Abstract
Wound healing has been extensively studied through the lens of inflammatory disorders and cancer, but limited attention has been given to hematophagy and arthropod-borne diseases. Hematophagous ectoparasites, including ticks, subvert the wound healing response to maintain prolonged attachment and facilitate blood-feeding. Here, we unveil a strategy by which extracellular vesicles (EVs) ensure blood-feeding and arthropod survival in three medically relevant tick species. We demonstrate through single cell RNA sequencing and murine genetics that wildtype animals infested with EV-deficient Ixodes scapularis display a unique population of keratinocytes with an overrepresentation of pathways connected to wound healing. Tick feeding affected keratinocyte proliferation in a density-dependent manner, which relied on EVs and dendritic epidermal T cells (DETCs). This occurrence was linked to phosphoinositide 3-kinase activity, keratinocyte growth factor (KGF) and transforming growth factor β (TGF-β) levels. Collectively, we uncovered a strategy employed by a blood-feeding arthropod that impairs the integrity of the epithelial barrier, contributing to ectoparasite fitness.
Keywords: Arthropod Vectors, Ticks, Tick-Borne Diseases, Skin, Wound Healing
Introduction
Ectoparasitic arthropods obtain their nourishment by feeding on a vertebrate host, providing an avenue for microbial transmission during hematophagy (Sonenshine & Roe, 2014). In North America, tick encounters account for approximately 77% of all arthropod-borne diseases, with most interactions attributed to Ixodes, Amblyomma and Dermacentor species (Eisen, 2022; Rosenberg et al., 2018). Notably, Ixodes scapularis is an arthropod vector of several human illnesses, including Lyme disease (Eisen, 2022). The Lone star tick Amblyomma americanum transmits bacteria that cause ehrlichiosis, while the American dog tick Dermacentor variabilis and Dermacentor andersoni carry pathogens associated with Rocky Mountain spotted fever and tularemia (Eisen, 2022; Rosenberg et al., 2018).
The unique microenvironment generated at the skin interface during a tick bite is conducive to arthropod hematophagy and pathogen transmission (Wikel, 2013). Unlike other blood-feeding arthropods, the development of hard ticks incorporates a series of events which necessitate long-term attachment to the skin (Sonenshine & Roe, 2014). This prolonged disruption of the host physical barrier poses a new challenge for tick survival as defense mechanisms are engaged. Tick saliva has been shown to be critical to subvert inflammation, blood coagulation, and nociception and antagonizes host immunity to enable attachment to the skin (Esteves et al., 2017; Francischetti et al., 2009; Kazimirova & Stibraniova, 2013; Kotal et al., 2015; Kotsyfakis et al., 2007; Kramer et al., 2011; Poole et al., 2013; Ribeiro et al., 1992; Ribeiro et al., 1985; Ribeiro et al., 1988; Simo et al., 2017; Valenzuela et al., 2002).
Observations across various species further demonstrate a conserved ability whereby tick effectors perturb pro-inflammatory mediators responding to ectoparasite feeding (Bakshi et al., 2019; Dickinson et al., 1976; Esteves et al., 2017; Karim & Ribeiro, 2015). Such antagonism of host responses has been implicated in the dissemination and persistence of vector-borne pathogens (Chen et al., 2014; Kotsyfakis et al., 2010; Oliva Chávez et al., 2021).
As the largest organ in the body, the skin serves as the first line of defense against arthropod infestation and pathogen transmission (Eyerich et al., 2018; Glatz et al., 2017; Kabashima et al., 2019). The skin is comprised of three primary layers: the outermost epidermis, the underlying dermis, and the hypodermis or subcutaneous fat (Eyerich et al., 2018). The complex architecture and specialized cell populations comprising the skin affords the mammalian host a protective barrier against environmental and microbial threats, in addition to aiding in thermoregulation and prevention of trans-epidermal water loss (Kabashima et al., 2019; Proksch et al., 2008). During injury, such as laceration of the skin by a tick hypostome, various immune and sentinel cells are activated and release soluble factors that prompts the highly complex and intricate process of wound healing (Singer & Clark, 1999). Proper wound healing requires a high degree of coordination to orchestrate a response to an insult, which is broadly comprised of four overlapping stages: hemostasis, inflammation, proliferation, and tissue remodeling (Peña & Martin, 2024).
Little attention has been paid to the latter phases of skin healing during tick hematophagy, including proliferation, which drives the process of re-epithelialization. Re-epithelialization, which is largely dependent on the proliferation of keratinocytes, culminates in the regeneration of the epidermal-dermal junction and restoration of barrier integrity (Pastar et al., 2014; Rousselle et al., 2019). Keratinocytes function as structural cells which serve as the outermost layer in mammals (Pastar et al., 2014; Rousselle et al., 2019). Keratinocytes have also been recognized as sentinels, facilitating crosstalk with immune cells, and partaking in the initiation of the wound healing response upon injury (Piipponen et al., 2020). Dysfunction in keratinocyte-mediated closure has been reported in chronic wounds indicating their crucial role in skin homeostasis (Pastar et al., 2014; Wikramanayake et al., 2014).
The current paradigm at the tick-skin interface is that salivary molecules are deposited within the dermis during feeding, where they actively regulate the cutaneous response to an insult (Bernard et al., 2020; Wikel, 2013). The impact of tick feeding on the epidermis, which interfaces with the external environment, has been mostly neglected. The significance of the epidermis in countering tick infestation was documented in the late 1970s wherein Langerhans cells were shown to respond to salivary antigens (Allen et al., 1979). We also implicated extracellular vesicles (EVs) originating from the tick I. scapularis in promoting tick fitness and generating distinct outcomes of pathogen transmission in mammals. This mechanism was accomplished through the tick SNARE protein Vamp33 and epidermal γδ T cells (Oliva Chávez et al., 2021).
In this article, we combined single cell RNA sequencing (scRNA-seq), murine genetics, intravital microscopy and flow cytometry to reveal that tick EVs disrupt intraepithelisal homeostasis. We discovered a unique population of keratinocytes in wildtype animals with an overrepresentation of pathways connected to wound healing during a bite from EV-deficient ticks. We further underpinned this biological network by demonstrating that tick EVs impacted epithelial proliferation through the disruption of phosphoinositide 3-kinase (PI3K) activity, keratinocyte growth factor (KGF) and transforming growth factor β (TGF-β). Collectively, we illustrate a tick-induced interference of wound healing via the skin epidermis, contributing to the process of arthropod hematophagy.
Results
Tick extracellular vesicles enable arthropod fitness.
We previously observed that EVs derived from I. scapularis enabled hematophagy (Oliva Chávez et al., 2021). We sought to corroborate our findings in other tick species of public health importance and assess the impact of EVs on arthropod fitness. Total genetic ablation in ticks remains beyond current technical capabilities because editing through clustered regularly interspaced short palindromic repeats (CRISPR) has only been applied to score morphological phenotypes, but not signaling pathways (Sharma et al., 2022). Thus, we silenced the expression of the vesicle associated membrane protein 33 (vamp33) through RNA interference (RNAi) to study the effect of tick EVs.
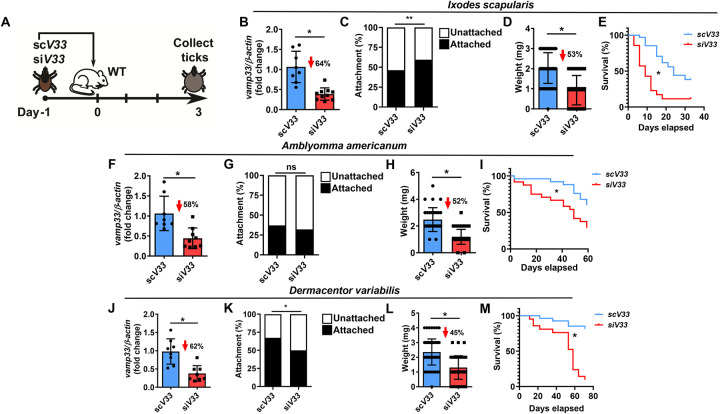
We designated arthropods that had reduced vamp33 gene expression as siV33, EV-deficient ticks, and the scramble control treatment as scV33, EV-sufficient ticks. SiV33 and scV33 microinjected nymphs were placed on C57BL/6 mice and allowed to feed for 3 days (Fig. 1A). On day 3, I. scapularis were assessed for efficiency of vamp33 silencing, attachment, and collected for weight and post-feeding survival (Fig. 1B–E). We observed a statistically significant difference in attachment between siV33 and scV33 nymphs (Fig. 1C) compared to our previous evaluation (Oliva Chávez et al., 2021). Diminished feeding was also measured for EV-deficient ticks as demonstrated by a 53% reduction in tick weight (Fig. 1D). Interrupted feeding in I. scapularis led to reduced survival post-detachment (Fig. 1E). An EV-associated fitness cost upon vamp33 silencing was observed in all three clinically relevant tick species (Fig. 1B–M). A notable exception was the lack of phenotypic differences in attachment for A. americanum compared to I. scapularis and D. variabilis (Fig. 1C, G, and K). Collectively, these findings offer the prospect of a cross-species integrated management for mammalian infestation despite the distinct tick phylogeny.
Figure 1: Tick EVs affect hematophagy and survival.
(A) Graphical illustration of experimental design. (B-M) Vamp33 siRNA (siV33) (red) or vamp33 scramble control (scV33) (blue) microinjected nymphs were placed on C57BL/6 mice and allowed to feed for 3 days. On day 3, ticks were harvested and assessed for fitness measurements. Efficiency of Vamp33 silencing and tick attachment, weight, and survival curves for (B-E) I. scapularis, (F-I) A. americanum and (J-M) D. variabilis. Graphs represent at least three independent experiments combined. Statistical significance shown as *p<0.05, **p<0.01, ns = not significant was assessed by t test (B, F, J); Fisher’s exact test (C, G, K); Mann Whitney test (D, H, L); and Log-rank (Mantel-Cox) test (E, I, M).
Tick EVs alter epidermal immune surveillance.
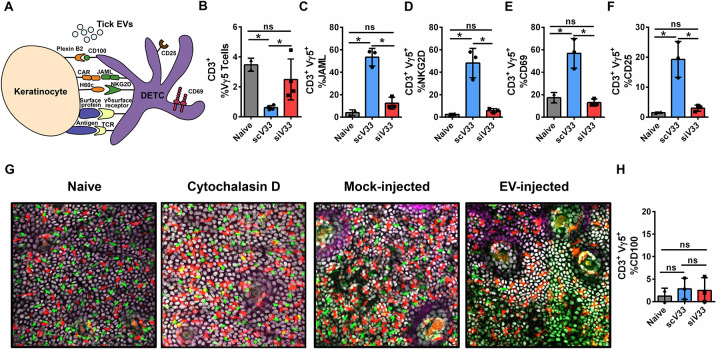
Recently, we reported that tick EVs within saliva affect the frequency of dendritic epidermal T cells (DETC) and alter the cytokine and chemokine milieu of the skin (Oliva Chávez et al., 2021). DETC surveillance of keratinocytes via various cell surface receptors is critical in a wounding response, leading to the activation and recruitment of immune cells, stimulation of keratinocytes for proliferation and survival, and anti-microbial responses (Jameson et al., 2002; Jameson et al., 2004; Keyes et al., 2016; Macleod & Havran, 2011; Sharp et al., 2005) (Fig. 2A). This crosstalk and surveillance between DETC and keratinocytes led us to reason that tick EVs might not solely impact DETCs, but also likely influence the most abundant epidermal cell, the keratinocyte. Hence, we allowed EV-deficient (siV33) and EV-sufficient (scV33) I. scapularis nymphs to feed on mice for 3 days and collected the skin biopsy for flow cytometry evaluation (Supplementary Fig. 1). We observed a decrease in DETC frequency during scV33 tick feeding on mice compared to naïve skin (Fig. 2B). Conversely, DETC frequency remained at homeostatic levels after impairment of tick EVs (siV33) and ectoparasite feeding on murine animals (Fig. 2B).
