Abstract
Hydrophobins are small (length, about 100 ± 25 amino acids), cysteine-rich, hydrophobic proteins that are present in large amounts in fungal cell walls, where they form part of the outermost layer (rodlet layer); sometimes, they can also be secreted into the medium. Different hydrophobins are associated with different developmental stages of a fungus, and their biological functions include protection of the hyphae against desiccation and attack by either bacterial or fungal parasites, hyphal adherence, and the lowering of surface tension of the culture medium to permit aerial growth of the hyphae. We identified and isolated a hydrophobin (fruit body hydrophobin 1 [Fbh1]) present in fruit bodies but absent in both monokaryotic and dikaryotic mycelia of the edible mushroom Pleurotus ostreatus. In order to study the temporal and spatial expression of the fbh1 gene, we determined the N-terminal amino acid sequence of Fbh1. We also synthesized and cloned the double-stranded cDNA corresponding to the full-length mRNA of Fbh1 to use it as a probe in both Northern blot and in situ hybridization experiments. Fbh1 mRNA is detectable in specific parts of the fruit body, and it is absent in other developmental stages.
Pleurotus ostreatus is an edible basidiomycete in which there has been increasing biotechnological interest due to its ability to degrade wood (12, 24) and chemicals related to lignin degradation products (3). Furthermore, this fungus produces secondary metabolites with pharmaceutical applications (4, 5, 19) and some proteins that may have industrial use (29, 30). P. ostreatus, as well as other higher fungi, can survive long desiccation periods, can attach to different substrates, and can survive attacks by some pathogens due to the unique properties of the hydrophobins present in their cell walls (38). Hydrophobins are members of a family of small hydrophobic proteins that have molecular masses around 10 kDa, are 100 ± 25 amino acids long, and are characterized by the conserved pattern of spacing of the eight cysteine residues present in their sequences (33). In vitro, hydrophobins spontaneously self-assemble at water-air interfaces into highly insoluble protein layers (rodlet layers) whose dissociation requires treatment with strong agents, such as performic acid (35) or trifluoroacetic acid (TFA) (8). In vivo, hydrophobins are synthesized as preproteins, processed, and exported out of the hyphae, where they polymerize into the rodlet layer, which confers a high degree of hydrophobicity (32).
The formation of the rodlet layer seems to have a critical morphogenetic role as mutants affected in hydrophobin production are drastically affected in the formation of aerial structures in Schizophyllum commune (35). In addition, several independent experiments performed to identify genes that are actively transcribed at the time of fruit body development have resulted in the isolation of different members of the hydrophobin family of proteins (7, 9, 22, 26). The findings indicate that expression of hydrophobin genes is developmentally regulated. The spatial expression of the hydrophobin genes, however, has not been extensively studied, mainly because of technical problems that make in situ detection of the hydrophobin transcripts difficult.
Although hydrophobins have been extensively studied in many basidiomycetes (see reference 33 for a review), little is known about these proteins in P. ostreatus. The goals of the present study were to identify fruit-body-specific hydrophobins of the edible mushroom P. ostreatus and to study the distribution of transcripts coding for this protein in the structures of the fruit body of this fungus. This study was part of the search for genes which are differently expressed in different organs and tissues of higher fungi.
MATERIALS AND METHODS
Fungal strains and culture conditions.
P. ostreatus var. florida strain N001 (mating type genotype A1A2B1B2) was the commercial strain used in this work. It was kindly supplied by Gurelan S. C., Pamplona, Spain. Two monokaryon strains (M1 and M5) with complementary mating types (A1B1 and A2B2, respectively) were isolated from a collection of monokaryons produced after germination of spores released by the dikaryotic strain. Monokaryotic and dikaryotic mycelia were grown on petri dishes containing 25 ml of the malt agar medium described by Eger (20 g of malt extract, 15 g of agar, 1 liter of H2O) (10) at 24°C in the dark. For the analysis of the mRNA present in monokaryon and dikaryon mycelia, mycelia were fragmented and inoculated onto soft malt agar medium (20 g of malt extract, 7 g of agar, 1 liter of H2O) and incubated for 4 to 6 days under the conditions described above. For small-scale production of fruit bodies, the dikaryon was grown on petri dishes containing a thin layer of malt agar medium (approximately 8 to 10 ml of culture medium per plate) under the conditions described above until the mycelia filled the plates. Then fruiting was induced by a cold shock (4°C, overnight) and incubation was continued with intermittent illumination (12 h of light, 12 h of darkness) at 17°C and 90% humidity for 7 to 10 days (10). For large-scale production of fruit bodies, dikaryotic mycelium was grown in 5-liter plastic bags filled with sterile hydrolyzed rye straw at 24°C in the dark. After 7 to 10 days, fruiting was induced and incubation was continued as described above for small-scale fruit body production. Fruit bodies were processed immediately after harvesting or were frozen in liquid nitrogen and stored at −80°C.
The Escherichia coli strain used in this work is described below. Bacterial cultures were grown on Luria-Bertani medium (10 g of Bacto Tryptone [Difco], 10 g of yeast extract, 5 g of NaCl, 1 liter of H2O) (21) with shaking (200 rpm) at 37°C. Ampicillin was obtained from Sigma Chemical Co. (St. Louis, Mo.).
Fbh1 purification and analysis.
