Abstract
Background:
Somatic loss of the tumour suppressor RB1 is a common event in tubo-ovarian high-grade serous carcinoma (HGSC), which frequently co-occurs with alterations in homologous recombination DNA repair genes including BRCA1 and BRCA2 (BRCA). We examined whether tumour expression of RB1 was associated with survival across ovarian cancer histotypes (HGSC, endometrioid (ENOC), clear cell (CCOC), mucinous (MOC), low-grade serous carcinoma (LGSC)), and how co-occurrence of germline BRCA pathogenic variants and RB1 loss influences long-term survival in a large series of HGSC.
Patients and methods:
RB1 protein expression patterns were classified by immunohistochemistry in epithelial ovarian carcinomas of 7436 patients from 20 studies participating in the Ovarian Tumor Tissue Analysis consortium and assessed for associations with overall survival (OS), accounting for patient age at diagnosis and FIGO stage. We examined RB1 expression and germline BRCA status in a subset of 1134 HGSC, and related genotype to survival, tumour infiltrating CD8+ lymphocyte counts and transcriptomic subtypes. Using CRISPR-Cas9, we deleted RB1 in HGSC cell lines with and without BRCA1 mutations to model co-loss with treatment response. We also performed genomic analyses on 126 primary HGSC to explore the molecular characteristics of concurrent homologous recombination deficiency and RB1 loss.
Results:
RB1 protein loss was most frequent in HGSC (16.4%) and was highly correlated with RB1 mRNA expression. RB1 loss was associated with longer OS in HGSC (hazard ratio [HR] 0.74, 95% confidence interval [CI] 0.66–0.83, P = 6.8 ×10−7), but with poorer prognosis in ENOC (HR 2.17, 95% CI 1.17–4.03, P = 0.0140). Germline BRCA mutations and RB1 loss co-occurred in HGSC (P < 0.0001). Patients with both RB1 loss and germline BRCA mutations had a superior OS (HR 0.38, 95% CI 0.25–0.58, P = 5.2 ×10−6) compared to patients with either alteration alone, and their median OS was three times longer than non-carriers whose tumours retained RB1 expression (9.3 years vs. 3.1 years). Enhanced sensitivity to cisplatin (P < 0.01) and paclitaxel (P < 0.05) was seen in BRCA1 mutated cell lines with RB1 knockout. Among 126 patients with whole-genome and transcriptome sequence data, combined RB1 loss and genomic evidence of homologous recombination deficiency was correlated with transcriptional markers of enhanced interferon response, cell cycle deregulation, and reduced epithelial-mesenchymal transition in primary HGSC. CD8+ lymphocytes were most prevalent in BRCA-deficient HGSC with co-loss of RB1.
Conclusions:
Co-occurrence of RB1 loss and BRCA mutation was associated with exceptionally long survival in patients with HGSC, potentially due to better treatment response and immune stimulation.
INTRODUCTION
Despite a high response rate to primary treatment, the progressive development of acquired drug resistance is common in tubo-ovarian high-grade serous carcinoma (HGSC), a histotype that is associated with approximately 70% of ovarian cancer deaths1. The frequent acquisition of resistance-conferring alterations in HGSC2–4 suggests that the development of drug resistance may be inevitable when curative surgery is not achieved in these patients. Countering that view, however, is the observation that a small subset of patients with HGSC advanced disease experience an exceptional response to treatment, survive well beyond a median of 3.4 years5, and in some cases, remain disease free6,7. Interest in studying long-term cancer survivors is growing as they may assist the discovery of prognostic biomarkers, novel treatments, and approaches to limit the development of resistance8.
Several clinical and molecular factors that influence treatment response and overall survival (OS) in HGSC have been described. Complete surgical debulking is associated with a more favourable outcome compared to patients left with residual disease9–11. Molecular subtypes defined by distinct gene expression patterns in primary HGSC are associated with different outcomes12, including the poor survival C1/mesenchymal subtype that is more often seen in patients where complete surgical tumour resection cannot be achieved13–15. By contrast, the C2/immunoreactive subtype is typified by extensive infiltration of intraepithelial T cells12, a feature known to be strongly associated with improved survival16,17. Tumours arising in individuals with germline or somatic alterations in BRCA1 or BRCA2 genes are typically more responsive to conventional chemotherapy and poly(ADP-ribose) polymerase (PARP) inhibitors, whereas those tumours with intact homologous recombination (HR) DNA repair are more often resistant to treatment18–20. Patients with germline BRCA1 or BRCA2 pathogenic variants show more favourable survival at five years post-diagnosis compared to non-carriers, with BRCA2 mutation carriers retaining a long-term (>10 year) survival advantage21–23. Although deleterious mutations in BRCA1, BRCA2 and other genes involved in HR DNA repair are associated with a favourable response to treatment, these are not sufficient alone to confer long-term survival and a large proportion of such patients experience a typical disease trajectory. A differential outcome in mutation carriers can in part be ascribed to alternative splicing24 or retention of the wild-type BRCA allele in tumours25, both of which appear to limit the effectiveness of chemotherapy.
We previously characterised a small series of HGSC exceptional survivors and found that co-occurring loss of function alterations in both BRCA and RB1 were associated with unusually favourable survival7,26. Disruption of the RB pathway is found in many cancer types but with variable impacts on patient outcome. For example, co-loss of RB1 and BRCA is associated with shorter survival in breast and prostate cancer, possibly due to lineage switching and resistance to hormonal therapy27–29. A transcriptomic signature of RB1 loss was recently described to be associated with poor outcomes across cancer types30. We have previously found that chromosomal breakage is the most common mechanism of RB1 inactivation in HGSC3, accounting for approximately 80% of all RB1 alterations. In addition to its crucial role in cell cycle regulation, RB1 is involved in non-canonical functions in a context- and tissue-dependent manner31–33, including HR mediated DNA repair. Loss of RB1 expression in HGSC has been associated with a survival benefit34, including in the context of abnormal block-like p16 staining35.
Factors underlying the association of RB1 loss with improved outcome in HGSC are unknown. Here, we contrast the pattern and clinical consequences of RB1 loss in HGSC with other epithelial ovarian cancer subtypes, investigate the relevance of co-occurring BRCA1 or BRCA2 mutations and RB1 loss in HGSC patients, and explore the functional effects of combined BRCA and RB1 impairment in HGSC cell lines.
PATIENTS AND METHODS
Patient cohorts
The study population consisted of 7436 patients diagnosed with invasive epithelial ovarian, peritoneal or fallopian tube cancer from 20 studies or biobanks participating in the Ovarian Tumor Tissue Analysis (OTTA) consortium36 (Supplementary Fig. S1). Written informed consent or IRB approved waiver of consent was obtained at each site for patient recruitment, sample collection, and study protocols (Supplementary Table S1).
Whole-genome sequence and matched transcriptome sequence data of primary HGSC tumours were available from 126 patients from the Multidisciplinary Ovarian Cancer Outcomes Group (MOCOG) study26 (Supplementary Fig. S1). This cohort consisted of 34 short-term survivors (OS <2 years), 32 moderate-term survivors (OS ≥2 and <10 years) and 60 long-term survivors (OS ≥10 years) with advanced stage (IIIC/IV) disease, enrolled in the Australian Ovarian Cancer Study (AOCS), the Gynaecological Oncology Biobank at Westmead Hospital (Sydney) or the Mayo Clinic Study.
Molecular analyses
RB1 protein expression was determined by immunohistochemistry (IHC) staining and scoring of tissue microarrays (TMAs) from formalin-fixed paraffin-embedded (FFPE) tumour samples, using our previously described protocol7 (RB1 antibody clone 13A10, Leica Biosystems; Supplementary Material). Subsets of HGSC patients had additional molecular or immune data available (Supplementary Fig. S1), including tumour p53 protein expression status previously classified37 as normal (wild-type) or abnormal (overexpression, complete absence, and cytoplasmic), germline BRCA1 and BRCA2 pathogenic variant status obtained from OTTA, RB1 mRNA tumour expression obtained using NanoString (ref34 and unpublished data), transcriptional subtypes of tumours using NanoString38 and CD8+ tumour infiltrating lymphocyte (TIL) density was previously classified39 based on the number of CD8+ TILs per high-powered field: negative (no TILs), low (<3 TILs), moderate (3–19 TILs) or high (≥20 TILs).