Figure 2: Tick EVs alter epidermal immune surveillance.
(A) Schematic representation of the DETC-keratinocyte crosstalk at the skin epidermis. (B-F, H) I. scapularis scV33 (blue) or siV33 (red) ticks were placed on C57BL/6 mice and allowed to feed for 3 days. On day 3, biopsies were taken from the skin at the bite site and compared to the naïve treatment (gray). (B) DETC (Vγ5), (C) JAML, (D) NKG2D, (E) CD69, (F) CD25, and (H) CD100 cells were assessed by flow cytometry. Graphs represent 1 of 3 independent experiments. (G) Epidermis containing Langerhans cells (red), DETCs (green), and keratinocytes (white) imaged on day 3 after injection with phosphate buffered saline (PBS - mock) or EV (4×107 particles) into the mouse ear. Cytochalasin D (100 μg) was applied topically on the mouse ear every 24 hours for 2 days to induce DETC rounding as a positive control. Langerhans cells, DETCs and epithelial cells were simultaneously visualized in the huLangerin-CreER; Rosa-stop-tdTomato; CX3CR1-GFP+/−; K14-H2B-Cerulean mouse strain. Cre expression was induced with an intraperitoneal injection of tamoxifen (2 mg). Images from one out of three independent experiments. Statistical significance shown as *p<0.05, ns = not significant. Data are presented as a mean with standard deviation. Significance was measured by One-way ANOVA followed by Tukey’s post hoc test.
DETCs exhibit a dendritic shape that allows for continuous surveillance of neighboring keratinocytes through various receptor-ligand interactions (Jameson et al., 2002; Witherden et al., 2012) (Fig. 2A). Upon tissue damage, stressed keratinocytes upregulate ligands and antigens that stimulate DETCs in a non-major histocompatibility complex (MHC)-restricted manner (Havran et al., 1991). Activated DETCs will then alter their morphology by retracting dendrites and assuming a rounded configuration to facilitate migration to the site of injury (Jameson et al., 2002; Nielsen et al., 2017). To determine the possible role of keratinocytes during tick feeding, we assessed the DETC co-stimulatory markers that facilitate immune surveillance. We observed an elevated co-receptor frequency among DETCs found at the skin interface where EV-sufficient ticks fed on mice, including the junctional adhesion molecule-like (JAML) and the C-type lectin-receptor NKG2D (also known as KLRK1) (Girardi et al., 2001; Whang et al., 2009) (Fig. 2C–D). Similar findings were also observed for the activation markers CD69 and CD25 (Fig. 2E–F). Conversely, JAML, NKG2D, CD69 and CD25 were not upregulated in the bite of EV-deficient ticks during murine feeding (Fig. 2C–F).
Morphologically, the hallmark of DETC activation is the conversion of a dendritic to a rounded morphology that facilitates intraepidermal migration, a phenomenon that is partially regulated by CD100 signaling (Thelen & Witherden, 2020; Witherden et al., 2012). To capture morphological changes in DETCs, we employed intravital microscopy of EV injection into the ear of a triple-reporter mouse model. Intravital microscopy of EV injection into the ear of this mouse model revealed that tick EVs did not promote rounding of DETCs, as compared to the positive control cytochalasin D (Fig. 2G). Supporting epidermal intravital imaging findings, expression of CD100 was not altered during a tick bite regardless of the EV status (Fig. 2H). Altogether, these findings provided evidence that tick EVs functionally alter immune surveillance of the epidermal niche by DETCs.
ScRNA-seq characterization of epidermal cells during tick feeding.
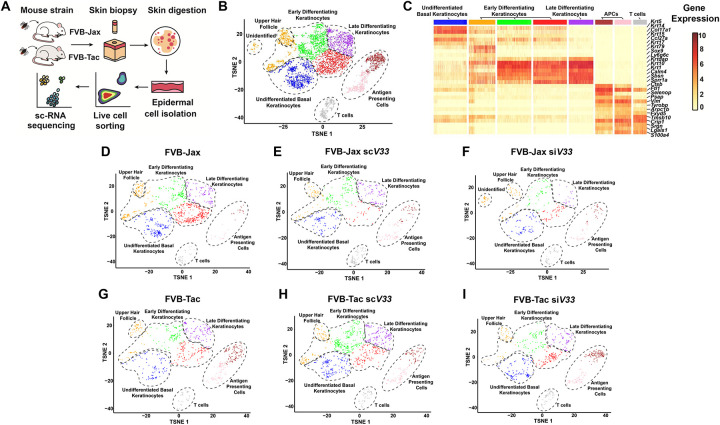
Given the functional perturbations in DETCs during tick feeding, and the well documented importance of the DETC-keratinocyte crosstalk during wounding, we hypothesized that the epidermal healing circuitry is likely being altered during tick feeding. To evaluate this hypothesis, we utilized scRNA-seq to analyze the impact of tick EVs on the epidermal immune environment in both DETC-deficient (FVB-Tac) and DETC-sufficient (FVB-Jax) mice three days after tick feeding. FVB-Tac mice are depleted of functional DETCs due to a failure of thymic selection because of a natural mutation of the skint1 gene (Barbee et al., 2011; Boyden et al., 2008; Lewis et al., 2006). Skin punch biopsies were obtained from the bite site, and the epidermis was enzymatically separated from the dermis. Live cells were sorted by fluorescence activation and libraries were generated for Illumina sequencing (Fig. 3A).
Figure 3: Epidermally-enriched scRNA-seq of the tick bite site.
(A) Overview of the experimental design. ScV33 and siV33 I. scapularis nymphs were placed on FVB-Jackson (FVB-Jax) or FVB-Taconic (FVB-Tac) mice and fed for 3 days. Skin biopsies at the bite site were digested with dispase and collagenase for epidermal cell isolation. Cells were sorted and prepared for scRNA-seq. (B) Composite tSNE plot of keratinocyte, T cell and antigen presenting cells in FVB-Jax and FVB-Tac mice in the presence or absence of I. scapularis nymphs microinjected with scV33 or siV33. tSNE plot represents 5,172 total cells following filtration as described in the materials and methods. (C) Heatmap depicting expression of the top 5 marker genes present in clusters from the epidermally enriched tSNE plot clusters (as shown in B). (D-I) Individual tSNE plots separated by mouse strain (FVB-Jax or FVB-Tac) in the presence or absence of I. scapularis nymphs microinjected with scV33 or siV33.
Our analysis encompassed approximately 20,640 cells, with an average of 88,027 reads. Our initial investigation resulted in 23 clusters (Supplementary Dataset 1). Next, we applied a fixed threshold to retain cells with more than 2500 UMIs (Supplementary Fig. 2A–B) and applied the DoubletFinder R package to predict doublets (Supplementary Fig. 2C–D). We identified 10 distinct groups of cells through an analysis of marker genes within each cluster relative to the entire dataset (Supplementary Dataset 2). Keratinocytes, T cells, fibroblasts and endothelial cells were observed in our scRNA-seq results (Supplementary Fig. 2D). The presence of dermal clusters in our study was likely due to an incomplete epidermal-dermal border separation during the enzymatic dissociation of skin biopsies. Thus, we subjected keratinocytes, T cells, and antigen-presenting cells (APCs) to a second round of clustering (Supplementary Dataset 3). This dataset revealed a total of 8 clusters visualized in t-distributed stochastic neighbor embedding (t-SNE) (Fig. 3B) for a total of 5,172 total cells with a median UMI count of 13,910 per cell.
Throughout the process of differentiation, keratinocytes express different types of keratins, including keratins (Krt) 1, 5, 10, and 14 (Fuchs, 1993). Elevated levels of Krt5 and Krt14 expression enabled the recognition of undifferentiated cells residing within the basal layer of the epidermis (Fig. 3C, Supplementary Dataset 4). Krt1, Krt10, and Involucrin were used to discern early and late-stage differentiation of keratinocytes (Fig. 3C, Supplementary Fig. 3, Supplementary Dataset 4). APCs and T cells were identified by the T cell receptor alpha constant (Trac), the T cell receptor delta constant (Trdc), and the histocompatibility class II antigen (H2-Aa) (Supplementary Dataset 3, Supplementary Table 2). The mouse epidermis harbors hair follicles with distinct physiological functions (Joost et al., 2018; Joost et al., 2016). Our dataset only accounted for compartments in anatomical proximity to the epidermis (Supplementary Fig. 4, Supplementary Table 2).
We then determined the percent distribution of interfollicular epidermal cells per treatment. In the skin biopsy where ticks fed on immune intact mice (FVB-Jax scV33 and FVB Jax siV33), we observed a decrease in keratinocytes and an overrepresentation of T cells and APCs compared to the naïve skin (Supplementary Fig. 5A–B, Supplementary Dataset 5). A similar effect was not observed when ticks fed on the skin of DETC-deficient mice, presumably due to the diminished wound healing capacity in FVB-Tac animals (Keyes et al., 2016). We confirmed the depletion of DETCs in the epidermis of FVB-Tac mice. Gene expression of Trdv4 in the T cell cluster, which encodes for the receptor Vδ1 in DETCs, was reduced in FVB-Tac compared to the FVB-Jax mouse strain (Supplementary Fig. 5C). Notably, partitioning of epidermal clusters by experimental conditions revealed an unidentified keratinocyte population found solely when EV-deficient ticks fed on FVB-Jax mice (Fig. 3D–I, Supplementary Dataset 3). The presence of this distinct cluster raised the hypothesis that EVs might exert an influence on keratinocytes within the context of DETCs, given its absence in FVB-Tac mice.
Tick EVs impact a keratinocyte population with a prominent wound healing signature.
