Abstract
The role of oxygen availability in determining the local physiological activity of Pseudomonas aeruginosa growing in biofilms was investigated. Biofilms grown in an ambient-air environment expressed approximately 1/15th the alkaline phosphatase specific activity of planktonic bacteria subjected to the same phosphate limitation treatment. Biofilms grown in a gaseous environment of pure oxygen exhibited 1.9 times the amount of alkaline phosphatase specific activity of air-grown biofilms, whereas biofilms grown in an environment in which the air was replaced with pure nitrogen prior to the inducing treatment did not develop alkaline phosphatase activity. Frozen cross sections of biofilms stained for alkaline phosphatase activity with a fluorogenic stain demonstrated that alkaline phosphatase activity was concentrated in distinct bands adjacent to the gaseous interfaces. These bands were approximately 30 μm thick with biofilms grown in air, 2 μm thick with biofilms grown in pure nitrogen, and 46 μm thick with biofilms grown in pure oxygen. Overall biofilm thickness ranged from approximately 117 to approximately 151 μm. Measurements with an oxygen microelectrode indicated that oxygen was depleted locally within the biofilm and that the oxygen-replete zone was of a dimension similar to that of the biologically active zone, as indicated by alkaline phosphatase induction. These experiments revealed marked spatial physiological heterogeneity within P. aeruginosa biofilms in which active protein synthesis was restricted by oxygen availability to the upper 30 μm of the biofilm. Such physiological heterogeneity has implications for microbial ecology and for understanding the reduced susceptibilities of biofilms to antimicrobial agents.
It is now generally accepted that bacteria have a marked tendency to attach to surfaces and initiate biofilm formation (8, 13). Biofilms lead to many undesired problems in industry, such as decreased heat transfer in cooling towers, regrowth in drinking water distribution systems, and deterioration of materials. In the medical area, biofilms are responsible for dental plaque and persistent infections on medical implants. It has long been observed that biofilms are much less susceptible to antimicrobial agents than are their planktonic counterparts (4, 7, 20, 25), but the underlying basis for this recalcitrance is not well established. One of the primary mechanisms suggested by many researchers is transport limitation of the antimicrobial agents into the biofilm community. This mechanism is related to the reaction or sorption of the antimicrobial agent within the biofilm, which establishes a diffusion barrier (6, 16, 18). However, transport limitation might not be sufficient to completely explain biofilm recalcitrance. Nichols (24) developed a mathematical model of the penetration of two antibiotics (the aminoglycoside tobramycin and the β-lactam compound cefsulodin) into biofilms of Pseudomonas aeruginosa and concluded that transport limitation did not adequately explain biofilm resistance to these antibiotics. Stewart (27) recently argued that, for most antibiotics, transport limitation was insufficient to explain the reduced susceptibilities of biofilms. Therefore, the roles of attachment-induced responses and physiological adaptation to nutrient limitation of bacteria in biofilms are receiving more attention (1, 14). Specifically, it has been speculated that the internal portions of biofilms experience starvation and slow growth due to nutrient limitation.
Several recent papers have examined the relationship between biofilm growth and susceptibility (2, 3, 11, 12). Growth rate limitation occurs in biofilms presumably as a result of restriction for a particular nutrient that fails to fully penetrate the biofilm. For example, Tresse et al. (28) reported that oxygen limitation contributed to enhanced antibiotic resistance of agar-entrapped Escherichia coli. Oxygen concentration gradients in biofilms have been experimentally demonstrated many times by microelectrode technology (10).