The MOCOG whole-genome and transcriptome sequencing dataset of 126 short-, moderate- and long-term survivors was uniformly processed as previously described26, and included detailed characterisation of each tumour sample for inactivating alterations in RB1 and HR pathway genes, including germline and/or somatic mutations in BRCA1, BRCA2, BRIP1, PALB2, RAD51C and RAD51D, or promoter methylation of BRCA1 and RAD51C. Homologous recombination deficiency (HRD) status was assessed using the CHORD (Classifier of Homologous Recombination Deficiency) method40, which uses specific base substitution, indel and structural rearrangement signatures detected in tumour genomes to generate BRCA1-type and BRCA2-type HRD scores. Primary tumours were classified as either BRCA1-HRD & RB1 altered; BRCA1-HRD & RB1 wild-type; BRCA2-HRD & RB1 altered; BRCA2-HRD & RB1 wild-type; homologous recombination proficient (HRP) & RB1 altered, or HRP & RB1 wild-type. For details on differential gene expression analyses, see Supplementary Material.
Cell culture
The AOCS patient-derived cell lines (AOCS1, AOCS3, AOCS7.2 AOCS9, AOCS11.2, AOCS14, AOCS16, AOCS22, AOCS30) were established from ascites drained from patients with HGSC, as previously described4. All AOCS cell lines were authenticated against matched patient germline DNA using short tandem repeat markers (STR, GenePrint10 System, Promega). Commercial cell lines OAW28 and CAOV3, categorised as likely HGSC41, were purchased from the American Type Culture Collection (ATCC), and JHOS2 and OVCAR4 were obtained from the National Cancer Institute Repository. Commercial lines were authenticated by comparing STR profiles (GenePrint10 System, Promega) to those published by online repositories (Cancer Cell Line Encyclopaedia, The Cancer Genome Atlas) before use in experiments. Cell lines were confirmed to be free of Mycoplasma by PCR at each revival and after finishing experiments. For details on cell growth conditions, CRISPR-mediated gene knockout, and molecular and functional cell line characterisation, see Supplementary Material.
Statistical analyses
Cox proportional hazards models were used to estimate hazard ratios (HRs) with 95% confidence intervals (CIs) using the ‘coxph’ function of the R package survival (v3.2–7). Final models were fitted using Cox regression adjusted for age at diagnosis and FIGO stage. A spline function was used for age at diagnosis with degree of freedom (df) 5 to account for the non-linear effect of the continuous variable. Regression models were fitted separately by histotype. The HGSC regression models were also stratified by site of participant recruitment, and sites with fewer than 10 events within the study period were excluded. The ENOC regression model was not stratified by site due to the limited number of overall patients per site. The OTTA survival dataset was right censored at 10 years from diagnosis to reduce the number of non-ovarian cancer related deaths. In the final Cox regression model, there was evidence for deviation from the proportional hazard assumption, but the degree of deviation was not substantial when considered alongside the large sample size and Schoenfeld residuals. The Kaplan–Meier method was used to estimate and plot progression-free and overall survival probabilities, and the log-rank (Mantel–Cox) test used to compare the survival duration between subgroups. In the Kaplan-Meier curves, the number of patients at risk on the date of diagnosis (time = 0) may be fewer than subsequent time intervals, owing to left truncation of follow-up resulting from delayed study enrolment at some OTTA sites. Differences in proportions of categorical features were assessed by either the chi-square or Fisher’s exact test as indicated. Differences in continuous variables were assessed by either a Wilcoxon Rank Sum Test or a Kruskal-Wallis test. All in vitro assays were performed across at least three independent experiments, and data are expressed as mean ± standard error of the mean (SEM) as indicated, from a minimum of three independent measurements. All statistical tests were two-sided and considered significant when P < 0.05. Statistical analyses were performed using either Prism (v9.3.1) or R (v3.6.3).
RESULTS
Loss of RB1 expression is most frequent in HGSC
RB1 protein expression was assessed by IHC in tumour samples from 7436 ovarian cancer patients using TMAs from 20 centres participating in the OTTA consortium (Supplementary Tables S1 and S2). RB1 tumour expression was classified as either retained or lost in 6564 samples, with 872 samples excluded that had either subclonal loss (n = 66), cytoplasmic (n = 17), or uninterpretable results (n = 789) due to either sample drop out or the absence of an internal positive control (Fig. 1A, Supplementary Material).
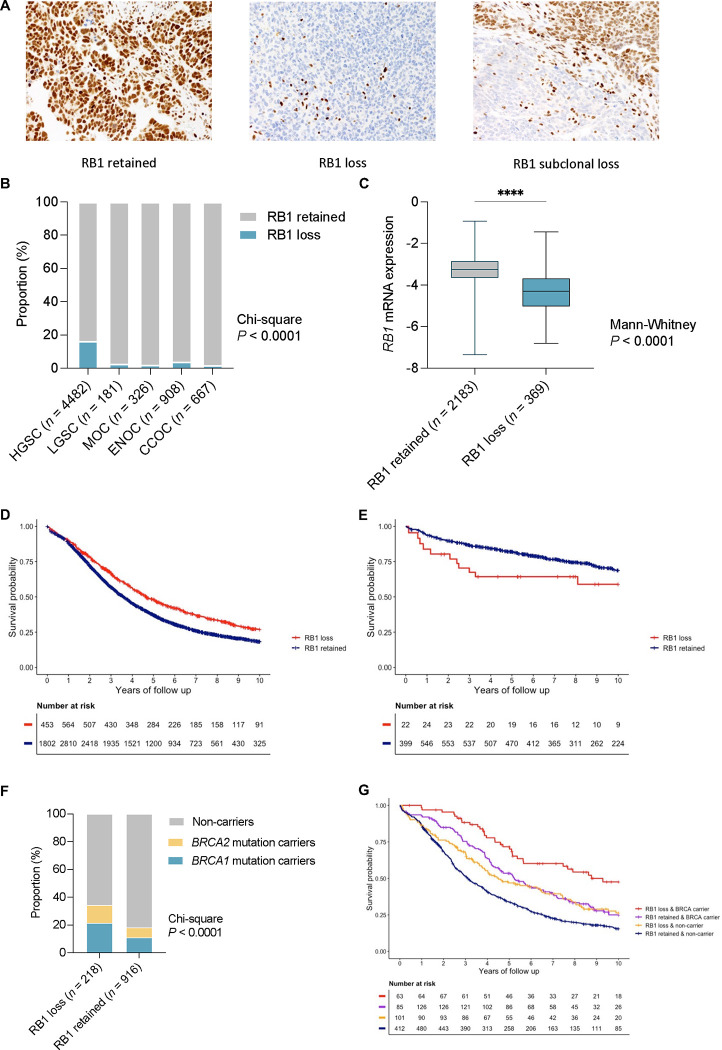
Figure 1. Expression of RB1 and survival associations across ovarian cancer histotypes.
(A) Representative images of immunohistochemical detection of RB1 expression in ovarian carcinoma tissues, showing examples of the three most common expression patterns: retained, lost and subclonal loss. (B) Proportion of patients with loss or retention of RB1 protein expression in tumour samples by ovarian cancer histotypes. Chi-square P value reported for difference in proportions across all histotypes. HGSC, tubo-ovarian high-grade serous carcinoma; LGSC, low-grade serous carcinoma; MOC, mucinous ovarian cancer; ENOC, endometrioid ovarian cancer; CCOC, clear cell ovarian cancer. (C) Boxplots show RB1 mRNA expression (NanoString) by RB1 protein expression status; lines indicate median and whiskers show range (Mann-Whitney test P value reported). Kaplan-Meier analysis of overall survival in patients diagnosed with HGSC (D) and ENOC (E) stratified by tumour RB1 expression. (F) Loss of RB1 tumour expression is more common in germline BRCA1 and BRCA2 mutation carriers than retained RB1 expression. Chi-square P value is reported. (G) Kaplan-Meier estimates of overall survival in HGSC patients by combined germline BRCA and tumour RB1 expression status.
