C | |
| 10.T1 | T | A1 | IIa | Moscow region | 1993 | C | |
| DP.1 | T | A2 | Ia | Odintzovo, Moscow region | 1993 | C | |
| I0.24 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K27 | P | A2 | IIa | Noginsk, Moscow region | 1991 | C | |
| K3.4 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.9 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.22 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.37 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.43 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.53 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.54 | P | A2 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K3.56 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K4.4 | P | A1 | IIa | Odintzovo, Moscow region | 1994 | C | |
| K4.5 | P | A1 | IIa | Odintzovo, Moscow region | 1994 | C | |
| K4.25 | P | A1 | Ia | Odintzovo, Moscow region | 1994 | C | |
| K4.26 | P | A1 | Ia | Odintzovo, Moscow region | 1994 | C | |
| K4.29 | P | A1 | Ia | Odintzovo, Moscow region | 1994 | C | |
| K4.31 | P | A1 | IIa | Odintzovo, Moscow region | 1994 | C | |
| K4.39 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| K4.42 | P | A1 | IIa | Odintzovo, Moscow region | 1993 | C | |
| KT2 | P | A2 | IIa | Taldom, Moscow region | 1991 | C | |
| KT5 | P | A2 | IIa | Taldom, Moscow region | 1991 | C | |
| KT16 | P | A2 | IIa | Taldom, Moscow region | 1991 | C | |
| KT17 | P | A2 | IIa | Taldom, Moscow region | 1991 | C | |
| KT19 | P | A2 | IIa | Taldom, Moscow region | 1991 | C | |
| KT21 | P | A1 | IIa | Taldom, Moscow region | 1991 | C | |
| KT23 | P | A1 | IIa | Taldom, Moscow region | 1991 | C | |
| KT33 | P | A1 | IIa | Taldom, Moscow region | 1991 | C | |
| PT.8 | T | A1 | Ia | Odintzovo, Moscow region | 1993 | C | |
| RYD138 | P | A1 | Ib | Belorussia | 1990 | C | |
| RYD151 | T | A2 | IIa | Moscow region | 1987 | C | |
| RYD55 | P | A1 | Ib | Moscow region | 1987 | C | |
| RYD3101 | T | A1 | IIa | Moscow region | 1992 | C | |
| RYD3210 | T | A1 | Ib | Moscow region | 1992 | C | |
| TA.33 | T | A1 | Ia | Adler, southern Russia | 1992 | C | |
| TA.61 | T | A1 | Ia | Adler, southern Russia | 1992 | C | |
| T0.K2 | P | A1 | IIa | Siberia, Tomsk region | 1994 | C | |
| T0.K3 | P | A1 | IIa | Siberia, Tomsk region | 1994 | C | |
| T0.K4 | P | A1 | IIa | Siberia, Tomsk region | 1994 | C | |
| T.11 | T | A2 | Ia | Odintzovo, Moscow region | 1993 | C | |
| Taiwan | Tai1 | T | A1 | Ib | Hsinche, Nantou Province | 1991 | D |
| TYH1 | T | A1 | Ib | Tainan | 1991 | D | |
| TYH2 | T | A1 | Ib | Tainan | 1992 | D | |
| TJCJul7 | T | A1 | Ib | Puli, Nantou | 1994 | D | |
| Tanzania | TZNW1 | T | A1 | Ib | Tengeru, Arusha | 1994 | E |
| United Kingdom | 78/06 | P | A1 | Ib | Llandegai, Gwynedd, Wales | 1978 | A |
| 78/18 | P | A1 | Ib | Wrington, Bristol, England | 1978 | A | |
| 81/2 | P | A1 | Ib | Llandegai, Gwynedd, Wales | 1981 | A | |
| 81/9 | P | A1 | Ia | Llandegai, Gwynedd, Wales | 1981 | A | |
| 83/5 | P | NK | Ia | Bangor, Gwynedd, Wales | 1983 | A | |
| 1756sz | P | NK | Ia | Long Ashton, Avon, England | 1985 | A | |
| 85/123 | P | A1 | Ia | Haskayne, Lancashire | 1985 | A | |
| 87/141/4 | P | A1 | IIa | Llangefni, Ynys Mon, Wales | 1987 | A | |
| 88/130/4 | P | A1 | Ia | Penmon, Ynys Mon, Wales | 1988 | A | |
| 91/03/1 | P | NK | Ia | ADAS, Bristol, England | 1991 | A | |
| 91/12/2 | T | NK | Ia | Epsom, Surrey, England | 1991 | A | |
| 92/8 | T | A1 | Ia | Llandegai, Gwynedd, Wales | 1992 | A | |
| 93/20/1 | P | A1 | IIa | Boston, Lincolnshire, England | 1993 | A | |
| 93/025/5 | P | A1 | Ia | Starcross, Devon, England | 1993 | A | |
| 93/028/8 | P | A1 | Ia | Biwmaris, Ynys Mon, Wales | 1993 | A | |
| 93/102 | P | A2 | Ia | Liskeard, Cornwall, England | 1993 | A | |
| United States | Ca29 | T | A1 | IIb | California | 1984 | F |
| Ca65 | T | A1 | IIb | California | 1984 | F | |
| F276 | T | A2 | Ia | Yolo County, California | 1993 | G | |
| F277 | T | A2 | Ia | Yolo County, California | 1993 | G | |
| F284 | T | A2 | IIb | Woodland, California | 1993 | G | |
| F286 | T | A2 | IIb | Saticoy, California | 1993 | G |
The original host plants are indicated as P (potato) and T (tomato).