The spatial heterogeneity of growth within a biofilm is a crucial issue in determining susceptibility to growth rate-dependent antimicrobial agents. The purpose of the work reported in this article was to test the dual hypotheses that physiological status varies spatially within the biofilm and that, for a P. aeruginosa model biofilm, physiological activity is controlled by oxygen availability. As a physiological indicator, we have used the expression of alkaline phosphatase (APase) upon exposure to phosphate starvation, which reflects the capacity for de novo protein synthesis. Oxygen delivery was controlled by varying the composition of the gaseous environment and measured directly with an oxygen microelectrode.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
A pure culture of P. aeruginosa ERC1 was used throughout. It was isolated from an industrial water system, identified with API NFT (bioMérieux Vitek, Inc., Hazelwood, Mo.), and retained in the culture collection of the Center for Biofilm Engineering. The 16S ribosomal DNA of the isolate was PCR amplified with 27F (sequence, 5′-AGA GTT TGA TCC TGG CTC AG-3′, which corresponds to E. coli 16S rRNA positions 8 to 27) as a forward primer and 1392R (sequence, 5′-ACG GGC GGT GTG TAC-3′, which corresponds to E. coli 16S rRNA positions 1392 to 1406) as a reverse primer and commercially sequenced at the University of Montana (Missoula, Mont.). When the sequence was aligned with the most similar Ribosomal Database Project (21) sequence with Genetic Data Environment, version 2.3, software, the percent similarity between P. aeruginosa NIH18 and ERC1 was 99.0%. MOPS (morpholinopropanesulfonic acid) minimal medium prepared as described by Neidhardt et al. (23) was used in both planktonic and biofilm experiments. High-phosphate medium contained 1 g of Na2HPO4 per liter, while low-phosphate medium contained 0.01 g of Na2HPO4 per liter. Glucose (1.0 g/liter of the planktonic culture medium and 0.1 g/liter of the biofilm culture medium) was used as the sole carbon source. All the experiments were carried out at room temperature, i.e., 22.2 ± 3.0°C.
Planktonic culture procedure.
P. aeruginosa overnight cultures were harvested by centrifugation at 7,500 rpm with a rotor 55-34, model RC5C centrifuge (Sorvall, Inc., Oklahoma City, Okla.) for 10 min, washed twice with low-phosphate medium, and resuspended in 100 ml of low-phosphate medium to induce phosphate starvation. The low-phosphate culture was stirred for 24 h. Two-milliliter aliquots were withdrawn every hour for APase and total protein assays. To test the effect of anoxic conditions on APase production, a low-phosphate culture was left in ambient air for 2.5 h, connected to pure nitrogen through a bacterial air vent for 3 h, and then changed back to ambient air for another 2.5 h. Two-milliliter aliquots were withdrawn every 30 min.
Biofilm culture procedure.
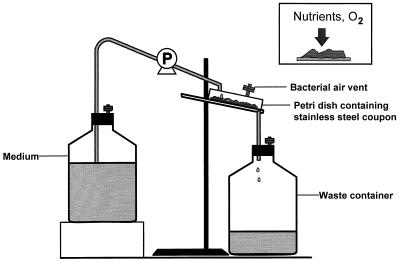
A drip-flow plate reactor was designed to cultivate biofilms (Fig. 1). Stainless steel slides in petri dishes were continuously bathed with medium that dripped onto the biofilm at a constant flow rate of 50 ml/h. After inoculation with an overnight culture (3 × 108 cells/ml in 0.1 g of glucose MOPS medium/liter) and incubation for 24 h, the reactor was fed for another 72 h with high-phosphate medium, which was then replaced with low-phosphate medium. The bacterial air vent of the reactor was either (i) connected to pure nitrogen (120 to 130 ml/min) or pure oxygen (120 to 130 ml/min) or (ii) exposed to ambient air to create different gaseous environments.
FIG. 1.
Schematic diagram of our drip-flow biofilm reactor.
APase activity and total protein assay.
In planktonic experiments, 2-ml aliquots were sampled, centrifuged, and then resuspended in 1 ml of TEP solution (10 mM Tris-Cl [pH 8.0], 1 mM EDTA [pH 8.0], 1 mM phenylmethylsulfonyl fluoride). In biofilm experiments, attached cells were scraped into 20 ml of phosphate-buffered saline buffer with a rubber policeman. After being treated with a homogenizer (Tissuemizer, type SDT 1810; Tekmar Co., Cincinnati, Ohio) with an output speed of 13,500 rpm in an ice bath for 3 min, 2-ml aliquots were suspended in 1 ml of TEP solution. Bacterial suspensions in TEP solution were disrupted by ultrasonic treatment with an ultrasonic cell disrupter (TORBEO, 36810 series; Cole-Parmer, Vernon Hills, Ill.) and then centrifuged. The supernatant was used for enzyme and total protein assays. APase activity was determined by the rate of hydrolysis of p-nitrophenyl phosphate to p-nitrophenol, measured by absorbance at 410 nm. The changes in colorimetric intensities were monitored over a 4-min interval. Total protein was determined with Sigma (St. Louis, Mo.) diagnostic kit no. 690, which uses a modified micro-Lowry method.
