Abstract
Resistance to the toxic compound potassium tellurite (Telr) has been employed as a selection marker built into a set of transposon vectors and broad-host-range plasmids tailored for genetic manipulations of Pseudomonas strains potentially destined for environmental release. In this study, the activated Telr determinants encoded by the cryptic telAB genes of plasmid RK2 were produced, along with the associated kilA gene, as DNA cassettes compatible with cognate vectors. In one case, the Telr determinants were assembled between the I and O ends of a suicide delivery vector for mini-Tn5 transposons. In another case, the kilA and telAB genes were combined with a minimal replicon derived from a variant of Pseudomonas plasmid pPS10, which is able to replicate in a variety of gram-negative hosts and is endowed with a modular collection of cloning and expression assets. Either in the plasmid or in the transposon vector, the Telr marker was combined with a 12-kb DNA segment of plasmid pWW0 of Pseudomonas putida mt-2 encoding the upper TOL pathway enzymes. This allowed construction of antibiotic resistance-free but selectable P. putida strains with the ability to grow on toluene as the sole carbon source through an ortho-cleavage catabolic pathway.
The use of recombinant Pseudomonas strains in bioremediation and biotransformation of organic chemicals (27) is sometimes limited by the paucity of genetic tools for constructing and tracing bacteria destined to perform under noncontained conditions (2, 4, 5, 12, 22). In many cases, having such strains carry antibiotic resistance markers is undesirable, while in other cases new traits introduced into a natural strain might be difficult to select for due to the natural resistance to multiple antibiotics that frequently exists in environmental isolates (1). In this context, a number of vectors tailored for construction of recombinant strains destined for environmental release have been designed in recent years (24, 25). One significant advance in this area was the combination of mini-Tn5 transposon vectors with nonantibiotic selection markers (6, 11). This permitted stable insertion of heterologous DNA segments into the chromosomes of many gram-negative eubacteria without the introduction of antibiotic resistance genes. While transposon vectors still remain the best choice for this purpose, the nonantibiotic selection markers (i.e., herbicide or heavy metal resistance) initially proposed by Herrero et al. (11) do present some problems in practical use. Resistance to herbicides, such as bialaphos (phosphinothricin tripeptide) or the monoisopropylamine salt of N-phosphonomethyl glycine (glyphosate) (25), is flawed by the high levels of tolerance and/or spontaneous mutation rates observed in gram-negative bacteria (11). Resistance to mercuric salts (encoded by the mer genes) is also hampered by the very narrow and varying ranges of concentrations of the agents at which selection is effective. Finally, tolerance to arsenite may not be easy to select for due to potential interference with the phosphate in the medium (11). Although these problems can be overcome in some cases by using nutritional markers (9) or excisable selection markers (19), none of the alternative resistance markers available previously was optimal for general use. In this work, we show that unlike other nonantibiotic determinants, resistance to tellurite salts (typically potassium tellurite, K2TeO3) is a potential marker for constructing strains destined for environmental release.
The toxicity of tellurite is believed to arise from its oxidative activity, and therefore tellurite has a broad spectrum of activity against many microorganisms. Resistance to tellurite (Telr) is found in both gram-positive and gram-negative bacteria, but in gram-negative bacteria it is frequently encoded by cryptic, nonexpressed genes borne by the chromosome (31) or by conjugative plasmids (33). The Telr marker used in this study originated from a variant of plasmid RK2 of the IncP-α group which actively expresses the Telr phenotype. Three genes borne by the plasmid (kilA, telA, and telB [28] or, alternatively, klaA, klaB, and klaC [8]) are required for tellurite resistance. The levels of the products of these genes are finely tuned, so even a low level of expression results in cells with high levels of resistance (27, 28). The mechanism of resistance is not fully known yet, but it systematically results in reduction of tellurite to elemental tellurium (15, 26, 31).
As shown below, we assembled the kilA and telAB genes either in minitransposon vectors or in a specialized mobilizable replicon derived from pPS10. Plasmid pPS10 is a natural plasmid that was originally isolated from a Pseudomonas syringae pv. savastanoi strain (18), whose replication mechanism is known in some detail. The basic replicon of this plasmid, which spans 1.8 kb, contains the origin of replication (oriV) and the gene of the initiator protein (repA) (18). Although wild-type pPS10 cannot be efficiently established in genera other than the genus Pseudomonas, certain repA variants expand the host range of the replicon to a variety of gram-negative bacteria (7). These variants allow propagation of the plasmid in Escherichia coli as a low-copy-number replicon and result in a moderately high copy number in Pseudomonas cells.
On the basis of the elements described above (resistance to tellurite, minitransposon vectors, and broadened-host-range replicons), we describe below the construction and performance of recombinant Pseudomonas putida strains designed for bioconversion of toluene and several alkyl and chloro- and nitro-substituted derivatives of toluene into the corresponding benzoates. This study was based on introduction into a plasmidless P. putida strain of the upper TOL operon of plasmid pWW0 borne by a Telr plasmid or a hybrid Telr minitransposon.