NK, not known.
SF, self-fertile.
Suppliers are indicated as follows: A, UW Bangor culture collection; B, G. Forbes (CIP, Quito, Ecuador); C, Y. Dyakov (Moscow State University, Russia); D, Y.-H. Huang (AVRDC, Tainan, Taiwan); E, R. Nono-Wondim (AVRDC, Arusha, Tanzania); F, M. Coffey (University of California, Riverside); G, B. Gabor (Petoseed Ltd., Woodland, Calif.).
Sources of host lesions.
Leaf and stem lesions of late blight were collected from naturally infected crops in the field (various cultivars) or were grown on detached leaflets of potato (cv. Maris Piper) or tomato (cv. FMX-93) inoculated with droplets of zoospores. Leaflets were incubated in moist chambers at 18°C for 3 to 4 days (potato) or 5 to 7 days (tomato). Some leaflet lesions were air dried by incubation in an open tray in the laboratory for 7 days.
Extraction of DNA.
DNA was extracted by using a variation of the method of Doyle and Doyle (9) with modifications devised by DuTeau and Leslie (11). Briefly, this involved cutting small slabs (ca. 1 cm2) of agar-containing mycelium from a culture grown on ryeA medium and subjecting these to rapid freezing twice with liquid nitrogen (poured into a microcentrifuge tube containing the slab and allowed to evaporate; no maceration), followed by incubation in 800 μl of modified CTAB extraction buffer (100 mM NaCl, 100 mM Tris-HCl [pH 8.0], 1.4 M NaCl, 2% CTAB [hexadecyltrimethylammonium bromide], 20 mM EDTA [sodium salt, pH 8.0]) for 60 min at 65°C. After removal of the agar slab with a sterile toothpick, 600 μl of water-saturated chloroform was added and the tube was vortex mixed for 10 s and microcentrifuged (17,000 × g) for 10 min. Six hundred microliters of the upper, aqueous layer was transferred to a fresh microcentrifuge tube, and 0.6 volume of isopropanol (360 μl) was added. The tubes were vortex mixed, left to stand at room temperature for 5 min, and microcentrifuged (17,000 × g) for 10 min. The liquid was decanted, and the DNA pellet was washed with 1 ml of 70% (vol/vol) ethanol. Tubes were vortexed and left at 65°C for 20 min before microcentrifugation (17,000 × g) for 10 min as before. After the tubes had been left open at 37°C for 30 min to remove the last traces of 70% ethanol, the DNA pellet was resuspended in 100 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) and stored at −20°C. Quality and concentration were checked by electrophoresis of 5 μl of the DNA solution on a 0.8% agarose gel.
DNA was extracted from late-blight lesions. Pieces of colonized leaf (5 by 5 mm), dried or fresh, were excised and ground in a microcentrifuge tube with fine sand (ca. 50 mg) and liquid nitrogen with a disposable polythene microcentrifuge grinder (Anachem Ltd.). Thereafter, the extraction procedure was the same as that described above. The purified DNA was dissolved in TE (500 μl); 1 μl was used for PCR.
PCR-RFLP procedure.
PCRs with a model PC2 PCR thermal cycler (Techne Ltd., Cambridge, United Kingdom) were optimized to maximize yield of the desired PCR product and reduce levels of nonspecific products (data not shown). Amplification was as follows for all primer combinations (final concentrations): deoxynucleoside triphosphates, 200 μM each; MgCl2, 2.75 mM; oligonucleotide primer (Cruachem Ltd., Glasgow, United Kingdom; see Table 2), 0.325 mM each; ethidium bromide, 0.2 μg ml−1; bovine serum albumin, 160 μg ml−1; 1× Thermo buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]); Taq DNA polymerase, 0.2 μl (1 U). All reagents were supplied by Promega Ltd. (Southampton, United Kingdom) unless stated otherwise. One to ten nanograms of total DNA was mixed with 20 μl of a master mix of the other PCR reactants in 0.5-ml microcentrifuge tubes (final volume, 25 μl) and overlaid with 50 μl of mineral oil. The PCR conditions were as follows: 1 cycle of 94°C for 90 s and 40 cycles of 94°C for 40 s, 55°C for 60 s, and 72°C for 90 s.
TABLE 2.