Fruit-body-specific hydrophobins of P. ostreatus were isolated by using protocols described elsewhere (8). The hydrophobins were isolated by obtaining the fraction of fruit bodies homogenized with an X-Press apparatus (AB Biox, Göteborg, Germany) which was insoluble in a boiling solution containing 10 g of sodium dodecyl sulfate (SDS) in 1 liter of 0.1 M sodium phosphate buffer (pH 7.0). In order to dissociate the insoluble hydrophobin aggregates, they were sonicated in TFA at 0°C, and the acid was then removed by flushing the dissociated fraction with a stream of nitrogen. The dried hydrophobin preparation (fruit body hydrophobin 1 [Fbh1]) was dissolved in water for further analysis.
For electrophoretic analysis of the hydrophobins, samples were prepared in the SDS sample buffer described by de Vries et al. (8) and were subjected to denaturing SDS-polyacrylamide gel electrophoresis (PAGE) in gels containing 15% polyacrylamide (20). The proteins resolved were revealed by using the silver staining method described by Merril (25). The protein molecular weight markers used were obtained from Gibco BRL (Life Technologies Ltd., Paisley, United Kingdom). For N-terminal sequencing, the proteins resolved by SDS-PAGE were electrotransferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore Corporation, Bedford, Mass.) by the semidry transfer method (Multiphor II electrophoresis system; Pharmacia, Uppsala, Sweden) at 0.8 mA cm−2, and the blotted proteins were sequenced by using a gas phase sequencer equipped with a phenylthiohydantoin analyzer (Applied Biosystems, Foster City, Calif.). For detection of carbohydrates, the proteins were transferred onto Immobilon P membranes as described above, and the membranes were incubated for 1 h at room temperature with a periodic acid solution (5 g of period acid, 1 liter of H2O). Then they were washed with dilute acetic acid (50 ml of acetic acid, 1 liter of H2O) and stained with Schiff reagent (Sigma).
Self-aggregation of hydrophobins at water-air interfaces was examined by electrobubbling (23). Briefly, 10 ml of a hydrophobin solution in water was placed in a glass tube (diameter, 1 cm), and two platinum electrodes were introduced opposite each other along the wall of the tube. Bubbling was accomplished by applying a constant current (20 mA) and was maintained until no more foam was formed. The protein aggregates present in the foam interfaces were collected by centrifugation.
Isolation, separation, and hybridization of RNA.
Total RNA was extracted by the hot phenol procedure, precipitated with lithium chloride, and denatured with formamide and formaldehyde (36). For Northern blot analysis the concentrations of RNA in the RNA samples were determined spectrophotometrically, and equal amounts of RNA were separated by electrophoresis on agarose gels (10 g of agarose per liter of 2.2 M formaldehyde) and transferred onto Hybond N+ membranes (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) (28). Hybridizations were performed at 65°C as described by Church and Gilbert (6).
cDNA clone isolation and DNA sequencing.
cDNA from total fruit body mRNA was synthesized by a reverse transcriptase (RT) reaction by using a 1st Strand cDNA synthesis kit for RT-PCR (avian myeloblastosis virus [AMV]) (Boehringer Mannheim, Mannheim, Germany) according to the supplier’s specifications. Each 50-μl reaction mixture contained 20 μg of total RNA (previously denatured at 65°C for 10 min), 50 U of AMV RT, 275 pmol of dT17 adapter primer (11), 300 mM dATP, 300 mM dCTP, 300 mM dGTP, 300 mM dTTP, 2.5 μl of RNase inhibitor, and 1× AMV RT buffer. The reaction mixture was incubated for 1 h at 50°C. To determine the sequence of the mRNA coding for Fbh1, a double-stranded DNA fragment was synthesized by a PCR in which 5 μl of the reverse transcription product was mixed with 200 mM dATP, 200 mM dCTP, 200 mM GTP, 200 mM dTTP, 2.5 U of Taq DNA polymerase (Promega, Southampton, United Kingdom), 100 pmol of a degenerate oligonucleotide primer corresponding to the possible coding sequence for the nine amino acids present in the amino-terminal sequence of mature Fbh1 (26-mer; 5′-ACNGARACNCCNGTNAAYCARTGYGG-3′), 30 pmol of dT17 adapter primer, each nucleotide at a concentration of 200 mM, and a buffer provided by the manufacturer. The PCR was carried out in a final volume of 50 μl. The cycling program started with a 5-min denaturation step at 94°C, which was followed by 30 cycles consisting of 1 min of annealing at 50°C, 1 min of extension at 72°C, and 1 min of denaturation at 94°C. To determine the sequence of the 5′ end of the mRNA coding for Fbh1, a 5′/3′ RACE kit (Boehringer Mannheim) was used with the following oligonucleotide primers based on the nucleotide sequence of the Fbh1 mRNA determined above: 5′-CAGGTTGACCCGCCCACTT-3′ and 5′-CTGGACATTGGTGAGGATG-3′. The amplified DNA fragments were cloned in pGEM-T vectors (Promega). Double-stranded DNA fragments were sequenced in both directions with a Thermo-Sequenase cycle sequencing kit (Amersham Ibérica, Madrid, Spain) by using a Vistra model 725 DNA sequencer (Molecular Dynamics Inc., Sunnyvale, Calif.).
Production of the hydrophobin Fbh1 in E. coli.