MATERIALS AND METHODS
Strains, media, and general procedures.
The bacterial strains and plasmids used in this study are described in Table 1. E. coli CC118λpir and S17-1λpir were used as hosts to propagate plasmids containing an R6K origin of replication (6). Solid and liquid Luria-Bertani (LB) media and M9 minimal medium (supplemented with 10 mM citrate as the sole carbon source [6]) were amended, when required, with ampicillin (100 μg/ml), piperacillin (40 μg/ml), kanamycin (75 μg/ml), chloramphenicol (30 μg/ml), rifampin (30 μg/ml), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml). Tellurite (K2TeO3; potassium tellurite hydrate; Aldrich Chemical Co.) was dissolved in water at a concentration of 40 mg/ml, and the preparation was filtered and kept frozen indefinitely at −20°C. The working concentrations of this salt in selective plates were 30 to 80 μg/ml. Growth on toluene as the sole carbon source was tested by patching the clones that were being investigated onto the surfaces of M9 mineral agar plates (16) without C and then saturating the plates with toluene vapor and incubating them for 2 to 4 days at 30°C. To amplify DNA segments by the PCR, 50 to 100 ng of a template was mixed in a 100-μl reaction mixture with 50 pmol of each primer used and 2.5 U of Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The reaction mixtures were then subjected to 25 cycles consisting of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C. Other gene cloning techniques were carried out by using previously published protocols (23). Mini-Tn5 vectors were inserted into the chromosome of P. putida by using the procedure described in detail by de Lorenzo and Timmis (6) and generally known as the pUT system. Triparental and biparental matings were performed to mobilize the transposon delivery plasmids from E. coli donors into the recipient Pseudomonas strain. The same mobilization strategy was used to transfer broadened-host-range derivatives of pPS10 from E. coli to Pseudomonas cells. The selection media used to identify exconjugants in all cases are indicated below. Expression of the luxAB luciferase genes of Vibrio harveyi was monitored by exposing the exponentially growing cultures being studied to n-decanal and measuring the emission of light with an LKB luminometer (13).
TABLE 1.
Bacteria and plasmids used
| Strain or plasmid(s) | Relevant characteristics | Reference or origin |
|---|---|---|
| Strains | ||
| E. coli CC118 | Δ(ara-leu) araD ΔlacX74 galE galK phoA thi-1 rpsE rpoB argE(Am) recA1 | 11 |
| E. coli CC118λpir | CC118 lysogenized with λpir phage | 11 |
| E. coli S17-1λpir | Tpr SmrrecA thi hsdRM+, λpir phage lysogen RP4::2-Tc::Mu::Km Tn7 | 6 |
| E. coli HB101 | SmrrecA thi pro leu hsdRM+ | Lab collection |
| P. putida KT2442 | Rifr CmrhsdR+, prototrophic | 6 |
| Plasmids | ||
| RK600 | Cmr ColE1oriV RK2mob+ tra+ | 6 |
| pDT1558 | Apr, pUC8 derivative containing at its SmaI site a 3.0-kb HincII fragment from RK2 containing kilA and telAB | 32 |
| pUC18Sfi | Apr, same as pUC18 but multiple cloning site flanked by SfiI-AvrII sites | 6 |
| pUC18Not | Apr, same as pUC18 but multiple cloning site flanked by NotI sites | 6 |
| pJMT4 | Apr, pUC18Sfi containing 3.2-kb HindIII-BamHI fragment from pDT1558 containing kilA and telAB | This study |
| pSHA1 | Apr, pUC18Not containing 3.2-kb HindIII-BamHI fragment from pDT1558 containing kilA and telAB | J. Benedí |
| pUTlacZ1 | Apr Kmr, delivery vector for mini-Tn5 lacZ1 NotI flanking a kanamycin resistance gene, SfiI-AvrII sites inserted with a promoterless lacZ | 6 |
| pJMT5 | Apr Kmr, pUTlacZ1 derivative in which the lacZ SfiI-AvrII insert has been replaced by the 3.0-kb AvrII fragment of pJMT4 containing kilA and telAB | This study |
| pJMT6 | Apr Telr, pUT/mini-Tn5 Tel (NotI site free), pJMT5 derivative with an internal NotI deletion | This study |
| pJMT9 | Apr Telr, pUT/mini-Tn5 Tel (SfiI-AvrII site free), pUTlacZ derivative in which the lacZ SfiI-AvrII insert has been deleted and the kanamycin NotI inset has been replaced by the 3.0-kb NotI fragment of pSHA1 containing kilA and telAB | This study |
| pUTluxAB | Apr Tcr, delivery vector for mini-Tn5 luxAB NotI flanking a promoterless 3.2-kb luxAB reporter | 6 |