RB1 loss was most frequent in HGSC (16.4%), followed by endometrioid ovarian cancer (ENOC; 4.1%, Chi-square P < 0.0001, Fig. 1B). Loss of RB1 expression was less frequent in all other histotypes (1.8% to 2.8%). RB1 mRNA expression was also assessed by NanoString in a subset of HGSC tumours (n = 2552) and was significantly associated with RB1 protein expression (Fig. 1C, P < 0.0001).
RB1 loss is associated with longer survival in HGSC
Loss of RB1 protein expression was associated with longer OS in patients with HGSC (HR 0.74, 95% CI 0.66–0.83, P = 6.8×10−7; Table 1) following multivariate analysis adjusting for stage and age at diagnosis and stratified by study. Patients with HGSC were comparable in terms of stage regardless of RB1 loss or retained expression (P = 0.9246), however those with RB1 loss had a younger age at diagnosis (median 59 years versus 61 years, P = 0.0003; Supplementary Table S3). Median OS was 4.7 years for patients with RB1 loss compared to 3.6 years for those with retained RB1 expression (Fig. 1D).
Table 1.
Multivariate analysis of molecular alterations and overall survival in patients with HGSC and ENOC
| Histotype | Feature | Category | No. patients (events, %) | HR (95% CI) | P | P for interaction |
|---|---|---|---|---|---|---|
|
| ||||||
| HGSCa,b | RB1 | Retained | 3453 (71.3) | 1 [Reference] | ||
| Loss | 686 (61.1) | 0.74 (0.66-0.83) | 6.8 × 10−7 | |||
| ENOCa | RB1 | Retained | 649 (22.7) | 1 [Reference] | ||
| Loss | 28 (39.3) | 2.17 (1.17-4.03) | 0.014 | |||
| HGSCa,b | RB1 and BRCA status | RB1 retained & non-carrier | 714 (76.3) | 1 [Reference] | 0.24 | |
| RB1 loss & non-carrier | 135 (60.7) | 0.74 (0.57-0.96) | 0.023 | |||
| RB1 retained & BRCA carrier | 159 (67.9) | 0.69 (0.55-0.86) | 0.001 | |||
| RB1 loss & BRCA carrier | 70 (42.9) | 0.38 (0.25-0.58) | 5.2 × 10−6 | |||
| ENOCa | RB1 and p53 | RB1 retained & p53 normal | 492 (17.5) | 1 [Reference] | 0.698 | |
| RB1 retained & p53 abnormal | 58 (36.2) | 2.26 (1.38-3.71) | 0.001 | |||
| RB1 loss & p53 normal | 11 (27.3) | 1.77 (0.56-5.65) | 0.332 | |||
| RB1 loss & p53 abnormal | 12 (58.3) | 5.34 (2.43-11.8) | <0.001 | |||
Adjusted for stage and age at diagnosis.
Stratified by study.
HR, hazard ratio, CI, confidence interval; HGSC, tubo-ovarian high-grade serous carcinoma; ENOC, endometrioid ovarian cancer.
In contrast to HGSC, loss of RB1 expression in tumours from patients with ENOC was associated with advanced stage (P = 0.0003) and poorer survival (HR 2.17, 95% CI 1.17–4.03, P = 0.0140; Table 1, Fig. 1E, Supplementary Table S4). RB1 loss and abnormal p53 protein expression, which is highly predictive of TP53 mutation42, were strongly correlated (chi-square P < 0.0001; Supplementary Fig. 2A). While TP53 mutation is known to be associated with inferior survival in patients with ENOC37,43, we note that combined RB1 loss and abnormal p53 expression were associated with the shortest patient survival (median OS 3.0 years; Supplementary Fig. 2B), suggesting that loss of RB1 and TP53 mutation have a compounding negative impact on survival in patients with ENOC.
Combined RB1 loss and germline BRCA mutation is associated with exceptionally good survival
We previously observed that co-occurrence of somatic RB1 protein loss and BRCA1 or BRCA2 alteration (somatic or germline) was associated with longer progression-free survival (PFS) and OS in HGSC7. Here, germline BRCA1 and BRCA2 status was available for 1134 HGSC patients for which we had RB1 IHC data (Supplementary Fig. S1). Consistent with having a younger age of diagnosis, patients with RB1 loss were more likely to have concurrent germline BRCA1 or BRCA2 mutations than those with retained RB1 expression (Fig. 1F, Chi-square P < 0.0001). Patients with both RB1 loss and a germline BRCA mutation had a 62% reduced risk of death compared with non-carriers with retained RB1 (HR 0.38, 95% CI 0.25–0.58, P = 5.2×10−6; Table 1). The median OS of BRCA germline carriers with RB1 loss was three times longer than non-carriers with RB1 retained tumours (median OS 9.3 years vs. 3.1 years, respectively), while median OS was 5.2 years for BRCA carriers with retained RB1 expression and 4.5 years for non-carriers with RB1 loss (Fig. 1G; Supplementary Table S5).
Enhanced response to chemotherapy in cells with impaired BRCA and RB1 function
To investigate whether co-occurrence of RB1 and BRCA alterations enhances sensitivity to standard-of-care ovarian cancer drugs, nine patient-derived HGSC cell lines with confirmed pathogenic TP53 mutation and known RB1 and BRCA status were treated with cisplatin, paclitaxel and olaparib (Supplementary Fig. S3A–C). AOCS14, the only cell line with a germline BRCA1 mutation and concomitant loss of RB1 expression, showed the best response to cisplatin and olaparib, and was the second most sensitive cell line to paclitaxel. In contrast AOCS11.2, a line with BRCA1 promoter methylation and loss of RB1 expression, was relatively resistant to paclitaxel and olaparib. Among cell lines with intact RB1 protein expression and BRCA wildtype background, AOCS3 was resistant to cisplatin, paclitaxel and olaparib.
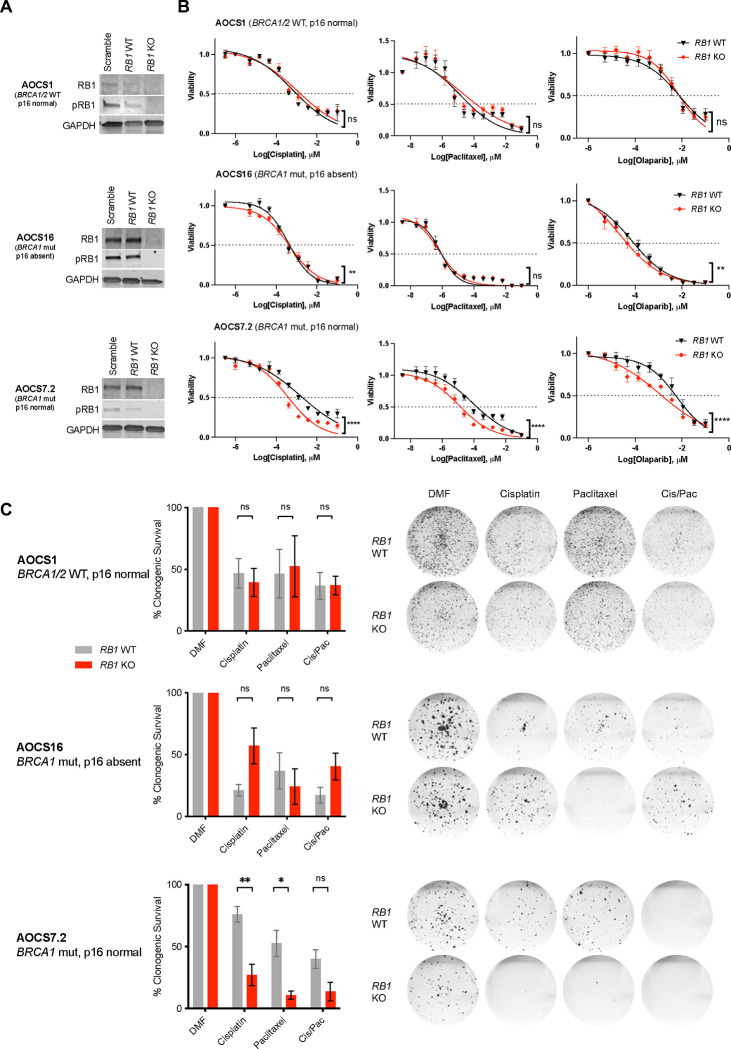
Except for the chemo-naïve cell lines AOCS30 and AOCS14, all other lines were derived from patients previously treated with chemotherapy. Since the evaluation of HGSC cell lines with existing RB1 mutations may have been confounded by their prior, differential exposure to chemotherapy we therefore characterised responses in isogenically matched lines deleted of RB1 and/or BRCA1. We first inactivated RB1 in two BRCA1-mutant (AOCS7.2, AOCS16) and one wild-type line (AOCS1) using CRISPR-Cas9 (Fig. 2A, Supplementary Fig. S4A). RB1 knockout clones of the BRCA1-mutant cell line AOCS7.2 had enhanced sensitivity to cisplatin and paclitaxel compared to RB1 wild-type clones, which was observed both in short-term drug assays (72 hours, Fig. 2B) and longer-term clonogenic survival assays (12 days, Fig. 2C). In this cell line, sensitivity to paclitaxel and olaparib was increased after RB1 knockout (paclitaxel IC50 92.0 nM versus 11.8 nM, P < 0.0001; olaparib IC50 6.1 versus 1.1 nM, P < 0.0001). Further, significantly fewer colonies grew in this BRCA1-mutant cell line after RB1 knockout upon treatment with cisplatin (P = 0.01), paclitaxel (P = 0.02) or a combination of both drugs (P = 0.067) in a clonogenic survival assay (n = 3). This effect was not apparent in the BRCA-wild-type line (AOCS1) or the other BRCA1-mutant line (AOCS16). Western blot and IHC analysis (Supplementary Fig. S4A) found that AOCS16 lacked expression of p16, which may functionally disrupt the RB1 pathway irrespective of an RB1 knockout44.