Oligonucleotide primers used in this investigation
| Primer | Sequence | Primer length (bp) and positiona |
|---|---|---|
| F1 | 5′-GCAATGGGTAAATCGGCTCAA-3′ | 21 23737–23757 |
| R1 | 5′-AAACCATAAGGACCACACAT-3′ | 20 24835–24854 |
| F2 | 5′-TTCCCTTTGTCCTCTACCGAT-3′ | 21 13619–13639 |
| R2 | 5′-TTACGGCGGTTTAGCACATACA-3′ | 22 14688–14667 |
| F3 | 5′-ATGGTAGAGCGTGGGAATCAT-3′ | 21 2892–2912 |
| R3 | 5′-AATACCGCCTTTGGGTCCATT-3′ | 21 4199–4179 |
| F4 | 5′-TGGTCATCCAGAGGTTTATGTT-3′ | 22 9329–9350 |
| R4 | 5′-CCGATACCGATACCAGCACCAA-3′ | 22 10292–10271 |
Primers were synthesized by Cruachem Ltd. The indicated positions refer to primer position on the P. infestans mitochondrial genome in relation to the origin (5′ end of the large subunit of the mitochondrial rRNA gene).
Three to four microliters of the amplified DNA was digested with the following restriction enzymes in a 20-μl volume restriction digest at 37°C for a period lasting between 1 h and overnight as follows: P1, CfoI; P2, MspI; P3, EcoRI; and P4, EcoRI. (EcoRI had greater activity in Promega Multi-Core buffer than in the manufacturer’s recommended buffer). The digested DNA samples were then mixed with 5 μl of gel-loading buffer (20% Ficoll, 0.02% bromophenol blue, 0.002% xylene cyanol), and 15 μl was loaded into a slot on a 2% agarose gel (Gibco BRL Ltd.) in 1× Tris-borate-EDTA (TBE) buffer (containing 0.1 μg of ethidium bromide ml−1). The gel was run at 10 V cm−1 for 60 to 90 min. Restriction patterns were visualized with a UV transilluminator at 254 nm, and the images were recorded by a gel documentation system (Appligene, Chester-le- Street, United Kingdom).
PCR amplification of longer fragments of the mitochondrial genome was achieved by a modification of the long PCR technique of Barnes (2) as follows (final concentrations): deoxynucleoside triphosphates, 325 μM each; 0.325 μM (each) oligonucleotide primer; ethidium bromide, 0.2 μg ml−1; 1× long PCR buffer (50 mM Tris-HCl [pH 9.2 at 25°C], 16 mM (NH4)2SO4, 1.75 mM MgCl2). A mixture of Taq DNA polymerase (Promega) and Tli polymerase (250 U of Taq, 1 U of Tli polymerase) was made, and 0.2 μl (i.e., 1 U of Taq plus 0.004 U of Tli polymerase) was added to each reaction in thin-walled 0.5-ml tubes. PCR cycling conditions were as follows: 1 cycle of 95°C for 1 s, then 94°C for 20 s; 20 cycles of 95°C for 1 s, 94°C for 10 s, 54°C for 60 s, and 67.5°C for 5 min; 8 cycles of 95°C for 1 s, 94°C for 10 s, and 67.5°C for 6 min.
RESULTS
Design of primers for analysis of mitochondrial haplotypes.
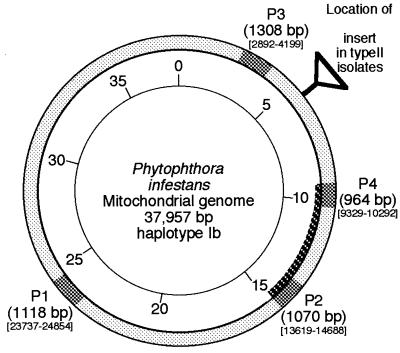
The complete mitochondrial genome of P. infestans (Ib mtDNA haplotype; 37,957 bp) has been sequenced, and parts of it have been published (5, 29; also http://megasun.bch.umontreal.ca/People/lang/species/phyti/phyti.html). Restriction site characteristics of the four haplotypes identified by Carter et al. (4; referred to here as Carter haplotypes) were located within the complete mitochondrial genome sequence (kindly supplied prior to full publication by B. F. Lang). Based on this sequence, pairs of oligonucleotide primers (listed in Table 2) were designed to amplify four regions of the mitochondrial genome in which polymorphisms were known to occur (Fig. 1). P2 is the site of the Ia/Ib polymorphism, while P1, P3, and P4 are sites of polymorphism between haplotype Ia/Ib and haplotype IIa/IIb. The primers were predicted to produce amplification products of 1,118 (P1), 1,070 (P2), 1,308 (P3), and 964 bp (P4).
FIG. 1.
Diagrammatic representation of the mitochondrial genome of P. infestans illustrating the location of the various amplification products (shaded). Sequence locations are according to the provisional sequence supplied by B. F. Lang. The position of a ca.-1.6-kb insert present in haplotype II isolates (Cornell haplotype B) is indicated. The darker shading indicates the extent of the longer F4-R2 PCR product. Sequence positions correspond to those published by Paquin et al. (29) (available at http://megasun.bch.umontreal.ca/People/lang/species/phyti/phytmtDNA.html).