| pJMT8 | Apr Telr, pJMT6 containing the 3.2-kb NotI fragment of pUTluxAB | This study |
| pCK04AxylR | Cmr, low-copy-number plasmid with a 12-kb NotI insert containing the upp TOL catabolic segment | 6 |
| pJMT7 | Apr Telr, pJMT6 containing the 12-kb NotI fragment of pCK04AxylR | This study |
| pGP704 | Apr, oriT RK2, oriV R6K | 17 |
| pNot18 | Apr, same as pUC18 but the α-lac fragment is flanked by NotI sites | M. Herrero |
| pNot19 | Apr, same as pUC19 but the α-lac fragment is flanked by NotI sites | M. Herrero |
| pVTR-A, -B, and -C | Cmr, pSC101 replicon-based vectors with excisable expression of lacIq/Plac NotI cassette | 21 |
| pPS10 | Kmr plasmid isolated from P. syringae pv. savastanoi | 18 |
| pMM141 | Kmr, pSP10 replicon, broadened host range, replicates in E. coli and Pseudomonas strains | 7 |
| pJPS6 | Kmr, selection marker cloned as a 1.7-kb NotI insert between the RK2 oriT segment (0.5 kb) and the pMM141 minimal replicon (1.8 kb), SfiI-AvrII site available for cloning | This study |
| pJPS8 | Telr, similar to pJPS6 but selection marker replaced by the 3.0-kb NotI insert of pSHA1, SfiI-AvrII site available for cloning | This study |
| pJPS9 | Smr, selection marker cloned as a 2.1-kb SfiI insert between RK2 oriT and pMM141 oriV, NotI site available for cloning | This study |
| pJPS10 | Telr, similar to pJPS9 but selection marker replaced by the 3.0-kb AvrII insert of pJMT4, NotI site available for cloning | This study |
| pJPS11 | Telr, pJPS10 containing at its NotI site the 12-kb NotI upp TOL catabolic segment of pCK04AxylR | This study |
Assembly of Telr transposon vectors.
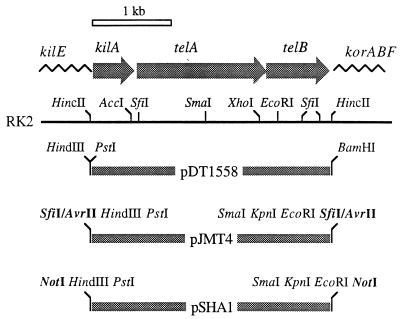
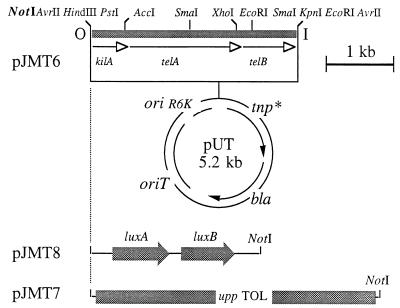
The genetic determinants for Telr, encoded by the kilA and telAB genes of plasmid RK2, were obtained as a 3.0-kb HindIII-BamHI fragment (32) from plasmid pDT1558 (kindly provided by D. Taylor, University of Alberta, Edmonton, Alberta, Canada). This DNA segment was cloned independently in the corresponding sites of vectors pUC18Sfi and pUC18Not in order to flank the Telr genes with rare restriction sites compatible with matching transposon or plasmid vectors. In one case (pJMT4) (Fig. 1), the kil and AtelAB genes were flanked by SfiI-AvrII sites, while in another case (pSHA1) (Fig. 1), the same genes were on a NotI segment. To assemble these segments as the selection markers of minitransposon vectors, the 3.0-kb AvrII insert of pJMT4 was exchanged with the AvrII segment of pUTlacZ1, resulting in pJMT5 (Table 1). This construction was then digested with NotI and religated to release the NotI inserts from the plasmid and to generate a minitransposon with Telr as the only selection marker (borne by pJMT6) (Fig. 2) and a free NotI site available for cloning within the mobile element. Similarly, the 3.0-kb NotI insert of plasmid pSHA1 was exchanged with the NotI segment of pUTlacZ1, digested with AvrII, and religated. This produced the delivery plasmid (pJMT9) of a second Telr mini-Tn5 transposon bearing a single AvrII site for insertion of heterologous cloned fragments of DNA. Insertion of the 3.2-kb NotI fragment of pUTluxAB (spanning the promoterless luxAB genes) into the single NotI site of pJMT6 resulted in pJMT8. In this construction, the luxAB genes were placed inside the minitransposon that was selectable through Telr. Similarly, plasmid pJMT7 (Fig. 2) was a derivative of pJMT6 containing a 12-kb NotI segment spanning the entire upper TOL pathway of plasmid pWW0 along with its cognate regulatory gene, xylR (19).
FIG. 1.
Telr resistance cassettes in recombinant plasmids and transposon vectors. The organization of the kilA locus of the RK2Telr plasmid, spanning the kilA telAB cistrons (29) (alternatively designated klaABC [8]), is shown at the top lined up with the neighboring regions of the plasmid. The location and orientation of each gene are indicated. The three genes appear to be transcribed from a single promoter upstream of kilA. telA and telB may be translationally coupled. The 3.0-kb RK2Telr HincII segments used to construct the tellurite resistance cassettes shown at the bottom are labeled with the designations of the plasmids.