In order to produce the hydrophobin Fbh1 in E. coli cells, the pET system (Novagen, Madison, Wis.) was used. The region coding for the mature Fbh1 protein was amplified by PCR from the fruit body cDNA mixture synthesized above. The PCR was carried out by using as primers oligonucleotides which contain sequences corresponding to the N-terminal and C-terminal ends of mature Fbh1 and BamHI and HindIII restriction sites. The sequences of the primers are as follows (the restriction sites are underlined): 5′-CGCGGATCCACTGAAACGCCGGTTA-3′ (primer corresponding to the N-terminal end) and 5′-GGGAAGCTTATAGCATTGAGCAAGT-3′ (primer corresponding to the C-terminal end). The amplified DNA was digested with EcoRI and BamHI and cloned downstream from the six-histidine sequence (His-Tag sequence) present in the pET28c expression vector; the translation frame was kept in order to generate a fusion protein (His-Tag-Fbh1). The resulting plasmid, pET28c-Fbh1, was transformed into the E. coli BL21 (F− ompT rB− mB−) lysogen of bacteriophage DE3 (Novagen). The construct was sequenced to verify the correct frame of the fusion protein.
To recover the His-Tag-Fbh1 fusion protein, expression of the fusion protein was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) to a final concentration of 1 mM to an early-exponential-phase culture of E. coli BL21(DE3) transformed with plasmid pET28c-Fbh1. After 3 h of induction, the bacteria were harvested by centrifugation, resuspended in column binding buffer containing 5 mM imidazole, 500 mM NaCl, and 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (pH 7.9), and disrupted by sonication (Soniprep 150 apparatus; MSE, Frisons PLC, Houston, Tex.) until the sample was no longer viscous. The debris was removed by centrifugation (12,000 × g, 10 min), and the protein was purified from the supernatant by metal affinity chromatography as recommended by the manufacturer (Novagen). Following elution with imidazole, the purified fusion protein was concentrated to the desired volume with Centriprep 3 filters (Amicon, Beverly, Mass.). The fusion protein was conserved at 4°C.
Preparation of anti-Fbh1 antibodies.
To prepare anti-Fbh1 antibodies, the Fbh1 purified from mycelia and the fusion His-Tag-Fbh1 protein were subjected to denaturing SDS-PAGE. The positions of the proteins were determined by comparison with parallel lanes which were silver stained (see above). The unstained regions of the polyacrylamide gel containing the proteins were excised, homogenized with an X-press apparatus at −20°C, and resuspended in phosphate-buffered saline (8 g of NaCl, 0.2 g of KCl, 1.45 g of Na2HPO4 · 2H2O, 0.23 g of NaH2PO4 · H2O, 1 liter of H2O; pH 7.6), and the resulting preparation was injected subcutaneously five times at 10-day intervals into rabbits. Blood was collected before immunization and 7 days after the last injection. For immunodetection of the proteins, the proteins were resolved by SDS-PAGE, transferred to PVDF membranes as described above, and detected by using anti-Fbh1 and anti-fusion protein polyclonal antibodies and the ECL chemiluminescence system (Amersham).
In situ detection of fbh1 mRNA.
The pattern of expression of the fbh1 gene was investigated by in situ detection of the mRNA coding for Fbh1 by hybridization with the appropriate single-stranded RNA probes in paraffin-embedded sections of mature fruit bodies. The digoxigenin-labeled RNA probes were prepared by using a DIG RNA labeling kit according to the instructions of the manufacturer (Boehringer Mannheim). A DNA fragment coding for Fbh1 (see above) was cloned into the polycloning site of the pGEM-T vector, and the sense and antisense RNA probes were generated by using one of the two promoters that flank the polycloning site. Hybridization was performed as previously described by Goday et al. (14). In all cases, no signal greater than the background signal was observed when control sense strand probes were used.
Nucleotide and protein sequence comparisons.
The GeneJockeyII package (Biosoft, Cambridge, United Kingdom) was used to predict the amino acid sequence of Fbh1 from the nucleotide sequence data, to align sequences and amino acids, and to generate hydrophobicity plots. The predicted amino acid sequence of Fbh1 was compared with translated sequences obtained from GenBank and EMBL by using the BLASTX and BLASTP programs (1, 13).
Nucleotide sequence accession number.
The sequence of Fbh1 cDNA has been deposited in the EMBL and GenBank nucleotide sequence databases under accession no. AJ004883.
RESULTS
Detection and purification of fruit-body-specific hydrophobins.
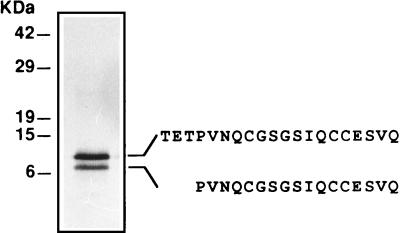
Fruit body hydrophobins of P. ostreatus were detected by extracting fruit body homogenates with hot SDS (8). After dissociation of the hot-SDS-insoluble aggregates with TFA and subsequent purification by 60% ethanol extraction (37), two protein bands at apparent relative molecular masses of 7 and 12 kDa were resolved by SDS-PAGE (Fig. 1). The amino acid sequences of the N-terminal ends of the proteins resolved by SDS-PAGE were determined by direct sequencing. The N-terminal sequence of the 12-kDa protein was TETPVNQ?GSGSIQ??ESVQ, and the N-terminal sequence of the 7-kDa protein was PVNQ?GSGSIQ??ESVQ (? indicates a probable cysteine residue). Both the pattern of putative cysteines [C-(Xaa)5/6-CC] of the purified proteins and the solubility properties were consistent with the properties expected for members of the hydrophobin protein family.