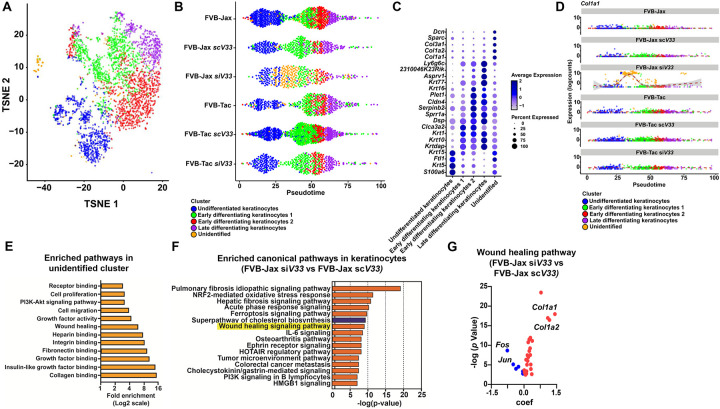
The emergence of this unique keratinocyte population responding to siV33 tick feeding prompted us to further investigate their role by subjecting these cells to a subsequent round of clustering. This examination revealed keratinocyte populations at various differentiated states and highlighted the presence of an unidentified epidermal population (Fig. 4A, Supplementary Dataset 6). Next, we relied on pseudotime to align keratinocytes along an inferred developmental trajectory based on their expression profile (Fig. 4B). Gene expression signatures mirrored the sequence of differentiation, starting with markers associated with undifferentiated basal states to terminally differentiated keratinocytes (Supplementary Fig. 6). We identified a unique keratinocyte population present along the pseudotime axis of the condition where EV deficient ticks fed on FVB-Jax mice (FVB-Jax siV33), setting it apart from the other treatments (Fig. 4B).
Figure 4: Impact of tick EVs on wound healing circuitry.
(A) Composite tSNE plot of keratinocytes in FVB-Jax and FVB-Tac mice in the presence or absence of I. scapularis nymphs microinjected with scV33 or siV33. (B) Cells colored by clusters originated from the keratinocyte tSNE plot (as shown in A) ordered across pseudotime (x-axis) for naïve, scV33-, and siV33-tick bites of FVB-Jax and FVB-Tac mice. (C) Dot plot of the top 5 marker genes present in the keratinocyte clusters (as shown in A). Average gene expression is demarked by the intensity of color. Percent of gene expression within individual clusters is represented by the dot diameter. (D) Expression of Col1a1 across treatments ordered across pseudotime (x-axis) for naïve, scV33-, and siV33-tick bites of FVB-Jax and FVB-Tac mice. (E) Enriched pathways in the unidentified cell cluster based on functional annotation in DAVID. Fold enrichment is indicated in a Log2 scale. *p value and false discovery rate (FDR)<0.05 were set as threshold. KEGG, GO and InterPro were used as reference annotation databases. (F) Ingenuity pathway analysis comparing keratinocytes of skin biopsies from FVB-Jax siV33 to FVB-Jax scV33. Blue denotes pathways predicted to be inhibited (negative z-score) whereas orange indicates pathways predicted to be activated (positive z-score) based on default parameters. Differential expression datasets were assessed for canonical pathway analysis. Results are shown in a −log (p-value) scale. *p value and FDR< 0.05 were set as threshold. (G) Volcano plot of genes representing the wound healing signaling pathway in keratinocytes of FVB-Jax siV33 compared to FVB-Jax scV33 datasets (highlighted in yellow; F). Blue denotes decrease whereas red indicates increase in the coefficient (coef) of expression.
The heterogeneity of keratinocytes is crucial for various functions, both during homeostasis and in response to external stimuli (Rice & Rompolas, 2020). Their transcriptional program has been recently explored to elucidate how different populations aid in the coordination of broader cellular circuits (Joost et al., 2020; Joost et al., 2018; Joost et al., 2016). Thus, we unraveled the transcriptional program employed by this unique keratinocyte population where EV deficient ticks fed on FVB-Jax mice. We computationally separated keratinocyte populations according to their respective experimental conditions (Supplementary Fig. 7). Then, we assessed marker genes in the keratinocyte population where EV deficient ticks fed on FVB-Jax mice, which revealed elevated expression of Col1a1, Col1a2, and Col3a1 (Fig. 4C). Evaluation of the marker gene Col1a1 across pseudotime further underscored the distinct transcriptional program of this unique keratinocyte subcluster (Fig 4D). Altogether, these results suggested an increase in the collagen production by this keratinocyte population in response to feeding of EV-deficient ticks.
Pathway enrichment analysis of all significant marker genes in the unidentified keratinocyte population revealed an overrepresentation of genes associated with the wound healing circuitry, including growth factor, collagen, fibronectin and heparin binding, and phosphoinositide 3-kinase (PI3K) activity (Fig. 4E, Supplementary Dataset 7). These molecules have been implicated in keratinocyte function during wound healing, primarily by enhancing keratinocyte proliferation and migration to support re-epithelialization and tissue repair (Bártolo et al., 2022; Matsuura-Hachiya et al., 2018; Misiura et al., 2020). Our findings suggested that a unique keratinocyte population with a prominent wound healing signature was selectively responding to EV-deficient ticks during hematophagy. To make a comparison between EV-deficient and EV-sufficient ticks in the murine skin, we subjected keratinocytes to a differential expression analysis and assessed enriched pathways through ingenuity pathway analysis (IPA). Notably, we observed a wound healing signature in the skin of DETC-sufficient mice fed with EV-deficient ticks, which was not detected in animals deficient for DETCs (Fig. 4F).
Further inspection of differentially expressed genes annotated for wound healing revealed a decrease in transcript levels for Fos and Jun and an increase of expression for Col1a1 and Col1a2 (Fig. 4G, Supplementary Dataset 8). Fos and Jun are subunits of AP-1, which is important for epithelial proliferation and differentiation (Angel et al., 2001; Li et al., 2003). Conversely, collagens have various roles during all stages of wound healing, aiding in the regulation of the wound healing response, reinforcing barrier integrity and facilitating the stratification of epidermal layers (Matsuura-Hachiya et al., 2018). Collectively, tick EVs impaired wound healing through specific molecular pathways in keratinocytes.
Tick EVs interfere with keratinocyte proliferation.
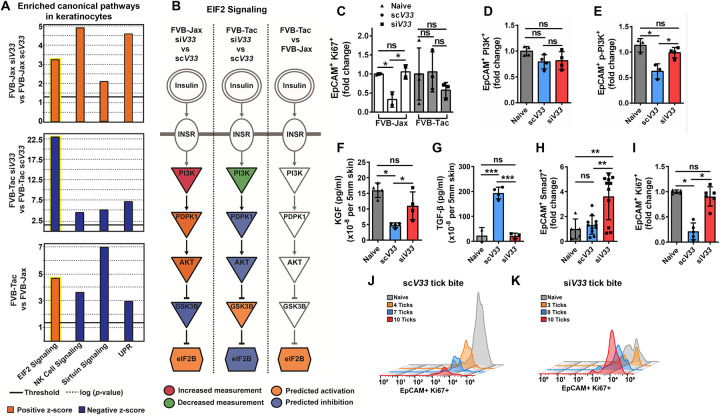
To understand how tick EVs influenced wound healing in keratinocytes, we then evaluated molecular networks altered in the epidermis of DETC-sufficient and DETC-deficient mice. We performed a similar analysis in naive animals to exclude confounding effects originated from genetic differences occurring between these two strains. Four pathways were identified: eukaryotic Initiation Factor 2 (EIF2), natural killer (NK) cell, sirtuin signaling, and the unfolded protein response (UPR) (Fig. 5A, Supplementary Dataset 9). The results obtained concerning NK cell, sirtuin signaling, and the UPR pathways were likely due to the skint1 deficiency in FVB-Tac mice. However, the EIF2 cascade was dependent on tick EVs because the computational prediction occurred regardless of the mouse genetic background (yellow highlight, Fig. 5A).
Figure 5: Tick EVs impact keratinocyte proliferation.
(A) Ingenuity pathway analysis derived from siV33 compared to the bite of scV33 ticks on FVB-Jax or FVB-Tac mice. Canonical pathways predicted to be inhibited (blue, negative z-score) or activated (orange, positive z-score) based on differential expression profile. The solid line indicates the p value significance threshold of 0.05 (−log=1.3). (B) The signaling cascade of EIF2 (highlighted in yellow, A), yielding (→) or inhibitory (┤) arrows. Orange indicates activation whereas blue shows inhibition according to the IPA prediction. Gene expression based on the scRNA-seq experiment is indicated in red (increased) or green (decreased). Gray – denotes no expression or prediction. (C) ScV33 (circle) or siV33 (square) injected I. scapularis nymphs were fed on FVB-Jax (white) or FVB-Tac (gray) mice for 3 days. Biopsies were taken from the skin at the bite site and assessed for EpCAM+ Ki67+ keratinocytes by flow cytometry. (D-H) ScV33 or siV33 ticks fed on C57BL/6 mice for 3 days. Biopsies were taken from the skin at the bite site and processed for flow cytometry analysis. (D) PI3K p85+, and (E) phospho-PI3K p85/p55+. (F) ELISA analysis of KGF levels normalized to total protein per 5 mm skin punch biopsy. (G) ELISA analysis of TGF-β levels normalized to total protein per 5 mm skin punch biopsy. (H) EpCAM+ Smad7+ keratinocytes assessed by flow cytometry. (I) EpCAM+ Ki67+ keratinocytes assessed by flow cytometry. Graph displays proliferation changes within the scV33 or siV33 treatments compared to the naïve skin. (C-I) Significance was measured by One-way ANOVA followed by Tukey’s post hoc test. (J-K) Flow cytometry histogram plots of EpCAM+ Ki67+ keratinocytes. (J) scV33 or (K) siV33 treatments displayed according to the number of ticks bitten per biopsy. X-axis shows fluorescence intensity, and the Y-axis indicates the count of events in the fluorescence channel. All experiments have statistical significance shown as ***p<0.001, **p<0.01, *p<0.05, ns = not significant. Data are presented as a mean with standard deviation.
A granular view of the EIF2 signaling pathway displayed PI3K as part of the biological circuit targeted by tick EVs (Fig. 5B, Supplementary Dataset 10). The PI3K/Akt pathway is important for skin development and wound healing, two processes dependent on keratinocyte proliferation and differentiation (Calautti et al., 2005). Upon injury, keratinocytes adjacent to the wound are quiescent, opting for a migratory phenotype that allows for the initiation of re-epithelialization (Dekoninck & Blanpain, 2019). Conversely, keratinocytes farther from the wound edge undergo a proliferative burst, allowing for the closure of the gap generated by migratory keratinocytes (Aragona et al., 2017). Given that the PI3K/Akt/mTOR pathway has been observed in the proliferative zone and correlates with accelerated wound closure, we reasoned that tick EVs interfered with keratinocyte proliferation. To evaluate this hypothesis, we used the protein Ki-67 and flow cytometry as a readout for proliferative keratinocytes (Supplementary Fig. 8). We observed a significant reduction in keratinocyte proliferation when EV-sufficient ticks fed on wildtype mice (Fig. 5C). However, the effect of keratinocyte proliferation was not observed in the absence of tick EVs (Fig. 5C). As noted above, the impact of tick feeding on keratinocyte proliferation was fully dependent on DETCs. In the absence of DETCs, the observed phenotype for keratinocyte proliferation in EV-sufficient ticks did not occur (Fig. 5C). Taken together, our reductionist approach orthogonally validated our scRNA-seq results, demonstrating that tick EVs decrease keratinocyte proliferation, which is a key step in wound healing.