FIG. 2.
Organization of Telr transposon vector mini-Tn5 Tel of pJMT6 and its derivatives. This plasmid is the delivery vector for the minitransposon shown at the top. The suicide donation system used (lower part of the figure) was the pUT system (11) and included the Tn5 transposase gene devoid of NotI sites (tnp*), an Apr selection marker (bla), an origin of transfer for RP4-mediated mobilization (oriT), and the origin of replication of plasmid R6K, which is dependent on the π protein encoded by the pir gene carried by specialized λpir E. coli hosts. The top part of the figure shows the Telr cassette of pJMT4 included in the mini-Tn5 transposon vector portion of the plasmid, including a single NotI site used for cloning heterologous DNA segments. The NotI insertions used included the luxAB genes (3.2 kb) in the case of pJMT8 and the upper TOL segment (upp TOL) (12 kb, not to scale) in pJMT7.
Construction of mobilizable, broadened-host-range plasmids derived from pPS10.
A minimal mobilizable replicon able to replicate in both E. coli and Pseudomonas cells was assembled as follows. A 545-bp segment of plasmid pGP704 spanning the oriT region of RK2 (17) was amplified by PCR with primers RP2 (5′AATTAGGCCTAGGCGGCCAGGAACGCAACCGCAGC3′) and RP1 (5′AATTAGCGGCCGCTCCTCAATCGCTCTTCGTTCGTC3′); these primers introduced SfiI-AvrII and NotI restriction sites, respectively, at the ends of the amplified fragment. Similarly, two other PCR primers, REPA1B (5′AATTAG GCCGCCTAGGCCCAGCTGGGTAGCGGAGCTATCCAACGGCTG3′) and REPA2B (5′AATTAGCGGCCGCGAAGGGTTGTTTCTGTAGAATGGG3′), which bear flanking SfiI and NotI sites as well, were used to amplify a 1.8-kb segment of plasmid pMM141. This plasmid is a variant of pPS10 (18) in which a conservative A32V mutation in the replication protein RepA broadened the host range of the plasmid so that it thrived in E. coli and other gram-negative hosts (7). The amplified segment of pMM141, which contained oriV and the mutated repA variant of the plasmid, was mixed with the segment containing oriT, digested with SfiI and NotI, and ligated to a 1.7-kb NotI fragment (from plasmid pUTlacZ1) (Table 1) encoding resistance to kanamycin. This resulted in plasmid pJPS6, which consisted of a basic replication-transference unit along with a kanamycin resistance selection marker. Plasmid pJPS6 was the basis for the following additional derivatives: pJPS8 (in which the Kmr NotI insert was replaced by the Telr marker of plasmid pSHA1), pJPS9 (in which the Kmr marker was deleted and an Smr marker of pUTSm [6] was added as an SfiI insert), and pJPS10 (in which the Kmr marker was deleted and the Telr determinants from pJMT4 were added as an SfiI-AvrII insert). All of these vectors contained three functional fragments (the resistance marker, the origin of transference, and the minimal replicon) and, depending on the combination, allowed insertion of various cloning and expression systems at either the single NotI site or the SfiI-AvrII sites (Table 1). In one case, the 12-kb NotI segment of pCK04AxylR (19) containing the upper pathway of the TOL plasmid and the regulatory gene xylR was cloned in the Telr plasmid vector pJPS10, giving rise to pJPS11.
Monitoring the stability of recombinant traits.
E. coli and P. putida clones carrying the Telr vectors were pregrown overnight at 37 and 30°C, respectively, in Luria broth amended with 80 μg of tellurite per ml in order to start with saturated cultures which retained 100% of the marker. The cultures were then diluted 1,000-fold, regrown to saturation levels in media with or without tellurite, and plated onto selective and nonselective agar plates. The procedure was repeated several times until the cells had gone through 80 generations. Stability was expressed as the percentage of cells that retained the Telr marker during growth. The results presented below are the averages from three independent experiments.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the DNA sequences used in this study are as follows: kilA and telAB genes, M62846 and M38697; oriT sequence of RK2, J04942; and pPS10 sequence, X58896, S80705, and X64048.
RESULTS AND DISCUSSION
Rationale for using Telr as a workable nonantibiotic selection marker.
