Abstract
Objective:
We examined interactions between genotype and a Dietary Approaches to Stop Hypertension (DASH) diet score in relation to systolic blood pressure (SBP).
Methods:
We analyzed up to 9,420,585 biallelic imputed single nucleotide polymorphisms (SNPs) in up to 127,282 individuals of six population groups (91% of European population) from the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium (CHARGE; n=35,660) and UK Biobank (n=91,622) and performed European population-specific and cross-population meta-analyses.
Results:
We identified three loci in European-specific analyses and an additional four loci in cross-population analyses at P for interaction < 5e-8. We observed a consistent interaction between rs117878928 at 15q25.1 (minor allele frequency = 0.03) and the DASH diet score (P for interaction = 4e-8; P for heterogeneity = 0.35) in European population, where the interaction effect size was 0.42±0.09 mm Hg (P for interaction = 9.4e-7) and 0.20±0.06 mm Hg (P for interaction = 0.001) in CHARGE and the UK Biobank, respectively. The 1 Mb region surrounding rs117878928 was enriched with cis-expression quantitative trait loci (eQTL) variants (P = 4e-273) and cis-DNA methylation quantitative trait loci (mQTL) variants (P = 1e-300). While the closest gene for rs117878928 is MTHFS, the highest narrow sense heritability accounted by SNPs potentially interacting with the DASH diet score in this locus was for gene ST20 at 15q25.1.
Conclusion:
We demonstrated gene-DASH diet score interaction effects on SBP in several loci. Studies with larger diverse populations are needed to validate our findings.
Keywords: DASH Diet, Association Study, Genome-Wide, Gene Environment Interaction, MTHFS, ST20, Systolic Pressure, CHARGE consortium, UK Biobank
Introduction
Nearly half of American adults (and 32% of the global population) have hypertension, an established risk factor for cardiovascular disease [1, 2]. The Dietary Approach to Stop Hypertension (DASH) diet promotes consumption of fruits, vegetables, whole grains, nuts and legumes, and low-fat dairy products, and limits the consumption of red and processed meats, sugar-sweetened beverages, and sodium [3]. The DASH diet is based on diets provided in a multicenter, randomized controlled trial (RCT), where the primary results successfully reduced blood pressure (BP) among individuals with prehypertension and hypertension and was subsequently shown in several additional RCTs [3–7]. For example, in the DASH-Sodium trial, the low sodium DASH diet reduced systolic BP by 7 mm Hg compared with a typical American diet [6]. In population-based studies, DASH diet scores have been developed to reflect an individual’s adherence to the DASH diet plan that was tested in the trials [8, 9]. Several independent studies showed that higher DASH diet scores were associated with a decreased risk of hypertension and cardiovascular disease [10–13].
The genetic architecture of hypertension has been studied extensively [14–16]. For example, a recent genome-wide association study (GWAS) reported that individuals in the high polygenic score quintile, calculated based on over 2,000 BP-associated loci, had a five-fold greater risk of hypertension compared to those in the lowest quintile with a low polygenic score [16]. Unhealthy lifestyle such as poor diet, smoking, and sedentary lifestyle is also a significant risk factor of hypertension and may interact with genetic factors. For example, an earlier study in UK Biobank showed an interaction between a polygenic score, built with 314 BP-associated loci, and a healthy lifestyle score for systolic BP. Compared to individuals with a low lifestyle score, systolic BP was 4.9 mm Hg lower in those with a high lifestyle score and a low genetic risk and 4.1 mm Hg lower in those with a high lifestyle score and a high polygenic score [17]. Gene-diet interaction has also been examined in other general populations [18–23], however, these studies are limited to investigating specific dietary components such as sodium intake [18, 19], different types of dietary fat [20, 21], or caloric intake [22]. Little is known about the interaction between genes and overall diet quality in relation to BP in the general population. To address this knowledge gap, the Gene-Lifestyle Interactions Working Group, part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, investigated genome-wide gene-DASH diet score interaction in relation to systolic BP [23–25].
Methods
Study Populations.
We analyzed six participating cohorts in the CHARGE consortium and the UK Biobank (Supplemental Appendix 1, Supplemental Table 1). Descriptions of the CHARGE consortium and the UK Biobank have been published previously [23, 26]. We included participants if they were aged 18 to 80 years and had no missing data on genotype, diet, systolic BP measurements, or covariates. The different populations in these cohorts included African population (AFR), Admixed American population (AMR), Central/South Asian population (CSA), East Asian population (EAS), European population (EUR), and Middle Eastern population (MID). In the UK Biobank, we excluded those who were related to other participants up to the third degree. All participants provided written informed consent. The present study protocol was approved by the Tufts University Institutional Review Board.
Study Design.
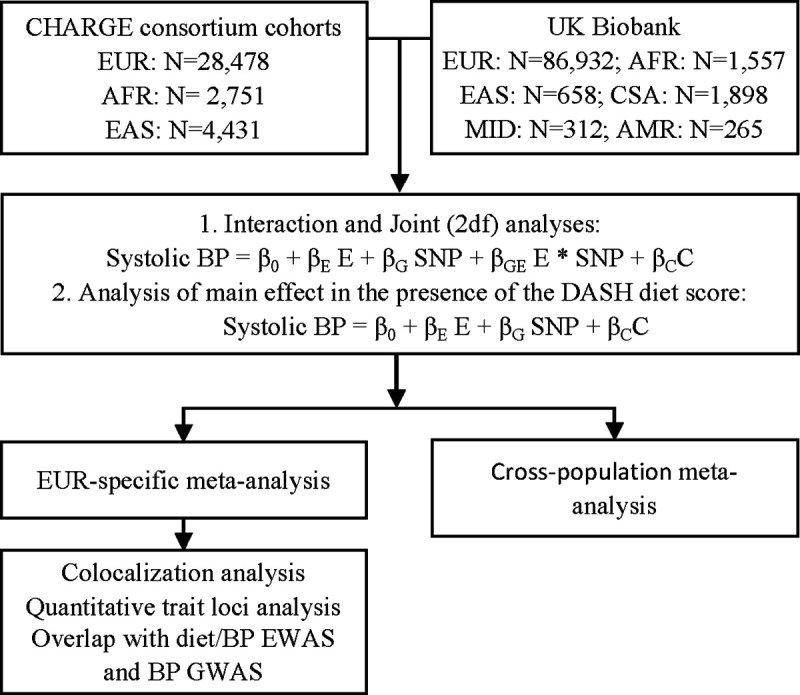
The study design is depicted in Figure 1. We developed the analytical protocol based on the method created by the Gene-Lifestyle Interactions Working Group [27]. Each cohort conducted population-specific analysis according to this protocol. Population-specific and cross-population meta-analyses were then conducted centrally.