FIG. 1.
SDS-PAGE of the SDS-insoluble TFA-soluble fraction of P. ostreatus fruit body proteins. The N-terminal amino acid sequences of the two proteins are shown.
In order to determine whether the purified protein was glycosylated, the protein was subjected to SDS-PAGE, electrotransferred onto an PVDF membrane, and treated with Schiff reagents. Hydrophobin SC3 from S. commune, in which glycosylation has been confirmed (2), was used as a positive control. No glycosylation was detected in the hydrophobins purified from P. ostreatus fruit bodies (data not shown).
Sequence of the fruit body hydrophobin mRNA.
In order to determine the sequence of the mRNA coding for the hydrophobin purified from P. ostreatus fruit bodies, total fruit body RNA was isolated and used as a template for the synthesis of the cDNA of the mRNA fraction. The cDNA coding for the hydrophobin purified from fruit bodies was amplified by PCR by using as primers a degenerate oligonucleotide corresponding to the possible sequences coding for the first nine amino acids of the 12-kDa protein and an oligo(dT)17 primer. This approach yielded a single DNA band at approximately 580 bp, which was cloned and sequenced. The sequence contained a 342-nucleotide open reading frame whose predicted amino acid sequence revealed an 88-amino-acid protein with a relative molecular mass of 8,893 and an isoelectric point of 4.77. The predicted protein contains two clusters of cysteines spaced like the clusters described previously for hydrophobins (33) (C-X6-CC-X31-C, C-X5-CC-X12-C). All of the data obtained suggest that the protein purified from the fruit bodies is the hydrophobin that is referred in this paper to as Fbh1. The 5′ end of the mRNA coding for Fbh1 was amplified by PCR by using a 5′/3′ RACE kit (see Materials and Methods) and an oligonucleotide based on the Fbh1 coding sequence determined as described above as the specific primer.
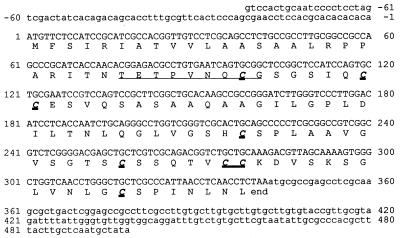
Figure 2 shows the complete cDNA sequence corresponding to the mRNA coding for Fbh1. This sequence contains an 82-bp 5′ untranslated region and a 342-bp open reading frame followed by a 155-nucleotide sequence before the start point of the polyadenylation tail. A putative polyadenylation signal (AATATT) is present 43 nucleotides before the actual start of the poly(A) tail. There is a unique ATG codon in frame with the expected amino acid sequence of mature Fbh1; this was thought to be the translation start point. The open reading frame coded for a 113-amino acid protein with a relative molecular mass of 11,247 Da and an isoelectric point of 7.95. The first 25 amino acids show a hydropathy profile compatible with a signal peptide that can be processed to yield the mature Fbh1 described above.
FIG. 2.
Fbh1 cDNA nucleotide and deduced amino acid sequences. The underlined sequence is the first nine amino acids of the mature protein. The cysteine residues are in boldface type.
Antibodies against Fbh1.
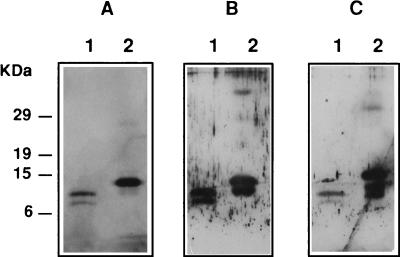
In order to confirm that the Fbh1 protein whose sequence was deduced from the cDNA sequence data was indeed the hydrophobin purified from the fruit bodies of P. ostreatus, the cDNA coding for mature Fbh1 was overexpressed in E. coli by using the pET system as described in Materials and Methods. After induction of protein expression and subsequent hydrophobin purification, a yield of 50 to 100 μg of pure recombinant protein per 100 ml of bacterial culture was obtained. The protein purified from the induced cultures was analyzed by SDS-PAGE, and a major protein band was produced at approximately 14 kDa (Fig. 3). In addition, when an aqueous solution of the hydrophobin-like protein purified from the induced E. coli cultures was subjected to electrobubbling, protein aggregates that produced a white foam were formed, as expected for a protein belonging to the hydrophobin family. This foam collapsed under a vacuum, and the insoluble fraction could be recovered by centrifugation. SDS-PAGE analysis of this material treated with TFA revealed a unique protein band corresponding to the major band present in the purified fraction from the induced cultures (data not shown).
FIG. 3.
SDS-PAGE of Fbh1 purified from P. ostreatus fruit bodies (lanes 1) or from transformed E. coli cells (lanes 2). (A) Silver staining. (B) Immunoreactions with antibodies raised against the protein purified from fruit bodies. (C) Immunoreactions with antibodies raised against the protein purified from E. coli transformed cells.
Polyclonal antibodies were raised against either the protein overexpressed in E. coli (fusion protein) or the protein purified from the P. ostreatus fruit bodies (natural protein). Both types of antibodies were unreactive with a total SDS-soluble extract from fruit bodies (data not shown). Figure 3 shows the results of detection of both the natural protein and the fusion protein with antibodies raised against either of the two proteins. These results confirmed that the mRNA detected in the fruit bodies whose sequence was obtained and is discussed above indeed coded for the hydrophobin protein purified from fruit bodies of P. ostreatus.