Once optimized, amplification of all four templates from relatively crudely extracted DNA from agar cultures was robust and generated a single band of the predicted size. The template DNA for P4 could be diluted to 150 pg and still provide suitable amplification (data not shown). Similar minimum quantities of template were required for amplification of the other products.
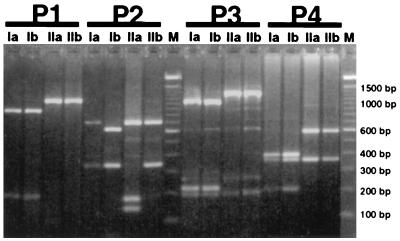
Restriction enzyme digestion of the four PCR products generally revealed the polymorphisms predicted by Carter et al. (4) (Fig. 2 and 3). A CfoI site on the P1 product was present in haplotypes Ia and Ib, while in both the P3 and P4 products, additional EcoRI sites were present in Ia and Ib haplotypes but not in types IIa and IIb. Digestion of the P1, P3, and P4 products provided the same information, but the P4 primers were used routinely because they amplified more reproducibly and produced a more distinctive restriction pattern. As predicted (4), an MspI digest of P2 yielded two fragments for haplotype Ia and IIb isolates and three fragments for haplotype Ib; however, haplotype IIa isolates unexpectedly also yielded three fragments due to an MspI site not identified previously due to the small size of the additional fragment (203 bp; Fig. 3). The reliability of the assay was confirmed with a double blind test in which a number of isolates, previously genotyped by the method of Carter et al. (4), were correctly identified.
FIG. 2.
Restriction enzyme digestions of PCR products amplified from P. infestans with primer pairs F1-R1 (cut with CfoI), F2-R2 (cut with MspI), F3-R3 (cut with EcoRI), and F4-R4 (cut with EcoRI). Amplifications were conducted with DNA from four isolates representing each of the four Carter (4) mitochondrial DNA haplotypes, namely Ia, 96.70; Ib, K1067; IIa, E14c2; and IIb, Ca65. Lanes marked M contain 100-bp ladders (Gibco BRL Ltd.).
FIG. 3.
Diagrammatic representation of the sizes of restriction fragments generated by digestion of the four PCR products from P. infestans. Restriction sites are indicated with an arrow, and the presence or absence of these sites is indicated by + or −.
The four PCR products were tested for new RFLPs by digestion with a further eight enzymes which cut the mtDNA frequently (AluI, DraI, EcoRI, EcoRV, HaeIII, HindIII, RsaI, and Sau3AI). However, among 12 geographically diverse isolates, which included all four Carter haplotypes, no further polymorphisms were detected, and all restriction fragments were as predicted from the sequence data.
Amplification of larger portions of the mtDNA was achieved with other combinations of the eight primers described in Table 2 and the modified long PCR amplification conditions of Barnes (2). Amplification with primers F4 and R2 yielded a PCR product, P2/4, 5,359 bp in length (including both the P2 and P4 products as well as the intervening region; Fig. 1). Digestion of P2/4 from six isolates, including all four haplotypes, with eight enzymes known to cut the predicted product at several positions (Fig. 4) did not reveal additional polymorphisms. This long PCR procedure was also successful in amplifying the 7,400-bp fragment of mtDNA between primers F3 and R4 (on either side of the 1.6-kb insert in haplotypes IIa and IIb) from a haplotype Ia isolate, but several smaller, nonspecific products were also amplified, and no restriction analysis was conducted (data not shown).
FIG. 4.
Long PCR amplification with F4-R2 primers of isolates Cer002t (haplotype Ia, lanes 1); GF266 (haplotype IIa, lanes 2); 1.T1 (haplotype Ib, lanes 3); and Tail (haplotype Ib, lanes 4) digested with HindIII, MspI, and RsaI. Lane M, 1-kb ladder (Gibco BRL Ltd.).
Isolates from 17 countries from both potato and tomato were screened by amplifying and digesting P2 (MspI digest; Fig. 5) and P4 (EcoRI digest). The presence of ethidium bromide in the PCR mixture allowed amplification to be verified by examining the PCR tubes with a UV transilluminator, making preliminary electrophoresis unnecessary. Products were digested most conveniently with microtiter plates.
FIG. 5.
F2-R2 PCR products amplified from various Russian isolates of P. infestans and digested with MspI. Further details about these isolates are given in Table 1. Lane M, 1-kb ladder (Gibco BRL Ltd.).
Haplotype IIb was found only in California and Canada (Vancouver). These isolates were mating types A1 and A2. Haplotypes Ia and IIa were both found in most countries. Haplotype Ia was most often associated with A1, and haplotype IIa was most often associated with A2 mating type. The Ib haplotype was detected in five countries (16 isolates in total) and tended to be from older collections.