Systolic BP.
Resting or sitting systolic BP (mm Hg) was measured according to standard protocol in each cohort [27]. When multiple BP readings (typically two or three readings) were available, the mean systolic BP of all readings was analyzed. For subjects taking anti-hypertensive medications, we adjusted their systolic BP by adding 15 mm Hg [14]. We winsorized systolic BP values that were more than 6 standard deviations (SD) from the mean.
DASH Diet Score.
Different versions of cohort-specific Food Frequency Questionnaires (FFQ) were used to assess dietary intakes largely during the same time for BP measurement in the CHARGE cohorts. Cohort specific dietary intake information is provided in Supplemental Table 2. In the UK Biobank, we analyzed at least two internet-based 24-hour dietary recalls collected within two years after the baseline BP measurement [28]. If a UK Biobank participant did not have a dietary recall within two years of the baseline BP measurement, but had BP measured during a follow-up visit, we analyzed their dietary recalls collected within two years of that follow-up BP measurement. Each cohort applied their own criteria for quality control (QC) of their dietary assessment tools, which is summarized in Supplemental Table 2. In each cohort, the DASH diet score was calculated using the same algorithm based on consumption of eight components: fruits, vegetables, whole grains, nuts and legumes, low-fat dairy, red and processed meats, sugar-sweetened beverages, and sodium [29]. Each component was computed and classified into cohort-specific quintiles. A score of 1 to 5 was then given for each component according to its quintile rank with higher scores for foods where high intake was favorable, and reverse quintile scores for the components where low intake was desired. The sum of the component scores resulted in a DASH diet score ranging from 8 to 40, with higher scores indicating better diet quality for BP control. Sodium intake was not included in the UK Biobank DASH diet score, as the nutrient component was not available when the analysis was run.
Association of the DASH diet score with systolic BP.
In the Framingham Heart Study (FHS) and the UK Biobank, we examined cross-sectional associations of the DASH diet score with systolic BP in a multivariate model with adjustment for age, age squared, sex, energy intake, and population (only in the UK Biobank) in the first model, and additionally adjusted for BMI in the second model.
Genotyping.
Genotyping was performed by each participating cohort using either Illumina or Affymetrix arrays. Specific details of genotyping platforms and imputation tools are described in Supplemental Table 3. SNP dosages were imputed in each cohort using reference panels including 1000 Genomes Phase 1 and Phase 3 panels and Haplotype Reference Consortium (HRC) panel [15, 30, 31]. In the CHARGE cohorts, we focused on biallelic variants with minor allele frequency (MAF) ≥0.01. Because of the larger sample size, we analyzed biallelic SNPs with MAF ≥0.005 in the UK Biobank. In all cohorts, we only included variants where the minor allele counts were >10. Only autosomal SNPs with imputation quality index ≥0.3 were considered in statistical analyses.
Statistical analysis.
Within each cohort, we first performed population-specific interaction analyses with the quantitative DASH diet score by examining the multiplicative interaction with SNP dosage. To explore whether the potential gene-diet interaction was driven by threshold effect, we also analyzed interactions with the DASH diet score dichotomized by its median or lower quartile. Linear regression models or linear mixed effect models (for familial data in the CHARGE cohorts) were run adjusting for age, age squared, sex, energy intake, BMI, field center (for multi-center studies), cohort-specific SNP-based principal components, and additional cohort-specific covariates, if any. Narrow sense heritability was approximated by the R2 derived from the regression models. The EasyQC R package [32] with 1000 Genomes data as reference [33] was used to conduct QC of summary statistics across all CHARGE cohorts. We followed the UK Biobank’s QC protocol [34] and only analyzed UK Biobank SNPs if these SNPs were also analyzed in the CHARGE cohorts. An inverse variance-weighted, fixed-effect meta-analysis was performed to combine cohort-specific results using METAL [35]. Genomic control was applied in the meta-analyses.
In secondary analyses, we examined associations for SNP-systolic BP (i.e., a main-effect model) and performed a 2-degree-of-freedom (2df) test [36] to jointly examine both SNP main effect and SNP-DASH diet interaction effect, with adjustment for the same covariates included in the interaction analysis (Figure 1). Similarly, population-specific analyses were conducted in each cohort, and meta-analyses were performed to combine cohort-specific findings. For the interaction analyses and the 2df tests, robust estimates of the standard errors (SE) and covariances were used in meta-analyses to protect against potential misspecification of the mean models [27]. In all analyses, heterogeneity across cohorts was determined based on Cochran’s Q-test. We considered SNPs with joint P <5e-8 that were present in at least two cohorts as significant. Novel loci were identified as SNPs with P-value < 5e-8 that are not in linkage disequilibrium (LD R2 ≥0.1; based on the 1000 Genomes data) or are ±500 kb from any previously validated BP–associated SNPs in the GWAS Catalog [37].
To further characterize SNPs significantly interacting with the DASH diet score, we conducted colocalization analysis to evaluate whether SNPs with an interaction effect were independent of those significant in the main effect GWAS. A Bayesian model, implemented with the Coloc R package, was used [38]. SNPs residing within 1 mb of the lead significant variants from the interaction analysis were included. Regression coefficients and squared SEs for the interaction term from the present meta-analysis and the corresponding values for the same SNPs from a large GWAS for systolic BP [15] were used. The colocalization analysis tested five hypotheses (H0-H5) using default priors, i.e., p1=1e-4, p2=1e-4, and p12=1e-5, where H0: no SNP has a genetic association in the region; H1: only interaction SNPs had a genetic association in the region; H2: only main effect SNPs had a genetic association in the region; H3: the two groups of SNPs are associated independently to the locus of interest, e.g., with different causal variants; H4: the two groups of SNPs are associated dependently to the locus of interest, e.g., they share a single causal variant. We considered the posterior probabilities ≥0.8 for H3 (i.e., two groups of SNPs are associated independently with the locus of interest) as significant evidence in support of two independent causal variants for systolic BP in one locus.