Staining procedures.
ELF-97 phosphatase substrate (Molecular Probes, Eugene, Oreg.) is a water-soluble faintly blue fluorescent stain. Upon cleavage by APase, the substrate yields a bright yellow-green intracellular fluorescent precipitate that exhibits excellent photostability, since photobleaching is insignificant. Therefore, bacterial cells with and without APase can be simultaneously visualized as yellow-green and blue fluorescence, respectively, by epifluorescence microscopy. The biofilms were stained ex situ in a homemade staining box with 5 ml of staining solution at 35°C for 45 min. For visual microscopic examination and photography, frozen biofilm sections were counterstained with 5 μl of 10-μg/ml propidium iodide to improve the contrast of APase-positive and -negative cells. After propidium iodide counterstaining, cells with APase activity exhibited yellow-green fluorescence while cells without APase activity were red through a U filter. For image analysis, biofilm sections were counterstained with 5 μl of 1-μg/liter tetramethylrhodamine.
Cryoembedding and cryosectioning.
Biofilm samples were cryoembedded with Tissue-Tek O.C.T. compound (Miles Inc., Elkhart, Ind.) as described previously (31). Embedded samples were sectioned with a model CM 1800 cryostat (Leica Inc., Deerfield, Ill.). The 5-μm-thick sections were mounted on Superfrost Plus microscopic slides (Fisher Scientific, Pittsburgh, Pa.).
Microscopy.
An Olympus (Lake Success, N.Y.) BH-2 microscope with epifluorescence illumination was used for the examination of the biofilm sections. After enzyme-labeled-fluorescence staining, faintly blue and intensely yellow-green fluorescence was visualized with an Olympus U filter cubic unit containing an excitation filter (wavelength, 334 to 365 nm), a dichroic mirror (model DM-400), and a barrier filter (model L-420).
Image analysis.
After being counterstained with tetramethylrhodamine, cells containing APase activity (green) and all cells (red) were selectively captured with an Olympus U and G filter cubic unit, respectively. The G filter cubic unit contained an excitation filter (model BP-545), a diachronic mirror (model DM-570), and a barrier filter (model O-590). The images captured at the same spot by different filters were digitalized by a cooled color charge-coupled device camera (Optronics, Goleta, Calif.) and saved as 8-bit gray-scale TIFF files. The fluorescence intensity was determined by MARK image analysis software (15).
Dissolved-oxygen profile measurement.
Oxygen profiles were measured with a Clark-type dissolved-oxygen microelectrode equipped with a guard cathode. The electrode was constructed as described by Revsbech (26). Microelectrode measurements were conducted with a micromanipulator (model M3301L; World Precision Instruments, New Haven, Conn.) equipped with a stepper motor (model 18503; Oriel, Stratford, Conn.). Custom data acquisition software was used to control the microelectrode movement. The microelectrode was introduced into a biofilm from the top, perpendicular to the substratum of stainless steel slides. Data was collected at 10-μm increments and 2-s intervals. Current produced by the electrode was collected and converted to oxygen concentration by the software.
RESULTS
Planktonic and biofilm APase specific activity.