Figure 2. Sensitivity to therapeutic agents in BRCA1-mutant cell lines with RB1 knockout.
(A) RB1 was knocked out using CRISPR/Cas9 in 3 patient-derived Australian Ovarian Cancer Study (AOCS) HGSC cell lines with either wild-type or mutant BRCA1 background. Representative Western Blots show protein levels of RB1 and phosphorylated RB1 (pRB1) compared to GAPDH loading control in single cell cloned, homozygous RB1 wildtype (WT) and knockout (KO) colonies in comparison to heterogeneous populations with a scramble single guide RNA (sgRNA). Independent blots were used for RB1 and pRB1. (B) Cell viability was compared between RB1 WT and KO clones following treatment with cisplatin (72 hours), paclitaxel (72 hours) or olaparib (120 hours). Nonlinear regression drug curves are shown; P values of a curve fit, extra sum-of squares F test (ns, not significant; ** P < 0.01; **** P < 0.0001; n = 3). Error bars indicate ± SEM; for some values error bars are shorter than the symbols and thus are not visible. (C) Proportion of surviving colonies following 16 days of treatment with cisplatin, paclitaxel or a combination of both (with half of the IC50 determined per drug and cell line respectively) relative to DMF vehicle control (n = 3 replicates). Data are presented as mean ± SEM. Mean values were compared by student’s t-test (ns, not significant; *P < 0.05; **P < 0.01). Representative scans of the fixed cell colonies stained with crystal violet are shown for each condition.
Given that RB1 plays a central role in the negative control of the cell cycle44,45, we tested whether the enhanced chemosensitivity of RB1 knockout AOCS 7.2 cells was associated with increased cell division. Live cell imaging showed similar growth rates of RB1 wildtype and knockout clones of all three isogenically matched HGSC cell lines (Supplementary Fig. S4B). In both BRCA wild-type and BRCA1 mutant cell lines, RB1 knockout did not alter cell cycle distribution at baseline or after 24 hours of cisplatin treatment (Supplementary Fig. S4C). Paclitaxel treatment resulted in a larger proportion of cells with a tetraploid DNA content in RB1 knockout cells compared to RB1 wild-type cells, indicating arrest in the G2 or M phase of the cell cycle. This effect was observed in all cell lines independent of BRCA or p16 status, however the arrest was more profound in the AOCS7.2 cell line (AOCS1, G2/M difference 8.59% ± 4.73%, P = 0.144; AOCS16, G2/M difference 8.13% ± 4.45%, P = 0.142; AOCS7.2: G2/M difference 14.49% ± 3.99%, P = 0.022; Supplementary Fig. S4C).
We extended our analysis of isogenically matched pairs by inactivating BRCA1 and/or RB1 in the chemo-naïve cell line AOCS30. While we were readily able to establish RB1 knockout lines, all BRCA1 targeted clones were hemizygous for BRCA1 deletion and retained BRCA1 expression (Supplementary Table S6), suggesting that engineered homozygous loss of BRCA1 was cell lethal, even in a tumour type where BRCA1 loss is frequently observed46.
Genomic and transcriptional landscape of HGSC with combined inactivation of BRCA and RB1
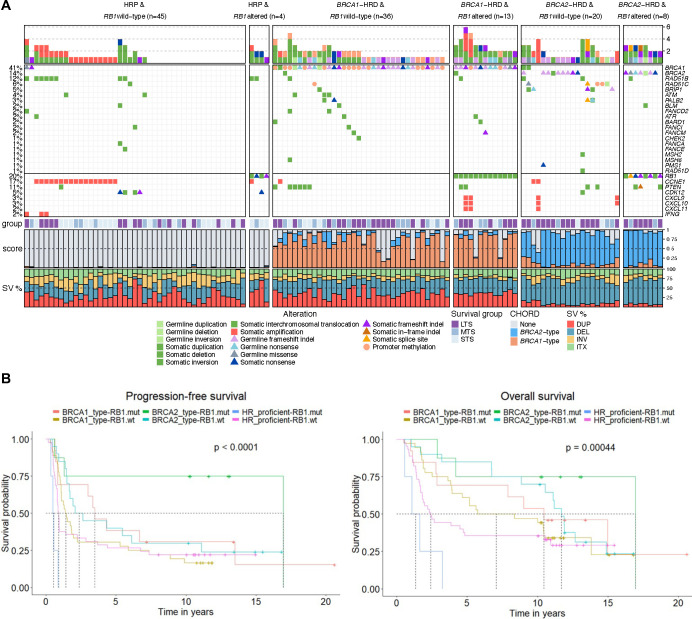
To further understand how RB1 loss may impact the biology of HGSC with co-loss of BRCA1 or BRCA2, we explored matched whole-genome and transcriptome data of primary HGSC tumours in the MOCOG cohort26 of 126 short- (OS <2 years), moderate- (OS ≥2 to <10 years) and long-term (OS ≥10 years) survivor patients (Supplementary Fig. S1). Each tumour genome was classified according to their HRD and RB1 status, resulting in 6 groups: BRCA1-HRD & RB1 altered (n = 13); BRCA1-HRD & RB1 wild-type (n = 36); BRCA2-HRD & RB1 altered (n = 8); BRCA2-HRD & RB1 wild-type (n = 20); HRP & RB1 altered (n = 4), or HRP & RB1 wild-type (n = 45; Fig. 3A).
Figure 3. Genomic landscape of high-grade serous ovarian tumours with co-occurring BRCA and RB1 alterations.
(A) Pathogenic germline and somatic alterations in homologous recombination (HR) and DNA repair genes detected by whole-genome sequencing and DNA methylation analysis of 126 primary HGSC samples26 are shown, as well as alterations in immune genes and CCNE1. Samples are grouped by HRD and RB1 status (wt, wild-type; mut, mutation). Bars at the top indicate the number of alterations in each listed gene per patient. Patients are annotated with survival group (LTS, long-term survivor, OS >10 years; MTS, mid-term survivor, OS 2–10 years; STS, short-term survivor, OS <2 years), tumour CHORD40 scores, and the proportion of structural variant (SV) type (DUP, duplication; DEL, deletion; INV, inversion; ITX, intra-chromosomal translocation). (B) Kaplan-Meier estimates of progression-free and overall survival of patients with according to HR status (BRCA1-type HRD, BRCA2-type HRD or homologous recombination proficient tumours) and RB1 status (mut, mutation; wt, wild-type).