Specific expression of fbh1 in fruit bodies.
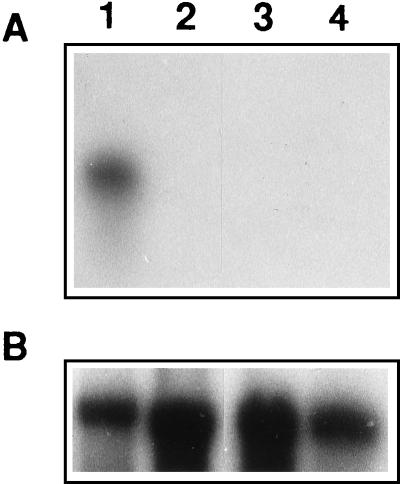
The expression of fbh1 in P. ostreatus fruit body, dikaryotic, and monokaryotic mycelia was studied by Northern blotting by using the cloned Fbh1 cDNA as a probe. Two monokaryons with compatible complementary mating type genes were used to eliminate the possibility of cosegregation of the cloned fbh1 allele with a given mating type allele. Figure 4 shows that hybridization could be detected only with RNA purified from fruit bodies, suggesting that fbh1 is expressed only in this developmental stage. In this experiment the amount of RNA was monitored by using as the control a fragment of P. ostreatus 28S rRNA.
FIG. 4.
Northern blot analysis showing specific expression of fbh1. (A) Hybridization in which Fbh1 cDNA was used as the probe. (B) Hybridization performed with a probe for P. ostreatus 28S rRNA as a control. Total RNAs were isolated from mature Florida N001 fruit bodies (lane 1), vegetatively growing Florida N001 dikaryotic mycelium (lane 2), the M1 monokaryotic strain (lane 3), and the M5 monokaryotic strain (lane 4).
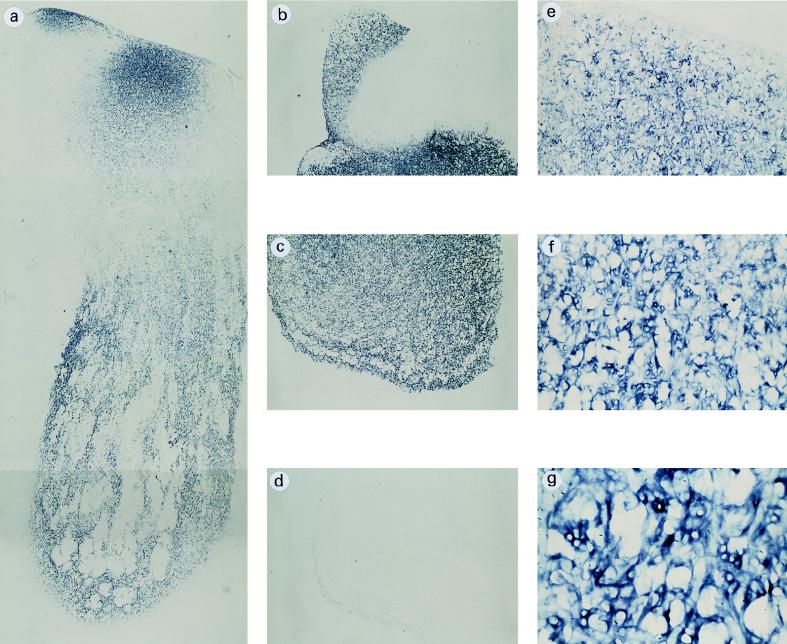
In order to analyze more precisely the parts of the fruit bodies that express the fbh1 gene, in situ mRNA hybridization experiments were performed with paraffin-embedded sections of P. ostreatus fruit bodies. The results indicated that fbh1 was expressed throughout the fruit body with the exception of the gills, which were completely unreactive to the fbh1 probe (Fig. 5). The levels of expression were not, however, homogeneous; greater reactivity was seen in the middle outer region of the stipe, whereas an irregular pattern of fbh1 expression was observed in the inner tissue of the pileus.
FIG. 5.
Localization of fbh1 mRNA in fruit bodies. (a) In situ hybridization in a longitudinal paraffin-embedded section of a mature fruit body hybridized with digoxigenin-labeled antisense fbh1 and viewed under a bright field, which gave a blue label. (b and c) Transverse sections from the upper part of panel a. (e through g) More detailed views of the upper part of panel a. (d) Control hybridized with the fbh1 sense probe. (a through d) Magnification, ×20. (e) Magnification, ×50. (f) Magnification, ×100. (g) Magnification, ×200.
Taken together, the results described above confirm that Fbh1 is a hydrophobin that is specific for fruit bodies in the oyster mushroom P. ostreatus.