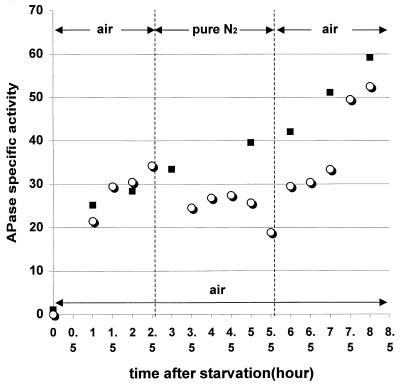
APase activity was below detectable levels in bacteria grown in planktonic suspensions in high-phosphate medium. After we induced phosphate starvation by transferring the culture to low-phosphate medium, APase was readily measured in planktonic bacteria. APase specific activity kept increasing during the first 8 h of phosphate starvation (Fig. 2). When pure nitrogen was introduced after a 2.5-h induction in air, APase production was immediately arrested. Accumulation of enzyme activity resumed immediately after the nitrogen was changed back to air (Fig. 2). When bacterial biofilm was subjected to the same duration of exposure to low-phosphate medium under ambient aerobic conditions, the level of APase specific activity was approximately 1/15th that of comparable planktonic bacteria (Table 1). Biofilm induction of APase was totally blocked when pure nitrogen was administered during the induction period, whereas induction of APase increased approximately twofold when pure oxygen was administered (Table 1).
FIG. 2.
APase specific activity of planktonic P. aeruginosa in response to phosphate starvation. APase specific activity is expressed as the change in the A410 mg of protein−1 min−1. ■, APase specific activity under ambient air without disruption for 8 h; ○, APase specific activity of a culture exposed to air, and then pure nitrogen, and finally air again.
TABLE 1.
APase specific activities under different conditions of planktonic and biofilm P. aeruginosa organisms
| Bacteria | Conditionsa | Enzyme sp act (ΔA410 mg of protein−1 min−1)b | Cell densityc |
|---|---|---|---|
| Planktonic | HP for 8 h | 0 | 1.52 × 109 |
| LP for 8 h | 59.19 | 2.6 × 108 | |
| Biofilm | LP + air for 8 h | 3.95 ± 0.56 | 4.26 ± 0.96 × 109 |
| LP + pure N2 for 8 h | 0.13 ± 0.079 | 4.43 ± 1.43 × 109 | |
| LP + pure O2 for 8 h | 7.49 ± 0.83 | 4.72 ± 0.53 × 109 |
HP, growth in high-phosphate medium; LP, growth in low-phosphate medium.
ΔA410, change in A410.
Cell densities are expressed as CFU per milliliter of planktonic bacteria and CFU per square centimeter of biofilm.
Patterns of APase expression in biofilms.
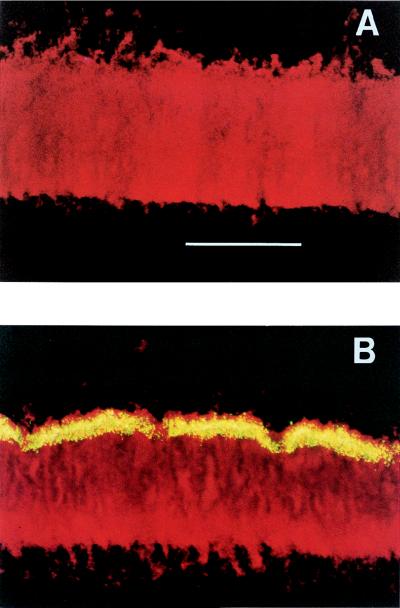
Staining with a fluorogenic phosphatase substrate revealed a distinct, spatially nonuniform pattern of APase expression within P. aeruginosa biofilm after induction by switching to low-phosphate medium. A control biofilm grown continuously with high-phosphate medium showed no APase activity (Fig. 3A). APase was expressed in a sharply delineated band adjacent to the biofilm-bulk fluid interface (Fig. 3B). Biofilm subjected to low-phosphate medium in a nitrogen atmosphere exhibited no visible phosphatase activity (image not shown). Conversely, when the reactor atmosphere was pure oxygen, not only was the zone of APase expression expanded but also localized sites of APase activity could be found much deeper inside the biofilm (image not shown).
FIG. 3.
Photomicrographs of P. aeruginosa biofilm cross sections stained for APase activity under different conditions, namely, high-phosphate medium with an ambient aerobic atmosphere (control) (A) and low-phosphate medium with an ambient aerobic atmosphere (B). The green-yellow color represents APase-positive cells, and the red color represents all cells. The images are oriented with the substrata at the bottoms of the pictures. Bar, 100 μm.