The genetic constitution of a mouse may lead to substantial alterations in phenotypic traits (Tanner & Lorenz, 2022; Woodworth et al., 2004). We therefore investigated the ability of I. scapularis to interfere with keratinocyte homeostasis in C57BL/6 mice, a more commonly used strain. We ascertained the keratinocyte PI3K status by flow cytometry due to its ability to assess protein expression on limited cell counts. Variation in the total PI3K comparing keratinocyte populations among treatments was not observed (Fig. 5D). However, a decrease in phospho-PI3K-positive keratinocytes was recorded when ticks deficient in EVs fed on C57BL/6 mice (Fig. 5E). Additionally, the bite of I. scapularis ticks reduced levels of the growth factor KGF and increased levels of TGF-β in the skin, compared to the EV-deficient treatment (Fig. 5F–G). These findings correlated to a significant decrease in the frequency of EpCAM+ keratinocytes expressing the negative regulator of TGF-β signaling, Smad7, in skin infested with scV33 ticks compared to siV33 ticks. (Fig. 5H; Supplementary Fig. 8). Next, we observed a significant decline in the frequency of EpCAM+ Ki67+ keratinocytes when EV-sufficient ticks fed on C57BL/6 mice (Fig. 5I). Remarkably, the ability of ticks to impair keratinocyte proliferation was observed in a density-dependent manner. As the number of ticks feeding on C57BL/6 mice increased, the capacity of keratinocytes to proliferate decreased (Fig. 5J). This observation was not recorded in mice infested with ticks deficient for EVs (Fig. 5K). In summary, we uncovered that tick EVs: (i) impacted keratinocyte proliferation; (ii) suppressed KGF and PI3K activity; and (iii) enhanced TGF- β levels, thereby, maintaining successful arthropod hematophagy.
Discussion
Ticks are ancient hematophagous arthropods that co-evolved with their hosts for millions of years (Sonenshine & Roe, 2014). Currently, we have a limited understanding of the mechanisms employed by these ectoparasites to feed on a mammal and enable pathogen transmission. Previously, we implicated Ixodes scapularis EVs in promoting fitness and generating distinct outcomes of pathogen transmission. This was accomplished through the SNARE protein Vamp33 and DETCs (Oliva Chávez et al., 2021). Here, we connect the mammalian wound healing circuitry as a target of tick-mediated host immunomodulation via salivary EVs. Our work connects in vivo proliferation of keratinocytes as a tick EV regulatory process during hematophagy.
We established the importance of EVs as a conserved strategy for arthropod fitness in three medically relevant tick species: I. scapularis, A. americanum, and D. variabilis. We demonstrated that I. scapularis EVs led to a decrease in DETCs at the bite site; yet DETCs present in the skin epidermis displayed upregulated co-stimulatory molecules. During skin injury, DETCs are activated whereby recognition of self-antigens on damaged or stressed keratinocytes allows for the orchestration of a host response (Jameson et al., 2004). Reduction of DETCs at the tick-skin interface may not be a result of cellular migration. DETC migration is facilitated by the conversion of a dendrite to a rounded morphology, which is partially regulated by CD100 signaling (Jameson et al., 2002; Witherden et al., 2012). To capture morphological changes, we employed intravital microscopy in an EV-injected triple-reporter mouse model (Park et al., 2021). Our results suggested that DETCs may not migrate during tick feeding due to the lack of cell rounding and CD100 upregulation. Importantly, DETCs regulate epidermal homeostasis and coordinate a wound healing response together with epidermal cells (Jameson et al., 2002). Collectively, our data suggest that tick EVs alter epidermal immune surveillance by restricting DETC presence and altering cell surface receptor expression, ultimately disrupting epidermal function and promoting hematophagy.
By employing a scRNA-seq approach in a mouse model naturally devoid of DETCs, we characterized the epidermal response to tick infestation. We revealed a unique keratinocyte population when EV-deficient ticks fed on DETC-sufficient mice. The absence of this unique cell population when EV-deficient ticks fed on DETC-deficient mice suggested that EVs may alter keratinocyte function within the context of intraepithelial γδ T cells. Sub clustering and pseudotime analysis of keratinocytes further emphasized the distinct nature of this unique population. Remarkably, this subcluster exhibited an overrepresentation of pathways associated with the wound healing circuitry, including growth factor, collagen and fibronectin binding, and cell proliferation. Moreover, specific biological signatures were associated with down regulation of AP-1 and upregulation of PI3K transcripts in EV-deficient tick fed on DETC-sufficient mice. These molecular circuits have been linked to epithelial proliferation and maintenance of barrier integrity in the skin epidermis (Angel et al., 2001; Jochum et al., 2001; Li et al., 2003; Matsuura-Hachiya et al., 2018).
Consistent with our systems level approach, EV-sufficient ticks fed on DETC-sufficient mice led to a decrease in keratinocyte proliferation. This observation was dependent on the role of DETCs as DETC-deficient mice did not exhibit a decrease in Ki67+ keratinocytes. Wound healing is marked by keratinocyte proliferation and migration to restore barrier function of the epidermis (Dekoninck & Blanpain, 2019). For instance, proliferation was deemed as a necessary step for proper wound closure at the leading edge in the murine tail (Aragona et al., 2017). Conversely, proliferation was judged dispensable for wound closure in the murine ear (Park et al., 2017). Our work was done using the natural site of tick infestation in mammals, the skin of the dorsal neck. Whether proliferation is necessary for migration during a tick bite remains to be determined.
KGF serves as a strong mitogenic factor for both mouse and human keratinocytes, and its overexpression can lead to a hyperproliferative state associated with skin disorders (Ni & Lai, 2020). Upon damage of the skin, activated DETCs secrete KGF to promote wound repair (Jameson et al., 2002). We observed that tick feeding on mice led to decreased KGF levels compared to the EV-deficient treatment. We also postulated that tick EVs interfered with other components of the wound healing biological program, including PI3K and TGF-β. We observed decreased levels of phosphorylated PI3K during EV-sufficient tick feeding on C57BL/6 mice, mirroring the findings from the scRNA-seq studies. Furthermore, we observed an increase in TGF-β release during tick feeding, which correlated to lower Smad7 levels compared to EV-deficient feeding at the bite site. Increased levels of TGF-β in the epidermis have been associated with the inhibition keratinocyte proliferation (Sellheyer et al., 1993). Moreover, TGF-β transduction is mediated by SMAD proteins, with Smad7 acting as a negative regulator of the TGF-β signaling network (Schmierer & Hill, 2007). Thus, we suggest that the observed reduction of DETCs during tick feeding may obstruct the necessary levels of KGF and TGF-β in the epidermis during wounding.
Extracts from tick salivary glands have shown their capability to impede cellular growth in vitro (Hajnicka et al., 2011). Accordingly, we demonstrated in vivo that tick EVs led to a significant reduction in the frequency of Ki67+ keratinocytes. Strikingly, the ability of ticks to impair epithelial cell proliferation was observed in a quantitative-dependent manner. An increase in the number of ticks fed simultaneously at a given skin site, resulted in a decrease of proliferative keratinocytes. In sum, this study unveiled the immunomodulatory effects of tick EVs in the epidermal layer, deviating from the established viewpoint that arthropod saliva mainly influences dermal responses.
Materials and Methods
Reagents and resources
All primers, reagents, resources, and software used in this study, together with their manufacturer’s information and catalog numbers are listed in Supplementary Tables 1 and 3.
Ticks
I. scapularis nymphs were obtained from two independent sources: (1) Dr. Ulrike Munderloh and Dr. Jonathan Oliver at the University of Minnesota; and the (2) tick rearing facility at Oklahoma State University. A. americanum and D. variabilis nymphs were obtained from the tick rearing facility at Oklahoma State University. Partially engorged I. scapularis adult ticks were obtained from Dr. Albert Mulenga and Dr. Adela Oliva Chavez at Texas A&M University. Upon arrival, ticks were maintained in a Percival I-30BLL incubator at 23°C with 85% relative humidity and a 12/10-hours light/dark photoperiod regimen.
Mice
Experiments were performed on C57BL/6, FVB/N Jax, and FVB/N Tac mice. Breeding pairs were purchased from the Jackson Laboratory except FVB/N Tac mice, which were purchased from Taconic Biosciences. All mouse strains were bred at the University of Maryland School of Medicine, unless otherwise indicated. Male mice (7–9 weeks) were used for all experiments. All mouse experiments were approved by the Institutional Biosafety (IBC-00002247) and Animal Care and Use (IACUC numbers 0119012 and 1121014) committees at the University of Maryland School of Medicine and complied with the National Institutes of Health (NIH) guidelines (Office of Laboratory Animal Welfare [OLAW] assurance number A3200–01). huLangerin-CreER;Rosa-stop-tdTomato;CX3CR1-GFP+/−;K14-H2B-Cerulean mice used for intravital microscopy imaging were housed at Michigan State University as described elsewhere (Park et al., 2021) (IACUC number PROTO202300065). To activate DETCs, Cytochalasin D (Sigma-Aldrich, C8273) was delivered topically as previously described (Park et al., 2021). Briefly, Cytochalasin D was dissolved in a 25 mg/ml stock solution in dimethyl sulfoxide (DMSO), and later, the stock solution was diluted 100 times in 100% petroleum jelly (Vaseline; final concentration is 250 μg/ml). One hundred micrograms of the mixture of Cytochalasin D and the petroleum jelly were spread evenly on the skin once every 24 hours for 2 days. A mixture of 100% DMSO in petroleum jelly (1:100) was used as a vehicle control.
RNA interference
siRNAs and scRNAs for vamp33 were designed as previously described (Oliva Chávez et al., 2021). Both siRNAs and scRNAs were synthesized according to the Silencer® SiRNA construction kit (Thermo Fisher Scientific). Primers are described in Supplementary Table 1. Unfed nymphs were microinjected with 60–80 ng of siRNA or scRNA using a Nanoject III (Drummond Scientific Company). Ticks recovered overnight at 23°C with saturated humidity before being placed on respective mice.
EV-depleted media
L15C300 medium was supplemented with 5% FBS (Millipore-Sigma), 5% tryptose phosphate broth (TPB) (BD), 0.1% lipoprotein concentrate (LPC) (MP Biomedicals), 0.25% sodium bicarbonate (Millipore-Sigma), and 25 mM HEPES (Millipore-Sigma). Media was cleared from EVs by ultracentrifugation at 100,000×g for 18 h at 4 °C in a LE-80 ultracentrifuge (Beckman Coulter) with a 60Ti rotor. EV-free media was then passed through a 0.22-μm Millipore Express® PLUS (Millipore-Sigma). The absence of EVs was confirmed by determining the particle size distribution with the NanoSight NS300 (Malvern Panalytical) for nanoparticle tracking analysis (NTA).