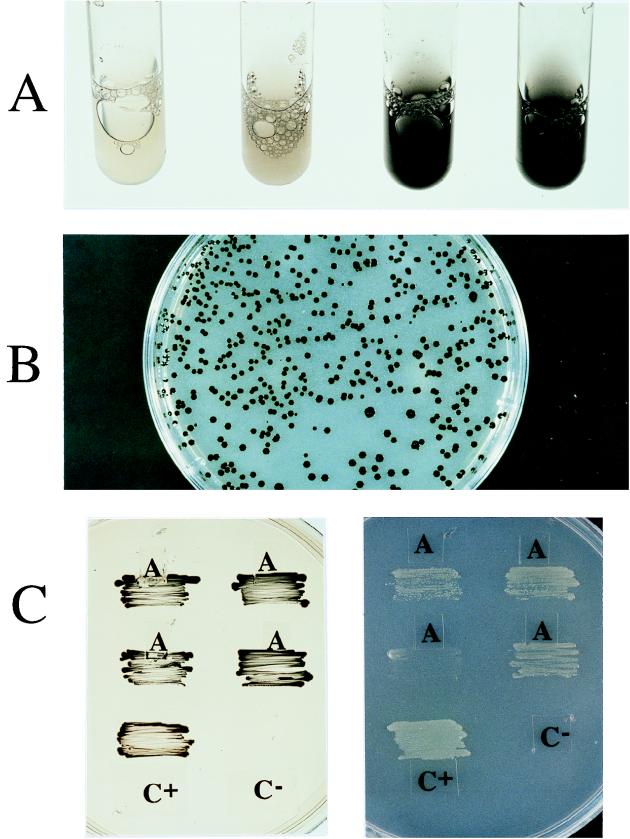
In this paper we describe a number of genetic tools tailored for pseudomonads and related genera in which genes for resistance to potassium tellurite are used as the selection phenotype. This resistance is an attractive marker, since potassium tellurite is toxic for most microorganisms (especially gram-negative bacteria) (28, 29); spontaneous tolerance is very infrequent, and resistance determinants are rarely expressed (33). Furthermore, the mechanism of tolerance to the oxianion makes Telr bacteria produce a characteristic black color when they are grown in selective medium containing the salt (Fig. 3A). Although Telr genes are available from different sources (8, 30, 32, 33), we chose the Telr genes present on a variant of the broad-host-range plasmid RK2 in which the kilA telAB cluster is effectively expressed by virtue of a mutation in kilB which activates expression of this set of otherwise cryptic genes (31). Finally, handling tellurite does not require the precautions necessary for handling mercuric or arsenic salts, tellurite-containing plates can be disposed of like any other antibiotic-containing cultures, and unlike resistance to herbicides, selection can be done in both minimal and rich media.
FIG. 3.
Phenotypes endowed by Telr in P. putida. (A) Expression of Telr in P. putida cells grown in a liquid culture. The cells were transformed with plasmid pJPS6 and grown in LB medium supplemented with different concentrations of potassium tellurite (6, 12, 50, and 80 μg/ml). Note the blackening of the culture medium during growth. (B) Selection of P. putida exconjugants containing mini-Tn5 Tel. Mating of E. coli S17-1λpir(pJMT6) and P. putida KT2442 was performed as indicated in the text, and the preparation was plated onto M9 medium containing citrate and tellurite. The P. putida cells which received the minitransposon in their chromosome gave rise to black colonies on the selection plate. (C) Selection of Telr P. putida clones expressing the upp TOL catabolic segment. The patches show the phenotypes resulting from expression of the upper TOL pathway in P. putida cells which received the corresponding DNA segment in Telr plasmid pJPS10 (top patches) or as mini-Tn5 Tel (upp TOL) (middle patches). The same clones were patched on M9 medium containing citrate and tellurite (left) and on M9 medium lacking a carbon source and then exposed to saturating toluene vapor (right). Positive (C+) and negative (C−) controls were included.
Performance of the Telr marker for selection of minitransposons.
The reference construct used to examine the utility of the kilA and telAB determinants as a selection marker was pJMT6 (Fig. 2). This plasmid is the suicide delivery plasmid for the transposon vector mini-Tn5 Tel bearing a free NotI site. Using previously described protocols (6), we randomly inserted this minitransposon into the chromosome of P. putida KT2442. In one case, a biparental mating was performed between donor strain E. coli S17-1λpir(pJMT6) and recipient strain P. putida KT2442, and the mating mixture was plated onto LB medium containing rifampin and tellurite to select for insertions in rich medium. Alternatively, a triparental mating was performed with E. coli CC118λpir(pJMT6), P. putida KT2442, and helper strain E. coli HB101(RK600), and the mating mixture was plated onto M9 minimal medium plates containing citrate and tellurite to select for insertions and to counterselect for the donor and helper cells. Regardless of the selection medium used and the form of mating, Telr exconjugants of Pseudomonas cells appeared at a frequency of 10−4 to 10−5 (6), which is within the range of frequencies of appearance described for other mini-Tn5 transposons (11), while the frequency of spontaneous tolerance to tellurite was negligible (<10−8). The Telr colonies had a distinct black color and were visible to the naked eye against the background used (Fig. 3B). Of the 100 Telr exconjugant colonies tested from each mating, 96 and 89 turned out to be sensitive to the β-lactam antibiotic piperacillin. This indicated that the Telr phenotype was due to authentic transposition of mini-Tn5 Tel into the chromosome of the target strain and not to cointegration of the whole pJMT6 delivery plasmid, which bears a β-lactamase gene (Fig. 2).