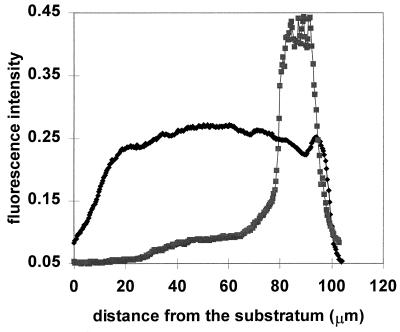
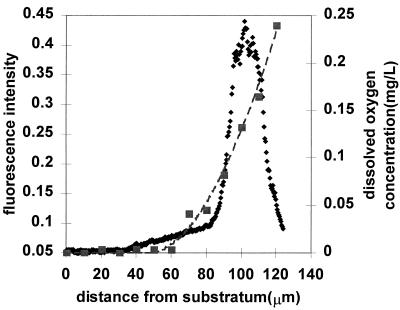
Image analysis was applied to images like those in Fig. 3 to quantitate the dimensions of the zones of APase expression under different gaseous environments. A representative image analysis result, showing a profile for phosphatase activity along with a profile for the entire biofilm, is shown in Fig. 4. The mean thicknesses of biofilms ranged from 117 to 151 μm (Table 2). The mean dimension of APase activity bands was approximately 30 μm when biofilms were grown in air. With a pure-oxygen environment, the zone of APase activity was expanded approximately 1.5-fold (Table 2).
FIG. 4.
Representative image analysis result of APase expression in P. aeruginosa biofilms with phosphate starvation under ambient aerobic condition. ⧫, biofilm cells stained with tetramethylrhodamine; ■, APase-positive cells.
TABLE 2.
Thicknesses of zones of APase expression under different gaseous conditions
| Atmosphere | Mean biofilm thickness ± SD (μm) | Mean APase band thickness ± SD (μm) |
|---|---|---|
| Ambient air | 117.13 ± 11.54 | 29.89 ± 4.39 |
| N2 | 142.04 ± 19.14 | 1.82 ± 3.11 |
| O2 | 150.59 ± 30.61 | 46.43 ± 5.72 |
Dissolved-oxygen measurement.
We measured the dissolved-oxygen profile within a biofilm under ambient aerobic conditions and superimposed image analysis data of APase activity with the dissolved-oxygen concentration profile (Fig. 5). The dissolved-oxygen concentration decreased from approximately 0.25 mg/liter at the biofilm-bulk fluid interface to essentially 0 (less than 0.01 mg/liter) at a point approximately 40 μm above the substratum. The band of APase expression in the upper region of the biofilm coincided with dissolved-oxygen concentrations of greater than 0.05 mg/liter.
FIG. 5.
Correlation of the dissolved-oxygen profile with the image analysis result of APase expression under aerobic conditions. ⧫, APase activity; ■, dissolved oxygen; ––, trend line representing dissolved-oxygen concentration.
DISCUSSION
APase was first discovered in P. aeruginosa by Hou et al. in 1966 (17). In E. coli it is induced under the control of a complex regulatory circuit and functions in scavenging phosphate from organic phosphate esters (22). APase is also easily detectable in planktonic suspensions of P. aeruginosa. In P. aeruginosa biofilms, the enzyme-labeled-fluorescence technique coupled with cryoembedding and cryosectioning enabled the visualization of the spatial pattern of APase expression (19). The pattern is distinct in P. aeruginosa biofilms in an atmosphere of ambient air. We hypothesized that this pattern is due to oxygen limitation because (i) oxygen is often limiting in aerobic biofilms because of its rapid consumption; (ii) in the drip-flow system, both nutrients and oxygen come from the top of the biofilm and cells near the bulk fluid-biofilm interface have greater access to oxygen and can consume it; (iii) oxygen is limiting deep within biofilms as suggested by de Beer et al. (10); and (iv) enzyme biosynthesis requires both nutrients and oxygen, since P. aeruginosa is an obligate aerobe under the experiment conditions used (no nitrate available).