DISCUSSION
In this paper we describe the isolation and characterization of a fruit-body-specific hydrophobin of the edible white rot fungus P. ostreatus Florida N001. When the hydrophobins present in fruit bodies of P. ostreatus Florida N001 were purified by using the previously described protocols (8), only two different proteins were recovered; these proteins had apparent relative molecular masses of 7 and 12 kDa. Amino-terminal microsequencing of these two proteins revealed that the 12-kDa protein contained a three-amino-acid amino-terminal extension (TET) that the 7-kDa protein did not contain. It is unlikely, however, that this is the only difference between the two proteins, although the electrophoretic behaviors of the highly hydrophobic proteins hamper accurate estimates of the actual size difference between the two proteins (18), and the possibility that there are chemical modifications that lead to proteolysis during the extraction protocol cannot be ruled out. Nevertheless, two additional explanations for the differences should be discussed. (i) The difference between the 7- and 12-kDa proteins in the three amino-terminal amino acids can be explained by alternative or inaccurate posttranslational processing of the proteins during maturation. Wessels et al. (34) described other examples in which different amino-terminal ends of a single hydrophobin are produced during the process of extracellular transport. (ii) The strain used (P. ostreatus Florida N001) is a dikaryon, and there could be two different fruit body hydrophobins encoded by different alleles. In this context, genetic analysis of the parents of strain Florida N001 has shown that this strain was derived from a cross between two monokaryons that appear to be very different from a genetic point of view (20a). Given the great variability found among hydrophobins (33), it seems unlikely that the two alleles of fbh1 present in Florida N001 are identical. Moreover, only one cDNA class coding for Fbh1 has been recovered from the total mRNA present in fruit bodies, although the 5′ primer oligonucleotide used in the PCR experiment should have allowed recovery of different variants having the same sequence at the 5′ end. Taken together, these data suggest that there is only one class of Fbh1 that is expressed in the fruit bodies of P. ostreatus and that the two protein variants obtained by denaturing PAGE are produced as a consequence of modification of a single class of proteins.
Glycosylation has been reported to be a posttranslational modification in hydrophobin SC3 of S. commune. This posttranslational modification can modify the hydropathy profile of the protein and affect its self-aggregation. We have been unable to find a glycosylation signal similar to that of SC3 in Fbh1. A major structural difference between SC3 and Fbh1 is the length of the amino acid sequence preceding the first cysteine residue. This sequence is very short in Fbh1 (seven amino acids in the 12-kDa form and four amino acids in the 7-kDa form), and putative sites for glycosylation in this region are scarce.
Based on their solubility properties, hydrophobins have been divided into two groups (32). Class I hydrophobins form very stable complexes that are insoluble in SDS and contain cysteine doublets followed by stretches of hydrophilic amino acids. Class II hydrophobins are soluble in SDS, and their cysteine doublets are immediately followed by hydrophobic residues. In the case of Fbh1, hydrophilic amino acids follow each cysteine doublet, indicating that this protein belongs to class II, which is consistent with the insolubility of Fbh1 aggregates in hot SDS.
Analysis of the sequence of the cDNA amplified by using as the primer an oligonucleotide whose sequence corresponded to the amino-terminal sequence of Fbh1 allowed prediction of the sequence of a 113-amino-acid protein with an isoelectric point of 7.95. The first 24 amino acids of the predicted protein have hydrophobicity and structural characteristics compatible with those of a signal peptide. When the signal peptide is cleaved off the protein, the mature hydrophobin is 88 amino acids long and has a predicted relative molecular mass of 8,624 Da and a pI of 4.77. The isoelectric point for the mature hydrophobin coincides with the expected external pH close to the hyphal membrane (17). The coincidence of the external pH and Fbh1 pI may facilitate polymerization of the hydrophobin in the external medium.
Comparisons of the Fbh1 sequence with sequences in the GenBank database revealed significant degrees of similarity with other hydrophobins purified from P. ostreatus, S. commune, Coprinus cinereus, and Agaricus bisporus. The sequence comparisons revealed a high level of identity (68% amino acid identity) between the signal peptide of Fbh1 and the signal peptide of hydrophobin POH1 from P. ostreatus var. ostreatus fruit bodies (1a), whereas the level of global identity between the two proteins is lower (51% amino acid identity). The low level of similarity between these two fruit body hydrophobins suggests either that they have different functions and are products of genes at different loci or that the variability among Fbh1 alleles is very high. If the latter is true, a similar situation would be expected for the two alleles of Fbh1 present in P. ostreatus var. florida N001, as discussed above. Other hydrophobins exhibiting high levels of similarity to Fbh1 were also purified from fruit bodies of other higher fungi; these hydrophobins included SC1 and SC4 (41 and 40% amino acid identity, respectively) of S. commune (34) and ABH1/hypA (7, 22) (39% amino acid identity) of A. bisporus. These findings demonstrated that the levels of amino acid sequence homology between hydrophobins from different (or related) species are low (data not shown).
The isolation of Fbh1 by biochemical methods and the PCR amplification of a cDNA coding for the protein from the total mRNA purified from fruit bodies raised a question about the actual relationship between the cDNA isolated and the purified protein. To answer this question, an indirect approach based on the cross-reactions of polyclonal antibodies with Fbh1 was used. This approach necessitated overexpression of Fbh1 in E. coli. Hydrophobin overexpression in bacteria had been unsuccessful previously. In the system described here, the modification of the protein caused by the histidine tag fused to the amino-terminal end of the protein very likely reduced the toxicity of the native protein for E. coli, allowing overexpression. The fusion protein, on the other hand, had many of the physicochemical properties of natural Fbh1 and could be used for induction of polyclonal antibodies that reacted positively against natural Fbh1. The positive reaction of natural Fbh1 with the antibodies raised against the His-Tag-Fbh1 fusion protein overexpressed in E. coli and the positive reaction of the overexpressed fusion protein with the antibodies raised against the natural Fbh1 protein justified the use of the cDNA present in the fusion construct as a probe for the studies on the spatial specificity expression of the protein.