Our observations suggested that the kilA and telAB genes can be used as an effective selection marker for P. putida. To ascertain whether the same marker could be used to produce insertions of heterologous DNA segments encoding nonselectable phenotypes, we repeated the same matings by using as donors λpir E. coli strains transformed with plasmid pJMT8. This plasmid is a pJMT6 derivative in which a promoterless luxAB reporter is cloned inside the Telr mini-transposon vector (Table 1). Telr exconjugants from the matings with the new donors appeared with the same ease of selection and with the same characteristics as exconjugants without the luxAB cassette appeared. Twenty Telr Pips colonies were reisolated in the absence of selection and were grown overnight in LB medium without tellurite. They were briefly exposed to n-decanal vapor, and the emission of light was measured with a luminometer as explained elsewhere (13). All of the clones tested emitted light at a detectable level, thus confirming the association of the Telr phenotype with the luxAB genes. The relative levels of luminiscence did vary, however, between approximately 500 and 12,000 U, suggesting that there were independent insertions in different locations of the chromosome. This was confirmed by a PCR and Southern blot analysis of selected exconjugants (data not shown). Taken together, our results confirmed that the Telr genes could be used in transposon vectors for Pseudomonas cells with the same efficiency, performance, and selection clarity as standard antibiotic markers. Although we used the Telr marker only for manipulations of P. putida, the nature of the resistance genes makes this marker potentially useful for a variety of gram-negative strains (28, 29).
Construction and performance of P. putida strains for degradation of toluene through an ortho-cleavage degradation pathway.
During construction of Pseudomonas biocatalysts for degradation of mixtures of Cl-toluenes (3, 14), the bacteria had to be able to completely metabolize toluene without any meta-cleavage of intermediate catechols. Since such a pathway is not naturally available, we set out to construct a P. putida strain that was able to convert toluene into benzoate and then further metabolize this compound through the chromosomally encoded benzoate degradation pathway involving the housekeeping ben/cat genes (10). To do this, we tried to insert the entire upper TOL pathway of plasmid pWW0 along with its cognate, toluene-responsive regulator, xylR, into the chromosome of P. putida KT2442. TOL plasmid pWW0 of P. putida mt-2 includes two operons for biodegradation of toluene and m- and p-xylenes. The first stage consists of sequential oxidation of the methyl group down to benzoate and toluates (10). The oxidative enzymes of the upper pathway (i.e., the enzymes that convert toluene into benzoate) have a broad substrate spectrum, so that a number of toluene derivatives can be converted into the corresponding acids (10, 19). Because of this, we excised the catabolic segment containing the upper TOL genes and xylR (the so-called upp TOL segment [19]) as a 12-kb NotI fragment from plasmid pCK04AxylR (Table 1) and cloned it at the corresponding site of pJMT6. This gave rise to the delivery plasmid (designated pJMT7) of transposon mini-Tn5 Tel (upp TOL), as shown in Fig. 2. This element was targeted to the chromosome of P. putida KT2442 through triparental mating of this strain with E. coli CC118λpir(pJMT7) and E. coli HB101(RK600), as explained above. The mating mixture was plated onto M9 selection medium containing citrate and tellurite. Like the outcome observed with minitransposons bearing smaller inserts, the transposition frequencies ranged from 10−4 to 10−5. Of the 50 Telr exconjugants examined further, 30 could grow well on a mineral medium containing toluene as the only carbon source (Fig. 3C), which confirmed the prediction made on the basis of the performance of the hybrid pathway.
That the acquired trait (growth on toluene) was stably inherited was shown by repeated transfer of one of the clones which grew the best on toluene into LB medium in the absence of selective pressure, as explained in Materials and Methods. After 1,000 generations, we could not detect any significant loss of either the Telr marker or the ability to grow on toluene, thus suggesting that the inserted genes were at least as stable as any other chromosomal DNA segment of the strain.
Application of Telr in broadened-host-range plasmids.
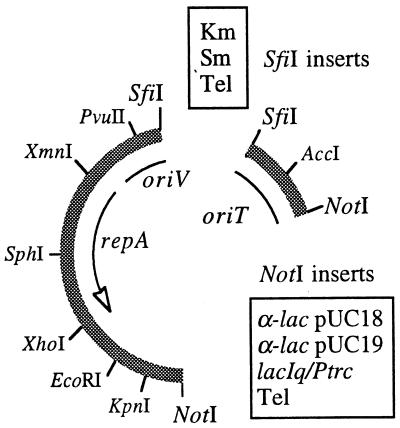
In a second type of vector, we combined the kilA and telAB genes with an artificially assembled mobilizable replicon tailored to match the marker. As shown in Fig. 4, this replicon consisted of a variant of Pseudomonas-specific plasmid pPS10 (18) with an ability to replicate in various gram-negative hosts (7) combined with the broad-range mobilization origin of plasmid RP4 (RK2). These two elements (which spanned as little as 2.3 kb) were constructed so that they could be combined with a variety of antibiotic or nonantibiotic selection markers (e.g., Telr) inserted at the available SfiI-AvrII site, as well as with cloned fragments of DNA that could be inserted as NotI fragments into the corresponding free site (Fig. 5). The result of these combinations was a series of cloning vectors which can replicate in a variety of gram-negative hosts and can be mobilized among strains through RP4-assisted conjugal transfer. On the other hand, the plasmids possess neither a stabilization system (20) nor a replication termination system, which results in a degree of long-term instability when the external selective pressure is removed.