Direct analysis of mitochondrial haplotype from late-blight lesions.
DNA was extracted from late-blight lesions on potato leaflets and was amplified with all four pairs of primers to yield amounts of product comparable to those amplified with DNA from pure cultures. A background smear or faint additional bands were frequently observed but did not obscure digestion products. P2 and P4 were reliably amplified from DNA from fresh lesions and from air-dried lesions of both tomato and potato. Four samples of air-dried lesions on tomato leaflets sent to Wales from Tanzania were found to be of the same haplotype (Ib) as a pure culture established in Tanzania (TNZW1; Table 1) from the same crop.
Amplification of mtDNA from other Phytophthora spp.
The sequences of mitochondrial genomes in Phytophthora spp. are believed to be moderately conserved (12, 29), meaning that the primers described above could be used to identify and characterize mtDNA of other species.
We tested DNA from nine Phytophthora spp. with the mitochondrial primers described above. At an annealing temperature of 55°C (optimal for P. infestans), amplification was generally poor for all primers. However, when a lower annealing temperature (53.5°C) was used, amplification was successful in the majority of cases as shown in Table 3. The P1 and P3 primers were the most suitable for the reliable amplification of mtDNA from these other species. The amount of P2 and P4 products were generally less than the amount from P. infestans.
TABLE 3.
Amplification of P1–P4 PCR products from nine Phytophthora species
| Species | Isolate | Origin (source, country) | Collectora | Primerb
|
|||
|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | ||||
| P. capsici | Pc34 | Tomato, Taiwan | A | ++ | + | ++ | ++ |
| P. citrophthora | BP265 | Cocoa, Brazil | B | ++ | + | ++c | + |
| P. colocasiae | PT265 | Taro, China | C | + | + | ++ | ++ |
| P. cryptogea | 77/6 | Tomato, United Kingdom | D | ++ | − | ++ | − |
| P. drechsleri | P208 | Capsicum, Mexico | E | ++ | + | ++ | + |
| P. fragariae var. rubi | R6 | Raspberry, United Kingdom | F | ++ | − | ++ | − |
| P. megasperma | 17 | Juniper, United States | G | ++ | − | ++ | − |
| P. nicotianae | BT113 | Tomato, United Kingdom | D | ++ | + | ++ | ++ |
| P. palmivora | 7298 | Soil, PNG | H | ++ | − | ++ | + |
Collectors are indicated as follows: A, Y.-H. Huang (AVRDC, Tainan, Taiwan); B, T. Dakwa (New Tafo, Ghana); C, K. M. Zhang (Hainan, China); D, P. Smith (GCRI, Littlehampton, United Kingdom); E. J. Galindo (Chapingo, Mexico); F, J. Duncan (SCRI, Dundee, United Kingdom); G, E. Hansen (Oregon State University, Oregon); H, F. Arentz (Bulolo, Papua New Guinea).
The amount of product generated at an annealing temperature of 53.5°C with each of the primer pairs is indicated as follows: ++, amount of amplification product similar to that obtained with P. infestans; +, clearly detectable PCR product but bands significantly weaker than for P. infestans; −, no detectable PCR product.
An additional band was also amplified.
There was some variation in the sizes of the undigested PCR products from the different species (data not shown), but more distinctive differences were seen following endonuclease digestion (Fig. 6). There were frequently similarities between the restriction patterns obtained from different species. Primers P2 and P4, routinely used for P. infestans, either failed to generate PCR product or gave a distinctive restriction pattern after digestion with the other species; on two occasions (data not shown), isolates from foliage blight of tomato from Taiwan produced restriction patterns quite different from those of P. infestans. These were subsequently identified as Phytophthora capsici on the basis of colony and sporangial morphology.
FIG. 6.
Amplification of DNA from various Phytophthora species with the P3 primers (at an annealing temperature of 53.5°C) followed by digestion with DraI. Lanes: 1, P. capsici (P. cap) (P139); 2, P. citrophthora (P. cit) (IMI362668); 3, P. colocasiae (P. col) IMI 368918); 4, P. drechsleri (P. dre) (P533); 5, P. fragariae var. fragariae (P. fra) (FVF24); 6, P. nicotianae (P. nic) (Nic1); 7, P. infestans (P. inf) (86.126.1a). Lanes M, contain a 100-bp ladder (Gibco BRL Ltd.). DNA was kindly supplied by David Cooke, SCRI, Dundee, Scotland.
DISCUSSION
Isolation of P. infestans into pure culture followed by the production of sufficient mycelium (typically >1 g) for Southern analysis or purification of mitochondrial DNA is a process which requires several weeks to complete. Methods for which only small (micrograms) quantities of pure P. infestans DNA are needed and methods not requiring the isolation of fungus into pure culture permit rapid and accurate monitoring of populations, which could help determine the most appropriate control strategy. For example, haplotype Ib has never become highly resistant to one of the most useful systemic and curative fungicides, metalaxyl (1, 6).