There are many previous reports discussing the developmental regulation of hydrophobins (see reference 32 for a review). In the case of Fbh1, the data presented in this paper show that this protein is specifically expressed in the fruit bodies and that transcription of the fbh1 gene cannot be detected in either monokaryotic or dikaryotic mycelia. Another hydrophobin whose expression appears to be developmentally regulated is A. bisporus ABH1/HypA (7, 22), whose expression appears to be associated with rapid expansion of the mushroom caps (7). A different pattern of expression control occurs in the SC4 and SC1 hydrophobins abundantly expressed in S. commune fruit bodies as these proteins are also produced in a growing dikaryon in the absence of fruiting (26). Fbh1 expression control in P. ostreatus seems to be more strict; in addition to the Fbh1 gene, we identified another hydrophobin gene that seems to be expressed in vegetatively growing mycelia but not in fruit bodies (26a). This expression specificity during different stages of development seems to indicate that variations in a basic structure are responsible for the adaptation of different hydrophobins to different functions.
A more detailed picture of the pattern of expression of fbh1 was obtained by in situ detection of the transcripts of this gene. In situ hybridization experiments are very often difficult to perform with higher fungi because of the low level of organization of fungal tissues and because of the chemical composition of fungal cell walls. However, we used a standard in situ hybridization protocol to study the distribution of fbh1 transcripts in the developing fruit body. Expression of fbh1 was distributed throughout the whole fruit body except the gills, where no hybridization signal was detected. Taking into account the fact that mushroom fungal hyphae are joined together in a rather loose mesh without tissue or organ differentiation (27), the higher levels of digoxygenin labeling detected in some areas of the pileus could be due to denser packaging of actively growing hyphae. Fruit body formation has been reported to occur in two phases (17); first a mycelial mass is produced by nontrophic extension of hyphae in a coordinated manner, and then the mycelial mass expands due to an increase in hyphal volume accompanied by uniform expansion of the hyphal walls. Thus, the signal detected in the pileus and the stipe could correspond to the hyphal wall expansion phase. The expansion phase has been reported to require protein synthesis (15), and Fbh1 synthesis could be part of this process.
The conserved eight-cysteine pattern that the proteins described here present resembles that of the lipid transfer plant proteins (LTPs) (31). The LTPs share obvious similarities with fungal hydrophobins (they are small extracellular basic proteins with a constant pattern of eight cysteines), and it has been proposed that they are involved in plant cuticle formation and in general plant defense mechanisms. The cuticle is a continuous layer of lipophilic material found on the outermost surfaces of the aerial parts of plants (16), and it could be considered functionally homologous to the hydrophobin layer in fungi. As the three-dimensional structures of some LTPs have been determined, the similarities discussed above suggest that LTP structure can be used to formulate a working hypothesis for elucidation of the tertiary structure of fungal hydrophobins.
ACKNOWLEDGMENTS
We thank N. Razkin for technical help, L. Lugones for helpful and enthusiastic discussions, and V. Muez and Gurelan S. C. for providing strain Florida N001 and for advice.
This work was supported by research project BIO94-0443 of the Comisión Nacional de Ciencia y Tecnología, by funds from the Universidad Pública de Navarra (Pamplona, Spain), and by the University of Groningen (Haren, The Netherlands). M.M.P. received a grant from the Ministerio de Educación y Ciencia, Programa de Formación de Profesores de Universidad (FPU), Spain.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 1a.Ásgeirsdóttir, S. Unpublished data.
- 2.Ásgeirsdóttir S A. Ph.D. thesis. Groningen, The Netherlands: Rijksuniversiteit Groningen; 1994. [Google Scholar]
- 3.Bezalel L, Hadar Y, Cerniglia C E. Enzymatic mechanisms involved in phenanthrene degradation by the white rot fungus Pleurotus ostreatus. Appl Environ Microbiol. 1997;63:2495–2501. doi: 10.1128/aem.63.7.2495-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobek P, Kuniak L, Ozdin L. The mushroom Pleurotus ostreatus reduces secretion and accelerates the fractional turnover rate of very-low-density lipoproteins in the rat. Ann Nutr Metab. 1993;37:142–145. doi: 10.1159/000177762. [DOI] [PubMed] [Google Scholar]
- 5.Bobek P, Ozdin L. The mushroom Pleurotus ostreatus accelerates plasma very-low-density lipoprotein clearance in hypercholesterolemic rat. Physiol Res. 1994;43:205–206. [PubMed] [Google Scholar]
- 6.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Groot P W J, Schaap P J, Sonnenberg A S M, Visser J, Van Griensven L J L D. The Agaricus bisporus HypA gene encodes a hydrophobin and specifically accumulates in peel tissue of mushroom caps fruit body development. J Mol Biol. 1996;257:1008–1018. doi: 10.1006/jmbi.1996.0219. [DOI] [PubMed] [Google Scholar]
- 8.de Vries O M H, Fekkes M P, Wösten H A B, Wessels J G H. Insoluble hydrophobin complexes in the walls of Schizophyllum commune and other filamentous fungi. Arch Microbiol. 1993;159:330–335. [Google Scholar]