FIG. 4.
Organization of the minimal replication-transfer segment of the pJPS plasmid series. The DNA sequence shown (length, 2,372 bp) includes a 1,815 bp SfiI-AvrII–NotI segment spanning the sequence of the repA gene and the target oriV of plasmid pMM141 (uppercase letters), as well as a 545-bp NotI–SfiI-AvrII fragment with the origin of transfer (oriT) of plasmid RK2 (lowercase letters). Depending on the specific construct (Fig. 5), these two segments may appear next to each other (as shown) or may be separated by NotI or SfiI-AvrII inserts.
FIG. 5.
Modular assembly of pJPS plasmids. The combinations of NotI and SfiI-AvrII inserts possible with the exchange of selection cassettes and cloned segments are shown. The NotI site is the preferred site for insertion of segments that originated in the previously reported cloning and expression vector pNot18, pNot19, and pVTR plasmid series (Table 1). The names of some of the combinations of these segments are indicated in the text.
To examine the performance of these plasmids when they were bearing catabolic segments, we constructed pJPS11. This plasmid is a derivative of the Telr vector pJPS10, in which the same upp TOL segment employed above was inserted at the free NotI site. This gave rise to a Telr replicon containing all of the genes required for conversion of toluene to benzoate. pJPS11 was mobilized to P. putida KT2442 through triparental mating of this recipient with E. coli CC118λpir and E. coli HB101(RK600), followed by selection on M9 medium containing citrate and tellurite. In this case, the frequency of transfer was approximately 10−1. Telr exconjugants were then examined for growth on mineral medium containing toluene as the carbon source. In this case, 100% of the exconjugants grew on the aromatic compound (Fig. 3C). However, when P. putida KT2442(pJPS11) was grown in the absence of selection, both the Telr marker and the ability to grow on toluene were lost at a rate of approximately 1 to 2% per generation. The new trait could therefore be maintained as long as the external conditions were favorable for its selective advantage.
ACKNOWLEDGMENTS
We are indebted to D. Taylor for the kind gift of pDT1558 and to S. Panke, M. Herrero, J. Benedí, A. Haro, S. Hernández, and I. Cases for some of the constructs described in this paper.
This work was funded by grant BIO95.788 from the Spanish Interministerial Commission for Science and Technology (CICYT) and by European Commission contracts BIO4-CT97-2040 and ENV4-CT95-0141.
REFERENCES
- 1.Bianco N, Neshat S, Poole K. Conservation of the multidrug resistance efflux gene oprM in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:853–856. doi: 10.1128/aac.41.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin R, Bellmare G, Dion P. Novel narrow host range vectors for direct cloning of foreign DNA in Pseudomonas. Curr Microbiol. 1994;28:41–47. doi: 10.1007/BF01575984. [DOI] [PubMed] [Google Scholar]
- 3.Brinkmann U, Reineke W. Degradation of chlorotoluenes by in vivo constructed hybrid strains: problems of enzyme specificity, induction and prevention of meta-pathway. FEMS Microbiol Lett. 1992;96:81–88. doi: 10.1016/0378-1097(92)90460-6. [DOI] [PubMed] [Google Scholar]
- 4.de Lorenzo V. Genetic engineering strategies for environmental applications. Curr Opin Biotechnol. 1992;3:227–231. doi: 10.1016/0958-1669(92)90097-3. [DOI] [PubMed] [Google Scholar]
- 5.de Lorenzo V. Designing microbial systems for gene expression in the field. Trends Biotechnol. 1994;12:365–371. doi: 10.1016/0167-7799(94)90037-X. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 7.Fernández-Tresguerres M E, Martín M, García de Viedma D, Giraldo R, Díaz-Orejas R. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol. 1995;177:4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goncharoff P, Saadi S, Chang C-H, Saltman L H, Figurski D H. Structural, molecular, and genetic analysis of the kilA operon of broad-host-range plasmid RK2. J Bacteriol. 1991;173:3463–3477. doi: 10.1128/jb.173.11.3463-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen L H, Sørensen S J, Sørensen L B. Chromosomal insertion of the entire Escherichia coli lactose operon into two strains of Pseudomonas using a modified mini-Tn5 delivery system. Gene. 1997;186:167–173. doi: 10.1016/s0378-1119(96)00688-9. [DOI] [PubMed] [Google Scholar]
- 10.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion from foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh N, Koide Y, Furuzawa H, Hirose S, Inukai T. Novel plasmid vectors for gene cloning in Pseudomonas. J Biochem. 1991;110:614–621. doi: 10.1093/oxfordjournals.jbchem.a123629. [DOI] [PubMed] [Google Scholar]