Restriction enzyme digestion of PCR products for RFLP analysis facilitates monitoring of genetic changes among field populations of P. infestans. We have shown that the screening process can be simplified by the direct analysis of genotypes from fresh or dried blight lesions and by the use of microtiter plates for digestion. One advantage of our method is that it includes both positive and negative controls; differences in the size of a positive product rather than a simple product/no product result are scored. By ribosomal DNA (rDNA)-based detection methods (32, 34) results could be false negative if a PCR fails due to sample contamination. A positive control for restriction digestion is also included, as one MspI site (P2) and one EcoRI site (P4) are present in all haplotypes; digestion failures are identified by uncut product.
Digestion of products P2 and P4, with MspI and EcoRI respectively, permits the unambiguous identification of the four Carter mitochondrial haplotypes. The simplicity of the process is such that 50 to 100 samples can be processed (i.e., DNA extraction, PCR amplification, restriction digestion, and gel electrophoresis) by a single person in less than 2 working days. This method might be further simplified by substituting a DNA extraction from late-blight lesions with NaOH (32, 34).
Some correlation has been made between the Carter haplotypes and the six mitochondrial haplotypes (A to F) designated by Goodwin (17), and it is known that the haplotype Ib isolates also have a nuclear DNA fingerprint (with probe RG57; 18) identical to the US-1 clonal lineage (6). Screening of isolates known to contain Goodwin’s haplotypes C to F (17) may reveal additional polymorphism within the PCR products described here.
In this study, haplotype Ib was detected in collections from most countries. It is now clear that this haplotype is almost always associated with A1 mating type, isozyme genotypes Gpi-1 86/100 and Pep-1 92/100 and multilocus RG57 fingerprint US-1. The four isolates from Taiwan were, as expected, haplotype Ib, as Taiwan is one of few countries in which this multilocus genotypes has been the only one detected (25). All older collections from Russia, Europe, and the eastern United States were only of this genotype (21); more recent collections show that this apparent clone has been or is being replaced by genotypes of haplotypes Ia or IIa, different isozyme alleles, and different RG57 fingerprints. It appears that the old genotype did not recombine with the new, highly variable genotypes which probably migrated from Mexico in the 1970s (21). An exception was detected in an English isolate from 1995 which was haplotype Ib but had a fingerprint quite different from US-1 (6); another isolate from Brittany in 1996 was Gpi-1 86/100 Pep-1 92/100 and haplotype Ib but again had a non-US-1 fingerprint (26).
In some countries only a single haplotype was detected (e.g., Costa Rica, 18 isolates all haplotype Ia; Bolivia, 10 isolates all haplotype IIa; data not shown), while in others (e.g., Russia, United Kingdom; Table 1) two or three haplotypes were detected. Lack of diversity may be due to the limited number of samples collected or sites sampled and/or to restricted sampling dates. In Western Europe, recent collections are predominantly of haplotype Ia, with much lower frequencies of IIa (6, 26). In this study, haplotype IIb was detected only in California and British Columbia (Vancouver), but it has since been detected in one isolate from The Netherlands (11a).
The amplification procedure described here works well with crudely prepared template DNA from both pure cultures and late-blight lesions, and is sensitive to as little as 150 pg of template DNA, which is equivalent to 100 to 200 nuclei of P. infestans (33). A PCR-based diagnostic assay devised by Trout et al. (34) to detect symptomless early infection detected as little as 10 pg of DNA of P. infestans but did not permit differentiation of genetically different isolates. The increased sensitivity of the latter assay is probably due to the higher copy number of the rDNA target sequences and the short 456-bp product which was generated. Like the method used in the present study, the rapid cellulose acetate electrophoresis method of Goodwin et al. (19) for isozyme analysis also does not require isolation of the fungus into pure culture. However, incubation of lesions may be required to induce development of sporangia. Our PCR-RFLP method has been tested successfully on small numbers of sporangia (26b).
The results shown in Fig. 4 demonstrate that modified long PCR amplification can be used as a preliminary search for restriction polymorphisms. The advantage of amplifying a longer fragment lies in the increased probability of locating polymorphic sites. Unfortunately, with the combinations of restriction enzymes and DNA samples tested here, polymorphisms were not detected. However, a similar method could be used to localize the mutations and/or insertions characteristic of mitochondrial haplotypes C to F described by Goodwin (17). We have also used a similar long PCR approach to amplify the 6-kb sequence of the InterGenic Spacer region (IGS) of the rDNA locus and digestion with RsaI to detect polymorphisms (data not shown).
Over the past decade, RFLPs have been exploited to analyze the mitochondrial genomes of various Phytophthora species (e.g., references 7, 8, 12, 13, 28). However, in all cases it was necessary to purify mtDNA by a time-consuming method of cesium chloride centrifugation. In the present study, PCR primers, originally designed to amplify mtDNA from P. infestans, were found to be effective for other Phytophthora species. In particular, P1 and P3 permitted amplification across a wide range of species. Although these primers were tested on only one isolate of each species here, it is anticipated that they will be of general use for diagnostic assays.