To understand the potential functions of SNPs significant in the interaction analysis, we carried out analyses to determine whether these loci are enriched with expression quantitative trait loci (eQTL) and DNA methylation quantitative trait (mQTL) variants. We used the eQTL and mQTL databases from FHS [39, 40]. Both cis- and trans-eQTL and mQTL variants residing within 1 mb regions of the lead SNPs significant in the interaction analysis were examined, with significance determined using a one-sided Fisher’s exact test and corrected for multiple testing as needed.
Results
Participant Characteristics.
We analyzed data from up to 35,660 individuals: 28,478 EUR, 2,751 AFR, and 4,431 EAS participants from cohorts participating in the CHARGE consortium and up to 91,622 unrelated UK Biobank participants comprising six population groups: EUR (N=86,932), AFR (N=1,557), EAS (N=658), CSA (N=1,898), MID (N=312), and AMR (N=265). Mean age ranged from 20 to 75 years in the CHARGE cohorts and 51 to 57 years in the UK Biobank. Both the CHARGE cohorts and the UK Biobank included more women than men, 59.1% and 54.2%, respectively. Demographic characteristics of participants are shown in Supplemental Table 1.
Association of the DASH diet score with systolic BP.
As shown in Supplemental Figure 1, higher DASH diet score was inversely associated with systolic BP. Systolic BP was 2.4±0.4 mm Hg lower in the FHS (P = 6.1e-8) and 1.1±0.2 mm Hg lower in the UK Biobank (P = 4.2e-12) per 10 units increase in the DASH diet score. Additional adjustment for BMI reduced the effect size to some extent: the inverse association became nonsignificant (0.2±0.2 mm Hg; P = 0.24) in UK Biobank, while the association remained significant in FHS (1.4±0.4 mm Hg; P = 7.6e-4).
Gene-DASH diet score interaction in relation to systolic BP in EUR population.
We examined 8,454,957 common biallelic SNPs available in at least two EUR cohorts. In the meta-analysis of all EUR individuals, we found potential interaction for the quantitative DASH score at three independent loci at 15q25.1 (lead SNP rs117878928, MTHFS, Pint =4e-8), 16q23.1 (lead SNP rs28562150, WWOX, Pint =3.9e-8), and 18q21.2 (lead SNP rs138826501, Pint =2.6e-8) (Table 1; Supplemental Figures 2 and 3). The direction of the interaction between rs117878928 (MTHFS at 15q25.1) and the quantitative DASH score was consistent in all study populations (Phet = 0.35; Supplemental Figure 4). The interaction effect size (mean and SE) for the lead SNP, rs117878928, was 0.42±0.09 (Pint = 9.4e-7) and 0.20±0.06 (Pint = 0.001) in the CHARGE cohorts and the UK Biobank, respectively. The other two loci were statistically significant in the CHARGE cohorts but not in the UK Biobank (Table 1).
Table 1.
Statistically Significantly interacting loci with the quantitative DASH diet score on systolic BP in meta-analysis of EUR participants.
| UKB | CHARGE | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||||
| SNP | Gene | Chr | Pos.38 | Region | EA | OA | EAF | Effect | SE | Pint | Phet | N | Effect | SE | P | N | Effect | SE | P |
|
| |||||||||||||||||||
| rs117878928 | MTHFS | 15 | 79841858 | 15q25.1 | A | C | 0.03 | 0.28 | 0.05 | 4e-8 | 0.35 | 86932 | 0.20 | 0.06 | 0.001 | 28507 | 0.42 | 0.09 | 9.4e-7 |
| rs28562150 | WWOX | 16 | 78530453 | 16q23.1 | A | G | 0.01 | 0.48 | 0.09 | 3.9e-8 | 1.85e-8 | 86932 | 0.21 | 0.11 | 0.05 | 13628 | 0.99 | 0.15 | 2.7e-11 |
| rs138826501 | 18 | 51789596 | 18q21.2 | T | C | 0.01 | 0.37 | 0.07 | 2.6e-8 | 0.003 | 86932 | 0.12 | 0.11 | 0.25 | 6945 | 0.54 | 0.09 | 4.2e-10 | |
Interaction analysis with dichotomized DASH diet scores.
To explore if the interaction analysis was influenced by a threshold effect, we analyzed the DASH diet score dichotomized by either the cohort-specific median or lower quartile (Supplemental Figure 2). In EUR participants, correlations of effect size and log10 Pint were slightly stronger in the analysis using the continuous DASH diet score compared to the median or lower quartile dichotomized DASH score (Supplemental Figure 5). For SNPs with Pint < 1e-3 in the quantitative DASH diet score analysis, their effect sizes and log10 Pint were correlated with that from analyses using the median dichotomized DASH score (Pearson r = 0.92 and 0.31, respectively; Supplemental Figure 6) and the lower quartile dichotomized DASH score (Pearson r = 0.74 and 0.16, respectively; Supplemental Figure 6).
Colocalization analysis.
We examined whether the lead interaction SNPs at the three loci colocalized with known systolic BP associated SNPs at the same loci (±1Mb region surrounding the lead SNPs). At the MTHFS locus (15q25.1), the colocalization analysis demonstrated that rs117878928 (MTHFS at 15q25.1) was a potential causal variant of systolic BP independent of the GWAS SNPs (i.e., SNPs with significant main effect in previous GWAS; posterior probably of H3 = 0.97; Table 2; Supplemental Figure 7). Because of low posterior probability of H1, H3, or H4, colocalization analysis did not support that the interaction SNPs identified in the other two loci (16q23.1 and 18q21.2) were causal variants to systolic BP (Table 2).
Table 2.
Colocalization analysis for the three statistically significant interaction loci in the EUR analysis.
| Lead SNP | rs117878928 | rs28562150 | rs138826501 |
| Number of SNPs tested | 5672 | 9885 | 5304 |
|
| |||
| PP.H0 | 0.00 | 0.57 | 0.00 |
| PP.H1 | 0.00 | 0.18 | 0.00 |
| PP.H2 | 0.03 | 0.19 | 0.91 |
| PP.H3 | 0.97 | 0.06 | 0.09 |
| PP.H4 | 0.00 | 0.01 | 0.00 |
Stratified analysis of lead SNP at 15q25.1.