Tick salivary gland culture
Salivary gland EVs were purified from ex vivo cultures that originated from partially engorged adult female ticks. Adult I. scapularis females were fed on New Zealand white rabbits for 5–6 days at either Dr. Albert Mulenga or Dr. Adela Oliva Chavez laboratories at Texas A&M University, as previously described (Oliva Chávez et al., 2021). Then, ticks were shipped to the University of Maryland School of Medicine. Partially-fed adult female ticks (90–120) were dissected 1–2 days post-removal. Briefly, midguts, Malpighian tubes, and other organs were removed. PBS was added to samples to avoid desiccation. Salivary glands were dissected and cultured in 24-well cell culture plates (Corning). 10 salivary glands from adult ticks were placed in each well, containing 500 μl of L15C300 EV-free medium supplemented with 1x penicillin/streptomycin (Corning) and 1x Amphotericin B (Gibco). Salivary glands were incubated for 24 h at 34 °C to allow EV secretion.
EV purification
Medium collected from salivary gland cultures were cleared of any live cells by centrifugation at 300 × g for 10 minutes at 4 °C. Dead cells were removed by a second centrifugation at 2,000 × g for 10 minutes at 4 °C. The supernatant was collected, and apoptotic bodies were removed by a third centrifugation at 10,000 × g for 30 minutes at 10°C. The supernatant was filtered through a 0.22-μm Millipore syringe filter (Millipore-Sigma) to reduce the number of EVs >200 nm in size. EVs were pelleted by ultracentrifugation (100,000 × g) for 18 hours at 4 °C. Supernatant was discarded and EVs were resuspended in PBS. EV concentration and sizes were determined using the NanoSight 300 machine (Malvern Panalytical) with the software versions 2.0 or 3.0. The mean of the size generated in the reports was used to calculate the average size of the EVs in each sample. The concentration of proteins in tick EVs was determined using the BCA assay (Thermo Scientific), following the manufacturer’s procedure.
Mouse capsule placement
Capsules made from the upper portion of a snap or screw top tube were adhered to the dorsal neck of each mouse to contain the ticks in one area. This technique is referred to as the capsule-feeding method and was adapted from a previous study (Schoeler et al., 1999). Briefly, capsule adhesive solution was made from 3 parts gum rosin (Sigma-Aldrich) and 1 part beeswax (FisherScience). Mice were anesthetized using isoflurane and shaved between the shoulder blades to the top of the cranium. Capsules were applied with the warmed adhesive and allowed to dry up for 24 hours prior to tick placement. Capsules were sealed with either a glued piece of mesh or a screw top after tick placement. Naïve groups consisted of capsule placement without ticks.
Tick feeding experiments
Microinjected ticks were placed on mice using either the free-feeding or capsule-feeding method and allowed to feed for 3 days. On day 3, ticks were collected, weighed, and either placed in a humidified chamber for survival analysis or frozen at −80°C for RNA purification. To purify the mRNA, ticks were flash-frozen in liquid nitrogen and crushed with small plastic pestles. TRIzol® reagent (200 μl) was added to the crushed tick and RNA was purified using the PureLink™ RNA mini kit. cDNA was synthesized from 50 to 200 ηg (5–10 μl) of RNA using the Verso cDNA synthesis kit (Thermo scientific).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
qRT-PCR was performed to measure gene expression. qRT-PCR was performed with the CFX96 Touch Real-Time PCR Detection 233 System (Biorad). No template controls were included to verify the absence of primer-dimers formation and/or contamination. Reactions on each sample and controls were run in duplicate. Gene expression was determined by relative quantification normalized to the tick actin, using the primers listed in Supplementary Table 1.
Flow cytometry of skin cell populations
I. scapularis nymphs fed on C57BL/6, FVB/N Jax, or FVB/N Tac male mice. On the third day of feeding, mice were euthanized with CO2. A 10- or 5-mm skin punch biopsy was taken while ticks were still attached. Skin samples from un-infested control mice were collected from matching locations. Single cell suspensions were prepared from each skin sample. Briefly, skin samples were cut into small pieces with sterile surgical scissors and placed into round-bottom tubes containing digestion buffer consisting of 90% RPMI-1640 (Quality Biological), 10% Liberase™ TL Research Grade (Roche), and 0.1% DNAse I (Millipore-Sigma). Digestions were carried out for 1 hour and 15 minutes at 37°C with constant shaking. Single cell suspensions were obtained by passing the digested tissues through a 40-μm cell strainer (Corning), homogenizing the tissue with a plunger and flushing cells with wash buffer consisting of PBS and 2 mM EDTA. Cells were centrifuged at 300 × g for 5 minutes at 4 °C, resuspended in 1 ml FACS buffer (PBS containing 1% BSA, 2 mM EDTA, and 0.05% NaN3) or FACS intracellular buffer (PBS containing 1% BSA and 0.05% NaN3). Cell suspensions were placed into a 96-well U-bottom plate and stained with respective antibody panels.
Live and dead cells were discriminated using Zombie Violet Fixable Live Dead stain (BioLegend). Cells were washed with FACS buffer. Cells were then blocked with anti-FcR (CD16-CD32) (BioLegend 156603), and subsequently stained with the respective antibody panel for 15 minutes at 4°C and washed with FACS buffer. Whenever appropriate, anti-rat IgM was added to the cells, incubated for 15 minutes at 4°C, and washed twice with the FACS buffer. Finally, cells were resuspended in 4% paraformaldehyde. For intracellular staining, cells were further processed following the instructions for the BioLegend’s FOXP3 Fix/Perm Buffer Set kit. Cells were measured with a LSRII flow cytometer (BD) at the Flow & Mass Cytometry Facility at the University of Maryland School of Medicine. Analysis was performed using the FlowJo software.
DETC populations in the murine skin were labeled with APC anti-CD45 (BioLegend 103111) or PE/Cyanine7 anti-CD45 (BioLegend 103114), FITC anti-CD3 (BioLegend 100203), BV60 anti-Vγ5 (BD 743241), APC anti-Thy1.2 (BioLegend 105312), and/or monoclonal antibody 17D1 (kindly provided by Dr. Adrian Hayday, King’s College London, and Dr. Robert Tigelaar, Yale University), and PE mouse anti-rat IgM (BD 553888). DETC costimulatory markers were measured with PE anti-JAML (BioLegend 128503), BV711 anti-CD100 (BD 745492), PE/Cyanine5 anti-CD44 (BioLegend 103010), APC/Cyanine7 anti-CD25 (BioLegend 102026), PerCP/Cyanine5.5 anti-CD69 (BioLegend 104522), and APC anti-CD314 (BioLegend 130212). Keratinocyte populations in the murine skin were labeled with BV711 anti-CD324 (BioLegend 118233), PE anti-CD200 (BioLegend 123807), PE/Cyanine5 anti-CD34 (BioLegend 119312), BV605 Sca1 (BioLegend 108133), and/or PE anti-CD49f (BioLegend 313612). Keratinocyte proliferation was labeled with the Alexa Fluor 700 anti-Ki-67 (BioLegend 652420). Smad7 was labeled with the anti-MADH7/SMAD7 polyclonal antibody (Abcam ab216428) and Alexa Fluor 405 goat anti-Rabbit IgG secondary antibody (Thermo Fischer Scientific A-31556).
Enzyme-linked immunosorbent assay
To determine levels of KGF, 5 mm skin biopsies were placed in 200μL of RPMI media tissue bath for one hour shaking at 32°C (150 revolutions per minute). KGF levels in tissue bath supernatant were determined using the R&D Systems KGF/FGF-7 Quantikine ELISA kit according to manufacturer instructions. To determine levels of TGF-β, 5mm skin biopsies were homogenized in lysis buffer containing 1X RIPA buffer (catalogue number 20–188, Millipore) with 1X protease-phosphatase inhibitor cocktail (catalogue number 78420, Thermo Scientific). TGF-β levels in tissue supernatant were determined using the R&D Systems TGF-β 1 Quantikine ELISA kit according to manufacturer instructions. Total protein in samples was determined using the Pierce BCA Protein Assay Kit (catalogue number 23227 Thermo Scientific). Sample concentration of KGF/TGF-β were normalized to the total protein in a sample.
Intravital microscopy
Epidermal intravital imaging studies were done in collaboration with Dr. Sangbum Park at Michigan State University. All in vivo imaging and analysis were performed, as described previously (Park et al., 2021). Simultaneous visualization of Langerhans cells, DETCs and epithelial cells was achieved by utilizing the huLangerin-CreER;Rosa-stop-tdTomato;CX3CR1-GFP+/−;K14-H2B-Cerulean mice.
Epidermal single-cell isolation, scRNA-seq library preparation and sequencing
I. scapularis nymphs were microinjected with vamp33 si or vamp33 sc and fed on FVB/N Jax or FVB/N Tac mice. On the third day of feeding, mice were euthanized with CO2. Partially fed ticks were removed and the sites where ticks bit were shaved followed by an application of a light layer of Nair depilatory lotion. A total of three 5-mm skin punch biopsies were obtained from the dorsal neck for each mouse. 5-mm skin punch biopsies were obtained from the same physiological site of naïve mice. Skin samples were incubated in dispase solution (4 U/mL dispase, 5mM MgCl2, and 0.4mM CaCl2 in PBS) for 2.5 hours at 37°C with constant shaking/stirring. Epidermal sheets were separated from the dermal layer using forceps. Epidermal sheets were then incubated in a digestion solution (2.5mg/mL collagenase D and 0.2mg/mL DNase in RPMI Medium) for 1 hour at 37°C with constant shaking/stirring.
Cells were resuspended using a wide-bore pipette tip and three samples per treatment per mouse were combined. Samples were passed through a 40 μM cell strainer and washed with RPMI +10% FBS. Cells were counted using the Countess II FL Automated Cell Counter, stained with 5 μl of 7-AAD per million cells, and incubated in the dark for 10 minutes at 4°C. Samples were then sorted at the CIBR Flow Cytometry Core Facility at the University of Maryland School of Medicine. Cells were sorted into a PBS in the absence of calcium and magnesium + 10% FBS collection buffer. They were then transported on ice to the Institute of Genome Sciences at the University of Maryland School of Medicine for library preparation and sequencing. Single cell libraries were generated with the 3’ NextGEM v3.1 kit targeting 3800–5000 cells. Libraries were sequenced with a NovaSeq 6000, S2 flowcell targeting 375M read pairs per sample.
Bioinformatics
All scRNA-seq reads were processed and mapped to the mouse mm10 reference genome using 10X Genomics’ Cell Ranger software. Approximately 20,640 total cells were profiled with 88,027 mean reads per cell across all conditions. A count matrix (gene-by-cell) generated by cell ranger count for each library was then aggregated into a single count matrix. Expression matrices were generated using the Bioconductor packages scater (v1.22.0) (Lun, McCarthy, et al., 2016) and scran (v1.22.1) (Lun, Bach, et al., 2016). Cells with less than 2,500 or greater than 60,000 UMIs were removed after calculating cell metrics using scater (v1.22.0). DoubletFinder (v2.0.1) (McGinnis et al., 2019) was applied removing 1,364 cells, which yielded a total of 10,715 cells. The remaining transcriptomes were normalized by first calculating size factors via the scran functions quickCluster and computeSumFactors. Then, we computed normalized counts for each cell with logNormCounts function in scran (v1.22.1).