Our aim in this study was to provide a simple, robust system whereby mtDNA haplotypes could be characterized according to a unified system which was easily comparable between laboratories. The simplicity of the method described combined with the wide availability of facilities for conducting PCR in most laboratories should mean that this and similar fingerprinting techniques can be used by those with little previous experience of molecular biology. Informal communication has already resulted in this method being used in a number of laboratories in several countries studying populations of P. infestans. We would be happy to comply with requests for moderate quantities of these primer pairs (along with a more detailed protocol).
ACKNOWLEDGMENTS
The financial support of the United Kingdom Government Overseas Development Administration (Department for International Development) is gratefully acknowledged.
Maintenance and culture of exotic isolates were conducted under the conditions of MAFF license no. PHF 1571/1156/84. We are grateful to the following people for supplying DNA or cultures of P. infestans and other species: Greg Forbes (CIP, Peru); Yuri Dyakov, Svetlana Bagirova, and Julia Maleeva (Moscow State University), Lowell Black and Yi-Hsiou Huang (AVRDC, Tainan, Taiwan), Remi Non-Wondim (AVRDC, Tanzania), K. M. Zhang, (Hainan, China), Vera Sanchez (CATIE, Costa Rica), and David Cooke (SCRI, Dundee, Scotland). We are grateful to Geoffrey Hall of the International Mycological Institute for assistance with importation of isolates, and we are indebted to B. F. Lang of the University of Montreal for access to unpublished sequence data for the P. infestans mitochondrial genome. Also, we thank the following persons for useful discussions: Jenny Day, Sue Whittaker, Nick Pipe, Richard Shattock, and David Cooke.
REFERENCES
- 1.Andrivon D, Beasse C, Laurent C. Characterization of isolates of Phytophthora infestans collected in northwestern France from 1988 to 1992. Plant Pathol. 1994;43:471–478. [Google Scholar]
- 2.Barnes W. PCR amplification of up to 35kb DNA with high fidelity and high yield from bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter D A, Archer S A, Buck K W, Shaw D S, Shattock R C. DNA polymorphism in Phytophthora infestans: the UK experience. In: Lucas J A, Shattock R C, Shaw D S, Cooke L R, editors. Phytophthora. Cambridge, United Kingdom: Cambridge University Press; 1991. pp. 272–294. [Google Scholar]
- 4.Carter D A, Archer S A, Buck K W, Shaw D S, Shattock R C. Restriction fragment length polymorphisms of mitochondrial DNA of Phytophthora infestans. Mycol Res. 1990;94:1123–1128. [Google Scholar]
- 5.Chesnick J M, Tuxbury K, Coleman A, Burger G, Lang B F. Utility of the mitochondrial nad4l gene for algal and protistan phylogenetic analysis. J Phycol. 1996;32:452–456. [Google Scholar]
- 6.Day J P, Shattock R C. Aggressiveness and other factors relating to displacement of populations of Phytophthora infestans in England and Wales. Eur J Plant Pathol. 1997;103:379–391. [Google Scholar]
- 7.de Cock A W A M, Karlovsky P, Jahnke K-D, Bahnweg G. Similarities in restriction fragment patterns of mitochondrial DNAs of Phytophthora species strongly depend on the restriction enzyme used due to heterogeneous base distribution and sequence conservation. Fungal Genet Biol. 1996;20:36–42. doi: 10.1006/fgbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 8.de Cock A W A M, Neuvel A, Bahnweg G, de Cock J C J M, Prell H H. A comparison of morphology, pathogenicity and restriction fragment patterns of mitochondrial DNA among isolates of Phytophthora porri Foister. Neth J Plant Pathol. 1992;98:277–289. [Google Scholar]
- 9.Doyle J J, Doyle J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 10.Drenth A, Goodwin S B, Fry W E, Davidse L C. Genotypic diversity of Phytophthora infestans in the Netherlands revealed by DNA polymorphisms. Phytopathology. 1993;83:1087–1092. [Google Scholar]
- 11.DuTeau N M, Leslie J F. A simple, rapid procedure for the isolation of DNA for PCR from Gibberella fujikuroi (Fusarium section Liseola) Fungal Genet Newsl. 1991;38:72. [Google Scholar]
- 11a.Flier, W. Personal communication.