- 9.Dons J J M, Springer J, de Vries S C, Wessels J G H. Molecular cloning of a gene abundantly expressed during fruiting body initiation in Schizophyllum commune. J Bacteriol. 1984;157:802–808. doi: 10.1128/jb.157.3.802-808.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eger G. Rapid method for breeding Pleurotus ostreatus. Mushroom Sci. 1976;9:567–576. [Google Scholar]
- 11.Frohman M A. RACE: rapid amplification of cDNA ends. In: Innis M A, Gelfrand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 28–38. [Google Scholar]
- 12.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidiomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 14.Goday A, Jensen A, Culiañez-Macià F A, Alba M M, Figueras M, Serratosa J, Torrent M, Pagès M. The maize abscisic acid responsive protein Rab17 is located in the nucleus and cytoplasm and interacts with nuclear localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gooday G W. Control of development of excised fruit bodies and stipes of Coprinus cinereus. Trans Br Mycol Soc. 1974;62:391–399. [Google Scholar]
- 16.Holloway P J. Structure and biochemistry of plant cuticular membranes: an overview. In: Cutler D F, Alvin K L, Price C E, editors. The plant cuticle. London, United Kingdom: Academic Press; 1982. pp. 1–32. [Google Scholar]
- 17.Jennings D H, Lysek G. Fungal biology: understanding the fungal lifestyle. Oxford, United Kingdom: BIOS Scientific Publishers Ltd.; 1996. [Google Scholar]
- 18.Kaufmann E, Geisler N, Weber K. SDS-PAGE strongly over-estimates the molecular masses of the neurofilament proteins. FEBS Lett. 1984;170:81–84. doi: 10.1016/0014-5793(84)81373-3. [DOI] [PubMed] [Google Scholar]
- 19.Kurashige S, Akuzawa Y, Endo F. Effects of Lentinus edodes, Grifola frondosa, and Pleurotus ostreatus administration on cancer outbreak, and activities of macrophages and lymphocytes in mice treated with a carcinogen, N-butyl-N-butanolnitrosamine. Immunopharmacol Immunotoxicol. 1997;192:175–183. doi: 10.3109/08923979709007657. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20a.Larraya, L. Unpublished data.
- 21.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 22.Lugones L, Bosscher J S, Scholtmeyer K, de Vries O M H, Wessels J G H. An abundant hydrophobin (ABHI) forms hydrophobic rodlet layers in Agaricus bisporus. Microbiology. 1996;142:1321–1329. doi: 10.1099/13500872-142-5-1321. [DOI] [PubMed] [Google Scholar]
- 23.Lugones L G, Wösten H A B, Wessels J G H. A hydrophobin (ABH3) secreted by the substrate mycelium of Agaricus bisporus (common white button mushroom). 1998. Microbiology, in press. [DOI] [PubMed] [Google Scholar]
- 24.Marzullo L, Cannio R, Giardina P, Santini M T, Sannia G. Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. J Biol Chem. 1995;270:3823–3827. doi: 10.1074/jbc.270.8.3823. [DOI] [PubMed] [Google Scholar]
- 25.Merril C R, Goldman D, Sedman S A, Ebert M H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981;211:1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- 26.Mulder G H, Wessels J G H. Molecular cloning of RNAs differentially expressed in monokaryons and dikaryons of Schizophyllum commune. Exp Mycol. 1986;10:214–227. [Google Scholar]
- 26a.Peñas, M. M. Unpublished data.
- 27.Ruiters M H J, Wessels J G H. In situ localization of specific RNAs in whole fruiting colonies of Schizophyllum commune. J Gen Microbiol. 1989;135:1747–1754. [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sarkar S, Martínez A T, Martínez M J. Biochemical and molecular characterization of a manganese peroxidase isoenzyme from Pleurotus ostreatus. Biochim Biophys Acta. 1997;1339:23–30. doi: 10.1016/s0167-4838(96)00201-4. [DOI] [PubMed] [Google Scholar]
- 30.Shin K-S, Oh I-K, Kim C-J. Production and purification of Remazol brilliant blue R decolorizing peroxidase from the culture filtrate of Pleurotus ostreatus. Appl Environ Microbiol. 1997;63:1744–1748. doi: 10.1128/aem.63.5.1744-1748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thoma S, Kaneko Y, Sommerville C. A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 1993;3:427–436. doi: 10.1046/j.1365-313x.1993.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- 32.Wessels J G H. Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol. 1994;32:413–427. [Google Scholar]
- 33.Wessels J G H. Hydrophobins: proteins that change the nature of fungal surface. Adv Microb Physiol. 1997;38:1–44. doi: 10.1016/s0065-2911(08)60154-x. [DOI] [PubMed] [Google Scholar]
- 34.Wessels J G H, de Vries O M H, Asgeirsdóttir S A, Schuren F H J. Hydrophobin genes involved in the formation of aerial hyphae and fruit bodies in Schizophyllum commune. Plant Cell. 1991;3:793–799. doi: 10.1105/tpc.3.8.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wessels J G H, de Vries O M H, Asgeirsdóttir S A, Springer J. The thn mutation of Schizophyllum commune which suppresses formation of aerial hyphae, affects expression of the Sc3 hydrophobin gene. J Gen Microbiol. 1991;137:2439–2445. doi: 10.1099/00221287-137-10-2439. [DOI] [PubMed] [Google Scholar]
- 36.Wessels J G H, Mulder G H, Springer J. Expression of dikaryon-specific and non-specific mRNAs of Schizophyllum commune in relation to environmental conditions and fruiting. J Gen Microbiol. 1987;133:2557–2561. [Google Scholar]
- 37.Wösten H A B, de Vries O M H, Wessels J G H. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell. 1993;5:1567–1574. doi: 10.1105/tpc.5.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wösten H A B, Wessels J G H. Hydrophobins, from molecular structure to multiple functions in fungal development. Mycoscience. 1997;38:363–374. [Google Scholar]