For downstream analysis, highly variable genes were selected using getTopHVGs before performing the Principal Component Analysis (PCA) and the tSNE projection. Clustering was conducted using kmeans function based on the calculated tSNE. Differential gene expression between clusters was calculated using find Markers function. Only identified epidermal cells of interest (Keratinocytes, T cells, and APCs) were further analyzed, resulting in a total of 5,172 cells with a median UMI count of 13,910 per cell. For pseudotime analysis, the Bioconductor matrix was imported into slingshot (v2.2.1) (Street et al., 2018). To compare the T cell receptor delta variable 4 (Trdv4) expression, normalized counts were used for visualization by the violin plot. The permutation test was applied to calculate the significance of the difference in the mean expression between two groups. A list of differentially expressed keratinocyte genes between treatments was generated by MAST (v1.24.0) (Finak et al., 2015) with significance testing under the Hurdle model for downstream analysis by the IPA.
Gene set enrichment analysis
Gene set enrichment analysis was performed using DAVID, version 2021. Default DAVID parameters were employed and included the following categories for the enrichment analysis: GOTERM_BP_DIRECT, GOTERM_CC_DIRECT and GOTERM_MF_DIRECT (from Gene_Ontology), KEGG_PATHWAY (from Pathways) and INTERPRO (from Protein_Domains). p value and FDR< 0.05 were set as a threshold.
Ingenuity pathway analysis
Differentially expressed keratinocyte genes from the following samples were analyzed in the IPA as independent datasets: 1) FVB-Tac Naïve versus FVB-Jax Naïve 2) FVB-Jax siV33 versus FVB-Jax scV33 and 3) FVB-Tac siV33 versus FVB-Tac scV33. Genes were considered differentially expressed if the p value and FDR were < 0.05. Dataset input criteria for the IPA included expression, p value, log ratio, FDR, and Ensemble ID codes. All datasets were examined for canonical pathway and upstream regulator analysis. FVB-Tac Naïve versus FVB-Jax Naïve dataset had 591 IDs, including 589 mapped and 2 unmapped IDs. FVB-Jax siV33 versus FVB-Jax scV33 dataset had 1207 IDs, including 1204 mapped and 3 unmapped IDs. FVB-Tac siV33 versus FVB-Tac scV33 had 732 IDs, including 728 mapped and 4 unmapped IDs. The IPA proprietary algorithm segments the network map between molecules into multiple networks and assigns scores for each network as described previously (Calvano et al., 2005). For the canonical pathway analysis, −log (P-value) >2 was taken as threshold and for the upstream regulator analysis, the p value of overlap <0.05 was set as the threshold. A positive Z-score was defined as the predicted activation, and a negative Z-score was defined as the predicted inhibition.
Statistical analysis
Statistical significance was assessed as follows: percent tick attachment was calculated by the Fisher’s exact test, tick weight by the t test or the Mann Whitney test, and survival curve by the Log-rank (Mantel-Cox) test. One-way ANOVA followed by Tukey’s post hoc test for multiple comparisons was also used. Kruskal-Wallis ANOVA was implemented if the dataset failed normality of residuals or displayed heterogeneity of variance. We used GraphPad PRISM® (version 9.1.0) for all statistical analyses. Outliers were detected by a GraphPad Quickcals program (https://www.graphpad.com/quickcalcs/Grubbs1.cfm). p values of < 0.05 were considered statistically significant.
Supplementary Material
Supplementary Figure 1: DETC flow cytometry gating strategy. 5 mm skin punch biopsies were obtained from the bite of ticks and compared to the naïve skin followed by flow cytometry analysis. Representative flow cytometry plots were gated for (A) DETCs (Vy5+) and (B) DETC co-receptors (JAML+, NKG2D+, CD25+, CD69+, CD100+ or CD44+).
Supplementary Figure 2: ScRNA-seq data filtration. Composite datasets of FVB-Jax and FVB-Tac samples included 20,640 cells (A) before filtration by scran (R package). (B) tSNE plot of fixed threshold filtration, set to 2500–60,000 UMIs. (C) Doublet finder (R package) of dataset. tSNE was colored by the doublet score. (D) tSNE plot after fixed threshold filtration and doublet finder analysis.
Supplementary Figure 3: Expression of keratinocyte-specific markers. (A) Graphical illustration of keratinocyte stratified layers with select marker genes. tSNE of keratinocyte clusters depicting gene expression of (B) Krt14, (C) Krt5, (D) Krt1, (E) Krt10 and (F) Ivl.
Supplementary Figure 4: Expression of hair follicle-specific markers. (A) Graphical illustration of hair follicle microanatomy with select marker genes. tSNE of keratinocyte clusters depicting gene expression of (B) Shh, (C) Krt75, (D) Lgr5, (E) Mgst1 and (F) Krt79.
Supplementary Figure 5: Epidermal cell type characterization. Cluster frequency of keratinocytes, antigen presenting and T cells in (A) FVB-Jax and (B) FVB-Tac mice in the presence or absence of I. scapularis nymphs microinjected with scV33 or siV33. (C) Violin plot displaying the expression of the TCR-Vδ1 gene, Trdv4, in the epidermal T cell cluster of naïve FVB-Jax and FVB-Tac mice. Significance shown as *p<0.05 based on a permutation test using R statistical packages.
Supplementary Figure 6: Keratinocyte-specific markers along pseudotime trajectory. (A) Krt14, (B) Krt1, and (C) Ivl gene expression along pseudotime values (x axis) for naïve, scV33-, or siV33-tick bites on FVB-Jax or FVB-Tac mice. Cells colored by clusters from keratinocyte tSNE plot (as shown in Figure 4D) ordered across the pseudotime (x-axis).
Supplementary Figure 7: Individual tSNE plots of keratinocyte clusters. Subclustering analysis of keratinocytes across samples: (A) FVB-Jax, (B) FVB-Jax scV33, (C) FVB-Jax siV33, (D) FVB-Tac, (E) FVB-Tac scV33, and (F) FVB-Tac siV33.
Supplementary Figure 8: Flow cytometry gating strategy in keratinocytes. 5 mm punch biopsies were obtained from the bite site of ticks or naïve skin and processed for flow cytometry. Representative flow cytometry plots were gated for (A) EpCAM+ keratinocytes, (B) Ki67+, (C) Anti-rabbit IgG+ for PI3K and Smad7, and (D) p-PI3K+ keratinocytes.
Acknowledgements
We acknowledge members of the Pedra laboratory for providing insightful discussions. We thank the rearing facility at Oklahoma State University for providing I. scapularis, A. americanum, and D. variabilis ticks; Xiaoxuan Fan, Bryan Hahn, Regina Harley, and Sean McGill (University of Maryland School of Medicine) for flow cytometry and sorting assistance; Adrian Hayday (King’s College London) and Robert Tigelaar (Yale University) for the monoclonal antibody 17D1; the Maryland Genomics Core at the Institute for Genome Sciences, University of Maryland School of Medicine for the services provided in next generation sequencing; the University of Maryland Greenebaum Comprehensive Cancer Center Flow Cytometry Shared Service core facility and the Flow & Mass Cytometry Facility at the University of Maryland School of Medicine for flow cytometry services; Cristiana Cairo, Nevil Singh, Nicholas Carbonetti (University of Maryland School of Medicine) and Jere McBride (University of Texas Medical Branch) for insightful advice. This work was supported by grants from the NIH to F31AI152215 (AJO), F31AI167471 (LRB), R01AI134696 (JHFP), R01AI116523 (JHFP), P01AI138949 (JHFP), T32AI162579 (HJL-Y), R01AR083086 (SP), the United States Department of Agriculture, National Institute of Food and Agriculture (USDA-NIFA) Hatch-Multistate Project to TEX0-1-7714 (AOC), and the Knipling-Bushland-Swahrf fellowship from the Department of Entomology at Texas A&M University (BL-G). The content is solely the responsibility of the authors and does not represent the official views of the NIH, the Department of Health and Human Services, the USDA-NIFA or the United States government.
Footnotes
Resource Availability
Further information and request for resources and reagents should be directed to and will be honored by the corresponding author: Joao HF Pedra (jpedra@som.umaryland.edu)
Data and Code Availability
All scRNA sequences are deposited into the NCBI Sequence Read Archive under the BioProject accession PRJNA905677. R codes for scRNA sequencing datasets were adapted from https://bioconductor.org/books/3.16/OSCA/ and specified R package vignettes. Tokens can be made available upon request.