The cohort had been selected for a long-term survivor study26 and hence was enriched for patients with very long survival. Among BRCA2-HRD patients, those with RB1 alterations had longer OS (median OS 17.0 years) compared with those without RB1 alterations (median OS 11.7 years, P = 0.0004; Fig. 3B). Similarly, BRCA1-HRD patients with RB1 alterations survived longer (median OS 10.4 years) than those with an intact RB1 gene (median OS 7.1 years). There were few HRP tumours with RB1 alterations, however these patients had a worse survival (median OS 1.4 years) compared to the HRP group with no RB1 alteration (median OS 2.4 years).
Examination of genomic features revealed relatively similar patterns within BRCA1-HRD and BRCA2-HRD groups, although there were a few discriminatory features identified between those with and without RB1 alterations (Supplementary Figs. S5 and S6). For example, the BRCA1-associated rearrangement signature Ovary_G47 was more enriched in BRCA1-HRD tumours with RB1 alterations compared to those without (P = 0.039). Among BRCA2-HRD tumours, the mutational signatures DBS6 (unknown etiology) and SBS3 (associated with HRD)48 were higher in RB1-altered tumours compared to non-altered tumours, although this was not significant (P = 0.082 and P = 0.1 respectively). Concordantly, the average BRCA1-type and BRCA2-type CHORD scores40 were highest in BRCA1- and BRCA2-HRD tumours with RB1 alterations respectively, indicating a higher probability of HRD. As described previously49, CCNE1 gene amplifications were absent in tumours with both HRD and RB1 alterations (P = 0.0006; Supplementary Fig. S7).
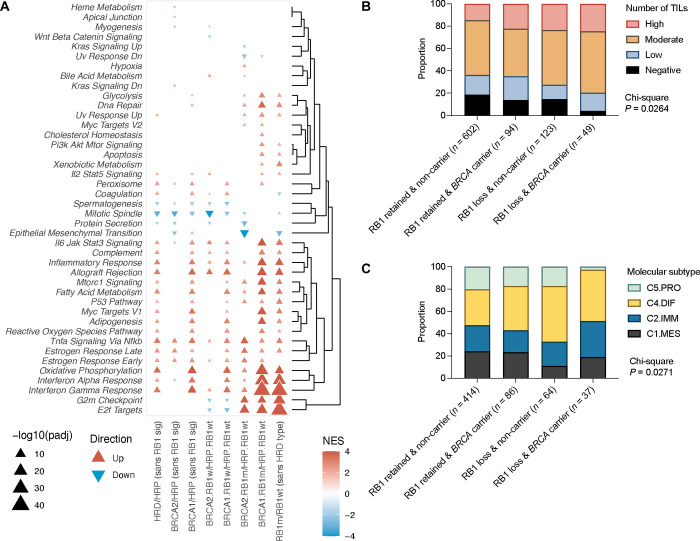
We hypothesised that tumours with combined HRD and RB1 loss may have unique transcriptional profiles. To explore this, we compared gene expression profiles between each HRD/RB1 group and the reference set of tumours that were HRP and RB1 wild-type (Supplementary Table S7, Supplementary Fig. S8). There was significant enrichment of MSigDB hallmark gene sets among genes differentially expressed in BRCA1-HRD tumours with RB1 alterations, the most prominent being interferon gamma response (up), interferon alpha response (up), oxidative phosphorylation (up), and E2F targets (up; adjusted P < 0.0001; Fig. 4A). The differentially expressed genes identified between BRCA2-HRD / RB1 altered tumours and the reference set were significantly enriched for the MSigDB hallmark gene sets: E2F targets (up), epithelial mesenchymal transition (down), G2M checkpoint (up), and TNF alpha signalling via NF-kB (up; adjusted P < 0.0001).
Figure 4. Characterisation of HGSC with co-loss of RB1 and BRCA.
(A) Gene set enrichment analysis indicating up- and downregulated pathways in tumours according to BRCA and RB1 status. HRP, homologous recombination proficient; HRD, homologous recombination deficient; RB1wt, RB1 wild-type; RB1m, RB1 altered. (B) Proportion of tumour infiltrating lymphocytes (TILs) in HGSC tumours grouped by RB1 expression and BRCA germline mutation status (Chi-square P value is indicated). (C) Proportion of tumours classified as each HGSC molecular subtype12 grouped by RB1 expression and BRCA germline mutation status (Chi-square P value is indicated; C5.PRO, C5/proliferative subtype; C4.DIF, C4/differentiated subtype; C2.IMM, C2/immunoreactive subtype; C1.MES, C1/mesenchymal subtype).
Since enhanced tumour cell proliferation has been associated with long-term survival in HGSC7,26, and loss of RB1 might accelerate proliferation31, we evaluated the expression of proliferation markers across the RB1 and BRCA subgroups. BRCA1-HRD tumours with RB1 alterations had significantly higher mRNA levels of the cell proliferation related genes PCNA (proliferating cell nuclear antigen) and MCM3 (minichromosome maintenance complex component 3) compared to BRCA1-HRD tumours without RB1 alterations (P < 0.0001, Supplementary Fig. S6). However, there were no significant differences in the proportion of Ki-67 positive cancer cell nuclei (P = 0.3297) across the subgroups (Supplementary Fig. S6), which was previously quantified by immunohistochemistry7 in a subset of primary tumours (n = 59).
Germline BRCA mutation carriers with somatic loss of RB1 tumour expression show elevated immune activity
Having observed that HGSC with combined RB1 loss and HRD have enrichment of transcriptional signatures associated with an enhanced immune response, we accessed existing immunohistochemical data39 to determine the prevalence of CD8+ TILs in HGSC samples that also had RB1 protein expression and BRCA germline mutation status (n = 868). BRCA carriers with RB1 loss had a significantly higher proportion of tumours (79.6%) with moderate and high densities of CD8+ TILs, compared to BRCA carriers with retained RB1 (64.9%), non-carriers with RB1 loss (72.4%) and non-carriers with retained RB1 (63.6%, P = 0.0264; Fig. 4B). Tumours with complete absence of CD8+ TILs were the least frequent in BRCA carriers with RB1 loss (4.1%) compared to the other groups (13.8 % of BRCA carriers with retained RB1 tumour expression, 14.6% of non-carriers with RB1 tumour loss, 18.8% of non-carriers with retained RB1 tumour expression).
Gene expression-based molecular subtypes12,38 also differed by RB1 and BRCA status (P = 0.0271, n = 601; Fig. 4C). As expected, there was enrichment for the C2/immunoreactive subtype, a subtype characterised by the presence of intratumoural CD8+ T cells and good survival, in germline BRCA carriers with RB1 loss (32.4%) compared to the other subgroups (between 19.8% and 23.4%). Additionally, tumours with RB1 loss were enriched for the C4/differentiated molecular subtype, a subtype characterised by cytokine expression and good survival, regardless of BRCA status (45.9% in BRCA carriers with RB1 loss, 50.0% in non-carriers with RB1 loss, 39.5% in BRCA carriers with retained RB1, 32.1% of non-carriers with retained RB1). BRCA carriers with RB1 loss also had the lowest proportion of the C5/proliferative molecular subtype (2.7% versus 17.2% to 20.3% in the other groups), a subtype associated with diminished immune cell infiltration and poor survival12,19.
DISCUSSION
Identifying the determinants of long-term patient survival, particularly in cancers with a generally unfavourable prognosis such as HGSC, may reveal novel therapeutic targets and inform personalised treatment strategies8. Improved survival associated with RB1 loss has been described previously in HGSC7,34,35,50 but the underlying factors contributing to this survival benefit have not been studied to date. We assessed tumour samples from a cohort of more than 7,000 women with ovarian cancer, including a subset with high resolution genomic data, to understand how RB1 loss may impact on therapeutic response and patient survival.
Alteration of the RB1 pathway is a frequent event in tumourigenesis, including loss of regulators such as p16, activation of D- and E-type cyclins and their associated cyclin dependent kinases, and loss of RB1 itself (reviewed in 51). Our study showed that RB1 loss is associated with longer survival in patients with advanced stage HGSC, but by contrast, loss of RB1 in ENOC was associated with a shorter survival, particularly in combination with p53 mutation. Similar to ENOC, in endocrine-driven breast and prostate cancer, RB1 loss is associated with poorer survival: early co-loss of BRCA2 and RB1 is associated with an aggressive, castration-resistant prostate cancer subtype (CRPC) characterised by epithelial-to-mesenchymal transition and shorter survival29. RB1 loss facilitates lineage plasticity and, with p53-comutation, leads to an androgen-independent phenotype52,53 and consequently resistance to anti-androgen therapy. In estrogen-receptor (ER) positive breast cancer, CDK4/6 inhibitor resistance is associated with RB1 loss and cyclin E2 activation54,55.