To test this hypothesis we designed a simple experiment using planktonic bacteria to see if we could switch APase production off and on by switching from anaerobic to aerobic conditions. The result shown in Fig. 2 supported our hypothesis. Pure nitrogen stopped APase production immediately, and APase production was resumed when the biofilm was exposed to air. We then exposed biofilms to different gaseous environments to see how the pattern of APase production would change in the attached population. With pure nitrogen, the expression of APase was totally blocked. This result showed that enzyme synthesis within the P. aeruginosa biofilm required oxygen. The experiment with a pure-oxygen environment led to a very interesting observation. The dimension of the band of APase expression increased 1.5-fold as measured by image analysis. Also, disperse green crystals, indicating isolated sites of APase activity, were found deep within the biofilm. Collective APase specific activity of the entire biofilm under ambient aerobic conditions was approximately 1/15 of that of comparable planktonic bacteria, which makes sense because only the upper 1/5 to 1/4 of the layer of the biofilm was expressing the enzyme. Also, 1.0 g of glucose per liter was used in planktonic culture to achieve enough cell mass. APase production of the whole biofilm was increased twofold under pure oxygen. This result supports our speculation that oxygen can penetrate deeper within the biofilm when pure oxygen is used, but the physiological heterogeneity of the community with oxygen-starved cells deep within a biofilm might lead to a different response. Hence, it was of interest to measure the penetration of dissolved oxygen. A good correlation was observed between the oxygen penetration profile, when the oxygen was measured under ambient aerobic conditions, and the band of APase expression. These findings also correspond somewhat to studies of microbial mats where changes in guild activity reflect changing chemical composition with increasing depth in the community (5, 9, 29).
The spatial patterns of expression of the phosphate starvation gene that we obtained in biofilms are distinct because we chose P. aeruginosa, which is an obligate aerobe, as our test microorganism to study the role of oxygen limitation in the expression of the phosphate starvation response. These results may also be related to the reduced susceptibilities of bacterial biofilms to antimicrobial agents. Specifically, it may be important to determine the role of limiting nutrients in the establishment of physiological gradients to understand mechanisms of recalcitrance. Since the spatial physiological patterns of biofilms of different microorganisms may not be the same as those of pure P. aeruginosa biofilms (19), that goal is further complicated in natural biofilm communities composed of both aerobes and anaerobes. However, the results reported here for P. aeruginosa biofilms give a representative picture and support the concept of physiological heterogeneity within biofilms (19, 30).
ACKNOWLEDGMENTS
This work was supported through cooperative agreement EEC-8907039 between the National Science Foundation and Montana State University and by the industrial associates of the Center for Biofilm Engineering.
We thank Betsey Pitts (Center for Biofilm Engineering, Montana State University, Bozeman, Mont.) for PCR amplifying the 16S ribosomal DNA of the isolate and Mary Bateson (Department of Microbiology, Montana State University) for assistance with the phylogenetic analysis.
REFERENCES
- 1.Anwar H, Strap J L, Costerton J W. Establishment of aging biofilms: possible mechanism of bacterial resistance to antimicrobial therapy. Antimicrob Agents Chemother. 1992;36:1347–1351. doi: 10.1128/aac.36.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashby M J, Neale J E, Knott S J, Critchley I A. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J Antimicrob Chemother. 1994;33:443–452. doi: 10.1093/jac/33.3.443. [DOI] [PubMed] [Google Scholar]
- 3.Brown M R W, Allison D G, Gilbert P. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J Antimicrob Chemother. 1988;22:777–780. doi: 10.1093/jac/22.6.777. [DOI] [PubMed] [Google Scholar]
- 4.Brown M R W, Gilbert P. Sensitivity of biofilms to antimicrobial agents. J Appl Bacteriol Symp Suppl. 1993;74:87S–97S. doi: 10.1111/j.1365-2672.1993.tb04345.x. [DOI] [PubMed] [Google Scholar]
- 5.Canfield D E, Des Marais D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Stewart P S. Chlorine penetration into artificial biofilm is limited by a reaction-diffusion interaction. Environ Sci Technol. 1996;30:2078–2083. [Google Scholar]