References
- Allen J. R., Khalil H. M., & Wikel S. K. (1979). Langerhans cells trap tick salivary gland antigens in tick-resistant guinea pigs. J Immunol, 122 (2), 563–565. [PubMed] [Google Scholar]
- Angel P., Szabowski A., & Schorpp-Kistner M. (2001). Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene, 20 (19), 2413–2423. 10.1038/sj.onc.1204380 [DOI] [PubMed] [Google Scholar]
- Aragona M., Dekoninck S., Rulands S., Lenglez S., Mascre G., Simons B. D., & Blanpain C. (2017). Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat Commun, 8, 14684. 10.1038/ncomms14684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi M., Kim T. K., Porter L., Mwangi W., & Mulenga A. (2019). Amblyomma americanum ticks utilizes countervailing pro and anti-inflammatory proteins to evade host defense. PLoS Pathog, 15 (11), e1008128. 10.1371/journal.ppat.1008128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee S. D., Woodward M. J., Turchinovich G., Mention J. J., Lewis J. M., Boyden L. M., Lifton R. P., Tigelaar R., & Hayday A. C. (2011). Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc Natl Acad Sci U S A, 108 (8), 3330–3335. 10.1073/pnas.1010890108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártolo I., Reis R. L., Marques A. P., & Cerqueira M. T. (2022). Keratinocyte growth factor-based strategies for wound re-epithelialization. Tissue Eng Part B Rev, 28 (3), 665–676. 10.1089/ten.TEB.2021.0030 [DOI] [PubMed] [Google Scholar]
- Bernard Q., Grillon A., Lenormand C., Ehret-Sabatier L., & Boulanger N. (2020). Skin interface, a key player for Borrelia multiplication and persistence in Lyme borreliosis. Trends in Parasitology, 36 (3), 304–314. https://doi.org/ 10.1016/j.pt.2019.12.017 [DOI] [PubMed] [Google Scholar]
- Boyden L. M., Lewis J. M., Barbee S. D., Bas A., Girardi M., Hayday A. C., Tigelaar R. E., & Lifton R. P. (2008). Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet, 40 (5), 656–662. 10.1038/ng.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calautti E., Li J., Saoncella S., Brissette J. L., & Goetinck P. F. (2005). Phosphoinositide 3-kinase signaling to Akt promotes keratinocyte differentiation versus death. J Biol Chem, 280 (38), 32856–32865. 10.1074/jbc.M506119200 [DOI] [PubMed] [Google Scholar]
- Calvano S. E., Xiao W., Richards D. R., Felciano R. M., Baker H. V., Cho R. J., Chen R. O., Brownstein B. H., Cobb J. P., Tschoeke S. K., Miller-Graziano C., Moldawer L. L., Mindrinos M. N., Davis R. W., Tompkins R. G., Lowry S. F., Inflamm, & host response to injury large scale collab. research program. (2005). A network-based analysis of systemic inflammation in humans. Nature, 437 (7061), 1032–1037. 10.1038/nature03985 [DOI] [PubMed] [Google Scholar]
- Chen G., Wang X., Severo M. S., Sakhon O. S., Sohail M., Brown L. J., Sircar M., Snyder G. A., Sundberg E. J., Ulland T. K., Olivier A. K., Andersen J. F., Zhou Y., Shi G. P., Sutterwala F. S., Kotsyfakis M., & Pedra J. H. (2014). The tick salivary protein sialostatin L2 inhibits caspase-1-mediated inflammation during Anaplasma phagocytophilum infection. Infect Immun, 82 (6), 2553–2564. 10.1128/IAI.01679-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekoninck S., & Blanpain C. (2019). Stem cell dynamics, migration and plasticity during wound healing. Nat Cell Biol, 21 (1), 18–24. 10.1038/s41556-018-0237-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R. G., O’Hagan J. E., Schotz M., Binnington K. C., & Hegarty M. P. (1976). Prostaglandin in the saliva of the cattle tick Boophilus microplus. Aust J Exp Biol Med Sci, 54 (5), 475–486. 10.1038/icb.1976.48 [DOI] [PubMed] [Google Scholar]
- Eisen L. (2022). Tick species infesting humans in the United States. Ticks and tick-borne diseases, 13 (6), 102025. https://doi.org/ 10.1016/j.ttbdis.2022.102025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves E., Maruyama S. R., Kawahara R., Fujita A., Martins L. A., Righi A. A., Costa F. B., Palmisano G., Labruna M. B., Sá-Nunes A., Ribeiro J. M. C., & Fogaça A. C. (2017). Analysis of the salivary gland transcriptome of unfed and partially fed Amblyomma sculptum ticks and descriptive proteome of the saliva. Frontiers in Cellular and Infection Microbiology, 7, 476. 10.3389/fcimb.2017.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S., Eyerich K., Traidl-Hoffmann C., & Biedermann T. (2018). Cutaneous barriers and skin immunity: differentiating a connected network. Trends Immunol, 39 (4), 315–327. 10.1016/j.it.2018.02.004 [DOI] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M., Linsley P. S., & Gottardo R. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol, 16, 278. 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti I. M., Sa-Nunes A., Mans B. J., Santos I. M., & Ribeiro J. M. (2009). The role of saliva in tick feeding. Front Biosci, 14 (6), 2051–2088. 10.2741/3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. (1993). Epidermal differentiation and keratin gene expression. Journal of Cell Science, 1993, 197–208. 10.1242/jcs.1993.Supplement_17.28 [DOI] [PubMed] [Google Scholar]
- Girardi M., Oppenheim D. E., Steele C. R., Lewis J. M., Glusac E., Filler R., Hobby P., Sutton B., Tigelaar R. E., & Hayday A. C. (2001). Regulation of cutaneous malignancy by γδ T cells. Science, 294 (5542), 605–609. https://doi.org/doi: 10.1126/science.1063916 [DOI] [PubMed] [Google Scholar]
- Glatz M., Means T., Haas J., Steere A. C., & Mullegger R. R. (2017). Characterization of the early local immune response to Ixodes ricinus tick bites in human skin. Exp Dermatol, 26 (3), 263–269. 10.1111/exd.13207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnicka V., Vancova-Stibraniova I., Slovak M., Kocakova P., & Nuttall P. A. (2011). Ixodid tick salivary gland products target host wound healing growth factors. Int J Parasitol, 41 (2), 213–223. 10.1016/j.ijpara.2010.09.005 [DOI] [PubMed] [Google Scholar]
- Havran W. L., Chien Y. H., & Allison J. P. (1991). Recognition of self antigens by skin-derived T cells with invariant gamma delta antigen receptors. Science, 252 (5011), 1430–1432. 10.1126/science.1828619 [DOI] [PubMed] [Google Scholar]
- Jameson J., Ugarte K., Chen N., Yachi P., Fuchs E., Boismenu R., & Havran W. L. (2002). A role for skin gammadelta T cells in wound repair. Science, 296 (5568), 747–749. 10.1126/science.1069639 [DOI] [PubMed] [Google Scholar]
- Jameson J. M., Cauvi G., Witherden D. A., & Havran W. L. (2004). A keratinocyte-responsive γδ TCR is necessary for dendritic epidermal T cell activation by damaged keratinocytes and maintenance in the epidermis. The Journal of Immunology, 172 (6), 3573–3579. 10.4049/jimmunol.172.6.3573 [DOI] [PubMed] [Google Scholar]
- Jochum W., Passegué E., & Wagner E. F. (2001). AP-1 in mouse development and tumorigenesis. Oncogene, 20 (19), 2401–2412. 10.1038/sj.onc.1204389 [DOI] [PubMed] [Google Scholar]
- Joost S., Annusver K., Jacob T., Sun X., Dalessandri T., Sivan U., Sequeira I., Sandberg R., & Kasper M. (2020). The molecular anatomy of mouse skin during hair growth and rest. Cell Stem Cell, 26 (3), 441–457.e447. 10.1016/j.stem.2020.01.012 [DOI] [PubMed] [Google Scholar]
- Joost S., Jacob T., Sun X., Annusver K., La Manno G., Sur I., & Kasper M. (2018). Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep, 25 (3), 585–597.e587. 10.1016/j.celrep.2018.09.059 [DOI] [PubMed] [Google Scholar]
- Joost S., Zeisel A., Jacob T., Sun X., La Manno G., Lönnerberg P., Linnarsson S., & Kasper M. (2016). Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst, 3 (3), 221–237.e229. 10.1016/j.cels.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K., Honda T., Ginhoux F., & Egawa G. (2019). The immunological anatomy of the skin. Nature Reviews Immunology, 19 (1), 19–30. 10.1038/s41577-018-0084-5 [DOI] [PubMed] [Google Scholar]
- Karim S., & Ribeiro J. M. (2015). An insight into the sialome of the lone star tick, Amblyomma americanum, with a glimpse on its time dependent gene expression. PLoS One, 10 (7), e0131292. 10.1371/journal.pone.0131292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazimirova M., & Stibraniova I. (2013). Tick salivary compounds: their role in modulation of host defences and pathogen transmission. Front Cell Infect Microbiol, 3, 43. 10.3389/fcimb.2013.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes B. E., Liu S., Asare A., Naik S., Levorse J., Polak L., Lu C. P., Nikolova M., Pasolli H. A., & Fuchs E. (2016). Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell, 167 (5), 1323–1338.e1314. 10.1016/j.cell.2016.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotal J., Langhansova H., Lieskovska J., Andersen J. F., Francischetti I. M., Chavakis T., Kopecky J., Pedra J. H., Kotsyfakis M., & Chmelar J. (2015). Modulation of host immunity by tick saliva. J Proteomics, 128, 58–68. 10.1016/j.jprot.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyfakis M., Horka H., Salat J., & Andersen J. F. (2010). The crystal structures of two salivary cystatins from the tick Ixodes scapularis and the effect of these inhibitors on the establishment of Borrelia burgdorferi infection in a murine model. Mol Microbiol, 77 (2), 456–470. 10.1111/j.1365-2958.2010.07220.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyfakis M., Karim S., Andersen J. F., Mather T. N., & Ribeiro J. M. (2007). Selective cysteine protease inhibition contributes to blood-feeding success of the tick Ixodes scapularis. J Biol Chem, 282 (40), 29256–29263. 10.1074/jbc.M703143200 [DOI] [PubMed] [Google Scholar]
- Kramer C. D., Poole N. M., Coons L. B., & Cole J. A. (2011). Tick saliva regulates migration, phagocytosis, and gene expression in the macrophage-like cell line, IC-21. Exp Parasitol, 127(3), 665–671. 10.1016/j.exppara.2010.11.012 [DOI] [PubMed] [Google Scholar]
- Lewis J. M., Girardi M., Roberts S. J., Barbee S. D., Hayday A. C., & Tigelaar R. E. (2006). Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol, 7 (8), 843–850. 10.1038/ni1363 [DOI] [PubMed] [Google Scholar]
- Li G., Gustafson-Brown C., Hanks S. K., Nason K., Arbeit J. M., Pogliano K., Wisdom R. M., & Johnson R. S. (2003). c-Jun is essential for organization of the epidermal leading edge. Dev Cell, 4 (6), 865–877. 10.1016/s1534-5807(03)00159-x [DOI] [PubMed] [Google Scholar]
- Lun A. T., Bach K., & Marioni J. C. (2016). Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol, 17, 75. 10.1186/s13059-016-0947-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A. T., McCarthy D. J., & Marioni J. C. (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res, 5, 2122. 10.12688/f1000research.9501.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod A. S., & Havran W. L. (2011). Functions of skin-resident γδ T cells. Cell Mol Life Sci, 68 (14), 2399–2408. 10.1007/s00018-011-0702-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Hachiya Y., Arai K. Y., Muraguchi T., Sasaki T., & Nishiyama T. (2018). Type IV collagen aggregates promote keratinocyte proliferation and formation of epidermal layer in human skin equivalents. Exp Dermatol, 27 (5), 443–448. 10.1111/exd.13328 [DOI] [PubMed] [Google Scholar]
- McGinnis C. S., Murrow L. M., & Gartner Z. J. (2019). DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst, 8(4), 329–337 e324. 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiura M., Baszanowska W., Ościłowska I., Pałka J., & Miltyk W. (2020). Prolidase stimulates proliferation and migration through activation of the PI3K/Akt/mTOR signaling pathway in human keratinocytes. Int J Mol Sci, 21 (23), 9243. 10.3390/ijms21239243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X., & Lai Y. (2020). Keratinocyte: a trigger or an executor of psoriasis? J Leukoc Biol, 108 (2), 485–491. 10.1002/JLB.5MR0120-439R [DOI] [PubMed] [Google Scholar]