Triple negative breast cancer (TNBC) provides an important contrast to the findings for RB1 loss in ER-positive breast cancer. In TNBC, RB1 loss is most common in the basal-like subtype, where BRCA1 mutation and promoter hypermethylation is associated with frequent RB1 gene disruption and RB1 loss28. RB1 loss alone, as well as co-occurrence with BRCA1 promoter hypermethylation, is associated with a favourable chemotherapy response and outcome27,56–58. Notably, TNBC and HGSC are more similar than the cancers that they are grouped with anatomically, sharing gene expression patterns, genetic drivers including BRCA1 and BRCA2, ubiquitous loss of TP53, extensive copy number variation, and susceptibility to platinum-based chemotherapy59,60. Taken together, the relationship between RB1 loss and patient survival appears to be dependent on cancer type and molecular context61.
Some, but not all TNBC and early metastatic prostate cancers are associated with germline variants in BRCA1, BRCA2 and other genes involved in HR DNA repair. However, previous tumour studies of RB1 expression have not also defined the HRD status of individual samples. A strength of this study was the known BRCA germline status of 1134 of the HGSC patients for which we also had RB1 protein expression, and this revealed the strong association of co-mutation in either BRCA1 or BRCA2 and RB1 with survival. In addition to germline mutations in BRCA1 or BRCA2, germline or somatic mutations, and promoter methylation of other genes involved in HR DNA repair, such as RAD51C, can result in a similar molecular phenotype, characterised by distinct genomic scarring26. Using whole-genome sequence data, we determined the likely tumour HRD status in a subset of 126 tumours using an algorithm that recognises genomic scarring associated with HRD (Fig. 3A), rather than simply designating BRCA mutation status, which does not account for all mechanisms of HR repair inactivation. Although the number of samples with RB1 loss and HR proficiency was small, the very poor outcome we observed with this group indicated that for RB1 to impart a survival benefit in HGSC, it must occur in an HRD background. Validation of this finding in a larger cohort may further inform how RB1 loss could favourably influence survival in certain histological and molecular contexts.
We have previously noted that enhanced proliferation in HGSC is associated with long-term survival7,26 and it is reasonable to suggest that RB1 loss may be imparting an effect through deregulating the cell cycle. However, data on the effect of RB1 loss on proliferation in HGSC tumours and cancer cell lines is inconsistent. RB1 knockout in our HGSC cell lines did not cause cell cycle alterations in the absence of treatment, and despite differences in proliferative markers at the mRNA level, there was no significant difference in the proportion of Ki-67 positive nuclei between tumours with or without RB1 protein expression. In a recent OTTA study, Ki-67 expression was not associated with survival in HGSC; however, there was strong correlation between loss of RB1 and the proliferative marker MCM362, which may provide a more accurate measure of tumour cell proliferation than Ki-6763.
In addition to its role in driving progression through the G1 stage of the cell cycle, RB1 has non-canonical functions. RB1 has been shown to participate in HR DNA repair through interactions with BRG1 and ATM64. A recent pan-cancer study65 found that combined loss of TP53 and RB1 was associated with a particularly high genome-wide loss-of-heterozygosity score, one of the key elements of genomic scarring associated with HRD. In our whole-genome analysis, HGSC tumours with dual loss of HRD and RB1 did not exhibit overall higher mutation burden; however, we did observe elevated levels of mutational signatures associated with HRD, which may be evidence of compounding DNA repair defects. It remains possible that the combined inactivation of RB1 and HR genes contribute to enhanced chemotherapy response and/or an impaired ability for tumour cells to develop therapy resistance.
When we evaluated a set of patient derived HGSC lines, those with germline BRCA1 mutation and RB1 alteration were most sensitive to cisplatin and olaparib. Knockout of RB1 in the AOCS 7.2 cell line which had a pre-existing BRCA1 mutation, resulted in an increase in chemosensitivity, consistent with the notion that co-mutation enhances chemotherapy response7. Unfortunately, despite considerable efforts, we were unable to generate a larger series of isogenically matched cell lines with combinations of conditional knockouts of RB1 and BRCA1 as all surviving clones retained at least one BRCA1 allele. BRCA1 loss is embryonic lethal and engineered loss in cell lines has been reported as lethal elsewhere including in the human haploid cell line, HAP146.
Our data provides evidence of an enhanced immunogenicity in HGSC with RB1 loss, with higher CD8+ TIL counts and upregulated expression of IFN-γ signalling pathways. RB1 has been shown to inhibit innate IFN-β production in immunocompetent mice66 and RB1 deficiency triggered an increased IFN-β and IFN-α secretion. Co-mutation of RB1 and TP53 was recently found to be associated with an enhanced response to the immune checkpoint inhibitor atezolizumab in metastatic urothelial bladder cancer67. Similarly, a case report described a complete response to atezolizumab in heavily pre-treated, RB1-negative TNBC68. This generates the hypothesis that RB1 loss could predict response to such therapies in HGSC, since this tumour type ubiquitously harbours TP53 mutations69. However, a recent biomarker study in ovarian cancer patients treated with atezolizumab or placebo and standard chemotherapy found that deleterious mutations in RB1 were prognostic for a better PFS, regardless of the addition of atezolizumab70. While it appears RB1 loss alone may not be predictive of response to the PD-L1 inhibitor atezolizumab, response rates to PD-1/PD-L1 pathway checkpoint inhibitors are generally quite low in HGSC, with the best objective response rates between 8% and 15%71. Our study has identified a subset of patients with combined RB1 and BRCA inactivation who demonstrate exceptional immune responses and may provide clues for the development of new immunotherapeutic strategies for HGSC that extend beyond targeting PD-L1/PD-1.
Our work highlights the importance of RB1 loss to treatment response and survival and focuses attention on other therapeutic opportunities in this subset of HGSC patients. Approximately 20 percent of HGSC patients have somatic loss of RB1 assessed using genomic data3,26, a figure that is consistent with the immunohistochemical results obtained in the large patient cohort described here. Both approaches indicate that RB1 loss is generally clonal, enhancing its value as a therapeutic target if selective inhibitors can be identified. Casein kinase 2 (CK2) inhibitors have been reported to enhance the sensitivity of RB1-deficient TNBC and HGSC cells to carboplatin and niraparib72. In addition, Aurora kinase A and B inhibition is synthetically lethal in combination with RB1 loss in breast and lung cancer cells73–75. Irrespective of HRD status, RB1 mutations correlate with sensitivity to WEE1 inhibition in TP53 mutant TNBC and HGSC patient-derived xenografts76, indicating additional treatment options that exploit RB1 inactivation in these tumours. In this study, the BRCA1-mutant cell line AOCS7.2 with induced RB1 knockout was more sensitive to olaparib suggesting that RB1 loss may also predict responses to PARP inhibitors in HGSC. RB1 staining of tumour tissue by IHC is a relatively low-cost pathology-based assay that could be used in prospective studies to test whether RB1 expression is predictive of responses to PARP inhibitors, either alone or in combination with approved HRD tests.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Beach and L. Bowes for their contributions to the study. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia (1186505 to DWG; 1092856, 1117044 and 2008781 to DDLB; 2009840 to SJR), the National Institutes of Health (NIH) / National Cancer Institute (R01CA172404 to SJR, P50 CA136393 to SHK) and the U.S. Army Medical Research and Materiel Command Ovarian Cancer Research Program (Award No. W81XWH-16-2-0010 and W81XWH-21-1-0401). DWG is supported by a Victorian Cancer Agency / Ovarian Cancer Australia Low-Survival Cancer Philanthropic Mid-Career Research Fellowship (MCRF22018). FAMS is supported by a Swiss National Foundation Early Postdoc Mobility Fellowship (P2BEP3-172246), a Swiss Cancer League grant BIL KFS-3942-08-2016 and a Prof. Max Cloëtta foundation grant. KIP is supported by a NHMRC CJ Martin Overseas Biomedical Fellowship (APP1111032). ELC is supported by a Victorian Cancer Agency Mid-Career Fellowship (MCRF21004). MW is supported by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme grant agreement No 742432 (BRCA-ERC). KS is supported by the Swedish Cancer Foundation. MSA is funded through a Michael Smith Health Research BC Scholar Award (18274) and the Janet D. Cottrelle Foundation Scholars program managed by the BC Cancer Foundation.