We conducted stratified analyses by rs117878928 genotype, CC (dosage ≤ 0.35, n = 8,1792), CA (dosage ≥ 0.75 and ≤ 1.25, n= 4,967), AA (dosage ≥ 1.65, n= 75) in the UK Biobank. CA and AA were combined due to small sample size (n = 5,042), and participants with ambiguous rs117878928 genotype were excluded. After adjusting for sex, age, age squared, energy intake, and BMI, we observed that one SD higher DASH diet score was associated with 0.15±0.07 (P = 0.02) mm Hg lower systolic BP in individuals with CC genotype, whereas one SD higher DASH diet score was associated with 0.78±0.27 (P = 0.004) mm Hg higher systolic BP in those with CA or AA genotype.
Expression and DNA methylation quantitative loci variants at the MTHFS locus.
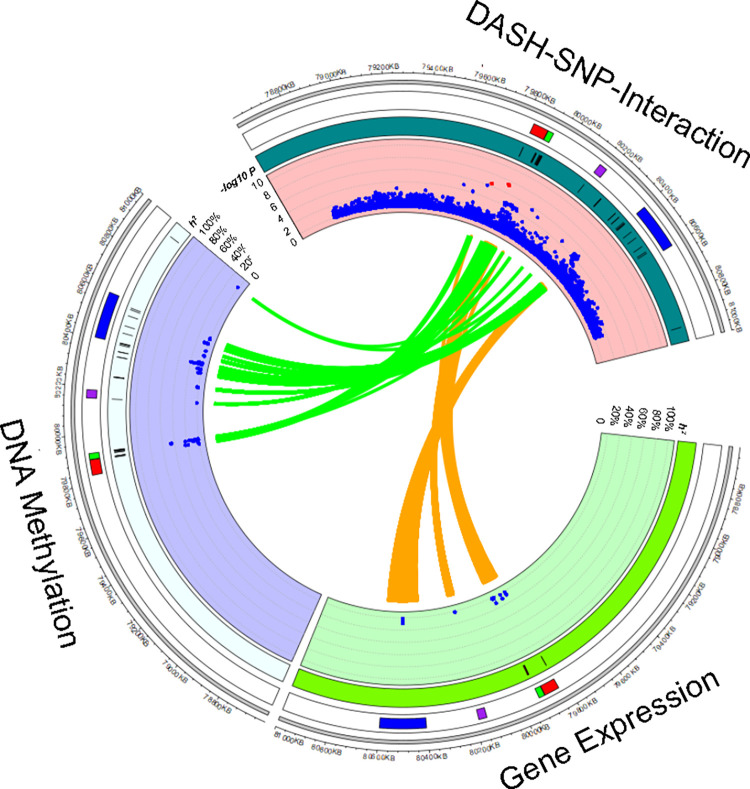
Low heterogeneity and the colocalization analysis suggest that, out of the significant three loci, rs117878928 may be causally interacting with the quantitative DASH score and associated with systolic BP. We further found that this locus was enriched with both cis-eQTL variants (Fisher exact test; P = 4e-273) and cis-mQTL variants (P = 1e-300). Figure 2 showed the link between SNPs with Pint < 1e-3 and cis-eQTL and cis-meQTL variants. In the 1 mb region around the lead SNP (rs117878928), we found 419 cis-eQTL variant-gene transcript pairs from 144 unique cis-eQTL variants and seven genes (including four protein coding genes; Supplemental Table 4) and 1,629 cis-mQTL variant-CpG pairs from 151 unique cis-mQTL variants and 32 CpGs (mapped to five protein coding genes; Supplemental Table 5) [39, 40]. For example, we found that a cis-eQTL variant, rs12915498 (Pint = 8.8e-4), accounted for 9.4% of heritability of expression levels of MTHFS and a cis-meQTL variant, rs11856431 (Pint = 6.8e-4), accounted for 22.7% of heritability of methylation levels of cg23855392 (a DNA methylation site ~6 kb away from the transcription start site of MTHFS). The highest heritability accounted by SNPs potentially interacting with the DASH diet score was for ST20 (a neighbor gene of MTHFS at 15q25.1). A cis-eQTL variant, rs35666771 (Pint = 5.2e-5), accounted for 11.1% of heritability of expression levels of ST20 and a cis-mQTL, rs3178646 (Pint = 9.9e-4), accounted for 46.5% of heritability of methylation levels of cg21315874 (a DNA methylation site ~6 kb away from the transcription start site of ST20).
We found that 139 cis-mQTL variants with Pint < 0.001 at the MTHFS locus were linked to four CpGs (cg13805518, cg02196730, cg26673396, and cg00225070) that were associated with a Mediterranean-style diet score at random-effect meta-analysis P < 0.05 [41]. The top cis-mQTL variants of the four diet-associated CpGs are presented in Supplemental Table 6. Furthermore, 81.2% of cis-mQTL variants associated with cg13805518 (annotated to ARNT2 at 15q25.1) were cis-mQTLs of cg13148921 (another CpG annotated to ARNT2; Supplemental Table 7), which was nominally associated with systolic BP in previous meta-analysis (P = 0.01) [42]. For example, rs11072902 (Pint = 2.6e-5) was associated with cg13805518 at P = 1e-46 and cg13148921 at P = 1e-8 [40]. However, none of the cis-mQTLs of cg13805518 and cg13148921 was in strong LD with the lead SNP (rs117878928) for gene-DASH diet score interaction, LD R2 < 0.01.
2df test in EUR participants.
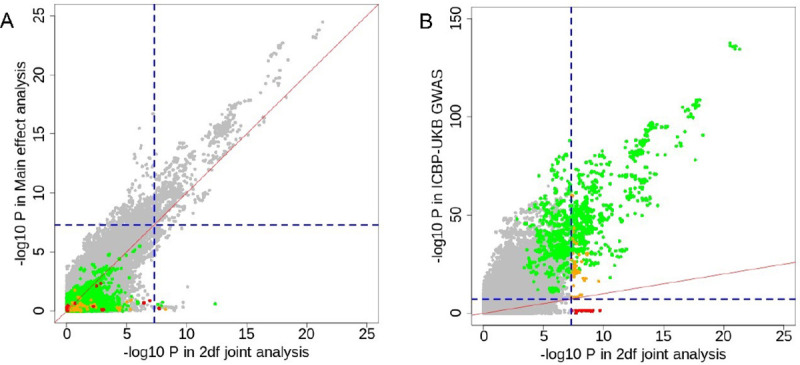
In secondary analysis, we ran a 2df test to jointly evaluate the SNP and diet-SNP interaction effect. Most SNPs (93%; n=1230) with Pjoint < 5e-8 in the 2df joint analyses were driven by their main genetic effect on systolic BP (Pmain < 5e-8; Figure 3A). We found that 11 loci reached Pjoint < 5e-8 with main effect Pmain ≥ 0.001 (Supplemental Table 8; Figure 3A). In these eleven loci, five loci (45%) had Pint < 0.001 (Supplemental Table 8). We further compared our 2df test statistics with that from the GWAS (i.e. main genetic effect) conducted by the International Consortium for Blood Pressure (ICBP) and the UK Biobank in N~750k EUR individuals [15]. As shown in Figure 3B, 98.7% (1230) SNPs with Pjoint < 5e-8 in the 2df test had P < 5e-8 in the previous GWAS for BP [15]. Among these, none of the SNPs with Pjoint < 5e-8 in the 2df test had Pint < 0.001, and 43 SNPs had Pint < 0.005. Five loci with Pint < 0.05 were shown in Supplemental Table 9.