- 12.Förster H, Kinscherf T G, Leong S A, Maxwell D P. Estimation of relatedness between Phytophthora species by analysis of mitochondrial DNA. Mycologia. 1988;80:466–478. [Google Scholar]
- 13.Förster H, Oudemans P, Coffey M D. Mitochondrial and nuclear DNA diversity within six species of Phytophthora. Exp Mycol. 1990;14:18–31. [Google Scholar]
- 14.Fry W E, Goodwin S B, Matuszak J M, Spielman L J, Milgroom M G. Population genetics and intercontinental migrations of Phytophthora infestans. Annu Rev Phytopathol. 1992;30:107–129. [Google Scholar]
- 15.Fry W E, Goodwin S B. Re-emergence of potato and tomato late-blight in the United States. Plant Dis. 1997;81:1349–1357. doi: 10.1094/PDIS.1997.81.12.1349. [DOI] [PubMed] [Google Scholar]
- 16.Gallegly M E, Galindo J. Mating types and oospores of Phytophthora infestans in nature in Mexico. Phytopathology. 1958;48:274–277. [Google Scholar]
- 17.Goodwin S B. DNA polymorphisms in Phytophthora infestans: the Cornell experience. In: Lucas J A, Shattock R C, Shaw D S, Cooke L R, editors. Phytophthora. Cambridge, United Kingdom: Cambridge University Press; 1991. pp. 256–271. [Google Scholar]
- 18.Goodwin S B, Drenth A, Fry W E. Cloning and genetic analyses of two highly polymorphic, moderately repetitive nuclear DNAs from Phytophthora infestans. Curr Genet. 1992;22:107–115. doi: 10.1007/BF00351469. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin S B, Schneider R E, Fry W E. Use of cellulose-acetate electrophoresis for rapid identification of allozyme genotypes of Phytophthora infestans. Plant Dis. 1995;79:1181–1185. [Google Scholar]
- 20.Goodwin S B, Spielman L J, Matuszak J M, Bergeron S N, Fry W E. Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology. 1992;82:955–961. [Google Scholar]
- 21.Goodwin S B. The population genetics of Phytophthora. Phytopathology. 1997;87:462–473. doi: 10.1094/PHYTO.1997.87.4.462. [DOI] [PubMed] [Google Scholar]
- 22.Griffith G W, Snell R, Shaw D S. Late blight (Phytophthora infestans) on tomato in the tropics. Mycologist. 1995;9:87–89. [Google Scholar]
- 23.Kato M, Mizubuti E S, Goodwin S B, Fry W E. Sensitivity to protectant fungicides and pathogenic fitness of clonal lineages of Phytophthora infestans in the United States. Phytopathology. 1997;87:973–978. doi: 10.1094/PHYTO.1997.87.9.973. [DOI] [PubMed] [Google Scholar]
- 24.Klimczak L J, Prell H H. Isolation and characterisation of mitochondrial DNA of the oomycetous fungus Phytophthora infestans. Curr Genet. 1984;8:323–326. doi: 10.1007/BF00419731. [DOI] [PubMed] [Google Scholar]
- 25.Koh Y J, Goodwin S B, Dyer A T, Cohen B A, Ogoshi A, Sato N, Fry W E. Migration and displacements of Phytophthora infestans populations in East Asian countries. Phytopathology. 1994;84:922–927. [Google Scholar]
- 26.Lebreton L, Laurent C, Andrivon D. Evolution of Phytophthora infestans populations in the two most important potato production areas of France during 1992–96. Plant Pathol. 1998;47:427–439. [Google Scholar]
- 27.Maleeva, J. Personal communication.
- 28.Möller E M, de Cock A W A M, Prell H H. Mitochondrial and nuclear DNA restriction enzyme analysis of the closely related Phytophthora species P. infestans, P. mirabilis and P. phaseoli. J Phytopathol. 1993;139:309–321. [Google Scholar]
- 29.Paquin B, Laforest M J, Forget L, Roewer I, Wang Z, Longcore J, Lang B F. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- 30.Sujkowski L S, Goodwin S B, Dyer A T, Fry W E. Increased genotypic diversity via migration and possible occurrence of sexual reproduction of Phytophthora infestans in Poland. Phytopathology. 1994;84:201–207. [Google Scholar]
- 31.Tooley P W. Use of uncontrolled freezing for liquid nitrogen storage of Phytophthora species. Plant Dis. 1988;72:680–682. [Google Scholar]
- 32.Tooley P W, Bunyard B A, Carras M M, Hatziloukas E. Development of PCR primers from internal transcribed spacer region 2 for detection of Phytophthora species infecting potatoes. Appl Environ Microbiol. 1997;63:1467–1475. doi: 10.1128/aem.63.4.1467-1475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tooley P W, Therrien C D. Cytophotometric determination of the nuclear DNA content of 23 Mexican and 18 non-Mexican isolates of Phytophthora infestans. Exp Mycol. 1987;11:19–26. [Google Scholar]
- 34.Trout C L, Ristaino J B, Madritch M, Wangsomboondee T. Rapid detection of Phytophthora infestans in late blight-infected potato and tomato using PCR. Plant Dis. 1997;81:1042–1048. doi: 10.1094/PDIS.1997.81.9.1042. [DOI] [PubMed] [Google Scholar]
- 35.Whittaker S L, Assinder S J, Shaw D S. Inheritance of mitochondrial DNA in Phytophthora infestans. Mycol Res. 1994;98:569–575. [Google Scholar]