BC’s Gynecological Cancer Research team (OVCARE) receives support through the BC Cancer Foundation and the VGH & UBC Hospitals Foundation. The Gynaecological Oncology Biobank at Westmead was funded by the NHMRC (ID310670, ID628903); the Cancer Institute NSW (12/RIG/1-17, 15/RIG/1-16); and acknowledges support from the Department of Gynaecological Oncology, Westmead Hospital, and the Sydney West Translational Cancer Research Centre (Cancer Institute NSW 15/TRC/1-01). The Women’s Cancer Research Program at Cedars-Sinai Medical Center (LAX) is supported by The National Center for Advancing Translational Sciences (NCATS) Grant UL1TR000124. The Study of Epidemiology and Risk Factors in Cancer Heredity (SEARCH) is funded by Cancer Research UK (C490/A10119 C490/A10124 C490/A16561) and the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The UKOPS study was funded by The Eve Appeal (The Oak Foundation) with contribution to authors’ salary through MRC core funding MC_UU_00004/01 and the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
The investigators also acknowledge generous contributions from the Border Ovarian Cancer Awareness Group, the Peter MacCallum Cancer Foundation, the Graf Family Foundation, Wendy Taylor, Arthur Coombs and family, and the Piers K Fowler Fund. The contents of the published material are solely the responsibility of the authors and do not reflect the views of the NHMRC, NIH, and other funders.
Footnotes
COMPETING INTERESTS
DDLB is an Exo Therapeutics advisor and has received research grant funding from AstraZeneca, Genentech-Roche and BeiGene for unrelated work. SF, NT, KA, and ADeF received grant funding from AstraZeneca for unrelated work. AGM and UM report funded research collaborations for unrelated work with industry: Intelligent Lab on Fiber, RNA Guardian, Micronoma and Mercy BioAnalytics. UM had stock ownership (2011–2021) awarded by University College London (UCL) in Abcodia, which held the licence for the Risk of Ovarian Cancer Algorithm (ROCA). UM reports research collaboration contracts with Cambridge University and QIMR Berghofer Medical Research Institute. UM holds patent number EP10178345.4 for Breast Cancer Diagnostics. UM is a member of Tina’s Wish Scientific Advisory Board (USA) and Research Advisory Panel, Yorkshire Cancer Research (UK). The remaining authors declared no conflicts of interest.
REFERENCES
- 1.Bowtell DD, Böhm S, Ahmed AA, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nature Reviews Cancer 2015; 15(11): 668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norquist B, Wurz KA, Pennil CC, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. Journal of Clinical Oncology 2011; 29(22): 3008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patch AM, Christie EL, Etemadmoghadam D, et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015; 521(7553): 489–94. [DOI] [PubMed] [Google Scholar]
- 4.Christie EL, Pattnaik S, Beach J, et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nature communications 2019; 10(1): 1295-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gockley A, Melamed A, Bregar AJ, et al. Outcomes of Women With High-Grade and Low-Grade Advanced-Stage Serous Epithelial Ovarian Cancer. Obstetrics and gynecology 2017; 129(3): 439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dao F, Schlappe BA, Tseng J, et al. Characteristics of 10-year survivors of high-grade serous ovarian carcinoma. Gynecologic Oncology 2016; 141(2): 260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garsed DW, Alsop K, Fereday S, et al. Homologous recombination DNA repair pathway disruption and retinoblastoma protein loss are associated with exceptional survival in high-grade serous ovarian cancer. Clinical Cancer Research 2018; 24(3): 569–80. [DOI] [PubMed] [Google Scholar]
- 8.Saner FAM, Herschtal A, Nelson BH, et al. Going to extremes: determinants of extraordinary response and survival in patients with cancer. Nature Reviews Cancer 2019; 19(6): 339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.du Bois A, Reuss A, Pujade-Lauraine E, Harter P, Ray-Coquard I, Pfisterer J. Role of surgical outcome as prognostic factor in advanced epithelial ovarian cancer: A combined exploratory analysis of 3 prospectively randomized phase 3 multicenter trials. Cancer 2009; 115(6): 1234–44. [DOI] [PubMed] [Google Scholar]
- 10.Wallace S, Kumar A, Mc Gree M, et al. Efforts at maximal cytoreduction improve survival in ovarian cancer patients, even when complete gross resection is not feasible. Gynecologic Oncology 2017; 145(1): 21–6. [DOI] [PubMed] [Google Scholar]
- 11.Harter P, Sehouli J, Vergote I, et al. Randomized Trial of Cytoreductive Surgery for Relapsed Ovarian Cancer. The New England journal of medicine 2021; 385(23): 2123–31. [DOI] [PubMed] [Google Scholar]
- 12.Tothill RW, Tinker AV, George J, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clinical Cancer Research 2008; 14(16): 5198–208. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Beach JA, Agadjanian H, et al. Suboptimal cytoreduction in ovarian carcinoma is associated with molecular pathways characteristic of increased stromal activation. Gynecologic Oncology 2015; 139(3): 394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Armasu SM, Kalli KR, et al. Pooled Clustering of High-Grade Serous Ovarian Cancer Gene Expression Leads to Novel Consensus Subtypes Associated with Survival and Surgical Outcomes. Clinical cancer research : an official journal of the American Association for Cancer Research 2017; 23(15): 4077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres D, Kumar A, Bakkum-Gamez JN, et al. Mesenchymal molecular subtype is an independent predictor of severe postoperative complications after primary debulking surgery for advanced ovarian cancer. Gynecologic Oncology 2019; 152(2): 223–7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T Cells, Recurrence, and Survival in Epithelial Ovarian Cancer. New England Journal of Medicine 2003; 348(3): 203–13. [DOI] [PubMed] [Google Scholar]
- 17.Hwang WT, Adams SF, Tahirovic E, Hagemann IS, Coukos G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: A meta-analysis. Gynecologic Oncology 2012; 124(2): 192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010; 28(15): 2512–9. [DOI] [PubMed] [Google Scholar]
- 19.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474(7353): 609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research 2014; 20(3): 764–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolton KL, Chenevix-Trench G, Goh C, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012; 307(4): 382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alsop K, Fereday S, Meldrum C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian ovarian cancer study group. Journal of Clinical Oncology 2012; 30(21): 2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Candido-dos-Reis FJ, Song H, Goode EL, et al. Germline mutation in BRCA1 or BRCA2 and ten-year survival for women diagnosed with epithelial ovarian cancer. Clinical cancer research 2015; 21(3): 652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Bernhardy AJ, Cruz C, et al. The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Research 2016; 76(9): 2778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell KN, Wubbenhorst B, Wenz BM, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nature communications 2017; 8(1): 319-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garsed DW, Pandey A, Fereday S, et al. The genomic and immune landscape of long-term survivors of high-grade serous ovarian cancer. Nature genetics 2022; 54(12): 1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson OA, Jonasson JG, Olafsdottir K, et al. CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 2011; 6(5): 638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jönsson G, Staaf J, Vallon-Christersson J, et al. The Retinoblastoma Gene Undergoes Rearrangements in BRCA1 -Deficient Basal-like Breast Cancer. Cancer Research 2012; 72(16): 4028–36. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty G, Armenia J, Mazzu YZ, et al. Significance of BRCA2 and RB1 co-loss in aggressive prostate cancer progression. Clinical Cancer Research 2020; 26(8): 2047–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WS, Alshalalfa M, Zhao SG, et al. Novel RB1-Loss Transcriptomic Signature Is Associated with Poor Clinical Outcomes across Cancer Types. Clinical Cancer Research 2019; 25(14): 4290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nature Reviews Cancer 2008; 8(9): 671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nature reviews Cancer 2008; 8(9): 714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vélez-Cruz R, Manickavinayaham S, Biswas AK, et al. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes and Development 2016; 30(22): 2500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millstein J, Budden T, Goode EL, et al. Prognostic gene expression signature for high-grade serous ovarian cancer. Annals of Oncology 2020; 31(9): 1240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milea A, George SHL, Matevski D, et al. Retinoblastoma pathway deregulatory mechanisms determine clinical outcome in high-grade serous ovarian carcinoma. Modern Pathology 2014; 27(7): 991–1001. [DOI] [PubMed] [Google Scholar]