Cross-population analysis.
In this analysis, we included 9,420,585 bi-allelic SNPs available in at least two cohorts with different populations. We found that five loci reached Pint < 5e-8, including one locus at 16q23.1 with statistical significance in EUR analysis. It should be noted that three of these five loci are driven by low frequency SNPs. (Table 3; Supplemental Figure 8 for Manhattan plot; Supplemental Figure 9 for regional plots). The lead SNPs in the five loci are intronic variants to THSD7B, SPATA5, UBE3D, GATA4, and WWOX, respectively. None of the SNPs with Pint < 0.05 at 1 mb region surrounding the five lead SNPs overlapped with SNPs associated with systolic BP (P < 1e-5) in the GWAS catalog [43].
Table 3.
Statistically significant SNPs in cross-population meta-analysis of gene-quantitative DASH diet score interaction.
| SNP | Chr | Pos.hg38 | Gene | Region | N | EA | OA | EAF | Beta | SE | Interaction P | Heterogeneity P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| rs145158769 | 2 | 137235853 | THSD7B | 2q22.1 | 4918 | A | T | 0.02 | −1.73 | 0.3 | 6e-9 | 1.4e-5 |
| rs180939244 | 4 | 122940020 | SPATA5 | 4q28.1 | 102530 | C | G | 0.71 | −0.44 | 0.08 | 9.5e-9 | 4.4e-16 |
| rs117137155 | 6 | 82895547 | UBE3D | 6q14.1 | 6038 | T | C | 0.99 | 2.06 | 0.28 | 1e-13 | 8.3e-11 |
| rs116170345 | 8 | 11689753 | GATA4 | 8p23.1 | 3477 | A | C | 0.99 | −1.26 | 0.21 | 1.6e-9 | 0.04 |
| rs28633096 | 16 | 78529552 | WWOX | 16q23.1 | 109405 | T | C | 0.31 | 0.41 | 0.08 | 5e-8 | 6.7e-6 |
Discussion
In this genome-wide interaction analysis in the CHARGE cohorts and the UK biobank, we showed that the association of DASH diet score with systolic BP was modified by multiple SNPs: at three loci in EUR analyses and an additional four loci in cross-population analysis. Our interaction SNP hits are independent of known BP loci from BP-GWAS. Furthermore, at the MTHFS locus (15q25.1), we demonstrated that the SNP-DASH diet score interaction may affect systolic BP through regulating levels of DNA methylation at this locus. While limitations exist in this study, our findings provide novel insights into gene-diet interactions in BP with respect to a better understanding of potential mechanisms of BP regulation and more personalized dietary advice.
Hypertension is the leading risk factor for cardiovascular disease [44]. Both environmental and genetic risk factors can lead to hypertension. A recent review summarized gene-lifestyle interactions in relation to hypertension and highlighted several genes that may interact with lifestyle factors to modify the risk of hypertension [45]. For example, several studies investigated interactions between genes and dietary components such as salt, alcohol, and fat intake [19, 46–48]. In a study of 4,414 individuals who participated in the Korean Genome and Epidemiology Study, an interaction was observed between rs3784789 (intronic to CSK and upstream to MIR4513) and estimated 24-hour urinary sodium to potassium ratio in relation to the risk of hypotension [46]. This SNP was nominally significant in the present interaction analysis, both in the EUR and cross-population analyses (P = 0.009 and 0.005, respectively). As also indicated in this previous review study [44], most studies had modest sample sizes, and the observed interactions were often different across studies. In the present study, we also observed that most of the loci with significant interactions had high heterogeneity, specifically the poor replication between observations in the CHARGE cohorts and the UK Biobank (Supplemental Figure 10). This high heterogeneity may be partly due to the diverse food habits that limit the ability of the DASH diet score to consistently reflect overall diet quality across cohorts or different dietary tools used in our participating cohorts. The present study adds novel evidence to the literature regarding interaction between genetic variants and overall diet quality, nonetheless, future analyses with larger sample sizes and better harmonized dietary information are needed to validate our findings.
At 15q25.1, the lead SNP, rs117878928 resides ~2 kb downstream of long noncoding RNA (LOC124903536) and ~2 kb upstream of a protein coding gene (MTHFS). MTHFS encodes methenyltetrahydrofolate synthase that catalyzes the conversion of 5-formyltetrahydrofolate to 5,10-methenyltetrahydrofolate. Activation of MTHFS may accelerate folate catabolism by modifying folate one-carbon forms, leading to impaired methylation reactions such as DNA methylation [49]. Folate metabolism has been implicated in the risk of hypertension [50, 51], although its role is not fully established. In an earlier study conducted in EUR participants from two participating cohorts of the present study, the Atherosclerosis Risk Communities study (ARIC) and the FHS, an intronic variant (rs6495446) of MTHFS has been linked to chronic kidney disease [52], a disease tightly associated with elevated BP. However, rs6495446 is not in LD with rs117878928 (R2 = 0.006).