- 13.Kristensen C, Eberl L, Sánchez-Romero J M, Giskov J M, Molin M S, de Lorenzo V. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J Bacteriol. 1995;177:52–58. doi: 10.1128/jb.177.1.52-58.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehning A, Happe B, Timmis K N, Pieper D. Metabolism of chlorotoluenes by Burkholderia sp. strain PS12 and toluene dioxygenase of Pseudomonas putida F1: evidence for monooxygenation by toluene and chlorobenzene dioxygenases. Appl Environ Microbiol. 1997;63:1974–1979. doi: 10.1128/aem.63.5.1974-1979.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd-Jones G, Osborn A M, Ritchie D A, Strike P, Hobman J L, Brown N L, Rouch D A. Accumulation and intracellular fate of tellurite in tellurite-resistant E. coli: a model for the mechanism of resistance. FEMS Microbiol Lett. 1994;118:113–120. doi: 10.1111/j.1574-6968.1994.tb06812.x. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nieto C, Giraldo R, Fernández-Tresguerres E, Díaz-Orejas R. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae pv. savastanoi. J Mol Biol. 1992;223:415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- 19.Panke S, Sánchez-Romero J M, de Lorenzo V. Engineering of quasi-natural Pseudomonas putida strains for toluene metabolism through an ortho-cleavage degradation pathway. Appl Environ Microbiol. 1998;64:748–751. doi: 10.1128/aem.64.2.748-751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pecota D C, Kim C S, Wu K, Gerdes K, Wood T K. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl Environ Microbiol. 1997;63:1917–1924. doi: 10.1128/aem.63.5.1917-1924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Martín J, de Lorenzo V. VTR expression cassettes for engineering conditional phenotypes in Pseudomonas: activity of the Pu promoter of the TOL plasmid under limiting concentrations of the XylR activator protein. Gene. 1996;172:81–86. doi: 10.1016/0378-1119(96)00193-x. [DOI] [PubMed] [Google Scholar]
- 22.Prosser J L. Molecular marker systems for detection of genetically engineered microorganisms in the environment. Microbiology. 1994;140:5–17. doi: 10.1099/13500872-140-1-5. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sánchez-Romero, J. M., and V. de Lorenzo. Genetic engineering of nonpathogenic Pseudomonas strains as biocatalysts for industrial and environmental processes. In J. Davies and G. Cohen (ed.), Manual of industrial biotechnology, 2nd ed., in press. American Society for Microbiology, Washington, D.C.
- 25.Sánchez-Romero, J. M., K. N. Timmis, and V. de Lorenzo. Transposon-vectors in microbial ecology and environmental biotechnology. FEMS Microbiol. Ecol., in press.
- 26.Suzina N E, Duda V I, Anisimova L A, Dmitriev V V, Boronin A M. Cytological aspects of resistance to potassium tellurite conferred on Pseudomonas cells by plasmids. Arch Microbiol. 1995;163:282–285. doi: 10.1007/BF00393381. [DOI] [PubMed] [Google Scholar]
- 27.Timmis K, Steffan R, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 28.Turner R J, Weiner J H, Taylor D. Characterization of the growth inhibition phenotype of the kilAtelAB operon from IncPα plasmid RK2Ter. Biochem Cell Biol. 1994;72:333–342. doi: 10.1139/o94-046. [DOI] [PubMed] [Google Scholar]
- 29.Turner R J, Weiner J H, Taylor D E. In vivo complementation and site-specific mutagenesis of the tellurite resistance determinant kilAtelAB from IncPα plasmid RK2Ter. Microbiology. 1994;140:1319–1326. doi: 10.1099/00221287-140-6-1319. [DOI] [PubMed] [Google Scholar]
- 30.Turner R J, Weiner J H, Taylor D E. Neither reduced uptake nor increased efflux is encoded by tellurite resistance determinants expressed in E. coli. Can J Microbiol. 1995;41:92–98. doi: 10.1139/m95-012. [DOI] [PubMed] [Google Scholar]
- 31.Turner R J, Weiner J H, Taylor D E. The tellurite resistance determinants tehAtehB and klaAklaBtelB have different biochemical requirements. Microbiology. 1995;141:3133–3140. doi: 10.1099/13500872-141-12-3133. [DOI] [PubMed] [Google Scholar]
- 32.Walter E G, Taylor D E. Comparison of tellurite resistance determinants from the IncPα plasmid RP4Ter and the IncHII plasmid pHH1508a. J Bacteriol. 1989;171:2160–2165. doi: 10.1128/jb.171.4.2160-2165.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter E G, Taylor D E. Plasmid-mediated resistence to tellurite: expressed and cryptic. Plasmid. 1992;27:52–64. doi: 10.1016/0147-619x(92)90006-v. [DOI] [PubMed] [Google Scholar]