- 36.Sieh W, Köbel M, Longacre TA, et al. Hormone-receptor expression and ovarian cancer survival: An Ovarian Tumor Tissue Analysis consortium study. The Lancet Oncology 2013; 14(9): 853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köbel M, Kang EY, Weir A, et al. p53 and ovarian carcinoma survival: an Ovarian Tumor Tissue Analysis consortium study. The Journal of Pathology: Clinical Research 2023; 9(3): 208–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talhouk A, George J, Wang C, et al. Development and Validation of the Gene Expression Predictor of High-grade Serous Ovarian Carcinoma Molecular SubTYPE (PrOTYPE). Clinical Cancer Research 2020; 26(20): 5411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ovarian Tumor Tissue Analysis (OTTA) Consortium Goode EL, Block MS, et al. Dose-Response Association of CD8+ Tumor-Infiltrating Lymphocytes and Survival Time in High-Grade Serous Ovarian Cancer. JAMA oncology 2017; 3(12): e173290–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen L W. M. Martens J, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nature Communications 2020; 11(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Communications 2013; 4(2126). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köbel M, Piskorz AM, Lee S, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. The Journal of Pathology: Clinical Research 2016; 2(4): 247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollis RL, Thomson JP, Stanley B, et al. Molecular stratification of endometrioid ovarian carcinoma predicts clinical outcome. Nature Communications 2020; 11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81(3): 323–30. [DOI] [PubMed] [Google Scholar]
- 45.Genovese C, Trani D, Caputi M, Claudio PP. Cell cycle control and beyond: emerging roles for the retinoblastoma gene family. Oncogene 2006; 25(38): 5201–9. [DOI] [PubMed] [Google Scholar]
- 46.Findlay GM, Daza RM, Martin B, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature 2018; 562(7726): 217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degasperi A, Amarante TD, Czarnecki J, et al. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nature Cancer 2020; 1(2): 249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature 2020; 578(7793): 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang EY, Weir A, Meagher NS, et al. CCNE1 and survival of patients with tubo-ovarian high-grade serous carcinoma: An Ovarian Tumor Tissue Analysis consortium study. Cancer 2022; 54(4): 538–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.da Costa AABA, do Canto LM, Larsen SJ, et al. Genomic profiling in ovarian cancer retreated with platinum based chemotherapy presented homologous recombination deficiency and copy number imbalances of CCNE1 and RB1 genes. BMC Cancer 2019; 19(1): 422-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandigo AC, Tomlins SA, Kelly WK, Knudsen KE. Relevance of pRB Loss in Human Malignancies. Clin Cancer Res 2022; 28(2): 255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017; 355(6320): 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mu P, Zhang Z, Benelli M, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53 - and RB1 -deficient prostate cancer. Science 2017; 355(6320): 84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palafox M, Monserrat L, Bellet M, et al. High p16 expression and heterozygous RB1 loss are biomarkers for CDK4/6 inhibitor resistance in ER(+) breast cancer. Nat Commun 2022; 13(1): 5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wander SA, Cohen O, Gong X, et al. The Genomic Landscape of Intrinsic and Acquired Resistance to Cyclin-Dependent Kinase 4/6 Inhibitors in Patients with Hormone Receptor-Positive Metastatic Breast Cancer. Cancer Discov 2020; 10(8): 1174–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Derenzini M, Donati G, Mazzini G, et al. Loss of Retinoblastoma Tumor Suppressor Protein Makes Human Breast Cancer Cells More Sensitive to Antimetabolite Exposure. Clinical Cancer Research 2008; 14(7): 2199–209. [DOI] [PubMed] [Google Scholar]
- 57.Treré D, Brighenti E, Donati G, et al. High prevalence of retinoblastoma protein loss in triple-negative breast cancers and its association with a good prognosis in patients treated with adjuvant chemotherapy. Annals of Oncology 2009; 20(11): 1818–23. [DOI] [PubMed] [Google Scholar]
- 58.Patel JM, Goss A, Garber JE, et al. Retinoblastoma protein expression and its predictors in triple-negative breast cancer. NPJ breast cancer 2020; 6(1): 19-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer 2010; 10(11): 803–8. [DOI] [PubMed] [Google Scholar]
- 60.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490(7418): 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Köbel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS medicine 2008; 5(12): e232–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang EY, Millstein J, Popovic G, et al. MCM3 is a novel proliferation marker associated with longer survival for patients with tubo-ovarian high-grade serous carcinoma. Virchows Archiv 2022; 480(4): 855–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y, Wang Y, Zhu F, Zhang J, Ma X, Zhang D. Gene expression profiling revealed MCM3 to be a better marker than Ki67 in prognosis of invasive ductal breast carcinoma patients. Clinical and Experimental Medicine 2020; 20(2): 249–59. [DOI] [PubMed] [Google Scholar]
- 64.Velez-Cruz R, Manickavinayaham S, Biswas AK, et al. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev 2016; 30(22): 2500–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westphalen CB, Fine AD, André F, et al. Pan-cancer Analysis of Homologous Recombination Repair–associated Gene Alterations and Genome-wide Loss-of-Heterozygosity Score. Clinical Cancer Research 2022; 28(7): 1412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng J, Liu X, Zhang P, et al. Rb selectively inhibits innate IFN-β production by enhancing deacetylation of IFN-β promoter through HDAC1 and HDAC8. Journal of Autoimmunity 2016; 73: 42–53. [DOI] [PubMed] [Google Scholar]
- 67.Manzano RG, Catalan-Latorre A, Brugarolas A. RB1 and TP53 co-mutations correlate strongly with genomic biomarkers of response to immunity checkpoint inhibitors in urothelial bladder cancer. BMC Cancer 2021; 21(432). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molinero L, Li Y, Chang C-W, et al. Tumor immune microenvironment and genomic evolution in a patient with metastatic triple negative breast cancer and a complete response to atezolizumab. Journal for ImmunoTherapy of Cancer 2019; 7(274). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. Journal of Pathology 2010; 221(1): 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Landen CN, Molinero L, Hamidi H, et al. Influence of Genomic Landscape on Cancer Immunotherapy for Newly Diagnosed Ovarian Cancer: Biomarker Analyses from the IMagyn050 Randomized Clinical Trial. Clinical Cancer Research 2023; 29(9): 1698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kandalaft LE, Odunsi K, Coukos G. Immune Therapy Opportunities in Ovarian Cancer. American Society of Clinical Oncology Educational Book 2020; 3(40): e228–e40. [DOI] [PubMed] [Google Scholar]
- 72.Bulanova D, Akimov Y, Senkowski W, et al. A synthetic lethal dependency on casein kinase 2 in response to replication-perturbing drugs in RB1-deficient ovarian and breast cancer cells. bioRxiv 2022: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong X, Du J, Parsons SH, et al. Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov 2019; 9(2): 248–63. [DOI] [PubMed] [Google Scholar]
- 74.Lyu J, Yang EJ, Zhang B, et al. Synthetic lethality of RB1 and aurora A is driven by stathmin-mediated disruption of microtubule dynamics. Nat Commun 2020; 11(1): 5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oser MG, Fonseca R, Chakraborty AA, et al. Cells Lacking the RB1 Tumor Suppressor Gene Are Hyperdependent on Aurora B Kinase for Survival. Cancer Discov 2019; 9(2): 230–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serra V, Wang AT, Castroviejo-Bermejo M, et al. Identification of a Molecularly-Defined Subset of Breast and Ovarian Cancer Models that Respond to WEE1 or ATR Inhibition, Overcoming PARP Inhibitor Resistance. Clinical Cancer Research 2022; 28(20): 4536–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.