Our observation regarding the enrichment of cis-mQTLs in this locus appears to be consistent with the functionality of MTHFS. In FHS, SNP rs117878928 is a cis-mQTL variant for cg21315874 (h2 = 0.015, P = 4.4e-15) [40], which is residing at 5’ untranslated region (UTR) of ST20. In the Genetics of DNA Methylation Consortium (GoDMC) [53], rs117878928 is also a cis-mQTL variant for other CpGs (e.g., cg22389121, P=2.3e-42) at 5’UTR or close the transcription start site of ST20. ST20 is adjacent to MTHFS, and it is a tumor suppressing gene involved in several processes such as apoptotic signaling pathway, cellular response to ultraviolet C, and negative regulation of cell growth [54]. ST20-MTHS readthrough transcript can be formed through splicing to produce a fusion protein that shares sequence identity from both genes, which are highly expressed in the liver and kidney (https://www.proteinatlas.org) [55]. Overall, our observations indicate that a DNA methylation related mechanism may be relevant to the gene-DASH diet score interaction observed in this region.
The joint analysis of SNP main effects and interaction effects has been shown to be more powerful than the analysis of SNP main effects or interaction effects alone, when the genetic effects are relatively weak and the interaction effects are moderate [36, 56]. Thus, the joint analysis is a promising approach to identify additional loci relevant to traits of interest. In the present study, we compared the joint analysis results with main effect statistics obtained from our study sample and a larger GWAS for systolic BP [15], and we identified several candidate loci for future validation (Supplemental Table 8). For example, the significant 2df test for an intronic SNP (rs140635454) of NUP93 was driven by its interaction with the DASH diet score. A recent study reported a significant 2df test for rs76976871 at this gene in a sleep-by-gene interaction analysis for high density lipoprotein cholesterol [57]. Although the interaction term was modest in the sleep study [57], our results and theirs combined highlight the importance of conducting a comprehensive joint analysis to facilitate identification of loci that are potentially modified by diet and other lifestyle factors.
There are several limitations in this study that should be discussed. First, the cross-sectional nature of the study does not allow us to account for reverse causation, which might have occurred if individuals with hypertension were advised to change their dietary patterns to help control their blood pressure. The DASH diet score was calculated using different versions of FFQs in the CHARGE cohorts and multiple 24-hour recalls in the UK Biobank. All these dietary assessments tools are based on self-reported dietary intake, which is subjective to both random and systematic bias [58]. In addition, different versions of FFQs have different food lists and different levels of detail, e.g., a 126-item FFQ was used in FHS, while the ARIC Study used a 66-item FFQ; therefore, some DASH diet score components may include a different numbers and types of food items. These combined may partly explain the high heterogeneity regarding gene-DASH diet score interaction across cohorts. The majority of our study participants (91%) were EUR; our cross-population analysis was therefore mainly driven by the EUR participants. Furthermore, the numerous associations detected in the lower quartile dichotomized DASH score are probably due to misclassification, therefore larger sample size to replicate the threshold analysis is necessary.
In conclusion, we demonstrated gene-DASH diet score interaction in several loci, particularly at the MTHFS locus (15q25.1). In addition, we showed that DNA methylation may be a relevant mechanism linking gene-diet interaction for systolic BP. Compared to large GWAS, the sample size of the present study is modest; therefore, studies with larger sample size and more diverse populations are needed to validate our findings and to facilitate the measure of inter-individual differences in dietary response, allowing for identification of high risk subgroups who would benefit the most from dietary modifications.
Acknowledgements
ARIC
The Atherosclerosis Risk in Communities study (ARIC) has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL059367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
CHS
This Cardiovascular Health Study (CHS) research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, 75N92021D00006; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and U01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
FHS
The authors thank the staff and participants of the Framingham Heart Study (FHS) study for their important contributions. The Framingham Heart Study was supported by NIH contracts N01-HC-25195, HHSN268201500001I, and 75N92019D00031.
NEO
The authors of the NEO study thank all individuals who participated in the Netherlands Epidemiology in Obesity study, all participating general practitioners for inviting eligible participants and all research nurses for collection of the data. We thank the NEO study group, Petra Noordijk, Pat van Beelen and Ingeborg de Jonge for the coordination, lab and data management of the NEO study. The genotyping in the NEO study was supported by the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze. The NEO study is supported by the participating Departments, the Division and the Board of Directors of the Leiden University Medical Center, and by the Leiden University, Research Profile Area Vascular and Regenerative Medicine. Raymond Noordam was supported by a grant from NHLBI (grant number R01 HL156991).
Raine Study
The Raine Study was supported by the National Health and Medical Research Council of Australia [Grant Numbers 572613, 403981, 1059711, 1027449, 1044840, 1021858], the Canadian Institutes of Health Research [Grant Number MOP-82893], and WA Health, Government of Western Australia (WADOH) [Future Health WA G06302]. Funding was also generously provided by Safe Work Australia. The authors are grateful to the Raine Study participants and their families, and to the Raine Study team for cohort coordination and data collection. The authors gratefully acknowledge the NHMRC for their long-term funding to the study over the last 30 years and also the following institutes for providing funding for Core Management of the Raine Study: The University of Western Australia (UWA), Curtin University, Women and Infants Research Foundation, Telethon Kids Institute, Edith Cowan University, Murdoch University, The University of Notre Dame Australia and The Raine Medical Research Foundation. This work was supported by resources provided by the Pawsey Supercomputing Centre with funding from the Australian Government and the Government of Western Australia.
SP & LVB
The Multi-Ethnic Cohort and Singapore Population Health Study 2012 studies (from which the Singapore Prospective Study Program [SP] and Living Biobank [LVB] are drawn from are supported by individual research and clinical scientist award schemes from the National Medical Research Council (NMRC) and the Biomedical Research Council (BMRC) of Singapore, the Singapore Ministry of Health, National University of Singapore and National University Health System, Singapore.
WHI
Women’s Health Initiative (WHI) funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: https://www-whi-org.s3.us-west-2.amazonaws.com/wp-content/uploads/WHI-Investigator-Long-List.pdf. Additional support to NF was provided by NIH DK117445 and MD012765.
UK Biobank
UKB (United Kingdom Biobank): The authors thank the staff and participants of the UK Biobank study for their important contributions. This research has been conducted using the UK Biobank Resource under the UK Biobank project number 56735 and application title “Interaction of Diet Quality Scores and Genetic Variants in Relation to Cardiometabolic Traits”.
Disclosed Funding and sources of support:
JM: Supported by K22-HL-135075; DCR: R01 HL118305 and R01 HL156991; PBM and HRW acknowledge the support of the NIHR Biomedical Research Centre, Queen Mary University of London; NR funding: R01-MD012765, R01-DK117445, R01-HL163972; This project was partly supported by the Intramural Research Program of the National Human Genome Research Institute of the National Institutes of Health through the Center for Research on Genomics and Global Health (CRGGH).