- Nielsen M. M., Witherden D. A., & Havran W. L. (2017). γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat Rev Immunol, 17 (12), 733–745. 10.1038/nri.2017.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva Chávez A. S., Wang X., Marnin L., Archer N. K., Hammond H. L., Carroll E. E. M., Shaw D. K., Tully B. G., Buskirk A. D., Ford S. L., Butler L. R., Shahi P., Morozova K., Clement C. C., Lawres L., Neal A. J. O., Mamoun C. B., Mason K. L., Hobbs B. E.,…Pedra J. H. F. (2021). Tick extracellular vesicles enable arthropod feeding and promote distinct outcomes of bacterial infection. Nature Communications, 12(1), 3696. 10.1038/s41467-021-23900-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Gonzalez D. G., Guirao B., Boucher J. D., Cockburn K., Marsh E. D., Mesa K. R., Brown S., Rompolas P., Haberman A. M., Bellaiche Y., & Greco V. (2017). Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat Cell Biol, 19(2), 155–163. 10.1038/ncb3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Matte-Martone C., Gonzalez D. G., Lathrop E. A., May D. P., Pineda C. M., Moore J. L., Boucher J. D., Marsh E., Schmitter-Sanchez A., Cockburn K., Markova O., Bellaiche Y., & Greco V. (2021). Skin-resident immune cells actively coordinate their distribution with epidermal cells during homeostasis. Nat Cell Biol, 23 (5), 476–484. 10.1038/s41556-021-00670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I., Stojadinovic O., Yin N. C., Ramirez H., Nusbaum A. G., Sawaya A., Patel S. B., Khalid L., Isseroff R. R., & Tomic-Canic M. (2014). Epithelialization in wound healing: a comprehensive review. Adv Wound Care 3 (7), 445–464. 10.1089/wound.2013.0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña O. A., & Martin P. (2024). Cellular and molecular mechanisms of skin wound healing. Nature Reviews Molecular Cell Biology 25 (8), 599–616. 10.1038/s41580-024-00715-1 [DOI] [PubMed] [Google Scholar]
- Piipponen M., Li D., & Landén N. X. (2020). The immune functions of keratinocytes in skin wound healing. Int J Mol Sci, 21 (22), 8790. 10.3390/ijms21228790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole N. M., Mamidanna G., Smith R. A., Coons L. B., & Cole J. A. (2013). Prostaglandin E2 in tick saliva regulates macrophage cell migration and cytokine profile. Parasites & Vectors, 6 (1), 261. 10.1186/1756-3305-6-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proksch E., Brandner J. M., & Jensen J.-M. (2008). The skin: an indispensable barrier. Experimental Dermatology, 17 (12), 1063–1072. https://doi.org/ 10.1111/j.1600-0625.2008.00786.x [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M., Evans P. M., MacSwain J. L., & Sauer J. (1992). Amblyomma americanum: characterization of salivary prostaglandins E2 and F2 alpha by RP-HPLC/bioassay and gas chromatography-mass spectrometry. Exp Parasitol, 74 (1), 112–116. 10.1016/0014-4894(92)90145-z [DOI] [PubMed] [Google Scholar]
- Ribeiro J. M., Makoul G. T., Levine J., Robinson D. R., & Spielman A. (1985). Antihemostatic, antiinflammatory, and immunosuppressive properties of the saliva of a tick, Ixodes dammini. J Exp Med, 161 (2), 332–344. 10.1084/jem.161.2.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro J. M., Makoul G. T., & Robinson D. R. (1988). Ixodes dammini: evidence for salivary prostacyclin secretion. J Parasitol, 74 (6), 1068–1069. [PubMed] [Google Scholar]
- Rice G., & Rompolas P. (2020). Advances in resolving the heterogeneity and dynamics of keratinocyte differentiation. Curr Opin Cell Biol, 67, 92–98. 10.1016/j.ceb.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R., Lindsey N. P., Fischer M., Gregory C. J., Hinckley A. F., Mead P. S., Paz-Bailey G., Waterman S. H., Drexler N. A., Kersh G. J., Hooks H., Partridge S. K., Visser S. N., Beard C. B., & Petersen L. R. (2018). Vital signs: trends in reported vectorborne disease cases - United States and Territories, 2004–2016. MMWR Morb Mortal Wkly Rep, 67(17), 496–501. 10.15585/mmwr.mm6717e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselle P., Braye F., & Dayan G. (2019). Re-epithelialization of adult skin wounds: cellular mechanisms and therapeutic strategies. Adv Drug Deliv Rev, 146, 344–365. 10.1016/j.addr.2018.06.019 [DOI] [PubMed] [Google Scholar]
- Schmierer B., & Hill C. S. (2007). TGFβ–SMAD signal transduction: molecular specificity and functional flexibility. Nature Reviews Molecular Cell Biology, 8(12), 970–982. 10.1038/nrm2297 [DOI] [PubMed] [Google Scholar]
- Schoeler G. B., Manweiler S. A., & Wikel S. K. (1999). Ixodes scapularis: effects of repeated infestations with pathogen-free nymphs on macrophage and T lymphocyte cytokine responses of BALB/c and C3H/HeN mice. Exp Parasitol, 92(4), 239–248. 10.1006/expr.1999.4426 [DOI] [PubMed] [Google Scholar]
- Sellheyer K., Bickenbach J. R., Rothnagel J. A., Bundman D., Longley M. A., Krieg T., Roche N. S., Roberts A. B., & Roop D. R. (1993). Inhibition of skin development by overexpression of transforming growth factor beta 1 in the epidermis of transgenic mice. Proc Natl Acad Sci U S A, 90 (11), 5237–5241. 10.1073/pnas.90.11.5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Pham M. N., Reyes J. B., Chana R., Yim W. C., Heu C. C., Kim D., Chaverra-Rodriguez D., Rasgon J. L., Harrell R. A. 2nd, Nuss A. B., & Gulia-Nuss M. (2022). Cas9-mediated gene editing in the black-legged tick, Ixodes scapularis, by embryo injection and ReMOT Control. iScience, 25 (3), 103781. 10.1016/j.isci.2022.103781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp L. L., Jameson J. M., Cauvi G., & Havran W. L. (2005). Dendritic epidermal T cells regulate skin homeostasis through local production of insulin-like growth factor 1. Nat Immunol, 6 (1), 73–79. 10.1038/ni1152 [DOI] [PubMed] [Google Scholar]
- Simo L., Kazimirova M., Richardson J., & Bonnet S. I. (2017). The essential role of tick salivary glands and saliva in tick feeding and pathogen transmission. Front Cell Infect Microbiol, 7, 281. 10.3389/fcimb.2017.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A. J., & Clark R. A. F. (1999). Cutaneous wound healing. New England Journal of Medicine, 341 (10), 738–746. 10.1056/nejm199909023411006 [DOI] [PubMed] [Google Scholar]
- Sonenshine D. E., & Roe R. M. (2014). Biology of Ticks (Vol. 1). Oxford University Press. [Google Scholar]
- Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., & Dudoit S. (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics, 19 (1), 477. 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S. M., & Lorenz R. G. (2022). FVB/N mouse strain regulatory T cells differ in phenotype and function from the C57BL/6 and BALB/C strains. FASEB Bioadv, 4(10), 648–661. 10.1096/fba.2021-00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen F., & Witherden D. A. (2020). Get in touch with dendritic epithelial T cells! Front Immunol, 11, 1656. 10.3389/fimmu.2020.01656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela J. G., Francischetti I. M. B., Pham V. M., Garfield M. K., Mather T. N., & Ribeiro J. M. C. (2002). Exploring the sialome of the tick Ixodes scapularis. Journal of Experimental Biology, 205 (18), 2843–2864. 10.1242/jeb.205.18.2843 [DOI] [PubMed] [Google Scholar]
- Whang M. I., Guerra N., & Raulet D. H. (2009). Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J Immunol, 182(8), 4557–4564. 10.4049/jimmunol.0802439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel S. (2013). Ticks and tick-borne pathogens at the cutaneous interface: host defenses, tick countermeasures, and a suitable environment for pathogen establishment. Front Microbiol, 4, 337. 10.3389/fmicb.2013.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake T. C., Stojadinovic O., & Tomic-Canic M. (2014). Epidermal differentiation in barrier maintenance and wound Healing. Adv Wound Care, 3(3), 272–280. 10.1089/wound.2013.0503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherden Deborah A., Watanabe M., Garijo O., Rieder Stephanie E., Sarkisyan G., Cronin Shane J. F., Verdino P., Wilson Ian A., Kumanogoh A., Kikutani H., Teyton L., Fischer Wolfgang H., & Havran Wendy L. (2012). The CD100 receptor interacts with its plexin B2 ligand to regulate epidermal γδ T cell function. Immunity, 37 (2), 314–325. https://doi.org/ 10.1016/j.immuni.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth C. D., Michael E., Smith L., Vijayachandra K., Glick A., Hennings H., & Yuspa S. H. (2004). Strain-dependent differences in malignant conversion of mouse skin tumors is an inherent property of the epidermal keratinocyte. Carcinogenesis, 25 (9), 1771–1778. 10.1093/carcin/bgh170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: DETC flow cytometry gating strategy. 5 mm skin punch biopsies were obtained from the bite of ticks and compared to the naïve skin followed by flow cytometry analysis. Representative flow cytometry plots were gated for (A) DETCs (Vy5+) and (B) DETC co-receptors (JAML+, NKG2D+, CD25+, CD69+, CD100+ or CD44+).
Supplementary Figure 2: ScRNA-seq data filtration. Composite datasets of FVB-Jax and FVB-Tac samples included 20,640 cells (A) before filtration by scran (R package). (B) tSNE plot of fixed threshold filtration, set to 2500–60,000 UMIs. (C) Doublet finder (R package) of dataset. tSNE was colored by the doublet score. (D) tSNE plot after fixed threshold filtration and doublet finder analysis.
Supplementary Figure 3: Expression of keratinocyte-specific markers. (A) Graphical illustration of keratinocyte stratified layers with select marker genes. tSNE of keratinocyte clusters depicting gene expression of (B) Krt14, (C) Krt5, (D) Krt1, (E) Krt10 and (F) Ivl.
Supplementary Figure 4: Expression of hair follicle-specific markers. (A) Graphical illustration of hair follicle microanatomy with select marker genes. tSNE of keratinocyte clusters depicting gene expression of (B) Shh, (C) Krt75, (D) Lgr5, (E) Mgst1 and (F) Krt79.
Supplementary Figure 5: Epidermal cell type characterization. Cluster frequency of keratinocytes, antigen presenting and T cells in (A) FVB-Jax and (B) FVB-Tac mice in the presence or absence of I. scapularis nymphs microinjected with scV33 or siV33. (C) Violin plot displaying the expression of the TCR-Vδ1 gene, Trdv4, in the epidermal T cell cluster of naïve FVB-Jax and FVB-Tac mice. Significance shown as *p<0.05 based on a permutation test using R statistical packages.
Supplementary Figure 6: Keratinocyte-specific markers along pseudotime trajectory. (A) Krt14, (B) Krt1, and (C) Ivl gene expression along pseudotime values (x axis) for naïve, scV33-, or siV33-tick bites on FVB-Jax or FVB-Tac mice. Cells colored by clusters from keratinocyte tSNE plot (as shown in Figure 4D) ordered across the pseudotime (x-axis).
Supplementary Figure 7: Individual tSNE plots of keratinocyte clusters. Subclustering analysis of keratinocytes across samples: (A) FVB-Jax, (B) FVB-Jax scV33, (C) FVB-Jax siV33, (D) FVB-Tac, (E) FVB-Tac scV33, and (F) FVB-Tac siV33.
Supplementary Figure 8: Flow cytometry gating strategy in keratinocytes. 5 mm punch biopsies were obtained from the bite site of ticks or naïve skin and processed for flow cytometry. Representative flow cytometry plots were gated for (A) EpCAM+ keratinocytes, (B) Ki67+, (C) Anti-rabbit IgG+ for PI3K and Smad7, and (D) p-PI3K+ keratinocytes.
Data Availability Statement
All scRNA sequences are deposited into the NCBI Sequence Read Archive under the BioProject accession PRJNA905677. R codes for scRNA sequencing datasets were adapted from https://bioconductor.org/books/3.16/OSCA/ and specified R package vignettes. Tokens can be made available upon request.