- 7.Costerton J W. The formation of biocide-resistant biofilms in industrial, natural and medical systems. Dev Ind Microbiol. 1984;25:363–372. [Google Scholar]
- 8.Costerton J W, Lewandoski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilm. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 9.D’Amelio E D, Cohen Y, Des Marais D J. Comparative functional ultrastructure of two hypersaline submerged cyanobacterial mats: Guerrero Negro, Baja California Sur, Mexico, and Solar Lake, Sinai, Egypt. In: Cohen Y, Rosenberg E, editors. Microbial mats: physiological ecology of benthic microbial communities. Washington, D.C: American Society for Microbiology; 1989. pp. 97–113. [Google Scholar]
- 10.de Beer D, Stoodley P, Lewandowski Z. Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 11.Evans D J, Allison D G, Brown M R W, Gilbert P. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms toward ciprofloxacin: effect of specific growth rate. J Antimicrob Chemother. 1991;27:177–184. doi: 10.1093/jac/27.2.177. [DOI] [PubMed] [Google Scholar]
- 12.Evans D J, Brown M R W, Allison D G, Gilbert P. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J Antimicrob Chemother. 1990;25:585–591. doi: 10.1093/jac/25.4.585. [DOI] [PubMed] [Google Scholar]
- 13.Geesey G G. Microbial exopolymers: ecological and economic considerations. ASM News. 1982;48:9–14. [Google Scholar]
- 14.Gilbert P, Collier P J, Brown M R W. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob Agents Chemother. 1990;34:1865–1868. doi: 10.1128/aac.34.10.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harkin G, Shope P. The Mark image analysis system. Technical report. Bozeman, Mont: Center for Biofilm Engineering, Montana State University; 1993. [Google Scholar]
- 16.Hodges N A, Gordon C A. Protection of Pseudomonas aeruginosa against ciprofloxacin and β-lactams by homologous alginate. Antimicrob Agents Chemother. 1991;35:2450–2452. doi: 10.1128/aac.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou C I, Gronlund A F, Campbell J J R. Influence of phosphate starvation on cultures of Pseudomonas aeruginosa. J Bacteriol. 1966;92:851–855. doi: 10.1128/jb.92.4.851-855.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyle B D, Alcantora J, Costerton J W. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob Agents Chemother. 1992;36:2054–2056. doi: 10.1128/aac.36.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C-T, Xu K D, McFeters G A, Stewart P S. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl Environ Microbiol. 1998;64:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeChevallier M W, Cawthon C D, Lew R G. Inactivation of biofilm bacteria. Appl Environ Microbiol. 1988;54:2492–2499. doi: 10.1128/aem.54.10.2492-2499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olson G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moat A G, Foster J W, editors. Microbial physiology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 196–197. [Google Scholar]
- 23.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols W W. Susceptibility of biofilms to toxic compounds. In: Characklis W G, Wilderer P A, editors. Structure and function of biofilms. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 321–331. [Google Scholar]
- 25.Nickel J C, Ruseska I, Wright J B, Costerton J W. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revsbech N P. An oxygen microsensor with a guard cathode. Limnol Oceanogr. 1989;34:474–478. [Google Scholar]
- 27.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tresse O, Jouenne T, Junter G-A. The role of oxygen limitation in the resistance of agar-entrapped, sessile-like Escherichia coli to aminoglycoside and β-lactam antibiotics. J Antimicrob Chemother. 1995;35:521–526. doi: 10.1093/jac/36.3.521. [DOI] [PubMed] [Google Scholar]
- 29.Ward D M, Bauld J, Castenholz R W, Pierson B K. Modern phototrophic microbial mats: anoxygenic, intermittently oxygenic/anoxygenic, thermal, eukaryotic, and terrestrial. In: Schopf J W, Klein C, editors. The proterozoic biosphere, a multidisciplinary study. New York, N.Y: Cambridge University Press; 1992. pp. 309–324. [Google Scholar]
- 30.Wentland E J, Stewart P S, Huang C T, McFeters G A. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol Prog. 1996;12:316–321. doi: 10.1021/bp9600243. [DOI] [PubMed] [Google Scholar]
- 31.Yu F P, Callis G M, Stewart P S, Griebe T, McFeters G A. Cryosectioning of biofilm for microscopic examination. Biofouling. 1994;8:85–91. [Google Scholar]