Melanie Guirette (Tufts University Friedman School of Nutrition Science and Policy)
Jessie Lan (Tufts University Friedman School of Nutrition Science and Policy)
Nicola Mckeown (Boston University)
Michael Brown (University of Texas Health Science Center at Houston School of Public Health)
Han Chen (The University of Texas Health Science Center at Houston School of Public Health)
Paul De Vries (University of Texas Health Science Center at Houston)
Hyunju Kim (University of Washington)
Casey Rebholz (Johns Hopkins Bloomberg School of Public Health)
Alanna Morrison (University of Texas Health Science Center at Houston)
Traci Bartz (University of Washington)
Amanda Fretts (University of Washington)
Xiuqing Guo (Harbor-UCLA Medical Center)
Rozenn Lemaitre (University of Washington)
Ching-Ti Liu (Boston University School of Public Health)
Raymond Noordam (Leiden University Medical Center)
Renée de Mutsert (Leiden University Medical Center)
Frits Rosendaal (Leiden University Medical Center)
Carol Wang (The University of Newcastle)
Lawrence Beilin (University of Western Australia)
Trevor Mori (Universty of Western Australia)
Wendy Oddy (University of Tasmania)
Craig Pennell (University of Newcastle)
Jin-Fang Chai (National University of Singapore and National University Health System)
Clare Whitton (Curtin University School of Population Health)
Rob van Dam (National University of Singapore)
Jianjun Liu (Genome Institute of Singapore)
E Shyong Tai (National University of Singapore and National University Health System)
Xueling Sim (National University of Singapore and National University Health System)
Marian Neuhouser (Fred Hutchinson Cancer Research Center)
Charles Kooperberg (Fred Hutchinson Cancer Center)
Lesley Tinker (Fred Hutchinson Cancer Research Center)
Nora Franceschini (University of North Carolina at Chapel Hill)
TianXiao Huan (National Heart, Lung, and Blood Institute)
Thomas Winkler (University of Regensburg)
Amy Bentley (National Institutes of Health)
W. James Gauderman (University of Southern California)
Luc Heerkens (Wageningen University)
Toshiko Tanaka (National Institute on Aging)
Jeroen Van Rooij (Erasmus Medical Center)
Patricia Munroe (Queen Mary University of London)
Helen Warren (Queen Mary University of London)
Trudy Voortman (Erasmus MC, University Medical Center)
Honglei Chen (Michigan State University)
D.C. Rao (Washington University in St. Louis, School of Medicine)
Daniel Levy (National Heart Lung and Blood Institute)
Jiantao Ma (Tufts University Friedman School of Nutrition Science and Policy)
References
- 1.Tsao C.W., et al. , Heart Disease and Stroke Statistics-2023 Update: A Report From the American Heart Association. Circulation, 2023. 147(8): p. e93–e621. [DOI] [PubMed] [Google Scholar]
- 2.Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet, 2021. 398(10304): p. 957–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appel L.J., et al. , A clinical trial of the effects of dietary patterns on blood pressure. DASH Collaborative Research Group. N Engl J Med, 1997. 336(16): p. 1117–24. [DOI] [PubMed] [Google Scholar]
- 4.Sacks F.M., et al. , Effects on blood pressure of reduced dietary sodium and the Dietary Approaches to Stop Hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N Engl J Med, 2001. 344(1): p. 3–10. [DOI] [PubMed] [Google Scholar]
- 5.Appel L.J., et al. , Effects of comprehensive lifestyle modification on blood pressure control: main results of the PREMIER clinical trial. JAMA, 2003. 289(16): p. 2083–93. [DOI] [PubMed] [Google Scholar]
- 6.Appel L.J., et al. , Effects of protein, monounsaturated fat, and carbohydrate intake on blood pressure and serum lipids: results of the OmniHeart randomized trial. JAMA, 2005. 294(19): p. 2455–64. [DOI] [PubMed] [Google Scholar]
- 7.Sacks F.M., et al. , Effects of high vs low glycemic index of dietary carbohydrate on cardiovascular disease risk factors and insulin sensitivity: the OmniCarb randomized clinical trial. JAMA, 2014. 312(23): p. 2531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djousse L., et al. , DASH Score and Subsequent Risk of Coronary Artery Disease: The Findings From Million Veteran Program. J Am Heart Assoc, 2018. 7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung T.T., et al. , Adherence to a DASH-style diet and risk of coronary heart disease and stroke in women. Arch Intern Med, 2008. 168(7): p. 713–20. [DOI] [PubMed] [Google Scholar]
- 10.Mertens E., et al. , Adherence to a healthy diet in relation to cardiovascular incidence and risk markers: evidence from the Caerphilly Prospective Study. Eur J Nutr, 2018. 57(3): p. 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folsom A.R., Parker E.D., and Harnack L.J., Degree of concordance with DASH diet guidelines and incidence of hypertension and fatal cardiovascular disease. Am J Hypertens, 2007. 20(3): p. 225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blumenthal J.A., et al. , Effects of the DASH diet alone and in combination with exercise and weight loss on blood pressure and cardiovascular biomarkers in men and women with high blood pressure: the ENCORE study. Arch Intern Med, 2010. 170(2): p. 126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y. and Wang J.G., Genome-Wide Association Studies of Hypertension and Several Other Cardiovascular Diseases. Pulse (Basel), 2019. 6(3–4): p. 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren H.R., et al. , Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet, 2017. 49(3): p. 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evangelou E., et al. , Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet, 2018. 50(10): p. 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren H., et al. , Genome-wide analysis in over 1 million individuals reveals over 2,000 independent genetic signals for blood pressure. Research Square, 2022. [Google Scholar]
- 17.Pazoki R., et al. , Genetic Predisposition to High Blood Pressure and Lifestyle Factors: Associations With Midlife Blood Pressure Levels and Cardiovascular Events. Circulation, 2018. 137(7): p. 653–661. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y., Ye Z., and Fang Y., Spatial analysis of the effects of PM2.5 on hypertension among the middle-aged and elderly people in China. Int J Environ Health Res, 2021. 31(6): p. 729–740. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., et al. , Neuropeptide Y promoter polymorphism modifies effects of a weight-loss diet on 2-year changes of blood pressure: the preventing overweight using novel dietary strategies trial. Hypertension, 2012. 60(5): p. 1169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tagetti A., et al. , Intakes of omega-3 polyunsaturated fatty acids and blood pressure change over time: Possible interaction with genes involved in 20-HETE and EETs metabolism. Prostaglandins Other Lipid Mediat, 2015. 120: p. 126–33. [DOI] [PubMed] [Google Scholar]
- 21.Goni L., et al. , Influence of fat intake and BMI on the association of rs1799983 NOS3 polymorphism with blood pressure levels in an Iberian population. Eur J Nutr, 2017. 56(4): p. 1589–1596. [DOI] [PubMed] [Google Scholar]
- 22.Htun N.C., et al. , Association of the catechol-O-methyl transferase gene Val158Met polymorphism with blood pressure and prevalence of hypertension: interaction with dietary energy intake. Am J Hypertens, 2011. 24(9): p. 1022–6. [DOI] [PubMed] [Google Scholar]
- 23.Psaty B.M., et al. , Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet, 2009. 2(1): p. 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Psaty B.M. and Sitlani C., The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium as a model of collaborative science. Epidemiology, 2013. 24(3): p. 346–8. [DOI] [PubMed] [Google Scholar]
- 25.Rao D.C., et al. , Multiancestry Study of Gene-Lifestyle Interactions for Cardiovascular Traits in 610 475 Individuals From 124 Cohorts: Design and Rationale. Circ Cardiovasc Genet, 2017. 10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudlow C., et al. , UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med, 2015. 12(3): p. e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao D.C., et al. , Multiancestry Study of Gene-Lifestyle Interactions for Cardiovascular Traits in 610 475 Individuals From 124 Cohorts: Design and Rationale. Circ Cardiovasc Genet, 2017. 10(3): p. e001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B., et al. , Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr, 2011. 14(11): p. 1998–2005. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y., et al. , Higher diet quality relates to decelerated epigenetic aging. Am J Clin Nutr, 2022. 115(1): p. 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentley A.R., et al. , Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat Genet, 2019. 51(4): p. 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature, 2018. 562(7726): p. 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler T.W., et al. , Quality control and conduct of genome-wide association meta-analyses. Nat Protoc, 2014. 9(5): p. 1192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C., et al. , A global reference for human genetic variation. Nature, 2015. 526(7571): p. 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westerman K.E., et al. , Genome-wide gene-diet interaction analysis in the UK Biobank identifies novel effects on hemoglobin A1c. Hum Mol Genet, 2021. 30(18): p. 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer C.J., Li Y., and Abecasis G.R., METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 2010. 26(17): p. 2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraft P., et al. , Exploiting gene-environment interaction to detect genetic associations. Hum Hered, 2007. 63(2): p. 111–9. [DOI] [PubMed] [Google Scholar]
- 37.Evangelou E., et al. , Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet, 2018. 50(10): p. 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giambartolomei C., et al. , Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet, 2014. 10(5): p. e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C., et al. , Whole genome DNA and RNA sequencing of whole blood elucidates the genetic architecture of gene expression underlying a wide range of diseases. Sci Rep, 2022. 12(1): p. 20167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma J., et al. , Elucidating the genetic architecture of DNA methylation to identify promising molecular mechanisms of disease. Sci Rep, 2022. 12(1): p. 19564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J., et al. , Whole Blood DNA Methylation Signatures of Diet Are Associated With Cardiovascular Disease Risk Factors and All-Cause Mortality. Circ Genom Precis Med, 2020. 13(4): p. e002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richard M.A., et al. , DNA Methylation Analysis Identifies Loci for Blood Pressure Regulation. Am J Hum Genet, 2017. 101(6): p. 888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sollis E., et al. , The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res, 2023. 51(D1): p. D977–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carey R.M., et al. , Prevention and Control of Hypertension: JACC Health Promotion Series. J Am Coll Cardiol, 2018. 72(11): p. 1278–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokubo Y., et al. , Gene and environmental interactions according to the components of lifestyle modifications in hypertension guidelines. Environ Health Prev Med, 2019. 24(1): p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park Y.M., et al. , Interaction between Single Nucleotide Polymorphism and Urinary Sodium, Potassium, and Sodium-Potassium Ratio on the Risk of Hypertension in Korean Adults. Nutrients, 2017. 9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simino J., et al. , Gene-alcohol interactions identify several novel blood pressure loci including a promising locus near SLC16A9. Front Genet, 2013. 4: p. 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feitosa M.F., et al. , Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS One, 2018. 13(6): p. e0198166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anguera M.C., et al. , Methenyltetrahydrofolate synthetase regulates folate turnover and accumulation. J Biol Chem, 2003. 278(32): p. 29856–62. [DOI] [PubMed] [Google Scholar]
- 50.McRae M.P., High-dose folic acid supplementation effects on endothelial function and blood pressure in hypertensive patients: a meta-analysis of randomized controlled clinical trials. J Chiropr Med, 2009. 8(1): p. 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forman J.P., et al. , Folate intake and the risk of incident hypertension among US women. JAMA, 2005. 293(3): p. 320–9. [DOI] [PubMed] [Google Scholar]
- 52.Kottgen A., et al. , Genome-wide association study for renal traits in the Framingham Heart and Atherosclerosis Risk in Communities Studies. BMC Med Genet, 2008. 9: p. 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Min J.L., et al. , Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet, 2021. 53(9): p. 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim T.E., et al. , Candidate tumor suppressor, HCCS-1, is downregulated in human cancers and induces apoptosis in cervical cancer. Int J Cancer, 2002. 97(6): p. 780–6. [DOI] [PubMed] [Google Scholar]
- 55.Uhlen M., et al. , Proteomics. Tissue-based map of the human proteome. Science, 2015. 347(6220): p. 1260419. [DOI] [PubMed] [Google Scholar]
- 56.Manning A.K., et al. , Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP x environment regression coefficients. Genet Epidemiol, 2011. 35(1): p. 11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noordam R., et al. , Multi-ancestry sleep-by-SNP interaction analysis in 126,926 individuals reveals lipid loci stratified by sleep duration. Nat Commun, 2019. 10(1): p. 5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bingham S.A., et al. , Comparison of dietary assessment methods in nutritional epidemiology: weighed records v. 24 h recalls, food-frequency questionnaires and estimated-diet records. Br J Nutr, 1994. 72(4): p. 619–43. [DOI] [PubMed] [Google Scholar]